Abstract
Developmental programs are executed by tightly controlled gene regulatory pathways. Here, we combined the unique sample retrieval capacity afforded by laser capture microscopy with analysis of mRNA abundance by CEL-Seq (cell expression by linear amplification and sequencing) to generate a spatiotemporal gene expression map of the Caenorhabditis elegans syncytial germline from adult hermaphrodites and males. We found that over 6000 genes exhibit spatiotemporally dynamic expression patterns throughout the hermaphrodite germline, with two dominant groups of genes exhibiting reciprocal shifts in expression at late pachytene during meiotic prophase I. We found a strong correlation between restricted spatiotemporal expression and known developmental and cellular processes, indicating that these gene expression changes may be an important driver of germ cell progression. Analysis of the male gonad revealed a shift in gene expression at early pachytene and upregulation of subsets of genes following the meiotic divisions, specifically in early and late spermatids, mostly transcribed from the X chromosome. We observed that while the X chromosome is silenced throughout the first half of the gonad, some genes escape this control and are highly expressed throughout the germline. Although we found a strong correlation between the expression of genes corresponding to CSR-1-interacting 22G-RNAs during germ cell progression, we also found that a large fraction of genes may bypass the need for CSR-1-mediated germline licensing. Taken together, these findings suggest the existence of mechanisms that enable a shift in gene expression during prophase I to promote germ cell progression.
Keywords: gene expression, germline, gametogenesis, oogenesis, spermatogenesis, meiosis, C. elegans
AT the core of metazoan sexual reproduction lies the development of haploid gametes, namely the sperm and the egg produced from diploid germline stem cells (L’Hernault 1997; Yoshida 2010; Sánchez and Smitz 2012; Chu and Shakes 2013; Ellis and Stanfield 2014; Tanaka 2014). This developmental process includes well-defined cellular steps, but the genetic program that drives them is largely unknown. During gametogenesis, the mitotically cycling stem cells must produce diploid cells that lose their stem cell identity, accumulate proteins required for entry into meiosis, and then proceed onto two consecutive rounds of meiotic cell divisions (meiosis I and II). A critical phase within gametogenesis occurs before the first meiotic division during prophase I. Specifically, homologous chromosomes find each other, pair, synapse, and exchange segments via homologous recombination in preparation for their segregation away from each other toward opposite poles of the meiosis I spindle. During prophase I of oogenesis in many organisms, the oocytes also accumulate transcripts required for early stages of embryogenesis [reviewed in Sánchez and Smitz (2012) and Kim et al. (2013)]. Yet, despite decades of studies providing descriptions of the gametogenic processes, we know very little about what drives and controls the development of sperm and oocytes. This stands in contrast to studies in yeast in which the availability of large quantities of synchronized meiotic cells have allowed transcriptome and ribosome profiling analysis of the budding yeast sporulation process (Chu et al. 1998; Primig et al. 2000; Brar et al. 2012).
The Caenorhabditis elegans gonad offers a unique opportunity for the study of progression from the undifferentiated proliferative germ cells through spermatogenesis or oogenesis (L’Hernault 1997; Dernburg 2001; Couteau et al. 2004; Greenstein 2005; Zetka 2009; Schvarzstein et al. 2010; Chu and Shakes 2013; Lui and Colaiácovo 2013; Nousch and Eckmann 2013; Ellis and Stanfield 2014). Nuclei in both the adult male and hermaphrodite tube-like gonads are arranged along the distal–proximal axis in a spatiotemporal fashion from the germ stem cell mitotic proliferative zone through the various stages of meiotic prophase, and then differentiate into mature sperm or oocytes (Kimble and Crittenden 2005; Shakes et al. 2009; Pazdernik and Schedl 2013; Ellis and Stanfield 2014) (Figure 1A and Figure 6A). Important contributions to our understanding of the genes required for germ cell progression were achieved through RNA-sequencing (RNA-seq), DNA microarray, and SAGE (serial analysis of gene expression) analyses of the entire C. elegans gonad (Reinke et al. 2000, 2004; Wang et al. 2009; Ortiz et al. 2014). However, due to the gonad’s syncytial nature, in which germ “cells” do not become fully cellularized until late diakinesis, and instead maintain a bridge or connection to a common core referred to as the rachis to which they contribute their cytoplasmic contents that flow into developing oocytes, it has so far been impossible to analyze the dynamics of the transcriptome through this process.
Figure 1.
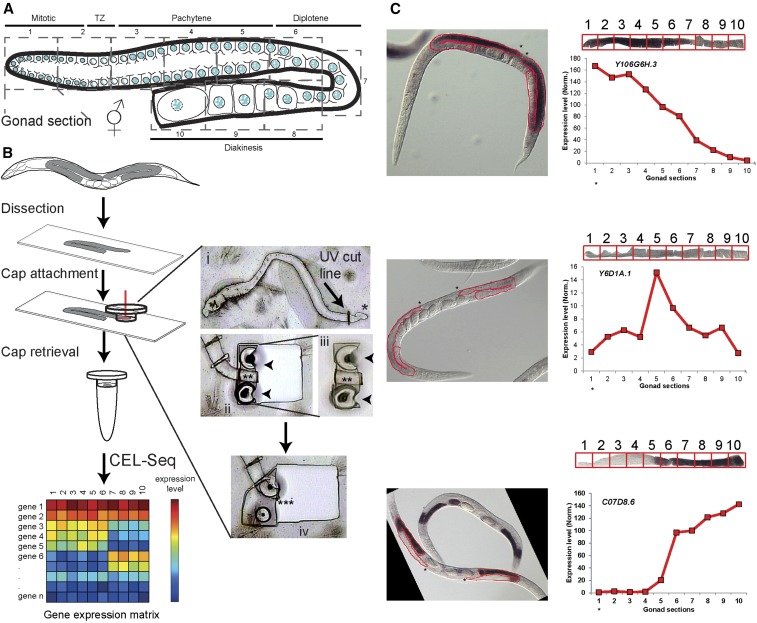
Methodology and verification of the spatiotemporal analysis of gene expression in the gonad. (A) Illustration of the C. elegans hermaphrodite gonad. The nuclei are arranged in a spatiotemporal pattern progressing from the distal mitotic tip (section 1) through the different stages of prophase I until the last (mature) oocyte (section 10). Oocytes become fully cellularized by late diakinesis. The numbers of the consecutive dissected sections and corresponding developmental stages are indicated. (B) Experimental work-through and representative images. Worms were dissected, and gonads were transferred to membrane slides. Sections equivalent to one-tenth of the total gonad length were cut via UV laser (red vertical line in cap represents the laser beam). (i) This inset shows the UV cut line on the dissected gonad, where * indicates the distal mitotic tip for orientation. (ii) Shows a section adhered to the cap by melting (arrowheads). (iii) Shows the cap from (ii) that was lifted to be examined at the QC (quality control) station (** indicates the UV dissected gonad section). (iv) Shows next section of the gonad adhered to cap by melting (*** indicates the corresponding empty area left behind after the previous section shown in (iii) was picked; note that no parts were left behind and no liquid was spilled). RNA from the caps was analyzed by CEL-Seq. (C) Verification of dynamic expression via comparison with NEXTDB RNA in situ images. Three representative images of early (Y106G6H.3), mid (Y6D1A.1), and late (C07D8.6) expression are shown for the indicated genes, as well as expression level (y-axis) along the different sections (x-axis), as found in our analysis with digitally linearized gonad images extracted from the NEXTDB images (the portions of the gonads that were linearized are outlined in red on the in situ images). Boxes on the linearized gonads, numbered 1 through 10, indicate the different sections for which expression levels were quantified. Asterisks on the NEXTDB images indicate the distal tip and are also placed under gonad section 1 on the x-axes of the expression analysis to orient the direction in which the sections are located along these gonads. TZ: transition zone.
Figure 6.
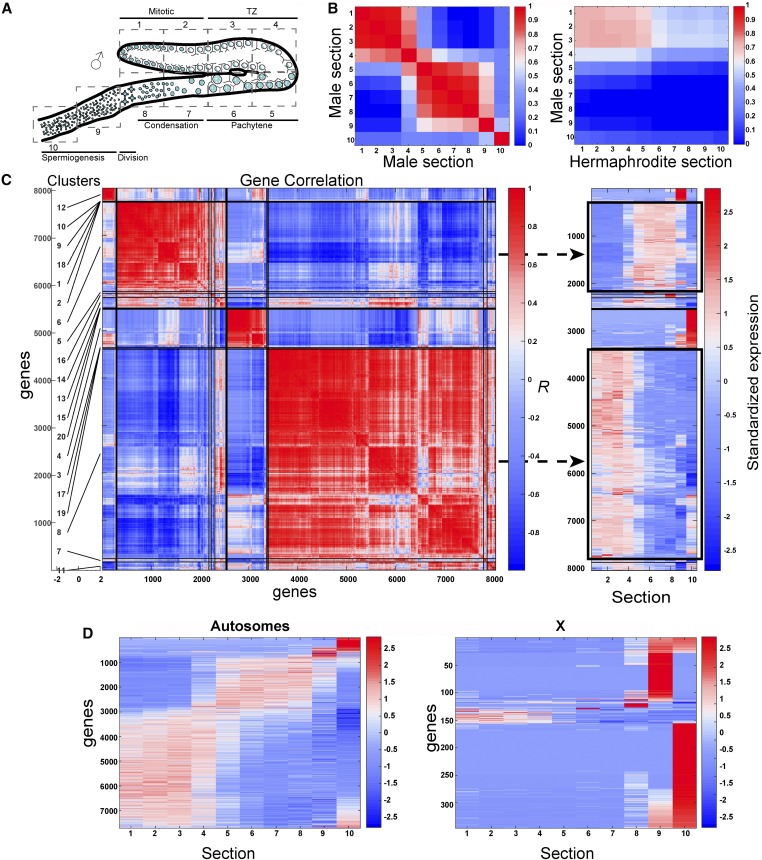
Dynamic expression of genes in the male gonad. (A) Illustration of the C. elegans male gonad. The nuclei are arranged in a spatiotemporal pattern from the distal mitotic tip (section 1) through the different stages of prophase I (from section 3 to early 8), the division zone (late section 8), and spermiogenesis (sections 9–10). (B) Pairwise profile correlation coefficients between male sections (left) and male vs. hermaphrodite sections (right). (C) Left: hierarchical grouping analyses of male germline gene expression profiles using Pearson’s correlation. The top 20 gene groups are numbered and demarcated to the left of the y-axis. Right: standardized gene expression levels. Gene expression levels were mean subtracted and divided by the SD. Genes are arranged in the same linear order of the clusters on the left (y-axis) and gonad sections (X). The two major groups are indicated by black rectangles. (D) Standardized gene expression level along the male gonad (sections 1–10) in autosomes (left) vs. the X chromosome (right). TZ: transition zone.
Recent advancements in high-throughput sequencing have enabled the analysis of mRNA abundances from small biological samples as small as individual cells (Tang et al. 2011; Hashimshony et al. 2012; Avital et al. 2014; Islam et al. 2014; Macaulay and Voet 2014). Among the available methods, CEL-Seq (cell expression by linear amplification and sequencing) is a multiplexed method that uses in vitro transcription for single-cell transcriptome analysis (Hashimshony et al. 2012). However, these methods have only recently started to be used to analyze transcriptome dynamics through an entire organ or microscopically defined syncytial tissue (Jan 2017).
Here, we report the development of a novel approach for transcriptome analysis of syncytial tissues by combining ARCTURUS Laser Capture Microscopy (LCM) for sample extraction and CEL-Seq analysis. With this approach, we analyzed the gene expression profiles in 10 sections of the adult C. elegans hermaphrodite and male germlines. Analysis of this unique data set revealed surprising dynamic changes in the transcription levels of over 6000 genes. The genes exhibiting dynamic changes in expression can be clustered into functional groups that differ in their annotated functional and cellular roles, expression profiles, and include dramatic differences between the autosomes and the X chromosome. We used these germline data sets to test different models for gene expression in the germline, and to analyze their spatiotemporal validity and sexual dimorphism.
Materials and Methods
Strains and alleles
The C. elegans N2 Bristol strain was used. Worms were cultured at 20° under standard conditions, as described in Brenner (1974).
Isolation of gonad segments
First, 20–24-hr post-L4 adult hermaphrodites or males were placed onto 30 μl of Egg Buffer (118 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2, and 0.025 mM HEPES pH 7.4) on glass slides, where gonads were dissected and transferred to polyethylene naphthalate (PEN) membrane slides (cat # LCM0522; Life Technologies) with a glass pipette. Next, 2 to 6 gonads were dissected per slide. Excess buffer was removed, and slides were frozen on dry ice and kept at −80°. To capture the sections, slides were thawed, loaded onto the ARCTURUS VERITAS LCM, and the distal—proximal axis of the gonad was measured from the distal tip to the end of the −1 oocyte or seminal vesicle for hermaphrodites and males, respectively (gonad axis or GA length). Nonspecific material was removed by loading it onto a CapSure HS LCM Cap (cat # LCM0214; Life Technologies), which was then discarded. New caps were used for loading each desired section and each section was equivalent to one-tenth of the GA total length. The cap was adhered to the membrane surrounding the desired section by two to three infrared (IR) laser melt points (power = 70–90 mW, pulse = 1000–3000 μsec, and intensity = 100 mV), and the section was isolated with an ultraviolet (UV) laser (laser power = 7–17). We verified under the microscope that: (1) the laser sealed the gonad along the incision line, (2) only the defined section was attached to the cap by examining the cap at the QC station, and (3) that all of the section was removed at the site. Caps were retrieved, immediately placed into 0.5 ml sterile tubes, and kept in dry ice.
CEL-Seq analysis
RNA was extracted from gonad sections using Trizol in the presence of LPA (linear polyacrylamide) to help RNA precipitation. ERCC (external RNA control consortium) spike-ins were added with the Trizol; 1 μl diluted 1:100,000 to each sample. RNA was resuspended in 5 μl water and CEL-Seq was performed as previously published (Hashimshony et al. 2012), with the exception that different primers were used containing a unique molecule identifier (UMI) and a shorter barcode.
Quantitative RT-PCR analysis
A Power SYBR Green RNA-to-Ct 1-Step Kit was used to perform the quantitative real-time RT-PCR analysis of gene expression levels in the gonad sections. Primer sequences for the genes are: C48B4.10 (Forward: 5′-TTTGCCTACGCCTTCATCTT-3′; Reverse: 5′-CTTGTGTGGTGTCCAAGTCG-3′), C01G5.2 (Forward: 5′-GAAAGGTGATCAACCGCCTA-3′; Reverse: 5′-CTTCTTCTCCCTTGCCATTG-3′), R04D3.3 (Forward: 5′-AAGCCACGGAAGACACAGAG-3′; Reverse: 5′-TCCAGGTCAGACCAGTCCTC-3′), and F08F3.6 (Forward: 5′-GACTTCCTCGAATGCCCATA-3′; Reverse: 5′-TCGTGCCAGACAACAAACAT-3′).
Ribosomal density profiling
Synchronized adult worms (24-hr post-L4) were washed with lysis buffer (20 mM Tris pH 8.0, 140 mM KCl, 5 mM MgCl2, 100 μg/ml cycloheximide, 0.5 mM DTT, and 1% Triton X-100) and flash frozen in liquid nitrogen. Frozen worm pellets were homogenized by cryogrinding with a BioSpec cryomill and suspended in the lysis buffer on ice. Lysates were cleared of debris by centrifugation for 20 min at 14,000 rpm and digested with RNAse I (Life Technologies) for 1 hr at room temperature. The monosome collection and ribosome footprint library were prepared as in Labunskyy et al. (2014). Briefly, digested lysates were loaded on linear 10–50% sucrose gradients and centrifuged for 3 hr at 35,000 rpm using a SW-41 Ti rotor. Gradients were fractionated and the fraction representing the monosome peak was collected. Ribosome-protected mRNA fragments were released with a release buffer [20 mM Tris pH 7.0, 2 mM EDTA, and 40 U/ml Superasein (Life Technologies)]. The obtained footprints were used to generate sequencing libraries with the ARTseq Ribosome Profiling Kit (Epicentre) by following the manufacturer’s instructions. High-throughput sequencing for ribosome profiling was performed using the Illumina platform. Reads were trimmed of the first nucleotide and the adapter sequence. Resulting reads > 23 nt were used for mapping. In-house Perl scripts were used to prepare reference databases. A total set of predicted/verified coding sequences from the WormBase WS246 release with 18 nt of 5′ and 3′ genomic flanking sequences were used as reference sequences. For transcripts with multiple isoforms, the longest isoform with the earliest start codon was selected. For transcripts encoding the same protein, one transcript was selected arbitrarily. Reads were mapped to the reference data with Bowtie v.1.1.1 (Langmead et al. 2009). Reads mapped to unique sites were reported with two mismatches allowed.
The same worm lysates used for ribosome profiling were used to isolate mRNA for RNA-seq analysis. Briefly, RNAs were first extracted by hot acid phenol:chloroform treatment and mRNAs were then isolated with a Dynabeads mRNA direct kit (Life Technologies) following the manufacturer’s instructions. Purified mRNA was fragmented and used to generate sequencing libraries. High-throughput sequencing was performed and reads were aligned to the same set of genes that were used for ribosome profiling mapping with Bowtie v.1.1.1. Reads mapped to the 3′-end 300 nt of the transcripts (WormBase WS246 release) were used to calculate mRNA abundances.
To obtain normalized intensity levels for different expression profiles, the ribosome densities were normalized to transcript expression levels and were averaged across two independent experiments.
Bioinformatics analyses
CEL-Seq initial analysis pipeline:
Transcript levels were obtained from sequencing data using a custom computational pipeline. Briefly, after trimming and filtering, barcoded reads were demultiplexed to different samples/gonad sections. For each sample, reads were mapped to the C. elegans genome release WS230 using bowtie2 (Langmead and Salzberg 2012) and counted using htseq-count (Anders et al. 2015), while correcting for read amplification bias using UMIs (Supplemental Material, Tables S1 and S9 for hermaphrodite and male, respectively). Transcript counts were then normalized to the total number of counted reads in a specific sample and multiplied by 105 (mean counted transcripts across sections) (Tables S2B and S10B for hermaphrodite and male, respectively). Finally, transcript levels were obtained by averaging expression across the two independent samples (Tables S2A and S10A, respectively).
Generation of expression profiles and profile-specific gene sets:
For all analyses, genes were defined as expressed if their transcript abundances at any gonad section were ≥ 2 and they were dynamically expressed with a fold difference across sections of ≥ 2. To identify specific expression profiles for in-depth analysis, we used hierarchical clustering of log2-transformed gene expression data identifying groups of coexpressed genes (Figure 2 and Table S3). In addition, we cross-referenced with publicly available in situ gene expression data for the hermaphrodite gonad (NEXTDB: nematode.lab.nig.ac.jp/, see main text). This procedure resulted in the definition of seven specific expression profiles with the following binary representations: “1000000000,” “1100000000,” “1111110000,” “0011110000,” “0011111111,” “0000011100,” and “0000001111” (1 indicates high expression, 0 indicates low expression). Four specific expression profiles for the male gonad were determined based on expression data clustering with the following binary representations: “1111000000,” “0000111100,” “0000000010,” and “0000000001.” Dynamically expressed genes were annotated to the binary profiles using Pearson’s correlation. Genes that did not correlate well (< 0.5) with any of the profiles were not associated with an expression profile.
Figure 2.
Dynamic expression of genes in the hermaphrodite gonad. (A) Left: hierarchical grouping analyses of dynamic germline gene expression profiles using Pearson’s correlation. The top 20 gene groups are numbered and demarcated along the left of the y-axis. Right: standardized gene expression levels (gene expression levels were mean subtracted and divided by the SD). Genes are arranged in the same linear order of the clusters on the left (y-axis) and gonad sections (X). The two major groups are indicated by black rectangles. Dynamically expressed genes are inversely correlated to each other (r < 0) if their profiles contain expression in opposite gonad sections. (B) Pairwise profile correlation coefficients among gonad sections. (C) Standardized gene expression levels along the gonad (sections 1–10) in autosomes compared to the X chromosome.
Gene ontology enrichment analysis:
For each of the determined expression profiles (see above) we tested for gene ontology (GO) term enrichments using the hypergeometric distribution with a P-value ≤10−5.
Ribosome loading and density analyses:
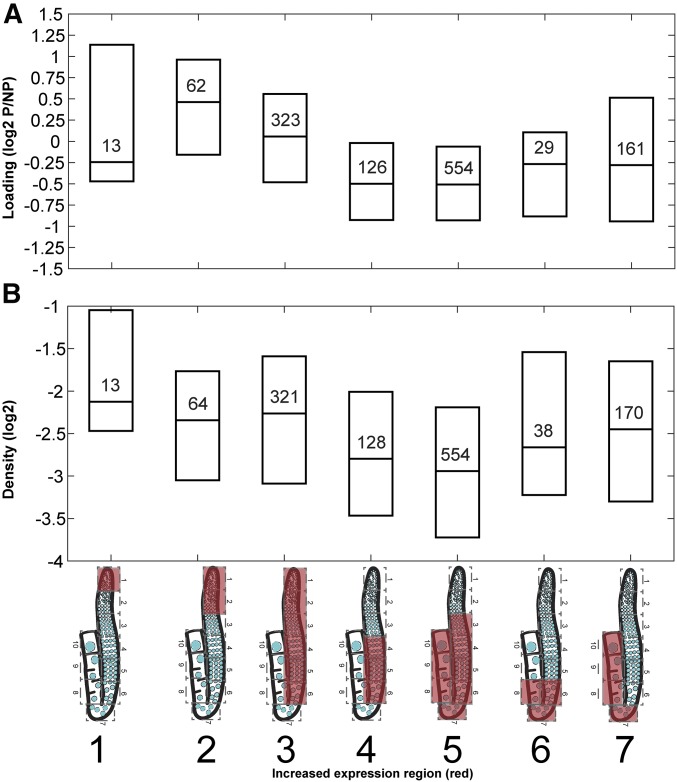
The fraction of transcript bound to polysomes was obtained using adult worms, as described in Nousch et al. (2014), and examined for germline-enriched genes only. Figure 4 depicts boxplots (25th, median, and 75th quantile) of the distribution of polysome loading (upper panel) and density (lower panel) for germline-enriched genes [“Figure 1Bi wt vs glp-4 enriched genes.txt” in Reinke et al. (2004)] associated with different expression profiles.
Figure 4.
Translational spatiotemporal changes. Polysome-associated vs. -free mRNAs of germline-enriched genes from (A) whole-worm polysome data published by Nousch et al. (2014), restricting our analysis to germline-enriched genes (Reinke et al. 2004), and (B) ribosome density (this study) in different expression profiles of hermaphrodite gonads. Boxplots indicate 25th, median, and 75th quantile. The number of genes is indicated within each box. Expression profiles are indicated as cartoons of the gonads with red covering the region of high gene expression. Gene cluster numbers are indicated below the cartoons.
Transcription factor analysis
We associated each of the C. elegans expressed genes with promoters containing a set of strong transcription factor (TF)-binding motifs present in the Cis-BP database (Narasimhan et al. 2015; FIMO (Find Individual Motif Occurrences) P-value ≤ 10−5 in the region −500 relative to the transcriptional start site). We then tested whether each of the 292 annotated TFs overlapped significantly with specific expression profile genes using the hypergeometric distribution (P-value ≤ 10−4).
Comparison of the UV- with the IR-based collection methods
Collection:
For both methods, 20–24-hr post-L4 adult hermaphrodites were placed onto 30 μl of Egg Buffer (118 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2, and 0.025 mM HEPES pH 7.4) on glass slides. Next, 2 to 6 gonads were dissected per slide and transferred to new slides (PEN membrane slides for UV and glass slides for IR). Excess buffer was removed, and slides were frozen on dry ice and kept at −80°. To capture the sections with IR, slides were thawed and dehydrated by using the following sequential washes: 2 min in 70% ethanol, 1 min in 95% ethanol, 1 min in 100% ethanol, and then 5 min in xylene. Dried slides were loaded onto the ARCTURUS VERITAS LCM, and the distal–proximal axis of the gonad was measured from the distal tip to the end of the −1 oocyte. CapSure HS LCM Cap (cat # LCM0214; Life Technologies) was loaded (for each desired section) and one-tenth of the gonad length was measured. The cap was adhered to the section by 2–8 IR laser melt points (power = 70–100 mW, pulse= 1000–3000 μsec, and intensity = 100–110 mV), and then that section was isolated (note that the UV laser is not used in the case of the IR method; for comparison see “Isolation of gonad segments” above). We verified under the microscope that the entire desired section was attached to the cap, and if any remnants bigger than 10% the size of that section were still visible, we repeated the process above to capture these remaining pieces on the same cap. Caps were retrieved, immediately placed into 0.5 ml sterile tubes, and kept in dry ice. Only one gonad was collected from each slide; two gonads (biological replicates) were used in the IR analysis and two for the UV analysis.
Bioinformatics analysis method:
To infer the quality of the IR collection method for each replicate, we measured the level of pairwise correlations among the 10 sections. While an overall high (Pearson’s r > 0.9) correlation was observed for adjacent gonad sections, two of the sections of the first replicate (# 4, 7) and two of the sections of the second replicate (# 1, 2) showed consistently lower levels of correlation with other sections, and were excluded from further analysis.
Transcript levels were obtained from sequencing data using the same protocol described for the UV-based collection method, except that for sections 1, 2, 4, and 7 data were generated using a single replicate rather than averaging across two.
Correlation between the two collection methods:
Supporting the high quality of our UV-based gonad data set, we also found high correlations with gonad sections obtained using a different method (IR method for gonad dissection discussed above). We found overall high correlations both when comparing gonad expression profiles across sections (Figure S3, B and C) and across genes (Figure S3D). In addition, we looked at several specific dynamic genes with broad expression ranges and found high correlations between their profiles using either method of gonad section isolation (Figure S3E).
Data availability
Strains and reagents are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and supplemental files. File S1 contains the list of the mapped reads number of all genes in each hermaphrodite gonad section. File S2 contains the normalized expression of all genes in each hermaphrodite gonad section. File S3 contains the normalized expression of dynamic genes in each hermaphrodite and male gonad section. File S4 contains clustered hermaphrodite and male genes in the defined expression profiles. File S5 contains the analysis of the comparison between NEXTDB in situ images to spatiotemporally resolved germline gene expression data. File S6 contains the list of X-linked genes expressed in the first five sections of the hermaphrodite gonad. File S7 contains a list of clusters with the illustrative spatiotemporal representation of high (red) and low (white) expression, binary designation, and main gametogenic processes. File S8 contains ribosome profiling data. File S9 contains a list of the mapped reads number of all genes in each section of the male gonad. File S10 contains normalized expression of all genes in each section of the male gonad. File S11 contains lists of the shared and specific hermaphrodite- and male germline-expressed genes. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6726293.
Results
A map of gene expression with spatiotemporal resolution for the C. elegans gonad
We dissected sequential sections from the C. elegans hermaphrodite gonad for gene expression analysis by using the ARCTURUS VERITAS LCM (Gallagher et al. 2012). We microdissected gonads from young adult worms (20–24-hr post-L4) and transferred them to PEN membrane slides. The slides were mounted on the LCM, and sequential segments, each equivalent to one-tenth of the gonad’s total length were cut from each gonad and sealed with a UV laser (hence sections 1–10). Although each section contains similar biomaterial volume, the number of nuclei varies from dozens in section 1 to a single nucleus in the last two sections due to the increase in oocyte volume during oogenesis (Greenstein 2005). Each section of the gonad was isolated or extracted by adhering the collection caps onto the membrane at specific positions with an IR laser (Figure 1B and see Materials and Methods).
RNA was extracted from each section and the levels of polyadenylated RNAs were analyzed by CEL-Seq (Hashimshony et al. 2012) (Table S1). Individual sections from duplicate biological samples showed a high level of correlation between samples and consecutive sections (Figure 2B, Figure S1, and Figure S2). We detected 7019 genes expressed in all 10 combined sections extracted from these gonads (Tables S1 and S2). Expression of four genes was validated by quantitative RT-PCR analysis for sections 2 (premeiotic region) and 9 (diakinesis) (Figure S4A). Specifically, all four genes exhibited the same expression patterns as detected by CEL-Seq, with two showing upregulated expression in the distal portion (C48B4.10 and C01G5.2) and two exhibiting upregulated expression in the proximal region of the gonads (F08F3.6 and R04D3.3). Next, to test if our data sets encompass a significant cohort of germline-expressed genes, we compared the genes in our list to those detected in previously reported transcriptome analyses of the entire gonad. In total, 60% of the genes expressed in our samples were also detected by Wang et al., who used SAGE to identify germline-expressed genes, while 10% of the genes detected in that study were not identified in ours (Wang et al. 2009). Comparison with the whole-gonad high-throughput sequencing analysis data from Ortiz et al. (2014) showed that 91% of our gene set was also detected in that study, while our segmented analysis did not identify 40% of the genes that they detected (Ortiz et al. 2014).
Our success in detecting a high level of mRNA in small biological samples encouraged us to use it for analyzing gene control during gametogenesis, while taking under consideration the inherent potential caveats of this methodology. Our method is built on utilizing small biological samples, and therefore low or transient changes in mRNA levels may be undetectable for some genes in the gonad, even if they do have important roles. This may be the case for sygl-1 and lin-41, which are required to maintain germline stem cell and oocyte growth (Kershner et al. 2014; Spike et al. 2014), and yet were not detected in our analysis. Also, the CEL-Seq method uses the poly-A tail for enrichment of the transcripts, and therefore the level of genes that undergo exceptional poly-A processing (e.g., gld-1) may also be misrepresented in our databases. Like previous gonad transcriptome projects (Reinke et al. 2000, 2004; Wang et al. 2009; Ortiz et al. 2014), it is also possible that our analysis includes transcripts that originate from the thin layer of the somatic gonadal sheath cells (Hall et al. 1999), yet due to the small volume of these somatic cells compared to the germline, we estimate that the number of these genes is limited to a small subset that have high expression in the somatic gonad, mostly at the proximal end (see Discussion).
To verify our results, we also collected gonad sections using a different approach (see Materials and Methods) in which the gonads were first completely dehydrated, and then the LCM caps were adhered to the various segments of the gonad using IR laser, therefore pulling the segments along with them when lifted. In this method, no ionizing UV laser is utilized. CEL-Seq analysis of 10 sequential segments showed very similar results to those obtained with the LCM/UV laser method of retrieval (see Materials and Methods, Figure S3). In particular, we found that the average correlation across segment replicates is 0.93 and that the median correlation coefficient between a gene’s profile across the two methods is 0.76. In addition, quantitative RT-PCR analysis comparing the expression of four genes between sections isolated by both methods revealed mostly similar levels of expression, regardless of the method of section isolation (Figure S4, A and B). Taken together, the comparisons with other studies examining germline gene expression, and the different retrieval methods tested, show that our novel approach is able to detect the expression of most genes in the gonad sections that we isolated. Since using the LCM/UV laser resulted in more accurate and reproducible results, all further results described herein were achieved by retrieving gonad sections via this method.
Most hermaphrodite germline-expressed genes show a dynamic expression that changes by late pachytene of meiotic prophase I
To determine whether there are dynamic changes in the transcriptome, we compared the relative mRNA levels of all the genes between the different sections isolated from the gonads. We found that 6054 of 7019 (86%) genes whose expression could be detected exhibit a dynamic pattern (more than a twofold change, herein referred to as dynamic expression) along the gonad axis (Figure 2A and Table S3). This result suggests that although hundreds of germline nuclei contribute their cytoplasmic content to a cytoplasmic core, most of the genes’ mRNAs are present at specific regions of the gonad at varying levels. To validate this result, we compared the top 105 most dynamic genes in our list exhibiting different expression profiles to in situ hybridization data from the nematode expression pattern database (NEXTDB: nematode.lab.nig.ac.jp/). Although the in situ signal is not quantitative, and the population used in that analysis was not age-matched to our samples in that their images show combined L4 and adult worms while we strictly analyzed young adults, we found that 82% (74/90) of the genes exhibiting changes in signal levels along the gonad based on our analysis also showed a germline-enriched change in the signal by in situ hybridization. Moreover, 76% of those showed a similar pattern to the one detected by our cluster designation (± 2 sections; Figure 1C, Figure S5, and Table S5).
We further validated our data set by comparing the expression patterns of genes on the X chromosome to those on the autosomes. Consistent with previous observations that unlike the autosomes, the X chromosomes lack histone post-translational modifications associated with active transcription until the end of pachytene (Kelly et al. 2002), we found that > 92% of the genes on the X chromosome exhibit negligible or low mRNA levels (defined as having less than two mapped reads) between the proliferative mitotic region (section 1) and the late pachytene region (section 5). Previous studies showed that de novo transcription drops during diakinesis (Gibert et al. 1984; Schisa et al. 2001; Kelly et al. 2002; Sheth et al. 2010), yet examples of RNA in situ data for several genes (including genes on the X chromosome) show that their transcripts are present at elevated levels in diakinesis (e.g., Kelly et al. 2002 and NEXTDB images of sec-3). These mRNAs were probably transcribed during pachytene and diplotene, and were either transported via cytoplasmic streaming or maintained during the later stages of prophase I (Wolke et al. 2007). This is in line with our findings showing that the relative expression levels of many genes located on the X chromosome increase at late pachytene and diplotene, and are maintained during diakinesis (Figure 2C). Surprisingly, we found that 19 of the 2868 X-specific genes (0.66%) were significantly expressed in sections 1–5 (Table S6), indicating that some genes may escape this chromosome-wide silencing. Thus, our comprehensive spatiotemporal analysis of germline gene expression shows extensive dynamic changes of mRNA levels throughout the C. elegans hermaphrodite syncytial gonad.
To find how the change in mRNA repertoire correlates with germ cell progression, we performed hierarchical grouping analyses of gene expression profiles using Pearson’s correlation coefficient. This unbiased grouping analysis revealed that most genes (> 70%) fall into two main groups with a reciprocal pattern of mRNA levels: the mitosis/early meiosis group and the diplotene/diakinesis group (groups 2 and 4, respectively, Figure 2A and Table S3). Genes in the first group show high levels of mRNA in the proliferative mitotic region (sections 1 + 2), the leptotene/zygotene region referred to as the transition zone (sections 2–3), and the pachytene region (sections 3–5), and their expression decreases as they progress toward the proximal end. Not surprisingly, genes involved in key steps taking place during early prophase, such as formation of the chromosome axes required for synaptonemal complex assembly (e.g., htp-1, htp-2, htp-3, rec-8, syp-1, and cra-1) and the DNA damage response (atl-1 and hus-1) fall into this group. The mRNA levels for the genes in the second group were low in sections 1–5, but increased toward late pachytene and diplotene (section 6), and remained elevated all the way through to the mature oocyte (section 10). This group included genes shown to be required for late meiotic events such as cell cycle progression (e.g., cyb-1, cyb-2.1, cyb-3, cdk-1, cdk-4, cyb-2.2, ify-1, and fzy-1), apoptosis (cep-1), chromosome segregation (air-2 and lab-1), meiotic spindle organization (mei-1, mei-2, and bub-3), polar body extrusion (ani-1), and yolk particle uptake (rme-2). Surprisingly, this group also included genes that are involved in DNA double-strand break repair from both the homologous recombination and nonhomologous end joining pathways (e.g., gen-1, msh-5, cku-80, mre-11, brc-2, and rpa-2), which suggests their requirement in late meiotic events. Other smaller groups with more distinct expression were consistent with specific oogonial roles. For example, among the genes in group 20, which included genes with high mRNA levels in section 6 (late pachytene–early diplotene), we found egl-1. Section 6 is the region in which the DNA damage checkpoint is activated (Ye et al. 2014), as underscored by our observations of elevated levels of egl-1, the most upstream core apoptotic machinery activator. The difference between the expression of cep-1 (group 2), which mediates cell cycle arrest and the early apoptotic commitment stage, and egl-1 (group 20), which initiates the activation cascade of CED-3, highlights the difference in the control of those two stages in the germ cell apoptosis pathway. This analysis suggests that gene expression elements act to shift most of the hermaphrodite germline-expressed genes such that they are either upregulated or downregulated toward late pachytene, and that a different control is exerted to regulate expression for some genes that are required for more specific roles.
Correlation between expression profile and cellular roles
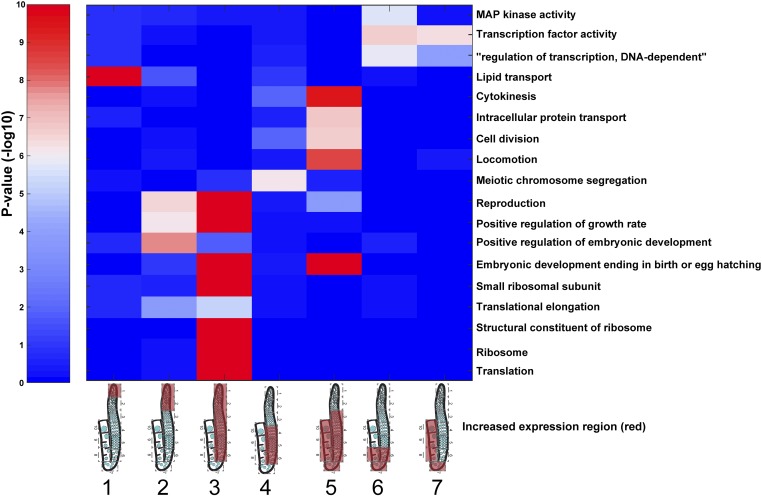
To better characterize gene expression dynamics across the hermaphrodite germline, we examined smaller clusters of genes with specific expression profiles. Based on the hierarchical clustering of genes and developmental stages, we designated seven profiles of expression and assigned their best-matching genes. Unlike the unbiased grouping (Figure 2), these clusters include genes exhibiting high expression in regions with defined biological processes: cluster 1: stem cells and mitosis; cluster 2: mitosis and very early stages of meiotic prophase I (leptotene/zygotene); cluster 3: mitosis, meiotic leptotene/zygotene, pachytene, and early diplotene; cluster 4: pachytene and early diplotene; cluster 5: pachytene, diplotene, and diakinesis; cluster 6: exit from late pachytene, diplotene, and early diakinesis; and cluster 7: late diplotene and all of diakinesis (Table S4 and Table S7, see Materials and Methods). We found that each of these clusters is enriched with specific GO terms (Figure 3 and Figure S6A). For example, a significant percentage of the genes with high mRNA levels in cluster 2 (sections 1 and 2), which encompasses mostly the mitotic proliferating region, were annotated as “positive regulation of embryonic development” unique to this profile, and with terms shared with genes expressed in cluster 3 (sections 1–6) such as reproduction and “positive regulation of growth rate.” In contrast, the genes exhibiting high expression in all meiotic prophase I sections (cluster 5, sections 3–10) were enriched for GO terms associated with late meiotic and embryonic processes (e.g., “cytokinesis,” “intracellular protein transport,” cell division, “locomotion,” and “embryonic development ending with birth”), indicating that the mRNA required for late stages of meiosis and as maternal contribution to the early embryo start to accumulate at the earliest stages of meiosis. The only GO term specifically enriched in cluster 1 (section 1) was “lipid transport,” and closer examination showed that the genes associated with this term are genes encoding for yolk proteins (VIT-1–6, Table S4). These vitellogenin proteins are highly expressed in the gut and transport lipids to the maturing oocyte (sections 9–10). A technical explanation for why we detected the vitellogenins’ mRNA in section 1 comes from our workflow procedure. We dissected adult worms in a small amount of buffer and transferred the gonads along with some buffer onto new slides. Most probably, these highly expressed mRNAs spilled from the gut during the dissections, and therefore were transferred with the residual carried over buffer and adhered to the membrane of the new slide. The first cap that was used to isolate section 1 may have also picked up this contamination, leading to this result. However, we believe that the influence of this contamination to our analyses is minor since: (1) we hardly detected vit-1-6 in the following sections and (2) most of the mRNAs we detected were also detected in previous whole-gonad analyses (see above).
Figure 3.
Gene ontology terms associated with genes expressed at specific regions of the hermaphrodite germline. For each of the indicated expression profiles (columns), the enrichment was computed for the genes expressed with that profile and the genes annotated to the indicated gene ontology categories (rows). For simplicity, only a subset of gene ontology terms with enrichment (hypergeometric distribution, see Materials and Methods) of less than P ≤ 10−5 for at least one profile are shown in the plot (full sets are shown in Figure S6). Profiles are indicated as cartoons of gonads, with red covering the high-expression region. Gene cluster numbers are indicated below the gonad cartoons. Color indicates the P-value of the enrichment with blue indicating low and red indicating high levels of enrichment.
Several enriched GO terms observed in the different profiles render further support to control mechanisms working in specific regions of the germline. The diplotene and diakinesis regions (cluster 6 and 7, sections 6–8 and 7–10, respectively) are enriched with genes annotated for TF activity, yet it is not clear if these are actively translated and act to change transcription. The exit from the late pachytene, diplotene, and early diakinesis region (cluster 6, sections 6–8) is also enriched with genes annotated for MAP kinase activity, which have been shown to control the execution of late meiotic stages (Hajnal and Berset 2002; Sundaram 2006; Hayashi et al. 2007; Lee et al. 2007b; Lopez et al. 2013). Interestingly, “meiotic chromosome segregation” is the only term enriched in the region encompassing pachytene and early diplotene (cluster 4, sections 4–6). This raises the question as to why regions of the germline where earlier stages of meiosis take place would be enriched for the expression of genes required for a step of meiosis that occurs after section 10? Closer examination revealed that this may be the result of erroneous annotation, since many of the genes included in this profile, and annotated with this term, are known to act in early meiosis where they are required to ensure the accurate chromosome segregation that ensues after section 10: syp-2, syp-3, htp-2, htp-3, zhp-3, him-8, him-5, him-17, rad-51, atl-1, and mre-11. Another explanation comes from the fact that de novo transcription is being shut down at late diplotene and diakinesis (Gibert et al. 1984; Schisa et al. 2001; Kelly et al. 2002; Sheth et al. 2010), and thus high levels of transcripts required for later stages (diakinesis) have to be produced at earlier stages (pachytene–diplotene). Taken together, the GO annotation analysis shows that different regions in the gonads are enriched with transcripts with distinct biological roles. Moreover, the annotations further support the quality and precision of the information that can be derived from the strategy applied in this current study, given the high concordance between the GO term enrichments observed for specific regions of the gonad and the mechanisms known to control germline processes at those regions.
Different levels of post-transcriptional regulation may be implemented throughout different regions of the germline
Previous studies suggesting that translational control plays an important role in oocyte development (e.g., Nousch et al. 2014), as well as the overrepresentation of ribosomal and translational GO terms identified for transcripts in cluster 3 (mitosis and meiotic leptotene/zygotene, pachytene, and early diplotene stages; Figure 3), prompted us to test how much translational control may contribute to the germline gene expression profiles that we observed. Therefore, we evaluated the level of the mRNA undergoing active translation analyzed in our samples. Since it is currently not possible to pull down sufficient polysomes for translation analysis from small biological samples, we first used the whole-worm polysome data published by Nousch et al. (2014), restricting our analysis to germline-enriched genes (Reinke et al. 2004). We examined the level of ribosome loading to transcripts of different expression profiles by sampling the ratio of polysome-bound to -free mRNAs (Nousch et al. 2014) (Figure 4A). Using this measure, it was previously suggested that germline-expressed genes are poorly translated (Nousch et al. 2014). Indeed, we found that the germline transcripts from most profiles exhibited low association with ribosomes (clusters 1 and 4–7, < 0), suggestive of lower translational activity. Cluster 3 (mitosis, meiotic leptotene/zygotene, pachytene, and early diplotene), and especially cluster 2 (mitosis and leptotene/zygotene of meiosis I), exhibited higher levels of association with polysomal fractions. Removing all transcripts associated with GO terms such as “small ribosomal subunit,” “structural constituent of ribosome,” “ribosome,” “translation,” and “translation elongation” from consideration did not change the polysome association ratio of cluster 2 (data not shown). These results suggest that although genes expressed in the germline have lower translational activity than in the soma (Nousch et al. 2014), in some regions of the gonad there may be a higher level of polysomal association.
Next, to further validate the polysome loading analysis utilizing previously published data, and to gain more insight into the level of active translation in the gonad, we performed polysome density analysis by doing ribosome profiling coupled with RNA-seq analysis for samples that we prepared from adult hermaphrodites (whole worms). We retrieved 4 million reads that aligned to ∼12,000 genes (Table S8), and by once again limiting our analysis to germline-enriched genes (Reinke et al. 2004) we found similar trends compared to the polysome loading analysis (Figure 4B). We also examined whether most genes in the same cluster have similar polysome binding and/or density. We found that most of the genes did not show high variability and that 50% of the genes exhibited a difference of less than an order of magnitude within the same cluster (Figure 4). Taken together, we found that despite the low level of translation in the germline, there are some minor spatiotemporal differences within the gonads as well as relatively limited variability within specific sections. However, it is unclear how much the region-specific translational control observed accounts for some of the dynamic changes in gene expression observed throughout the germline.
Specific TF targets are spatiotemporally coexpressed in the gonad
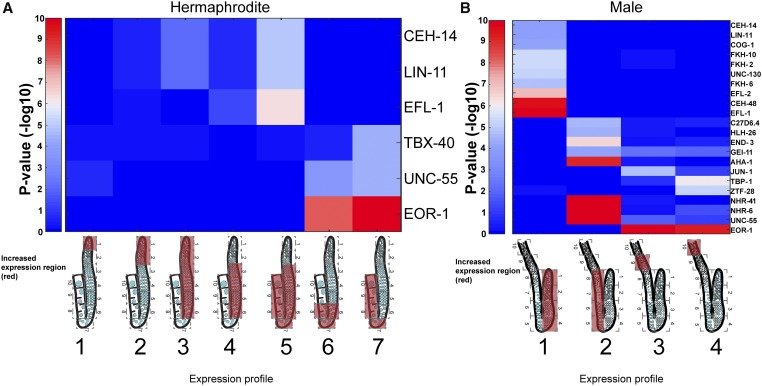
Our observation of a transcriptional shift occurring in the C. elegans gonad led us to ask how such boundaries are established and maintained. Some reports suggested that gene expression in the gonad is mostly governed by post-transcriptional control (e.g., Merritt et al. 2008). Although our data set does not contradict this model, it can be used to search for evidence of specific transcriptional control elements that may take part in driving germ cell progression. Studies in yeast suggest that a series of TFs drive meiosis and sporulation in that organism (Chu et al. 1998; Primig et al. 2000). To explore if some TFs are also involved in driving meiosis in C. elegans, we checked whether known TF targets are expressed in the same cluster in the gonad. Specifically, we used the characterized recognition motifs of 292 C. elegans TFs (Narasimhan et al. 2015) to test whether transcripts that are expected to be controlled by these motifs are enriched in our defined expression clusters (Figure 5A). We found an enrichment of the predicted targets for several TFs in specific expression clusters. Among these TFs, the strongest correlation with spatiotemporal expression was with EFL-1 and EOR-1. EFL-1 targets were enriched with genes that are expressed throughout meiotic prophase I (cluster 5), but they were not enriched in profiles with earlier expression or more limited spatiotemporal expression (Figure 5A). This result is not surprising given the known roles of EFL-1 in oogenesis and early embryogenesis (Chi and Reinke 2006), as well as the phenotypes observed upon the depletion of efl-1 by RNA interference (RNAi), which include embryonic lethality, a phenotype commonly associated with meiotic defects, as well as fewer germ cells, gonad development variation, and sterility (Lin and Reinke 2008). The predicted targets for EOR-1 consisted of genes that were highly enriched in diplotene through diakinesis (clusters 6 and 7, sections 6–10). Depletion of eor-1 in mutants for various other genes was reported to result in sterility and embryonic lethality (Lehner et al. 2006). Nevertheless, we cannot rule out the possibility that expression of these genes arises from the somatic gonad (see Discussion). Taken together, these results suggest that TFs may act during oogenesis to upregulate gene expression at different stages and patterns, but unlike the sporulation transcription pattern of yeast (Chu et al. 1998; Primig et al. 2000), we could not detect a simple consecutive TF program.
Figure 5.
Transcription factor targets are enriched at specific germline regions. Significant enrichments (P-value ≤ 10−4) of expression of transcription factor targets within the germline gene expression profiles. Scale: 10 indicates high expression, 0 indicates low expression. (A) Hermaphrodite. (B) Male. Expression profiles are indicated as cartoons of the gonads with red covering the regions of high gene expression. Gene cluster numbers are indicated below the cartoons.
The male gonad shows a shift in mRNA abundance by entrance into pachytene of meiotic prophase I
We next examined the male gonad, which is also mostly syncytial, with nuclei that are laid in a linear fashion from the mitotic proliferative region into meiotic stages and finally differentiate into sperm (L’Hernault 1997; Shakes et al. 2009; Chu and Shakes 2013) (Figure 6A). The first developmental stages of the male gonad are similar to the hermaphrodite gonad, but their relative physical length is different (Shakes et al. 2009; Chu and Shakes 2013; Figure 6A). Also, nuclei following pachytene in the male gonad undergo dramatically distinct differentiation stages to produce haploid spermatozoa [Figure 6A and Figure 8, reviewed in Chu and Shakes (2013)]. We dissected male gonads and analyzed gene expression levels along 10 sequential sections, similar to our hermaphrodite analysis. Once again, individual sections from duplicate biological samples showed a high level of correlation between samples and the consecutive sections (Figure 6B and Figure S1B), yet these correlations were lower than observed for the hermaphrodite samples. This could be a result of it being technically simpler to detect the end of the −1 oocyte in the hermaphrodite gonad compared to the more challenging detection of the end of the vas deferens in males. We detected 8052 genes expressed in all 10 combined sections (Table S3, Table S9, Table and S10). Comparison between the genes in our list and the data set of Ortiz et al. (2014) showed that 88% of the genes that we found expressed in our samples were also detected in that data set, while we did not detect 33% of the genes detected in that study (Ortiz et al. 2014). These tests show that the spatiotemporal analysis of male-expressed genes, similar to our analysis of gene expression in the hermaphrodite germline, is able to detect the majority of the mRNA levels along the gonad.
Figure 8.
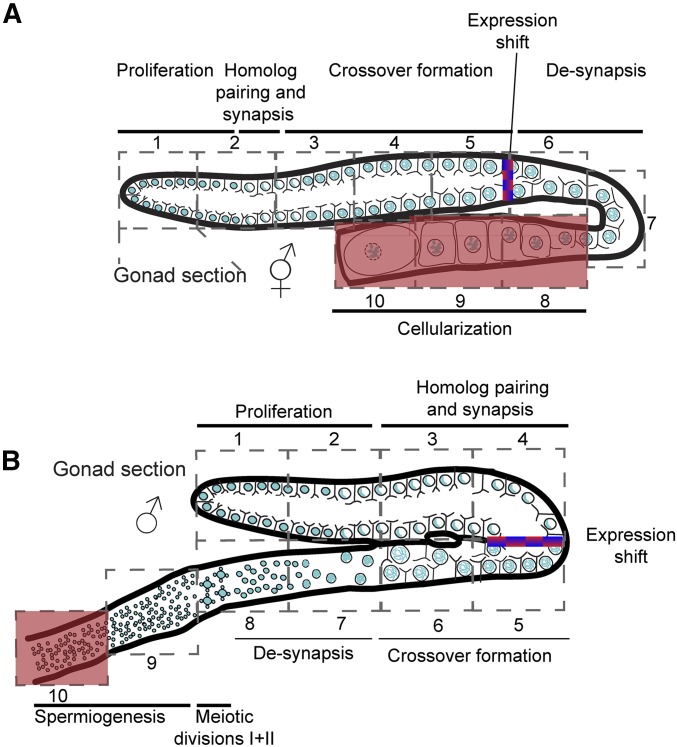
Spatiotemporal processes in the male and hermaphrodite gonads. Illustrations of the hermaphrodite (A) and male (B) gonads. Sections and corresponding major process are indicated. The area in which most of the mRNA levels shift is indicated as a red/blue gradient. The region in which de novo transcription was reported to decrease (Kelly et al. 2002) is highlighted in red.
To identify both the shared and specific genes operating during germ cell progression in the two sexes, we compared all the genes for which we detected expression in our hermaphrodite and male gonads and found 5633 shared, 1386 oogenesis-specific, and 2419 spermatogenesis-specific genes (Table S11). We also correlated individual sections from our male and hermaphrodite analyses, taking into account that the same developmental stages are not always present in the same cut between these two sexes, given that meiotic progression unfolds differently throughout these gonads and the presence of two postmeiotic differentiation sections in the male gonad. We found that the gene expression profiles for the first three sections in the male gonad (mitosis and early meiosis) highly correlate with the gene expression profiles observed for the first five sections of the hermaphrodite gonad, and to a lesser extent with the last five sections of the hermaphrodite gonad. This result shows that the proliferative zone and early meiotic steps of both the hermaphrodite and male germlines are similar at the mRNA level. In contrast, the last six sections of the male gonad (sections 5–10) are markedly different from any section in the hermaphrodite gonad (Figure 6B). These results show that the mRNA cohort of the distal parts of the male gonad is similar to the entire cohort of the hermaphrodite gonad, but after that point it diverges and a different gene expression pattern is adopted.
Comparing the expression pattern and unbiased grouping of genes in the male gonad showed that, similar to the hermaphrodite gonad, most genes fall into two main groups: genes with high expression in mitotic and early meiotic sections (sections 1–4, group 8 in Figure 6C), and genes with high expression from pachytene to the end of prophase I (sections 5–8, group 6 in Figure 6C). Two smaller groups included genes with high expression in early spermatids (section 9, group 12 in Figure 6C) and in the late spermatid region (section 10, group 4 in Figure 6C). A previous study suggested that gene expression changes also occur during pachytene in spermatocytes, as well as in spermatids and even spermatozoa in mice testes (Soumillon et al. 2013), indicating that these shifts may be evolutionarily conserved. The four major expression profiles we found in the worm male gonad were also enriched with specific GO term annotated groups (Figure S6B). In agreement with previous reports, we found that genes mapping to the X chromosome were mostly silent in the male gonad (Figure 6D and Kelly et al. 2002), yet > 300 genes encoded on the X chromosome had significant expression in at least one section. Most strikingly, the expression of > 200 genes on the X chromosome was restricted only to sections 9 and 10, albeit these could also be transcribed from the somatic gonadal vas deferens (see Discussion).
High expression of specific TF targets was again enriched among the four major expression profiles (Figure 5B). Similar to the hermaphrodite gonad, EOR-1 and EFL-1 targets were also expressed in the male gonads, but exhibited spatiotemporal differences in expression compared to the hermaphrodite gonads. While EFL-1 targets are expressed throughout all meiotic regions during oogenesis, they were limited to the mitotic/early meiotic cluster in the male gonad. While EOR-1 targets are expressed in mid–late prophase I regions during oogenesis, they are enriched postmeiotically in the male (compare Figure 5, A and B). Thus, our results suggest that some TFs are needed for general germ cell progression, but that the way the specific male or hermaphrodite gene expression programs are executed probably depends on other regulatory elements that affect both transcription and translation. Interestingly, the murine homolog of efl-1, E2F1, was shown to be expressed in spermatogonia and early meiotic stages in mouse testes (Rotgers et al. 2015), once again pointing toward evolutionary conservation of our findings. These results show that a shift in gene expression occurs by either early or late pachytene in the male and hermaphrodite gonads, respectively, and that the male also employs tight mRNA control in the spermiogenesis region, leading to sharp transitions in mRNA cohort profiles.
Spatiotemporal expression of genes corresponding to CSR-1-associated 22G RNAs
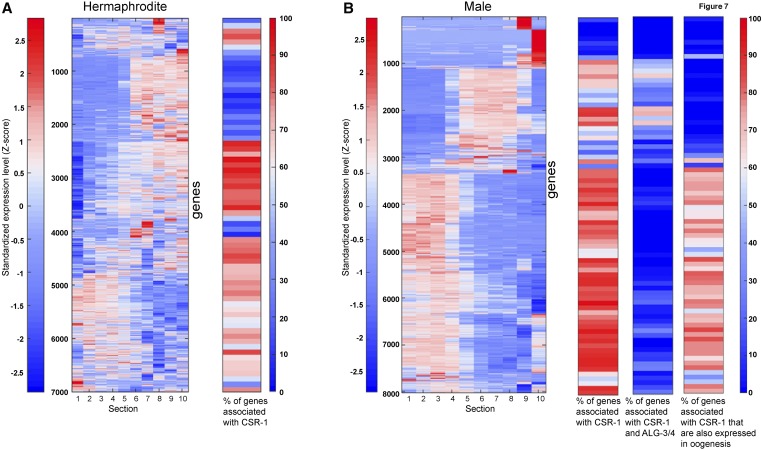
CSR-1 (Chromosome Segregation and RNAi-deficient-1) is an Argonaute protein that binds 22G RNAs that are antisense to genes exhibiting elevated germline expression [reviewed in Wedeles et al. (2014)]. Although CSR-1 has in vitro slicer activity (Aoki et al. 2007), it was shown that the expression of the genes that correspond to the CSR-1-associated 22G RNAs is downregulated in csr-1 mutants (Claycomb et al. 2009). Therefore, it was suggested that CSR-1 plays a direct role in licensing or protecting the transcription of germline-expressed genes via the 22G RNAs (Claycomb et al. 2009; Seth et al. 2013; Wedeles et al. 2013). Our spatiotemporal analysis of germline gene expression allowed us to revisit the possibility that genes that correspond to CSR-1-associated 22G RNAs are silenced in a localized fashion within the gonad, which may have escaped the whole-worm analysis (Claycomb et al. 2009), and that there is a requirement for CSR-1 for germline gene expression. Therefore, we compared the expression of 22G corresponding genes to our spatiotemporal germline gene expression profiles. We found that although CSR-1 is expressed throughout the gonad (Claycomb et al. 2009), most of the associated genes were mainly expressed in either the early or late large germline groups (Figure 7A). This result shows that although CSR-1 may license transcription in the germline, other factors also exist to spatiotemporally limit this transcription. Most strikingly > 2000 genes with significant expression in the gonad are not associated with CSR-1 22G RNAs. These are mostly late expression genes encoded from the X chromosome and genes with very limited spatiotemporal expression patterns (Figure 7A), indicating that other control mechanisms can license germline gene expression. Thus, our results show that although CSR-1 may license gene expression in the germline, possibly through binding to genomic sites and increasing transcription (Seth et al. 2013; Wedeles et al. 2013), it is neither sufficient for robust expression nor exclusively required for germline gene expression.
Figure 7.
CSR-1-associated genes are enriched in specific groups. (A) Left: standardized expression of the hermaphrodite genes along the gonad sections. Genes were hierarchically clustered according to their expression profiles. Right: histogram-plot (each bin representing 100 clustered genes) showing the frequency of the genes associated with CSR-1. (B) Left: standardized expression of the male genes along the gonad sections. Genes were hierarchically clustered according to their expression profiles. Right: histogram plots (each bin representing 100 clustered genes) showing the frequency of the genes that are either associated with CSR-1 (left), both CSR-1 and ALG-3/4 (middle), or CSR-1 oogenesis genes (right).
CSR-1 22G RNA expression licensing has been more thoroughly explored in the male gonad (Conine et al. 2013). There, the Argonaute proteins ALG-3/4 engage 26G RNAs to both positively and negatively regulate hundreds of mRNAs, and many of the positively regulated mRNAs also correspond to 22G RNAs bound by CSR-1. This and other data led to a model in which a feedback loop between the two classes sustains this positive expression mechanism, and licenses the expression of the targeted genes in both the male gonad and the next generation (Conine et al. 2013). When we tested the expression profile of genes corresponding to the CSR-1 male 22G RNAs, we found that most did not have significant expression throughout all sections of the male gonad. We found that > 83% of the genes that correspond to CSR-1 22G RNAs belonged to the two major dynamic groups (sections 1–4 or 5–8, Figure 7B). Genes with expression in sections 9 and/or 10 were largely not represented in the 22G CSR-1-associated RNAs, although these may come from the somatic cells (see Discussion). Surprisingly, the cohort of genes that corresponds to both CSR-1- and ALG-3/4-associated RNAs are mostly expressed in sections 5–8, even though ALG-3/4 are expressed in the male gonad until the mature sperm (Conine et al. 2010, 2013) (Figure 7B). As expected from our comparison of gene expression levels for different sections between male and hermaphrodite gonads (Figure 6B), the genes that correspond to CSR-1 22G RNAs and are also expressed in the hermaphrodite were expressed in sections 1–4, and were mostly not shared with the ALG-3/4 genes (Figure 7B). Our results suggest that CSR-1 may only license expression in sections 1–8 in the male gonad, and if a feedback loop with ALG-3/4 is present, it exhibits very limited or specific spatiotemporal expression.
Discussion
Dynamic changes in mRNA levels within a syncytium
Multinucleated cells exist in many multicellular organisms. In some of these syncytial cells (e.g., muscle fibers) the nuclear morphology is generally uniform (Bruusgaard et al. 2003), while in others like the human placenta, the nuclei are observed with different chromatin structures and the transcription levels vary between nuclei (Burton and Jones 2009; Ellery et al. 2009). Although during oogenesis in both Drosophila melanogaster and C. elegans the nuclei do not float in a shared cytoplasm, the oocyte develops while it is connected to a core of cytoplasm, whose contents flow and are shared with other nuclei. This scenario raises a critical question: how can a nucleus differentiate into a specific fate while other nuclei that share mRNA with it are present at a different state?
RNA-seq of biological samples as small as single cells has advanced our understanding of differences in transcription between cells, but applying these methods to microscopically defined syncytial tissues has been challenging. By combining microscopic laser dissection, and adhering techniques with linear amplification and RNA sequencing, we were able to bridge this gap. Using this method, we were able to analyze mRNA levels with spatiotemporal resolution along the axis of the C. elegans gonad and provide a comprehensive view of dynamic gene expression for an entire syncytial organ, as well as complete oogenesis and spermatogenesis differentiation processes. Our analysis provides evidence that clusters of mRNA levels change within the cytoplasmic core at specific regions throughout the germline.
Two nonmutually exclusive models can explain how mRNAs that diffuse within the shared cytoplasm and/or reach an equilibrium via cytoplasmic flow (Wolke et al. 2007) can still be present at different levels at different regions: (1) post-transcriptional control via mRNA degradation could reduce the level of specific transcripts at specific zones of the gonad and (2) constant production in only a specific region can create a gradient of the mRNA levels. The robust prevalence of dynamic expression found in this research and the sharp changes observed in a small subset of those transcripts lead us to suggest that both of these models act within the gonad, with the former model acting to quickly remove a few select transcripts, while more prevalent gradual changes are driven by transcription acting on most of the dynamic genes.
We found that the male gonad shows sharper contrasts in its mRNA cohort among groups of sections (compare Figure 2B to Figure 6B). Also, the mRNA cohort of sections 1–3 in the male is similar to all sections of the hermaphrodite (Figure 6B). We can envision two explanations for these results. It is possible that cytoplasmic flow (Wolke et al. 2007) is stronger in the germlines of hermaphrodites compared to males, and thus the mRNAs from the early stages are present throughout the hermaphrodite, but not in the male, gonad. It is also possible that spermatogenesis, which proceeds faster than oogenesis (Jaramillo-Lambert et al. 2007), requires sharper changes in gene expression, and thus early transcripts have to be eliminated in the male for progression of the developmental program. It would be interesting to test these models to have a deeper understanding of these two similar, yet different, programs.
Analyzing oogenesis through germline gene expression
Similar to other transcriptomic studies of the C. elegans gonads (Reinke et al. 2000, 2004; Wang et al. 2009; Ortiz et al. 2014), our data combine transcripts from both the germline and the somatic gonads. Although, for the most part, the volume of the somatic cells is negligible compared to the germline (Hall et al. 1999), it is possible that some of the genes we detected as enriched in certain regions were actually transcribed in the soma. For example, inx-8/9, which were both grouped into the late diakinesis cluster (Table S4), are known as sheath cell components (Starich et al. 2014). Expression of other genes present in this late diakinesis cluster (i.e., myo-3) could also arise from the sheath cells and not only as part of the maternal contribution. In a similar fashion, expression of genes in the postmeiotic divisions of the male gonad could arise from the somatic gonadal vas deferens. For example, K09C8.2 and R03H10.4, which have high expression in late stages in our data set, were shown to be expressed in the somatic vas deferens (Thoemke et al. 2005). This somatic expression could also explain the specific expression from the X chromosome that we found in sections 9 and 10. Nevertheless, the large difference in volume between the germline and somatic tissues, and the presence of many known germline transcripts, suggest that most transcripts we detected are part of the germline. Future studies applying our method to, for example, glp-4 mutants, in which worms reach adulthood lacking germline nuclei (Beanan et al. 1992), would provide a handle on somatic gonad-expressed genes.
A large body of work has been devoted to understanding the mitotic to meiotic switch [reviewed in Kimble (2011)]. Indeed, these studies defined a set of proteins that together control this irreversible decision. However, our results also show that a dramatic change in gene expression occurs within the first half of meiotic prophase I in both oogenesis and spermatogenesis (Figure 8). This midprophase I shift occurs both as a sharp transition as well as a gradual increase/decrease in germline gene expression (compare meg-1 to rmd-1 in Figure S5).
Why would so many genes have to be upregulated or downregulated at mid–late pachytene in hermaphrodites? One answer comes from a “no return” decision that takes place shortly after pachytene during oogenesis. This is the last point in which quality control mechanisms can direct a defective nucleus into an apoptotic fate (Gartner et al. 2008). Thus, any nucleus that enters diakinesis would need a large cohort of transcripts that would facilitate the subsequent meiotic divisions and early embryogenesis. Indeed, it is not surprising that either at or shortly after the end of pachytene in the hermaphrodite gonad, genes associated with cell cycle progression, chromosome segregation, apoptosis, and development are upregulated, leading to the oocyte maturation stages (this study). Yet, although this model would explain the switch in oogenesis, it would not explain our observations for the male gonad, where no apoptotic germ cell quality control is set in place (Gartner et al. 2000; Jaramillo-Lambert et al. 2010), and there is no clear evidence for paternal contribution. Rather, it would seem that in the male gonad, this switch represents the beginning of the cell differentiation stage, which would probably require a whole new set of gene functions.
What drives the midprophase I transition? One possibility is that it involves signaling by the MAP kinase (MAPK) pathway, which has been previously implicated in regulating meiotic progression [reviewed in Sundaram (2006)]. This is also supported by our study, which shows an enrichment for genes annotated with MAPK activity exhibiting elevated mRNA levels in diplotene (sections 6–8) (Figure 3). However, if MAPK is driving the midway transition, it must act rapidly since the shift we detected in germline gene expression occurs between sections 5 and 6, concomitant with the rise in activated MAPK (Hayashi et al. 2007; Lee et al. 2007a,b; Arur et al. 2011). Another possibility is that transcription from genes on the X chromosome, which is turned on at section 6 in the hermaphrodite germline (Figure 2C), drives this transition, but there are no robust transcripts from the X in sections 5–8 in the male (Figure 6D). Thus, it remains to be determined how the changes in germline gene expression we uncovered are regulated, and we suggest that our data sets provide a starting point for subsequent studies aiming to address this issue.
Controlling transcript levels throughout the germline
Previous studies suggested that gene regulation in the C. elegans germline is primarily executed post-transcriptionally by 3′-UTR elements (e.g., Merritt et al. 2008). Several lines of evidence presented in this research argue that transcript levels play a major role in the germline’s gene expression through either mRNA biogenesis or degradation. First, we found a very dynamic profile of germline gene expression, where genes on the X chromosome show differences in expression throughout the gonad and, contrary to what has been postulated before (e.g., Nousch and Eckmann 2013), transcript levels do not simply rise gradually from distal to proximal regions, but exhibit very complex patterns (Figure 2A). Second, we found dozens of TFs with dynamic patterns of expression (Table S3). Third, some gene clusters, especially with high expression in mid–late meiosis, are enriched for transcripts annotated with GO terms associated with transcription. Fourth, several predicted TF targets are expressed with similar profiles, suggesting activation by transcription (Figure 5). Fifth, we found that most transcripts do not show strong variability of association to polysomes, arguing against silent vs. active populations (Figure 4). Finally, we found that hundreds of genes corresponding to CSR-1-bound 22G RNAs, which are presumed to act via transcriptional activation (Claycomb et al. 2009), are present throughout most regions during germline progression for both sexes (Figure 7). Altogether, these observations suggest that the active pattern of transcription may be combined with patterns of post-transcriptional control to enable faster changes in gene expression within the gonad.
Ways to exploit the spatiotemporal profiles of germline gene expression
The data sets provided here are useful resources for many fields of study. Using these data sets, we show dramatic changes in gene expression at specific stages during prophase I in both spermatogenesis and oogenesis (Figure 8). Recently, computational integration of cytological and RNA-seq analyses was used to analyze the transcriptome of spermatogonia and meiotic prophase substages in juvenile mice (Ball et al. 2016). This work found a transcriptome shift during pachytene, as was previously suggested (Waldman Ben-Asher et al. 2010; Soumillon et al. 2013), indicating that this transcriptome shift may be evolutionarily conserved in metazoans. Specific TFs may be required to execute these shifts in gene expression at specific meiotic stages, as suggested by our analysis (Figure 5). Although most genes residing on the X chromosome are silenced from the onset of germ cell progression until the end of the divisions in spermatogenesis, we found that some genes escape this whole-chromosome control in the male germline. It will be interesting to identify the molecular mechanisms that facilitate this control. Although the methodology we employed cannot detect small RNAs (see Materials and Methods), it can detect the dynamic nature of genes that may be targeted by the endogenous small RNA machinery. Focusing on genes that correspond to CSR-1-associated 22G RNAs, we found indications for CSR-1’s mode of operation. Genes corresponding to CSR-1-associated 22G RNAs are expressed broadly in the gonads of both males and hermaphrodites, but do not exhibit sharp spatiotemporal transitions in expression. This supports a model by which CSR-1 can license transcription in the germline, but we found almost no support for robust positive feedback between ALG-3/4 and CSR-1, as the shared genes are mostly limited to very specific patterns.
Finally, our data set (Tables S1–S11) can be used to find new paths for the control of germline development, gametogenesis, and the different layers of gene regulation at the chromosomal and genome-wide levels. Recently, transcriptome profiling of the C. elegans somatic gonad precursor cells has been published (Kroetz and Zarkower 2015). As data sets for germline precursor cells, and the transcriptomes for different single-cell germline developmental stages become available, for C. elegans and other organisms, it will also be possible to assess the evolutionary conservation of these findings, and use these comparisons to find both common as well as unique strategies by which gene regulation is used to control germline proliferation and differentiation.
Acknowledgments
We thank Charles R. Vanderburg of the Harvard NeuroDiscovery Center and the Technion Genome Center for technical assistance, and Yuji Kohara for kindly granting us permission to use in situ data from the NEXTDB database (version 4.0). The N2 worms were provided by the Caenorhabditis Genetics Center, which is funded by the U.S. National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD-010440). This work was supported by Israel Science Foundation grants 1283/15 and 2090/15 to Y.B.T., a European Research Council grant (EvoDevoPaths) and a European Molecular Biology Organization Young Investigator Program to I.Y., and NIH grant R01 GM-072551 to M.P.C.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6726293.
Communicating editor: J. Engebrecht
Literature Cited
- Anders S., Pyl P. T., Huber W., 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K., Moriguchi H., Yoshioka T., Okawa K., Tabara H., 2007. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26: 5007–5019. 10.1038/sj.emboj.7601910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arur S., Ohmachi M., Berkseth M., Nayak S., Hansen D., et al. , 2011. MPK-1 ERK controls membrane organization in C. elegans oogenesis via a sex-determination module. Dev. Cell 20: 677–688. 10.1016/j.devcel.2011.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avital G., Hashimshony T., Yanai I., 2014. Seeing is believing: new methods for in situ single-cell transcriptomics. Genome Biol. 15: 110 10.1186/gb4169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball R. L., Fujiwara Y., Sun F., Hu J., Hibbs M. A., et al. , 2016. Regulatory complexity revealed by integrated cytological and RNA-seq analyses of meiotic substages in mouse spermatocytes. BMC Genomics 17: 628 10.1186/s12864-016-2865-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beanan M., Strome S., 1992. Characterization of a germline proliferation mutation in Caenorhabditis elegans. Development 116: 755–766. [DOI] [PubMed] [Google Scholar]
- Brar G. A., Yassour M., Friedman N., Regev A., Ingolia N. T., et al. , 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557. 10.1126/science.1215110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruusgaard J. C., Liestøl K., Ekmark M., Kollstad K., Gundersen K., 2003. Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J. Physiol. 551: 467–478. 10.1113/jphysiol.2003.045328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton G. J., Jones C. J., 2009. Syncytial knots, sprouts, apoptosis, and trophoblast deportation from the human placenta. Taiwan. J. Obstet. Gynecol. 48: 28–37. 10.1016/S1028-4559(09)60032-2 [DOI] [PubMed] [Google Scholar]
- Chi W., Reinke V., 2006. Promotion of oogenesis and embryogenesis in the C. elegans gonad by EFL-1/DPL-1 (E2F) does not require LIN-35 (pRB). Development 133: 3147–3157. 10.1242/dev.02490 [DOI] [PubMed] [Google Scholar]
- Chu D. S., Shakes D. C., 2013. Spermatogenesis. Adv. Exp. Med. Biol. 757: 171–203. 10.1007/978-1-4614-4015-4_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., et al. , 1998. The transcriptional program of sporulation in budding yeast. Science 282: 699–705. 10.1126/science.282.5389.699 [DOI] [PubMed] [Google Scholar]
- Claycomb J. M., Batista P. J., Pang K. M., Gu W., Vasale J. J., et al. , 2009. The argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134. 10.1016/j.cell.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine C. C., Batista P. J., Gu W., Claycomb J. M., Chaves D. A., et al. , 2010. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 107: 3588–3593. 10.1073/pnas.0911685107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine C. C., Moresco J. J., Gu W., Shirayama M., Conte D., Jr., et al. , 2013. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell 155: 1532–1544. 10.1016/j.cell.2013.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., Goodyer W., Zetka M., 2004. Finding and keeping your partner during meiosis. Cell Cycle 3: 1014–1016. 10.4161/cc.3.8.1077 [DOI] [PubMed] [Google Scholar]
- Dernburg A. F., 2001. Here, there, and everywhere: kinetochore function on holocentric chromosomes. J. Cell Biol. 153: F33–F38. 10.1083/jcb.153.6.F33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellery P. M., Cindrova-Davies T., Jauniaux E., Ferguson-Smith A. C., Burton G. J., 2009. Evidence for transcriptional activity in the syncytiotrophoblast of the human placenta. Placenta 30: 329–334. 10.1016/j.placenta.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. E., Stanfield G. M., 2014. The regulation of spermatogenesis and sperm function in nematodes. Semin. Cell Dev. Biol. 29: 17–30. 10.1016/j.semcdb.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher R. I., Blakely S. R., Liotta L. A., Espina V., 2012. Laser capture microdissection: arcturus(XT) infrared capture and UV cutting methods. Methods Mol. Biol. 823: 157–178. 10.1007/978-1-60327-216-2_11 [DOI] [PubMed] [Google Scholar]
- Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M. O., 2000. A conserved checkpoint pathway mediates DNA damage–induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5: 435–443. 10.1016/S1097-2765(00)80438-4 [DOI] [PubMed] [Google Scholar]
- Gartner A., Boag P. R., Blackwell T. K., 2008. Germline survival and apoptosis (December 28, 2005), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.145.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibert M. A., Starck J., Beguet B., 1984. Role of the gonad cytoplasmic core during oogenesis of the nematode Caenorhabditis elegans. Biol. Cell 50: 77–85. 10.1111/j.1768-322X.1984.tb00254.x [DOI] [PubMed] [Google Scholar]
- Greenstein D., 2005. Control of oocyte meiotic maturation and fertilization (September 4, 2008), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.53.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnal A., Berset T., 2002. The C. elegans MAPK phosphatase LIP-1 is required for the G(2)/M meiotic arrest of developing oocytes. EMBO J. 21: 4317–4326. 10.1093/emboj/cdf430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H., Winfrey V. P., Blaeuer G., Hoffman L. H., Furuta T., et al. , 1999. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev. Biol. 212: 101–123. 10.1006/dbio.1999.9356 [DOI] [PubMed] [Google Scholar]
- Hashimshony T., Wagner F., Sher N., Yanai I., 2012. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep. 2: 666–673. 10.1016/j.celrep.2012.08.003 [DOI] [PubMed] [Google Scholar]
- Hayashi M., Chin G. M., Villeneuve A. M., 2007. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 3: e191 10.1371/journal.pgen.0030191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S., Zeisel A., Joost S., La Manno G., Zajac P., et al. , 2014. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11: 163–166. 10.1038/nmeth.2772 [DOI] [PubMed] [Google Scholar]
- Jan S. Z., Vormer T.L., Jongejan A., Rolling M.D., Silber de Rooij D.G., et al. , 2017. Unraveling transcriptome dynamics in human spermatogenesis. Development 144: 3659–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Ellefson M., Villeneuve A. M., Engebrecht J., 2007. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev. Biol. 308: 206–221. 10.1016/j.ydbio.2007.05.019 [DOI] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Harigaya Y., Vitt J., Villeneuve A., Engebrecht J., 2010. Meiotic errors activate checkpoints that improve gamete quality without triggering apoptosis in male germ cells. Curr. Biol. 20: 2078–2089. 10.1016/j.cub.2010.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Schaner C. E., Dernburg A. F., Lee M. H., Kim S. K., et al. , 2002. X-chromosome silencing in the germline of C. elegans. Development 129: 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershner A. M., Shin H., Hansen T. J., Kimble J., 2014. Discovery of two GLP-1/notch target genes that account for the role of GLP-1/notch signaling in stem cell maintenance. Proc. Natl. Acad. Sci. USA 111: 3739–3744. 10.1073/pnas.1401861111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Spike C., Greenstein D., 2013. Control of oocyte growth and meiotic maturation in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 277–320. 10.1007/978-1-4614-4015-4_10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., 2011. Molecular regulation of the mitosis/meiosis decision in multicellular organisms. Cold Spring Harb. Perspect. Biol. 3: a002683 10.1101/cshperspect.a002683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., Crittenden S., 2005. Germline proliferation and its control (August 15, 2005), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.13.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroetz M. B., Zarkower D., 2015. Cell-specific mRNA profiling of the Caenorhabditis elegans somatic gonadal precursor cells identifies suites of sex-biased and gonad-enriched transcripts. G3 (Bethesda) 5: 2831–2841. 10.1534/g3.115.022517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labunskyy V. M., Gerashchenko M. V., Delaney J. R., Kaya A., Kennedy B. K., et al. , 2014. Lifespan extension conferred by endoplasmic reticulum secretory pathway deficiency requires induction of the unfolded protein response. PLoS Genet. 10: e1004019 10.1371/journal.pgen.1004019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L., 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9: 357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Hook B., Pan G., Kershner A. M., Merritt C., et al. , 2007a Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 3: e233 10.1371/journal.pgen.0030233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Ohmachi M., Arur S., Nayak S., Francis R., et al. , 2007b Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics 177: 2039–2062. 10.1534/genetics.107.081356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B., Crombie C., Tischler J., Fortunato A., Fraser A. G., 2006. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 38: 896–903. 10.1038/ng1844 [DOI] [PubMed] [Google Scholar]
- L’Hernault S. W., 1997. Spermatogenesis, in C. elegans II, edited by Riddle D. L., Blumenthal T., Meyer B. J., Priess J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Lin B., Reinke V., 2008. The candidate MAP kinase phosphorylation substrate DPL-1 (DP) promotes expression of the MAP kinase phosphatase LIP-1 in C. elegans germ cells. Dev. Biol. 316: 50–61. 10.1016/j.ydbio.2007.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez A. L., III, Chen J., Joo H. J., Drake M., Shidate M., et al. , 2013. DAF-2 and ERK couple nutrient availability to meiotic progression during Caenorhabditis elegans oogenesis. Dev. Cell 27: 227–240. 10.1016/j.devcel.2013.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui D. Y., Colaiácovo M. P., 2013. Meiotic development in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 133–170. 10.1007/978-1-4614-4015-4_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay I. C., Voet T., 2014. Single cell genomics: advances and future perspectives. PLoS Genet. 10: e1004126 10.1371/journal.pgen.1004126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C., Rasoloson D., Ko D., Seydoux G., 2008. 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol. 18: 1476–1482. 10.1016/j.cub.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimhan K., Lambert S. A., Yang A. W., Riddell J., Mnaimneh S., et al. , 2015. Mapping and analysis of Caenorhabditis elegans transcription factor sequence specificities. Elife 4: e06967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousch M., Eckmann C. R., 2013. Translational control in the Caenorhabditis elegans germ line. Adv. Exp. Med. Biol. 757: 205–247. 10.1007/978-1-4614-4015-4_8 [DOI] [PubMed] [Google Scholar]
- Nousch M., Yeroslaviz A., Habermann B., Eckmann C. R., 2014. The cytoplasmic poly(A) polymerases GLD-2 and GLD-4 promote general gene expression via distinct mechanisms. Nucleic Acids Res. 42: 11622–11633. 10.1093/nar/gku838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M. A., Noble D., Sorokin E. P., Kimble J., 2014. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 (Bethesda) 4: 1765–1772. 10.1534/g3.114.012351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazdernik N., Schedl T., 2013. Introduction to germ cell development in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 1–16. 10.1007/978-1-4614-4015-4_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primig M., Williams R. M., Winzeler E. A., Tevzadze G. G., Conway A. R., et al. , 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26: 415–423. 10.1038/82539 [DOI] [PubMed] [Google Scholar]
- Reinke V., Smith H. E., Nance J., Wang J., Van Doren C., et al. , 2000. A global profile of germline gene expression in C. elegans. Mol. Cell 6: 605–616. 10.1016/S1097-2765(00)00059-9 [DOI] [PubMed] [Google Scholar]
- Reinke V., Gil I. S., Ward S., Kazmer K., 2004. Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131: 311–323. 10.1242/dev.00914 [DOI] [PubMed] [Google Scholar]
- Rotgers E., Nurmio M., Pietila E., Cisneros-Montalvo S., Toppari J., 2015. E2F1 controls germ cell apoptosis during the first wave of spermatogenesis. Andrology 3: 1000–1014. 10.1111/andr.12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez F., Smitz J., 2012. Molecular control of oogenesis. Biochim. Biophys. Acta 1822: 1896–1912. 10.1016/j.bbadis.2012.05.013 [DOI] [PubMed] [Google Scholar]
- Schisa J. A., Pitt J. N., Priess J. R., 2001. Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 128: 1287–1298. [DOI] [PubMed] [Google Scholar]
- Schvarzstein M., Wignall S. M., Villeneuve A. M., 2010. Coordinating cohesion, co-orientation, and congression during meiosis: lessons from holocentric chromosomes. Genes Dev. 24: 219–228. 10.1101/gad.1863610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth M., Shirayama M., Gu W., Ishidate T., Conte D., Jr., et al. , 2013. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev. Cell 27: 656–663. 10.1016/j.devcel.2013.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakes D. C., Wu J. C., Sadler P. L., Laprade K., Moore L. L., et al. , 2009. Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet. 5: e1000611 10.1371/journal.pgen.1000611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth U., Pitt J., Dennis S., Priess J. R., 2010. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development 137: 1305–1314. 10.1242/dev.044255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon M., Necsulea A., Weier M., Brawand D., Zhang X., et al. , 2013. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 3: 2179–2190. 10.1016/j.celrep.2013.05.031 [DOI] [PubMed] [Google Scholar]
- Spike C. A., Coetzee D., Eichten C., Wang X., Hansen D., et al. , 2014. The TRIM-NHL protein LIN-41 and the OMA RNA-binding proteins antagonistically control the prophase-to-metaphase transition and growth of Caenorhabditis elegans oocytes. Genetics 198: 1535–1558. 10.1534/genetics.114.168831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starich T. A., Hall D. H., Greenstein D., 2014. Two classes of gap junction channels mediate soma-germline interactions essential for germline proliferation and gametogenesis in Caenorhabditis elegans. Genetics 198: 1127–1153. 10.1534/genetics.114.168815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram M. V., 2006. RTK/Ras/MAPK signaling (February 11, 2006), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.80.1, http://www.wormbook.org. [Google Scholar]
- Tanaka M., 2014. Vertebrate female germline–the acquisition of femaleness. Wiley Interdiscip. Rev. Dev. Biol. 3: 231–238. 10.1002/wdev.131 [DOI] [PubMed] [Google Scholar]
- Tang F., Lao K., Surani M. A., 2011. Development and applications of single-cell transcriptome analysis. Nat. Methods 8: S6–S11. 10.1038/nmeth.1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoemke K., Yi W., Ross J. M., Kim S., Reinke V., et al. , 2005. Genome-wide analysis of sex-enriched gene expression during C. elegans larval development. Dev. Biol. 284: 500–508. 10.1016/j.ydbio.2005.05.017 [DOI] [PubMed] [Google Scholar]
- Waldman Ben-Asher H., Shahar I., Yitzchak A., Mehr R., Don J., 2010. Expression and chromosomal organization of mouse meiotic genes. Mol. Reprod. Dev. 77: 241–248. [DOI] [PubMed] [Google Scholar]
- Wang X., Zhao Y., Wong K., Ehlers P., Kohara Y., et al. , 2009. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics 10: 213 10.1186/1471-2164-10-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeles C. J., Wu M. Z., Claycomb J. M., 2013. Protection of germline gene expression by the C. elegans Argonaute CSR-1. Dev. Cell 27: 664–671. 10.1016/j.devcel.2013.11.016 [DOI] [PubMed] [Google Scholar]
- Wedeles C. J., Wu M. Z., Claycomb J. M., 2014. Silent no more: endogenous small RNA pathways promote gene expression. Worm 3: e28641 10.4161/worm.28641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolke U., Jezuit E. A., Priess J. R., 2007. Actin-dependent cytoplasmic streaming in C. elegans oogenesis. Development 134: 2227–2236. 10.1242/dev.004952 [DOI] [PubMed] [Google Scholar]
- Ye A. L., Ragle J. M., Conradt B., Bhalla N., 2014. Differential regulation of germline apoptosis in response to meiotic checkpoint activation. Genetics 198: 995–1000. 10.1534/genetics.114.170241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S., 2010. Stem cells in mammalian spermatogenesis. Dev. Growth Differ. 52: 311–317. 10.1111/j.1440-169X.2010.01174.x [DOI] [PubMed] [Google Scholar]
- Zetka M., 2009. Homologue pairing, recombination and segregation in Caenorhabditis elegans. Genome Dyn. 5: 43–55. 10.1159/000166618 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and reagents are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and supplemental files. File S1 contains the list of the mapped reads number of all genes in each hermaphrodite gonad section. File S2 contains the normalized expression of all genes in each hermaphrodite gonad section. File S3 contains the normalized expression of dynamic genes in each hermaphrodite and male gonad section. File S4 contains clustered hermaphrodite and male genes in the defined expression profiles. File S5 contains the analysis of the comparison between NEXTDB in situ images to spatiotemporally resolved germline gene expression data. File S6 contains the list of X-linked genes expressed in the first five sections of the hermaphrodite gonad. File S7 contains a list of clusters with the illustrative spatiotemporal representation of high (red) and low (white) expression, binary designation, and main gametogenic processes. File S8 contains ribosome profiling data. File S9 contains a list of the mapped reads number of all genes in each section of the male gonad. File S10 contains normalized expression of all genes in each section of the male gonad. File S11 contains lists of the shared and specific hermaphrodite- and male germline-expressed genes. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6726293.