Abstract
Boundaries (insulators) in the Drosophila bithorax complex (BX-C) delimit autonomous regulatory domains that orchestrate the parasegment (PS)-specific expression of the BX-C homeotic genes. The Fab-7 boundary separates the iab-6 and iab-7 regulatory domains, which control Abd-B expression in PS11 and PS12, respectively. This boundary is composed of multiple functionally redundant elements and has two key functions: it blocks cross talk between iab-6 and iab-7 and facilitates boundary bypass. Here, we show that two BEN domain protein complexes, Insensitive and Elba, bind to multiple sequences located in the Fab-7 nuclease hypersensitive regions. Two of these sequences are recognized by both Insv and Elba and correspond to a CCAATTGG palindrome. Elba also binds to a related CCAATAAG sequence, while Insv does not. However, the third Insv recognition sequences is ∼100 bp in length and contains the CCAATAAG sequence at one end. Both Insv and Elba are assembled into large complexes (∼420 and ∼265–290 kDa, respectively) in nuclear extracts. Using a sensitized genetic background, we show that the Insv protein is required for Fab-7 boundary function and that PS11 identity is not properly established in insv mutants. This is the first demonstration that a BEN domain protein is important for the functioning of an endogenous fly boundary.
Keywords: Abdominal B, BEN DNA-binding domain, bithorax, boundary, chromatin, Elba, Fab-7, Insensitive, insulator, parasegment identity
SPECIAL elements called chromatin boundaries, or insulators, form looped domains and play important roles in gene regulation (Chetverina et al. 2014; Matzat and Lei 2014; Ma et al. 2016). Their known genetic functions include blocking the action of enhancers and silencers, and an ability to mediate long-distance regulatory interactions (Holdridge and Dorsett 1991; Kellum and Schedl 1991, 1992; Geyer and Corces 1992; Sigrist and Pirrotta 1997; Muller et al. 1999; Cai and Shen 2001; Muravyova et al. 2001; Kyrchanova et al. 2008, 2013; Li et al. 2011; Fujioka et al. 2013). Genome-wide ChIP (chromatin immunoprecipitation) experiments with known insulator proteins, together with chromatin conformation capture experiments, have shown that boundaries are pervasive components of eukaryotic chromosomes, demarcating distinct chromatin and regulatory domains and helping mediate distant regulatory interactions (Cléard et al. 2006; Holohan et al. 2007; Cuddapah et al. 2009; Jiang et al. 2009; Smith et al. 2009; Nègre et al. 2010; Schwartz et al. 2012; Vietri Rudan and Hadjur 2015; Ali et al. 2016; Merkenschlager and Nora 2016). Studies in vertebrates on DNA-binding proteins essential for the architectural functions of boundary elements have focused almost exclusively on a single protein, CTCF (Vietri Rudan and Hadjur 2015; Ali et al. 2016; Merkenschlager and Nora 2016). In contrast, experiments in flies have implicated more than a dozen DNA-binding proteins in boundary function, and this number is likely to double or triple (Chetverina et al. 2014; Cuartero et al. 2014; Maksimenko and Georgiev 2014; Zolotarev et al. 2016; Fedotova et al. 2017). The properties of fly boundaries are equally complex. Endogenous boundaries span DNA sequences of > 200 bp and their activities depend upon unique assemblages of associated proteins. In the cases that have been studied in detail, the cis- and trans-acting elements are often functionally redundant with no single element being absolutely essential (Cuartero et al. 2014; Zolotarev et al. 2016; Kyrchanova et al. 2017). This means that a mutation in the recognition sequence for a boundary protein or in the protein itself may have no obvious phenotypic consequences.
One example of a boundary that is composed of multiple functionally redundant elements is Fab-7 from the Drosophila bithorax complex (BX-C) (Schweinsberg et al. 2004; Wolle et al. 2015). BX-C contains three homeotic genes, Ultrabithorax (Ubx), abdominal-A (abd-A), and Abdominal-B (Abd-B). These genes are responsible for specifying the parasegments (PS5–13) that make up the posterior two-thirds of the fly (Kyrchanova et al. 2015; Maeda and Karch 2015). Parasegment-specific expression of the three homeotic genes is orchestrated by a ∼300-kb regulatory region that is organized into three gene-specific transcriptionally associated regulatory domains (TARDs). For example, the Abd-B TARD contains four parasegment-specific cis-regulatory domains—iab-5, iab-6, iab-7, and iab-8—and these direct Abd-B expression in PS10, PS11, PS12, and PS13, respectively (Figure 1A) (Lewis 1978; Sánchez-Herrero et al. 1985).
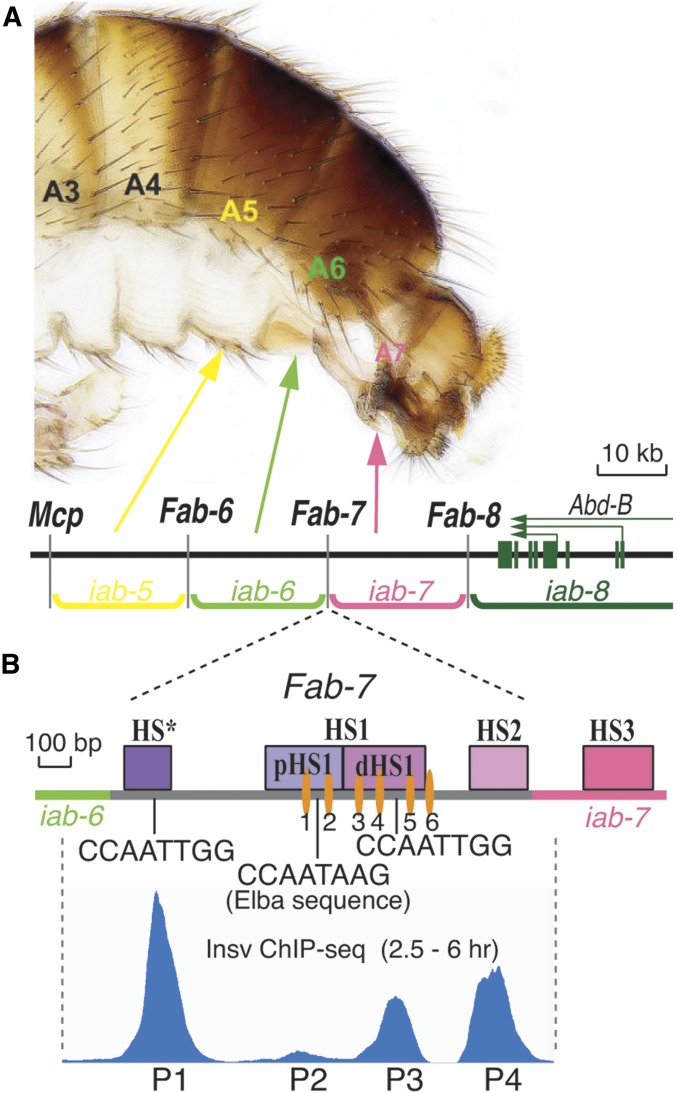
Figure 1.
The Fab-7 boundary. (A) Map of the Abd-B region of the bithorax complex. The relative location of the Abd-B regulatory domains—iab-5, iab-6, iab-7, and iab-8—and the segments that they specify in the adult male fly are indicated in the panel. Also shown are the positions of the boundary elements—Mcp, Fab-6, Fab-7, and Fab-8—and the Abd-B transcription unit. (B) Map of the Fab-7 nuclease hypersensitive sites—HS*, HS1, and HS2—and location of the binding motifs (GAGAG) for the GAGA factor (GAF) (orange ovals) and Insv. Also shown is a diagram of the Insv chromatin immunoprecipitation (ChIP)-sequencing peaks spanning the Fab-7 boundary and the adjacent iab-7 PRE (polycomb response element), adapted from Dai et al. (2015). The peaks are aligned with position of the Fab-7 hypersensitive sites.
Individual PS-specific cis-regulatory domains have to function autonomously to properly specify PS identity, and autonomous activity is conferred by the boundaries (Fabs and Mcp; Figure 1A) that bracket each cis-regulatory domain. The Fab-7 boundary, which is located between the iab-6 and iab-7 cis-regulatory domains, is the most thoroughly characterized BX-C boundary (Figure 1B) (Gyurkovics et al. 1990; Galloni et al. 1993; Hagstrom et al. 1996; Zhou et al. 1996; Mihaly et al. 1997). It maps to a region that contains four nuclease-hypersensitive sites: HS*, HS1, HS2, and HS3 (Hagstrom et al. 1996; Zhou et al. 1996; Mihaly et al. 1997). A combination of P-element excision-induced deletions and transgene assays map the sequences required for boundary activity to a 1.2-kb DNA segment that spans HS*, HS1, and HS2. The fourth hypersensitive site, HS3, has Polycomb silencing activity, and corresponds to the iab-7 PRE (polycomb response element). When Fab-7 is deleted (Mihaly et al. 1997), cross talk between regulatory elements in iab-6 and iab-7 either ectopically activates or silences the fused domain in PS11. As a consequence, a complex mixture of gain- (PS12 identity: GOF) and loss-of-function (PS10 identity: LOF) phenotypes are observed in PS11 (Mihaly et al. 1997). If the HS3 iab-7 PRE is also deleted (class I deletions), the fused domain is always activated, completely transforming PS11 into a duplicate copy of PS12.
Though Fab-7 appears to have boundary function throughout development, this constitutive activity depends upon cis-acting subelements that have stage- or tissue/cell type-specific activity. Transgene enhancer-blocking assays showed that sequences on the proximal side of HS1 (pHS1) have insulator activity in early embryos during the initiation phase of BX-C regulation, but not later in development (Schweinsberg and Schedl 2004; Schweinsberg et al. 2004). In contrast, sequences on the distal side of HS1 (dHS1) have reduced activity in early embryos, but are fully functional in older embryos and adults when BX-C regulation has switched to the maintenance phase. Two factors, Elba and LBC, which have boundary activity during the initiation and maintenance phase, respectively, have been identified (Aoki et al. 2008, 2012; Wolle et al. 2015). The Elba factor binds to a conserved 8-bp sequence in pHS1: CCAATAAG. It is detected in early 0–6-hr embryonic nuclear extracts while it is absent from older 6–18-hr embryonic nuclear extracts. The heterotrimeric Elba factor is composed of two proteins, Elba1 and Elba2, that have C-terminal BEN DNA-binding domains and a third protein, Elba3, that functions to link Elba1 and Elba2 together (Aoki et al. 2012). The ∼1000-kDa LBC binds to an ∼140-bp sequence in dHS1 and its temporal profile is the opposite of the Elba factor: there is little LBC activity in 0–6-hr extracts, while the LBC is enriched in older 6–18-hr nuclear extracts (Wolle et al. 2015).
Elba and LBC are not the only factors that bind to Fab-7 boundary sequences. The constitutively expressed C2H2 zinc finger protein Pita binds to two sites in HS2 (Maksimenko et al. 2015; Kyrchanova et al. 2017). Because of functional redundancy, mutations in these Pita sites in a Fab-7 boundary replacement that has HS* + HS1 + HS2 (but not HS3) have no effect on boundary function. However, in a genetically sensitized replacement, consisting of only HS1 + HS2, mutations in the HS2 Pita-binding sites eliminate boundary function (Kyrchanova et al. 2017).
Another factor that is known to associate with Fab-7 in vivo is Insensitive (Insv). Insv was initially identified as a corepressor for the Suppressor of Hairless [Su(H)] protein in the Notch signaling pathway (Duan et al. 2011). However, subsequent studies (Dai et al. 2015) revealed that it is associated with many chromatin boundaries including Fab-7. Intriguingly, Insv is a close relative of the Fab-7 boundary factors Elba1 and Elba2, and, like these two proteins, it has a C-terminal BEN domain. Also, like the Elba complex proteins, high levels of Insv are present throughout blastoderm- and early gastrula-stage embryos. While the Elba proteins disappear during gastrulation, Insv expression resolves into 14 stripes, predominantly in cells from the CNS and PNS (Duan et al. 2011; Bonchuk et al. 2015). The predicted Insv recognition sequence from genome-wide ChIP experiments is a palindrome, CCAATTGG, which differs by only two bases from the Elba recognition sequence in Fab-7 pHS1, CCAATAAG. The crystal structures of the Insv and Elba1 BEN domains bound to this palindrome sequence have been determined, and they share many similarities (Dai et al. 2013, 2015). Both bind to DNAs as dimers and each monomer interacts with both strands of the DNA helix. They also share amino acid–nucleotide contacts. There is also suggestive evidence that the biological activities of Insv may closely overlap those of the Elba complex. Genome-wide ChIPs show that Insv colocalizes with several known boundary proteins, including CTCF, Mod(mdg4), BEAF, and CP190 at relatively high frequencies. Moreover, in the case of CP190, this colocalization is not coincidental as these two proteins were found to co-immunoprecipitate (Dai et al. 2015). While the colocalization of Insv with known boundary factors is suggestive, the evidence for an architectural function is only correlative. In fact, previous in vivo assays have pointed to a quite different role, namely, in repressing transcription, and not to an architectural function (Duan et al. 2011; Dai et al. 2013).
In the studies reported here, we have used a combination of genetic and molecular approaches to explore the biological properties of the Insv protein. We have identified Insv-binding sequences in Fab-7 and shown that two of these are also recognized by the Elba factor. We also show that a third Insv-binding “site” spans a sequence of 100 bp and includes the Elba sequence, CCAATAAG, at one end. Next, we used two genetically sensitized backgrounds to demonstrate that Insv contributes to Fab-7 boundary function in vivo.
Materials and Methods
Insv rescue transgene
The genomic rescue construct, insv+3.67, contains a wild-type 3.67-kb fragment from the insv genomic region. It was amplified according to Duan et al. (2011). The rescue construct was injected into preblastoderm embryos containing an attP site at cytogenetic location 86F (y1M{vas-int.Dm}ZH-2A w*; M{3xP3-RFP.attP}ZH-86Fb; Bloomington stock RRID:BDSC_24749) (Bischof et al. 2007).
Fly stocks and genetic crosses
All flies were maintained at 25° on standard medium. The Fab-7GAGA1–5 and HS1 + HS2 replacements were generated using the Fab-7attP50 landing platform described previously (Wolle et al. 2015). The null mutation of insv gene, insv23B, was provided by Eric Lai (Department of Developmental Biology, Sloan-Kettering Institute, New York) (Reeves and Posakony 2005; Duan et al. 2011). Oregon-R was used as wild-type. In the rescue experiment, the genomic rescue transgene was recombined with Fab-7GAGA1–5. The recombinant was verified by PCR and then introduced into an insv23B mutant background.
Cuticle preparations and immunostaining
Adult abdominal cuticles of homozygous eclosed 3–4-day-old flies were prepared essentially as described in Mihaly et al. (1997) and mounted in Hoyer’s solution.
Electrophoretic mobility shift assays
The probe sequences used in this work are shown in Supplemental Material, Table S1. Short probes (≤ 32 bp) were obtained by annealing the synthesized oligo DNAs, while long probes (> 32 bp) were generated by PCR reactions followed by extractions from 3% agarose/TBE gels. For labeling the probe, 1 pmol of probe was 5′-end labeled with [γ-32P] ATP (MP Biomedicals) using T4 Polynucleotide Kinase (New England Biolabs, Beverly, MA) in a reaction mix of 50 μl. The reaction was incubated for 45 min at 37°. Labeled probe was separated from free ATP using columns packed with Sephadex G-50, fine gel (Amersham, Piscataway, NJ). The eluted volume was adjusted to 100 μl using deionized water for a final concentration of 10 fmol/μl labeled probe. For binding reactions, a 20-μl volume consisting of 25 mM Tris-Cl (pH 7.4), 100 mM KCl, 1 mM EDTA, 0.1 mM DTT, 0.1 mM PMSF, 0.03 mg/ml BSA, 10% glycerol, 0.5% Triton X-100, 0.25 mg/ml poly(dI-dC), 0.5 μl labeled probe, and 1 μl of nuclear extract (corresponding to ∼2.5 mg of embryos) was used. Nuclear extracts from Oregon R embryos were prepared as in Aoki et al. (2008, 2012). In samples containing unlabeled competitor DNA, the DNA was included so that the final concentration of the competitor was at 100-fold excess. The reaction mixture described above was incubated for 30 min at room temperature and loaded onto a 4% acrylamide/bis-acrylamide (37.5/1) gel (0.5× TBE and 2.5% glycerol). The 36–64 lane gels (Triple-wide gel system; C.B.S. Scientific Company, Del Mar, CA) were electrophoresed at 60 V for 1∼2 hr at 4° with 0.5× TBE and 2.5% glycerol running buffer, and 20-lane gels (7.5 inches long) were electrophoresed at 180 V for 3∼4 hr at 4° with the same running buffer. Gels were dried on 3MM chromatography paper (Whatman) and imaged using a Typhoon 9410 scanner and Image Gauge software and/or X-ray film.
Size-exclusion chromatography
For size estimations, 200 μl of the nuclear extracts was loaded onto a 24-ml bed volume Superdex 200 10/300 size-exclusion column (GE Healthcare) assembled in an Akta system (GE Healthcare). Next, 250 μl fractions were collected from 4 to 22 ml. Fractions (5.6 μl each) were analyzed by electrophoretic mobility shift assays (EMSAs) to determine DNA-binding activity. Size-exclusion standards (Bio-Rad, Hercules, CA) ranging from 1.35 to 670 kDa were used to calculate the partition coefficient and estimate the size of the protein complexes.
Antibodies
Rabbit Elba and Insv antibodies used for EMSA and ChIP experiments were previously described in Aoki et al. (2014).
ChIP
The methods used for ChIP experiments are described in Aoki et al. (2014). The Insv ChIP-seq (ChIP-sequencing) diagrams were generated from the BEDgraph data of the ChIP-seq experiments of Dai et al. (2015).
Data availability
Reagents (antibodies and strains) used for the experiments in this paper are available upon request. Results presented in figures and supplemental figures support the conclusions in this paper. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.6932915.
Results
Both Insv and Elba bind to CCAATTGG palindromes in HS* and HS1
ChIP-seq data from Dai et al. (2015), reproduced in Figure 1B, indicate that there are four Insv peaks in Fab-7. The first, P1, maps to the CCAATTGG palindrome in HS*. P2 and P3 map to HS1; the former spans the Elba recognition sequence in pHS1 while the latter maps to the CCAATTGG palindrome in dHS1. Finally, P4 maps to HS2. To further document the in vivo association of Insv with these sites in Fab-7, we cross-linked 2–5-hr embryos using either the standard formaldehyde cross-linking procedure or a procedure that uses a combination of formaldehyde and the bifunctional cross-linker disuccinimidyl glutarate (DSG), which we developed for ChIP experiments on the Elba factor (Aoki et al. 2014). Figure S1, A and B show that the four Insv sites in the Fab-7 boundary identified by Dai et al. (2015) can be detected using a different Insv antibody.
To confirm these findings and map the actual Insv-binding sequences, we used EMSAs and nuclear embryonic extracts prepared from early (0–6 hr) and late (6–12 or 6–20 hr) embryos. Based on the pattern of Insv expression (Bonchuk et al. 2015; Dai et al. 2015), we expected to detect Insv DNA-binding activity at both developmental stages with probes spanning the P1 and P3 palindromes. Figure 2A shows EMSA experiments with early (0–6 hr) and late (6–18 hr) wild-type nuclear extracts using a 32-bp probe spanning the CCAATTGG palindrome in P3. This probe gives a complex pattern of shifts that differ in early and late nuclear extracts. A similar set of early and late shifts are observed for a probe spanning the P1 palindrome (data not shown).
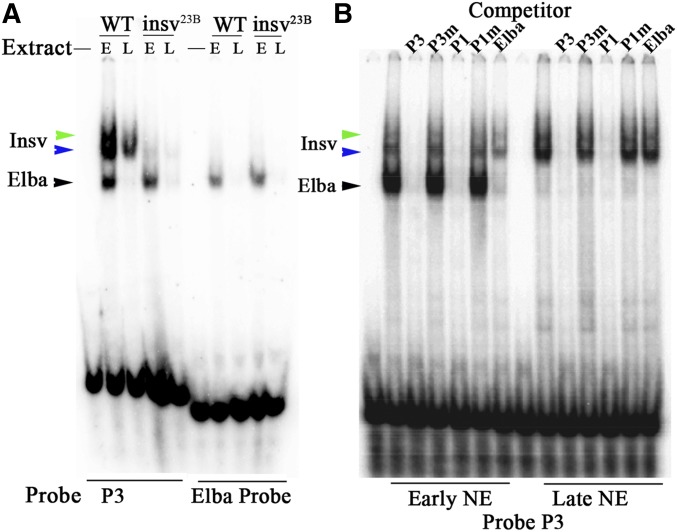
Figure 2.
Shifts generated by Insv and Elba. (A) Nuclear extracts (NEs) were prepared from 0 to 6-hr and 6–18-hr wild-type (WT) and insv23B embryos. The NEs were then used for electrophoretic mobility shift assay (EMSA) experiments with either the P3 (left side) or Elba (right side) probes. With the P3 probe, Elba and Insv shifts are observed in WT 0–6-hr extracts. The two sets of shifts are present in roughly similar yields in WT. While the Elba shift is also detected in the 0–6-hr insv23B extracts, the 0–6-hr Insv shift is greatly reduced in yield. However, there is a residual Insv shift in 0–6-hr insv23B extracts that we attribute to a low level of contamination with WT flies (red eye flies were found in our insv23B population cages). Like the 0–6-hr NEs, the yield of the Insv shift in 6–18-hr insv23B is also greatly reduced comparative to the Insv shift in 6–18-hr WT NEs. The Elba probe was used for shifts on the right side. In this case, shifts are observed in 0–6-hr nuclear extracts from both WT and isnv23B embryos, but not in extracts from 6 to 18-hr embryos. E, early (0–6 hr); L, late (6–18hr). (B) Competition experiments. EMSA of the P3 probe incubated with NEs from 0 to 6 hr (E) and 6–18 hr (L) embryos, either without (−) or in the presence of excess (100-fold) cold competitor, as indicated: P3, P3m (mutant), P1, P1m (mutant), or the 27-bp Elba probe. Sequences of probes are in Table S1. Positions of the Elba and Insv shifts are indicated.
Since the Elba recognition sequence, CCAATAAG, is closely related to the predicted Insv-binding sequence, it seemed likely that Elba might also bind to the CCAATTGG palindrome. In this case, the shifts that are present in early, but not in late, nuclear extracts would be expected to be generated by the Elba factor, not Insv. To explore this possibility, we used early and late nuclear extracts in EMSA experiments with a 27-bp probe spanning the Elba CCAATAAG sequence. As can be seen in Figure 2A, early extracts generate a prominent shift with the Elba probe (black arrowhead), while late extracts do not. The early shift with the Elba probe comigrates with the most rapidly migrating P3 (or P1) shift. On the other hand, the set of more slowly migrating shifts (blue and green arrowheads) that are detected with the P3 (or P1) probes are not observed with the Elba probe. We can draw several inferences from these findings. First, the set of more slowly migrating shifts (blue and green arrowheads) with the P3 or P1 probes in early and late extracts are likely to correspond to the Insv protein. In contrast, the more rapidly migrating shift (black arrowhead), which is enriched in early extracts, corresponds to the Elba factor. Second, the Elba factor recognizes both the CCAATAAG and CCAATTGG palindromes, while Insv only recognizes the CCAATTGG palindrome. Below, we have tested these inferences.
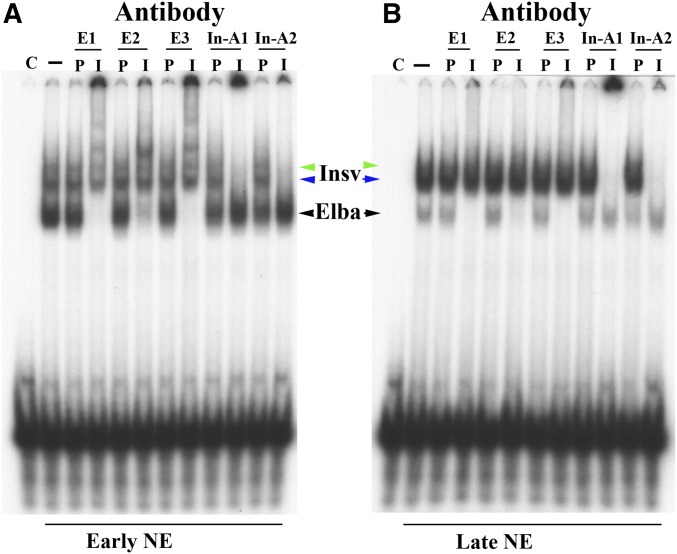
Antibody supershift experiment: Figure 3A shows antibody supershift experiments with early extracts. Antibodies against the three Elba proteins—Elba1 (E1), Elba2 (E2), and Elba3 (E3)—supershift the rapidly migrating band (black arrowhead), but not the collection of more slowly migrating shifts (blue and green arrowheads), while the corresponding preimmune serums (P) do not affect the shifts. A different result is obtained for two independent Insv antibodies, In-A1 and In-A2. These antibodies supershift the collection of more slowly migrating shifts (blue and green arrowheads), but have no effect on the Elba shift (black arrow). Again, the preimmune controls for these two Insv antibodies do not alter the pattern of shifts. Essentially, the same results are obtained for the P3 shifts generated in late nuclear extracts [note that in this particular late nuclear extract preparation, the Elba shift is readily apparent; the Elba factor is typically present at much lower levels in late nuclear extracts (see Figure 2)]. The slowly migrating bands are supershifted by Insv antibodies, while the more rapidly migrating band is shifted by the three Elba antibodies.
Competition experiments: Figure 2B shows competition experiments with wild-type and mutant versions of the P3 and P1 probes, and the 27-bp Elba probe. Excess cold P3 and P1 DNA competes with both the Elba (black arrowhead) and Insv (blue and green arrowheads) shifts in early nuclear extracts. In contrast, mutant versions of the P3 and P1 probes fail to compete with either Elba or Insv shifts. The Elba shift is competed by the cold 27-bp Elba probe, while the Insv shift is not. In late nuclear extracts, the Insv shifts are competed by wild-type versions of P1 and P3, but not by mutant probes or by the Elba probe. To further confirm that Elba and Insv recognize the CCAATTGG palindrome, we introduced 3-bp mutations in 1-bp increments across the entire 32-bp P3 and P1 sequences. We found that mutations in the palindrome sequence disrupted DNA binding by Elba and Insv, whereas mutations outside of the palindrome had little if any effect.
Nuclear extracts from insv25B mutant embryos: To provide additional evidence that the cluster of slowly migrating shifts observed in embryonic nuclear extracts with the P3 (and P1) probe is generated by the Insv protein, we prepared nuclear extracts from flies carrying the insv23B deletion. Shifts with early and late insv23B nuclear extracts are shown in Figure 2A. As can be seen in the figure, the cluster of Insv shifts is substantially reduced compared to wild-type, while the yield of Elba shift is similar to wild-type. While the yield of the Insv is substantially reduced, a weakly labeled Insv-like shift can be detected at both stages. Since the insv23B mutation deletes most of the insv gene, an Insv shift would not be expected. However, we suspect that this residual Insv shift is observed because the insv23B fly cages we prepared for the nuclear extracts had a few contaminating wild-type (w+) flies (∼0.5%). insv23B have reduced fecundity and the presence of even a small percentage of wild-type females could contribute a sufficient number of embryos to generate a detectable Insv shift.
Figure 3.
Antibody supershift experiments using staged embryonic nuclear extracts (NEs). (A and B) Supershift experiments with antibodies against Elba1 (E1), Elba2 (E2), Elba3 (E3), and two different anti-Insv rabbit polyclonal antibodies, In1 and In2. P corresponds to the preimmune serum, while I corresponds to immune serum. Stages of the NEs are indicated below.
In vitro-translated Elba and Insv proteins bind to the CCAATTGG palindrome
In previous studies, we found that the two BEN domain Elba proteins, Elba1 and Elba2, were unable to bind DNA on their own or when mixed together. Binding to the CCAATAAG sequence was only observed when Elba3 was included in the reaction mix (Aoki et al. 2012). These findings suggested that Elba3 functions to link the Elba1 and Elba2 BEN domains together. Consistent with this suggestion, we found that the Elba1:Elba2 combination is able to bind to the CCAATAAG sequence in the absence of Elba3 when both proteins have an N-terminal GST moiety, which can mediate dimerization. Similar results were obtained when the C-terminal BEN domains of Elba1 and Elba2 were fused to an N-terminal GST. Interestingly, in our experiments with the GST fusions, shifts were only observed when both GST-Elba fusions were included in the reaction mix. This finding indicated that Elba1 and Elba2 homodimers could not bind to the CCAATAAG sequence. These results fit with our antibody supershift experiments, which showed that the Elba shift of the CCAATAAG probe in nuclear extracts is generated by all three Elba proteins. The supershift experiments with Elba antibodies in Figure 2 indicate that Elba also binds to the palindrome CCAATTGG sequence as a heterotrimeric (Elba1 + ElBa2 + Elba3) complex. However, while we did not detect any shifts with nuclear extracts that might correspond to the independent binding of either Elba1 or Elba2 homodimers to the palindrome, Dai et al. (2015) were able to generate a crystal structure of Elba1 alone bound to the palindrome.
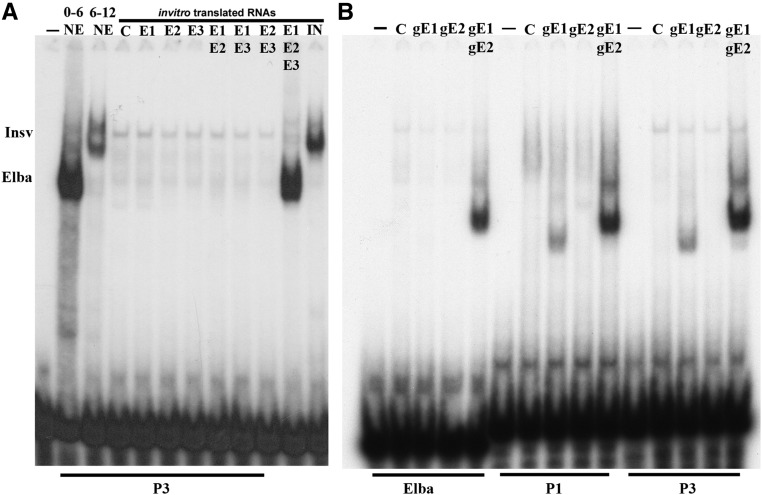
To explore this issue further, we generated the three Elba proteins by in vitro translation in rabbit reticulocyte extracts. Figure 4A shows the shifts generated by the individual translated Elba proteins and by different combinations of these proteins. Like the Elba sequence CCAATAAG, all three Elba proteins must be included in the reaction mix to generate a shift with the palindrome. We also tested whether in vitro-translated Insv protein is able to bind on its own to the CCAATTGG palindrome. In contrast to the two Elba BEN domain proteins, Insv alone is sufficient to generate the Insv shift. This result indicates that, in spite of the protein sequence similarity between Elba1, Elba2, and Insv, Insv does not require a coupling protein like Elba3 for its DNA-binding activity.
Figure 4.
In vitro-translated Elba and Insv proteins. (A) Electrophoretic mobility shift assay (EMSA) of probe P3 using staged nuclear extracts or in vitro-translated RNAs, as indicated. (−) no nuclear extracts, 0–6 and 6–12: staged nuclear extracts. Rabbit reticulocyte translation mix with no added mRNA (C) or with RNAs encoding the proteins as indicated: E1 (Elba1), E2 (Elba2), E3 (Elba3), and Insv (Insensitive), either alone or in different combinations. (B) EMSA experiments with in vitro-translated GST-Elba1/Elba2 BEN domains and the Elba, P1, and P3 probes. –, no reticulocyte translation mix; C, reticulocyte translation mix, no RNAs; gE1, reticulocyte translation mix primed with an RNA encoding the GST-Elba1CBEN protein; gE2, reticulocyte translation mix primed with an RNA encoding the GST-Elba2CBEN protein; gE1/gE2, reticulocyte translation mix primed with an RNA encoding the GST-Elba1CBEN and GST-Elba2CBEN proteins. Note that gE1 (GST-Elba1CBEN) can bind to the P1 and P3 probes, but not to the Elba probe on its own.
We also tested whether GST-mediated dimerization would enable Elba1 and Elba2 to bind to the palindrome, either on their own or when combined with each other. For this purpose, we used in vitro translation to generate C-terminal BEN domain proteins fused to an N-terminal GST. As found previously, neither GST-Elba1CBEN (gE1) nor GST-Elba2CBEN (gE2) are able to bind to the CCAATAAG sequence on their own, while the combination of two proteins (gE1 gE2) generates a prominent shift (Figure 4B). A different result is obtained for the CCAATTGG palindrome. GST-Elba1CBEN shifts both the P1 and P3 palindrome probes, though the yield of the shift is less than that observed for the GST-Elba1CBEN/GST-Elba2CBEN combination (gE1 gE2). In contrast, at most, only a very weak shift is generated by GST-Elba2CBEN (gE2) on its own.
Insv and Elba are assembled into large complexes in nuclear extracts
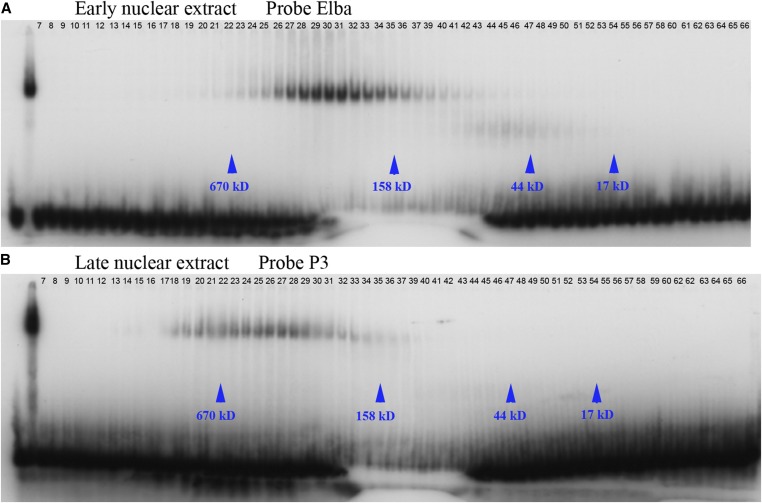
To further characterize the Insv and Elba DNA-binding activity in nuclear extracts, we fractionated early and late wild-type extracts by gel filtration on a Superdex 200 size-exclusion column. In the experiment presented in Figure 5, we simplified the shift patterns by using the Elba probe for early nuclear extracts and the P3 probe for late nuclear extracts. The former is expected to give the Elba shift while the latter should give the Insv shift. The peak fractions for Elba in the size-exclusion column are between 22 and 27, and correspond to an estimated size of ∼265–290 kDa (Figure 5A). The three Elba proteins are each about 41.6–43.3 kDa and assemble into a tripartite complex. If each Elba protein were present in the complex in only one copy, the predicted size of the complex would only be about ∼130 kDa. One explanation for this discrepancy is that other proteins are associated with the Elba factor in nuclear extracts. While we cannot exclude this possibility, no other proteins were present in significant yield when we purified the complex from nuclear extracts (Aoki et al. 2012). Thus, an alternative explanation is that Elba is a hexamer with two copies of Elba1, Elba2, and Elba3.
Figure 5.
Insv and Elba are assembled into large complexes in nuclear extracts. Early (0–6 hr) (A) and late (6–18 hr) (B) nuclear extracts were fractionated on a Superdex 200. The column fractions were then used in electrophoretic mobility shift assay experiments with P3. Column fractions loaded as indicated in each panel. Lanes on left are no extract, plus nuclear extract followed by the column fractions. Lanes on far left of (A) are nuclear extract (58) and no extract (59).
The peak fractions for Insv elute even earlier from the gel filtration column than Elba (Figure 5B), and we estimate that Insv is assembled into a complex of ∼420 kDa. Like the three Elba proteins, an Insv monomer is ∼42 kDa. Thus, if this complex consists of only the Insv protein, there would be ∼10 Insv monomers. Since a dimer is required for DNA binding (Dai et al. 2013), a complex of this composition would have five DNA-binding Insv BEN dimers. As such, it could interact simultaneously with five different recognition motifs, potentially tethering distant boundary elements in close proximity. Alternatively, since Insv has been shown to interact with CP190 and Su(H) (Duan et al. 2011; Dai et al. 2015), it is also possible that these proteins might be components of the Insv complex in nuclear extracts.
The P2 Insv recognition sequence is unusually large
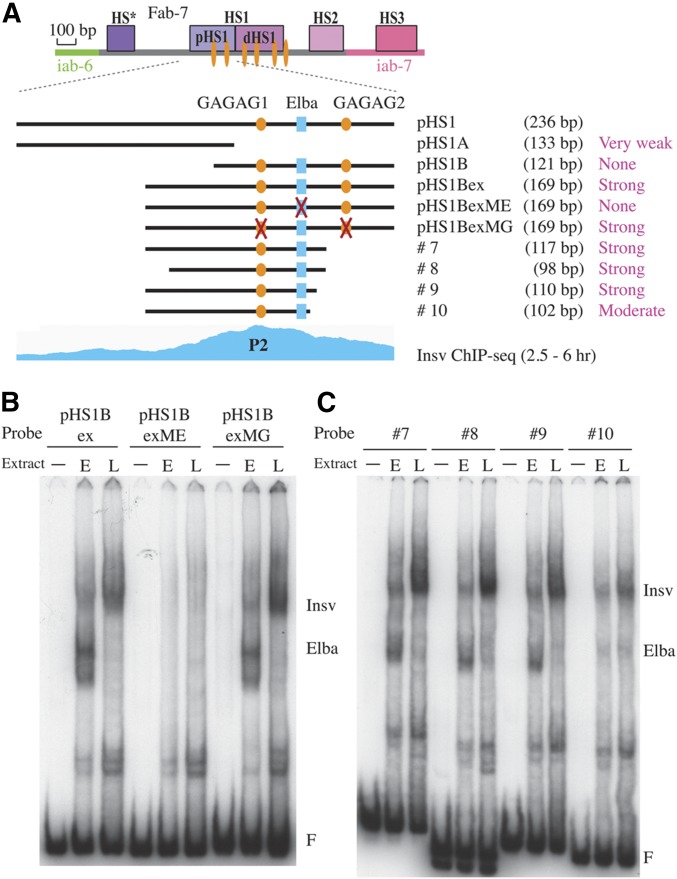
We also attempted to identify the sequences responsible for the P2 and P4 Insv ChIP peaks. Unlike P1 and P3, the ChIP peaks for P2 (pHS1) and P4 (HS2) map to sequences that do not contain the CCAATTGG palindrome. The pHS2 does include the Elba motif, CCAATAAG; however, a 27-bp probe containing this sequence is not shifted by Insv (see Figure 2A). Since Insv is assembled into a large complex in nuclear extracts that could potentially contain multiple dimeric BEN DNA-binding domains, it seemed possible that the complex might be able to interact with an extended sequence. Consistent with this idea, the P2 ChIP peak is quite broad. For this reason, we tested a series of larger fragments spanning the pHS1 region of HS1 (Figure 6A).
Figure 6.
Electrophoretic mobility shift assay (EMSA) of probes spanning P2. (A) Diagram of probes spanning P2. Indicated are the extent of the probes, their length, and relative binding affinity for Insv. Also indicated are recognition sequences for GAF (GAGAG, orange ovals) and Elba (CCAATAAG, blue rectangle). The Insv chromatin immunoprecipitation-sequencing (ChIP-seq) diagram of early embryos (Dai et al. 2015) is aligned on the bottom. (B) EMSA of wild-type pHS1Bex (A) and mutated versions of pHS1Bex. pHS1BexME, mutation of the Elba sequence; pHS1BexMG, mutation of both GAGAG sequences). (C) EMSA of probes #7, #8, #9, and #10. The location and extent of these four probes is indicated in (A). (F) free probe. Positions of the Elba and Insv shifts are indicated. − no extract; E, 0–6 hr “early” embryo nuclear extract; L, 6–18 hr “late” embryo extract.
As indicated in the diagram in Figure 6A, an Insv-like shift is observed when the entire 236-bp pHS1 sequence is used as a probe (see Figure S2A). When we divided pHS1 into proximal (pHS1A) and distal (pHS1B) halves, the former gave a weak Insv-like shift while the latter did not (Figure S2A). However, when we extended pHS1B proximally by 47 bp (pHS1Bex), a strong Insv-like shift was observed with both early and late nuclear extracts (Figure 6B and Figure S2A). Note that, as expected, the early extract also gives an Elba shift, while the late extract does not.
We used a combination of competition and supershift experiments to determine whether the Insv-like shift observed with the 169-bp pHS1Bex probe corresponds to Insv. If Insv is responsible for the slowly migrating shift with the pHS1Bex probe, binding should be competed by excess cold 32-bp P1 probe. Figure S2A shows that the pHS1Bex shift in late nuclear extracts is indeed competed by the P1 probe. In contrast, a mutant version of the P1 probe (that does not bind Insv: see Figure 2B) fails to compete the pHS1Bex shift. Also, as expected, the pHS1Bex shift is not competed by the short 27-bp Elba-binding sequence (or a mutant version of this sequences). Antibody supershift experiments with late nuclear extracts provide a further demonstration that the pHS1Bex shift is generated by Insv (Figure S2B). Both of the Insv rabbit polyclonal antibodies supershift the pHS1Bex shift, while the preimmune serum does not. In addition, antibodies against the three Elba subunits have no effect on the pHS1Bex shift.
To further define the minimal sequences required for Insv binding, we generated a series of deletions in the pHS1Bex probe (Figure 6, A and C). These deletion mutants indicate that Insv binding requires a sequence of between 63 and 91 bp (the combination of pHS1B and probe #9). The sequence required for Insv binding begins just distal to the Elba-binding site and extends proximally between 63 (pHS1B) and 91 bp (probe #9) (Figure 6). In addition to the Elba-binding site, this sequence contains one of the proximal GAGAG motifs. To test whether either of these motifs are needed for Insv binding, we generated mutations in the GAGAG motifs and Elba-binding site in the pHS1Bex probe. Figure 6B shows that the GAGAG motifs are not required for Insv binding, while the Elba site is.
Unlike the P2 site, we failed to detect any Insv-containing shifts with probes from HS2 (the P4 peak), even when we used DNA fragments of up to 200 bp in length. This observation suggests that Insv association with HS2 in ChIP experiments involves yet another mechanism. For example, Insv binding could be stabilized through interactions with another DNA-binding protein, like Pita, which binds to sequences in HS2. Alternatively, Insv protein bound to sequences in HS* and HS1 could be cross-linked to a protein associated with HS2. Further studies will be required to clarify the mechanism responsible for the P4 ChIP peak.
Insv contributes to Fab-7 boundary activity
The findings in the previous sections confirm that Insv binds to multiple sites in Fab-7. To extend this analysis, we tested whether Insv contributes to Fab-7 boundary function. In a null insv23B background, there are no obvious transformations in PS11 (A6) parasegment identity; however, this is not all that surprising because cis-acting elements in Fab-7 are known to be functionally redundant (Schweinsberg et al. 2004; Kyrchanova et al. 2017). For this reason, we tested the effects of the insv23B mutation in genetic backgrounds in which Fab-7 activity in BX-C is sensitized. For this purpose, we used two different Fab-7 replacements. The first is an HS1 + HS2 replacement. This replacement retains these two Fab-7 hypersensitive sites, but lacks HS* (which has P1: Figure 1) and hypersensitive site HS3, which corresponds to the iab-7 PRE. In the second replacement, Fab-7GAGA1–5, sequences spanning all four hypersensitive sites (HS* + HS1 + HS2 + HS3) are present; however, the GAGA factor (GAF) motifs GAGA1, GAGA2, GAGA3, GAGA4, and GAGA5 in HS1 are mutant.
The HS1 + HS2 replacement displays tissue-specific defects in boundary function (Kyrchanova et al. 2017). As illustrated in the male fly shown in Figure 7B, the A6 sternite on the ventral side is missing. This phenotype arises from a GOF transformation of A6 (PS11) into a duplicate copy of A7 (PS12), and is characteristic of deletions that remove both the Fab-7 boundary and iab-7 PRE. Although most HS1 + HS2 males lack a sternite, a small patch of ventral cuticle is observed in a few (∼10%). While boundary function is disrupted in ventral tissues, it is largely retained in the dorsal tergite. In most HS1 + HS2 male flies, the tergite is nearly the same size as in wild-type. Moreover, judging from the characteristic trichome pattern, the A6 tergite is also properly specified. A similar tissue-specific effect on boundary function is observed in female flies (data not shown). When we combine the HS1 + HS2 replacement with the insv23B mutation, boundary function in dorsal tissues is largely abrogated. Figure 7C shows that the entire A6 segment is almost absent in insv23B; HS1 + HS2 males, indicating that A6 (PS11) is transformed into a duplicate copy of A7 (PS12). The same GOF A6←→A7 transformation is also observed in insv23B; HS1 + HS2 females (data not shown).
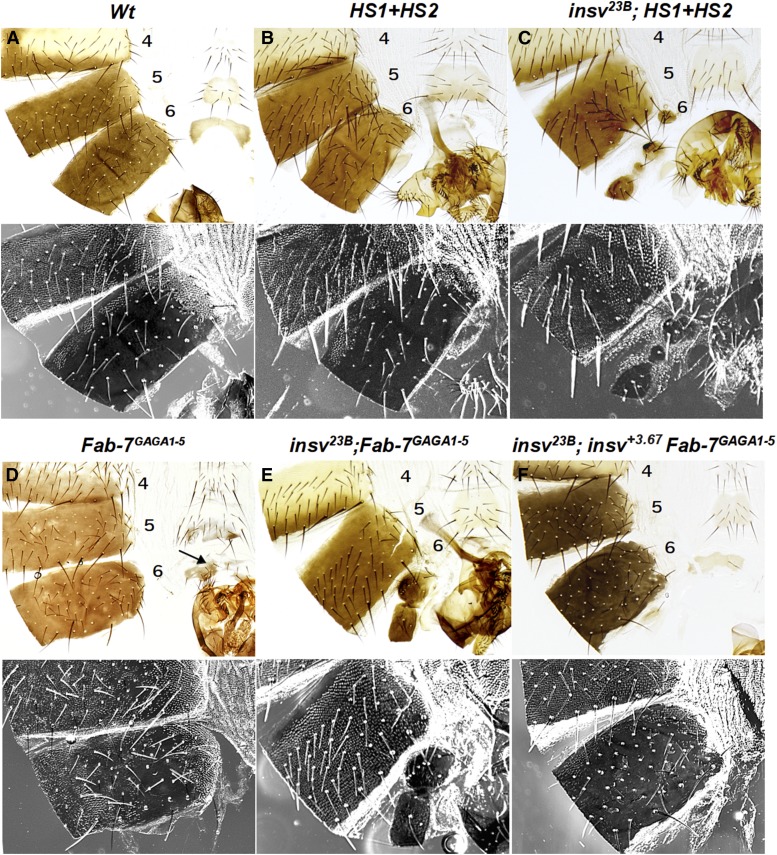
Figure 7.
Insv is required for Fab-7 function in vivo. (A) Adult abdominal cuticle preparations of a wild-type (Wt) male. The fifth and sixth tergites are pigmented, the A6 sternite is recognizable by the absence of bristles and a specific form. Trichomes are visible in the dark field and cover all the surface of the A5 tergite, and only a thin stripe along the anterior and ventral edges of the A6 tergite. (B and D) Homozygous males with a sensitized genetic background of Fab-7: HS1 + HS2 (B) and Fab-7GAGA1–5 (D) (see the text). (C) In the absence of the endogenous Insv, HS1 + HS2 males show a strong gain-of-function (GOF) transformation of A6 to A7, as revealed by the marked reduction of the A6 tergite and the absence of the sternite. (E): insv23B; Fab-7GAGA1–5 homozygous males have a mixed GOF and loss-of-function (LOF) phenotype. A6 is partially transformed into A7 (GOF transformation), but at the same time the residual A6 cuticle has morphological features characteristic of A5 (trichomes visible in the dark field) (LOF transformation). (F) Rescue of the mutant insv23B; Fab-7GAGA1–5 phenotype with a genomic rescue construct insv+3.67. The genomic Insv fragment restores the boundary function of Fab-7GAGA1–5. The A6 tergite is only marginally smaller than in Wt, and the trichomes are usually limited to small patches. The numbers 4, 5, and 6 indicate A5, A6, and A7 abdominal segments, respectively. Arrows in (D) show extra bristles on the A6 sternite.
The Fab-7GAGA1–5 replacement was modeled after the GAGA site mutations that had previously been tested in enhancer-blocking transgene assays (Schweinsberg et al. 2004). These studies indicated that GAGA sites 1 and 2 (Figure 1) contribute to the early boundary activity of pHS1 (perhaps facilitating the binding or functioning of Elba and Insv) (Schweinsberg et al. 2004; Aoki et al. 2008). GAGA sites 3, 4, and 5 were found to be critical for LBC binding and, consequently, are important for the late boundary activity of the dHS1 sequence (Wolle et al. 2015). In transgene enhancer-blocking assays, the GAGA1–5 mutations weakened but did not eliminate boundary activity. This is also true in the context of BX-C. The Fab-7GAGA1–5 replacement retains boundary function and ∼15% of the mutant flies resemble wild-type. For the remaining Fab-7GAGA1–5 flies, the alterations in segment morphology are quite weak and can differ, often substantially, between flies or even within the same fly. As illustrated in Figure 7D, the A6 tergite in most male flies is marginally reduced in size, as would be expected for a GOF transformation. At the same time, there are also small patches of trichomes in regions of the A6 tergite that should be devoid of these hairs. This is characteristic of a loss-of-function transformation of A6 (PS11) into A5 (PS10). The sternites also show a mixture of weak GOF and LOF.
Combining the insv23B mutation with the Fab-7GAGA1–5 replacement disrupts boundary activity, giving a phenotype that resembles class II Fab-7 deletions, which retain HS3 (Mihaly et al. 1997). A typical insv23B; Fab-7GAGA1–5 male is shown in Figure 7E. The weak mixture of GOF and LOF phenotypes seen in Fab-7GAGA1–5 is replaced by a much stronger parasegmental transformation. As expected for a PS11←→PS12 transformation, the sternite is almost completely absent, while the size of the tergite is substantially reduced and quite irregular in shape. The two connected patches of residual tissue in the cuticle in Figure 7E are covered in trichomes, indicating that the surviving PS11 cells have assumed a PS10 identity. Likewise, in animals that have a residual sternite it is misshapen and has one or two bristles, as would be expected for PS10 identity.
To confirm that the insv23B mutation is responsible for the loss of Fab-7 boundary activity when combined with Fab-7GAGA1–5, we used a ϕC31 integration platform to introduce a genomic DNA fragment spanning the insv transcription unit. We found that the genomic insv fragment (insv+3.67) restores the boundary function of Fab-7GAGA1–5. Unlike their insv23B; Fab-7GAGA1–5 counterparts, insv23B; insv+3.67 Fab-7GAGA1–5 flies exhibit the same range and frequency of weak GOF and LOF transformations as seen in Fab-7GAGA1–5 (Figure 7F). The rescuing activity of the ϕC31 insv integrant is most clearly illustrated in the size of the tergite. In insv23B; Fab-7GAGA1–5 flies, there is a substantial reduction in the size of the A6 tergite and the residual tissue is typically covered in trichomes, as expected for a PS10 identity (Figure 7E). In contrast, in insv23B; insv+3.67 Fab-7GAGA1–5 flies, the tergite is typically only marginally smaller than wild-type and the PS10-like trichome hairs are usually limited to small patches (Figure 7F).
Discussion
Genome-wide ChIP experiments have shown that most Insv sites in vivo overlap sites for known fly architectural proteins, including BEAF, CP190, and CTCF (Dai et al. 2015). Among these sites, several correspond to boundaries in the fly homeotic complexes, including Fab-7 and Fab-8. However, this association was only a correlation, and there was no experimental evidence that Insv actually has an architectural function in flies. Indeed, there are no obvious homeotic transformations evident in insv23B mutant flies. On the other hand, the absence of phenotypic effects is not unusual, as fly boundaries typically utilize multiple factors and can in many instances tolerate the loss of any single factor (Cleard et al. 2017; Kyrchanova et al. 2017). Additionally, in the case of Insv, both its sequence recognition properties and its pattern of expression resemble another, closely related BEN domain boundary factor, Elba.
To determine whether Insv has architectural activities like the Elba factor, we focused on the Fab-7 boundary. ChIP experiments by Dai et al. (2015) indicate that Insv associates with four sites in the Fab-7 boundary. Two of these, P2 and P3, are located in the large nuclease hypersensitive site HS1, while the other two, P1 and P4, are located in hypersensitive sites HS* and HS2, respectively. The P1 and P3 peaks include the consensus Insv recognition motifs in HS* and HS1, the CCAATTGG palindrome, deduced from the ChIP experiments (Dai et al. 2015). We found that Insv binds to short probes spanning the P1 and P3 palindromes, and that mutations in these palindromes abrogate Insv binding. Like Insv, the Elba factor also binds to the two Fab-7 CCAATTGG palindromes, and in nuclear extracts from 0 to 6 hr embryos, the P1 and P3 probes are shifted by both Insv and Elba. Expression of the Elba factor is restricted to blastoderm and early gastrula stages, and Elba-binding activity disappears thereafter. Typically, there is little Elba activity detected in 6–18-hr nuclear extracts, and the P1 and P3 shifts are generated by Insv.
While the P1 and P3 peaks span the Insv palindrome, the two other peaks, P2 and P4, map to regions of Fab-7 that do not contain obvious Insv recognition sequences. P2 is located on the proximal side of the large hypersensitive region, HS1 (Figure 1). The P2 region contains the Elba recognition sequence; however, Insv does not bind to short probes containing the Elba motif (CCAATAAG). Instead, Insv binding requires a sequence of at least 60 bp. This Insv recognition sequence includes the Elba motif at its distal end and an internal GAF-binding sequence, GAGAG. We found that the Elba motif is important for Insv binding while the GAGAG motif is not. The crystal structure of the Insv BEN domain shows that it binds to its recognition sequence (the CCAATTGG palindrome) as a dimer. However, it is not clear how a dimerized Insv BEN domain would interact with a recognition sequence that has the Elba motif at one end and extends ≥ 60 bp away. Likely relevant to Insv interactions with this large sequences is the fact that it is found in a large complex in nuclear extracts. While an Insv dimer should be ∼80 kDa, Insv in nuclear extracts fractionates as a ∼420-kDa complex. It is possible that this complex contains as many as five Insv dimers, or has some additional protein(s) that mediate binding to this large recognition sequence. One plausible candidate would be CP190, which has been shown to interact directly with Insv (Dai et al. 2015). The other region of Fab-7 that is associated with Insv in vivo is located in HS2. However, this sequence does not contain the CCAATTGG palindrome and we were unable to detect Insv binding to probes spanning the HS2 region in our EMSA experiments. Given the unusual size of the P2 recognition sequence, it is possible that our probe design did not include all of the elements needed for Insv binding.
To assay for insv chromosome architectural activities, we used two Fab-7 replacements in which boundary function is partially compromised. In the first replacement, HS1 + HS2, DNA sequences for HS* and iab-7 PRE (HS3) are deleted, leaving the Insv/Elba-binding sites in HS1 and HS2. The HS1 + HS2 boundary is unusual in that it is nearly, if not fully, functional in cells giving rise to the adult tergite, while it has only minimal activity in cells that form the sternite. When combined with insv23B, a GOF transformation of A6 to A7 equivalent to that of class I deletions (which remove the Fab-7 boundary and iab-7 PRE) is observed. In the second, Fab-7GAGA1–5, GAGA sites 1 and 2 in pHS1 and 3–5 in dHS1 are mutant. The GAGA1–2 site mutations partially compromise boundary function during the initiation phase of BX-C regulation, while the GAGA3–5 mutations abrogate LBC binding and compromise boundary function during the maintenance phase. The mutant boundary retains all of the Insv/Elba- and Pita-binding sites and the iab-7 PRE (HS3), and in an otherwise wild-type background gives a mixture of weak GOF and LOF phenotypes in A6. However, in an insv23B background, boundary activity is completely lost and the phenotypic effects are the same as those observed in class II boundary deletions, which retain the iab-7 PRE. Confirming that insv is responsible, we found that a genomic insv rescue transgene restores Fab-7GAGA1–5 boundary function to that seen in an otherwise wild-type background.
It is worth noting that the Elba complex is developmentally restricted and contributes to Fab-7 boundary activity during the initiation phase of BX-C regulation, but not during the maintenance phase. As Elba and Insv bind to the same (the CCAATTGG palindrome) or overlapping (P2) sequences in Fab-7 (and presumably elsewhere in the genome), and both are ubiquitously expressed at the blastoderm stage, this raises questions about their respective roles. For example, ChIP and EMSA experiments localize Elba and Insv to the same sites in Fab-7; however, there is no indication of cooccupancy in our EMSA experiments with 0–6 hr nuclear extracts. In the case of the partially compromised Fab-7 boundaries, we have shown that Insv is essential for their residual function. This could mean that the Elba complex cannot substitute for Insv in these particular Fab-7 boundary mutants. In this respect, the differences in the developmental expression profile of Insv and the Elba complex proteins might be important. Alternatively, a combination of both Insv and Elba might be required for the functioning of these mutant boundaries.
In either case, it is clear that Insv is dispensable when the Fab-7 boundary is wild-type. This highlights a reoccurring feature of boundaries in flies: there are typically multiple backup mechanisms that ensure functionality. Yet another example would be the Pita sites in HS2. In an otherwise wild-type background, they are not needed for boundary function. Only when the Fab-7 boundary activity is compromised by mutations in binding sites for other proteins do they become important. Redundancy also extends to many other fly DNA-binding architectural proteins, including BEAF and highly conserved dCTCF. Flies can survive to the adult stage in the absence of either BEAF or dCTCF proteins (Mohan et al. 2007; Roy et al. 2007; Bonchuk et al. 2015). Curiously, while dCTCF is one among many in flies, its vertebrate counterpart is widely believed to be the only globally important DNA-binding architectural factor. This is quite surprising. As redundancy in flies appears to play a central role in helping to ensure functional robustness, one might think that the demands for robustness in the subdivision and higher-order organization of the very large, and genetically much more complex, chromosomes of vertebrates would require even more functional redundancy and backup mechanisms than in flies. It is also possible that mammalian boundaries, like those in flies, utilize other proteins besides dCTCF. In this regard, it may be of interest that mutations in one of the mammalian BEN domain proteins, NAC-1, has a segmentation defect in vertebra patterning in which the sixth lumbar vertebra (L6) is transformed into a sacral identity (Yap et al. 2013). This transformation is reminiscent of the parasegmental transformations observed for deletions of BX-C boundaries, or as is found here in sensitized Fab-7 backgrounds for insv mutations.
One must also wonder why Elba and Insv are deployed in early embryos, and then dispensed with in most tissues/cell types later in embryonic development. One idea is that they are not “general” boundary factors like BEAF or dCTCF, but rather have dedicated boundary activities that specifically target a special class of regulatory interactions that might, for example, be especially prevalent in early embryos. In the context of BX-C, this would be blocking cross talk between parasegment-specific initiation elements in neighboring cis-regulatory domains during the initiation phase of BX-C regulation (Kyrchanova et al. 2015; Maeda and Karch 2015). In a model invoking a “dedicated” function, neither factor would be able to block cross talk between, for example, PcG and Trx elements during the maintenance phase of BX-C regulation. At this point, it is not possible to answer this question; however, it clearly will be of interest to test whether Insv (or Elba) have a “dedicated” boundary function that is either relevant or especially important during early embryogenesis. Needless to say, their developmentally limited patterns of expression mean that their activities are de facto restricted by stage and tissue types.
Acknowledgments
This study was performed using the equipment of the Institute of Gene Biology (IGB) Russian Academy of Sciences facilities supported by the Ministry of Science and Education of the Russian Federation. We especially thank E. Lai and Q. Dai for the insv mutant flies, valuable discussions, and for sharing unpublished results. This study was supported by the Russian Science Foundation [project number 14-24-00166 (to P.G.)], the National Institutes of Health (NIH) [to P.S. (GM-043432 and GM-126975), and by grants from the Donation Claraz, the State of Geneva, and the Swiss National Fund for Research to F.K. P.S. acknowledges support from a grant to the IGB by the Russian Federation Ministry of Education and Science (14.B25.31.0022). C.C. and D.W. were supported by an NIH training grant (T32-GM-007388).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.6932915.
Communicating editor: M. Kuroda
Literature Cited
- Ali T., Renkawitz R., Bartkuhn M., 2016. Insulators and domains of gene expression. Curr. Opin. Genet. Dev. 37: 17–26. 10.1016/j.gde.2015.11.009 [DOI] [PubMed] [Google Scholar]
- Aoki T., Schweinsberg S., Manasson J., Schedl P., 2008. A stage-specific factor confers Fab-7 boundary activity during early embryogenesis in Drosophila. Mol. Cell. Biol. 28: 1047–1060. 10.1128/MCB.01622-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T., Sarkeshik A., Yates J., Schedl P., 2012. Elba, a novel developmentally regulated chromatin boundary factor is a hetero-tripartite DNA binding complex. Elife 1: e00171 10.7554/eLife.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T., Wolle D., Preger-Ben Noon E., Dai Q., Lai E. C., et al. , 2014. Bi-functional cross-linking reagents efficiently capture protein-DNA complexes in Drosophila embryos. Fly (Austin) 8: 43–51. 10.4161/fly.26805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F., Basler K., 2007. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104: 3312–3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonchuk A., Maksimenko O., Kyrchanova O., Ivlieva T., Mogila V., et al. , 2015. Functional role of dimerization and CP190 interacting domains of CTCF protein in Drosophila melanogaster. BMC Biol. 13: 63 10.1186/s12915-015-0168-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H. N., Shen P., 2001. Effects of cis arrangement of chromatin insulators on enhancer-blocking activity. Science 291: 493–495. 10.1126/science.291.5503.493 [DOI] [PubMed] [Google Scholar]
- Chetverina D., Aoki T., Erokhin M., Georgiev P., Schedl P., 2014. Making connections: insulators organize eukaryotic chromosomes into independent cis-regulatory networks. Bioessays 36: 163–172. 10.1002/bies.201300125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cléard F., Moshkin Y., Karch F., Maeda R. K., 2006. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat. Genet. 38: 931–935. 10.1038/ng1833 [DOI] [PubMed] [Google Scholar]
- Cleard F., Wolle D., Taverner A. M., Aoki T., Deshpande G., et al. , 2017. Different evolutionary strategies to conserve chromatin boundary function in the bithorax complex. Genetics 205: 589–603. 10.1534/genetics.116.195586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuartero S., Fresán U., Reina O., Planet E., Espinàs M. L., 2014. Ibf1 and Ibf2 are novel CP190-interacting proteins required for insulator function. EMBO J. 33: 637–647. 10.1002/embj.201386001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S., Jothi R., Schones D. E., Roh T.-Y., Cui K., et al. , 2009. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 19: 24–32. 10.1101/gr.082800.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q., Ren A., Westholm J. O., Serganov A. A., Patel D. J., et al. , 2013. The BEN domain is a novel sequence-specific DNA-binding domain conserved in neural transcriptional repressors. Genes Dev. 27: 602–614. 10.1101/gad.213314.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q., Ren A., Westholm J. O., Duan H., Patel D. J., et al. , 2015. Common and distinct DNA-binding and regulatory activities of the BEN-solo transcription factor family. Genes Dev. 29: 48–62. 10.1101/gad.252122.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H., Dai Q., Kavaler J., Bejarano F., Medranda G., et al. , 2011. Insensitive is a corepressor for suppressor of hairless and regulates Notch signalling during neural development. EMBO J. 30: 3120–3133. 10.1038/emboj.2011.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedotova A. A., Bonchuk A. N., Mogila V. A., Georgiev P. G., 2017. C2H2 zinc finger proteins: the largest but poorly explored family of higher eukaryotic transcription factors. Acta Naturae 9: 47–58. [PMC free article] [PubMed] [Google Scholar]
- Fujioka M., Sun G., Jaynes J. B., 2013. The Drosophila eve insulator Homie promotes eve expression and protects the adjacent gene from repression by polycomb spreading. PLoS Genet. 9: e1003883 10.1371/journal.pgen.1003883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloni M., Gyurkovics H., Schedl P., Karch F., 1993. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 12: 1087–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer P. K., Corces V. G., 1992. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 6: 1865–1873. 10.1101/gad.6.10.1865 [DOI] [PubMed] [Google Scholar]
- Gyurkovics H., Gausz J., Kummer J., Karch F., 1990. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 9: 2579–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K., Muller M., Schedl P., 1996. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 10: 3202–3215. 10.1101/gad.10.24.3202 [DOI] [PubMed] [Google Scholar]
- Holdridge C., Dorsett D., 1991. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell. Biol. 11: 1894–1900. 10.1128/MCB.11.4.1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan E. E., Kwong C., Adryan B., Bartkuhn M., Herold M., et al. , 2007. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 3: e112 10.1371/journal.pgen.0030112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Emberly E., Cuvier O., Hart C. M., 2009. Genome-wide mapping of boundary element-associated factor (BEAF) binding sites in Drosophila melanogaster links BEAF to transcription. Mol. Cell. Biol. 29: 3556–3568. 10.1128/MCB.01748-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellum R., Schedl P., 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64: 941–950. 10.1016/0092-8674(91)90318-S [DOI] [PubMed] [Google Scholar]
- Kellum R., Schedl P., 1992. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 12: 2424–2431. 10.1128/MCB.12.5.2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Chetverina D., Maksimenko O., Kullyev A., Georgiev P., 2008. Orientation-dependent interaction between Drosophila insulators is a property of this class of regulatory elements. Nucleic Acids Res. 36: 7019–7028. 10.1093/nar/gkn781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Maksimenko O., Stakhov V., Ivlieva T., Parshikov A., et al. , 2013. Effective blocking of the white enhancer requires cooperation between two main mechanisms suggested for the insulator function. PLoS Genet. 9: e1003606 10.1371/journal.pgen.1003606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Mogila V., Wolle D., Magbanua J. P., White R., et al. , 2015. The boundary paradox in the Bithorax complex. Mech. Dev. 138: 122–132. 10.1016/j.mod.2015.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrchanova O., Zolotarev N., Mogila V., Maksimenko O., Schedl P., et al. , 2017. Architectural protein Pita cooperates with dCTCF in organization of functional boundaries in Bithorax complex. Development 144: 2663–2672. 10.1242/dev.149815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B., 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- Li H.-B., Müller M., Bahechar I. A., Kyrchanova O., Ohno K., et al. , 2011. Insulators, not Polycomb response elements, are required for long-range interactions between Polycomb targets in Drosophila melanogaster. Mol. Cell. Biol. 31: 616–625. 10.1128/MCB.00849-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Li M., Roy S., Liu K. J., Romine M. L., et al. , 2016. Chromatin boundary elements organize genomic architecture and developmental gene regulation in Drosophila Hox clusters. World J. Biol. Chem. 7: 223–230. 10.4331/wjbc.v7.i3.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda R. K., Karch F., 2015. The open for business model of the bithorax complex in Drosophila. Chromosoma 124: 293–307. 10.1007/s00412-015-0522-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimenko O., Georgiev P., 2014. Mechanisms and proteins involved in long-distance interactions. Front. Genet. 5: 28 10.3389/fgene.2014.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksimenko O., Bartkuhn M., Stakhov V., Herold M., Zolotarev N., et al. , 2015. Two new insulator proteins, Pita and ZIPIC, target CP190 to chromatin. Genome Res. 25: 89–99. 10.1101/gr.174169.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzat L. H., Lei E. P., 2014. Surviving an identity crisis: a revised view of chromatin insulators in the genomics era. Biochim. Biophys. Acta 1839: 203–214. 10.1016/j.bbagrm.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M., Nora E. P., 2016. CTCF and cohesin in genome folding and transcriptional gene regulation. Annu. Rev. Genomics Hum. Genet. 17: 17–43. 10.1146/annurev-genom-083115-022339 [DOI] [PubMed] [Google Scholar]
- Mihaly J., Hogga I., Gausz J., Gyurkovics H., Karch F., 1997. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 124: 1809–1820. [DOI] [PubMed] [Google Scholar]
- Mohan M., Bartkuhn M., Herold M., Philippen A., Heinl N., et al. , 2007. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 26: 4203–4214. 10.1038/sj.emboj.7601851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M., Hagstrom K., Gyurkovics H., Pirrotta V., Schedl P., 1999. The mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 153: 1333–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muravyova E., Golovnin A., Gracheva E., Parshikov A., Belenkaya T., et al. , 2001. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291: 495–498. 10.1126/science.291.5503.495 [DOI] [PubMed] [Google Scholar]
- Nègre N., Brown C. D., Shah P. K., Kheradpour P., Morrison C. A., et al. , 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6: e1000814 10.1371/journal.pgen.1000814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves N., Posakony J. W., 2005. Genetic programs activated by proneural proteins in the developing Drosophila PNS. Dev. Cell 8: 413–425. 10.1016/j.devcel.2005.01.020 [DOI] [PubMed] [Google Scholar]
- Roy S., Gilbert M. K., Hart C. M., 2007. Characterization of BEAF mutations isolated by homologous recombination in Drosophila. Genetics 176: 801–813. 10.1534/genetics.106.068056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Herrero E., Vernós I., Marco R., Morata G., 1985. Genetic organization of Drosophila bithorax complex. Nature 313: 108–113. 10.1038/313108a0 [DOI] [PubMed] [Google Scholar]
- Schwartz Y. B., Linder-Basso D., Kharchenko P. V., Tolstorukov M. Y., Kim M., et al. , 2012. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 22: 2188–2198. 10.1101/gr.138156.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg S., Hagstrom K., Gohl D., Schedl P., Kumar R. P., et al. , 2004. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 168: 1371–1384. 10.1534/genetics.104.029561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg S. E., Schedl P., 2004. Developmental modulation of Fab-7 boundary function. Development 131: 4743–4749. 10.1242/dev.01343 [DOI] [PubMed] [Google Scholar]
- Sigrist C. J., Pirrotta V., 1997. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics 147: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. T., Wickramasinghe P., Olson A., Loukinov D., Lin L., et al. , 2009. Genome wide ChIP-chip analyses reveal important roles for CTCF in Drosophila genome organization. Dev. Biol. 328: 518–528. 10.1016/j.ydbio.2008.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri Rudan M., Hadjur S., 2015. Genetic tailors: CTCF and cohesin shape the genome during evolution. Trends Genet. TIG 31: 651–660. 10.1016/j.tig.2015.09.004 [DOI] [PubMed] [Google Scholar]
- Wolle D., Cleard F., Aoki T., Deshpande G., Schedl P., et al. , 2015. Functional requirements for Fab-7 boundary activity in the bithorax complex. Mol. Cell. Biol. 35: 3739–3752. 10.1128/MCB.00456-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K. L., Sysa-Shah P., Bolon B., Wu R. C., Gao M., et al. , 2013. Loss of Nac1 expression is associated with defective bony patterning in the murine vertebral axis. PLos One 8: e69099 10.1371/journal.pone.0069099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Barolo S., Szymanski P., Levine M., 1996. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 10: 3195–3201. 10.1101/gad.10.24.3195 [DOI] [PubMed] [Google Scholar]
- Zolotarev N., Fedotova A., Kyrchanova O., Bonchuk A., Penin A. A., et al. , 2016. Architectural proteins Pita, Zw5,and ZIPIC contain homodimerization domain and support specific long-range interactions in Drosophila. Nucleic Acids Res. 44: 7228–7241. 10.1093/nar/gkw371 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Reagents (antibodies and strains) used for the experiments in this paper are available upon request. Results presented in figures and supplemental figures support the conclusions in this paper. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.6932915.