Abstract
Tumor-specific delivery of therapeutics is challenging. One of the major hurdles for successfully delivering targeted agents by nanovectors is the filtering role of the liver in rapidly sequestering nanovectors from the circulation. Exosomes, a type of endogenous nanoparticle, circulate continuously in the peripheral blood and play a role in intercellular communication. The aim of this study was to determine whether the level of endogenous exosomes has an effect on nanovector delivery efficiency of targeted agents.
Methods: Exosomes were isolated from peripheral blood and intravenously (I.V.) injected into tumor-bearing mice. Subsequently, 1,1-dioctadecyl-3,3,3'3'-tetramethylindotricarbocyanine-iodide (DiR) fluorescent dye-labeled nanoparticles, including grapefruit nanovectors (GNV) and standard liposomes, were I.V. injected in the mice. The efficiency of redirecting GNVs from liver to other organs of injected mice was further analyzed with in vivo imaging. The concentration of chemo drugs delivered by GNV was measured by HPLC and the anti-lung metastasis therapeutic effects of chemo drugs delivered by GNVs in mouse breast cancer and melanoma cancer models were evaluated.
Results: We show that tail vein-injected exosomes isolated from mouse peripheral blood were predominately taken up by liver Kupffer cells. Injection of peripheral blood-derived exosomes before I.V. injection of grapefruit-derived nanovector (GNV) decreased the deposition of GNV in the liver and redirected the GNV to the lung and to the tumor in breast and melanoma tumor-bearing mouse models. Enhanced therapeutic efficiency of doxorubicin (Dox) or paclitaxel (PTX) carried by GNVs for lung metastases was demonstrated when there was an I.V. injection of exosomes before therapeutic treatment. Furthermore, we found that CD36 and IGFR1 receptor-mediated pathways played a critical role in the exosome-mediated inhibitory effect of GNV entry into liver macrophages.
Conclusions: Collectively, our findings provide a foundation for using autologous exosomes to enhance therapeutic vector targeted delivery to the lung.
Keywords: Blood exosomes, nanovector uptake, liver Kupffer cells, CD36 and IGFR1, outer nuclear membrane cluster, lung metastasis
Introduction
Despite their many potential advantages for therapeutic delivery, nanoparticle-based delivery systems must overcome many hurdles such as eliminating the induction of cytotoxic effects due to off-targeting. Unlike the situation where nanovectors are synthesized artificially, nano-sized exosome-like nanoparticles from edible plants have been utilized for encapsulating drugs 1, siRNA 2, DNA expression vectors, and antibody to treat diseases in mouse models without causing side effects. Although the use of edible plant exosome-like nanovectors in therapeutic delivery holds great promise, effective and efficient delivery of the target remains to be developed.
The liver is the major site for removing circulating macromolecules including exosomes 3 and nano-sized exosome-like nanovectors, such as GNV made from grapefruit-derived lipids 4. The rapid sequestration of I.V.-injected nanovectors from the blood by Kupffer cells is one of the major problems for efficient delivery of targeted drug carriers to a desired cell population and for prevention of liver toxicity 5-8. From a clinical application standpoint, whether pre-injection of autologous exosomes blocks therapeutic nanovector uptake by liver macrophages has not been tested.
In this study, we hypothesized that uptake of circulating exosomes leads to a reduction of liver uptake of subsequently I.V.-administered therapeutic nanovectors. Here, we demonstrate that autologous exosomes are predominately taken up by liver Kupffer cells, and the autologous exosomes are critical for regulating the distribution of I.V.-injected nanovectors. Increasing the level of peripheral blood-derived exosomes by I.V. injection blocked accumulation of subsequently injected nanovectors in the liver. Uptake of peripheral blood-derived exosomes lead to redirection of I.V.-injected GNV from the liver to the lungs under “normal” physiological conditions and to the tumor in breast and melanoma tumor-bearing mouse models. The CD36- and IGFR1-mediated pathways play a role in exosome-mediated distribution of injected nanovectors. We also found that injection of exosomes isolated from circulating blood enhanced the amount of therapeutic agent delivered by GNV to the tumor and prevented lung metastasis. These findings suggest that an increased level of circulating exosomes is capable of enhancing the efficiency of targeted delivery of therapeutic agents to lungs where most human tumor metastases occur 9-12. This procedure could be done by isolating exosomes from the peripheral blood of a patient. Isolated exosome would then be perfused back into the same patient before the patient is I.V.-administered therapeutic nanovectors, such as GNVs, as demonstrated in mouse models in this study. Furthermore, these findings not only have implications for personalized therapy of lung cancer and lung metastasis, but also support the use of this strategy to increase delivery of therapeutic agents to primary tumor and further studying the effect of a delivered agent on the tumor microenvironment.
Results
Exosomes isolated from peripheral blood enhanced accumulation of grapefruit nanovectors (GNV) and liposomes in the lungs by inhibiting uptake of GNVs by liver macrophages
Despite the many potential advantages of nanovectors 13 and liposomes as therapeutic agent delivery systems, most of the nanovectors end up in the liver and spleen; thus, delivery of targeted therapeutic agents to the appropriate tissue is prevented. The rapid sequestering of nanovectors from the circulation in the liver presents a huge challenge for effective therapy. Exosomes are released from many different types of cells and circulate continuously in the blood. Although circulating exosomes provide a promising approach to assess biomarkers in human disease 14, their role in terms of modulating the route of therapeutic nanovectors injected I.V. is not known. The overall goal of this study was to determine whether circulating exosomes have an effect on the targeted delivery of therapeutic agents and whether these exosomes may improve the therapeutic efficiency and effectiveness of nanovectors in treatment of various diseases. We first sought to determine whether circulating exosomes home to the same tissue as the nanovectors we tested. In vivo imaging analysis indicated that the majority of the nanovectors homed to the liver and spleen within 12 h after tail vein injection (Figure S1A-B). Confocal imaging data further demonstrated that Kupffer cells took up the injected nanoparticles including the GNVs 4 and IGNVs 15 we made and commercial liposomes (Figure S1C). Furthermore, the data generated from depletion of Kupffer cells experiments (Figure S2) indicates that depletion of Kupffer cells led to a redirection of GNVs from the liver and spleen to the lungs. Enhanced GNV signals in the lungs correlates with a reduction of GNV signals in the liver and spleen (Figure S3A-C). Collectively, these data suggest that I.V.-injected nanovectors are taken up by Kupffer cells and depletion of Kupffer cells redirects the nanovectors to the lungs. However, from a clinical application standpoint, pre-depletion of a patient's Kupffer cells to prevent liver uptake of therapeutic nanovectors is not acceptable. Therefore, we tested if injection of autologous exosomes back into the patient before delivery of the therapeutic nanovector would block subsequent Kupffer cell uptake. Western blot analysis (Figure 1A) showed that exosomes were present in the blood sample and shared a number of exosomal protein markers including CD63, CD9, and CD81.
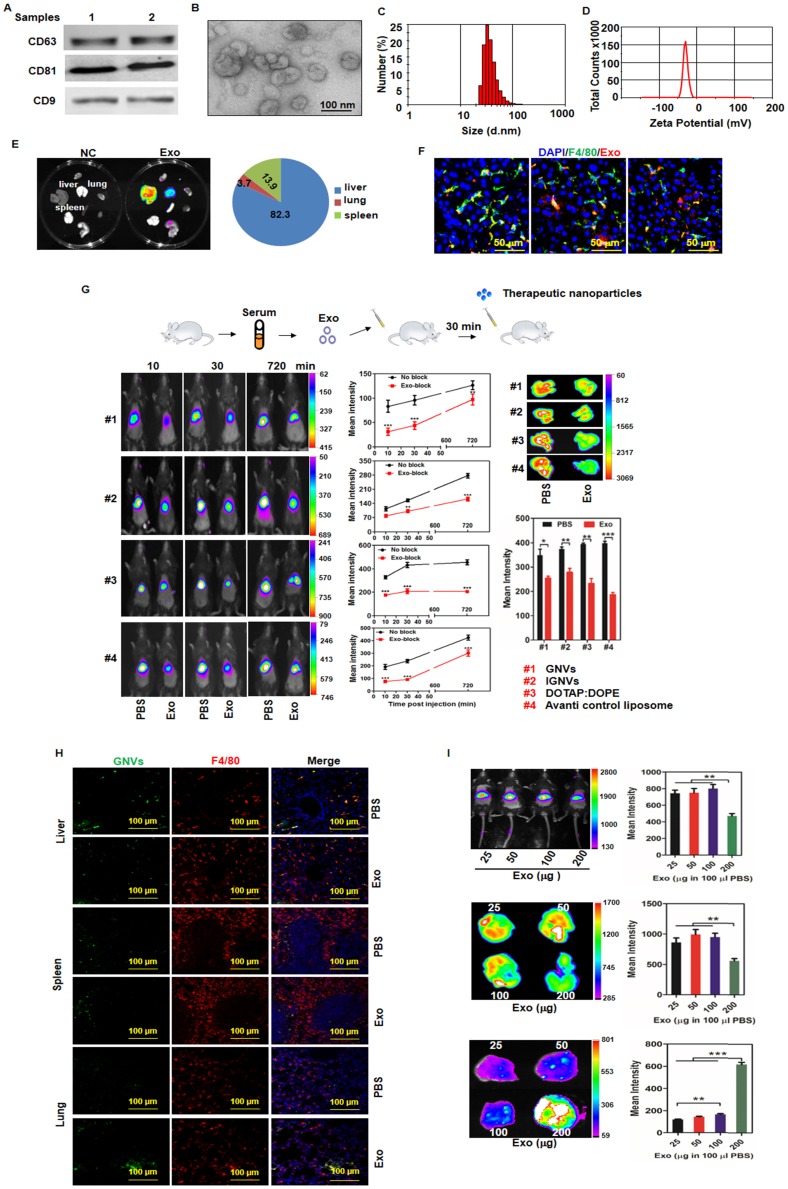
Figure 1.
The majority of circulating exosomes are taken up by liver F4/80 macrophages and pre-injection of exosomes leads to redirection of subsequently injected nanovectors from the liver to the lungs. Exosomes from normal mouse plasma were isolated using the PureExo® Exosomes Isolation kit. (A) Representative samples of Western blotting for exosome CD63, CD81, CD9 are shown. (B) The morphology of exosomes was examined and imaged using transmission electron microscopy (TEM). The size distribution (C) and surface Zeta potential (D) of exosomes were measured using a ZetaSizer. (E) Distribution of DiR-labeled exosomes in normal mice. Mice were I.V. injected with 200 μg of exosomes and DiR signals in the liver, lung, spleen, kidney, heart, thymus, brain and stomach were analyzed by scanning using a Kodak Image System. (F) Livers from mice were removed over a 24-72 h period after I.V. injection and liver tissue sections were stained with anti-mouse F4/80 antibody. Representative images of DiR-labeled exosomes from mice and F4/80-stained liver section. (G) Exosomes (200 μg) were isolated from plasma of normal mice and injected I.V. into mice. DiR dye-labeled nanovectors including grapefruit lipid-derived GNVs (#1), lymphocyte membrane-coated GNVs-IGNVs (#2), DOTAP:DOPE liposomes (#3) and liposomes from Avanti (#4) were I.V. injected into mice 30 min after an injection of exosomes. Accumulation of nanovectors in mouse liver was examined in living mice (left panel) and ex vivo livers (right panel). Representative images of DiR-labeled nanovectors from mice (left panel) and livers (right panel), followed by bar charts of the mean net intensity (sum intensity/area, n=5). For statistics, see method section “Statistics”. (H) PKH67-labeled GNVs (200 nmol) were I.V. injected into mice that were previously I.V. injected with exosomes (200 μg/mouse). Liver, lung and spleen were removed and PKH67-GNVs in tissues sections stained with anti-F4/80 antibody were imaged using confocal microscopy. Representative images are shown. (I) Inhibition of liver accumulation of nanovectors. Different doses of C57BL/6 mice serum-derived exosomes (25, 50, 100 and 200 μg) were I.V. injected into C57BL/6 mice. 30 min after injection of exosomes, mice were injected with 200 nmol DiR dye-labeled GNVs. DiR dye signals in living mice (top panel), liver (middle panel) and lung (bottom panels) were quantitatively analyzed using a Kodak Image System. The data are presented as the mean net intensity (sum intensity/area, n=5). Data are presented as mean ± SD, **p < 0.01, ***p < 0.001. Error bars represent SD. The data (A-F, H) shown are representative of at least 3 independent experiments (n=5).
Exosomal morphology, size distribution and Zeta potential were further evaluated using electron microscopy (Figure 1B) and Zetasizer Nano ZS analysis (Figure 1C-D), respectively. After tail vein injection, exosomes primarily trafficked to the liver (Figure 1E) and were taken up by Kupffer cells (Figure 1F). Remarkably, 30 min after I.V. injection of exosomes, the intensity of the liver signal of all 4 different types of nanovectors we tested was reduced significantly (Figure 1G); whereas, the number of positive GNVs in the lungs was increased (Figure 1H, bottom panel). The significantly increased signal of injected GNVs in the lungs was evident when 200 μg of GNVs/mouse were I.V. injected (Figure 1I). No abnormality was observed in terms of body weight of mice I.V. injected with autologous exosomes (200 μg in 100 μL PBS) compared with PBS alone (Figure S4).
Injection of peripheral blood-derived exosomes inhibited lung metastasis by enhancing the accumulation of therapeutic GNVs in the tumor and lungs
Most cancer deaths result from metastasis. The lungs are the most common clinically relevant site of cancer metastases including breast cancer, where metastatic breast cancer remains a therapeutic challenge. As proof-of-concept, GNVs were used to determine whether injection of circulating exosomes leads to enhanced accumulation of GNVs in tumor and lungs. The metastatic mouse mammary carcinoma 4T1 model was used. The mammary pads of mice were injected with 4T1 cells, and at day 14 post-injection, mice were I.V. injected with exosomes purified from circulating blood of BALB/c mice and PBS was used as a control. Thirty min later, mice were I.V. injected with DiR dye-labeled GNVs. Live mouse imaging data indicates that pre-injection of exosomes significantly enhanced the GNV signals detected in circulating blood (Figure 2A) and breast tumor (Figure 2B). This result was corroborated by quantitative analysis of the GNVs signals in the tumor and lungs (Figure 2C) and the delivery of the chemo drug PTX by GNVs (Figure 2D). More than 80% of the chemo drugs, including Dox and PTX, were efficiently coupled with GNVs (Figure S5).
Figure 2.
Exosomes redirect GNVs from the liver to the lungs and tumor. (A) DiR dye-labeled GNVs were I.V. injected into 6-week-old female BALB/c mice pretreated with exosomes or clodrosomes to deplete Kupffer cells, or PBS as a control (Normal). Peripheral blood was collected into anticoagulant tubes 30, 60, and 180 min after injection. The DiR dye signals in blood were quantified by scanning using a Kodak Image System. The data are presented as the mean net intensity (sum intensity/area, n=5). (B) DiR dye-labeled GNVs were I.V. injected into 4T1 tumor-bearing mice pretreated with exosomes or PBS as a control. 4T1 tumor-bearing mice without any treatment were used as a negative control (NC). Representative images collected at 1 h, 3 h, 6 h and 20 h after injection. The data are presented as the mean net intensity (sum intensity/area, n=5). (C) Mice were sacrificed, tumors and organs (liver, lung, spleen, heart, thymus, kidney, and lymph node) were removed, scanned using a Kodak Image System and quantitatively analyzed. The data are presented as the mean net intensity (sum intensity/area, n=5). (D) Distribution of paclitaxel in B16F10 tumor-bearing mice. B16F10 tumor-bearing mice were I.V. injected with paclitaxel-loaded GNV (GNV-PTX) 3 times. The concentration of paclitaxel in mouse tumor, liver, lung and spleen was analyzed using HPLC. (n=3). Data are presented as mean ± SD, *p < 0.05, **p < 0.01, ***p < 0.001. Error bars represent SD.
Next, we tested whether observing an enhanced GNV signal in the lung leads to better therapeutic effects regarding inhibition of 4T1 breast tumor metastasis. On day 35 post tumor cell injection, mice with 4T1 tumor cells succumbed to significant lung metastasis (Figure 3A). In contrast, mice pre-injected with autologous exosomes followed by I.V. administration of GNVs carrying Dox had a decreased number of macro- (Figure 3A) and micro- (Figure 3B) metastatic tumor nodules when compared to mice pre-injected with PBS as a control. Pre-injection with exosomes followed by I.V. administration of GNV carrying Dox significantly prolonged the survival rate of 4T1-bearing mice with lung metastases (Figure 3C).
Figure 3.
Pre-injection of blood-derived exosomes enhances anti-tumor metastasis of therapeutic agents delivered by GNVs. 1×105 4T1 cells were injected into a mammary fat pad of female BALB/c mice. On day 5 after the injection, mice were tail vein injected every 3 days for a total of 10 times with PBS, Dox, GNV-Dox, Exo/GNV-Dox. (A) Mice were sacrificed and lungs were imaged and the pulmonary metastatic nodules were quantified. (B) Lung tissue sections were stained with H&E. (C) Representative images of lung and sectioned lung tissue (n=5), and survival rates of mice were recorded. B16F10 cells (5×104) were I.V. injected into C57BL/6 mice. 5 days later, mice were tail vein injected every 3 days for a total of 10 times with PBS, PTX, GNV-PTX or Exo/GNV-PTX. Lungs were removed, imaged and the metastatic nodules in lungs were quantitative analyzed (D), tissue sections were stained with H&E (E), and survival rates of mice were recorded (F). Data are presented as mean ± SD, *p<0.05, p**<0.01 and p***<0.001. Error bars represent SD. The data (A-D) shown are representative of at least 3 independent experiments (n=5).
Collectively, these results indicate that pre-injection of autologous, peripheral blood exosomes enhanced the anti-lung metastasis therapeutic effects of Dox delivered by GNVs.
To test whether the enhanced GNV signal in the lung and breast tumor was applicable to other types of cancer, murine melanoma B16F10 was used since the current therapy for lung metastasis of melanoma is disappointing. The therapeutic effects of PTX on preventing the growth of I.V.-injected B16F10 tumor cells in the lung were determined.
To determine whether pre-injection of exosomes isolated from peripheral blood would result in reduced lung metastatic potential, 5 days after tail vein injection of tumor cells, mice were treated with GNV carrying PTX every 3 days for 30 days. Despite the fact that this route bypasses several of the steps occurring during metastasis, it enabled us to focus on the potential effect of exosomes injected at the final stages of metastasis. Injection of exosomes and GNV-PTX resulted in decreased numbers of macro lung metastases in the mice injected with B16F10 cells. The results indicate that mice pre-injected with exosomes followed by I.V.-administered GNV carrying PTX had fewer lung macro- and micro-metastatic tumor nodules than mice that were not injected with exosomes or injected with PBS as a control (Figure 3D). Confirmation of the tumors as melanomas in mice pre-injected with exosomes was confirmed by histological analysis of H&E-stained sections (Figure 3E). This result is also supported by the fact that the lowest mortality was observed in the group of mice pre-injected with exosomes, followed by I.V. administration of GNVs carrying PTX (Figure 3F).
CD36 and IGFR1 receptor-mediated pathways play a crucial role in exosomes-mediated prevention of GNV nanovector uptake
Collectively, the data suggest that exosomes are rapidly sequestered by liver macrophages and prevent subsequent macrophage uptake of GNV. Next, we determined whether preventing the uptake of GNVs is a macrophage receptor-mediated event. CD36 is known as FAT (fatty acid translocase) and binds ligands 16,17. To determine whether macrophage CD36 plays an in vivo role in exosomes entry, we performed confocal imaging analysis of the localization of GNVs with liver macrophages that were F4/80+CD36+. Our data indicate that F4/80+CD36+ liver macrophages took up GNVs 30 min after I.V. injection (Figure 4A). To determine whether pre-injection of exosomes had an effect on the location of GNVs in CD36+ liver macrophages, mouse liver F4/80+ macrophages were isolated from mice pre-injected with exosomes or PBS as a control. In the control group of mice I.V. injected with PBS, immunohistological staining revealed CD36 is clustered at the outer nuclear membrane and is co-localized with GNVs (Figure 4B, top panel). In contrast, when exosomes were pre-injected, we did not observe CD36 clustered at the outer nuclear membrane and there was a much weaker GNV signal on the outside of the nucleus (Figure 4B, bottom panel). The role of macrophage CD36 in the uptake of GNVs was further demonstrated by in vivo image analysis. In wild-type B6 mice used as a control, the liver GNV signal was reduced significantly (n=5) when mice were pre-injected with exosomes, compared with the liver signal of mice pre-injected with PBS (Figure 4C). CD36 KO lead to a reduced difference in GNV signal intensity between mice pre-injected with exosomes and mice pre-injected with PBS (Figure 4C). These data indicate that uptake of exosomes leads to blocking of CD36/GNV cluster formation at the outer membrane of the nucleus. The exosome-mediated reduction of cluster formation is associated with the subsequent inhibition of GNV entry into liver macrophages.
Figure 4.
Pre-injection of exosomes prevents co-localization of CD36 and GNVs and knockout of CD36 leads to canceling of exosomes-mediated inhibition of liver uptake of GNVs. Six-week-old B6 mice were I.V. injected with DiR-GNVs (2 nmol/100 μL). 30 min after the injection, mice were sacrificed and liver sections were immuno-stained with CD36 and F4/80. (A) Representative images of sectioned liver tissue (n=5). Six-week old B6 mice were I.V. injected with exosomes (200 μg/mouse) isolated from peripheral blood or PBS as a control. 30 min after the injection, DiR-GNVs (200 nmol) was administered intravenously. The mice were sacrificed and F4/80-positive cells were FACS-sorted and stained with CD36 antibody. (B) Representative confocal images of F4/80 positive cells isolated from liver (n=5). Wild-type B6 mice and age/sex-matched CD36 KO mice were I.V. injected with C57BJ/6 plasma exosomes (200 μg/mouse, n=5) followed by DiR-labeled GNVs (200 nmol) at 30 min intervals. (C) The DiR signals in living mice (top panel) and liver tissue (bottom panel) were imaged 30 min after the GNV injection.
It has been reported that the resulting formation of CD36 clusters initiates signal transduction and internalization of receptor-ligand complexes and tyrosine family kinases are required for CD36 clustering 18. How exosomes regulate kinase(s) to prevent subsequent GNV entry into macrophages is not known. From a clinical application standpoint, human monocytes were used to address this question. FACS analysis of U937 human monocytes pre-treated with/without exosomes isolated from peripheral blood of healthy subjects indicates that exosomes inhibit U937 uptake of GNVs as early as 30 min after exosome treatment (Figure 5A). Using siRNA technology, the results generated from a human tyrosine kinase siRNA library screening assay indicated that knockdown of LTK, IGFR1, and FYN genes completely reversed the exosome-mediated inhibition of GNV entry (Figure 5B, left panel). The efficiencies of LTK and FYN siRNA knockdown were highly variable (data not shown) although we did not determine the cause of the variation. However, the efficiency of IGFR1 siRNA knockdown was highly reproducible (Figure 5B, right panel).
Figure 5.
siRNA knockdown of IGFR1 reversed exosomes-mediated inhibition of GNV uptake by human monocytes. (A) U937 human monocytes were incubated for 30 min with exosomes (2 μg/100 culture media) isolated from healthy subjects. Treated cells were then incubated with PKH26-labeled GNVs (2.0 nmol) for additional 0-120 min and the cells were subsequently FACS analyzed. Representative FACS images of GNV-positive cells (n=5). (B) 48 h siRNA-transfected U937 human monocytes were incubated with/without exosomes for 30 min. Then, PKH26-labeled GNVs were added to the treated cells and incubated for an additional 60 min before cells were harvested for FACS analysis of PKH26-positive cells. Representative FACS images of GNV-positive cells (left panel) and the percentages of GNVs+ U937 cells as a result of siRNA knockout of IGFR1 are presented as mean ± SD (right panel), *p < 0.05. Error bars represent SD.
Discussion
In this study, we discovered the utility of blood-derived exosomes in enhancing targeted delivery of therapeutic agents carried by nanovectors for treatment of lung metastasis.
We demonstrated that I.V. injection of autologous exosomes results in a decreased capacity of Kupffer cells to take up subsequently injected GNV and redirects the GNVs from the liver to the lungs. The therapeutic utility of these results was further demonstrated by the inhibition of breast and melanoma lung metastasis in murine models. This finding provides a foundation for further studying the regulatory role of circulating exosomes in terms of response to circulating foreign nanoparticles including nanovectors in general. In addition, this approach has the potential of directly translating into clinical applications for treatment of lung-related diseases using autologous exosomes. Unlike tumor cell-derived exosomes, which are important in facilitating cancer progression and spread 9,19, no abnormality was observed in terms of body weight of mice I.V. injected with autologous exosomes. Although other nanoparticles could potentially be used for blocking liver Kupffer cell uptake, one common feature of all other nanoparticles is that they are made of “foreign” material, which the host immune system will recognize and respond to as foreign material. Therefore, the safety and side effects of those materials in an individual are unpredictable. Those issues are of no concern for autologous exosomes and no pharmaceuticals are needed to make them, which eliminates cost as an issue.
Furthermore, this study demonstrates that there is a relationship between CD36 and IGFR1-mediated signaling pathways. Our findings point to the molecular pathway underlying exosome-mediated inhibition of entry of GNVs into Kupffer cells. Our data show that this inhibitory effect is CD36 and IGFR1-dependent, since knockout of the CD36 gene encoding for CD36 receptor or siRNA knockdown of IGFR1 negates the inhibitory effect. These observations, together with earlier findings that CD36 can associate with and activate tyrosine family kinases 18, provide a defined pathway whereby the receptor triggers internalization of nanovectors, like GNVs, into Kupffer cells. Both CD36 and IGFR1-mediated pathways may work in an independent manner or via cross-talk with each other to control the level of nanovectors or nanoparticles taken up by Kupffer cells. Exosomes circulating in the peripheral blood may serve as an inter-pathway communicator for this cross-talk. However, further investigation into whether CD36 cross-talks with the IGFR1 pathway through exosomes that regulate the uptake of nanoparticles could be useful for learning more about the role of circulating exosomes in the context of cleaning up foreign particles in the circulatory system.
Liver macrophages take up circulating particles via a number of pathways. We speculate that at the exosome level, injecting (I.V.) less than 200 μg of exosomes could be under the threshold for endogenous exosome-mediated activation of CD36. GNV may continuously be taken up by liver macrophages via other pathways that are not affected by uptake of endogenous exosomes. When the necessary or required exosome level is reached to where the CD36 pathway is activated, the uptake of GNVs is inhibited. Under what conditions circulating exosomes can reach more than 200 μg of exosomal protein in the peripheral blood is not known and requires further investigation. In addition, whether the exosome-mediated inhibitory effect on the uptake of other types of nanovectors is applicable to other types of foreign particles needs to be further studied. Similarly, GNVs can be taken up by liver macrophages via a number of pathways. Our results presented in this study suggest that the CD36 pathway activated by exosomes is an inhibitory pathway for the uptake of GNVs. How the CD36-mediated inhibitory pathway interacts with the pathways that regulate GNV uptake needs to be further investigated. Furthermore, the fact that I.V. injection of exosomes partially inhibits the uptake of GNVs suggests that other pathways that may not be affected by elevated exosomes also play a role in GNV uptake.
The depletion of macrophages using clodrosomes leads to more GNVs in circulation and longer circulation in peripheral blood (Figure 2A) than does injection of exosomes isolated from peripheral blood. The longer circulation time of the nanovectors without being eliminated from the bloodstream is significant for their accumulation in tumor tissues through the enhanced permeability and retention (EPR) effect 20,21. As demonstrated in mouse models 22, the depletion of macrophages using clodrosomes has a better therapeutic effect on prevention of lung metastasis; however, using clodrosomes is not applicable in human patients. Collectively, our results and others 22 provides a rationale for future studies of whether combination approaches, such as autologous exosomes pretreatment, followed by antibody-mediated depletion of macrophages will further enhance targeted delivery efficiency for treatment of lung disease.
Although the exact nature of the GNV-induced outer nuclear membrane cluster inhibition by exosomes remains to be defined, the fact that the exosome treatment leads to blocking of the outer nuclear cluster formation induced by GNVs is significant. Proteins gain entry into the nucleus through the nuclear envelope (NE). The NE consists of concentric outer and inner membranes. The NE has important functions in regulating membrane rigidity, gene expression, and chromosome organization 23. Dysfunctions in the NE impair NE architecture and cause human diseases such as rapid aging and cancers 24. Liposome-like GNVs induce transient formation of the outer nuclear membrane and endogenous exosomes can inhibit the GNV-induced formation of the outer nuclear membrane cluster. This knowledge provides a foundation for further investigations of whether exosomes play a regulatory role in NE rigidity, gene expression, and chromosome organization.
Although this study focuses on clinical applications for treatment of lung metastasis, the findings in this study provide a foundation for further investigations of whether other microvesicles 25 may also contribute to blocking of nanovector uptake by liver macrophages. We also realize that the purity of exosomes isolated with the method described in this study is lower than the purity achieved using the sucrose gradient method 26. However, from a clinical therapeutic viewpoint, our method is superior to the sucrose cushion method. Our method can be accomplished: 1) within 1-2 h instead of more than 8 h by the sucrose gradient method; 2) with fewer steps, which reduces potential contamination during the purification process; and 3) better yield since fewer exosomes are lost at each step. Nevertheless, we don't currently have technology to isolate and purify exosomes without “contamination” 27. Even within a single exosome population, many distinct subtypes of exosomes exist; therefore, the development of new, more selective isolation techniques is urgently needed to achieve good purity in each subset of vesicle population for clinical applications.
The tumor microenvironment has an effect on tumor metastasis 9,28. Our data show that not only does the autologous exosome pretreatment lead to an enhanced therapeutic effect on lung metastasis, but also increases the GNV signal in the primary tumor. Therefore, the current study provides a foundation for further studying whether the treatments as described in this study have effects on the tumor microenvironment including cellular composition, cytokine profiles and other molecular profiles that have been demonstrated to have an effect on tumor metastasis.
Methods
Mice
C57BL/6j, BALB/c and CD36 knockout mice, 6-8 weeks of age were obtained from Jackson Laboratories. All animal procedures were approved by the University of Louisville Institutional Animal Care and Use Committee.
Reagents and antibodies
Mouse monoclonal anti-CD36 and rat anti-F4/80 were purchased from Abcam (Cambridge, MA, USA). Primary antibodies were detected by Alexa Fluor 488-, 594- or 647-conjugated goat anti-mouse, anti-rabbit IgG and anti-rat antibodies (1:600, Invitrogen, Waltham, MA, USA). Tissues were counterstained with DAPI and images were captured on a Zeiss LSM 510 confocal microscope equipped with a digital image analysis system (Pixera, San Diego, CA, USA).
Near-infrared lipophilic carbocyanine dye 1,1-dioctadecyl-3,3,3'3'-tetramethylindotricarbocyanine-iodide (DiR) was purchased from Invitrogen (Carlsbad, CA, USA). PKH26-GL and PKH67 (Sigma-Aldrich, St. Louis, MO, USA), PureExoR Exosome Isolation Kit for serum (101 Bio, Palo Alto, CA, USA) and clodrosomes were purchased (Encapsula NanoSciences LL (Brentwood, TN)). Human ON-TARGETplus Smartpool-Tyrosine Kinase library-black plates (version 2.0, Dharmacon) were ready to use at a final concentration of 50 nM.
The murine melanoma cell line B16F10, the murine breast tumor cell line 4T1 and human U937 monocytes were purchased (ATCC) and cultured according to the instructions provided (ATCC).
Exosomes isolation and usage
Exosomes from mouse plasma were isolated according to the instructions provided in the PureExo® Exosomes Isolation kit. In brief, debris in plasma was removed by centrifugation at 2000 ×g for 10 min. Then, the supernatant was transferred to a new glass tube and mixed with pre-prepared isolation solution, vortexed for 30 s and incubated at 4 °C for 2 h. The middle “fluff” layer was transferred onto a PureExo column without disturbing the top and bottom layers. The column was spun at 2,000 ×g for 5 min and the cloudy top layer was collected by flow-through.
To ensure injected exosomes and nanovectors used in this study were not aggregated into large particles, the stock of purified exosomes or nanovectors was always centrifuged at 10,000 ×g for 20 min just before use to pellet microparticles but not the exosomes or nanovectors. 200 μg of exosomes or 200 nmol of GNVs from the supernatants were used for injection.
Western blot
Exosomes were lysed with RIPA buffer. Lysates were subjected to SDS/PAGE (10% gel) and the proteins were transferred onto a polyvinylidine fluoride membrane (Life Technologies, Grand Island, NY, USA). After blocking with 5% BSA, membranes were incubated with rat anti-mouse CD9, goat anti-mouse CD63 and goat anti-mouse CD81 antibodies overnight at 4 °C (Santa Cruz Biotechnology, Inc., Dallas, TX, USA). The reaction product was finally visualized with an Amersham ECL Western Blotting system (GE Healthcare Life Sciences, Chalfont, UK).
Electron microscopy examination of isolated exosomes
Isolated exosomes in PBS were fixed in 2% paraformaldehyde (Electron Microscopy Science, PA) in PBS for 2 h at 22 °C followed by 1% glutaraldehyde (Electron Microscopy Science, PA) for 30 min at 22 °C. 15 µL of fixed samples were put on a 2% agarose gel with formvar/carbon-coated nickel grids on top and allowed to adsorb for 5-10 min. The grids with adherent exosomes were fixed in 2% paraformaldehyde in PBS for 10 min followed by extensive washing in PBS. Negative contrast staining was performed with 1.9% methyl cellulose and 0.3% uranyl acetate for 10 min. The grids with negatively stained exosomes were dried before observation under a Zeiss EM 900 electron microscope.
Size distribution and Zeta potential analysis
Size distribution and Zeta potential of exosomes were analyzed at a flow rate of 0.03 mL/min using a Zetasizer Nano ZS (Malvern Instrument, UK). Briefly, exosomes were washed in ddH2O by centrifugation at 100,000 ×g for 45 min, resuspended with 1 mL ddH2O and transferred into cuvettes for analysis. Protein concentration of exosomes was determined using a Bio-Rad Protein Quantitation Assay kit with bovine serum albumin as a standard.
Macrophage depletion
Macrophages were depleted by clodrosomes as previously reported 29. In brief, 150 μL (700 μg) of clodrosomes were I.V. injected into BALB/c mice. The presence of macrophages in mouse liver after the clodrosome treatment was checked by staining with anti-mouse F4/80 antibody.
Fluorescent labeling of exosomes and nanovectors
For exosomes labeling, 5 μL of DiR, at a concentration of 200 µg/mL in ethanol, was mixed with 200 µg exosomes in 100 µL PBS for 1 h. Ethanol and the unincorporated DiR was removed by a conditioned Sepharose™ CL-4B column.
For labeling 200 nmol of GNVs, IGNVs, DOTAP:DOPE and liposomes from Avanti were incubated with DiR dye (200 µg/mL) at 37 °C for 1 h. The unincorporated DiR was removed by centrifugation at 100,000 ×g for 30 min.
In vivo images
To check the distribution of particles in mice, DiR dye-labeled vectors including GNVs, IGNVs, DOTAP:DOPE (1:1, W/W) and liposomes (N-(carbonyl-ethoxypolyethylene glycol 2000)-1,2-distearoyl-sn-glycero-3-phosphoethanolamine sodium salt (MPEG-DSPE), 3.19 mg/mL; fully hydrogenated soy phosphatidylcholine (HSPC), 9.58 mg/mL; cholesterol, 3.19 mg/mL, ammonium sulfate, approximately 2 mg/mL) from Avanti were prepared as follows. 200 nmol of grapefruit lipids or DOTAP:DOPE (1:1, w/w) were dried in glass vials and DiR dye was added with ddH2O. Then the nanovectors were prepared according to the protocol as described in our previous publications 4,15. Free DiR dye was removed by centrifugation at 100,000 ×g for 1 h. The DiR dye-labeled nanovectors were injected into mice via the tail vein and images of living mice were obtained 0.5, 1, 6, 12 h after injection. DiR dye signals in organs were quantified by scanning mice using a Kodak Images Station.
To verify the distribution of exosomes, exosomes (200 μg) isolated from plasma of healthy BALB/c or C57BL/6j mice were labeled with DiR dye and injected into mice. Organs were removed and DiR dye signals in each organ were quantified after injection.
To demonstrate the re-distribution of particles in mice treated with clodrosomes (700 μg) or exosomes (200 μg), DiR dye-labeled GNVs were I.V. injected into mice (24 h after clodrosomes injection and 0.5 h after exosomes injection), respectively. DiR dye signals in living mice and organs were quantified using a Kodak Image System.
To exam the re-distribution of the nanovectors in 4T1 tumor-bearing mice, 200 μg of mouse blood-derived exosomes or DiR dye-labeled GNVs were I.V. injected into mice and DiR signals in living mice, 4T1 tumor tissue, liver, lung, spleen, kidney, thymus, heart and lymph node were collected.
Monitoring GNVs in peripheral blood
DiR dye labeled GNVs (200 nmol) were I.V. injected into mice receiving clodrosome or exosome treatment. Next, 100 μL of anticoagulated blood was collected at different time points (30, 60 and 180 min) and the DiR signals were quantified using a Kodak Image System.
Immunofluorescent staining
For Kupffer cell staining, the mice injected with clodrosomes were sacrificed at different time points (24, 48 and 72 h). Liver tissue was removed and fixed with Periodate-Lysine-Paraformaldehyde (PLP) fixative at 22 °C for 2 h, dehydrated with 30% sucrose solution at 4 °C overnight, embedded in OCT and cut into 8 μm sections. The tissue sections were blocked with 5% BSA at 22 °C for 45 min and then incubated with rat anti-mouse F4/80 antibody (1:100) at 37 °C for 2 h. After washing 3x, tissue sections were stained with Alexa 488- or 647-conjugated anti-rat secondary antibody (1:800) at 37 °C for 30 min and DAPI for 90 s. The tissue slides were mounted and viewed using a confocal microscope equipped with a digital image analysis system (Pixera, San Diego, CA, USA).
To check the distribution of nanovectors in normal mice with or without clodrosome or exosome injection, PKH67 or PKH26 dye labeled GNVs (200 nmol) were I.V. injected into mice. Mice were sacrificed 12 h after the injection; tissues including liver, lung, and spleen were fixed, dehydrated and cut into 8 μm sections. The tissue sections were stained with DAPI at 22 °C for 90 s.
To verify the cell target of nanovectors in lungs having 4T1 tumor metastases, 4T1 tumor-bearing mice were treated with 200 μg exosomes first and then PKH26-labeled GNVs (200 nmol) were I.V. injected into mice. 12 h after injection, lung tissue was removed, fixed, dehydrated and cut into 8 μm sections. The tissue sections were blocked with 5% BSA at 22 °C for 45 min, incubated with anti-mouse F4/80 at 37 °C for 2 h and then stained with Alexa Fluor® secondary antibody at 37 °C for 30 min. The co-localization of GNVs with cells was examined by confocal microscopy.
Measurement of the concentration of paclitaxel in mice tissues
B16F10 melanoma mice were I.V. injected with free paclitaxel or GNV-paclitaxel. Exosome (200 μg) pre-treated B16F10-bearing mice were intravenously administrated GNV-paclitaxel three times and the paclitaxel in liver, lung, tumor and spleen tissues was quantitatively analyzed using high performance liquid chromatography (HPLC) as described 30.
For GNV-Dox or GNV-PTX preparation, doxorubicin (Dox) or paclitaxel (PTX) were mixed with total lipids from grapefruit dissolved in chloroform and dried under nitrogen to obtain a thin and dried lipids-complex film. The film was reconstituted in PBS buffer, followed by sonication for 30 min, allowing the lipids to self-assemble into Dox or PTX-loaded nanoparticles. Then, GNV-Dox or GNV-PTX were purified by ultracentrifugation at 100,000 ×g for 1 h, the supernatants were collected and the residual Dox or PTX content was measured using a CARY 100 Bio UV-Visible Spectrophotometer at a wavelength of 486 or 265 nm, respectively. The loading efficiency was calculated as follows: Loading efficiency = (Amount of total drug - Amount of drug in the supernatant)/Amount of total drug× 100%.
4T1 and B16F10 tumor models
Six-week-old female BALB/c mice were injected in a mammary fat pad with murine breast tumor 4T1 cells (1×105 cells/ mouse in 50 μL PBS). 5 days later, mice were treated every 3 days for a total of ten times with PBS, Dox, GNV-Dox or exosomes and GNV-Dox (Exo/GNV-Dox). Lung metastases were imaged and the survival rate of the mice was calculated.
In the second set of experiments, 5×104 B16F10 cells were I.V. injected into C57BL/6 mice (six-week-old females). 5 days later, mice were treated intravenously every 3 days for a total of ten times with PBS, free paclitaxel (PTX), GNV-PTX or Exo/GNV-PTX.
Hematoxylin and Eosin (H&E) staining
Lungs from 4T1 and B16F10-bearing mice were fixed in 2% PLP fixative at 22 °C for 2 h, dehydrated in 30% sucrose solution overnight at 4 °C, embedded in OCT and cut into 8 μm sections. The tissue sections were stained with H&E.
Human peripheral blood and exosomes isolation
Blood samples were obtained from consenting healthy volunteers. Blood was drawn into 10 mL purple tubes (BD Vacutainer®, Becton Dickinson, Franklin Lakes, NJ) and all draws were performed using venipuncture. For the separation of plasma, blood samples were centrifuged at 22 °C at 1600 ×g for 10 min. The plasma layer was carefully removed without disturbing the buffy coat, transferred to a new tube and then centrifuged at 22 °C at 16000 ×g for 10 min to remove residual cells, cell debris, apoptotic bodies, and nuclei, and then used for isolation of exosomes with a standard differentiation centrifugation protocol as described 31.
Cell culture, transfection and FACS analysis
U937 cells (ATCC) were plated at 9.3×103 per well in black-walled 96-well plates (Costar) in antibiotic-free growth medium (Invitrogen) 16 h before transfection. Transfections were performed using a siPORT Amine agent (ThermoFisher Scientific) with siRNAs (final concentration 50 nM). Transfections were performed in duplicate and quadruplicate if knockdown was evaluated.
Transfected cells were incubated for 48 h to allow target knockdown. 30 min after exosomes were isolated from the peripheral blood of healthy subjects, the exosomes were added to each siRNA-transfected well. PKH26-labeled GNVs were added for an additional 0-2 h incubation at 37 °C. The treated cells were then washed and PKH26-positive cells were FACS analyzed using a previously described method 32. FlowJo was used for analysis.
Statistical analysis
All statistical analyses in this study were performed with SPSS 16.0 software. Data are presented as mean ± SD. The significance of mean values between two groups was analyzed by Student's t-test. Differences between individual groups were analyzed by one- or two-way analysis of variance tests. Differences were considered significant when the P-value was less than 0.05, 0.01 or 0.001 as indicated in the text.
Supplementary Material
Supplementary figures.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (NIH) (UH3TR000875, R01AT008617, RO1AT004294). Huang-Ge Zhang is supported by a Research Career Scientist (RCS) Award. Yun Teng is partially supported by a pilot project funded through the 1P30GM106396 Molecular Targets Phase III COBRE grant. Research reported in this publication was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM113226-6174 (Deng). National Natural Science Foundation of China (81772585, 81572977), Qilong Wang is partially supported by Natural Science Foundation of Jiangsu Province (BK20150421) and Huai'an key laboratory of Esophageal Cancer Biobank Project (HAP201605). We thank Dr. Jerald Ainsworth for editorial assistance.
Authors' contributions
QW, XZ, and H-GZ designed the research, analyzed and interpreted data, and drafted the manuscript; MS, JM, YT, ZD, LZ, KS, and AK performed experiments and interpreted data; JY and DM interpreted the findings.
Abbreviations
- DAPI
4',6-diamidine-2-phenylindole dihydrochloride
- DIR
1,1-dioctadecyl-3,3,3'3'-tetramethylindotricarbocyanine-iodide
- DOX
Doxorubicin
- EPR
enhanced permeability and retention
- FAT
fatty acid translocase
- GNV
grapefruit nanovectors
- IGFR1
insulin-like growth factor 1
- IGNVs
grapefruit-derived nanovectors (GNV) coated with inflammatory-related receptor enriched membranes of activated leukocytes
- I.V.
intravenously
- KO
knockout
- LTK
leukocyte tyrosine kinase
- NE
nuclear envelope
- PLP
periodate-Lysine-paraformaldehyde
- PTX
paclitaxel.
References
- 1.Zhuang X, Xiang X, Grizzle W. et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol Ther. 2011;19(10):1769–79. doi: 10.1038/mt.2011.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez-Erviti L, Seow Y, Yin H. et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–5. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 3.Imai T, Takahashi Y, Nishikawa M. et al. Macrophage-dependent clearance of systemically administered B16BL6-derived exosomes from the blood circulation in mice. J Extracell Vesicles. 2015;4:26238. doi: 10.3402/jev.v4.26238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Q, Zhuang X, Mu J. et al. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat Commun. 2013;4:1867. doi: 10.1038/ncomms2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen H, Sun T, Ferrari M. Nanovector delivery of siRNA for cancer therapy. Cancer Gene Ther. 2012;19(6):367–73. doi: 10.1038/cgt.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tavares AJ, Poon W, Zhang YN. et al. Effect of removing Kupffer cells on nanoparticle tumor delivery. Proc Natl Acad Sci U S A. 2017;114(51):E10871–E10880. doi: 10.1073/pnas.1713390114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang YN, Poon W, Tavares AJ. et al. Nanoparticle-liver interactions: Cellular uptake and hepatobiliary elimination. J Control Release. 2016;240:332–48. doi: 10.1016/j.jconrel.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Liu T, Li L, Fu C. et al. Pathological mechanisms of liver injury caused by continuous intraperitoneal injection of silica nanoparticles. Biomaterials. 2012;33(7):2399–407. doi: 10.1016/j.biomaterials.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Hoshino A, Costa-Silva B, Shen TL. et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–35. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rak J. Cancer: Organ-seeking vesicles. Nature. 2015;527(7578):312–4. doi: 10.1038/nature15642. [DOI] [PubMed] [Google Scholar]
- 11.Mantovani A. Cancer: Inflaming metastasis. Nature. 2009;457(7225):36–7. doi: 10.1038/457036b. [DOI] [PubMed] [Google Scholar]
- 12.Alderton GK. Metastasis: Directions to metastatic sites. Nat Rev Cancer. 2015;15(12):696–7. doi: 10.1038/nrc4046. [DOI] [PubMed] [Google Scholar]
- 13.Parodi A, Quattrocchi N, van de Ven AL. et al. Synthetic nanoparticles functionalized with biomimetic leukocyte membranes possess cell-like functions. Nat Nanotechnol. 2013;8(1):61–8. doi: 10.1038/nnano.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alderton GK. Diagnosis: Fishing for exosomes. Nat Rev Cancer. 2015;15(8):453. doi: 10.1038/nrc3990. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, Ren Y, Mu J. et al. Grapefruit-Derived Nanovectors Use an Activated Leukocyte Trafficking Pathway to Deliver Therapeutic Agents to Inflammatory Tumor Sites. Cancer Res. 2015;75(12):2520–9. doi: 10.1158/0008-5472.CAN-14-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pepino MY, Kuda O, Samovski D. et al. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu Rev Nutr. 2014;34:281–303. doi: 10.1146/annurev-nutr-071812-161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abumrad NA, Goldberg IJ. CD36 actions in the heart: Lipids, calcium, inflammation, repair and more. Biochim Biophys Acta. 2016;1861(10):1442–9. doi: 10.1016/j.bbalip.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heit B, Kim H, Cosío G. et al. Multimolecular signaling complexes enable Syk-mediated signaling of CD36 internalization. Dev Cell. 2013;24(4):372–83. doi: 10.1016/j.devcel.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa-Silva B, Aiello NM, Ocean AJ. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17(6):816–26. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin SY, Zhang AQ, Cheng SX. et al. Drug self-delivery systems for cancer therapy. Biomaterials. 2017;112:234–47. doi: 10.1016/j.biomaterials.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 21.Bazak R, Houri M, Achy SE. et al. Passive targeting of nanoparticles to cancer: A comprehensive review of the literature. Mol Clin Oncol. 2014;2(6):904–8. doi: 10.3892/mco.2014.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohara Y, Oda T, Yamada K. et al. Effective delivery of chemotherapeutic nanoparticles by depleting host Kupffer cells. Int J Cancer. 2012;131(10):2402–10. doi: 10.1002/ijc.27502. [DOI] [PubMed] [Google Scholar]
- 23.Hetzer MW, Wente SR. Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell. 2009;17(5):606–16. doi: 10.1016/j.devcel.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burke B, Stewart CL. Functional architecture of the cell's nucleus in development, aging, and disease. Curr Top Dev Biol. 2014;109:1–52. doi: 10.1016/B978-0-12-397920-9.00006-8. [DOI] [PubMed] [Google Scholar]
- 25.van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–28. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 26.Webber J, Clayton A. How pure are your vesicles. J Extracell Vesicles; 2013. p. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simonsen JB. What Are We Looking At? Extracellular Vesicles, Lipoproteins, or Both. Circ Res. 2017;121(8):920–2. doi: 10.1161/CIRCRESAHA.117.311767. [DOI] [PubMed] [Google Scholar]
- 28.DVF T, Palomo-Ponce S, Stork D. et al. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature. 2018;554(7693):538–43. doi: 10.1038/nature25492. [DOI] [PubMed] [Google Scholar]
- 29.Teng Y, Mu J, Hu X. et al. Grapefruit-derived nanovectors deliver miR-18a for treatment of liver metastasis of colon cancer by induction of M1 macrophages. Oncotarget. 2016;7(18):25683–97. doi: 10.18632/oncotarget.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng Z, Rong Y, Teng Y. et al. Broccoli-Derived Nanoparticle Inhibits Mouse Colitis by Activating Dendritic Cell AMP-Activated Protein Kinase. Mol Ther. 2017;25(7):1641–54. doi: 10.1016/j.ymthe.2017.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng Z, Cheng Z, Xiang X. et al. Tumor cell cross talk with tumor-associated leukocytes leads to induction of tumor exosomal fibronectin and promotes tumor progression. Am J Pathol. 2012;180(1):390–8. doi: 10.1016/j.ajpath.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 32.Deng Z, Mu J, Tseng M. et al. Enterobacteria-secreted particles induce production of exosome-like S1P-containing particles by intestinal epithelium to drive Th17-mediated tumorigenesis. Nat Commun. 2015;6:6956. doi: 10.1038/ncomms7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.