Abstract
Purpose
HOX transcript antisense RNA (HOTAIR) plays important roles in carcinogenesis of various kinds of malignant tumors, including lung cancer. Single nucleotide polymorphisms (SNPs) in HOTAIR were reported to be associated with susceptibility of several kinds of cancers. The present study assessed the associations between three SNPs (rs4759314, rs12826786, and rs920778) and lung cancer susceptibility, as well as gene–environment interaction between smoking exposure and the polymorphisms.
Patients and methods
A case–control study including 551 patients and 543 healthy controls was performed. The associations between SNPs and lung cancer susceptibility were assessed by logistic regression model.
Results
rs4759314 was observed to increase the susceptibility of lung cancer, lung adenocarcinoma, squamous lung cancer, and small cell lung cancer statistically significantly (OR of 4.048 for lung cancer; 3.584 for lung adenocarcinoma; 4.671 for squamous lung cancer; 4.502 for small cell lung cancer). In stratified analysis for sex and smoking exposure, rs4759314 GG and AG genotype was also observed to increase the risk of lung cancer statistically significantly (OR of 5.221 for male; 3.491 for female; 3.653 for nonsmoking individuals; 4.458 for smoking individuals). Results of gene–environment interaction analysis showed that there was no interaction between smoking exposure and rs4759314 on additive scale. Results of logistic regression model suggested that the interaction between smoking and rs4759314 was statistically significant on multiplicative scale. rs12826786 CT genotype carriers and T allele could decrease the risk of developing lung cancer (OR of 0.751 for CT carriers; 0.785 for T allele), and in dominant model, TC and TT genotype carriers also have a 0.249-fold decrease risk compared with CC genotype carriers. In stratified analysis for smoking exposure, TC and TT have a 0.432-fold decreased risk compared with CC genotype carriers.
Conclusion
HOTAIR rs4759314 and rs12826786 were associated with lung cancer susceptibility in Chinese Han population.
Keywords: lung cancer, lncRNAs, HOTAIR, single nucleotide polymorphism, susceptibility, interaction
Introduction
Lung cancer is acknowledged to be the most lethal of all malignant tumors, and lung cancer is the number one cause of death due to malignant tumors worldwide. The incidence of lung cancer in China is increasing rapidly in recent decades and statistical data showed that lung cancer was the most common incident cancer in China in 2015.1 Due to lack of clinical symptoms during early stages of lung cancer, a great majority of patients are diagnosed in intermediate and advanced stages, and the prognosis is so unfavorable that the overall 5 year survival was as low as below 16% in the past 30 years.2 Lung cancer is such a devastating disease that has led to enormous economic loss and a heavy burden for people all over the world.
Screening and protecting the high-risk individuals of lung cancer is indispensable to prevention and control of lung cancer in population; early diagnosis and treatment of lung cancer is the key to prolong the survival time of the patients. Nonetheless, practical and effective screening methods and molecular biomarkers were seldom developed for distinguishing the high-risk individuals for lung cancer so that the early diagnosis of lung cancer is extremely difficult. Carcinogenesis is a complicated process in which environmental risk factors and genetic predisposition play important roles. Although tobacco smoking is well established as the strongest environmental risk factor for development of lung cancer, yet it was reported that 25% of the lung cancer patients were nonsmokers which indicated that genetic risk factors may play important roles in carcinogenesis of lung cancer.3,4
Long noncoding RNAs (lncRNAs), a class of RNAs more than 200 nucleotides in length with no protein-coding function, expressed uniquely in different tissues, were reported to exert effects in gene expression regulation. lncRNAs were reported to be aberrantly expressed in different kinds of cancers and play a pivotal role in initiation and progression of cancer.5 Genome-wide association studies identified enormous number of single nucleotide polymorphisms (SNPs) which were associated with disease or traits, and more than one-third of these genetic variants reside in noncoding regions.6 Accumulating evidence demonstrated that SNPs that lie within the lncRNAs were associated with risk and prognosis of various kinds of malignant tumors such as breast cancer, colorectal cancer, and gastric cancer.
HOTAIR was reported to play important roles in development of various kinds of malignant tumors such as breast cancer, gastric cancer, colorectal cancer, and lung cancer. In addition, accumulating studies showed that SNPs in HOTAIR were associated with susceptibility of various kinds of cancers.7–18 Based on the oncogenic role HOTAIR plays in development of lung cancer, as well as the gene expression regulation effect of SNPs exerted on function of lncRNAs, we hypothesized that these SNPs in HOTAIR may modify the risk of lung cancer, and hence we performed this case–control study to investigate the associations of the three SNPs (rs4759314, rs920778, and rs12826786) that reside in HOTAIR with risk of developing lung cancer.
Material and methods
Subjects and data collection
There were 551 patients and 543 healthy controls included in the present study. All patients who were diagnosed with lung cancer and confirmed by pathological examination were enrolled from Liaoning Cancer Hospital and First Affiliated Hospital of China Medical University. All patients donated 5 mL of venous blood before subsequent therapeutic measures such as radiotherapy or chemotherapy were taken. Controls were recruited from the medical examination center of the same hospital mentioned above and donated 5 mL of venous blood during the same period. All subjects were unrelated Han Chinese and signed an informed consent. This study was approved by the ethics committee of China Medical University.
SNP selection
By using the Haploview 4.2 software and reading previous studies of HOTAIR polymorphism on cancer risk, we picked up the three SNPs (rs920778, rs4759314, and rs12826786). In using Haploview software, genotype data of Chinese Han in Beijing from 1000 genomes project were used (Phase 3), and threshold for analysis was set as r2>0.8 and minor allelic frequencies >0.05.
Data collection and SNP genotyping
Demographic data of studying subjects such as sex and age were collected from medical records of the hospitals. We collected data such as smoking status by interview with the subjects. Smoking status was defined as follows: smoking more than 100 cigarettes in the lifetime was defined as a smoker, otherwise a nonsmoker. Genomic DNA samples of all subjects were extracted from the venous blood by phenol–chloroform method and was stored at –20°C; subsequently, genotyping was performed by the TaqMan allelic discrimination method on Applied Biosystems 7500 fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA); TaqMan probe and primers were designed and manufactured by Invitrogen (Applied Biosystems); 10% of the samples were randomly selected for repeat genotyping for the sake of quality control and all the results of duplicate sets were concordant.
Statistical analysis
Normality test for continuous variables such as age was conducted by using Shapiro–Wilk test; for data not conforming to normal distribution, nonparametric test Mann–Whitney U-test was conducted. Student’s t-test was performed to examine the differences of continuous variables; Pearson’s chi-squared test was performed to examine the differences of categorical variables such as constituent ratio of sex and age composition between case and control groups. Hardy– Weinberg’s equilibrium (HWE) for each SNP in control group was calculated by the goodness-of-fit chi-squared test. The ORs and their 95% CIs which were used to assess the relationship of three SNPs with lung cancer risk were calculated by unconditional logistic regression analyses. Power analysis to calculate the power of the sample size was conducted by using Quanto software version 1.2.4 (University of Southern California, Los Angeles, CA, USA). Cross-over study was performed to evaluate the interaction of SNP and smoking status (additive interaction) and subjects with both the protective genotype and nonsmoking exposure were used as reference group. Logistic regression model was performed to evaluate the multiplicative interaction of SNPs and smoking status. According to research by Andersson et al,19 three estimation measures were calculated to assess the interaction: the synergy index (S), the attributable proportion due to interaction (AP), and the relative excess risk due to interaction (RERI). All statistical tests are two-sided; P<0.05 was considered statistically significant. All the statistical analysis was performed using Statistical Product and Service Solutions software 19.0 (IBM Corporation, Armonk, NY, USA).
Results
The baseline demographic data of the subjects are shown in Table 1. There were 551 cases and 543 controls included in the present study. The mean (±SD) age was 57.99±11.509 for the cases and 56.69±15.659 for the controls; there were no statistically significant differences in age (P=0.117) and sex (P=0.127) between cases and controls which indicated that the proportionality in two groups was appropriate. The result of normality test showed that age of the case and control groups didn’t conform to the normal distribution, so subsequently Mann–Whitney U-test was performed to compare the median age of the two groups, where the results showed that there were no differences in median age between the case and control groups (Mann–Whitney U-test P=0.640). Compared with the control group, there were more smokers in the case group as expected (29.9% among cases vs 20.4% among controls, P<0.001). For histological type of lung cancer in cases, there were 304 adenocarcinoma, 131 squamous lung cancer (SLC), 7 large cell lung cancer, and 109 small cell lung cancer (SCLC; Table 1).
Table 1.
Basic data of study subjects
| Variables | Case (n=551) | Control (n=543) | P |
|---|---|---|---|
|
| |||
| Mann–Whitney U-test | 0.640 | ||
| Age (mean±SD) | 57.99±11.509 | 56.69±15.659 | 0.117 |
| <57, n (%) | 232 (42.1) | 243 (44.8) | 0.377 |
| >57, n (%) | 319 (57.9) | 300 (55.2) | |
| Sex, n (%) | |||
| Male | 192 (34.8) | 165 (30.4) | 0.127 |
| Female | 359 (65.2) | 378 (69.6) | |
| Smoking, n (%) | |||
| No | 386 (70.1) | 432 (79.6) | <0.001* |
| Yes | 165 (29.9) | 111 (20.4) | |
| Pathological type | |||
| Adenocarcinoma, n (%) | 304 (53.5) | ||
| Squamous lung cancer, n (%) | 131 (20.5) | ||
| Large cell lung cancer, n (%) | 7 (11.7) | ||
| Small cell lung cancer, n (%) | 109 (14.3) | ||
Note: More smokers were statistically significant in cases than in controls.
Indicates statistical significance (P<0.05).
The observed genotypic frequencies of the three SNPs in the control group are all consistent with HWE which indicated that the controls in the present study were representative for the general studying population appropriately. The genotype distributions of three SNPs between the case and control group are shown in Table 2. The results showed that rs12826786 and rs4759314 were statistically significantly associated with lung cancer risk. Compared with rs12826786 CC genotype carriers, carriers of CT genotype had a 0.243-fold decreased risk for lung cancer (CT vs CC: adjusted OR=0.757, 95% CI=0.584–0.980, P=0.035), and in dominant genetic inheritance model, compared with wild-type CC genotype, carriers with variant genotype of rs12826786 (CT+TT) had a 0.249-fold decreased risk for developing lung cancer (CT+TT vs CC: adjusted OR=0.751, 95% CI=0.584–0.966, P=0.026). Subjects carrying AG genotype of rs4759314 had a 3.048-fold increased risk for lung cancer compared with AA genotype carriers (AG vs AA: adjusted OR=4.048, 95% CI=2.302–7.120, P<0.001) and a statistically significant association was also observed in the dominant genetic inheritance model (GG+AG vs AA: adjusted OR=3.869, 95% CI=2.230–6.714, P<0.001). The allelic frequencies of G allele of rs4759314 in the case group were 5.7%, which was significantly higher than that in the control group, G allele carriers had a 2.598-fold increased risk of developing lung cancer than A allele carriers (G vs A: adjusted OR=3.598, 95% CI=2.116–6.117, P<0.001). There were no statistically significant results for associations between rs920778 and lung cancer risk (Table 2).
Table 2.
Distributions of the three SNPs and alleles in two groups and their relationship with susceptibility of lung cancer
| SNP | Lung cancer
|
|||
|---|---|---|---|---|
| Cases, n (%) | Controls, n (%) | ORa (95% CI) | Pb | |
|
| ||||
| rs920778 | ||||
| TT | 324 (59.7) | 326 (59.2) | 1 (ref) | |
| TC | 197 (36.3) | 203 (36.8) | 1.026 (0.800–1.317) | 0.838 |
| CC | 22 (4.1) | 22 (4.0) | 0.989 (0.537–1.823) | 0.973 |
| TC+CC vs TT | 1.023 (0.803–1.302) | 0.856 | ||
| CC vs TT+TC | 0.980 (0.536–1.792) | 0.947 | ||
| T allele | 845 (77.8) | 855 (77.6) | ||
| C allele | 241 (22.2) | 247 (22.4) | 1.013 (0.828–1.239) | 0.901 |
| rs12826786 | ||||
| CC | 340 (62.6) | 381 (69.1) | 1 (ref) | |
| CT | 185 (34.1) | 156 (28.3) | 0.757 (0.584–0.980) | 0.035* |
| TT | 18 (3.3) | 14 (2.5) | 0.696 (0.341–1.422) | 0.321 |
| CT+TT vs CC | 0.751 (0.584–0.966) | 0.026* | ||
| TT vs CC+CT | 0.762 (0.375–1.548) | 0.452 | ||
| C allele | 865 (79.7) | 918 (83.3) | ||
| T allele | 221 (20.3) | 184 (16.7) | 0.785 (0.632–0.974) | 0.028* |
| rs4759314 | ||||
| AA | 526 (96.9) | 489 (88.7) | 1.00 (ref) | |
| AG | 16 (2.9) | 61 (11.1) | 4.048 (2.302–7.120) | <0.001* |
| GG | 1 (0.2) | 1 (0.2) | 1.023 (0.064–16.440) | 0.987 |
| GG+AG vs AA | 3.869 (2.230–6.714) | <0.001* | ||
| GG vs AA+AG | 0.931 (0.058–14.965) | 0.960 | ||
| A allele | 1,068 (98.3) | 1,039 (94.3) | ||
| G allele | 18 (1.7) | 63 (5.7) | 3.598 (2.116–6.117) | <0.001* |
Notes:
OR adjusted for age;
P-value adjusted for age
Indicates statistical significance (P<0.05).
Abbreviations: ref, reference; SNP, single nucleotide polymorphism.
Subsequently, we performed a stratification analysis for sex, smoking exposure, and pathological type of lung cancer. For rs4759314 in nonsmokers, carriers of genotype GG or AG had a 2.653-fold increased risk for developing lung cancer compared with AA genotype carriers (GG+AG vs AA: adjusted OR=3.653, 95% CI=1.918–6.959, P<0.001). Simultaneously, in smokers, subjects with GG and AG genotype could increase the risk of developing lung cancer compared with AA genotype carriers (GG+AG vs AA: adjusted OR=4.458, 95% CI=1.467–13.543, P=0.008). In stratified analysis for sex, there were also statistically significant associations between rs4759314 and lung cancer risk in male and female subgroups. In male subjects, rs4759314 GG or AG genotype carriers had a 4.221-fold increased risk of developing lung cancer compared with AA genotype carriers (GG+AG vs AA: adjusted OR=5.221, 95% CI=1.917–14.216, P<0.001). In female subjects, compared with AA genotype carriers, carriers of GG and AG genotype had a 2.491-fold increased risk of developing lung cancer (GG+AG vs AA: adjusted OR=3.491, 95% CI=1.783–6.837, P<0.001; Table 3). In stratified analysis for smoking status, rs12826786 TT and TC genotype carriers had a 0.432-fold decreased risk of developing lung cancer compared with CC genotype carriers (TT+TC vs CC: adjusted OR=0.568, 95% CI=0.340–0.950, P=0.031; Table 4). For rs920778, there were no statistically significant associations in the stratified analysis for smoking exposure and sex (Table 5).
Table 3.
Associations of rs4759314 with susceptibility of lung cancer, stratified by sex and smoking exposure
| Variables | rs4759314 (GG+AG/AA)
|
|||
|---|---|---|---|---|
| Cases (n) | Controls (n) | ORa (95% CI) | Pb | |
|
| ||||
| Sex | ||||
|
| ||||
| Male | 26/166 | 5/160 | 5.221 (1.917–14.216) | 0.001* |
| Female | 36/323 | 12/366 | 3.491 (1.783–6.837) | <0.001* |
|
| ||||
| Smoking status | ||||
|
| ||||
| No | 39/347 | 13/419 | 3.653 (1.918–6.959) | <0.001* |
| Yes | 23/142 | 4/107 | 4.458 (1.467–13.543) | 0.008* |
Notes:
OR adjusted for age;
P-value adjusted for age;
Indicates statistical significance (P<0.05).
Table 4.
Associations of rs12826786 with susceptibility of lung cancer, stratified by sex and smoking exposure
| Variables | rs12826786 (TT+TC/CC)
|
|||
|---|---|---|---|---|
| Cases | Controls | ORa (95% CI) | Pb | |
|
| ||||
| Sex | ||||
|
| ||||
| Male | 58/134 | 61/104 | 0.745 (0.473–1.173) | 0.204 |
| Female | 112/247 | 142/236 | 0.749 (0.552–1.017) | 0.064 |
|
| ||||
| Smoking status | ||||
|
| ||||
| No | 121/265 | 155/277 | 0.815 (0.609–1.091) | 0.169 |
| Yes | 49/116 | 48/63 | 0.568 (0.340–0.950) | 0.031* |
Notes:
OR adjusted for age;
P-value adjusted for age;
Indicates statistical significance (P<0.05).
Table 5.
Associations of rs920778 with susceptibility of lung cancer, stratified by sex and smoking exposure
| Variables | rs920778 (CC+TC/TT)
|
|||
|---|---|---|---|---|
| Cases | Controls | ORa (95% CI) | Pb | |
|
| ||||
| Sex | ||||
|
| ||||
| Male | 80/112 | 65/100 | 1.128 (0.729–1.745) | 0.587 |
| Female | 145/214 | 154/224 | 0.985 (0.734–1.322) | 0.921 |
|
| ||||
| Smoking status | ||||
|
| ||||
| No | 155/231 | 168/264 | 1.054 (0.796–1.396) | 0.713 |
| Yes | 70/95 | 51/60 | 0.885 (0.539–1.453) | 0.885 |
Notes:
OR adjusted for age;
P-value adjusted for age.
Results of stratified analysis by pathologic type of lung cancer are shown in Tables 6–8. In lung adenocarcinoma subgroup, rs4759314 AG genotype carriers also had a 2.584-fold increased risk of developing lung adenocarcinoma than AA genotype carriers (AG vs AA: adjusted OR=3.584, 95% CI=1.919–6.694, P<0.001) and the allelic frequency of rs4759314 G was 5.3% in cases, which was significantly higher than the control group (1.7%); the G allele elevated the risk of developing lung adenocarcinoma by 2.296-fold (G vs A: adjusted OR=3.296, 95% CI=1.834–5.924, P<0.001). In dominant model, rs4759314 GG and AG genotype carriers increased the risk of developing lung adenocarcinoma by 2.483-fold compared with AA genotype carriers (GG+AG vs AA: adjusted OR=3.483, 95% CI=1.893–6.411, P<0.001; Table 6). For SLC subgroup, it was shown that rs4759314 AG genotype could increase the risk of developing SLC by 3.671-fold compared with AA genotype (AG vs AA: adjusted OR=4.671, 95% CI=2.279–9.572, P<0.001). In dominant genetic model, rs4759314 GG and AG genotype carriers had a 3.614-fold increased risk for developing SLC (GG+AG vs AA: adjusted OR=4.614, 95% CI=2.286–9.311, P<0.001). The allelic frequency of rs4759314 G in cases (6.5%) was significantly higher than that in controls (1.7%), which indicated that G allele increased the risk of developing SLC by 3.117-fold (G vs A: adjusted OR=4.117, 95% CI=2.092–8.104, P<0.001; Table 7). In SCLC subgroup, rs12826786 CT genotype carriers had a 0.388-fold decreased risk of developing SCLC compared with CC genotype (CT vs CC: adjusted OR=0.612, 95% CI=0.379–0.987, P=0.044). For rs4759314, AG genotype carriers had a 3.502-fold increased risk of developing SCLC compared with wild-type AA genotype (AG vs AA: adjusted OR=4.502, 95% CI=2.095–9.673, P<0.001), and in dominant genetic model, GG and AG genotype increased the risk of developing SCLC by 3.253-fold compared with the wild-type genotype AA (GG+AG vs AA: adjusted OR=4.253, 95% CI=1.997–9.058, P<0.001). The allelic frequency of rs4759314 G was significantly higher in SCLC patients than that in the control group, which increased the risk of developing SCLC by 2.763-fold (G vs A: adjusted OR=3.763, 95% CI=1.815–7.799, P<0.001; Table 8). We failed to find any statistically significant associations between rs920778 and lung cancer risk in all analyses. We also performed a cross-over analysis to investigate whether there is additive interaction of smoking exposure and rs4759314 polymorphisms on lung cancer susceptibility. Results of the cross-over analysis showed that compared with carriers of genotype AA who had no smoking exposure, those carriers of GG and AG genotype had a 2.622-fold increased risk of developing lung cancer. An elevated risk was also observed in AA genotype carriers with smoke exposure and the group combining GG and AG genotype carriers (Table 9). Results of three measures were not statistically significant, which indicated that there was no additive interaction between smoking exposure and rs4759314 (RERI=2.718, 95% CI=−5.011–10.447; AP=0.391, 95% CI=−0.335–1.118; S=1.843, 95% CI=0.439–7.729; Table 10).
Table 6.
Distributions of the three SNPs and alleles in two groups and their relationship with susceptibility of lung adenocarcinoma
| SNP | Adenocarcinoma
|
|||
|---|---|---|---|---|
| Controls, n (%) | Cases, n (%) | ORa (95% CI) | Pb | |
|
| ||||
| rs920778 | ||||
| TT | 324 (59.7) | 180 (59.1) | 1.00 (ref) | |
| TC | 197 (36.3) | 113 (37.0) | 1.036 (0.772–1.392) | 0.812 |
| CC | 22 (4.1) | 12 (3.9) | 0.976 (0.472–2.018) | 0.947 |
| TC+CC vs TT | 1.030 (0.774–1.371) | 0.838 | ||
| CC vs TT+TC | 0.963 (0.469–1.974) | 0.917 | ||
| T allele | 845 (77.8) | 473 (77.5) | 1.00 (ref) | 0.899 |
| C allele | 241 (22.2) | 137 (22.5) | 1.013 (0.800–1.288) | |
| rs12826786 | ||||
| CC | 340 (62.6) | 207 (68.1) | 1.00 (ref) | |
| CT | 185 (34.1) | 90 (29.6) | 0.803 (0.591–1.090) | 0.159 |
| TT | 18 (3.3) | 7 (2.3) | 0.640 (0.263–1.558) | 0.325 |
| CT+TT vs CC | 0.788 (0.585–1.061) | 0.117 | ||
| TT vs CC+CT | 0.687 (0.284–1.665) | 0.406 | ||
| C allele | 865 (79.7) | 504 (82.9) | 1.00 (ref) | 0.104 |
| T allele | 221 (20.3) | 104 (17.1) | 0.808 (0.624–1.045) | |
| rs4759314 | ||||
| AA | 526 (96.9) | 273 (89.8) | 1.00 (ref) | |
| AG | 16 (2.9) | 30 (9.9) | 3.584 (1.919–6.694) | <0.001* |
| GG | 1 (0.2) | 1 (0.3) | 1.876 (0.117–30.164) | 0.657 |
| GG+AG vs AA | 3.483 (1.893–6.411) | <0.001* | ||
| GG vs AA+AG | 1.730 (0.108–27.810) | 0.699 | ||
| A allele | 1,068 (98.3) | 576 (94.7) | 1.00 (ref) | <0.001* |
| G allele | 18 (1.7) | 32 (5.3) | 3.296 (1.834–5.924) | |
Notes:
OR adjusted for age;
P-value adjusted for age;
Indicates statistical significance (P<0.05).
Abbreviations: ref, reference; SNP, single nucleotide polymorphism.
Table 7.
Distributions of the three SNPs and alleles in two groups and their relationship with susceptibility of squamous lung cancer
| SNP | Squamous lung cancer
|
|||
|---|---|---|---|---|
| Controls, n (%) | Cases, n (%) | ORa (95% CI) | Pb | |
|
| ||||
| rs920778 | ||||
| TT | 324 (59.7) | 76 (58) | 1.00 (ref) | |
| TC | 197 (36.3) | 51 (38.9) | 1.165 (0.785–1.730) | 0.449 |
| CC | 22 (4.1) | 4 (3.1) | 0.751 (0.250–2.250) | 0.609 |
| TC+CC vs TT | 1.122 (0.762–1.650) | 0.560 | ||
| CC vs TT+TC | 0.707 (0.239–2.095) | 0.532 | ||
| T allele | 845 (77.8) | 203 (77.5) | 1.00 (ref) | 0.909 |
| C allele | 241 (22.2) | 59 (22.5) | 1.019 (0.738–1.408) | |
| rs12826786 | ||||
| CC | 340 (62.6) | 91 (69.5) | 1.00 (ref) | |
| CT | 185 (34.1) | 38 (29.0) | 0.786 (0.516–1.198) | 0.262 |
| TT | 18 (3.3) | 2 (1.5) | 0.406 (0.092–1.790) | 0.234 |
| CT+TT vs CC | 0.751 (0.497–1.135) | 0.174 | ||
| TT vs CC+CT | 0.439 (0.100–1.923) | 0.275 | ||
| C allele | 865 (79.7) | 220 (84.0) | 1.00 (ref) | 0.113 |
| T allele | 221 (20.3) | 42 (16.0) | 0.747 (0.520–1.073) | |
| rs4759314 | ||||
| AA | 526 (96.9) | 114 (86.9) | 1.00 (ref) | |
| AG | 16 (2.9) | 17 (13.1) | 4.671 (2.279–9.572) | <0.001* |
| GG | 1 (0.2) | 0 (0.0) | – | – |
| GG+AG vs AA | 4.614 (2.286–9.311) | <0.001* | ||
| GG vs AA+AG | – | – | ||
| A allele | 1,068 (98.3) | 245 (93.5) | 1.00(ref) | <0.001* |
| G allele | 18 (1.7) | 17 (6.5) | 4.117 (2.092–8.104) | |
Notes:
OR adjusted for age;
P-value adjusted for age;
Indicates statistical significance (P<0.05); – indicates the results cannot be calculated.
Abbreviations: ref, reference; SNP, single nucleotide polymorphism.
Table 8.
Distributions of the three SNPs and alleles in two groups and their relationship with susceptibility of small cell lung cancer
| SNP | Small cell lung cancer
|
|||
|---|---|---|---|---|
| Controls, n (%) | Cases, n (%) | ORa (95% CI) | Pb | |
|
| ||||
| rs920778 | ||||
| TT | 324 (59.7) | 66 (58.9) | 1.00 (ref) | |
| TC | 197 (36.3) | 40 (35.7) | 0.995 (0.647–1.531) | 0.982 |
| CC | 22 (4.1) | 6 (5.4) | 1.347 (0.525–3.451) | 0.535 |
| TC+CC vs TT | 1.030 (0.681–1.558) | 0.888 | ||
| CC vs TT+TC | 0.995 (0.982–1.008) | 0.471 | ||
| T | 845 (77.8) | 172 (76.8) | 1.00 (ref) | 0.992 |
| C | 241 (22.2) | 52 (23.2) | 1.06 (0.753–1.492) | |
| rs12826786 | ||||
| CC | 340 (62.6) | 91 (69.5) | 1.00 (ref) | |
| CT | 185 (34.1) | 38 (29.0) | 0.612 (0.379–0.987) | 0.044* |
| TT | 18 (3.3) | 2 (1.5) | 1.209 (0.436–3.357) | 0.715 |
| CT+TT vs CC | 0.665 (0.423–1.043) | 0.076 | ||
| TT vs CC+CT | 1.401 (0.509–3.859) | 0.514 | ||
| C | 865 (79.7) | 182 (83.5) | 1.00 (ref) | 0.194 |
| T | 221 (20.3) | 36 (16.5) | 0.774 (0.526–1.140) | |
| rs4759314 | ||||
| AA | 526 (96.9) | 96 (88.1) | 1.00 (ref) | |
| AG | 16 (2.9) | 13 (11.9) | 4.502 (2.095–9.673) | <0.001* |
| GG | 1 (0.2) | 0 (0.0) | – | – |
| GG+AG vs AA | 4.253 (1.997–9.058) | <0.001* | ||
| GG vs AA+AG | – | – | ||
| A | 1,068 (98.3) | 205 (94.0) | 1.00 (ref) | <0.001* |
| G | 18 (1.7) | 13 (6.0) | 3.763 (1.815–7.799) | |
Notes:
OR adjusted for age;
P-value adjusted for age;
Indicates statistical significance (P<0.05); – indicates the results cannot be calculated.
Abbreviations: ref, reference; SNP, single nucleotide polymorphism.
Table 9.
Effect of interaction between rs4759314 and smoking exposure on lung cancer susceptibility
| SNP | Smoking exposure | Controls, n (%) | Cases, n (%) | ORa (95% CI) | Pb |
|---|---|---|---|---|---|
|
| |||||
| rs4759314 | |||||
| AA | Non-exposure | 419 (77.2) | 347 (63.0) | 1 | |
| GG+AG | Non-exposure | 13 (2.4) | 39 (7.0) | 3.622 (1.903–6.895) | <0.001* |
| AA | exposure | 107 (19.7) | 142 (25.8) | 1.602 (1.201–2.138) | <0.001* |
| GG+AG | exposure | 4 (0.7) | 23 (4.2) | 6.943 (2.378–20.268) | <0.001* |
Notes:
OR adjusted for age; bP-value adjusted for age;
Indicates statistical significance (P<0.05).
Abbreviation: SNP, single nucleotide polymorphism.
Table 10.
Interaction measures between rs4759314 and smoking exposure on lung cancer susceptibility
| Lung cancer
| ||
|---|---|---|
| Measure | Estimate | 95% CI |
|
| ||
| RERI | 2.718 | −5.011–10.447 |
| AP | 0.391 | −0.335–1.118 |
| S | 1.843 | 0.439–7.729 |
Abbreviations: AP, attributable proportion due to interaction; RERI, relative excess risk due to interaction; S, synergy index.
Results of multiplicative interaction analysis showed that the interaction of rs4759314 and smoke exposure was statistically significant, ORs (95% CIs) and P-values of interaction terms were 1.704 (1.333–2.179), P<0.001.
Discussion
Lung cancer is the leading cause of cancer-related deaths because early stage patients have almost no discomfort and clinical symptoms are often ignored, meaning the best treatment opportunity time is missed by most patients. Nearly 50% of the patients diagnosed with the stage I non-small-cell lung cancer (NSCLC) died in 10 years, so effective screening methods for early stage of lung cancer are urgently needed to be developed.20
Cancer was traditionally considered to be a genetic disease; yet, recent research has demonstrated that epigenetic regulation such as DNA methylation, histone deacetylation, chromatin remodeling, gene imprinting, and noncoding RNA (ncRNA) regulation also play indispensable roles in cancer development. lncRNAs which express in tissues and disease specificity model have attracted increasing attention. Thanks to the advancement of the high-throughput sequencing and tilling microarrays technology in elucidating human genomes and transcriptomes; ncRNAs which are actively transcribed constitute more than 75% of the human genome.
Deregulation in expression of lncRNAs was observed by numerous studies on different kinds of malignant tumors, which indicated that lncRNAs may exert an effect of oncogene or tumor suppressor gene, and lncRNAs could be potential biomarkers for diagnosis, prognosis, and therapeutics of cancers. Previous research reported that HOTAIR was upregulated in breast, gastric, esophageal squamous cell carcinoma, hepatocellular carcinoma, colorectal cancer, gallbladder cancer, pancreatic cancer, nasopharyngeal carcinoma, and cervical cancer.21–36 The overexpression of HOTAIR in these cancers may affect the proliferation, apoptosis, invasion, aggression, and metastasis which are involved in the process related with cancer initiation and progression.
HOTAIR was reported to play important roles in development of lung cancer, by some researchers. In a study on NSCLC, patients with high expression of HOTAIR in tumor tissues than adjacent normal tissues were more probably in bad prognosis, more advanced stage, lymph-vascular invasion, lymph node metastasis, and short disease-free survival. In vitro, upregulation of HOTAIR expression in A549 cell line could promote migration behavior and anchorage-independent cell growth of the cells.37 In another study on NSCLC, overexpression of HOTAIR was correlated with NSCLC advanced pathological stage and lymph node metastasis in which HOTAIR might have affected the metastatic and invasive potential of NSCLC cells by regulating expression of MMPs and HOXA5. Patients with high expression of HOTAIR have a poor prognosis compared with those who have low expression. In vitro, knockdown of HOTAIR by RNAi in NSCLC cell line SPC-A-1 decreased the migration and invasion of the NSCLC cells. In vivo, knockdown of HOTAIR in SPC-A-1 cells which were injected into athymic mice xenograft model resulted in reduction in the number of metastatic nodules, which indicates that HOTAIR can promote NSCLC cell metastasis.38 In addition, HOTAIR also plays an oncogenic role in the development of SCLC. A study on SCLC showed that patients with high HOTAIR expression may have significantly more lymphatic invasion and more relapse compared with group of low HOTAIR expression. In vitro, knockdown of HOTAIR in SCLC cell line SBC-3 cells decreased the potential of proliferation and invasiveness.39
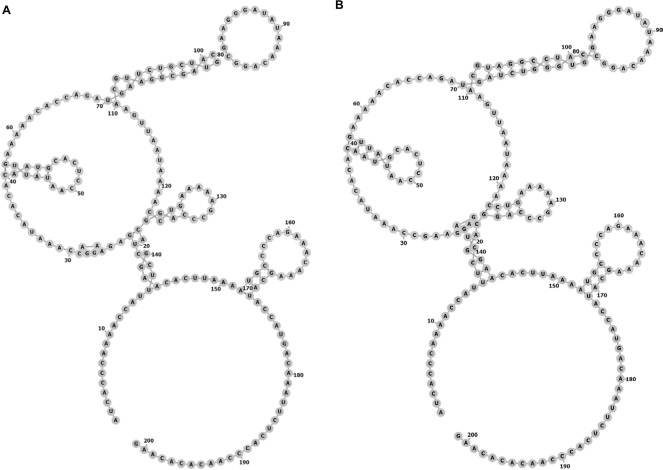
SNPs reside in the coding or noncoding genes and may exert extensive effects on gene expression in ways such as affecting the binding affinity of transcription factor and altering the RNA splicing, subsequently modulating the function of genes. Thus, we performed this case–control study to elucidate the relationship of three SNPs (rs920778, rs12826786, and rs4759314) in lncRNA HOTAIR with lung cancer risk. In the present study, rs4759314 was found associated with risk of developing lung adenocarcinoma, SLC, and SCLC. A previous study on gastric cancer in Chinese population demonstrated that carriers of rs4759314 G allele can significantly increase the risk of developing gastric cancer compared with rs4759314 A allele and the expression level of HOTAIR in AG genotype carriers was significantly higher than that of AA genotype carriers.11 Guo et al reported that rs12826786 polymorphism was associated with susceptibility of gastric cardia adenocarcinoma (GCA) and rs12826786 can affect the expression level of HOTAIR in normal tissues and GCA tumor tissues.8 To the best of our knowledge, we are the first to report the associations of HOTAIR rs4759314 and rs12826786 with lung cancer susceptibility. In our research, it was found that rs4759314 AG genotype could increase the risk of developing lung cancer, and in stratified analyses for pathologic type of lung cancer, sex, and smoking status, the associations between rs4759314 polymorphism and lung cancer risk were still statistically significant and consistent. The rs4759314 polymorphism resides in the first intron of HOTAIR, and hence we used SNP Function Prediction module of SNPinfo (https://snpinfo.niehs.nih.gov/cgi-bin/snpinfo/snpfunc.cgi) to predict the potential function of rs4759314 and the result showed that rs4759314 resided in transcription factor binding sites which indicated that rs4759314 may affect the expression level of HOTAIR. It is biologically plausible that rs4759314 polymorphism exerts effects on HOTAIR expression level that could promote the tumorigenesis of the lung. RNAfold web server was used to predict the effect of rs4759314 on RNA secondary structure and minimum free energy of HOTAIR (Figure 1A and B). We did not find any statistically significant associations between rs920778 and lung cancer risk; in addition, the results of function prediction of rs920778 showed that rs920778 resides in no transcription factor binding sites, maybe this polymorphism site could not have effects on the transcription of HOTAIR. Results of power analysis showed that power for rs4759314, rs12826786, and rs920778 were 0.9814, 0.60, and 0.0539, respectively. The power of sample size for rs920778 was too low due to the value of OR close to 1. According to the calculation of sample size before we conducted the study, we set prospective OR as 1.5 or 0.5 for the unverified SNPs in previous studies, so the sample size of the present study was calculated to be sufficient. Yet, after we carried out the genotyping, the anticipant sample size seems to be too small. Maybe relationship of rs920778 with lung cancer susceptibility could be verified in a study with a larger sample in the future.
Figure 1.
The centroid secondary structure of HOTAIR sequences.
(A) Structure of HOTAIR containing the A allele of rs4759314. The minimum free energy (MFE) of the centroid secondary structure is –29.74 kcal/mol. (B) Structure of HOTAIR containing the G allele of rs4759314. The MFE of the centroid secondary structure is –29.44 kcal/mol.
There are several limitations in the present study that should not be ignored. First, the present research was a hospital-based case–control study and all study subjects were enrolled from the three hospitals in northeast China which could lead to selection bias and may not be approximately representative for the overall population in China. Second, some important data of environmental risk factors of lung cancer such as secondhand smoke exposure and air pollution exposure could not be obtained, so these factors may confound the associations between the SNP and lung cancer susceptibility. Third, all the associations in our results were only statistically significant in which the underlying mechanisms need to be elucidated in future studies.
Conclusion
In the present study, we demonstrated that HOTAIR rs4759314 and rs12826786 polymorphism were associated with susceptibility of lung cancer in Chinese Han population.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No 81673261).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Carr SR, Akerley W, Hashibe M, Cannon-Albright LA. Evidence for a genetical contribution to non-smoking-related lung cancer. Thorax. 2015;70(11):1033–1039. doi: 10.1136/thoraxjnl-2014-206584. [DOI] [PubMed] [Google Scholar]
- 3.Subramanian J, Govindan R. Lung cancer in never smokers: a review. Journal of Clinical Oncology. 2007;25(5):561–570. doi: 10.1200/JCO.2006.06.8015. [DOI] [PubMed] [Google Scholar]
- 4.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers — a different disease. Nat Rev Cancer. 2007;7(10):778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 5.Verhaegh GW, Verkleij L, Vermeulen SH, et al. Polymorphisms in the H19 gene and the risk of bladder cancer. Eur Urol. 2008;54(5):1118–1126. doi: 10.1016/j.eururo.2008.01.060. [DOI] [PubMed] [Google Scholar]
- 6.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayram S, Sumbul AT. Dadas E. A functional HOTAIR rs12826786 C>T polymorphism is associated with breast cancer susceptibility and poor clinicopathological characteristics in a Turkish population: a hospital-based case-control study. Tumour Biol. 2016;374:5577–5584. doi: 10.1007/s13277-015-4430-y. [DOI] [PubMed] [Google Scholar]
- 8.Guo W, Dong Z, Bai Y, et al. Associations between polymorphisms of HOTAIR and risk of gastric cardia adenocarcinoma in a population of north China. Tumor Biology. 2015;36(4):2845–2854. doi: 10.1007/s13277-014-2912-y. [DOI] [PubMed] [Google Scholar]
- 9.Bayram S, Sümbül AT, Batmacı CY, Genç A. Effect of HOTAIR rs920778 polymorphism on breast cancer susceptibility and clinicopathologic features in a Turkish population. Tumour Biol. 2015;36(5):3863–3870. doi: 10.1007/s13277-014-3028-0. [DOI] [PubMed] [Google Scholar]
- 10.Guo L, Lu X, Zheng L, Liu X, Hu M. Association of Long Non-Coding RNA HOTAIR Polymorphisms with Cervical Cancer Risk in a Chinese Population. PLoS One. 2016;11(7):e0160039. doi: 10.1371/journal.pone.0160039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.du M, Wang W, Jin H, et al. The association analysis of lncRNA HOTAIR genetic variants and gastric cancer risk in a Chinese population. Oncotarget. 2015;6(31):31255–31262. doi: 10.18632/oncotarget.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayram S, Ülger Y, Sümbül AT, et al. A functional HOTAIR rs920778 polymorphism does not contributes to gastric cancer in a Turkish population: a case–control study. Fam Cancer. 2015;14(4):561–567. doi: 10.1007/s10689-015-9813-0. [DOI] [PubMed] [Google Scholar]
- 13.Jin H, Lu X, Ni J, et al. HOTAIR rs7958904 polymorphism is associated with increased cervical cancer risk in a Chinese population. Sci Rep. 2017;7(1):3144. doi: 10.1038/s41598-017-03174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi Q, Wang J, Huang B, et al. Association of HOTAIR polymorphisms rs4759314 and rs920778 with cancer susceptibility on the basis of ethnicity and cancer type. Oncotarget. 2016;7(25):38775–38784. doi: 10.18632/oncotarget.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue Y, Gu D, Ma G, et al. Genetic variants in lncRNA HOTAIR are associated with risk of colorectal cancer. Mutagenesis. 2015;30(2):303–310. doi: 10.1093/mutage/geu076. [DOI] [PubMed] [Google Scholar]
- 16.Pan W, Liu L, Wei J, et al. A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol Carcinog. 2016;55(1):90–96. doi: 10.1002/mc.22261. [DOI] [PubMed] [Google Scholar]
- 17.Xavier-Magalhães A, Oliveira AI, de Castro JV, et al. Effects of the functional HOTAIR rs920778 and rs12826786 genetic variants in glioma susceptibility and patient prognosis. J Neurooncol. 2017;132(1):27–34. doi: 10.1007/s11060-016-2345-0. [DOI] [PubMed] [Google Scholar]
- 18.Yan R, Cao J, Song C, et al. Polymorphisms in lncRNA HOTAIR and susceptibility to breast cancer in a Chinese population. Cancer Epidemiol. 2015;39(6):978–985. doi: 10.1016/j.canep.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 19.Andersson T, Alfredsson L, Källberg H, Zdravkovic S, Ahlbom A. Calculating measures of biological interaction. Eur J Epidemiol. 2005;20(7):575–579. doi: 10.1007/s10654-005-7835-x. [DOI] [PubMed] [Google Scholar]
- 20.Rosell R, Karachaliou N. Lung cancer: Maintenance therapy and precision medicine in NSCLC. Nat Rev Clin Oncol. 2013;10(10):549–550. doi: 10.1038/nrclinonc.2013.152. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z, Zhou L, Wu LM, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18(5):1243–1250. doi: 10.1245/s10434-011-1581-y. [DOI] [PubMed] [Google Scholar]
- 22.Wu ZH, Wang XL, Tang HM, et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;321:395–402. doi: 10.3892/or.2014.3186. [DOI] [PubMed] [Google Scholar]
- 23.Sørensen KP, Thomassen M, Tan Q, et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat. 2013;142(3):529–536. doi: 10.1007/s10549-013-2776-7. [DOI] [PubMed] [Google Scholar]
- 24.Ma MZ, Li CX, Zhang Y, et al. Long non-coding RNA HOTAIR, a c-Myc activated driver of malignancy, negatively regulates miRNA-130a in gallbladder cancer. Mol Cancer. 2014;13:156. doi: 10.1186/1476-4598-13-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G, Zhang H, Wan X, et al. Long noncoding RNA plays a key role in metastasis and prognosis of hepatocellular carcinoma. Biomed Res Int. 2014;2014:780521–8. doi: 10.1155/2014/780521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 27.Hajjari M, Behmanesh M, Sadeghizadeh M, Zeinoddini M. Upregulation of HOTAIR long non-coding RNA in human gastric adenocarcinoma tissues. Med Oncol. 2013;30(3):670. doi: 10.1007/s12032-013-0670-0. [DOI] [PubMed] [Google Scholar]
- 28.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39(6):2119–2128. doi: 10.1177/147323001103900608. [DOI] [PubMed] [Google Scholar]
- 30.Ge XS, Ma HJ, Zheng XH, et al. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104(12):1675–1682. doi: 10.1111/cas.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endo H, Shiroki T, Nakagawa T, et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One. 2013;8(10):e77070. doi: 10.1371/journal.pone.0077070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding C, Cheng S, Yang Z, et al. Long non-coding RNA HOTAIR promotes cell migration and invasion via down-regulation of RNA binding motif protein 38 in hepatocellular carcinoma cells. Int J Mol Sci. 2014;15(3):4060–4076. doi: 10.3390/ijms15034060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen FJ, Sun M, Li SQ, et al. Upregulation of the long non-coding rna hotair promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52(11):908–915. doi: 10.1002/mc.21944. [DOI] [PubMed] [Google Scholar]
- 34.Nie Y, Liu X, Qu S, et al. Long non-coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci. 2013;1044:458–464. doi: 10.1111/cas.12092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim K, Jutooru I, Chadalapaka G, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32(13):1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang L, Liao LM, Liu AW, et al. Overexpression of long noncoding RNA HOTAIR predicts a poor prognosis in patients with cervical cancer. Arch Gynecol Obstet. 2014;290(4):717–723. doi: 10.1007/s00404-014-3236-2. [DOI] [PubMed] [Google Scholar]
- 37.Nakagawa T, Endo H, Yokoyama M, et al. Large noncoding RNA HOTAIR enhances aggressive biological behavior and is associated with short disease-free survival in human non-small cell lung cancer. Biochem Biophys Res Commun. 2013;436(2):319–324. doi: 10.1016/j.bbrc.2013.05.101. [DOI] [PubMed] [Google Scholar]
- 38.Liu XH, Liu ZL, Sun M, et al. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi: 10.1186/1471-2407-13-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ono H, Motoi N, Nagano H, et al. Long noncoding RNA HOTAIR is relevant to cellular proliferation, invasiveness, and clinical relapse in small-cell lung cancer. Cancer Med. 2014;3(3):632–642. doi: 10.1002/cam4.220. [DOI] [PMC free article] [PubMed] [Google Scholar]