Abstract
Background
Acyl-CoA dehydrogenase long chain (ACADL) was revealed to have a correlation with malignant progression of cancer. However, whether ACADL plays a role in clinical therapy remains unclear. The clinicopathological role of ACADL in esophageal squamous cell carcinoma (ESCC) will be discussed in this study.
Materials and methods
The expression of ACADL was analyzed via real-time PCR and Western blotting to assess mRNA and protein levels in ESCC cell lines and normal esophageal epithelial cells (NEECs), in six paired ESCC tumors and relative normal tissues. Furthermore, immunohistochemical staining was performed on 135 paraffin-embedded ESCC specimens to assess ACADL expression. The clinicopathological significance of ACADL expression was further investigated via survival analysis and Cox regression analysis.
Results
ACADL was found to be markedly upregulated in ESCC cell lines when compared with NEECs. Moreover, various experiments such as quantitative real-time PCR, Western blot, and immunohistochemical analyses all revealed that ACADL expression was increased in all six paired ESCC tumors and matched normal tissues. Furthermore, immunohistochemical analysis revealed an increased level of ACADL protein expression in all 135 paraffin-embedded samples from ESCC patients, which increased with disease progression.
Conclusion
We demonstrated that ACADL is overexpressed in ESCC, both in cell lines and clinical specimens. ACADL is found to be a vital regulator in ESCC progression and can predict a worse outcome for ESCC patients, suggesting that ACADL might be a valuable molecule to be targeted for clinical therapy of ESCC treatment.
Keywords: ESCC, ACADL, prognosis, IHC, target
Background
Esophageal cancer is a common malignancy worldwide with high mortality1,2 and can be divided into two major types, which are squamous cell carcinoma and adenocarcinoma. In most parts of the world, esophageal squamous cell carcinoma (ESCC) is the predominant type.3,4 Specifically, in People’s Republic of China, the morbidity and mortality of esophageal cancer are both ranked the fourth.5 Although there have been developments in ESCC therapy in recent years, ESCC prognosis remains poor due to a lack of obvious early symptoms and detection markers.6,7 Lymph node metastasis is the most common death-related event in esophageal cancer. Recurrence and metastasis are the major causes of esophageal cancer-related death. Therefore, identifying diagnostic markers in a subpopulation of ESCC patients is necessary in predicting disease progression, prognosis, recurrence, and metastasis, and furthermore in contributing to the development of new treatments. Acyl-CoA dehydrogenase long chain (ACADL) is one member of the acyl-CoA dehydrogenase family; mitochondrial β-oxidation of straight chain fatty acids initiation could be affected by the members in this family.8 ACADL is responsible for lipid and energy metabolism9–15 and plays a crucial role in normal metabolism. Defects in this gene correlate with multiple human diseases, including cardiovascular disease, central obesity, nonketotic hypoglycemia, diabetes, and World Health Organization-defined metabolic syndrome.15–21 ACADL expression is characterized within specific organs and tissues, which could be used to explore the molecular constituents of human biology and disease.22 It has been reported that ACADL is markedly expressed in several human cancers such as breast cancer, prostatic carcinoma, and melanoma,23,24 and is involved in malignant progression of prostate cancer.24 These previous studies indicate that ACADL might be a key molecule in cancer promotion. However, whether ACADL plays a role in ESCC is still not clear.
In this study, we detected the expression of ACADL in ESCC cell lines, six paired ESCC sections, and matched adjacent normal ones, and 135 paraffin-embedded ESCC samples. We revealed that ACADL was significantly upregulated at both the mRNA and protein levels via quantitative real-time PCR, Western blotting and immunohistochemistry (IHC) analysis. Related clinical patients’ information and detailed statistical analyses furthermore provided evidence that higher ACADL expression level is correlated with ESCC progression, involving advanced clinical stage and a worse outcome. Altogether, we concluded that ACADL could serve as a molecular marker for predicting high-grade tumors and clinical outcomes in ESCC patients.
Materials and methods
Cell culture
As previously described, normal esophageal epithelial cells (NEECs) were constructed from fresh samples of adjacent relative normal esophageal tissue, 5 cm far away from the cancerous section.25 Professors SW Tsao and G Srivastava (University of Hong Kong) kindly provided us with some ESCC cell lines such as Eca109 and EC18. The remaining cell lines used in this study, including KYSE-520, KYSE-410, KYSE-30, KYSE-140, and KYSE-510, were all purchased from Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany), German Resource Center for Biological Material.26 Complete Dulbecco’s Modified Eagle’s Medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA) was used to culture the cells. The use of all cell lines was approved by the Yantai Yuhuangding Hospital Institution Review Board.
Patient information and tissue specimens
One hundred and thirty-five paraffin-embedded ESCC tumor specimens were collected for this study, from patients who had been histopathologically diagnosed with ESCC at the Affiliated Yantai Yuhuangding Hospital of Qingdao University from 2005 to 2010. Table 1 shows the detailed clinical information of the patient samples. Prior patient consent and approval have been obtained from the Ethics Committee of the Yantai Yuhuangding Hospital for the use of these clinical specimens for the purpose of research.
Table 1.
Clinicopathological characteristics of patient samples and expression of ACADL in ESCC
| Characteristics | Number of cases (%) |
|---|---|
|
| |
| Gender | |
| Female | 55 (40.7) |
| Male | 80 (59.3) |
| Age (years) | |
| >60 | 69 (51.1) |
| ≤60 | 66 (48.9) |
| Clinical stage | |
| I | 14 (10.4) |
| II | 80 (59.3) |
| III | 30 (22.2) |
| IV | 11 (8.1) |
| T classification | |
| T1 | 16 (11.9) |
| T2 | 52 (38.5) |
| T3 | 52 (38.5) |
| T4 | 15 (11.1) |
| N classification | |
| N0 | 91 (67.4) |
| N1 | 44 (32.6) |
| M classification | |
| No | 124 (91.9) |
| Yes | 11 (8.1) |
| Vital status (at follow-up) | |
| Alive | 72 (53.3) |
| Dead | 63 (46.7) |
| Histological differentiation | |
| Well | 36 (26.7) |
| Moderate | 74 (54.8) |
| Poor | 25 (18.5) |
| Expression of ACADL | |
| Low expression | 81 (60.0) |
| High expression | 54 (40.0) |
Abbreviations: ACADL, acyl-CoA dehydrogenase long chain; ESCC, esophagus squamous cell carcinoma.
RNA extraction, RT-qPCR
Trizol reagent (Invitrogen) was used to harvest the RNA of ESCC cell lines. Then, the extracted RNA was pretreated with RNase-free DNase, and 2 µg RNA was used for cDNA synthesis, with random hexamers acting as the primers. The RT-qPCR primers for ACADL and GAPDH were designed online, and the sequences used were as follows: forward primer for ACADL 5′-GGAAGCAAGGTGTTCATC AG-3′; reverse primer for ACADL 5′-TCTCCAAGTA GGGCACTAGC-3′; forward primer for GAPDH 5′-ACAT CCCCTCACCAATAACAAC-3′; and reverse primer for GAPDH: 5′-TAGCCAAATCATACTGCTCGTC-3′. Fast-Start Universal SYBR Green Master (ROX; Roche, Toronto, ON, Canada) was used in this experiment. Expression data were then normalized to housekeeper gene GAPDH and calculated as 2–[(Ct ofgene) − (Ct ofGAPDH)]. All experiments were repeated at least three times.
Western blotting
Cell proteins were harvested in sample buffer containing protease inhibitor when the cell confluence reached about 70%–80%. Bicinchoninic acid (BCA) assay (Pierce, Rockford, IL, USA) was used to examine the concentration of the harvested protein. Equal amounts (20 µg) of protein were used in Western blotting analysis. ACADL was detected with an anti-ACADL rabbit antibody (1:1,000; Millipore, Burlington, MA, USA). The α-tubulin expression was used as an equal loading control. The secondary antibodies were goat anti-rabbit/mouse IgG-HRP (1:50,000 dilution; Pierce).
IHC
135 paraffin-embedded ESCC specimens were used to assess the expression of ACADL via IHC. The process has been previously described.27 Goat monoclonal antibody (1:150) was used to target anti-ACADL antibody. The samples that were not incubated with any antibodies were regarded as control. The staining proportion and intensity were reviewed by two expert pathologists who were unaware of the related clinical information. The positive staining tumor cell proportions could be divided into four grades as follows: 1 (<20%), 2 (20%–40%), 3 (40%–70%), and 4 (>70%). The staining intensity was graded as follows: 0 (negative); 1 (weak), 2 (moderate), and 3 (strong). The staining index (SI) was calculated as follows: SI = staining intensity × proportion of positively stained cells. Via this assessment, 0, 1, 2, 3, 4, 6, 8, 9, or 12 would be the final SI. Samples with SI ≥6 were defined as high expression, while SI <6 was recognized as low expression. Finally, the Olympus BX51 image analysis system (Olympus Corporation, Tokyo, Japan) and the CellSens Dimension 1.5 Imaging software (Olympus Corporation) were both used to quantitate the protein level. Briefly, stained sections were analyzed at 200× magnification, and the mean optical density (MOD) was analyzed through ten representative staining fields for each section, which represents the strength of staining signals as measured per positive pixels. The independent samples t-test was used to analyze the statistic differences among different tissues, and P<0.05 was considered statistically significant.
Statistical analysis
IBM SPSS 16.0 statistical software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Correlation analysis were taken to evaluate the relationship between ACADL level and various clinical parameters including gender, age, TNM grading, vital status, tumor stage, and histological differentiation. Kaplan–Meier analysis and log-rank test were used to plot the survival curves. Univariate and multivariate Cox regression analyses were furthermore performed. A P-value <0.05 was considered statistically significant, and <0.01 was considered highly statistically significant.
Results
ACADL is remarkably upregulated in ESCC
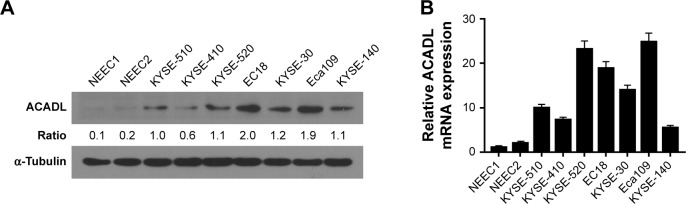
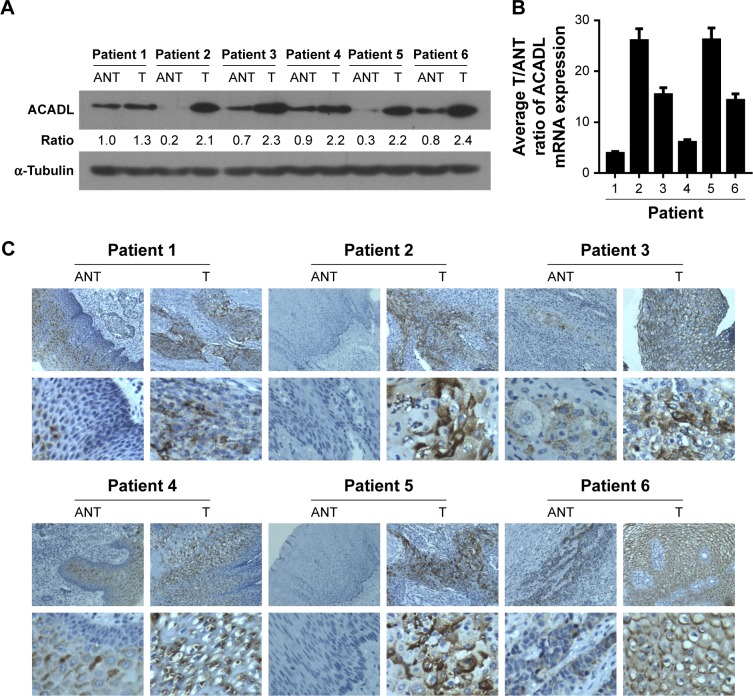
We detected ACADL expression via qPCR and Western blotting analysis in NEECs and ESCC cell lines, which include KYSE-510, KYSE-410, KYSE-520, EC18, KYSE-30, KYSE-109 and KYSE-140. Both mRNA and protein levels of ACADL were markedly upregulated in ESCC cell lines when compared to NEECs (Figure 1A and B). We furthermore validated ACADL expression in six paired normal primary samples. When comparing to the relatively normal tissues, ACADL protein expression was significantly upregulated in the matched ESCC ones (Figure 2A). As expected, the mRNA expression of ACADL was consistent with the protein expression (Figure 2B). Furthermore, IHC analysis also supported that ACADL expression was increased in ESCC tissues compared with the normal control (Figure 2C). Collectively, these results suggested a significant overexpression of ACADL in ESCC both in mRNA and protein levels.
Figure 1.
Expression pattern of ACADL in NEEC and ESCC cell lines.
Notes: Expression of ACADL in ESCC cell lines (KYSE-510, KYSE-410, KYSE-520, EC18, KYSE-30, KYSE-109 and KYSE-140) and NEEC were examined via Western blotting (A) and qPCR (B). α-Tubulin and GAPDH act as housekeeper gene in the experiments, respectively. Columns indicate mean from three parallel experiments; bars represent SD.
Abbreviations: ACADL, acyl-CoA dehydrogenase long chain; ESCC, esophageal squamous cell carcinoma; NEEC, normal esophageal epithelial cell; qPCR, quantitative PCR.
Figure 2.
ACADL is upregulated in ESCC specimens.
Notes: Western blotting (A) and RT-PCR (B) analysis were used to detect the protein and mRNA expression pattern in six paired ESCC tissues (T) and matched adjacent relative normal ones (N) respectively. IHC analysis was furthermore undertaken to validate ACADL expression in clinical samples (C).
Abbreviations: ACADL, acyl-CoA dehydrogenase long chain; ESCC, esophageal squamous cell carcinoma; IHC, immunohistochemistry; RT-PCR, real-time PCR.
ACADL is overexpressed in clinical ESCC specimens
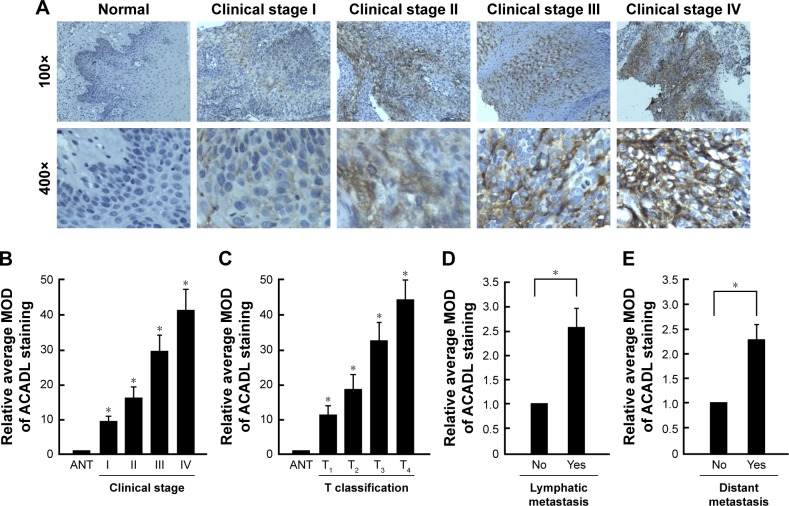
To investigate whether ACADL is essential to the progression of ESCC, the clinical significance of ACADL was further validated in 135 paraffin-embedded, archived clinical ESCC samples, of which 14 cases belonged to stage I, 80 cases belonged to stage II, 30 cases belonged to stage III, and 11 cases belonged to stage IV. Among all samples, 54 samples showed a high ACADL protein expression (40.0%) and 81 samples showed weak or no staining (60.0%, Table 1). As can be seen Figure 3A, IHC staining showed that ACADL expression is correlated with clinical ESCC stage. The MOD values of ACADL staining were higher in primary ESCC when compared to control ones. Tumor grades and T classification could both be influence factors to MOD (P<0.001, Figure 3B and C) in ESCC. Interestingly, there was a significant difference (positively relation) of MOD values of ACADL staining in lymphatic metastasis group (P<0.01, Figure 3D) and distant metastasis group when compared to vector ones (P<0.01, Figure 3E).
Figure 3.
IHC and statistical analysis of ACADL upregulation pattern in archived paraffin-embedded ESCC tissue sections.
Notes: Representative images of ACADL expression in normal esophagus tissues and ESCC sections of various clinical stages from IHC analyses (A). The average MOD of ACADL staining was analyzed between relative normal esophagus tissues and ESCC sections of various clinical stages (B). The average MOD of ACADL staining was analyzed between relative normal ones and ESCC samples from various T grades (C). The average MOD of ACADL staining was analyzed between nonlymphatic metastasis group and lymphatic metastasis group (D). The average MOD of ACADL staining was analyzed between nondistant metastasis group and distant metastasis group (E). *P<0.05.
Abbreviations: ACADL, acyl-CoA dehydrogenase long chain; ESCC, esophageal squamous cell carcinoma; IHC, immunohistochemistry; MOD, mean optical density.
ACADL expression is associated with clinicopathology of ESCC
To reveal the specific role of ACADL in ESCC, clinical patients’ information was taken into consideration to validate the vital role of ACADL in the progression of ESCC. As shown in Table 2, statistic analysis revealed that high expression of ACADL was positively correlated with tumor stage (P<0.001; Table 2), T/N classification (P<0.001), M classification (P=0.021), as well as pathological differentiation (P=0.003). To ensure these results, correlation analysis was used to explore ACADL expression levels with different clinical parameters including tumor stages (0.453; P<0.001), T classification (0.421; P<0.001), N classification (0.400; P<0.001), M classification (0.199; P=0.021), and pathological differentiation (0.277; P=0.001), as is shown in Table 3. These results showed that ACADL expression positively correlated with clinical stage, TNM classification, and poor pathological differentiation in ESCC.
Table 2.
Clinicopathological characteristics of patient samples and expression of ACADL in ESCC and correlation between ACADL expression and clinicopathological characteristics of ESCC patients
| Characteristics | Total | ACADL
|
χ2 test P-value | Fisher’s exact test P-value | |
|---|---|---|---|---|---|
| Low expression 60.0% | High expression 40.0% | ||||
|
| |||||
| Gender | 0.283 | 0.186 | |||
| Female | 55 | 36 (65.5) | 19 (34.5) | ||
| Male | 80 | 45 (56.3) | 35 (43.7) | ||
| Age (years) | 0.058 | 0.079 | |||
| >60 | 69 | 36 (52.2) | 33 (47.8) | ||
| ≤60 | 66 | 45 (68.2) | 21 (31.8) | ||
| Clinical stage | <0.001 | <0.001 | |||
| I | 14 | 14 (100.0) | 0 (0.0) | ||
| II | 80 | 55 (68.8) | 25 (31.2) | ||
| III | 30 | 9 (30.0) | 21 (70.0) | ||
| IV | 11 | 3 (27.3) | 8 (72.7) | ||
| T classification | <0.001 | <0.001 | |||
| T1 | 16 | 15 (93.8) | 1 (6.2) | ||
| T2 | 52 | 38 (73.1) | 14 (26.9) | ||
| T3 | 52 | 25 (48.1) | 27 (51.9) | ||
| T4 | 15 | 3 (20.0) | 12 (80.0) | ||
| N classification | <0.001 | <0.001 | |||
| N0 | 91 | 67 (73.6) | 24 (26.4) | ||
| N1 | 44 | 14 (31.8) | 30 (68.2) | ||
| M classification | 0.021 | 0.027 | |||
| Yes | 11 | 3 (27.3) | 8 (72.7) | ||
| No | 124 | 78 (62.9) | 46 (37.1) | ||
| Histological differentiation | 0.003 | 0.003 | |||
| Well | 36 | 27 (75.0) | 9 (25.0) | ||
| Moderate | 74 | 46 (62.2) | 28 (37.8) | ||
| Poor | 25 | 8 (32.0) | 17 (68.0) | ||
| Status | <0.001 | <0.001 | |||
| Alive | 72 | 62 (86.1) | 10 (13.9) | ||
| Dead | 63 | 19 (30.2) | 44 (69.8) | ||
Abbreviations: ACADL, acyl-CoA dehydrogenase long chain; ESCC, esophageal squamous cell carcinoma.
Table 3.
Spearman correlation analysis between ACADL and clinicopathologic factors
| Variables | ACADL expression level
|
P-value |
|---|---|---|
| Spearman correlation | ||
|
| ||
| Clinical staging | 0.453 | <0.001 |
| T classification | 0.421 | <0.001 |
| N classification | 0.400 | <0.001 |
| M classification | 0.199 | 0.021 |
| Status | 0.570 | <0.001 |
| Histological differentiation | 0.277 | 0.001 |
| Survival time | −0.602 | <0.001 |
Abbreviation: ACADL, acyl-CoA dehydrogenase long chain.
Upregulation of ACADL might predict a poor ESCC prognosis
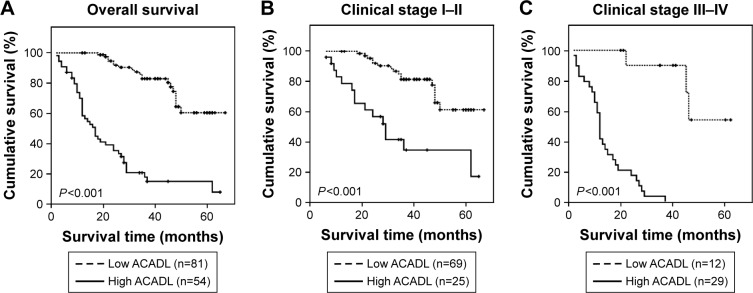
We revealed that increased ACADL expression could predict a shorter survival time in patients with ESCC (-0.602, P<0.001, Table 3), and that there is a close connection between them. Furthermore, Kaplan–Meier analysis and log-rank tests were undertaken to indicate the relationship between ACADL level and several clinicopathologic parameters including clinical stage, T/N/M classification, as well as pathological differentiation and status to predict patients’ outcome. The result showed higher ACADL expression could predict a lower cumulative 5-year survival rate, and lower ACADL expression could predict a longer one. In the low ACADL expression group, >60% of patients had better 5-year survival time, but the percent who had a better a 5-year survival time in the high ACADL expression group was <20% (Figure 4A).
Figure 4.
Survival curves and univariate analyses were assessed to evaluate the potentiality of ACADL as an independent prognostic marker for ESCC patients.
Notes: ACADL-low group and ACADL-high group were compared to predict the overall survival of patients who were diagnosed with ESCC. In the low-ACADL group, the rate of cumulative 5-year survival was 60.1% (95% CI, 44.2%, 76.0%), whereas it was only 14.4% (95% CI, 4.0%, 24.8%) in the high-ACADL group (A). The statistical significance of survival time for patients was compared both in clinical stage I–II subgroup (B) and III–IV subgroup based on the expression of ACADL (C). Log-rank tests were taken to calculate the P-values.
Abbreviations: ACADL, acyl-CoA dehydrogenase long chain; ESCC, esophageal squamous cell carcinoma.
We stratified patients based on their T/N/M classification and clinical stages and analyzed the value and possibility of being a molecular target for patients who are diagnosed with ESCC, we examined whether it could function as a marker both in stage I–II subgroup and stage III–IV subgroup. And we validated its vital role in the progression of ESCC. Low ACADL patients presented a longer survival time while high ACADL patients presented a shorter survival time. This was observed in both clinical stage I–II subgroup (P<0.001, Figure 4B) and III–IV subgroup (P<0.001, Figure 4C). Consistently, the survival time in pT1–2/pT3–4/pN−/pN+ pM−/pM+ subgroups was much shorter in ESCC patients with higher ACADL expression (Figure S1) and much longer in ESCC patients with lower ACADL expression. Collectively, these results indicate that ACADL will be a valuable marker in ESCC patients.
The value of ACADL expression as a prognostic factor for ESCC patient was evaluated by univariate and multivariate Cox regression analysis. The results identified that ACADL expression was also an independent marker to predict patients’ outcome together with clinical stage, and they are both recognized (Table 4). It is a useful biomarker for predicting progression of ESCC and patients’ prognosis.
Table 4.
Univariate and multivariate analyses of various prognostic parameters in patients with ESCC Cox regression analysis
| Variables | Univariate analysis
|
Multivariate analysis
|
||||
|---|---|---|---|---|---|---|
| Number of patients | P-value | Regression coefficient (SE) | P-value | Relative risk | 95% CI | |
|
| ||||||
| ACADL | <0.001 | 1.903 (0.297) | <0.001 | 6.707 | 3.746–12.008 | |
| Low expression | 69 | |||||
| High expression | 66 | |||||
| Clinical stage | <0.001 | 0.782 (0.177) | <0.001 | 2.185 | 1.543–3.093 | |
| I | 38 | |||||
| II | 113 | |||||
| III | 46 | |||||
| IV | 15 | |||||
Notes: ACADL protein expression level in ESCC significantly correlated with patient survival time (P<0.001); the correlation coefficient was −0.602 indicating that higher levels of ACADL expression correlated with shorter survival time. Five-year survival low expression 60.1% (44.2%–76.0%). High expression 14.4% (4.0%–24.8%).
Abbreviations: ACADL, acyl-CoA dehydrogenase long chain; ESCC, esophageal squamous cell carcinoma.
Discussion
ACADL is one of the acyl-CoA dehydrogenases involved in lipid and energy metabolism, which exhibits tissue-specific and spatial expression patterns, acts as a major regulator in the process of metabolism, and could affect various aspects of the process.22 Defects in this gene would cause long-chain acyl-CoA dehydrogenase deficiency, resulting in dysregulation of the gene in development and disease, and bringing about a false metabolic pattern in human body.28 Upregulation of ACADL correlates with lipolysis and glucose metabolism, resulting in a range of disorders such as fatty liver disease, central obesity, metabolic syndrome, which are defined by the World Health Organization, and type 2 diabetes.15,20 However, owing to its metabolic capabilities, the role of ACADL, an important mitochondrial flavoenzyme, in tumor progression, development, and therapeutic responses have been under investigation only recently, and many aspect of its function are still unclear and need further exploration.29 Studies have shown ACADL protein expression is downregulated in solid pseudopapillary tumor of the pancreas, which is a rare disease with favorable prognosis.30 Increased ACADL mRNA expression, along with TNFAIP8 and HIF-1α, is reported to regulate cell survival and cancer progression.23 Xie et al,24 found that ACADL contributes to the progression of prostate carcinoma, enhancing the malignant phenotypes of prostate cancer cells; however, Li et al31 showed that it is negatively correlated with methylation of breast cancer. These studies highlight the potentiality of ACADL as a biomarker in clinic.
In this study, we first discovered that the mRNA and protein expression levels of ACADL were both upregulated in ESCC cell lines and tumor samples when compared to the respective control. Meanwhile, our IHC analysis indicated that high ACADL protein expression in histological sections had a close relationship with clinical stage, TNM grading, pathological differentiation, and survival time in ESCC. Statistical analysis further confirmed that ACADL upregulation could give rise to a worse outcome for ESCC patients. These results reveal that upregulation of ACADL might enhance and promote the progression of ESCC.
Previous studies have demonstrated elevated ACADL expression in various tumors; our results are consistent with those studies, highlighting its important role in tumorigenesis. Particularly, we evaluated the potentiality of ACADL to function as an effective molecular therapeutic target for patients who are diagnosed with ESCC However, further investigations are needed to determine whether it is the upregulation of ACADL that leads to the malignant phenotype and properties in ESCC or reversely if it is the initiation and progression of the tumor that contributes to ACADL overexpression. The specific molecular mechanism by which ACADL gives rise to malignancy of ESCC also needs to be deciphered.
Conclusion
In this study, we demonstrate that elevated ACADL expression has a close relationship with histological grades and could be an effective factor that affects ESCC patients’ outcome, which gives light on the possibility that ACADL is a vital gene in the process of tumorigenesis. However, the specific role of ACADL in ESCC needs further investigation.
Supplementary material
Survival curves for ESCC patients in cases with ACADL expression and other clinical parameters into consideration.
Notes: The statistical significance of ACADL-high patients and ACADL-low patients was analyzed within subgroups of T grading of 1–2 and T grading of 3–4 (A, B); within subgroups of lymph node metastasis negative or positive (C, D); within subgroups of metastasis negative or positive (E, F). Log-rank tests were taken to calculate the P-values.
Abbreviations: ACADL, acyl-CoA dehydrogenase long chain; ESCC, esophageal squamous cell carcinoma; MOD, mean optical density; qPCR, quantitative PCR.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Peng X, Xue H, Lu L, Shi P, Wang J, Wang J. Accumulated promoter methylation as a potential biomarker for esophageal cancer. Oncotarget. 2017;8(1):679–691. doi: 10.18632/oncotarget.13510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Ren QG, Yang SL, Hu JL, Li PD, Chen YS, Wang QS. Evaluation of HO-1 expression, cellular ROS production, cellular proliferation and cellular apoptosis in human esophageal squamous cell carcinoma tumors and cell lines. Oncol Rep. 2016;35(4):2270–2276. doi: 10.3892/or.2016.4556. [DOI] [PubMed] [Google Scholar]
- 4.Hsieh MS, Yang PW, Wong LF, Lee JM. The AXL receptor tyrosine kinase is associated with adverse prognosis and distant metastasis in esophageal squamous cell carcinoma. Oncotarget. 2016;7(24):36956–36970. doi: 10.18632/oncotarget.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Y, Totsuka Y, He Y, et al. Epidemiology of esophageal cancer in Japan and China. J Epidemiol. 2013;23(4):233–242. doi: 10.2188/jea.JE20120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleinberg L, Forastiere AA. Chemoradiation in the management of esophageal cancer. J Clin Oncol. 2007;25(26):4110–4117. doi: 10.1200/JCO.2007.12.0881. [DOI] [PubMed] [Google Scholar]
- 7.Han X, Lu N, Pan Y, Xu J. Nimotuzumab Combined with Chemotherapy is a Promising Treatment for Locally Advanced and Metastatic Esophageal Cancer. Med Sci Monit. 2017;23(1):412–418. doi: 10.12659/MSM.902645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurtz DM, Tolwani RJ, Wood PA. Structural characterization of the mouse long-chain acyl-CoA dehydrogenase gene and 5′ regulatory region. Mamm Genome. 1998;9(5):361–365. doi: 10.1007/s003359900770. [DOI] [PubMed] [Google Scholar]
- 9.Ashmore T, Roberts LD, Morash AJ, et al. Nitrate enhances skeletal muscle fatty acid oxidation via a nitric oxide-cGMP-PPAR-mediated mechanism. BMC Biol. 2015;13(1):110. doi: 10.1186/s12915-015-0221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu S, Park S, Lim Y, Shin S, Han SN. Korean pine nut oil replacement decreases intestinal lipid uptake while improves hepatic lipid metabolism in mice. Nutr Res Pract. 2016;10(5):477–486. doi: 10.4162/nrp.2016.10.5.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee AR, Han SN. Pinolenic Acid Downregulates Lipid Anabolic Pathway in HepG2 Cells. Lipids. 2016;51(7):847–855. doi: 10.1007/s11745-016-4149-6. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki S, Awai K, Ishihara A, Yamauchi K. Cold temperature blocks thyroid hormone-induced changes in lipid and energy metabolism in the liver of Lithobates catesbeianus tadpoles. Cell Biosci. 2016;6(1):19. doi: 10.1186/s13578-016-0087-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park S, Shin S, Lim Y, et al. Korean Pine Nut Oil Attenuated Hepatic Triacylglycerol Accumulation in High-Fat Diet-Induced Obese Mice. Nutrients. 2016;8(1):59. doi: 10.3390/nu8010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao S, Liu Y, Sun W, et al. Genome-Wide Expression Profiling of Anoxia/Reoxygenation in Rat Cardiomyocytes Uncovers the Role of MitoKATP in Energy Homeostasis. Oxid Med Cell Longev. 2015;2015(1):756576. doi: 10.1155/2015/756576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun H, Liu D. Hydrodynamic delivery of interleukin 15 gene promotes resistance to high fat diet-induced obesity, fatty liver and improves glucose homeostasis. Gene Ther. 2015;22(4):341–347. doi: 10.1038/gt.2014.114. [DOI] [PubMed] [Google Scholar]
- 16.Kim SJ, Chae S, Kim H, et al. A protein profile of visceral adipose tissues linked to early pathogenesis of type 2 diabetes mellitus. Mol Cell Proteomics. 2014;13(3):811–822. doi: 10.1074/mcp.M113.035501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu X, Duan Z, Hu H, et al. Proteomic profile of carbonylated proteins in rat liver: exercise attenuated oxidative stress may be involved in fatty liver improvement. Proteomics. 2013;13(10–11):1755–1764. doi: 10.1002/pmic.201200522. [DOI] [PubMed] [Google Scholar]
- 18.Lin X, Hamilton-Williams EE, Rainbow DB, et al. Genetic interactions among Idd3, Idd5.1, Idd5.2, and Idd5.3 protective loci in the nonobese diabetic mouse model of type 1 diabetes. J Immunol. 2013;190(7):3109–3120. doi: 10.4049/jimmunol.1203422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirkov S, Myers JL, Ramírez J, Liu W. SNPs affecting serum metabolomic traits may regulate gene transcription and lipid accumulation in the liver. Metabolism. 2012;61(11):1523–1527. doi: 10.1016/j.metabol.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banasik K, Justesen JM, Hornbak M, et al. Bioinformatics-driven identification and examination of candidate genes for non-alcoholic fatty liver disease. PLoS One. 2011;6(1):e16542. doi: 10.1371/journal.pone.0016542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Illig T, Gieger C, Zhai G, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42(2):137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fagerberg L, Hallström BM, Oksvold P, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13(2):397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Day TF, Mewani RR, Starr J, et al. Transcriptome and proteome analyses of TNFAIP8 knockdown cancer cells reveal new insights into molecular determinants of cell survival and tumor progression. Methods Mol Biol. 2017;1513(1):83–100. doi: 10.1007/978-1-4939-6539-7_7. [DOI] [PubMed] [Google Scholar]
- 24.Xie BX, Zhang H, Wang J, et al. Analysis of differentially expressed genes in LNCaP prostate cancer progression model. J Androl. 2011;32(2):170–182. doi: 10.2164/jandrol.109.008748. [DOI] [PubMed] [Google Scholar]
- 25.Andl CD, Mizushima T, Nakagawa H, et al. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J Biol Chem. 2003;278(3):1824–1830. doi: 10.1074/jbc.M209148200. [DOI] [PubMed] [Google Scholar]
- 26.Shimada Y, Imamura M, Wagata T, Yamaguchi N, Tobe T. Characterization of 21 newly established esophageal cancer cell lines. Cancer. 1992;69(2):277–284. doi: 10.1002/1097-0142(19920115)69:2<277::aid-cncr2820690202>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 27.Li J, Gong LY, Song LB, et al. Oncoprotein Bmi-1 renders apoptotic resistance to glioma cells through activation of the IKK-nuclear factor-kappaB Pathway. Am J Pathol. 2010;176(2):699–709. doi: 10.2353/ajpath.2010.090502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurtz DM, Rinaldo P, Rhead WJ, et al. Targeted disruption of mouse long-chain acyl-CoA dehydrogenase gene reveals crucial roles for fatty acid oxidation. Proc Natl Acad Sci U S A. 1998;95(26):15592–15597. doi: 10.1073/pnas.95.26.15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Justus CR, Sanderlin EJ, Yang LV. Molecular Connections between Cancer Cell Metabolism and the Tumor Microenvironment. Int J Mol Sci. 2015;16(5):11055–11086. doi: 10.3390/ijms160511055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y, Xu H, Chen H, et al. Proteomic analysis of solid pseudopapillary tumor of the pancreas reveals dysfunction of the endoplasmic reticulum protein processing pathway. Mol Cell Proteomics. 2014;13(10):2593–2603. doi: 10.1074/mcp.M114.038786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Heng J, Yan J, et al. Integrated analysis of gene expression and methylation profiles of 48 candidate genes in breast cancer patients. Breast Cancer Res Treat. 2016;160(2):371–383. doi: 10.1007/s10549-016-4004-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Survival curves for ESCC patients in cases with ACADL expression and other clinical parameters into consideration.
Notes: The statistical significance of ACADL-high patients and ACADL-low patients was analyzed within subgroups of T grading of 1–2 and T grading of 3–4 (A, B); within subgroups of lymph node metastasis negative or positive (C, D); within subgroups of metastasis negative or positive (E, F). Log-rank tests were taken to calculate the P-values.
Abbreviations: ACADL, acyl-CoA dehydrogenase long chain; ESCC, esophageal squamous cell carcinoma; MOD, mean optical density; qPCR, quantitative PCR.