Significance
Chemical cross-linking mass spectrometry is a powerful method to identify protein interaction partners. The cross-links also provide approximate interresidue distances, which can help model the structures of complexes. However, current cross-linking reagents react with only a limited set of amino acid residues and their very high reactivity can induce cross-links between sites that are distant in the native state of the interrogated proteins. We therefore developed a “plant-and-cast” strategy, in which a heterobifunctional cross-linker is “planted” onto surface Lys residues using a highly reactive succinimide ester. The half-reacted cross-linker then “casts” a much less reactive sulfonyl fluoride across the proximal surface resulting in cross-links to neighboring Ser, Thr, Tyr, His, or Lys sidechains in a proximity-enhanced reaction.
Keywords: chemical cross-linker, sulfur–fluoride exchange, cross-linking mass spectrometry, proximity-enhanced reactivity, protein–protein interaction
Abstract
Chemical cross-linking mass spectrometry (CXMS) is being increasingly used to study protein assemblies and complex protein interaction networks. Existing CXMS chemical cross-linkers target only Lys, Cys, Glu, and Asp residues, limiting the information measurable. Here we report a “plant-and-cast” cross-linking strategy that employs a heterobifunctional cross-linker that contains a highly reactive succinimide ester as well as a less reactive sulfonyl fluoride. The succinimide ester reacts rapidly with surface Lys residues “planting” the reagent at fixed locations on protein. The pendant aryl sulfonyl fluoride is then “cast” across a limited range of the protein surface, where it can react with multiple weakly nucleophilic amino acid sidechains in a proximity-enhanced sulfur-fluoride exchange (SuFEx) reaction. Using proteins of known structures, we demonstrated that the heterobifunctional agent formed cross-links between Lys residues and His, Ser, Thr, Tyr, and Lys sidechains. This geometric specificity contrasts with current bis-succinimide esters, which often generate nonspecific cross-links between lysines brought into proximity by rare thermal fluctuations. Thus, the current method can provide diverse and robust distance restraints to guide integrative modeling. This work provides a chemical cross-linker targeting unactivated Ser, Thr, His, and Tyr residues using sulfonyl fluorides. In addition, this methodology yielded a variety of cross-links when applied to the complex Escherichia coli cell lysate. Finally, in combination with genetically encoded chemical cross-linking, cross-linking using this reagent markedly increased the identification of weak and transient enzyme–substrate interactions in live cells. Proximity-dependent cross-linking will dramatically expand the scope and power of CXMS for defining the identities and structures of protein complexes.
Chemical cross-linking mass spectrometry (CXMS) offers the unique ability to decipher protein interaction networks and to derive tertiary structural information of proteins, and thus is increasingly used to study large and transient protein assemblies and intrinsically disordered proteins that are challenging for classic protein structural analysis techniques (1–5). In CXMS, a bifunctional chemical reagent is applied to proteins to cross-link pairs of amino acid residues, which are identified by tandem MS. The identity and distance constraints obtained for amino acids then afford information of protein interactions and tertiary structures. The versatility and throughput of CXMS in combination with X-ray crystallography, NMR, or cryoelectron microscopy is advancing structural biology and interactomics in great strides.
The chemistry of the cross-linker is critical for acquiring abundant and accurate information in CXMS. Currently the most widely used cross-linkers consist of homobifunctional N-hydroxysuccinimidyl (NHS) esters to react with Lys (4, 5). Cross-linkers targeting Glu and Asp have also been developed (6–8), yet they require the carboxylic acid residue to be activated before cross-linking under reaction conditions that can distort protein structure (5). Alternatively, disulfides can be formed between Cys residues after treatment with reagents such as I2, which create highly reactive intermediates (9). However, these methods have limitations that have kept CXMS from reaching its full potential. The restricted repertoire of residues, Lys, Glu, Asp, Cys, limits the number and types of restraints that can be obtained. Moreover, the high reactivity of the intermediates often leads to spurious cross-links between residues that are far apart in the native structures of proteins, but brought into proximity by rare thermal fluctuations that are then trapped due to the high reactivity of the cross-linking chemistry. Thus, the Cα-Cα distances between cross-linked residues are often far greater than the combined distance of the sidechains plus the cross-linking moiety (10). This ambiguity decreases the precision with which interresidue distances can be specified, complicating their use as structural restraints for molecular modeling of complexes.
Besides these residue-specific chemistries, photoactivated cross-linkers such as diazirines target virtually all amino acids nonselectively (11). However, the possibility of excessive cross-linking dramatically complicates the data analysis, which remains a daunting challenge for CXMS, and high-density cross-linking may artificially distort protein tertiary structure as well (4, 5). In addition, photo–cross-linkers generally have short half-lives and thus limited use in studying weak and transient protein–protein interactions (PPIs) (12). Clearly, new chemical cross-linking strategies that are able to target a wide range of amino acid residues specifically with defined cross-linking sites could have a large impact on CXMS.
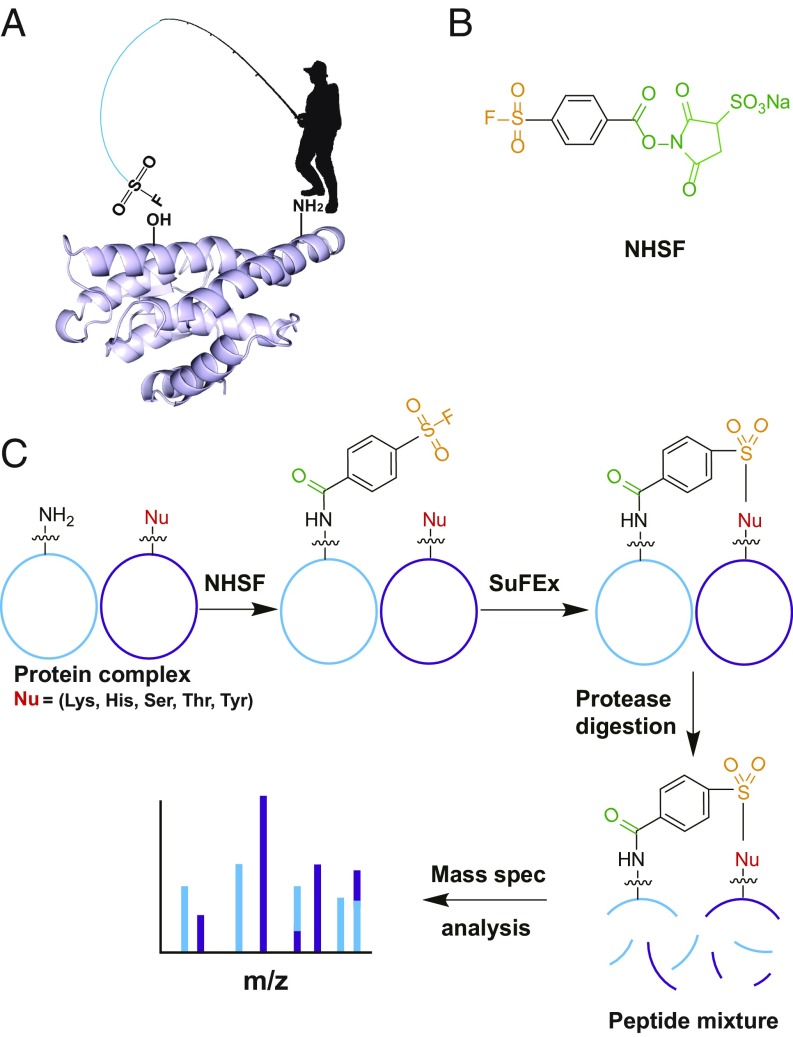
To address this need, we introduce here a plant-and-cast strategy in which highly reactive and weakly reactive electrophiles are combined into a single cross-linker (Fig. 1A). The stronger electrophile, in this case a succinimide ester, reacts rapidly with Lys sidechains, placing the weakly reactive electrophile on the surface of the protein. The flexibility of the Lys sidechain allows the half-reacted cross-linker to cast about along the protein surface, where its proximity and very high local concentration will facilitate reaction with a variety of sidechains that are generally not accessed by traditional cross-linking agents.
Fig. 1.
A plant-and-cast strategy for developing specific, multitargeting NHSF cross-linker. (A) The plant-and-cast strategy. (B) Structure of NHSF. (C) NHSF cross-links Lys with various nucleophilic residues via proximity-enhanced SuFEx reaction for CXMS.
We report a proximity-enhanced chemical cross-linker that is able to target multiple amino acid residues with high specificity and efficiency for CXMS. A heterobifunctional cross-linker containing N-hydroxysulfosuccinimide and aryl sulfonyl fluoride moieties (NHSF, Fig. 1B) was designed and shown to form cross-links between Lys and neighboring nucleophilic residues including His, Lys, Ser, Thr, and Tyr on proteins. Importantly, we found high structural compatibility of the identified cross-linking sites by NHSF, highlighting the potential of NHSF for accurate structural studies of proteins. We further showed that NHSF could cross-link complex Escherichia coli whole-cell lysate revealing cross-linked peptides undetectable with existing reagents. Finally, we showed that this approach can enhance the identification of weak and transient protein interactions when combined with genetically encoded chemical cross-linking (GECX) (12).
Results
A Plant-and-Cast Strategy for Developing Specific, Multitargeting Cross-Linker.
Reactivity and selectivity are two opposing demands, which are difficult to achieve simultaneously when designing chemical cross-linkers––especially when the reaction needs to be compatible with proteins and their milieu. Recently, we developed proximity-enabled bioreactivity (13–16), which allows unnatural amino acids (Uaas) bearing biocompatible functional groups to react with specific natural residues of proteins selectively by bringing the two residues into proximity (17). This methodology has enabled us to capture weak PPIs and transient enzyme–substrate interactions (12). In particular, a sulfonyl-fluoride–containing Uaa is able to react with Lys and His (18), and a fluorosulfate-containing Uaa FSY reacts with Lys, His, and Tyr, both via sulfur–fluoride exchange (SuFEx) reactions (19). Proximity-enhanced reactivity is a recently appreciated approach to direct and control reactivity, with wide-ranging applications in chemical biology (20). Also, sulfonyl fluorides have gained much attention recently in chemical proteomics and covalent drug discovery (21, 22), in which sulfonyl fluorides form noncovalent complex with target proteins and subsequently modify the protein covalently with high specificity toward multiple nucleophilic residues.
Since aryl sulfonyl fluorides have low intrinsic reactivity with nucleophilic residues at physiological conditions (21), we reasoned that a heterobifunctional cross-linker containing NHS and aryl sulfonyl fluoride groups (NHSF, Fig. 1B) would enhance the cross-linking efficiency over a homobifunctional cross-linker containing aryl sulfonyl fluoride group only. An initial rapid second-order reaction of the highly reactive NHS moiety with exposed Lys residues will plant the aryl sulfonyl fluoride in close proximity to nucleophilic residues, and the resultant proximity would enhance a subsequent first-order SuFEx reaction between aryl sulfonyl fluoride and a range of nucleophilic residues to achieve efficient cross-linking (Fig. 1C). The high effective concentration of the sulfonyl fluoride would assure reaction with weakly nucleophilic sidechains (e.g., Ser, Thr) that ordinarily would be unreactive. This type of heterobifunctional cross-linker has been previously attempted for stepwise cross-linking of two proteins with known interactions (23), but no single cross-linked residue was identified and its application for the powerful CXMS was not explored.
Reaction of NHSF with Model Peptide.
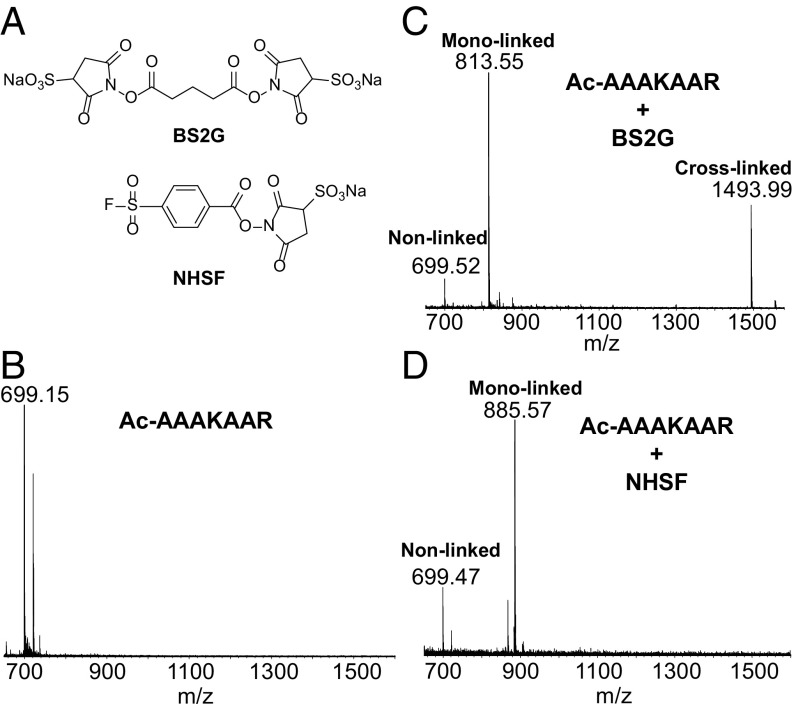
We first tested the reactivity of NHSF with a model peptide (Ac-AAAKAAR, 7KR) with a single Lys as a reactive group and compared its reactivity toward BS2G [bis(sulfosuccinimidyl) 2,2,4,4-glutarate-d4, Fig. 2A], a commercial NHS-based homobifunctional cross-linker of similar length. With the peptide in the same molar ratio of BS2G, we detected both a monolinked peptide as well as the cross-linked dimer (Fig. 2C). In contrast, when NHSF was used, the reaction stops at the monoadduct created by reaction of the succinimide ester group with the Lys sidechain. Only trace amounts of the corresponding sulfonamide were formed (Fig. 2D), consistent with the much lower reactivity of SF compared with the succinimide ester. Owing to the low reactivity of sulfonyl fluorides and the lack of reactive groups in 7KR, additional reaction was not observed with the Arg sidechain. Also the tethered sulfonyl fluoride in the monoadduct was resistant to spontaneous hydrolysis. This experiment suggests the ability to plant and stably cast the sulfonyl-fluoride group across a protein surface.
Fig. 2.
Reaction of NHSF with peptide 7KR. (A) Structures of BS2G and NHSF. Mass spectra of peptide 7KR (B), BS2G-treated 7KR (C), and NHSF-treated 7KR (D).
NHSF Cross-Links BSA.
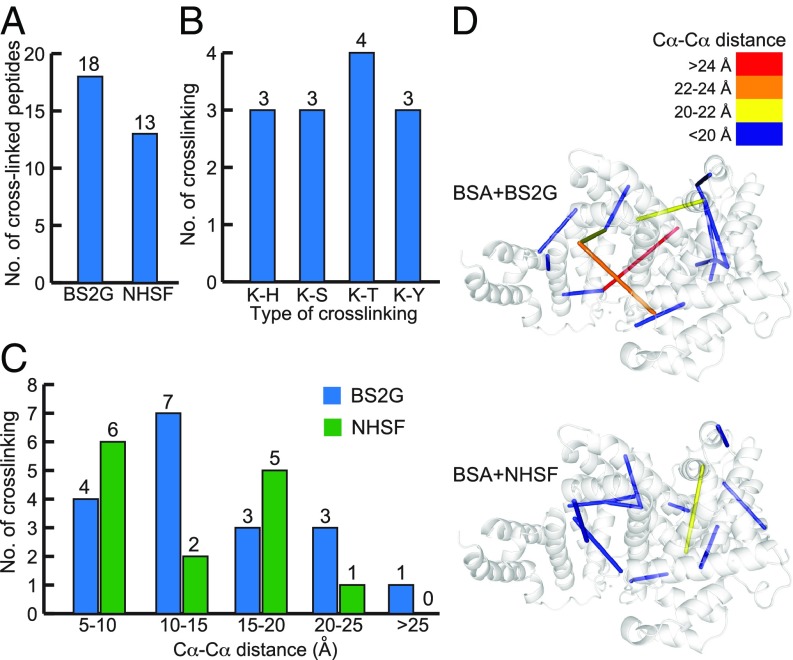
To determine the ability of NHSF to cross-link proteins, BSA was cross-linked by BS2G and NHSF, separately. A total of 18 interpeptide cross-links of BS2G sample and 13 interpeptide cross-links of NHSF sample were identified by MS (Fig. 3A and SI Appendix, Tables S1 and S2). While only Lys-Lys or N-terminal NH2-Lys cross-linking sites were identified in the BS2G sample, multiple cross-linking sites of Lys-His, Lys-Ser, Lys-Thr, and Lys-Tyr were detected in the NHSF sample (Fig. 3B), confirming the ability of NHSF to cross-link Lys with multiple weakly nucleophilic residues via proximity-enhanced SuFEx reaction as designed. Interestingly, no Lys-Lys cross-links were observed when NHSF was used. Thus, the information obtained from NHSF cross-linking was complementary to that of BS2G. The maximum Cα-Cα distance of Lys-Lys cross-linking site is ∼20 Å for BS2G, and 17–22 Å for NHSF cross-linking based on the combined structures of the amino acid sidechains plus the covalently connected cross-linking agent. We therefore used a limit of 20 Å for reasonable cross-linking distances, and evaluated the Cα-Cα distances for all cross-linked residues in the BSA structure (Fig. 3 C and D): There was only one cross-linked peptide slightly exceeding this limit for the NHSF sample, which measured a distance of 21.2 Å. In contrast, there were four cross-linked peptides exceeding the distance limit for the BS2G sample.
Fig. 3.
CXMS analyses of BSA protein cross-linked with NHSF or BS2G. (A) Total number of identified cross-linked peptides from BS2G or NHSF cross-linked BSA. (B) Types of cross-linking sites identified from the NHSF cross-linked BSA sample, with numbers indicated on top. (C) Distribution of Cα-Cα distance of cross-linked residues from BSA samples. (D) Identified cross-links mapped onto the crystal structures of BSA (Protein Data Bank ID code 3V03). The Cα-Cα distances of cross-links are color-coded.
NHSF Cross-Links GST.
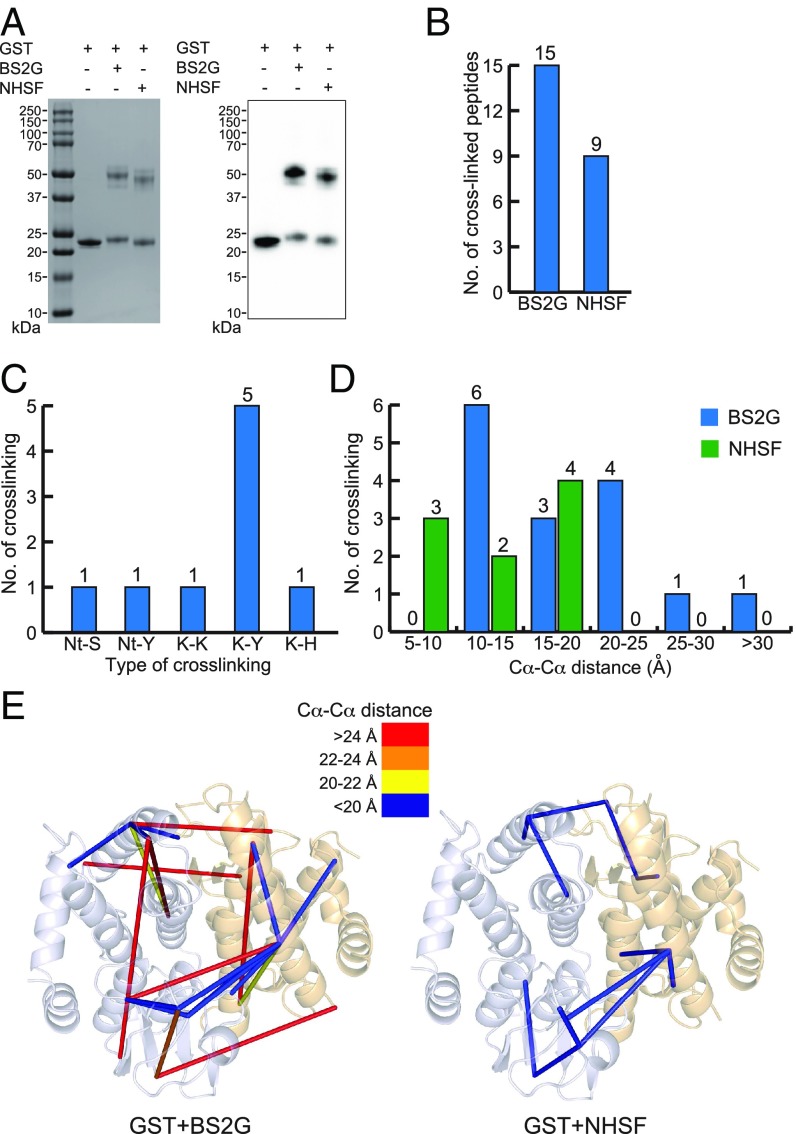
To further validate the cross-linking specificity of NHSF, the homodimeric GST was cross-linked by NHSF or BS2G. As shown by SDS/PAGE and Western blot run under reducing and denatured conditions, GST was successfully cross-linked in the dimeric form by both cross-linkers (Fig. 4A). MS identified 15 cross-linked peptides in the BS2G sample and 9 cross-linked peptides in the NHSF sample (Fig. 4B and SI Appendix, Tables S3 and S4). Again, only N-terminal NH2-Lys or Lys-Lys cross-links were identified for BS2G-treated GST, but multiple different types of cross-linked sites were identified in the NHSF cross-linked GST sample, including 1 N-terminal NH2-Ser, 1 N-terminal NH2-Tyr, 5 Lys-Tyr, and 1 Lys-His (Fig. 4C), further confirming NHSF’s unique ability to cross-link these residues that are infeasible to target with existing chemical cross-linkers. The one Lys-Lys cross-link observed for the NHSF sample was also observed with the BS2G as a cross-linker at a reasonable interresidue distance (Cα-Cα distance = 16.7 Å). All other Cα-Cα distances of cross-linked sites of NHSF cross-linking were highly compatible with the crystal structure of GST. By contrast, 6 out 15 cross-links of BS2G cross-linking were incompatible with the GST structure (Fig. 4 D and E). In summary, the NHSF cross-linker primarily identifies cross-links that are orthogonal to the set identified by BS2G, showing the complementarity of the two approaches. Moreover, the distance information obtained with NHSF is more compatible with the native structures of BSA and GST.
Fig. 4.
CXMS analyses of GST protein cross-linked with NHSF or BS2G. (A) SDS/PAGE (Left) and Western blot (Right) analyses of GST cross-linking. (B) Total number of identified cross-linked peptides from BS2G or NHSF cross-linked GST. (C) Types of cross-linking sites identified from the NHSF cross-linked GST sample, with numbers indicated on top. Nt represents N-terminal NH2. (D) Distribution of Cα-Cα distance of cross-linked residues from GST samples. (E) Identified cross-links mapped onto the crystal structures of GST (Protein Data Bank ID code 1N2A). The Cα-Cα distances of cross-links are color-coded.
NHSF Cross-Links E. coli Whole-Cell Lysate Demonstrates the Applicability to Complex Mixtures and Defines the Chemoselectivity of the Cross-Linking Reaction.
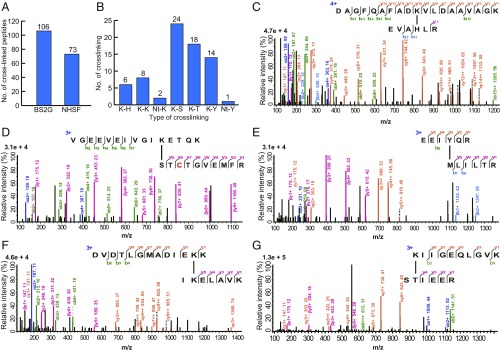
To determine whether NHSF could be used in complex biological samples to generate novel cross-links for MS identification, we applied BS2G and NHSF on E. coli whole-cell lysate. Consistent with the results from model proteins, we obtained large and comparable number of interlinked peptides by BS2G (106) and NHSF (73) (Fig. 5A and SI Appendix, Tables S5 and S6). Six types of cross-links were identified by MS (Fig. 5B). Each type of cross-link was unambiguously supported by high-quality mass spectra (Fig. 5 C–G).
Fig. 5.
CXMS analysis of E. coli cell lysate with NHSF or BS2G. (A) Total number of identified cross-linked peptides. (B) Types and numbers of cross-links identified from the NHSF-treated sample. Nt represents N-terminal NH2. (C–G) Representative tandem mass spectra for each cross-link generated by NHSF.
Remarkably, 86% of the cross-links involved the sidechains of Ser, Thr, Tyr, and His, which are inaccessible using other commonly employed chemical cross-linking reagents. Thus, NHSF and its chemoselectivity and distance-dependent reactivity bodes well for its use as a complement to existing technologies. We attribute the relatively low abundance of Lys-Lys cross-links to the relatively low reactivity of the sulfonyl-fluoride group. It is possible the dearth of Lys-Lys cross-links with NHSF reflects the high intrinsic reactivity of the succinimide ester, which effectively blocks the Lys sidechains toward further reaction, as seen in the above work with the model peptide. Once planted, the remaining sulfonyl-fluoride group is free to react with the remaining sidechains at a slower rate. The discovery of high-frequency cross-linking at Ser and Thr sidechains was rather unexpected, and speaks to the large rate acceleration that can be achieved from proximity-enhanced reactions. Finally, we note that the relative rates of the initial reaction with the succinimide ester are second order (first order in NHSF and first order in deprotonated Lys sidechains), while the second step is first order. Thus, the relative rates of the two reactions can be easily manipulated by changing the pH and the concentration of NHSF, which can be used in future applications to effect different product distributions.
NHSF in Combination with GECX to Identify Enzyme–Substrate Interaction.
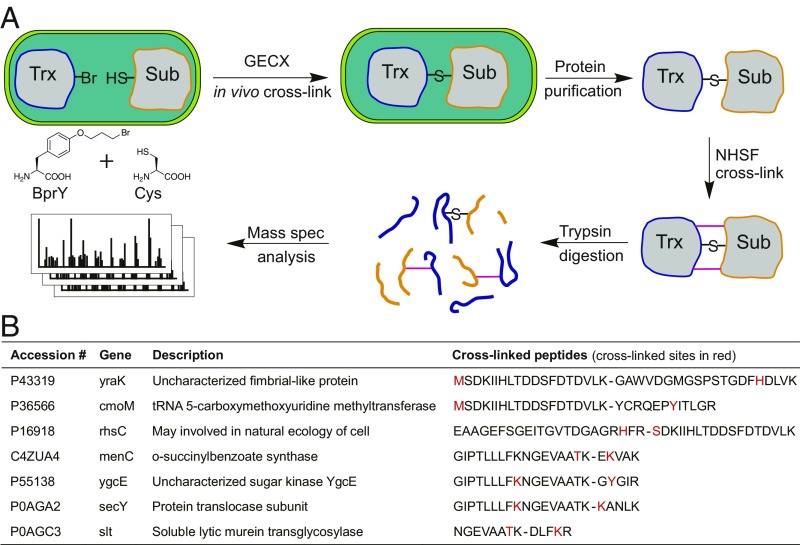
An outstanding challenge for studying PPIs and their networks is to identify weak and transient protein interactions. We previously developed GECX, which uses a bioreactive Uaa to capture such interactions in situ for subsequent MS identification (12). However, as a single cross-linked peptide is generated for each interacting protein in GECX, which may or may not be identifiable by tandem MS, the number of identified proteins with direct cross-linked peptides remains low. Since NHSF targets multiple residues, we reasoned it could be combined with GECX to increase the identifiable cross-linked peptides (Fig. 6A).
Fig. 6.
GECX followed by NHSF cross-linking increases the number of identifiable cross-linked proteins. (A) Scheme showing the combined procedures for identifying Trx-interacting proteins in E. coli cells. (B) Seven additional Trx-interacting proteins identified with NHSF cross-linking. Note His of rhsC cross-linked with the α-amine of the N-terminal Ser of Trx.
Using GECX, we genetically incorporated Uaa O-(3-bromopropyl)-l-tyrosine (BprY, Fig. 6A) into thioredoxin (Trx) to capture Trx-interacting proteins in E. coli cells (12, 15). Whenever a substrate protein of Trx interacts with Trx, BprY reacts with the Cys residue of the substrate protein via proximity-enhanced reactivity, thus covalently cross-linking the substrate protein with Trx in vivo. The cross-linked Trx complex was purified via the His6 tag appended at the C terminus of Trx, further treated with or without NHSF, and then subjected to MS analysis. In the absence of NHSF treatment, seven cross-linked peptides of Trx and interacting proteins were identified in tandem mass spectra (12). With NHSF treatment, an additional seven pairs of peptides of Trx and interacting proteins cross-linked by NHSF were identified (Fig. 6B). These results suggest that GECX followed by NHSF cross-linking indeed increased the number of identifiable cross-linked proteins that interact with thioredoxin.
Discussion
In summary, we developed a plant-and-cast approach to chemical cross-linking that relies on the use of a cross-linker with two groups of graded reactivity toward nucleophilic sidechains. In this approach, the more reactive residue plants the reagent in place, leaving the less reactive group free to cast over the protein surface, ultimately forming cross-links via proximity-enhanced reactivity. This work represents our first attempt to reduce this concept to practice; given the success described herein, we expect that it should be broadly applicable. We report a heterobifunctional chemical cross-linker NHSF capable of targeting multiple amino acid residues including Lys, His, Ser, Thr, and Tyr for CXMS via proximity-enhanced SuFEx reactivity. Existing CXMS chemical cross-linkers target Lys, Cys, Asp, and Glu only; the ability to cross-link His, Ser, Thr, and Tyr has not been feasible before and thus will dramatically expand the diversity of proteins amenable to CXMS with increasing multiplicity of cross-links. In particular, Tyr residues are often enriched at protein–protein interfaces (24). In addition, we demonstrated that NHSF shows no nonspecific cross-linking and provides distance constraints highly compatible with crystal structures, which will afford more accurate structural information to simplify the complexity and to improve the accuracy of structural modeling of large protein assemblies. This feature of NHSF should also be invaluable for the validation of structures obtained with cryoelectron microscopy. Moreover, CXMS is unable to address weak and transient protein interactions. Here we further demonstrated that GECX in combination with NHSF cross-linking markedly increased the identification of weak and transient enzyme–substrate interactions in live cells, which will find broad applications in the identification of unknown protein interactions. Future developments of NHSF will include MS-cleavable modification to advance simplified MS workflows and isotopic labeling to enable quantitative cross-linking MS. Lastly, aryl sulfonyl fluoride has been reported to react with catalytic Ser residues but is generally considered inactive toward unactivated Ser residues under physiological conditions (21, 25). Recently, small molecule probes concentrated in mitochondria were found to label Ser and Thr residues of proteins (26). Our results of NHSF cross-linking provide robust and abundant examples that aryl sulfonyl fluoride is able to react with noncatalytic Ser and Thr via proximity-enhanced reactivity, which will be valuable for designing reactive probes and covalent inhibitors in chemical biology and molecular pharmacology.
Materials and Methods
Chemical Synthesis of NHSF.
The synthesis of NHSF follows a previously published method with slight modifications (23). Briefly, a mixture of 4-(fluorosulfonyl)benzoic acid (0.6 g, 2.8 mmol), N-hydroxysulfosuccinimide (0.44 g, 2.0 mmol), and dicyclohexylcarbodiimide (0.6 g, 2.8 mmol) in 10 mL dry dimethylformamide was stirred under N2. The mixture was allowed to react on ice for 2 h and then overnight at room temperature (RT). After reaction, the dicyclohexylurea precipitate was removed by filtration. The filtrate was then added to 25 mL ethyl acetate and diethyl ether mixture (vol/vol, 3:2) to afford white precipitates. The crude material was further purified by HPLC and lyophilized to give final product as a white solid. NMR: 1H NMR (D2O, 800 MHz): δ 8.493 (d, J = 9.2 Hz, 2H), 8.255 (d, J = 9.2 Hz, 2H), 4.507 (m, 1H), 3.4 (m, 1H), 3.229 (dd, J = 2.9, 19.0 Hz, 1H). 13C NMR (DMSO-d6, 200 MHz): δ 169.1, 165.5, 132.3, 130.0, 56.9, 31.6. High-resolution mass spectrometry: Calculated for C11H7FNO9S2 [M-H]− m/z 379.9541, found 379.9544.
Peptide and Protein Cross-Linking.
Peptide synthesis, protein expression and purification, and E. coli cell lysate preparation are described in SI Appendix. Cross-linking of 7KR peptide: In a 20 μL reaction, 10 μM 7KR peptide (in PBS buffer, pH 7.5) was cross-linked at RT for 1 h with 10 μM BS2G or 10 μM NHSF. Reactions were acidified by formic acid at final concentration of 5% and desalted with Stagetip. BSA cross-linking: In a 20 μL reaction, 10 μM BSA (69 kDa, in PBS buffer, pH 7.5) was cross-linked at RT for 1 h with 1 mM BS2G or 1 mM NHSF, which corresponded to a 1:100 protein:cross-linker molar ratio. BS2G cross-linking reaction was terminated at RT by adding 20 mM ammonium bicarbonate and incubating for 20 min. NHSF cross-linking reaction was terminated at RT by adding 10 mM DTT and incubating for 20 min. GST cross-linking: In a 20 μL reaction, 10 μM GST (23 kDa, in PBS buffer, pH 7.5) was cross-linked at RT for 1 h with 0.5 mM BS2G or 0.5 mM NHSF, which corresponded to a 1:50 protein:cross-linker molar ratio. The cross-linking reactions were similarly terminated as described in BSA cross-linking above. E. coli cell lysate cross-linking: 20 μL lysate (10 mg/mL protein, 50 mM Hepes, pH 8.3, 150 mM NaCl) was incubated with 40 mM BS2G or 40 mM NHSF at RT for 2 h. BS2G cross-linking reaction was terminated at RT by adding 100 mM ammonium bicarbonate and incubating for 20 min. NHSF cross-linking reaction was terminated at RT by adding 100 mM DTT and incubating for 20 min. Thioredoxin sample: The cloning of thioredoxin, in vivo cross-linking via GECX, and purification were carried out as described before (12). The His-tag pull-down sample of thioredoxin (20 μL, 3.45 mg/mL protein, 50 mM Hepes, pH 8.3, 150 mM NaCl) was incubated with 20 mM NHSF at RT for 1 h. NHSF cross-linking reaction was terminated at RT by adding 100 mM DTT and incubating for 20 min.
Protein Digestion.
All protein samples were precipitated by six volumes of acetone at −20 °C for 30 min. Precipitated proteins were dried in air and resuspended in 8 M urea, 100 mM Tris, pH 8.5. After reduction with 2 mM DTT for 20 min and alkylation with 10 mM iodoacetamide for 15 min in the dark, samples were diluted to 2 M urea with 100 mM Tris, pH 8.5, and digested with trypsin (at 50:1 protein:enzyme ratio) at 37 °C for 16 h. Digestion was stopped by adding formic acid to 5% final concentration, and digested peptides were desalted with Stagetip.
Tandem MS Analysis.
MS experiments were performed using an Orbitrap Fusion Lumos instrument (ThermoFisher) coupled with an UltiMate 3000 nano LC. Mobile phase A and B were water and acetonitrile, respectively, with 0.1% formic acid. Protein digests were loaded directly onto a C18 PepMap EASYspray column (part number ES803; ThermoFisher Scientific) at a flow rate of 300 nL/min. E. coli whole-cell lysate digests were separated at 300 nL/min using a linear gradient of 2–35% B over 115 min. All other samples (except thioredoxin pull-down samples) were separated using a linear gradient of 2–40% B over 38 min. Survey scans of peptide precursors were performed from 375 to 1500 m/z at 60,000 FWHM resolution with a 4 × 105 ion count target and a maximum injection time of 50 ms. The instrument was set to run in top-speed mode with 3-s cycles for the survey and the MS/MS scans. After a survey scan, tandem MS was then performed on the most abundant precursors exhibiting a charge state from 2 to 7 (3 to 8 for E. coli samples) of greater than 5 × 104 intensity by isolating them in the quadrupole at 1.6 Da. Higher energy collisional dissociation (HCD) fragmentation was applied with 30% collision energy and resulting fragments detected in the Orbitrap detector at a resolution of 30,000. The maximum injection time limited was 50 ms and dynamic exclusion was set to 60 s with a 10-ppm mass tolerance around the precursor.
Measurement of thioredoxin His-tag pull down samples were performed using an Orbitrap Fusion Lumos instrument (ThermoFisher) coupled with an EasyLC1200 (ThermoFisher). Mobile phase A and B were water and 80% acetonitrile, respectively, with 0.1% formic acid. Protein digests were loaded directly onto a PicoFrit emitter (New Objective) self-packed to a 20-cm C18 column with 1.9-μm Reprosil PUR beads (Dr.Maisch GmbH HPLC) running at a flow rate of 300 nL/min. Digested peptides were separated at 300 nL/min using a linear gradient of 5–37% B over 120 min. Survey scans of peptide precursors were performed from 350 to 1650 m/z at 120,000 FWHM resolution with a 2 × 105 ion count target and a maximum injection time of 100 ms. The instrument was set to run in top-speed mode with 3-s cycles for the survey and the MS/MS scans. After a survey scan, tandem MS was then performed on the most abundant precursors exhibiting a charge state from 2 to 7 of greater than 5 × 104 intensity by isolating them in the quadrupole at 1.2 m/z. HCD fragmentation was applied with 28% collision energy and resulting fragments detected in the Orbitrap detector at a resolution of 30,000. Automatic gain control (AGC) target was set at 8 × 104 and the maximum injection time limited was 50 ms (the AGC target is allowed to be exceeded if there is available parallelizable time). Both MS1 and MS2 data are recorded at Profile mode. The dynamic exclusion was set to 30 s with a 10-ppm mass tolerance around the precursor from reselection.
Data Analysis.
Cross-linked peptides were identified using pLink 2 software. pLink search parameters: precursor mass tolerance 20 parts per million (ppm), fragment mass tolerance 20 ppm, peptide length minimum 6 amino acids and maximum 60 amino acids per chain, peptide mass minimum 600 and maximum 6,000 Da per chain, fixed modification C 57.02146, enzyme trypsin, three missed cleavage sites per chain. The E. coli protein sequences were downloaded from Uniprot. Other protein sequences (such as GST, BSA) were also downloaded from Uniprot. Data of thioredoxin sample was searched with slightly modified parameters set (peptide mass minimum 300 and maximum 2,500 Da per chain, peptide length minimum 3 amino acids and maximum 25 amino acids per chain).
Supplementary Material
Acknowledgments
We acknowledge the support of NIH Grants 5TL1TR001871 (to H.W.), GM122603 and P01AG002132 (to W.F.D.), and R01GM118384 and RF1MH114079 (to L.W.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813574115/-/DCSupplemental.
References
- 1.Young MM, et al. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. Proc Natl Acad Sci USA. 2000;97:5802–5806. doi: 10.1073/pnas.090099097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herzog F, et al. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337:1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y, et al. A strategy for dissecting the architectures of native macromolecular assemblies. Nat Methods. 2015;12:1135–1138. doi: 10.1038/nmeth.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu C, Huang L. Cross-linking mass spectrometry: An emerging technology for interactomics and structural biology. Anal Chem. 2018;90:144–165. doi: 10.1021/acs.analchem.7b04431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sinz A. Cross-linking/mass spectrometry for studying protein structures and protein-protein interactions: Where are we now and where should we go from here? Angew Chem Int Ed Engl. 2018;57:6390–6396. doi: 10.1002/anie.201709559. [DOI] [PubMed] [Google Scholar]
- 6.Novak P, Kruppa GH. Intra-molecular cross-linking of acidic residues for protein structure studies. Eur J Mass Spectrom (Chichester) 2008;14:355–365. doi: 10.1255/ejms.963. [DOI] [PubMed] [Google Scholar]
- 7.Leitner A, et al. Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proc Natl Acad Sci USA. 2014;111:9455–9460. doi: 10.1073/pnas.1320298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutierrez CB, et al. Developing an acidic residue reactive and sulfoxide-containing MS-cleavable homobifunctional cross-linker for probing protein-protein interactions. Anal Chem. 2016;88:8315–8322. doi: 10.1021/acs.analchem.6b02240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bass RB, Butler SL, Chervitz SA, Gloor SL, Falke JJ. Use of site-directed cysteine and disulfide chemistry to probe protein structure and dynamics: Applications to soluble and transmembrane receptors of bacterial chemotaxis. Methods Enzymol. 2007;423:25–51. doi: 10.1016/S0076-6879(07)23002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merkley ED, et al. Distance restraints from crosslinking mass spectrometry: Mining a molecular dynamics simulation database to evaluate lysine-lysine distances. Protein Sci. 2014;23:747–759. doi: 10.1002/pro.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suchanek M, Radzikowska A, Thiele C. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat Methods. 2005;2:261–267. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- 12.Yang B, et al. Spontaneous and specific chemical cross-linking in live cells to capture and identify protein interactions. Nat Commun. 2017;8:2240. doi: 10.1038/s41467-017-02409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiang Z, et al. Adding an unnatural covalent bond to proteins through proximity-enhanced bioreactivity. Nat Methods. 2013;10:885–888. doi: 10.1038/nmeth.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen XH, et al. Genetically encoding an electrophilic amino acid for protein stapling and covalent binding to native receptors. ACS Chem Biol. 2014;9:1956–1961. doi: 10.1021/cb500453a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang Z, et al. Proximity-enabled protein crosslinking through genetically encoding haloalkane unnatural amino acids. Angew Chem Int Ed Engl. 2014;53:2190–2193. doi: 10.1002/anie.201308794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoppmann C, Maslennikov I, Choe S, Wang L. In situ formation of an azo bridge on proteins controllable by visible light. J Am Chem Soc. 2015;137:11218–11221. doi: 10.1021/jacs.5b06234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L. Genetically encoding new bioreactivity. N Biotechnol. 2017;38:16–25. doi: 10.1016/j.nbt.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoppmann C, Wang L. Proximity-enabled bioreactivity to generate covalent peptide inhibitors of p53-Mdm4. Chem Commun (Camb) 2016;52:5140–5143. doi: 10.1039/c6cc01226d. [DOI] [PubMed] [Google Scholar]
- 19.Wang N, et al. Genetically encoding fluorosulfate-L-tyrosine to react with lysine, histidine, and tyrosine via SuFEx in proteins in vivo. J Am Chem Soc. 2018;140:4995–4999. doi: 10.1021/jacs.8b01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsukiji S, Miyagawa M, Takaoka Y, Tamura T, Hamachi I. Ligand-directed tosyl chemistry for protein labeling in vivo. Nat Chem Biol. 2009;5:341–343. doi: 10.1038/nchembio.157. [DOI] [PubMed] [Google Scholar]
- 21.Narayanan A, Jones LH. Sulfonyl fluorides as privileged warheads in chemical biology. Chem Sci (Camb) 2015;6:2650–2659. doi: 10.1039/c5sc00408j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong J, Krasnova L, Finn MG, Sharpless KB. Sulfur(VI) fluoride exchange (SuFEx): Another good reaction for click chemistry. Angew Chem Int Ed Engl. 2014;53:9430–9448. doi: 10.1002/anie.201309399. [DOI] [PubMed] [Google Scholar]
- 23.Woltjer RL, Weclas-Henderson L, Papayannopoulos IA, Staros JV. High-yield covalent attachment of epidermal growth factor to its receptor by kinetically controlled, stepwise affinity cross-linking. Biochemistry. 1992;31:7341–7346. doi: 10.1021/bi00147a019. [DOI] [PubMed] [Google Scholar]
- 24.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee H, et al. A study of the reactivity of S(VI)-F containing warheads with nucleophilic amino-acid side chains under physiological conditions. Org Biomol Chem. 2017;15:9685–9695. doi: 10.1039/c7ob02028g. [DOI] [PubMed] [Google Scholar]
- 26.Yasueda Y, et al. A set of organelle-localizable reactive molecules for mitochondrial chemical proteomics in living cells and brain tissues. J Am Chem Soc. 2016;138:7592–7602. doi: 10.1021/jacs.6b02254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.