Significance
Virtually all organisms require vitamin B1, including bacterioplankton that impact nutrient cycling and productivity in aquatic systems and Earth’s climate. Here, we show that B1 auxotrophy, the need for exogenous B1 or precursors for survival, is widespread among wild bacterioplankton. Genetic analyses of wild bacterioplankton revealed that most are B1 auxotrophs and the abundance of several B1-related genotypes changes temporally at an estuarine monitoring station, suggesting that B1/precursor availability influences bacterioplankton succession. Complementarily, in-field nutrient-amendment experiments and bioassays indicate that B1/precursor bioavailability periodically limits bulk growth of bacterioplankton. Together the presented data highlight the prevalent reliance of bacterioplankton upon exogenous B1/precursors and suggest a hitherto overlooked influence of B1/precursor availability on aquatic biochemical cycling.
Keywords: vitamin B1, thiamin, bacterioplankton, metagenomics, growth limitation
Abstract
Vitamin B1 (B1 herein) is a vital enzyme cofactor required by virtually all cells, including bacterioplankton, which strongly influence aquatic biogeochemistry and productivity and modulate climate on Earth. Intriguingly, bacterioplankton can be de novo B1 synthesizers or B1 auxotrophs, which cannot synthesize B1 de novo and require exogenous B1 or B1 precursors to survive. Recent isolate-based work suggests select abundant bacterioplankton are B1 auxotrophs, but direct evidence of B1 auxotrophy among natural communities is scant. In addition, it is entirely unknown if bulk bacterioplankton growth is ever B1-limited. We show by surveying for B1-related genes in estuarine, marine, and freshwater metagenomes and metagenome-assembled genomes (MAGs) that most naturally occurring bacterioplankton are B1 auxotrophs. Pyrimidine B1-auxotrophic bacterioplankton numerically dominated metagenomes, but multiple other B1-auxotrophic types and distinct uptake and B1-salvaging strategies were also identified, including dual (pyrimidine and thiazole) and intact B1 auxotrophs that have received little prior consideration. Time-series metagenomes from the Baltic Sea revealed pronounced shifts in the prevalence of multiple B1-auxotrophic types and in the B1-uptake and B1-salvaging strategies over time. Complementarily, we documented B1/precursor limitation of bacterioplankton production in three of five nutrient-amendment experiments at the same time-series station, specifically when intact B1 concentrations were ≤3.7 pM, based on bioassays with a genetically engineered Vibrio anguillarum B1-auxotrophic strain. Collectively, the data presented highlight the prevalent reliance of bacterioplankton on exogenous B1/precursors and on the bioavailability of the micronutrients as an overlooked factor that could influence bacterioplankton growth and succession and thereby the cycling of nutrients and energy in aquatic systems.
Vitamin B1 (thiamin, called “B1” herein) is a cofactor for enzymes involved in amino acid synthesis and central carbon metabolism; as a result B1 is required by virtually all cells (1). Canonical bacterial B1 biosynthesis is well studied and involves several core enzymes that generate pyrimidine and thiazole precursors and ultimately fuse them to yield the B1 molecule (1). In addition, several known transport systems are considered key to importation of exogenous B1 and/or B1 precursors by bacteria (1–3).
B1 is intriguing with respect to bacterial ecology, as B1-prototrophic (de novo synthesizers of the vitamin) and B1-auxotrophic populations (those incapable of de novo B1 biosynthesis) occur in nature, with the latter lacking one or more core B1-biosynthesis genes and requiring exogenous B1 or B1 precursors to survive (1, 4, 5). Surveys of isolate growth or genomes indicate that most (∼70–80%) bacterial isolates from estuarine/marine waters are B1 prototrophs (4–7). In notable contrast, isolates representing abundant marine and freshwater bacterioplankton lineages, such as the SAR11 and SAR86 clades and Actinobacteria, are B1 auxotrophs (8–10). Additionally, an analysis of metagenomes from a Sargasso Sea station noted a deficiency in the core B1-biosynthesis gene thiC encoding a pyrimidine synthase (8), which suggests a prevalence of B1 auxotrophy. Together these findings suggest B1 auxotrophy may be more prevalent among wild bacterioplankton communities than expected based on isolate surveys; however, direct evidence of this is limited and lacks temporal resolution that would help address the predominance of B1 auxotrophy over time.
Exogenous B1 and B1 precursors are bioavailable in aquatic systems (11–14) and presumably sustain B1-auxotrophic bacterioplankton, including different B1-auxotrophic types which are discussed to a degree in recent work (7, 8). Chemical techniques recently confirmed that B1, phosphorylated B1, and 4-amino-5-aminomethyl-2-methylprimidine (HMP) molecules occur in seawater (7, 15, 16). Nevertheless, it is unknown if exogenous B1 and/or precursors are always bioavailable at sufficient concentrations to sustain maximum growth of bacterioplankton in nature. Some experimental evidence suggests that increased B1 bioavailability can stimulate phytoplankton biomass or primary productivity in coastal marine waters (17–19). Also, recent work reported significant correlations between B1 concentrations and phytoplankton biomass (chlorophyll a) in temperate estuarine waters (20). Analogous tests of B1/precursor limitation of bacterioplankton growth have not been performed. To our knowledge, only a single marine experiment reports B1 and B12 colimitation of bacterial production rather than B1 limitation alone (21). B1 deficiency is of current interest as a factor influencing the productivity of aquatic ecosystems. For example, B1 is of concern in the Baltic Sea due to recent links between declines in bird and fish stocks and B1 dietary deficiency (22, 23). The Baltic Sea is a representative of large, relatively long-residence-time, temperate estuaries that are found worldwide and are of broad interest and concern since they contribute significantly to regional productivity, elemental cycling, and economics (24–27). Although B1 deficiency can be a critically important factor for higher organisms, microbial B1 deficiency in the Baltic Sea or parts of the world ocean is an unexplored issue.
Here we surveyed metagenomic datasets from fresh, estuarine, and marine waters to test the hypothesis that B1 auxotrophy is commonplace among wild bacterioplankton communities and, furthermore, that the prevalence of different auxotrophic types as well as transport and B1-salvaging strategies change over time. Motivated by metagenomic survey results from a Baltic Sea time-series monitoring station, we performed nutrient-enrichment experiments testing a second hypothesis that bacterial production is periodically limited by B1 and/or precursor bioavailability at the same station. Primarily the results reveal an extensive reliance of diverse and numerically dominant wild bacterioplankton upon exogenous B1 and B1 precursors while suggesting that B1/precursor bioavailability influences bacterioplankton succession and productivity, along with other bottom-up and top-down controls.
Results
Most Wild Bacterioplankton Are B1-Auxotrophic.
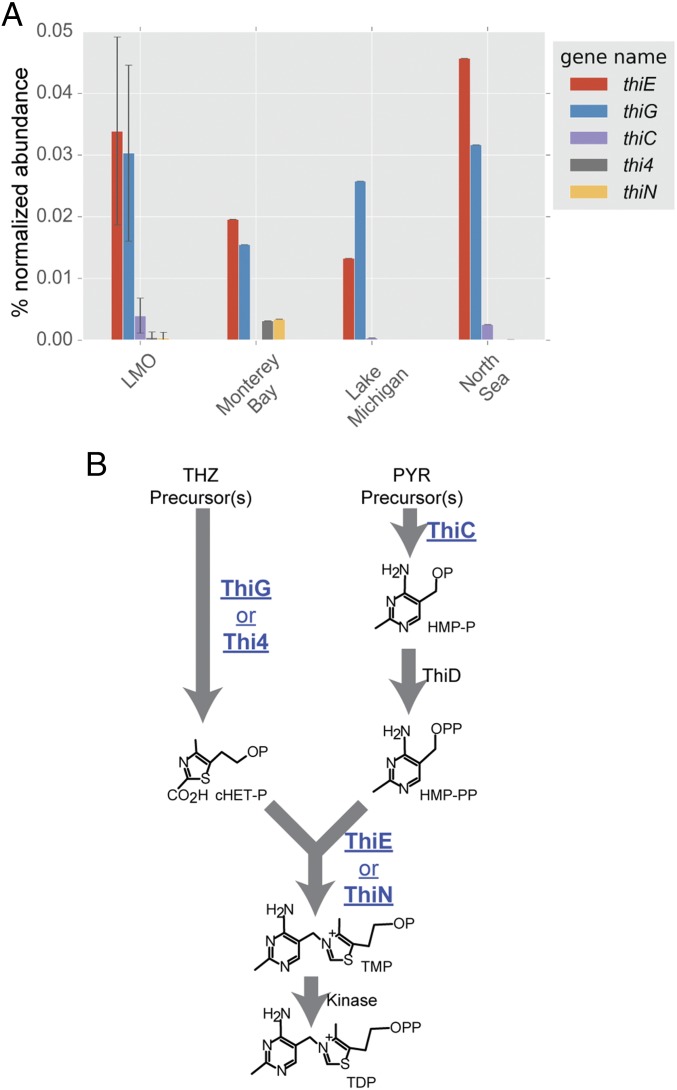
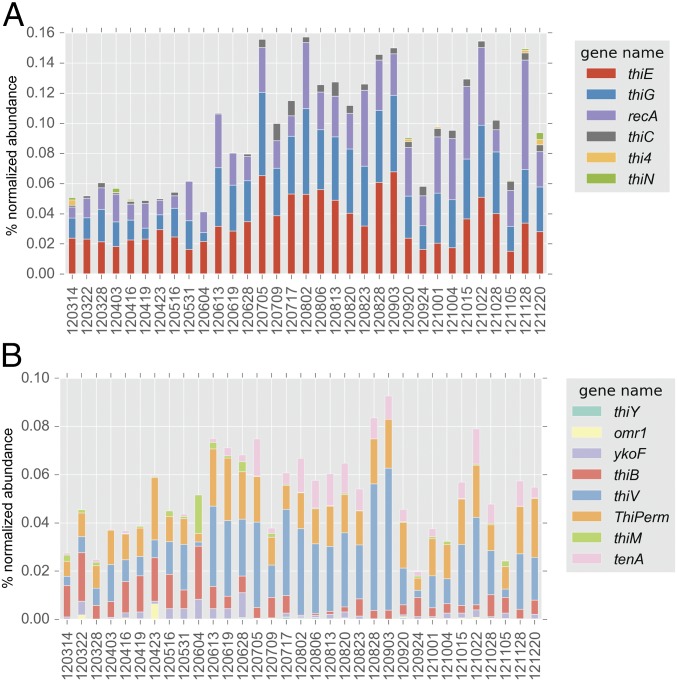
Metagenomes from freshwater, estuarine, and marine sites, specifically eastern-central Lake Michigan, the Baltic Sea proper at the Linnaeus Microbial Observatory (LMO) (28), the Kabeltonne station offshore of Helgoland in the North Sea (northeastern Atlantic Ocean), and mooring M0 in Monterey Bay, CA (eastern Pacific Ocean), were surveyed for essential core B1 biosynthesis genes (thiC, thiG, and thiE) in bacterioplankton, and all were deficient in thiC relative to the other two genes (Fig. 1A and SI Appendix, Table S1). The thiC, thiG (thi4 in Archaea), and thiE (thiN in Archaea) genes encode phosphomethylpyrimidine synthase, thiazole synthase, and thiamine monophosphate synthase, respectively, in canonical bacterial B1 biosynthesis (Fig. 1B) (1). The overt deficiency in thiC across habitats suggests that most wild bacterioplankton are pyrimidine B1 auxotrophs (Fig. 1A) and therefore obligately or in part rely on exogenous pyrimidine precursors, e.g., HMP (8), to survive.
Fig. 1.
(A) Abundance (percentage of normalized abundance; see Methods) of multiple B1 biosynthesis genes central to B1 biosynthesis in LMO metagenomes and in marine and freshwater metagenomes. Notably, all metagenomes exhibit a deficiency in thiC genes. For LMO data, columns represent mean values and error bars represent the SD (n = 37). (B) A metabolic map of core B1-biosynthesis reactions facilitated by proteins (and respective encoding genes) surveyed in this study (shown in blue bold type and underlined). cHET-P, 2-(2-carboxy-4-methylthiazol-5-yl)ethyl phosphate; HMP-P, 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate; HMP-PP, 4-amino-5-hydroxymethyl-2-methylpyrimidine diphosphate; P (in chemical compounds), PO4; PYR, pyrimidine; TDP, thiamin diphosphate; THZ, thiazole; TMP, thiamin monophosphate.
For comparison with the LMO (Baltic Sea) metagenomic survey results, 25 isolates readily cultivatable on nutrient-rich agar from Baltic Sea surface water and representing multiple bacterial classes (29–31) were tested for B1 auxotrophy by traditional growth surveys. Only eight (32%) of the surveyed isolates were B1 auxotrophs (SI Appendix, Table S2), notably less than the proportion derived from LMO metagenomes (Fig. 1A).
Multiple B1-Related Auxotrophic Types, Transporters, and Salvage Strategies Among Natural Bacterioplankton.
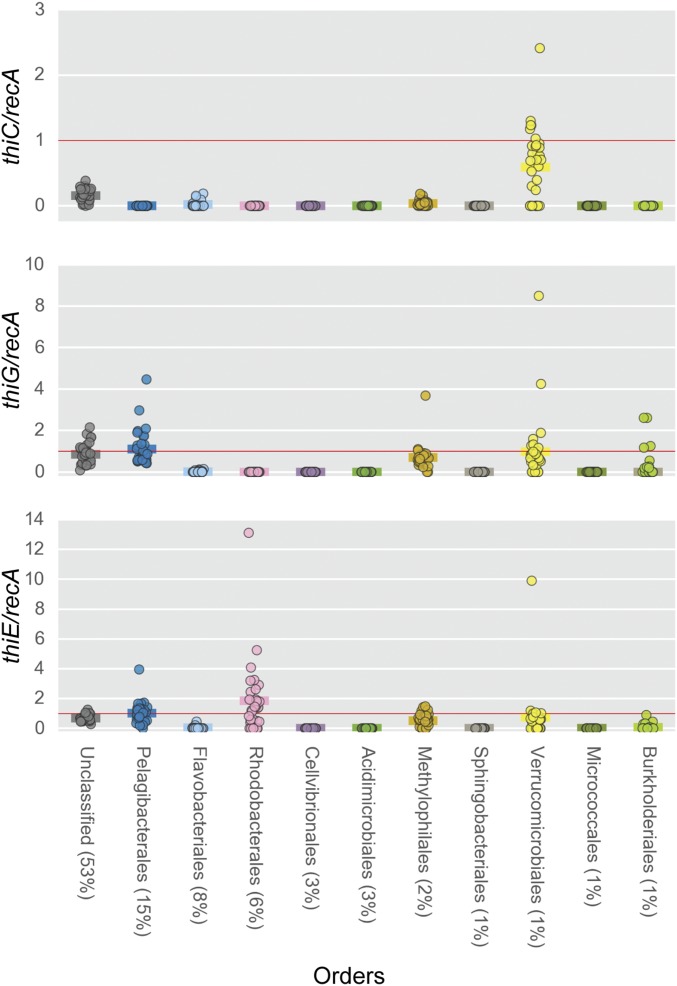
Surveying all the LMO metagenomes at a taxa-specific level (Order) revealed more evidence of multiple types of B1 auxotrophs, e.g., pyrimidine and thiazole (dual) auxotrophs, intact B1 auxotrophs, and pyrimidine B1 auxotrophs, existing among relatively abundant wild bacterioplankton (Fig. 2). Abundances of core B1-biosynthesis genes normalized to recA gene abundance, i.e., thiC/recA, thiG/recA, and/or thiE/recA ratios, were often below 1 across multiple numerically dominant bacterial orders in LMO metagenomes, which indicates a predominance of B1-auxotrophic populations across taxa (Fig. 2). The recA gene, encoding Recombinase A, occurs as a single-copy gene in most prokaryotic genomes; thus its abundance is frequently used as a proxy for genome numbers (32, 33).
Fig. 2.
recA-normalized abundances of B1 biosynthesis genes (thiC, thiG, thiE) for the most abundant bacterioplankton taxa across LMO metagenomes (based on the mean percentage of total prokaryotic sequences). Genes classifiable at the class level but not at the order level are in the unclassified category. Solid horizontal lines belonging to each order represent average abundance values.
Pyrimidine B1 auxotrophs lack only thiC of the core B1-biosynthesis genes (thiC, thiG, and thiE; as described above) and thus are represented by thiC/recA and thiG/recA values of 0 and ≥1, respectively. Unclassified orders, Pelagibacterales, Methylophilales, and Burkholderiales were largely composed of pyrimidine B1 auxotrophs, as their respective thiC/recA and thiG/recA values were ∼0 and ∼1, respectively (Fig. 2).
Thiazole and pyrimidine (dual) B1 auxotrophic populations lack pyrimidine synthase (thiC) and thiazole synthase (thiG; thi4 in Archaea) but possess thiamin monophosphate synthase (thiE; thiN in Archaea), resulting in thiC/recA and thiG/recA values of 0 and thiE/recA values ≥1. A strong signature of dual B1 auxotrophy was identified within the Rhodobacterales [encompassing the important Roseobacter clade (34, 35)], underscoring that many LMO rhodobacters are reliant on both exogenous pyrimidine and thiazole precursors to help meet their B1 demands. The dual B1-auxotrophy type has garnered little consideration in recent studies (7, 36); however, our surveying of publically available complete bacterioplankton genomes [obtained from the Integrated Microbial Genomes Joint Genome Institute (IMG JGI), https://img.jgi.doe.gov/] (Methods) identified a considerable number of isolates (41 of 330) as dual B1 auxotrophs (Dataset S1).
Intact B1 auxotrophs lack thiamin monophosphate synthase (ThiE or ThiN) and accordingly exhibit thiE/recA (or thiN/recA) values of 0. Many abundant LMO bacterial orders exhibited thiE/recA values of approximately 0 (as well as >1) (Fig. 2), which stands in stark contrast with the rarity of intact B1 auxotrophy among surveyed bacterioplankton isolate genomes (14 of 330; 4%) (SI Appendix, Table S3).
Also noted were thiC/recA, thiG/recA, or thiE/recA values >1 within select LMO bacterial orders, e.g., Rhodobacterales thiE/recA ∼13 (Fig. 2). Numerous bacterioplankton isolate genomes (109 of 330) possess more than one copy of thiE and/or thiN (Dataset S2), including at least two isoforms of the ThiE enzyme (SI Appendix, Fig. S1). A smaller set of isolate genomes (4 of 330) contain more than one copy of thiG or thi4 (SI Appendix, Table S4). Thereby, multiple copies of thiG and thiE per genome can explain thiE/recA and thiG/recA values >1 among LMO orders. Notably, the ecological advantage of bacteria (in general) harboring multiple copies of B1 biosynthesis genes is entirely unknown.
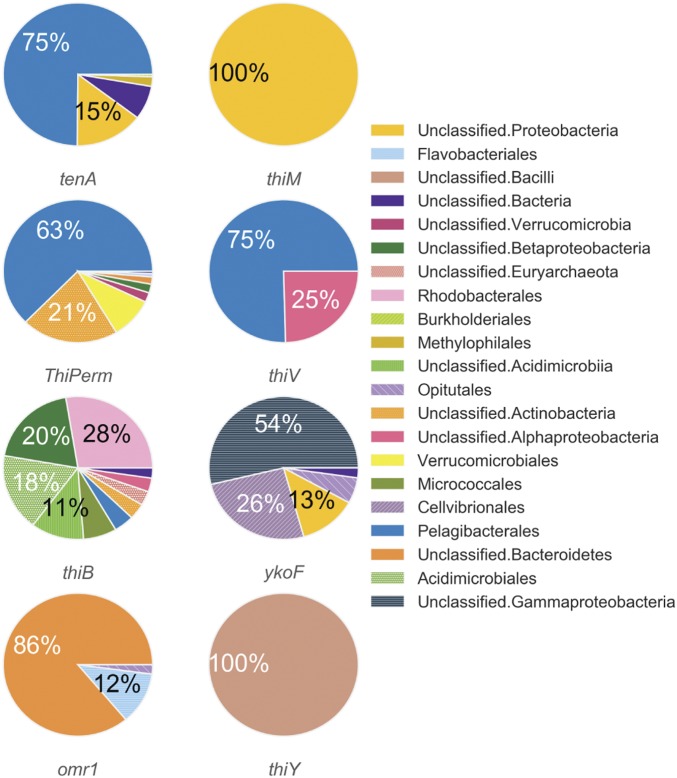
Genes attributed to salvage of B1 from pyrimidine and thiazole B1-degradation products, tenA and thiM (1, 37, 38), as well as several B1/B1 precursor transport systems (SI Appendix, Table S5), were also identified in LMO metagenomes. These genes generally affiliated with distinct sets of taxa; for example, at least nine different bacterial orders possessed thiB [a crucial substrate-binding component of an ATP-dependent B1 transport system (39)], while thiV [a putative HMP sodium symporter (8)] sequences affiliated only with Pelagibacterales and unclassified Alphaproteobacteria (Fig. 3). Further, four other B1/precursor transport genes (ThiPerm, ykoF, omr1, and thiY) were identified in LMO metagenomes (Fig. 3). Considered altogether, a noteworthy variety of B1-related genotypes persists among LMO bacterioplankton.
Fig. 3.
Taxonomic affiliation of protein-encoding genes linked to B1-related transport or salvage identified from all LMO metagenomes (n = 37).
B1 Genotypes and Auxotrophy in Baltic Sea and World Ocean Metagenome-Assembled Genomes.
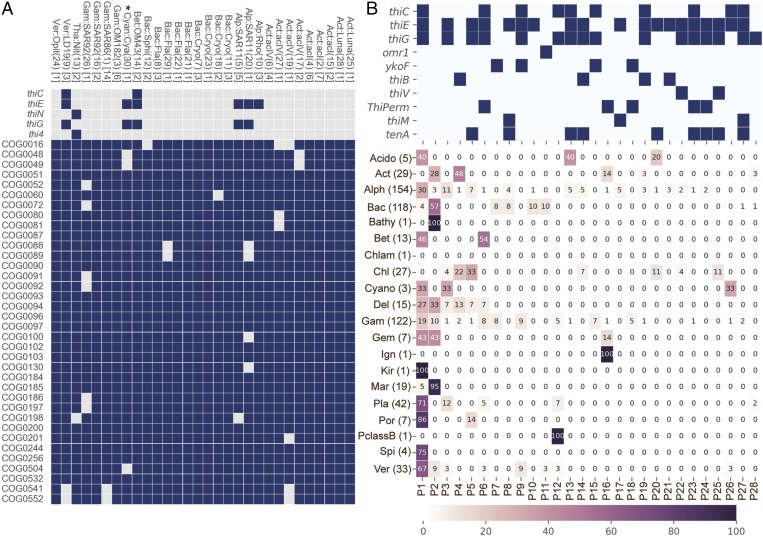
To investigate the prevalence of B1-related genotypes among diverse and abundant bacterioplankton lineages, we analyzed metagenome-assembled genome (MAG) clusters from the Baltic Sea and the Tara Oceans global sampling expedition. Surveying LMO MAG clusters generated from weekly samples (March to December 2012) from the LMO Baltic Sea site (28) for B1-related genes (as above) (SI Appendix, Table S1) notably showed B1 auxotrophy was prevalent. A total of 27 of 30 MAGs were putative B1 auxotrophs based on their lack of thiC, thiG (thi4 in Archaea), or thiE (thiN in Archaea) (Fig. 4A). Two MAGs affiliated with the ubiquitous SAR11 bacterioplankton clade lacked only thiC, which is indicative of pyrimidine B1 auxotrophy and is congruent with recent findings that several marine SAR11-affiliated isolates are pyrimidine B1 auxotrophs (8). Strikingly, however, most LMO MAG clusters (23 of 30, ∼77%) lacked thiE (or thiN) (Fig. 4A), making them intact B1 auxotrophs without thiamin monophosphate synthase and completely reliant on exogenous intact B1 or phosphorylated B1 to survive (4, 6). Adding to the genotypic variability, the intact B1 auxotrophs possessed one or more of three different known B1/B1 precursor transport systems with different modes of action and substrate specificity (SI Appendix, Fig. S2 and Table S5). Additionally, the rhodobacter-affiliated LMO MAG cluster Alph:Rho (Fig. 4A) (10) lacked thiC and thiG but possessed thiE, which reflects dual B1 auxotrophy and an ability to jointly use exogenous pyrimidine and thiazole precursors to meet cellular B1 demands (6).
Fig. 4.
(A) The presence (dark blue boxes) or absence (light grey boxes) of B1 biosynthesis genes (with the “Thi” prefix) and a set of genes expected in all bacteria (with the “COG” prefix) (105) in 30 LMO MAG clusters. The number of MAGs within a MAG cluster is shown in brackets at the right of its taxonomic ID. The MAG cluster (BACL) ID number is given within parenthesis and the taxonomic affiliations are as described earlier (28). acI, Actinobacteria clade acI; acIV, Actinobacteria clade acIV; Act, Actinobacteria; Alp, Alphaproteobacteria; Bac, Bacteroidetes; Bet, Betaproteobacteria; Cryo, Cryomorphaceae; Cya, Cyanobium; Cyan, Cyanobacteria; Fla, Flavobacteriaceae; Gam, Gammaproteobacteria; LD19, Verrucomicrobia subdivision LD19; Luna, Actinobacteria clade Luna; Nit, Nitrosopumilus; Opit, Opitutacae; Rho, Rhodobacteraceae; SAR11, SAR11 clade; SAR86, SAR86 clade; SAR92, SAR92 clade; Sphi, Sphingobacteriales; Tha, Thaumarchaeota; Ver, Verrucomicrobia. The Cyanobium MAG cluster (Cya) (30) is marked with an asterisk indicating it was not counted as a B1-auxotrophic cluster because it consists of a single MAG with 79.2% estimated completeness, and B1 auxotrophy is uncommon in cyanobacteria (7). (B) Twenty-eight different B1-related genotype profiles (unique repertoires of B1-related genes denoted P1–28 at the bottom of the figure) occurred in at least 3 of 603 near-complete (≥90%) Tara Oceans MAGs. The top grid indicates the presence (dark boxes) or absence (white boxes) of genes within each unique profile. All B1-related genes were surveyed using protein models (SI Appendix, Table S1), but only those detected are shown. In the lower grid, the taxa-specific prevalence (%) of each B1 profile is represented by color and by number within the grid. Phylum/class abbreviations are as in A or as follows: Bathy, Bathyarchaeota; Chl, Chloroflexi; Chlam, Chlamydiae; Cyano, Cyanobacteria; Del, Deltaproteobacteria; Gem, Gemmatimonadetes; Ign, Ignavibacteriae; Kir, Kiritimatiellaeota; Mar, Marinimicrobia; PclassB, Proteobacteria novel Class B; Pla, Planctomycetes; Por, Poribacteria; Spi, Spirochaetes. The number of MAGs within a MAG cluster is shown in parentheses at the right of its taxonomic ID.
Questioning whether diverse B1 genotypes exist among abundant bacterioplankton beyond the Baltic Sea, we surveyed for B1-related genes in 603 (≥90% complete) MAGs recently reconstructed from the Tara Oceans global sampling expedition dataset (40). The survey uncovered a high frequency of B1 auxotrophy, with ∼62% lacking at least one core B1-biosythesis gene (thiC, thiG, or thiE), similar to Baltic Sea MAGs, but also a notable variety of B1-related genotypic profiles (28 total) reflecting different B1 auxotrophic and prototrophic types and different transport and salvage capacities (Fig. 4B). Some pronounced lineage-specific differences were evident, with Actinobacteria, Bacteroidetes, and Chloroflexi MAGs being exclusively B1 auxotrophic (e.g., profiles 2, 4, and 16), Betaproteobacteria and Spirochaetes being exclusively B1 prototrophic, and Alphaproteobacteria composed of a variety of auxotrophic and prototrophic profiles (Fig. 4B). Thus, a sizable assortment of B1-related genotypes and hence different B1-related lifestyles (SI Appendix, Table S6) exist among abundant wild bacterioplankton in the global ocean, not only in the Baltic Sea (Fig. 4).
Temporal Dynamics in the Abundance of B1-Related Genes.
Pronounced temporal shifts in the abundance of B1-related core, transport, and salvage genes occurred across LMO time-series metagenomes derived from weekly (March to December 2012) samplings at the Baltic Sea station (Fig. 5). Sequence library- and size-normalized abundances of core B1-biosynthesis genes ranged from <0.001 to >0.06% (Fig. 5). Specifically, thiC, thiG, and thiE gene abundances peaked in midsummer (July to September) and again in the fall (October) with notably diminished abundances throughout the spring and into the early summer (March to June) (Fig. 5A). The ratio of thiC (pyrimidine synthesis) to thiG+thi4 (thiazole synthesis) genes remained well below 1 (0.06–0.28), indicative of persistent thiC deficiency and a predominance of pyrimidine B1 auxotrophs (SI Appendix, Fig. S3). Peak abundances of pyrimidine uptake and salvage genes thiV and tenA occurred in midsummer and fall during peaks in thiG and thiE abundances (Fig. 5B). Notably, the abundances of these genes showed a significant positive correlation with temperature (and cyanobacterial biomass) (e.g., for thiE and temperature, Spearman rho = 0.63, P = 8 × 10−4), but excluding thiC (Spearman rho = 0.38, P = 0.19) (SI Appendix, Fig. S5). In contrast, the highest abundances of thiB and ykoF genes (B1 transport) occurred in spring and early summer (March–June) (Fig. 5B) and exhibited either a significant negative or no correlation with temperature (SI Appendix, Fig. S4). These patterns point out seasonal changes in the reliance of numerically dominant bacterioplankton upon exogenous B1/precursors and especially the elevated reliance upon exogenous pyrimidine precursor(s) in summer and fall.
Fig. 5.
Abundances (% normalized abundance) of B1 biosynthesis genes (A) and of B1-related salvaging or transporter genes (B) fluctuate across LMO metagenomes. As mentioned in the text, thi4 and thiN are archaeal-associated functional analogs of thiG and thiE in bacteria. Sampling dates (in two digit year, month, and day format) linked to respective metagenomes are presented along the x axes.
Short-term (weekly) changes in the abundance of B1-related genes were also identified, highlighting periods of rapid change in reliance upon exogenous B1/precursors within the ecosystem. For example, from June 28 to July 5 thiE and thiG abundances doubled and tripled, respectively, with minimal change in thiC abundance (Fig. 5A), a sign of an increased need for exogenous pyrimidine precursor(s). Further, from May 31 to June 4 Proteobacteria-affiliated thiM gene abundance, linked to the use of exogenous thiazole B1 precursor(s) (13, 38, 41), increased by ∼10-fold (SI Appendix, Fig. S5). Diatom abundances notably increased by ∼10-fold at the same time, but the two variables did not covary significantly across the dataset (SI Appendix, Figs. S4 and S5), suggesting periodic increases in diatoms may increase the bioavailability of exogenous thiazole B1 precursor(s), as indicated in recent culture-based experiments (13). Cumulatively these results reveal previously unseen seasonal and weekly changes in the reliance of wild bacterioplankton on exogenous B1/precursors.
Experimental Evidence of B1 and B1 Precursor Limitation of Bacterial Production.
Given the prevalence of B1 auxotrophic bacterioplankton and temporal shifts in the prevalence of B1-related genotypes (B1-auxotrophic types) and also uptake and salvage strategies, which are indicative of temporal shifts in B1 and precursor bioavailability (Figs. 1, 4, and 5), we hypothesized that B1 or B1 precursor availability periodically limits bacterial production in LMO surface waters. Indeed, short-term (24–48 h) enrichment experiments with surface seawater revealed that the addition of B1 or B1 precursors (1 nM final concentration) stimulated bacterial production in three of five experiments (Table 1). In the experiments in which stimulation was observed, mean bacterial production rates were significantly stimulated, by 1.2- to 2.6-fold relative to controls (two-tailed Welch-corrected t tests) (Table 1).
Table 1.
Results of nutrient-amendment experiments at the LMO site and concentrations of bioavailable intact B1 in initially incubated LMO water based on V. anguillarum PF430-3 ∆thiE bioassays
| Observation | May 12, 2015 | May 26, 2015 | June 24, 2015 | August 4, 2015 | June 1, 2016 |
| Limitation | Yes (24 h) | No* | Yes (48 h) | Yes (48 h) | No |
| Stimulating amendment | All | — | All | B1 only | — |
| Intact B1, pM | 2.2 ± 0.76 | 4.6 ± 1.6 | 2.4 ± 0.69 | 3.7 ± 0.14 | 11 ± 0.88 |
Data from bacterial production limitation tests and V. anguillarum PF430-3 ∆thiE bioassays are provided in SI Appendix, Tables S6 and S10, respectively. Bold text highlights experiments in which bacterial production was stimulated due to amendment of B1 or precursor (HMP, HET, HMP + HET).
HET amendment elevated bacterial production rates compared with controls, but the mean rate for the amendment was not considered significantly elevated as its respective P value equaled the cutoff value of 0.05 (P = 0.046).
We developed a bioassay method using a genetically engineered Vibrio anguillarum strain PF430-3 deletion mutant lacking thiE (∆thiE) to determine bioavailable intact B1 in seawater at the start of enrichment experiments and put responses in nutrient-amendment experiments into context with intact B1 bioavailability. The PF430-3 ∆thiE mutant thus lacks a functional ThiE protein and is an intact B1 auxotroph that grows only on supplied B1 or phosphorylated B1 (SI Appendix, Fig. S6 and Tables S7 and S8). Further, PF430-3 ∆thiE exhibited a half-saturation growth constant (Km) of only 0.5 and 1.3 pM for B1 and thiamin diphosphate (TDP) in batch cultures (SI Appendix, Table S8), benefitting intact B1 detection in bioassays but also direct evidence that wild-type PF430-3, a B1-prototrophic (de novo B1-synthesizing) bacterium, is capable of high-affinity uptake of intact B1. Based on PF430-3 ∆thiE bioassays, intact B1 concentrations in LMO water used in enrichment experiments ranged from 2.2–11 pM (Table 1). Notably, stimulation of bacterial production by the addition of B1 or B1 precursor in enrichment experiments occurred when intact B1 concentrations were ≤3.7 pM (Table 1).
Discussion
Results from our bioinformatic and experimental efforts build upon knowledge of B1-related bacterioplankton ecology deduced from isolate-based studies (8, 10, 36, 42) and limited Sargasso Sea metagenomic data (8) in several important ways. First, most bacterioplankton are “cheaters” for B1 and/or precursors (Figs. 1A and 4), relying on exogenous pools to survive, including slow-growing oligotrophs (e.g., Pelagibacterales spp.) (43, 44) and fast-responding copiotrophs (e.g., several Gammaproteobacteria groups) (45, 46). It has long been known that select plankton (bacteria and protists) can use exogenous B1 and/or precursors to meet their B1 demands (6, 47); nonetheless, the reliance of wild bacterioplankton on these extracellular micronutrients remained unknown. Our results show B1-auxotrophic bacterioplankton numerically dominate natural waters. Markedly among them are multiple genotypes, including several largely overlooked in recent studies [specifically, dual (pyrimidine and thiazole) B1 auxotrophs and intact B1 auxotrophs, the latter receiving some recent consideration (8, 36)] that now are clearly common within certain major bacterial orders (e.g., Rhodobacterales) in the wild (Figs. 2 and 4). Additionally, there are populations with more than one B1 and/or precursor transporter (Fig. 4B, SI Appendix, Fig. S2, and Dataset S3), which presumably have an advantage over populations with a single B1 or precursor transporter, and B1-prototrophic populations with transporters capable of high-affinity B1 uptake (Fig. 4B, SI Appendix, Table S8, and Dataset S4), an ability thus far best described in model enteric B1-prototrophic bacteria, e.g., Escherichia coli (48). Combined, the analyses of metagenomes from freshwater, estuarine (including the Baltic Sea), and global ocean stations presented here directly show the ubiquity of B1 auxotrophy and reliance on exogenous B1/precursors in general. Thus, it is environmentally relevant to consider the costs/benefits of multiple B1-related lifestyles, particularly if we are to fully understand the niches of natural bacterioplankton and how their lifestyles (SI Appendix, Table S6) impact the biochemistry and productivity of aquatic systems.
The widespread cheating on the biochemical costs of B1 (rather than making it de novo) by bacterioplankton is further notable because most often they are viewed as vitamin producers/providers, particularly to algae (49–57). Arguably, the impact of bacterioplankton vitamin consumption or modification has received less attention (6, 8, 13, 36). Additionally, the metabolic capabilities of vitamin-consuming bacterioplankton versus purely de novo B1 synthesizers are important to consider in the future, especially in the context of climate change. For example, at the LMO site temperature and filamentous cyanobacterial biomass positively correlated with the prevalence of pyrimidine B1 auxotrophs, which is indicated by temperature being negatively correlated with thiC abundance and positively correlated with thiG and thiE and also positively correlated with tenA [linked to salvage of B1 from pyrimidines resulting from B1 degradation (37)] and thiV [linked putatively to HMP uptake (8)] (SI Appendix, Fig. S4). Increased temperature (and also light) promotes B1 degradation in seawater (58, 59). Notably, large filamentous cyanobacterial blooms occur in the Baltic Sea, especially in summer months (60, 61), and are rich in B1 (62); furthermore, cyanobacteria are B vitamin (including B1) prototrophs, with very few exceptions [e.g., Synechococcus sp. PCC7002 (63) and symbiotic Candidatus Atelocyanobacterium thalassa sp. (7, 64)], and are known releasers of vitamins (65). Thus, filamentous cyanobacteria are considered significant sources of B1 and precursors in the Baltic Sea microbiome. Multiple factors—temperature, light, and phytoplankton community—are poised to synergistically elevate the importance of exogenous pyrimidine B1 precursors under warmer, more stratified, and more light-exposed conditions, e.g., summertime at the LMO site (Fig. 5 and SI Appendix, Fig. S4). Given the predicted increase in Baltic Sea surface temperatures (66) and the increased dominance of cyanobacteria under warmer conditions (67, 68), pyrimidine precursor availability is expected to rise and to promote pyrimidine B1 auxotrophs and exogenous pyrimidine precursor consumers (possibly including B1 prototrophs). The ramifications of such community shifting are unknown but may impact community growth and metabolism, especially organic carbon processing, as populations more reliant on exogenous pyrimidines are poised to save on the energetic/elemental costs of de novo B1 biosynthesis and to repurpose those resources to support other metabolic activities, putatively increased respiration or growth.
Results of several recent studies suggest microbial interactions and metabolite exchanges (e.g., vitamin exchange) play a significant role in the diversification and interconnectedness among plankton in nature, especially bacterioplankton (56, 69–73). The pronounced temporal shifts observed suggest that the bioavailability of intact B1 and/or B1 precursors changes notably over time and plays a role in the succession of bacterioplankton communities on week to seasonal time scales (Fig. 5 and SI Appendix, Figs. S3 and S4). While details regarding the circulation and exchange of these micronutrients remain elusive, it is anticipated that both intimate and at-a-distance microbial interactions significantly influence the bioavailability of B1 and its precursors, alongside abiotic and enzymatic degradation (58, 59, 74).
Here we present experimental evidence that B1/precursors can periodically limit bacterial production (Table 1). This finding indicates that exogenous B1/precursor availability requires future consideration as a bottom-up control of bacterioplankton growth, along with organic carbon, phosphorous, nitrogen, and/or iron, which are commonly viewed as key nutrients limiting bulk bacterioplankton growth (75–79).
The simplest explanation for elevated bacterial production following B1/precursor amendment in our experiments is that the supply of B1/precursor stimulated one or more metabolic processes mediated by B1-requiring enzymes and in turn promoted bulk bacterioplankton growth. Since many B1-requiring enzymes occur in bacterioplankton, some with key roles in central (amino acid and carbon) metabolism [e.g., decarboxylation of alpha-ketoglutarate generating an energy-rich reductant in the tricarboxylic citric acid (TCA) cycle (7)] but also in more specialized biochemical pathways [e.g., sulfoacetaldehyde acetyltransferase (7)], there are many points in cellular metabolism at which limited B1 availability could stall growth.
We noted with interest that intact B1 concentrations at the start of nutrient-enrichment experiments were not abnormally low (∼2.2–11 pM) (Table 1) relative to B1 concentrations previously reported from diverse coastal waters using either chemical or bioassay measures (19, 80–82). We calculated whether a B1-auxotrophic bacterioplankton cell is expected to become B1-limited in LMO surface waters based on cell demands (quota), bioavailable intact B1 in LMO water (Table 1), and molecular diffusion. Based on a maximum B1 cell quota for V. anguillarum PF430-3 ∆thiE (∼530 B1 molecules per cell, converting from picomoles of B1 per cell) (SI Appendix, Table S6) and a diffusive flux of 1.06 × 106 B1 molecules/d for a 2-µm diameter V. anguillarum cell [cells are typically rod-like with a maximum length between 1.4 and 2.6 µm (83)], the B1-auxotrophic cell will encounter ∼4,000 times the number of B1 molecules it needs to divide at a rate of one division/d (see calculations in SI Appendix). Nevertheless, bacterial production stimulated by the addition of B1 or precursor amendment occurred when initial intact B1 concentrations were below ∼3.8 pM (Table 1), suggesting that this concentration is a tipping point for B1 limitation in the upper water column of the system. Whether this tipping point applies to other aquatic systems, e.g., the pelagic surface ocean, is unknown; nonetheless, it is useful for contextualizing results from nutrient-addition experiments and represents an important line of future research. Considering all these points together, we hypothesize the observed stimulation of bacterial production reported here (Table 1) resulted from one or more of the following: (i) higher B1 quotas in natural bacterioplankton than in V. anguillarum PF430-3 ∆thiE; (ii) the reliance of select bacterioplankton upon low-affinity B1 or precursor transporters requiring relatively high exogenous concentrations of B1/precursor for efficient uptake; and (iii) the influence of binding proteins in natural waters on the bioavailability of B1/precursors to specific taxa (e.g., the bioassay organism V. anguillarum PF430-3 can access protein-bound B1, but others in the natural bacterioplankton community cannot). Interestingly, regarding the last point, extracellular vitamin B12-binding proteins are produced by phytoplankton cultures and exhibit high affinity for dissolved B12 at environmentally relevant picomolar concentrations (84, 85). Likewise, research suggests that water-soluble B1-binding proteins can impact B1 bioavailability (86). Thus, it is conceivable that free dissolved protein binders of B1 and/or precursors may influence bioavailability of B1 (and potentially B1 precursors) to certain bacterioplankton; however, further research is needed to directly demonstrate this phenomenon.
Conclusion
Our results reveal that globally and naturally occurring bacterioplankton have a greater reliance on exogenous B1 and its precursors than previously realized. Specifically, (i) analyses of metagenomes from around the world show that most bacterioplankton in nature, including diverse genotypes and B1 auxotrophs and select B1 prototrophs, are reliant upon exogenous B1 and/or B1 precursor pools; (ii) Baltic Sea time-series metagenomes suggest that the reliance on exogenous B1 and precursor bioavailability changes substantially on weekly to seasonal time scales; and (iii) nutrient-addition experiments in the Baltic Sea show that B1 and/or B1 precursor bioavailability can periodically limit bulk growth of bacterioplankton. Collectively our results show the need for future investigations into B1/precursor dependency and exchange across microbial populations in aquatic systems.
At a broad aquatic ecosystem view, B1 availability is currently of greatest concern to megafauna, as multiple Northern Hemisphere animal species are declining in numbers as well as being metabolically and behaviorally impaired due to B1 deficiency (22, 23, 87). Our results suggest B1 deficiency at the picofaunal (bacterioplankton) level merits attention as well. Bacterioplankton are the foundations for microbial food webs, influencers of productivity, and performers of key ecosystem and global-level chemical transformations, e.g., nutrient regeneration through remineralization and respiration of organic carbon to carbon dioxide (88–90). Moving forward, the bioavailability of B1 and precursors as factors influencing bacterioplankton metabolism, growth, and succession deserves consideration alongside other bottom-up and top-down controls in the bigger ecological picture.
Methods
Bacterioplankton Isolate Surveys.
Bacterioplankton strains previously obtained from waters off eastern Sweden (29–31) were used in B1-auxotrophy surveys. Strains were randomly selected from five different classes. Colonies grown on marine broth 2216 (91) agar plates were transferred to liquid marine broth 2216 and were grown to turbid densities. Cells were pelleted (6,000 × g), were washed three times with charcoal-filtered, microwaved Baltic Sea surface water (cFMSW) depleted in vitamins (92), and were resuspended in cFMSW and transferred (1 µL to 1 mL) to triplicate sterile polystyrene tubes (4.5 mL; Greiner Bio One) containing the B1-deplete survey medium. B1 was added to select cultures at a final concentration of 500 pM. The B1-deplete medium consisted of cFMSW plus inorganic and organic nutrients (SI Appendix, Table S9). Inorganic and organic nutrients for isolate growth surveys were obtained from Sigma Aldrich, as was thiamin (≥99% purity). Growth was quantified as 590-nm absorbance using a FLUOstar Optima Plate Reader (BMG-LABTECH) and clear 96-well plates. Turbidity was measured after cultures were incubated for two or more weeks at room temperature in the dark. Strains exhibiting significantly higher growth versus unamended controls (determined by Welch- corrected t test; GraphPad Prism) in vitamin-amended tubes were marked as B1 auxotrophs; strains exhibiting no significant difference in growth were marked as B1 prototrophs.
Bioinformatic Analyses.
B1-related biosynthesis, salvage, and transporter proteins present in genomic and metagenomic datasets were surveyed using representative HMM profiles in the TIGRFAM (v. 15) and PFAM (v. 28) databases; in addition, custom HMM profiles were created for Omr1, ThiV, ThiY, and a decoy version of false-positive ThiY proteins (SI Appendix, Table S1). PFAM models PF09084 (SSUA/Thi5/NMT1) and PF00474 (SSSP) returned numerous nonspecific hits to isolate genomes and hence were not used in our analyses.
The custom profiles were created by first aligning seed proteins (publically available at https://figshare.com/articles/Custom_HMM_files/5928151) using MUSCLE (v3.8.1551) (93) with default settings. Alignments were then trimmed using the Gblocks (94) web server with parameters for less stringent selection: Allow smaller final blocks, gap positions within the final blocks, and less strict flanking positions. HMM profiles were generated from the trimmed alignments using hmmbuild from the hmmer (v. 3.1b2, hmmer.org) toolbox with default settings. Trusted cutoffs (TCs) for the custom profiles were determined by searching the seed proteins with the corresponding HMM and setting the cutoff to the lowest score in the output.
Protein sequences from metagenomes, MAGs, or isolate genomes were searched using hmmsearch from hmmer with per-domain output. Hits scoring above TCs for profiles were kept, and the highest-scoring, nonoverlapping HMMs were assigned to each protein sequence. Sequences identified as ThiE were those hit by the ThiE TIGRFAM model TIGR00693 or TMP-TENI PFAM model PF02581 above their respective TC values. Sequences identified as ThiB were hit by TIGRFAM model TIGR01254 or TIGR01276.
Metagenomic Surveys and Reference Metagenomes.
Four metagenomic datasets were surveyed. Three were obtained from the IMG database: AOAMet1_03_M0_10 (Project ID 1038530), COG_06 (Project ID 1016719), and LauGreDrversion2_2 (Project ID 1062136). Protein sequences and contig coverage values (which were used as proxies for protein abundance values) were downloaded from the corresponding folder at the JGI website. The fourth metagenomic dataset originates from time-series sampling of surface (2 m) microbial communities in the central Baltic Sea proper, specifically at the LMO station (56°55.851N, 17°03.640E) (28). This dataset included a coassembly of 37 samples annotated as described previously (28). Gene abundance in the LMO coassembly was normalized using transcript per million (TPM) calculations (95). For all datasets, gene abundances (TPM or contig coverage values) were divided by respective total abundances for all genes (total TPM or contig coverage values) for comparison across datasets and were converted to a percentage (described as percent normalized abundance).
Reference Genomes and MAGs.
Protein sequences from 330 genome-sequenced prokaryotic isolates (272 bacterial and 58 archaeal) were downloaded from the IMG database (https://img.jgi.doe.gov/) (96) and were used in protein sequence searches (Dataset S5). Additionally, a set of 83 MAGs reconstructed from LMO metagenomic assemblies and clustered into 30 Baltic Sea clusters (BACL) based on sequence similarity (28) and a set of 603 MAGs reconstructed from the Tara Oceans sampling expedition dataset with ≥90% completeness (40) were also surveyed.
Taxonomic Assignments.
Protein sequences in all metagenomic datasets were annotated taxonomically using similarity searches against the UniProt nonredundant (nr) database (2017-02-02) and two lowest common ancestor (LCA) steps. First, homologs of proteins were found in the nr database using DIAMOND (v. 0.8.26) (97), with the top 10 hits kept. Taxonomy was assigned to query sequences using the script ClassifyContigNR.py. This script calculates normalized weights (nw) for each hit with a minimum %identity ≥40% (default setting) by dividing the alignment length of the query by the total length of the query and then multiplying that by the %identity of the hit: nw = aLi100, where a = the alignment length of the query, L = length (in amino acids) of query, and i = percent identity of the hit. This nw forced to range between 0 and 1. For each query protein, the script then collates hits at each taxonomic rank and adds taxa weights (tw) to the taxonomy corresponding to hits at that rank. These taxa weights are calculated as tw = (nw − tr)/(1 − tr), where tr is a rank-specific threshold which by default is set to 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, and 0.95 for the ranks superkingdom, phylum, class, order, family, genus, and species, respectively. Only taxa that obtain a tw >0 are included in the assignments. Finally, the script attempts to assign a taxonomy for progressively higher levels in the taxonomy hierarchy, starting with species and moving up to genus, family, order, class, phylum, and finally superkingdom. For each rank, tws are normalized to the sum of tws at that rank, and the taxa with the highest normalized tw above 0.5 (default setting) is assigned to the query protein at that rank. If the script successfully assigns a taxonomy at a certain rank, the higher ranks (if any) obtain the corresponding taxonomy and the lower ranks (if any) are classified as the taxa with an “unclassified” prefix. Contigs are assigned taxonomy in a similar manner by collating all hits for query sequences encoded on a contig and normalizing the tws across all taxa.
Whole-Water Incubations.
Water samples were collected from ∼2 m depth at the LMO site for vitamin-amendment experiments on five occasions between May 2015 and June 2016 (Table 1). Upon return to shore, 0.5 L of LMO water was dispensed into acid-washed 1-L polycarbonate incubation bottles. Triplicate bottles received B1, HMP, HET (4-methyl-5-thiazoleethanol), or HMP + HET amendments (1-nM final concentration of each) or no addition (negative control), were incubated in the dark at ambient temperature, and were sampled for bacterial production after 24 h and 48 h. HET (≥95% purity) used in experiments was purchased from Sigma Aldrich, and HMP (>95% purity) was purchased from Enamine Ltd. Aliquots of vitamins and B1 precursors were freshly prepared in autoclaved Milli-Q water (Millipore) and were kept frozen; aliquots were kept on ice in the dark before use in experiments. Bacterial production was estimated by 3H-thymidine incorporation (20 nmol⋅L−1 final concentration; Perkin-Elmer) (98) using microcentrifugation (99), assuming 1.4 × 1018 cells per mole of thymidine (100) and 2.0 × 10−14 gC per cell (101). Outlier replicate data points were identified using a Grubbs’ test (0.05 significance level, two-sided; https://www.graphpad.com/quickcalcs/Grubbs1.cfm) and were excluded before averages and SDs were calculated across replicate bottles. Three samples from experiment 3 (after 24 h) with signals below kills were excluded from calculations. Unpaired Welch-corrected, two-tailed t tests (GraphPad Prism) were used to identify significant differences between treatments and controls. Data used in t tests are provided in Dataset S6.
Quantification of Intact B1 via Bioassays with an Engineered V. anguillarum PF430-3 Mutant.
A B1-auxotrophic V. anguillarum PF430-3 mutant requiring intact B1 was engineered by constructing an in-frame deletion of the gene encoding thiamin monophosphate synthase protein (ThiE) by allelic exchange, as previously described for the deletion of other genes in V. anguillarum NB10 (102) and PF430-3 (103). The thiE-specific suicide vector was constructed as follows: DNA sequences flanking the thiE gene were amplified from wild-type V. anguillarum PF430-3 by PCR using primers PE1 (5′-TTTAGATCTGGAGCACAAGTGCGCGATAATG-3′) and PE2 (5′-ACTACTTGGTAATCGCTAATTACTCCTCGCTAACGACTGACTG-3′) for the upstream region and primers PE3 (5′-GGAGTAATTAGCGATTACCAAGTAGTGCTGAGGAGGTATGTG-3′) and PE4 (5′-TTTGAGCTCGCTGACGACTGTGGATGTTATCC-3′) for the downstream region. Primer PE1 introduced a BglII restriction site, and primer PE4 introduced a SacI restriction site. The two PCR fragments, containing a 26-nt overlap region, were mixed as the template for a final PCR using primers PE1 and PE4. The final PCR product was purified, digested with BglII and SacI, and cloned into the BglII and SacI sites of pDM4 (102), creating pDM4thiE.
PF430-3 ∆thiE was confirmed to be B1-auxotrophic in culture as it reached high densities on liquid B1-deplete medium (cFMSW-VIB: cFMSW as above without any organic micronutrients and only C, N, P, and trace metals) supplemented with B1 but not in the same medium without B1 (SI Appendix, Fig. S7). Maximum cell yields of B1-limited cultures PF430-3 ∆thiE increased linearly with B1 addition; also, B1 and TDP half-saturation growth constants for PF430-3 ∆thiE were ca. 1 pM (0.510 ± 0.125 and 1.38 ± 0.286, respectively) (SI Appendix, Table S8), confirming the strain as a good candidate organism for quantification bioassays in which maximum cell yields indicate bioavailable B1 or phosphorylated B1 in a sample.
LMO water samples collected for bioassay quantification (on the same dates as samples for enrichment incubation experiments) were 0.2-µm filtered via sterile syringe filtration and were stored at −20 °C. On the day of bioassays, frozen samples were thawed in the dark, filtered aseptically through a 0.2-µm sterile syringe filter, and supplied nutrients in proportions equivalent to that of the cFMSW-VIB medium. The nutrient-enriched sample was combined with B1-deplete cFMSW-VIB medium (1:2 or 1:5), aliquoted into five sets of triplicate sterile polystyrene tubes (4.5 mL; Greiner Bio One), and amended with 0, 1, 2, 3, or 5 pM B1 for quantifying vitamin in the initial sample (0 pM addition) via an internal standard curve. Triplicate negative control tubes (cFMSW-VIB medium only) were run in parallel.
Early stationary-phase PF430-3 ΔthiE cells grown in marine broth 2215 (91) were centrifuged, washed, and resuspended at a dilution of 1:2,000 and then were added to the tubes and incubated in the dark at ∼16 °C, with shaking at 100 rpm. Abundances of PF430-3 ∆thiE cells in all tubes were determined by flow cytometric (FACS CANTO II flow cytometer; Becton Dickinson) counting of SYBR Green I-stained (104) cells fixed with 2% formaldehyde. Maximum abundances were determined via sampling on days 1, 2, 3, 5, 7, and 9 of incubation; initial monitoring of PF430-3 ∆thiE growth under incubation conditions revealed the stationary phase was reached by day 2 or 3.
Maximum PF430-3 ∆thiE cell yields from internal B1 additions were used to calculate the picomoles of B1 per PF430-3 ∆thiE cell for each bioassayed sample from a linear regression; this value was used to calculate concentrations in the initial undiluted water sample (0 pM addition). The maximum yields of negative controls were subtracted from the 0 pM internal standard (water sample) before calculations (SI Appendix, Table S10).
Supplementary Material
Acknowledgments
We thank Sachia Jo Traving and Elisabeth Münster Happel for assistance with whole-water incubations; Emil Skov Schlüter for assistance with auxotrophy screening of Baltic Sea isolates; Emil Fridolfsson, Carina Bunse, and Kristofer Bergström for laboratory assistance at Linnaeus University; Johannes Alneberg for bioinformatics support; and Hans Paerl for comments on the manuscript. This research was supported by the Joint Baltic Sea Research and Development Programme (BONUS) Blueprint project (Art 185) funded jointly by the European Union’s Seventh Programme for Research, Technological Development, and Demonstration, The Danish Council for Strategic Research (L.R.), and the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS) (J.P. and A.F.A.). The study was also supported by Swedish Research Council FORMAS Grant 215-2012-1319.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genomic and metagenomic data that support the findings of this study are publicly accessible via the Joint Genomic Institute (JGI) Genome Portal (https://genome.jgi.doe.gov/portal/) or the National Center for Biotechnology Information (NCBI) Sequence Read Archive (accession nos. SRR2053273–SRR2053308). Contigs for each MAG are available on the NCBI Whole Genome Shotgun database (accession nos. LIAK00000000–LIDO00000000, and centrally accessible via NCBI BioProject ID: PRJNA273799). Protein sequence alignments and custom HMM profiles generated and used in this study have been deposited in Figshare (https://figshare.com/articles/Custom_HMM_files/5928151).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806425115/-/DCSupplemental.
References
- 1.Jurgenson CT, Begley TP, Ealick SE. The structural and biochemical foundations of thiamin biosynthesis. Annu Rev Biochem. 2009;78:569–603. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J Biol Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 3.Jaehme M, Slotboom DJ. Diversity of membrane transport proteins for vitamins in bacteria and archaea. Biochim Biophys Acta. 2015;1850:565–576. doi: 10.1016/j.bbagen.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Burkholder PR, Lewis S. Some patterns of B vitamin requirements among neritic marine bacteria. Can J Microbiol. 1968;14:537–543. doi: 10.1139/m68-091. [DOI] [PubMed] [Google Scholar]
- 5.Berland BR, Bonin DJ, Durbec JP, Maestrini SY. Bactéries hétérotrophes aérobies prélevés devant le delta du rhône. IV. Besoins en vitamines et liberation de ces substances. Hydrobiologia. 1976;50:167–172. [Google Scholar]
- 6.Burkholder PR. Some nutritional relationships among microbes of sea sediments and water. In: Oppenheimer CH, editor. Symposium on Marine Microbiology. CC Thomas; Springfield, IL: 1963. pp. 133–150. [Google Scholar]
- 7.Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA. The role of B vitamins in marine biogeochemistry. Annu Rev Mar Sci. 2014;6:339–367. doi: 10.1146/annurev-marine-120710-100912. [DOI] [PubMed] [Google Scholar]
- 8.Carini P, et al. Discovery of a SAR11 growth requirement for thiamin’s pyrimidine precursor and its distribution in the Sargasso Sea. ISME J. 2014;8:1727–1738. doi: 10.1038/ismej.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupont CL, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012;6:1186–1199. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia SL, et al. Auxotrophy and intrapopulation complementary in the ‘interactome’ of a cultivated freshwater model community. Mol Ecol. 2015;24:4449–4459. doi: 10.1111/mec.13319. [DOI] [PubMed] [Google Scholar]
- 11.Natarajan KV, Dugdale RC. Bioassay and distribution of thiamine in the sea. Limnol Oceanogr. 1966;11:621–629. [Google Scholar]
- 12.Carlucci AF, Silbernagel SB. Bioassay of seawater. II. Methods for the determination of concentrations of dissolved vitamin B1 in seawater. Can J Microbiol. 1966;12:1079–1089. doi: 10.1139/m66-147. [DOI] [PubMed] [Google Scholar]
- 13.Paerl RW, et al. Use of plankton-derived vitamin B1 precursors, especially thiazole-related precursor, by key marine picoeukaryotic phytoplankton. ISME J. 2017;11:753–765. doi: 10.1038/ismej.2016.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carlucci AF, Bowes PM. Determination of vitamin B12, thiamine, and biotin in Lake Tahoe waters using modified marine bioassay techniques. Limnol Oceanogr. 1972;17:774–777. [Google Scholar]
- 15.Heal KR, et al. Determination of four forms of vitamin B12 and other B vitamins in seawater by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2014;28:2398–2404. doi: 10.1002/rcm.7040. [DOI] [PubMed] [Google Scholar]
- 16.Suffridge C, Cutter L, Sañudo-Wilhelmy SA. A new analytical method for direct measurement of particulate and dissolved B-vitamins and their congeners in seawater. Front Mar Sci. 2017;4:175. [Google Scholar]
- 17.Gobler CJ, Norman C, Panzeca C, Taylor GT, Sañudo-Wilhelmy SA. Effect of B-vitamins (B1, B12) and inorganic nutrients on algal bloom dynamics in a coastal ecosystem. Aquat Microb Ecol. 2007;49:181–194. [Google Scholar]
- 18.Natarajan KV. Distribution and significance of vitamin B12 and thiamine in the subarctic Pacific Ocean. Limnol Oceanogr. 1970;15:655–659. [Google Scholar]
- 19.Carlucci AF. The ecology of the plankton off La Jolla, California, in the period April through September, 1967. Part II: Vitamin B12, thiamine, and biotin. In: Strickland JD, editor. Bulletin of the Scripps Institution of Oceanography. University of California Press; Berkeley, CA: 1970. pp. 23–31. [Google Scholar]
- 20.Gómez-Consarnau L, et al. Mosaic patterns of B-vitamin synthesis and utilization in a natural marine microbial community. Environ Microbiol. 2018;25:3389. doi: 10.1111/1462-2920.14133. [DOI] [PubMed] [Google Scholar]
- 21.Panzeca C, et al. B vitamins as regulators of phytoplankton dynamics. Eos Trans Am Geophys Union. 2006;87:593–596. [Google Scholar]
- 22.Balk L, et al. Wild birds of declining European species are dying from a thiamine deficiency syndrome. Proc Natl Acad Sci USA. 2009;106:12001–12006. doi: 10.1073/pnas.0902903106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bengtsson B-E, et al. Reproductive disturbances in Baltic fish: A synopsis of the FiRe project. Ambio. 1999;28:2–8. [Google Scholar]
- 24.Nixon SW. Estuaries and Nutrients. Humana Press; New York: 1981. Remineralization and nutrient cycling in coastal marine ecosystems; pp. 111–138. [Google Scholar]
- 25.Ducklow HW, Purdie DA, Williams PJL, Davies JM. Bacterioplankton: A sink for carbon in a coastal marine plankton community. Science. 1986;232:865–867. doi: 10.1126/science.232.4752.865. [DOI] [PubMed] [Google Scholar]
- 26.Bianchi TS. Biogeochemistry of Estuaries. Oxford Univ Press; Oxford: 2007. [Google Scholar]
- 27.Montgomery MT, et al. 2,4,6-trinitrotoluene mineralization and bacterial production rates of natural microbial assemblages from coastal sediments. Environ Pollut. 2011;159:3673–3680. doi: 10.1016/j.envpol.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 28.Hugerth LW, et al. Metagenome-assembled genomes uncover a global brackish microbiome. Genome Biol. 2015;16:279. doi: 10.1186/s13059-015-0834-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riemann L, et al. The native bacterioplankton community in the central baltic sea is influenced by freshwater bacterial species. Appl Environ Microbiol. 2008;74:503–515. doi: 10.1128/AEM.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farnelid H, Harder J, Bentzon-Tilia M, Riemann L. Isolation of heterotrophic diazotrophic bacteria from estuarine surface waters. Environ Microbiol. 2014;16:3072–3082. doi: 10.1111/1462-2920.12335. [DOI] [PubMed] [Google Scholar]
- 31.Boström KH, Riemann L, Kühl M, Hagström A. Isolation and gene quantification of heterotrophic N2-fixing bacterioplankton in the Baltic Sea. Environ Microbiol. 2007;9:152–164. doi: 10.1111/j.1462-2920.2006.01124.x. [DOI] [PubMed] [Google Scholar]
- 32.Howard EC, et al. Bacterial taxa that limit sulfur flux from the ocean. Science. 2006;314:649–652. doi: 10.1126/science.1130657. [DOI] [PubMed] [Google Scholar]
- 33.Rusch DB, et al. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simon M, et al. Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 2017;11:1483–1499. doi: 10.1038/ismej.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo H, Moran MA. Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev. 2014;78:573–587. doi: 10.1128/MMBR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gómez-Consarnau L, et al. Proteorhodopsin light-enhanced growth linked to vitamin-B1 acquisition in marine Flavobacteria. ISME J. 2016;10:1102–1112. doi: 10.1038/ismej.2015.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenkins AL, Zhang Y, Ealick SE, Begley TP. Mutagenesis studies on TenA: A thiamin salvage enzyme from Bacillus subtilis. Bioorg Chem. 2008;36:29–32. doi: 10.1016/j.bioorg.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizote T, Nakayama H. The thiM locus and its relation to phosphorylation of hydroxyethylthiazole in Escherichia coli. J Bacteriol. 1989;171:3228–3232. doi: 10.1128/jb.171.6.3228-3232.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webb E, Claas K, Downs D. thiBPQ encodes an ABC transporter required for transport of thiamine and thiamine pyrophosphate in Salmonella typhimurium. J Biol Chem. 1998;273:8946–8950. doi: 10.1074/jbc.273.15.8946. [DOI] [PubMed] [Google Scholar]
- 40.Tully BJ, Graham ED, Heidelberg JF. The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans. Sci Data. 2018;5:170203. doi: 10.1038/sdata.2017.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paerl RW, et al. Carboxythiazole is a key microbial nutrient currency and critical component of thiamin biosynthesis. Sci Rep. 2018;8:5940. doi: 10.1038/s41598-018-24321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang I, Kim S, Islam MR, Cho J-C. The first complete genome sequences of the acI lineage, the most abundant freshwater Actinobacteria, obtained by whole-genome-amplification of dilution-to-extinction cultures. Sci Rep. 2017;7:42252. doi: 10.1038/srep42252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carini P, Steindler L, Beszteri S, Giovannoni SJ. Nutrient requirements for growth of the extreme oligotroph ‘Candidatus Pelagibacter ubique’ HTCC1062 on a defined medium. ISME J. 2013;7:592–602. doi: 10.1038/ismej.2012.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- 45.Ivars-Martinez E, et al. Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J. 2008;2:1194–1212. doi: 10.1038/ismej.2008.74. [DOI] [PubMed] [Google Scholar]
- 46.Tada Y, et al. Differing growth responses of major phylogenetic groups of marine bacteria to natural phytoplankton blooms in the western North Pacific Ocean. Appl Environ Microbiol. 2011;77:4055–4065. doi: 10.1128/AEM.02952-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lwoff A. Some aspects of the problem of growth factors for protozoa. Annu Rev Microbiol. 1947;1:101–114. [Google Scholar]
- 48.Kawasaki T, Miyata I, Esaki K, Nose Y. Thiamine uptake in Escherichia coli. I. General properties of thiamine uptake system in Escherichia coli. Arch Biochem Biophys. 1969;131:223–230. doi: 10.1016/0003-9861(69)90125-8. [DOI] [PubMed] [Google Scholar]
- 49.Grant MAA, Kazamia E, Cicuta P, Smith AG. Direct exchange of vitamin B12 is demonstrated by modelling the growth dynamics of algal-bacterial cocultures. ISME J. 2014;8:1418–1427. doi: 10.1038/ismej.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 51.Buchan A, LeCleir GR, Gulvik CA, González JM. Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol. 2014;12:686–698. doi: 10.1038/nrmicro3326. [DOI] [PubMed] [Google Scholar]
- 52.Haines KC, Guillard RRL. Growth of vitamin B12-requiring marine diatoms in mixed laboratory cultures with vitamin B12-producing marine bacteria. J Phycol. 1974;10:245–252. [Google Scholar]
- 53.Kazamia E, et al. Mutualistic interactions between vitamin B12 -dependent algae and heterotrophic bacteria exhibit regulation. Environ Microbiol. 2012;14:1466–1476. doi: 10.1111/j.1462-2920.2012.02733.x. [DOI] [PubMed] [Google Scholar]
- 54.Wagner-Döbler I, et al. The complete genome sequence of the algal symbiont Dinoroseobacter shibae: A hitchhiker’s guide to life in the sea. ISME J. 2010;4:61–77. doi: 10.1038/ismej.2009.94. [DOI] [PubMed] [Google Scholar]
- 55.Wienhausen G, Noriega-Ortega BE, Niggemann J, Dittmar T, Simon M. The exometabolome of two model strains of the Roseobacter group: A marketplace of microbial metabolites. Front Microbiol. 2017;8:1985. doi: 10.3389/fmicb.2017.01985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durham BP, et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci USA. 2015;112:453–457. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karl DM. Nutrient dynamics in the deep blue sea. Trends Microbiol. 2002;10:410–418. doi: 10.1016/s0966-842x(02)02430-7. [DOI] [PubMed] [Google Scholar]
- 58.Carlucci AF, Silbernagel SB, McNally PM. Influence of temperature and solar radiation on persistence of vitamin B12, thiamine and biotin in seawater. J Phycol. 1969;5:302–305. doi: 10.1111/j.1529-8817.1969.tb02618.x. [DOI] [PubMed] [Google Scholar]
- 59.Gold K. Some factors affecting the stability of thiamine. Limnol Oceanogr. 1968;13:185–188. [Google Scholar]
- 60.Niemi A. Blue-green algal blooms and N:P ratio in the Baltic Sea. Acta Bot Fenn. 1979;110:57–61. [Google Scholar]
- 61.Kononen K, Nômmann S. Marine Pelagic Cyanobacteria: Trichodesmium and Other Diazotrophs. Springer; Heidelberg: 1992. Spatio-temporal dynamics of the cyanobacterial blooms in the Gulf of Finland, Baltic Sea; pp. 95–113. [Google Scholar]
- 62.Fridolfsson E, Lindehoff E, Legrand C, Hylander S. Thiamin (vitamin B1) content in phytoplankton and zooplankton in the presence of filamentous cyanobacteria. Limnol Oceanogr. 2018 doi: 10.1002/lno.10949. [DOI] [Google Scholar]
- 63.Wilhelm SW, Trick CG. Effects of vitamin B12 concentration on chemostat cultured Synechococcus sp. strain PCC 7002. Can J Microbiol. 1995;41:145–151. [Google Scholar]
- 64.Tripp HJ, et al. Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature. 2010;464:90–94. doi: 10.1038/nature08786. [DOI] [PubMed] [Google Scholar]
- 65.Bonnet S, et al. Vitamin B12 excretion by cultures of the marine cyanobacteria Crocosphaera and Synechococcus. Limnol Oceanogr. 2010;5:1959–1964. [Google Scholar]
- 66.Andersson A, et al. Projected future climate change and Baltic Sea ecosystem management. Ambio. 2015;44:345–356. doi: 10.1007/s13280-015-0654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wasmund N. Occurrence of cyanobacterial blooms in the Baltic Sea in relation to environmental conditions. Int Revue Ges Hydrobiol Hydrogr. 1997;82:169–184. [Google Scholar]
- 68.Paerl HW, Huisman J. Climate. Blooms like it hot. Science. 2008;320:57–58. doi: 10.1126/science.1155398. [DOI] [PubMed] [Google Scholar]
- 69.Amin SA, Parker MS, Armbrust EV. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev. 2012;76:667–684. doi: 10.1128/MMBR.00007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amin SA, et al. Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature. 2015;522:98–101. doi: 10.1038/nature14488. [DOI] [PubMed] [Google Scholar]
- 71.Lima-Mendez G, et al. Tara Oceans coordinators Ocean plankton. Determinants of community structure in the global plankton interactome. Science. 2015;348:1262073. doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- 72.Needham DM, Fuhrman JA. Pronounced daily succession of phytoplankton, archaea and bacteria following a spring bloom. Nat Microbiol. 2016;1:16005. doi: 10.1038/nmicrobiol.2016.5. [DOI] [PubMed] [Google Scholar]
- 73.Vergin KL, et al. High-resolution SAR11 ecotype dynamics at the Bermuda Atlantic Time-series Study site by phylogenetic placement of pyrosequences. ISME J. 2013;7:1322–1332. doi: 10.1038/ismej.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu M, Papish ET, Begley TP. Mechanistic studies on thiaminase I: Identification of the product of thiamin degradation in the absence of the nucleophilic cosubstrate. Bioorg Chem. 2000;28:45–48. [Google Scholar]
- 75.Sala MM, et al. Seasonal and spatial variations in the nutrient limitation of bacterioplankton growth in the northwestern Mediterranean. Aquat Microb Ecol. 2002;27:47–56. [Google Scholar]
- 76.Kuparinen J, Heinänen A. Inorganic nutrient and carbon controlled bacterioplankton growth in the Baltic Sea. Estuar Coast Shelf Sci. 1993;37:271–285. [Google Scholar]
- 77.Elser JJ, Stabler LB, Hassett RP. Nutrient limitation of bacterial growth and rates of bacterivory in lakes and oceans: A comparative study. Aquat Microb Ecol. 1995;9:105–110. [Google Scholar]
- 78.Kirchman DL, et al. Carbon versus iron limitation of bacterial growth in the California upwelling regime. Limnol Oceanogr. 2000;45:1681–1688. [Google Scholar]
- 79.Pinhassi J, et al. Seasonal changes in bacterioplankton nutrient limitation and their effects on bacterial community composition in the NW Mediterranean Sea. Aquat Microb Ecol. 2006;44:241–252. [Google Scholar]
- 80.Koch F, et al. Vitamin b(1) and b(12) uptake and cycling by plankton communities in coastal ecosystems. Front Microbiol. 2012;3:363. doi: 10.3389/fmicb.2012.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sañudo-Wilhelmy SA, et al. Multiple B-vitamin depletion in large areas of the coastal ocean. Proc Natl Acad Sci USA. 2012;109:14041–14045. doi: 10.1073/pnas.1208755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barada LP, et al. The distribution of thiamin and pyridoxine in the western tropical North Atlantic Amazon River plume. Front Microbiol. 2013;4:25. doi: 10.3389/fmicb.2013.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baumann P, Furniss AL, Lee JV. Genus I. Vibrio. In: Krieg NR, Holt JG, editors. Bergey’s Manual of Systematic Bacteriology. 1st Ed. The Williams & Wilkins Co.; Baltimore: 1984. pp. 518–538. [Google Scholar]
- 84.Davies AG, Leftley JW. Vitamin B12 binding by microalgal ectocrines: Dissociation constant of the vitamin-binder complex determined using an ultrafiltration technique. Mar Ecol Prog Ser. 1985;21:267–273. [Google Scholar]
- 85.Droop MR. Vitamin B12 and marine ecology. IV. The kinetics of uptake, growth and inhibition in Monochrysis lutheri. J Mar Biol Assoc UK. 1968;48:689–733. [Google Scholar]
- 86.Itokawa Y, Kimura M, Nishino K. Thiamin-binding proteins. Ann N Y Acad Sci. 1982;378:327–336. doi: 10.1111/j.1749-6632.1982.tb31207.x. [DOI] [PubMed] [Google Scholar]
- 87.Balk L, et al. Widespread episodic thiamine deficiency in Northern Hemisphere wildlife. Sci Rep. 2016;6:38821. doi: 10.1038/srep38821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Azam F, Fenchel T, Field JG, Gray JS. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser. 1983;10:257–263. [Google Scholar]
- 89.Arnosti C. Microbial extracellular enzymes and the marine carbon cycle. Annu Rev Mar Sci. 2011;3:401–425. doi: 10.1146/annurev-marine-120709-142731. [DOI] [PubMed] [Google Scholar]
- 90.Gattuso J-P, Frankignoulle M, Wollast R. Carbon and carbonate metabolism in coastal aquatic ecosystems. Annu Rev Ecol Evol Syst. 1998;29:405–434. [Google Scholar]
- 91.ZoBell CE. Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J Mar Res. 1941;4:42–75. [Google Scholar]
- 92.Paerl RW, Bertrand EM, Allen AE, Palenik B, Azam F. Vitamin B1 ecophysiology of marine picoeukaryotic algae: Strain-specific differences and a new role for bacteria in vitamin cycling. Limnol Oceanogr. 2015;60:215–228. [Google Scholar]
- 93.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 95.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 96.Chen IA, et al. IMG/M: Integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 2017;45:D507–D516. doi: 10.1093/nar/gkw929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 98.Fuhrman JA, Azam F. Bacterioplankton secondary production estimates for coastal waters of British Columbia, Antarctica, and California. Appl Environ Microbiol. 1980;39:1085–1095. doi: 10.1128/aem.39.6.1085-1095.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smith DC, Azam F. A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Microb Food Webs. 1992;6:107–114. [Google Scholar]
- 100.Dinasquet J, et al. Cascading effects of the ctenophore Mnemiopsis leidyi on the planktonic food web in a nutrient-limited estuarine system. Mar Ecol Prog Ser. 2012;460:49–61. [Google Scholar]
- 101.Lee S, Fuhrman JA. Relationships between biovolume and biomass of naturally derived marine bacterioplankton. Appl Environ Microbiol. 1987;53:1298–1303. doi: 10.1128/aem.53.6.1298-1303.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Milton DL, O’Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tan D, Svenningsen SL, Middelboe M. Quorum sensing determines the choice of antiphage defense strategy in Vibrio anguillarum. MBio. 2015;6:e00627. doi: 10.1128/mBio.00627-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brussaard CPD, Payet JP, Winter C, Weinbauer MG. Manual of Aquatic Viral Ecology. Am Soc Limnol Oceanogr; Waco, TX: 2010. Quantification of aquatic viruses by flow cytometry; pp. 102–109. [Google Scholar]
- 105.Raes J, Korbel JO, Lercher MJ, von Mering C, Bork P. Prediction of effective genome size in metagenomic samples. Genome Biol. 2007;8:R10. doi: 10.1186/gb-2007-8-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.