Significance
Somatic mutations in some splicing factor genes are frequently found in myelodysplastic syndromes (MDS) and MDS-related acute myeloid leukemia (AML), blood cancers with few effective treatment options. However, the pathophysiological effects of these mutations remain poorly characterized. Here, we report the establishment of mouse models to study a common splicing factor mutation, U2AF1(S34F). Production of the mutant protein in the murine hematopoietic compartment disrupts hematopoiesis in ways resembling human MDS. We further identified deletion of the Runx1 gene and other known oncogenic mutations as changes that might collaborate with U2af1(S34F) to give rise to frank AML in mice. However, the U2af1(S34F) mutation was absent in two of the three AML cases, raising the possibility that this mutant protein plays a dispensable role in tumor maintenance.
Keywords: U2AF1, splicing factor, S34F, myelodysplastic syndromes, leukemia
Abstract
Mutations affecting the spliceosomal protein U2AF1 are commonly found in myelodysplastic syndromes (MDS) and secondary acute myeloid leukemia (sAML). We have generated mice that carry Cre-dependent knock-in alleles of U2af1(S34F), the murine version of the most common mutant allele of U2AF1 encountered in human cancers. Cre-mediated recombination in murine hematopoietic lineages caused changes in RNA splicing, as well as multilineage cytopenia, macrocytic anemia, decreased hematopoietic stem and progenitor cells, low-grade dysplasias, and impaired transplantability, but without lifespan shortening or leukemia development. In an attempt to identify U2af1(S34F)-cooperating changes that promote leukemogenesis, we combined U2af1(S34F) with Runx1 deficiency in mice and further treated the mice with a mutagen, N-ethyl-N-nitrosourea (ENU). Overall, 3 of 16 ENU-treated compound transgenic mice developed AML. However, AML did not arise in mice with other genotypes or without ENU treatment. Sequencing DNA from the three AMLs revealed somatic mutations homologous to those considered to be drivers of human AML, including predicted loss- or gain-of-function mutations in Tet2, Gata2, Idh1, and Ikzf1. However, the engineered U2af1(S34F) missense mutation reverted to WT in two of the three AML cases, implying that U2af1(S34F) is dispensable, or even selected against, once leukemia is established.
Myelodysplastic syndromes (MDS) are neoplastic diseases characterized principally by deficiencies of normal cells in myeloid, erythroid, and/or megakaryocytic lineages, mono- or multilineage dysplasia, clonal dominance of abnormal immature cells, and variable risks of developing secondary acute myeloid leukemia (AML) (1–3). In over half of MDS patients, the malignant clone carries a mutation in one of four genes (U2AF1, SRSF2, SF3B1, or ZRSR2) encoding factors critical for correct splicing of premessenger RNA (pre-mRNA) (4). Mutations in these “splicing factor genes” are presumed to be causative events in MDS because of their frequency, their recurrence at a few positions in coding sequences, their high allelic ratios, and their association with clinical outcomes (4–7). Still, despite intensive studies, the mechanisms by which such mutations contribute to the initiation or maintenance of neoplasias have not been identified.
We have been studying the most common mutation observed in U2AF1, the gene encoding an RNA-binding protein that helps direct the U2 small nuclear ribonucleoprotein particle (U2 snRNP) to the 3′ splice-acceptor site in pre-mRNA (8–10). This mutation replaces serine at position 34 of U2AF1 with phenylalanine (S34F) (4, 6), yielding a neomorphic splicing factor that changes splicing patterns for many RNAs and does not preserve enough normal splicing to permit cell survival in the absence of a WT allele (11). In addition, U2AF1(S34F) is present at high allelic frequencies (nearly 50%) in MDS (6), seems to predispose MDS patients to secondary AML (6, 12), and is also found, albeit at lower frequencies, in a variety of other neoplastic diseases, including solid tumors (13–15).
Progress toward understanding the oncogenic effects of U2AF1(S34F) has been impeded by a lack of appropriate biological models. Here we describe our development of genetically engineered mouse models in which U2af1(S34F) is assembled at the endogenous U2af1 locus by the action of the Cre recombinase. By activating Cre in the hematopoietic compartment of these mice to produce U2af1(S34F), the mice develop impairments of blood cells with MDS-like features accompanied by abnormal splicing patterns resembling those previously observed in human cells expressing this mutant splicing factor. In an effort to model U2AF1(S34F)-associated leukemia, we deprived the U2af1 mutant mice of a hematopoietic transcription factor, Runx1, often commutated in human MDS and leukemias (16, 17), and treated them with a chemical mutagen, N-ethyl-N-nitrosourea (ENU). Under those circumstances, clones of AML developed in 3 of 16 mice but not in mice lacking any of the three factors. Although AML did not occur often enough to conclude that the mutant splicing factor is required for leukemogenesis in this model, whole-exome sequencing (WES) of AML cells revealed somatic mutations in genes often mutated in human AML. In summary, we report knock-in mouse lines with conditional U2af1(S34F) alleles, demonstrate the effects of mutant U2af1 on mouse hematopoiesis, and identify additional mutations that may cooperate with U2af1(S34F) and Runx1 deficiency during leukemogenesis.
Results
Establishing Mice Carrying Conditional Knock-In S34F Alleles of U2af1.
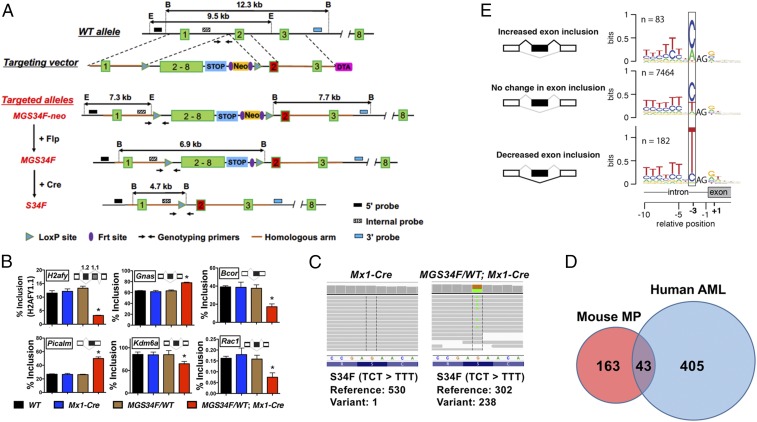
We used targeting vectors (termed “MGS34F” and “IES34F”) (Fig. 1A and SI Appendix, Fig. S2A) to create two conditional S34F mutant alleles at the endogenous U2af1 locus of B6/129 mice, and we documented the successful introduction of the targeting vectors at the appropriate sites by restriction mapping and Sanger sequencing (Materials and Methods and SI Appendix, Figs. S1A and S2B). Mice carrying either of the altered U2af1 alleles express GFP in all tissues because the mouse embryonic stem cell line used to generate the mutant mice carries a GFP transgene driven by the ubiquitously active human ubiquitin C (UBC) promoter (18), and the transgene is located on the same chromosome as the U2af1 locus, with a cross-over frequency of 1.95% (SI Appendix, Fig. S1H and Table S1).
Fig. 1.
Conditional expression of U2af1(S34F) from the mouse endogenous locus alters splicing similar to that in human cells expressing U2AF1(S34F). (A) Diagrams of the endogenous U2af1 locus, the MGS34F targeting vector, and modified alleles, with sites used for Southern blotting- and PCR-based genotyping. Numbers in boxes indicate exons or exonic sequences in cDNA; lines represent introns; STOP denotes a 3× transcriptional stop signal from SV40; B, BamHI site; E, EcoRI site. The red version of exon 2 encodes the S34F mutation (TCT-to-TTT). A more detailed description of the targeting vector is in SI Appendix, Supplemental Methods and Materials. (B) U2af1(S34F) changes the inclusion levels of indicated exons (or portions of exons; see inserted cartoons) found at six loci of genes (named in boxes) in total bone marrow cells from mice with the indicated genotypes (n = 3). The mice were treated with poly (IC) and were killed 2 wk later for RNA extraction. As noted in the text, alternative splicing of human homologs of these mRNAs was previously reported to be affected by U2AF1(S34F). Asterisks indicate statistically significant changes compared with other genotypes by t test (*P < 0.05). Error bars represent SEM. (C) Representative read coverage of U2af1 cDNA by RNA-seq, which was performed on MPs from mice of the indicated genotypes 4 wk after poly (IC) treatment. Each gray line represents a sequencing read. The reference DNA (noncoding strand) and protein sequences are shown below the sequencing reads, and the numbers of reference (WT) and variant (S34F) alleles are quantified below the graphs. (D) The Venn diagram indicates the numbers of orthologous genes from mouse and human datasets that show at least 10% change in cassette exon inclusion levels in the presence of mutant U2AF1. The mouse dataset is presented in SI Appendix, Table S3. The human dataset was published previously (21), using AML samples from The Cancer Genome Atlas (63). Genes with low levels of expression (median transcripts per million sequenced RNAs <10) or that have no mouse or human ortholog were excluded from the analysis. (E) U2af1(S34F) recognizes similar consensus sequences at 3′ splice sites in the mouse genome as U2AF1(S34F) does in the human genome. mRNA from MPs with or without U2af1(S34F) was sequenced to determine nucleotides at the 3′ splice-acceptor sites of cassette exons and was displayed as sequence logos according to whether inclusion of the exon in mRNA was increased, decreased, or unaffected by U2af1(S34F). The resemblance of these logos to those previously determined with human materials is discussed in the text. Additional molecular characterization of the established mouse lines (including another conditional allele, IES34F) and their cell derivatives is presented in SI Appendix, Figs. S1 and S2.
To assess whether these conditional alleles can be rearranged correctly, we crossed the two mouse strains with mice carrying a UBC-driven transgene encoding a tamoxifen-dependent Cre recombinase (UBC-CreERT2) (19). Mouse embryo fibroblasts (MEFs) derived from embryos with MGS34F; CreERT2 or IES34F; CreERT2 were treated with 4-hydroxyl-tamoxifen (4OHT) in culture; efficient appearance of the expected configurations of U2af1(S34F) was confirmed by Southern blotting (SI Appendix, Figs. S1B and S2C), and the anticipated changes in U2af1(S34F) mRNA were ascertained with allele-specific TaqMan assays (SI Appendix, Figs. S1C, S2D, and S3 C–E). MGS34F appeared to be recombined by Cre more efficiently than the IES34S allele and was used in all follow-up studies.
We next crossed mice heterozygous for the conditional MGS34F allele with mice carrying an Mx1-Cre transgene that is expressed in the blood lineage upon administration of poly (IC) (20). Two weeks after poly (IC) treatment of the resulting bitransgenic mice (MGS34F/WT; Mx1-Cre), Cre-mediated recombination was nearly complete in nucleated bone marrow cells, as revealed by Southern blotting (SI Appendix, Fig. S1E) and by quantitative analysis of PCR-generated fragments (SI Appendix, Fig. S3 A and B). The WT and mutant alleles appear to be expressed at equivalent levels, judging from the ∼1:1 ratio of mutant and WT U2af1 mRNA (SI Appendix, Fig. S1F).
Effects of U2af1(S34F) on Pre-mRNA Splicing in Mouse Cells Resemble the Effects of U2AF1(S34F) in Human Cells.
U2AF1(S34F) is known to affect pre-mRNA splicing and possibly other events in a variety of human cells (11, 21–27). We used cells and tissues from mice carrying the U2af1(S34F) allele to determine whether similar alterations occur in mice after expression of U2af1(S34F) at physiological levels. As an initial assessment, we measured changes in spliced products of pre-mRNA from six mouse genes (H2afy, Gnas, Bcor, Picalm, Kdm6a, and Rac1) homologous to human genes whose transcripts undergo changes in alternative splicing (11, 21, 27) in the presence of U2AF1(S34F). We also examined RNA products of one mouse gene, Atg7, whose transcripts were reported to show changes in use of polyadenylation sites (25). Isoform-sensitive primers were designed to target regions of the mRNAs that are predicted to undergo changes based on previous results in human or mouse cells (SI Appendix, Table S2). We found that similar alterations in pre-mRNA splicing occurred in the six homologous murine genes expressed in total bone marrow cells (Fig. 1B). However, the previously reported changes in Atg7 RNA polyadenylation sites were not observed in mouse bone marrow cells expressing U2af1(S34F) (SI Appendix, Fig. S1G).
We used whole-cell mRNA-sequencing methods to make a more extensive survey of U2af1(S34F)-induced alterations in pre-mRNA splicing, looking specifically for the changes in frequency with which so-called “cassette exons” are sometimes included in the spliced products, since these are the most prominent consequences of the S34F mutant in human cells (11, 21–24, 27). In addition, we performed these analyses in mouse myeloid progenitors (MPs) isolated from the bone marrow of MG(S34F)/WT; Mx1-Cre mice (and from Mx1-Cre mice as a control) 4 wk after poly (IC) treatment based on their immunophenotype, Lin−Sca-1−c-Kit+ (Lin, Lineage), which expressed U2af1(S34F) mRNA, as expected (Fig. 1C).
Consistent with previous reports, alterations in the use of cassette exons are the most frequent changes in MPs expressing U2af1(S34F) (268 of 389 events) (SI Appendix, Table S3). Overall, we found significant changes in the alternative splicing of cassette exons from 206 mouse genes; 43 of those genes (about 20%) are homologs of human genes similarly affected by U2AF1(S34F) or U2AF1(S34Y) in AML cells (Fig. 1D). Furthermore, the consensus nucleotide sequences preceding the invariant AG dinucleotide at the 5′ end of the affected cassette exons match those previously identified as determinants of altered splicing patterns in human cells expressing U2AF1(S34F) (11, 21–24, 27). For example, the first nucleotide position 5′ of the AG dinucleotide is frequently occupied by a C or A nucleotide upstream of exons that are more frequently included in mRNA and is often occupied by a T nucleotide upstream of cassette exons that are less frequently included (Fig. 1E). Similar changes were seen in MEFs expressing U2af1(S34F) (SI Appendix, Figs. S1D and S2E). Based on these results, we conclude that physiological expression of U2af1(S34F) causes changes in pre-mRNA splicing similar but not identical to those observed by us and others in human cells expressing U2AF1(S34F) (11, 21–24, 27).
U2af1(S34F) Affects Hematopoiesis in Mice with Features That Are Shared with Human MDS.
We next examined the impact of U2af1(S34F) on hematopoiesis in our genetically engineered mice. Blood samples were taken before and after administration of poly (IC) to induce Cre and were subjected to complete blood cell counts and flow cytometry. The cellular profiles of blood from WT mice and from mice carrying only the Mx1-Cre transgene or only the MGS34F allele were similar and normal, as expected. However, MGS34F/WT; Mx1-Cre mice showed mild but persistent changes in RBCs (reduced RBC count, hemoglobin concentration, and hematocrit and increased mean RBC volume) (Fig. 2 A and B and SI Appendix, Fig. S4 A and B). Platelet numbers were normal (SI Appendix, Fig. S4C), but the numbers of WBCs were reduced persistently by nearly 50% compared with results in mice with the control genotypes after poly (IC) treatment (Fig. 2C). Decreases were observed in all major WBC lineages as measured by flow cytometry (SI Appendix, Fig. S4 D–G), with the most marked reduction seen in B220+ B cells (Fig. 2 D–G). Despite the reduction in nearly all mature blood cell types upon activation of U2af1(S34F), bone marrow cellularity and spleen size were normal (SI Appendix, Fig. S4 I and J), with no evidence of pathological extramedullary hematopoiesis in mice with U2af1(S34F).
Fig. 2.
U2af1(S34F) causes multilineage cytopenia, macrocytic anemia, and reduction in the HSC and progenitor cell populations. (A–G) Multilineage cytopenia and macrocytic anemia in mice expressing U2af1(S34F). Blood samples taken from mice of the indicated genotypes (see keys in A) before and up to 24 wk after poly (IC) treatment were subjected to complete blood cell counts to determine numbers of RBCs (A), mean RBC volume (B), and WBCs (C) and were used for flow cytometry (D–G) to determine percentage of cells with the indicated lineage markers. Asterisks indicate significant changes for mice with the MGS34F/WT; Mx1-Cre genotype compared with mice of any other genotype by multiple t test (false-discovery rate <0.05). *P < 0.05. (H–L) U2af1(S34F) reduces the percentage of HSCs and progenitor cells in the bone marrow. Bone marrow cells from mice of the indicated genotypes were harvested 36 wk after poly (IC) treatment, and the percentages of HSC and progenitor cells were determined by flow cytometry using the stated markers. (H) Representative results of flow cytometry with numbers indicating the percentages of live nucleated cells in the boxed areas. (I–L) Bar heights indicate the mean percentages of cells with the designated phenotypes (vertical axes) in the bone marrow. Dots indicate the percentage of cells in a mouse. Asterisks denote significant changes by t test (*P < 0.05). N.S., not significant. Error bars represent the SEM. See SI Appendix, Fig. S4 for additional characterization of these mice. WT/WT, n = 10; WT/WT; Mx1-Cre, n = 10; MGS34F/WT, n = 10; MGS34F/WT; Mx1-Cre, n = 11.
We sought to determine whether these long-term changes were due to effects of the mutant splicing factor on blood stem and progenitor cells. We examined the abundance and function of hematopoietic stem cells (HSCs) and progenitors in mutant and control mice 36 wk after poly (IC) treatment. Bone marrow cells from mice with or without U2af1(S34F) were stained with antibodies against markers for HSCs and for various blood cell progenitors and were analyzed by flow cytometry. While no differences were observed in the percentages of Lin− cells (Fig. 2I) and MPs (Lin−Sca-1−c-Kit+) (Fig. 2 H and J) from mutant and control animals, the HSC- and progenitor-enriched LSK cells (marked by Lin−Sca-1+c-Kit+) were decreased in the bone marrow of U2af1(S34F) mice (Fig. 2 H and K). The decrease in the LSK cells was due to the reduction of short-term and long-term HSCs (marked by CD48−CD150− and CD48−CD150+, respectively, within the LSK cells) (Fig. 2 H and L and SI Appendix, Fig. S4K) but not the multipotent progenitors (marked by CD48+CD150− within the LSK cells) (SI Appendix, Fig. S4L) in the bone marrow of U2af1(S34F) mice. These findings suggest that reductions in HSCs likely account for the persistence of multilineage cytopenia observed in the peripheral blood of mice expressing mutant U2af1.
To determine whether the effects of U2af1(S34F) on mouse hematopoiesis were cell autonomous, we transplanted bone marrow cells from MGS34F/WT; Mx1-Cre mice and from control (MGS34F/WT) mice noncompetitively into lethally irradiated recipient mice (F1 progeny of a B6 × 129 cross) (SI Appendix, Supplemental Methods and Materials). Four weeks after transplantation, poly (IC) was used to induce production of Cre in the transplanted cells. Five weeks later, macrocytic anemia, multilineage cytopenia, and reduction in LSK cells were observed in the recipient mice (compare results in SI Appendix, Fig. S5 with those in Fig. 2 and SI Appendix, Fig. S4). Although the numbers of LSK cells were reduced, a slightly higher fraction of these cells was actively cycling, as shown by a BrdU incorporation assay (SI Appendix, Fig. S5Q), suggesting a compensatory mechanism to overcome the loss of these cells. Moreover, bone marrow cells from the U2af1(S34F) mice formed fewer myeloid colonies, especially after replating (SI Appendix, Fig. S5R), consistent with the reduced numbers of LSK cells observed by flow cytometry. Overall, these results demonstrate that the effects of U2af1(S34F) on hematopoiesis are cell autonomous.
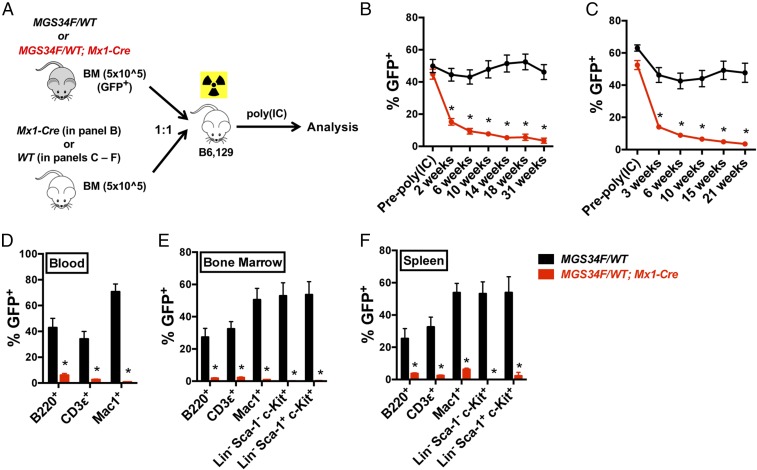
To further characterize the HSC defects attributed to U2af1(S34F), we performed competitive transplantations using bone marrow cells from MGS34F/WT; Mx1-Cre or MGS34F/WT mice (test cells) mixed with bone marrow cells from WT or Mx1-Cre mice (competitor cells). Equal numbers of test and competitor cells were mixed and transplanted into lethally irradiated recipient mice, and the recipients received poly (IC) 4 wk later (Fig. 3A). Mice that received test cells from MGS34F/WT; Mx1-Cre animals displayed a deficiency of U2af1(S34F)-expressing cells in all measured mature and immature cell lineages in the peripheral blood, bone marrow, and spleen as early as 2 wk after poly (IC) treatment (Fig. 3 B–F). In contrast, immature and mature blood cells from MGS34F/WT mice remained at levels of nearly 50% at all times. These results show that HSCs expressing U2af1(S34F) are defective in repopulating the hematopoietic compartment in a competitive transplant setting.
Fig. 3.
HSCs and progenitor cells with U2af1(S34F) are eliminated in a competitive transplant assay with cells lacking mutant U2af1. (A) Assay design. Equal numbers (0.5 million nucleated cells) of donor (MGS34F/WT or MGS34F/WT; Mx1-Cre, both GFP+) and competitor (WT or Mx1-Cre alone, GFP−) bone marrow cells were mixed and transplanted into lethally irradiated WT recipient mice. Recipient mice were treated with poly (IC) 4 wk after transplant. (B–F) Contributions of donor cells to hematopoietic compartments of recipient mice. (B and C) U2af1(S34F)-containing bone marrow cells from MGS34F/WT; Mx1-Cre mice, but not bone marrow cells from MGS34F/WT mice, constituted less of the WBCs with bone marrow competitors from Mx1-Cre (B) or WT (C) mice as measured by percentages of GFP+ cells in peripheral blood of recipient mice in a competitive transplantation. (D–F) All measured cell lineages showed competitive disadvantages of U2af1(S34F)-containing cells in the indicated cell lineages in the blood (D), bone marrow (E), and spleen (F) 21 wk after poly (IC) treatment. n = 5–10 for each group of mice. Asterisks indicate significant changes by t test. Error bars represent SEM. *P < 0.05.
Using light microscopy to seek the morphological features of myelodysplasia, we occasionally observed binucleated erythroid cells and hypo-segmented neutrophils in the bone marrow cells from U2af1(S34F)-expressing mice (SI Appendix, Fig. S6 A and B). Moreover, megakaryocytes from these mice occasionally formed clusters (SI Appendix, Fig. S6 C–E), even though platelet production was normal or only slightly diminished (SI Appendix, Figs. S4C and S5C). These dysplastic features typically affected less than 1% of bone marrow cells and were not present in the peripheral blood; therefore, they did not meet the diagnostic criteria for human MDS. Nevertheless, the multilineage cytopenia and dysplasia found in the U2af1(S34F) mutant mice are hallmarks of human MDS.
Generation of Mice with Conditional Alleles of both Runx1 and U2af1.
Although U2af1(S34F) affects hematopoiesis in our genetically engineered mice, these mice were relatively healthy (e.g., with normal weight gain) (SI Appendix, Figs. S4H and S5M), and they lived a normal life span. We reasoned that additional genetic changes might produce more severe hematologic defects or neoplasias dependent on U2af1(S34F). Therefore, we combined our MGS34F allele and the Mx1-Cre transgene with a Runx1 allele carrying LoxP sites that flank exon 4, encoding the DNA-binding Runt domain (28). When bred to homozygosity at the floxed Runx1 locus (Runx1F/F) in the presence of Mx1-Cre and MGS34F, Cre recombinase inactivates Runx1 and allows the production of the mutant splicing factor in the same cells after administration of poly (IC). Since loss-of-function mutations of Runx1 often co-occur with U2AF1 mutations in human myeloid neoplasms (16, 17), this seemed to be a promising genetic combination for eliciting pathological phenotypes, including leukemias.
The experimental cohort used for these purposes consisted of the compound-engineered mice (MGS34F/WT; Runx1F/F; Mx1-Cre, hereafter “URC mice”), mice in which poly (IC) activates U2af1(S34F) or inactivates Runx1 (MGS34F/WT; Mx1-Cre or Runx1F/F; Mx1-Cre), and control WT mice (or WT-equivalent mice lacking the Mx1-Cre transgene). After induction of Cre, URC mice developed the hematological abnormalities associated with either U2af1(S34F) (e.g., multilineage cytopenia, low-level myeloid, and erythroid dysplasia) or Runx1 deficiency alone (e.g., thrombocytopenia, increased percentage of myeloid cells, and increased myeloid colony formation, refs. 28 and 29), as shown in SI Appendix, Fig. S7 A–D. We also examined the effects of U2af1(S34F) and Runx1 deletion on gene expression and pre-mRNA splicing in MP cells from poly (IC)-treated URC mice and compared the findings with those obtained in mice with conditional alleles at either the U2af1 or Runx1 locus (SI Appendix, Fig. S7 E–G and Tables S3 and S4). As anticipated from its role as a gene encoding a transcription factor, Runx1 deficiency mainly affected gene expression, whereas U2AF1(S34F) predominantly changed mRNA splicing. As a result, MP cells with U2af1(S34F) and without functional Runx1 exhibited many changes in both mRNA splicing and gene expression, as illustrated in SI Appendix, Fig. S7 F and G.
URC Mice Develop Frank AML or AML Clones After ENU Mutagenesis.
Although URC mice showed many changes in hematopoiesis and in gene expression in MP cells (SI Appendix, Fig. S7), they had a normal life span and did not develop frank leukemia within 1.5 y after poly (IC) treatment. To seek additional genetic alterations that might cooperate with U2af1(S34F) and Runx1 deficiency to cause more profound pathology, including leukemias, we performed a forward genetic screen by treating URC mice (6–16 wk old) with the mutagen ENU 1 wk after inducing programmed genetic changes with poly (IC) (Fig. 4A). We used a relatively low dose of ENU (100 mg/kg) that, based on our previous experience (30), did not produce myeloid leukemia in otherwise normal animals. Mice that developed T cell malignancies and thymoma (known consequences of ENU) were excluded from further analysis.
Fig. 4.
Leukemogenesis in ENU-treated URC mice. (A) Strategy for testing leukemogenesis. Mice treated with poly (IC) and 1 wk later with ENU (100 mg/kg) were monitored until they were moribund or up to 1.5 y after receiving poly (IC). Splenic or bone marrow cells from these mice without frank AML were transplanted into sublethally irradiated WT recipient mice. Three to five recipient mice were used for each transplantation and were monitored for up to 1 y for the development of AML. (B) Summary of AML incidence. Mice with the control genotype lackwd either U2af1(S34F) or Runx1 deletions and were not used for transplant. NA, not available. Mice with T cell lymphoma or thymoma were censored from the study cohort. In total, 3 of 16 URC mice developed AML (P value = 0.13, based on four separate genotypes as shown; P value = 0.03 based on URC mice vs. all other genotypes, by Fisher’s exact test). (C) A case of primary AML in 1 of 16 poly (IC)- and ENU-treated URC mice (case 1). (Left) A high percentage of c-Kit+ cells was present in the peripheral blood of this mouse by flow cytometry, which coincided with infiltration of morphologically defined myeloblasts (arrowheads) in a Wright–Giemsa–stained blood smear (Center) and an H&E-stained section of the bone marrow (Right). (D) A case of late AML. Representative results are shown for the case 3 donor (Upper Row) and one of transplant recipients when it was moribund (Lower Row). (Left) Percentages of c-Kit+ cells were markedly increased in the peripheral blood of the recipient mouse compared with that of the donor. (Center) Abnormally nucleated RBCs (arrows) were observed in the blood smears from both donor and recipient, while myeloblasts (arrowheads) were observed only in blood from the recipient. (Right) Blast cells also filled the bone marrow of the recipient mouse. (Scale bars in C and D: 50 µm.) (E) Survival curves of sublethally irradiated mice after transplantations of bone marrow (black trace) or spleen (blue trace) cells from the primary donor mice of cases 1–3. The day count started with the day of transplantation. See SI Appendix, Fig. S9 for additional characterization of these AML cases.
All mice lacking Runx1 died prematurely [within 15 mo after poly (IC) treatment], regardless of U2af1 status, after receiving ENU (SI Appendix, Fig. S8A). However, 10 mo after administration of ENU, one of the 16 URC mice (case 1)—but none of the mice with Runx1 deletion alone or any other genotypes—developed AML (which we call “primary” or “1°” AML) (Fig. 4B). Morphologically defined myeloblasts were abundant in the peripheral blood, bone marrow, and spleen, and they infiltrated multiple organs, including the liver (Fig. 4C and SI Appendix, Fig. S9A). The malignant cells expressed the c-Kit receptor but not mature lineage markers, including Mac1 (Fig. 4C). When the bone marrow or spleen cells were transplanted from the case 1 mouse to sublethally irradiated mice, the recipients died with AML in 3 mo (Fig. 4E).
All the other ENU-treated Runx1F/F; Mx1-Cre mice, with or without the U2AF1(S34F) mutation, died prematurely with myeloid pathology: increased percentages of myeloid cells in the peripheral blood, enlarged spleen (SI Appendix, Fig. S8B), and extramedullary hematopoiesis. Low-grade dysplasia in the erythroid and neutrophil lineages was occasionally observed in the peripheral blood and bone marrow of moribund URC mice, consistent with the multilineage dysplasia associated with U2af1(S34F). These results suggest that ENU-treated mice with deletions of Runx1 develop lethal forms of myeloproliferative neoplasm (MPN) with occasional dysplastic features in the presence of U2af1(S34F).
We speculated that the animals with lethal forms of MPN but not frank AML might harbor malignant clones capable of producing myeloid leukemias if introduced into healthy recipient mice. To test this idea, spleen and/or bone marrow cells from moribund URC mice with MPN were transplanted to sublethally irradiated mice, and the recipients were monitored for up to 1 y after transplantation for development of AML (Fig. 4A). None of the animals receiving transplanted cells from Runx1F/F; Mx1-Cre mice (n = 11) developed AML or other hematopoietic malignancies. However, all recipients of spleen and/or bone marrow cells from 2 of the 11 ENU-treated URC donors (cases 2 and 3 in Fig. 4 B and D and SI Appendix, Fig. S9) died with AML 12 or 42 wk after transplantation (Fig. 4E). Several hematopoietic compartments, including blood, bone marrow, and spleen, as well as other tissues such as the liver, were filled with immature c-Kit+ hematopoietic precursors in the recipient mice but not in the donor mice (Fig. 4D and SI Appendix, Fig. S9 B and C). The c-Kit+ cells were GFP+ (SI Appendix, Fig. S9 D and E), confirming that they were derived from the donor mice. Results from secondary transplants using spleen cells from recipient mice confirmed that the malignancies were transplantable (SI Appendix, Fig. S9 F and G). We refer to the AMLs derived from cases 2 and 3 as “late AMLs” to distinguish them from the 1° AML observed in one ENU-treated URC mouse (case 1).
In summary, we observed a total of three cases of AML arising from the bone marrow of URC mice after ENU treatment (Fig. 4B). In contrast, AML did not appear in any of the ENU-treated mice with one gene alteration [i.e., only U2af1(S34F) or only deletion of exon 4 of Runx1], in recipients of bone marrow transplants from such animals, or in mice that did not receive ENU. These findings are consistent with the idea that initiation of AML in our cohort requires U2af1(S34F), Runx1 deficiency, and treatment with ENU. However, due to the low incidence of AML (in 3 of 16 URC mice), a larger cohort would be necessary to establish the requirement of all three of these factors, including mutant U2af1, for leukemogenesis in a statistically significant fashion.
Identification of Probable Leukemogenic ENU-Induced Mutations by WES of DNA from Cases 1–3.
We performed WES on DNA from GFP+Lin−c-Kit+ (LK) cells from the spleens of cases 1, 2, and 3 mice to seek additional genetic changes caused by ENU. DNA from GFP+CD3ε+ or GFP+CD19+ lymphocytes from each of the three affected URC mice was used to determine a reference sequence. In each leukemic sample, we identified more than 1,000 somatic variants by WES (SI Appendix, Table S5). The most common variants were single-nucleotide substitutions, among which G-to-A (or C-to-T) transitions, A-to-G (or T-to-C) transitions, and G-to-T (or C-to-A) transversions were the most frequent (SI Appendix, Fig. S10A). These changes are consistent with previously reported effects of ENU (SI Appendix, Table S6).
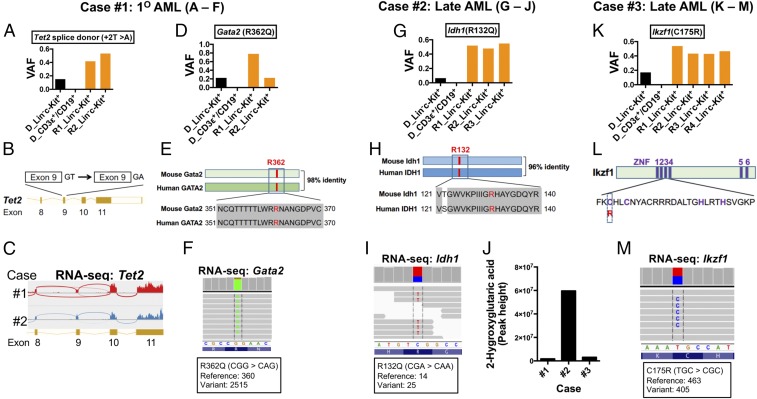
Case 1: By reviewing the variants for likely participants in leukemogenesis in primary AML cells from case 1, we identified a splice donor site mutation in Tet2 that is predicted to disrupt correct splicing and a missense mutation in Gata2 homologous to the GATA2(R362Q) mutation found in human AML (Fig. 5 A–F).
Fig. 5.
Acquired somatic mutations in AML cells in cases 1–3 affecting mouse homologs of human cancer genes. DNA was prepared from donor-derived (i.e., GFP+) Lin−c-Kit+ cells and lymphocytes (CD3ε+ or CD19+) from the spleens of donor mice (cases 1–3) and matched transplant-recipient mice and was used for WES to identify somatic mutations. RNA-seq and metabolic analysis from some samples were performed to confirm the WES findings. (A–F) Acquired mutations in Tet2 and Gata2 in AML cells from case 1. (A and D) VAFs for Tet2 and Gata2 mutations in the indicated cells from the donor (D) and transplant recipient (R) mice. (B and C) The T-to-A mutation in Tet2 disrupts a splice donor site at the 3′ end of exon 9, producing transcripts in which exon 9 is omitted as shown by RNA-seq in the Sashimi plot in C. (E) Comparison of mouse Gata2 and human GATA2 proteins, highlighting in red the conserved arginine residue that was changed to a glutamine residue by the missense mutation. (F) Confirmation of the Gata2(R362Q) mutation by mRNA sequencing from AML cells from a recipient mouse. (G–J) An Idh1(R132Q) mutation in AML cells from case 2. (G) VAFs for the R132Q allele in DNA from donor and recipient mice. (H) Position of the mutated amino acid residue in mouse Idh1 and human IDH1 proteins. (I) Confirmation of Idh1(R132Q) by sequencing of RNA from AML in a recipient mouse. (J) AML cells with Idh1(R132Q) had elevated levels of 2-hydroxyglutarate, as measured by LC-MS/MS. (K–M) An Ikzf1(C175R) mutation in case 3. (K) VAFs of the C175R allele in donor and recipient mice. (L) Diagram illustrating the position of C175 in the third zinc finger (ZNF) domain of Ikzf1. (M) Confirmation of the Ikzf1(C175R) mutation by sequencing RNA from AML cells from a recipient mouse. Additional characterizations of the acquired mutations are presented in SI Appendix, Fig. S10 and Tables S5 and S7.
Case 2: In the leukemic cells from case 2, we found a missense point mutation in Idh1 homologous to human IDH1(R132Q) (Fig. 5 G–J).
Case 3: In the leukemia cells from case 3, we found a missense mutation affecting a critical cysteine residue in a C2H2 zinc finger domain of the transcription factor Ikzf1 (C175R) (Fig. 5 K–M).
In general, the variant allele frequencies (VAFs) of these potentially pathogenic variants were low (0.2 or less) in the donor LK cells, absent in the donor’s normal lymphocytes, and near or even above 0.5 in the LK cells recovered from the recipient mice that had developed AML (Fig. 5 A, D, G, and K).
We next validated the presence of these variants and others by deep sequencing of several DNA samples from leukemic cells from cases 1–3, using a focused panel of cancer-related genes (SI Appendix, Table S7). This approach detected additional variants at low VAFs that are likely to be subclonal (SI Appendix, Fig. S10 B and C). We further confirmed the expression or functional consequences of these variants by mRNA sequencing. For example, the splice-site mutation in Tet2 resulted in exon skipping (Fig. 5C); the Gata2(R362Q), Idh1(R132Q), and Ikzf1(C175R) mutations were observed in mRNA (Fig. 5 F, I, and M); and Idh1(R132Q)-expressing cells had high levels of 2-hydroxyglutarate (Fig. 5J). These identified variants are likely drivers of leukemogenesis (see Discussion for details).
The U2af1(S34F) Allele Is Absent in AML Cells Derived from Cases 2 and 3.
Encountering mutations in these mouse AMLs that are identical or similar to mutations frequently reported in human AML was surprising, but analysis of the DNA-sequencing results revealed one additional surprise: U2af1(S34F) was absent in the DNA of LK cells from the recipients of transplants from cases 2 and 3 that developed late AML (SI Appendix, Fig. S10 F and G, Top). This was true although Cre-mediated recombination was complete in the late AML cells and there was no evidence of focal deletions by PCR (SI Appendix, Fig. S10 F and G, Bottom and SI Appendix, Fig. S10H). The LK cells from these recipients were also GFP+ (SI Appendix, Fig. S9 D and E) and showed complete deletion of the floxed exon 4 in the Runx1F alleles (SI Appendix, Fig. S10D), further confirming that these cells originated from the donors. The absence of U2af1(S34F) was also confirmed by mRNA sequencing (SI Appendix, Fig. S11A). In contrast, U2af1(S34F) was present in LK cells from the donor case 2 and 3 mice (albeit at a lower VAF), in matched lymphocytes, and in the LK cells from the case 1 mouse with primary AML (SI Appendix, Fig. S10 E–G). Moreover, the AML cells from case 1, but not those from cases 2 and 3, exhibited U2af1(S34F)-associated splicing changes in the consensus sequences preceding the altered cassette exons (SI Appendix, Fig. S11B), providing functional evidence for the presence or absence of U2af1(S34F) in these samples. These findings are considered further in Discussion.
Discussion
Here we report the establishment of genetically modified mouse strains that express a commonly observed mutant allele, U2af1(S34F), from the endogenous U2af1 locus in a Cre-dependent manner (Fig. 1A and SI Appendix, Fig. S2A). We show that production of U2af1(S34F) from one of the two U2af1 loci in heterozygous (S34F/WT) mice alters pre-mRNA splicing in mouse cells in ways similar to those observed in U2AF1(S34F)-expressing human cells, as previously reported by us and others (Fig. 1 B–E and SI Appendix, Figs. S1D and S2E and Table S3) (11, 21–24, 27). In the heterozygous mice, hematopoiesis is impaired, producing multilineage cytopenia and dysplasia (Figs. 2 and 3 and SI Appendix, Figs. S4–S6); these features are hallmarks of human MDS, a class of disorders in which mutant U2AF1, including U2AF1(S34F), or other mutant splicing factors are often found.
We also observed myeloid leukemogenesis in 3 of 16 mice in which U2af1(S34F) is combined with homozygous deletion of Runx1 and exposure to a mutagen, ENU, but not in mice lacking any one of those three factors (Fig. 4 and SI Appendix, Fig. S9). Sequencing of DNA from the three AMLs identified additional mutations associated with human AML (Fig. 5 and SI Appendix, Fig. S10 and Tables S5 and S7). However, the U2af1(S34F) missense mutation was absent in two of the three AML cases (SI Appendix, Figs. S10 F–H and S11), suggesting that U2af1(S34F) may be involved in the initiation but not the maintenance of AML.
U2af1(S34F) Changes RNA Processing in Ways both Common to and Different from Changes Observed in Human Cells Carrying U2AF1(S34F).
Specific alterations in pre-mRNA splicing are the most widely characterized consequences of expressing U2AF1(S34F) in human cells (11, 21–24, 27). In this report, we show that U2af1(S34F) causes similar but not identical changes in mRNA splicing in mouse fibroblasts and MP cells. Less than 5% of cassette exons show at least a 10% change in rates of inclusion in mouse cells expressing U2af1(S34F) (Fig. 1E and SI Appendix, Figs. S1D and S2E), numbers similar to those we previously observed in human cells expressing U2AF1(S34F) (11, 21). Moreover, the consensus 3′ splice-site sequences that precede the affected exons in mouse and human cells are very similar or identical. These similarities likely reflect the highly conserved sequence of U2AF1 protein and other components of the spliceosome machinery. For example, human and mouse U2AF1 proteins are nearly (more than 99%) identical, and the S34F mutations probably have similar effects on the two homologous proteins, recognizing highly similar 3′ splice sites in mouse and human transcriptomes.
Despite the similar effects of S34F mutations on sequence recognition and genes of altered splicing, the mouse mRNAs altered by U2af1(S34F) are not necessarily the homologs of the human mRNAs altered by U2AF1(S34F) (Fig. 1 B and D). Similar observations have been made previously in transgenic mice expressing U2AF1(S34F) (27) and in mice expressing other mutant splicing factors (31–34). Despite many differences (roughly 20% of genes with mRNAs significantly affected by the S34F mutation in mouse and human cells are homologs), a few of the genes with altered mRNAs in both species are often mutated in human MDS and AML or are partners in gene fusions in myeloid disorders (PICALM, GNAS, KDM6A, and BCOR) (35–38). Another gene (RAC1) plays an important role in HSCs (39, 40), and another (H2AFY) encodes isoforms reported to affect erythroid and granulomonocytic differentiation (41). It remains to be determined, however, whether any of the changes in spliced mRNAs from these genes has a role in the hematologic abnormalities we have observed in our U2af1(S34F) mouse model.
Mutant U2AF1 has also been claimed to cause neoplasia by compromising mitophagy through alternative polyadenylation (APA) of ATG7 mRNA (25) or by increasing the formation of R-loops (26). While we observed no evidence of effects of U2af1(S34F) on Atg7 APA selection in mouse bone marrow cells (SI Appendix, Fig. S1G), it remains to be determined whether R-loop formation is enhanced in mouse cells containing U2af1(S34F).
U2af1(S34F) Confers MDS-Like Features on Mouse Hematopoiesis but Does Not Cause Full-Fledged MDS.
We have found that expression of U2af1(S34F) from the endogenous locus in our mice impairs hematopoiesis, with features reminiscent of human MDS (Figs. 2 and 3 and SI Appendix, Figs. S4–S6). We also note similarities and differences in the hematopoietic phenotypes in our mice and the phenotypes in a previously reported transgenic mouse model (27), in which U2AF1(S34F) is driven by a tetracycline-responsive element in the Col1a1 locus. Cytopenia is a common feature of both models, but we observed cytopenia in all leukocyte lineages, whereas in the transgenic model, the affected cell populations are restricted to monocytes and B cells. Both models manifest defective HSC function, since they compete poorly with cotransplanted normal HSCs in irradiated recipients. However, we observed decreased numbers of LSK and HSC cells in our mice, while the number of LSK cells was increased in the transgenic model. Furthermore, our mice developed mild macrocytic anemia and low-grade myelodysplasia, which were not present in the transgenic model. Factors contributing to these differences might include the levels of expression of the mutant proteins (mutant and WT proteins are at equal levels in our model, but amounts may differ in the transgenic model) and the methods for inducing the mutant protein [transient treatment with poly (IC) to activate the Mx1-Cre transgene in our mice as opposed to the addition of doxycycline to activate the reverse tetracycline transactivator (rtTA) transcriptional regulator in the transgenic model].
Mice engineered to produce mutant splicing factors other than mutant U2AF1 (31–34) display similar features, including cytopenia and a competitive disadvantage of mutant-containing HSCs in repopulation assays (reviewed in refs. 42 and 43). Therefore, different mutant splicing factors may elicit effects on hematopoiesis that are both common to splicing factor mutations and specific for certain factors.
Regardless of the hematopoietic phenotypes produced by different mutant splicing factors, none of the mouse models (including ours) recapitulates all aspects of human MDS. These results suggest that a mutant splicing factor is insufficient to drive human MDS on its own. Similarly, expression of an MDS-associated SRSF2 mutant in human induced pluripotent stem cells did not affect the production of hematopoietic progenitor cells and only mildly affected hematopoiesis (44). In contrast, a much more drastic phenotype was produced by deletion of chromosome 7q, another common but much more extensive genetic alteration in MDS. These observations are consistent with findings that mutant U2AF1 is often associated with other genetic alterations, such as deletion of chromosome 20q and mutations in ASXL1, DNMT3A, TET2, and RUNX1 in human MDS (4, 6, 7, 12, 16, 17, 45).
Beyond MDS, the same mutations in U2AF1 or other splicing factor genes are found in apparently healthy individuals with so-called “clonal hematopoiesis of indeterminate potential” (CHIP) (46). HSCs from individuals with CHIP often carry loss-of-function mutations in DNMT3A and TET2 (47, 48). The combination of U2af1(S34F) and loss of Dnmt3A or Tet2 might produce hematopoietic phenotypes in mice that more precisely resemble human diseases. The occurrence of a Tet2 splice-site mutation in a case of AML in our mouse model (Fig. 5 A–C) may reflect such cooperativity between mutant U2af1 and loss of Tet2.
Does U2af1(S34F) Promote the Initiation of Myeloid Leukemia?
We have pursued the hypothesis that additional genetic changes are necessary to produce severe, progressive hematological disease in mice expressing U2af1(S34F) in the blood lineage. Although a larger cohort is necessary to reach statistical significance and confirm these findings, our results are consistent with the idea that U2af1(S34F) plays a necessary but not sufficient role in leukemogenesis in this setting. The results are also consistent with clinical observations that somatic mutation of U2AF1 is a risk factor for AML in MDS patients (6, 12), in individuals with age-related clonal hematopoiesis (49), and in apparently healthy individuals (50).
We were surprised to find that in two cases of AML—the two detected after bone marrow transplantation—the leukemic cells contained only U2af1 RNA with a WT coding sequence (SI Appendix, Figs. S10 F–H and S11). These cells retained the Cre-recombined MGS34F allele (SI Appendix, Fig. S10 F–H), but codon 34 was composed of TCT (serine) rather than TTT (phenylalanine). A T-to-C transition might have occurred during exposure to ENU treatment or randomly during the leukemogenic process and conferred a selective advantage of the kind demonstrated in the competitive transplantation experiments shown in Fig. 3. Regardless of the mechanism and significance of the loss of U2af1(S34F), our results suggest that U2af1(S34F) is dispensable for the maintenance of murine AML in this setting. This is consistent with our previous observation that U2AF1(S34F) is dispensable for the growth of established human lung adenocarcinoma cell lines carrying this mutation (11).
ENU-Induced Mutations That Likely Cooperate with U2af1(S34F) and Runx1 Deletion in Leukemogenesis.
Exome sequencing of leukemic cells from the three cases of AML derived from our mouse model identified at least four somatic mutations that are likely to have contributed functionally to leukemogenesis (Fig. 5):
-
i)
The Tet2 variant in AML case 1 affects a splice donor site of a constitutive exon (exon 9, as shown in Fig. 5B) so that exon 9 is skipped during splicing (Fig. 5C). Loss of exon 9 (138 bp in length) does not change the reading frame in the Tet2 coding sequence, but loss of the 2-oxoglutarate-Fe(II)–dependent dioxygenase domain, partly encoded by exon 9, likely eliminates normal Tet2 function.
-
ii)
The murine Gata2 variant in case 1 AML is equivalent to GATA2(R362Q) (Fig. 5E), which is one of the recurrent GATA2 mutations in AML [Catalog of Somatic Mutations in Cancer (COSMIC) ID: COSM87004] (refs. 51 and 52). GATA2 encodes a transcription factor that plays a critical role during hematopoiesis.
-
iii)
The Idh1 mutation identified in one of the late AML cases (case 2) is equivalent to human IDH1(R132Q) (Fig. 5H). R132 mutants of IDH1, including IDH1(R132Q), produce the onco-metabolite 2-hydroxyglutarate (Fig. 5J) that is commonly elevated in several human cancers including AML (53).
-
iv)
The variant allele Ikzf1(C175R), found in one case of late AML (case 3), affects a conserved cysteine in a zinc finger domain of the transcription factor it encodes (Fig. 5L) and thus likely compromises (or alters) the DNA-binding capacity of Ikzf1. IKZF1, a known tumor-suppressor gene for lymphoid neoplasms (54), is also recurrently deleted in MPN (55) and pediatric AML (56). In combination with our results, it seems likely that IKZF1 is a tumor-suppressor gene for myeloid cancers as well.
These likely driver mutations from the three AML cases in our mice occurred largely independently, but some appear to be functionally related, implying that they may affect hematopoietic cell functions that must be altered to produce murine AML. For example, mutant IDH1 is known to inhibit TET2 enzyme activity (57), and Gata2 and Ikzf1 (as well as Runx1) are transcription factors critical for hematopoietic lineage specification and/or HSC function. Mutant U2AF1 might cooperate with alterations of the epigenetic modifiers and lineage transcription factors to initiate leukemogenesis.
Materials and Methods
Generation of Mice, MEFs, and Related Procedures.
Details for generating the U2af1(S34F) conditional knock-in mice and related MEFs can be found in SI Appendix. All alleles except Runx1F were used in a heterozygous (or hemizygous) state in this study. Mice carrying the U2af1-targeted alleles were all backcrossed to B6 mice for at least three generations before they were used for the reported experiments and were considered to have a mixed B6,129 background. Littermates of animals with experimental genotypes were used as controls in all described studies. Mice carrying the MGS34F allele are available at The Jackson Laboratory as stock no. 032638.
All procedures involving mice were approved by the Institutional Animal Care and Use Committees at the National Human Genome Research Institute (NHGRI) (protocols G-13-1 and G-95-8) and at Weill Cornell Medicine (WCM) (protocol 2015-017). A brief description of these procedures can be found in SI Appendix.
Cell Culture, Reagents, and Assays.
MEFs.
Subconfluent cells were treated with 4OHT (Sigma-Aldrich) at the indicated doses for 24 h to induce Cre activity. The drug was then removed, and cells were cultured for an additional 48 h before harvest. In other instances, the same treatment was repeated before harvest to improve the efficacy of Cre-mediated recombination.
Colony-forming cell assays.
Thirty thousand nucleated bone marrow cells were seeded in MethoCult GF M3434 medium (STEMCELL Technologies) in a 35-mm dish in triplicate, according to the vendor’s instruction. The number of colonies, including erythroid (BFU-E), granulocyte-monocyte (CFU-GM), and multipotential (CFU-GEMM) colonies, were counted 14 d later (or 7 d later when secondary plating was performed). When secondary plating was performed, primary colony-forming cell (CFC) cultures from each set of triplicate cultures were harvested on day 8, pooled, and washed with Iscove’s Modified Dulbecco’s Medium (STEMCELL Technologies) containing 2% FBS. Thirty thousand or ten thousand nucleated cells were again seeded in a 35-mm dish with MethoCult GF M3434 medium in triplicate. The colonies were counted 12 d later. CFC assays were performed either as a contracted service (STEMCELL Technologies) or by the investigators.
RNA and RT-qPCR.
RNA and RT-qPCR were performed as previously described (11). A brief description of these procedures can be found in SI Appendix.
Cell-surface and intracellular staining and flow cytometry.
Cell-surface staining was conducted as previously described (58). RBCs in blood, bone marrow, or spleen cell suspensions were lysed using ACK lysing buffer (Quality Biological). Descriptions of the antibodies and procedures can be found in SI Appendix.
Metabolite quantification by LC-MS/MS.
Two million live splenocytes from case 1–3 recipient mice at the moribund stage were used to quantify soluble metabolite by LC-MS/MS at the Proteomics and Metabolics core laboratory (WCM) as a paid service.
High-Throughput Nucleotide Sequencing.
mRNA sequencing.
mRNA sequencing (RNA-seq) was conducted at either the Sequencing Facility (National Cancer Institute) or the Genome Resources Core Facility (WCM). RNA quality was assessed by a Bioanalyzer (Agilent). Total RNA with good integrity values (RNA integrity number >9.0) was used for poly(A) selection and library preparation using the Illumina TruSeq RNA library preparation kit. The MEF samples were run on a HiSeq 2500 instrument and were sequenced to the depth of 100 million paired-end 101-bp reads per sample. All other samples were run on a HiSeq 4000 instrument and were sequenced to the depth of 50 million paired-end 51-bp reads per sample. These RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE112174).
WES.
WES was performed in the WCM Genome Resources Core Facility. DNA quality was assessed with a Bioanalyzer (Agilent). A prehybridization library was prepared using the KAPA LTP library preparation kit (Kapa Biosystems). Targets were enriched by NimbleGen SeqCap EZ Exome Library v2.0 (Roche NimbleGen). Up to 18 samples were sequenced in one HiSeq 4000 lane to generate paired-end 101-bp reads.
Bioinformatics.
Differential gene-expression and splicing patterns were analyzed as described (11, 21). A brief description of these procedures can be found in SI Appendix.
For the analysis of WES data, raw sequencing reads were edited to remove low-quality bases, and adapter sequences were identified using cutadapt (59). The processed reads were then mapped to the mouse GRCm38 reference genome using Burrows–Wheeler Aligner (60). The alignments were further processed for local realignment and base quality score recalibration using Gene Analysis Toolkit (GATK) software (Broad Institute) (61). Somatic single-nucleotide variants and small insertions and deletions (indels) were detected using Varscan2 (62).
Statistics.
Statistical significance was determined by two-tailed Student’s t test using GraphPad Prism 6 or as otherwise stated. In all analyses, P values ≤ 0.05 are considered statistically significant.
Supplementary Material
Acknowledgments
We thank Sukanya Goswami, Shao Ning Yang, and Guoan Zhang (WCM); Hayley Motowski, Jackie Idol, Danielle Miller-O’Mard, Ursula Harper, Amalia Dutra, Evgenia Pak, Stacie Anderson, Martha Kirby, David Bodine, Shelley Hoogstraten-Miller, Guadalupe Lopez, Cecilia Rivas, and other members of the transgenic mouse core at the NHGRI for exceptional technical assistance; Nancy Speck (University of Pennsylvania) for providing the conditional Runx1-knockout mice; Devin Koestler (University of Kansas Medical Center) for consultation on statistical analysis; and members of the H.V. and the P.L. laboratories, Timothy A. Graubert, Pavankumar N. G. Reddy, and Borja Saez (Massachusetts General Hospital), Matthew J. Walter and Cara Lunn Shirai (Washington University in St. Louis), and Dan Larson, Murali Palangat, and Markus Hafner (NIH) for helpful discussions during the course of the study. H.V. and D.L.F. were supported by the Intramural Program at the NIH and are now supported by the Meyer Cancer Center at WCM and the Edward P. Evans Foundation. P.L., T. Zhen, and L.G. are supported by the Intramural Program of NHGRI, NIH.
Footnotes
The authors declare no conflict of interest.
Data deposition: RNA-sequencing data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE112174).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812669115/-/DCSupplemental.
References
- 1.Catenacci DVT, Schiller GJ. Myelodysplasic syndromes: A comprehensive review. Blood Rev. 2005;19:301–319. doi: 10.1016/j.blre.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Troy JD, Atallah E, Geyer JT, Saber W. Myelodysplastic syndromes in the United States: An update for clinicians. Ann Med. 2014;46:283–289. doi: 10.3109/07853890.2014.898863. [DOI] [PubMed] [Google Scholar]
- 3.Arber DA, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 5.Papaemmanuil E, et al. Chronic Myeloid Disorders Working Group of the International Cancer Genome Consortium Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graubert TA, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet. 2011;44:53–57. doi: 10.1038/ng.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damm F, et al. Groupe Francophone des Myélodysplasies Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood. 2012;119:3211–3218. doi: 10.1182/blood-2011-12-400994. [DOI] [PubMed] [Google Scholar]
- 8.Merendino L, Guth S, Bilbao D, Martínez C, Valcárcel J. Inhibition of msl-2 splicing by sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 9.Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 10.Zorio DA, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
- 11.Fei DL, et al. Wild-type U2AF1 antagonizes the splicing program characteristic of U2AF1-mutant tumors and is required for cell survival. PLoS Genet. 2016;12:e1006384. doi: 10.1371/journal.pgen.1006384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thol F, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119:3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- 13.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550, and erratum Nature (2014) 2622. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waterfall JJ, et al. High prevalence of MAP2K1 mutations in variant and IGHV4-34-expressing hairy-cell leukemias. Nat Genet. 2014;46:8–10. doi: 10.1038/ng.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu L, et al. Genomic landscape of CD34+ hematopoietic cells in myelodysplastic syndrome and gene mutation profiles as prognostic markers. Proc Natl Acad Sci USA. 2014;111:8589–8594. doi: 10.1073/pnas.1407688111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adema V, et al. U2AF1 mutations in S34 and Q157 create distinct molecular and clinical contexts. Blood. 2016;128:3155. [Google Scholar]
- 18.Schaefer BC, Schaefer ML, Kappler JW, Marrack P, Kedl RM. Observation of antigen-dependent CD8+ T-cell/dendritic cell interactions in vivo. Cell Immunol. 2001;214:110–122. doi: 10.1006/cimm.2001.1895. [DOI] [PubMed] [Google Scholar]
- 19.Ruzankina Y, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 21.Ilagan JO, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res. 2015;25:14–26. doi: 10.1101/gr.181016.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okeyo-Owuor T, et al. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia. 2015;29:909–917. doi: 10.1038/leu.2014.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brooks AN, et al. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS One. 2014;9:e87361. doi: 10.1371/journal.pone.0087361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Przychodzen B, et al. Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood. 2013;122:999–1006. doi: 10.1182/blood-2013-01-480970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park SM, et al. U2AF35(S34F) promotes transformation by directing aberrant ATG7 pre-mRNA 3′ end formation. Mol Cell. 2016;62:479–490. doi: 10.1016/j.molcel.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen L, et al. The augmented R-loop is a unifying mechanism for myelodysplastic syndromes induced by high-risk splicing factor mutations. Mol Cell. 2018;69:412–425.e6. doi: 10.1016/j.molcel.2017.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shirai CL, et al. Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell. 2015;27:631–643. doi: 10.1016/j.ccell.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Growney JD, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichikawa M, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10:299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 30.Castilla LH, et al. The fusion gene Cbfb-MYH11 blocks myeloid differentiation and predisposes mice to acute myelomonocytic leukaemia. Nat Genet. 1999;23:144–146. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- 31.Kim E, et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27:617–630. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kon A, et al. Physiological Srsf2 P95H expression causes impaired hematopoietic stem cell functions and aberrant RNA splicing in mice. Blood. 2018;131:621–635. doi: 10.1182/blood-2017-01-762393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obeng EA, et al. Physiologic expression of Sf3b1(K700E) causes impaired erythropoiesis, aberrant splicing, and sensitivity to therapeutic spliceosome modulation. Cancer Cell. 2016;30:404–417. doi: 10.1016/j.ccell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mupo A, et al. Hemopoietic-specific Sf3b1-K700E knock-in mice display the splicing defect seen in human MDS but develop anemia without ring sideroblasts. Leukemia. 2017;31:720–727. doi: 10.1038/leu.2016.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Caudell D, Aplan PD. The role of CALM-AF10 gene fusion in acute leukemia. Leukemia. 2008;22:678–685. doi: 10.1038/sj.leu.2405074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bejar R, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jankowska AM, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118:3932–3941. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Damm F, et al. BCOR and BCORL1 mutations in myelodysplastic syndromes and related disorders. Blood. 2013;122:3169–3177. doi: 10.1182/blood-2012-11-469619. [DOI] [PubMed] [Google Scholar]
- 39.Gu Y, et al. Hematopoietic cell regulation by Rac1 and Rac2 guanosine triphosphatases. Science. 2003;302:445–449. doi: 10.1126/science.1088485. [DOI] [PubMed] [Google Scholar]
- 40.Cancelas JA, et al. Rac GTPases differentially integrate signals regulating hematopoietic stem cell localization. Nat Med. 2005;11:886–891. doi: 10.1038/nm1274. [DOI] [PubMed] [Google Scholar]
- 41.Yip BH, et al. The U2AF1S34F mutation induces lineage-specific splicing alterations in myelodysplastic syndromes. J Clin Invest. 2017;127:3557. doi: 10.1172/JCI96202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inoue D, Abdel-Wahab O. Modeling SF3B1 mutations in cancer: Advances, challenges, and opportunities. Cancer Cell. 2016;30:371–373. doi: 10.1016/j.ccell.2016.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saez B, Walter MJ, Graubert TA. Splicing factor gene mutations in hematologic malignancies. Blood. 2017;129:1260–1269. doi: 10.1182/blood-2016-10-692400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang C-J, et al. Dissecting the contributions of cooperating gene mutations to cancer phenotypes and drug responses with patient-derived iPSCs. Stem Cell Reports. 2018;10:1610–1624. doi: 10.1016/j.stemcr.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papaemmanuil E, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627; quiz 3699. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steensma DP, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaiswal S, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Genovese G, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abelson S, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559:400–404. doi: 10.1038/s41586-018-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desai P, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat Med. 2018;24:1015–1023. doi: 10.1038/s41591-018-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luesink M, et al. High GATA2 expression is a poor prognostic marker in pediatric acute myeloid leukemia. Blood. 2012;120:2064–2075. doi: 10.1182/blood-2011-12-397083. [DOI] [PubMed] [Google Scholar]
- 52.Yan X-J, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 53.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mullighan CG, et al. Children’s Oncology Group Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jäger R, et al. Deletions of the transcription factor Ikaros in myeloproliferative neoplasms. Leukemia. 2010;24:1290–1298. doi: 10.1038/leu.2010.99. [DOI] [PubMed] [Google Scholar]
- 56.de Rooij JDE, et al. Recurrent deletions of IKZF1 in pediatric acute myeloid leukemia. Haematologica. 2015;100:1151–1159. doi: 10.3324/haematol.2015.124321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhen T, et al. Chd7 deficiency delays leukemogenesis in mice induced by Cbfb-MYH11. Blood. 2017;130:2431–2442. doi: 10.1182/blood-2017-04-780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. [Google Scholar]
- 60.Li H. March 16, 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2.
- 61.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koboldt DC, et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ley TJ, et al. Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.