Significance
Although MARF1 (meiosis regulator and mRNA stability factor 1) is an ancient protein, identification of its function in mammalian female germ cell development and fertility is recent. It is crucial for the progression of oocyte meiosis and defense against the ravages of retrotransposons, which can cause damage to the oocyte’s genome. These processes are dependent upon the ability of MARF1 to act alone both to bind RNA and to function as a ribonuclease during oogenesis. Here we reveal the molecular structure and functional mechanisms that enable MARF1 activity and provide insight into the complex posttranscriptional processes that shape the oocyte transcriptome.
Keywords: MARF1, RNase, oocyte meiosis, crystal structure, female infertility
Abstract
Producing normal eggs for fertilization and species propagation requires completion of meiosis and protection of the genome from the ravages of retrotransposons. Mutation of Marf1 (meiosis regulator and mRNA stability factor 1) results in defects in both these key processes in mouse oocytes and thus in infertility. MARF1 was predicted to have ribonuclease activity, but the structural basis for the function of MARF1 and the contribution of its putative ribonuclease domain to the mutant oocyte phenotype was unknown. Therefore, we resolved the crystal structures of key domains of MARF1 and demonstrated by biochemical and mutagenic analyses that the ribonuclease activity of MARF1 controls oocyte meiotic progression and retrotransposon surveillance. The N-terminal NYN domain of MARF1 resembles the nuclease domains of Vpa0982, T4 RNase H, and MCPIP1 and contains four conserved aspartate residues, D178, D215, D246, and D272. The C-terminal LOTUS domain of MARF1 adopts a winged helix-turn-helix fold and binds ssRNA and dsRNA. Purified MARF1 cleaved ssRNAs in vitro, but this cleavage activity was abolished by mutations of conserved aspartates in its NYN domain and truncation of the LOTUS domain. Furthermore, a point mutation in the D272 residue in vivo caused a female-only infertile phenotype in mice, with failure of meiotic resumption and elevation of Line1 and Iap retrotransposon transcripts and DNA double-strand breaks in oocytes. Therefore, the ribonuclease activity of MARF1 controls oocyte meiosis and genome integrity. This activity depends upon conserved aspartic residues in the catalytic NYN domain and the RNA-binding activity of the LOTUS domain.
The objective of germline development in mammals is to produce normal gametes for fertilization and stable transmission of genetic information to the next generation. This requires that germ cells complete meiosis to accomplish recombination and segregation of homologous chromosomes and maintain the integrity of their genome. The success of these key gametogenic processes is crucial for species propagation, survival, and diversification. Errors or failure in these processes can lead to infertility, miscarriage, birth defects, and genetic diseases. However, germ cells face a severe challenge from the derepression of retrotransposons during their early development when genome-wide DNA demethylation reprogramming takes place (1). To meet this challenge, germ cells evolved sophisticated defensive networks to protect their genome from retrotransposon-mediated DNA damage while also establishing the mechanisms of meiotic division. In mammalian species, a female germ cell-specific pathway governed by MARF1 (meiosis regulator and mRNA stability factor 1; formerly known as “meiosis arrest female 1”) enables completion of meiosis and retrotransposon silencing (2, 3). Alternatively, these key gametogenic processes in males require the evolutionarily conserved PIWI protein and the associated PIWI-interacting RNA (piRNA) pathway (4–19). Interruption of the PIWI–piRNA pathway by mutation of genes encoding PIWI protein and their interacting partners produces infertility in males but not in females (20–35). In contrast, mutation of Marf1 in mice causes female infertility without affecting males (3). Infertility of Marf1-mutant females is caused by ovulation of immature germinal vesicle (GV)-stage oocytes arrested at prophase I of the first meiotic division (3). Moreover, Marf1 mutations abrogate the normal silencing of retrotransposons Line1 and Iap, unleashing their attack on the oocyte genome (3). Therefore, at least in mice, the MARF1-mediated pathway in oocytes may function as a female counterpart to the male PIWI–piRNA pathway in spermatocytes.
The functional similarity between oocyte MARF1 and the spermatocyte PIWI–piRNA pathway might be based on potential structural analogies between the protein domains of MARF1 and those of PIWI proteins and their interacting partners, TDRD5/7. MARF1 contains three major domains, the N-terminal NYN domain belonging to PIN nuclease superfamily, two RRM domains, and a C-terminal repeat of LOTUS domains (Fig. 1A). Both the NYN and LOTUS domains are recently identified protein domains that are well conserved across eukaryotes and bacteria (36, 37). The NYN domain is predicted to be one major group of the PIN domain-like nucleases, which could resemble the RNA-slicing activity of the PIWI domain in PIWI proteins (37, 38). The LOTUS domain (also known as “OST-HTH”) is named after the germline proteins Limkain B1 (now MARF1), Oskar, and Tudor domain-containing proteins (TDRD) 5 and 7 owing to its presence in these proteins (36, 39). This LOTUS domain is predicted to bind to dsRNA or to stems of folded structures in RNAs, particularly those formed by small noncoding RNAs (snRNAs) after hybridizing with their RNA targets (36, 39). This RNA-binding property of the LOTUS domain equips it to be an “adapter” able to recruit targets for the effectors. Based on the copresence of the NYN RNase-like domain and the LOTUS RNA-binding domains in MARF1 protein, it has been hypothesized that MARF1 in oocytes may function as both an adaptor, like the TDRD5/7 in male spermatocytes, to recruit specific RNA targets, including those for retrotransposons Line 1 and Iap, and an effector, similar to the PIWI domain in PIWI proteins to catalyze the specific cleavage of target RNA (2). According to this hypothesis, the MARF1 protein alone in oocytes may be capable of performing the regulatory role played by multiple factors in the PIWI–piRNA pathway of spermatocytes. This model could explain the sexual dimorphic control of mammalian germ cell meiosis and retrotransposon surveillance. However, this notion has remained conjecture because the molecular properties and the function of the NYN and LOTUS domain in MARF1 were unclear. Moreover, there was no direct experimental evidence showing that the oocyte-expressed MARF1 has RNase activity or that the absence of this activity contributes to the described phenotype of mutant Marf1 oocytes. Here, we resolved the crystal structures of the NYN and LOTUS domains of MARF1 and demonstrated through biochemical and mutagenic analyses that MARF1 is indeed an RNase that controls oocyte meiotic progression and retrotransposon surveillance.
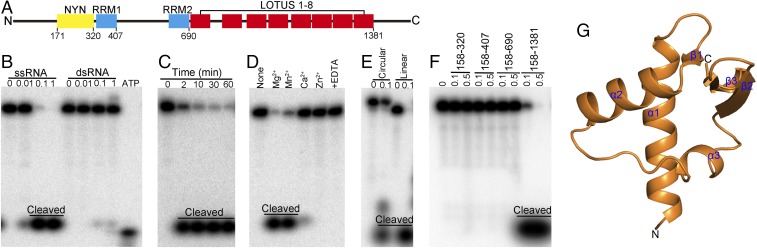
Fig. 1.
In vitro RNase activity assay of MARF1 and the crystal structure of the MARF1-LOTUS domain. (A) Schematic illustration of the domain architecture of M. musculus MARF1. It contains an N-terminal NYN domain (residues 171–320), two central RRM domains (residues 329–407 and 609–690 for RRM1 and -2, respectively), and eight C-terminal repeats of the LOTUS/OST-HTH domain (residues 691–1381). (B) Dose-dependent cleavage of ssRNA but not dsRNA by MARF1. The 5′ 32P-labeled 49-nt ssRNA or dsRNA was incubated with various doses (0.00–1.00 μM) of MARF1 at 37 °C for 5 min. Free [32P]ATP was loaded in the far right lane as a marker. (C) Time-dependent cleavage of ssRNA by MARF1. The 49-nt 32P-ssRNA was incubated with MARF1 (0.05 μM) at 37 °C for 1–60 min. (D) Effect of divalent cations and their chelator on the cleavage activity of MARF1. The 49-nt 32P-ssRNA was incubated with MARF1 (0.05 μM) in solution containing different bivalent cations and/or EDTA at 37 °C for 5 min. (E) The cleavage activity of MARF1 toward linear and circular ssRNA. The 49-nt 32P-ssRNA or circular ssRNA was incubated with or without 0.1 μM of MARF1 at 37 °C for 5 min. (F) RNase activity of different domains of MARF1.The 49-nt 32P-ssRNA was incubated with 0.1 or 0.5 μM of different domains of MARF1 at 37 °C for 5 min. (G) The structure of first LOTUS domain of MARF1.
Results and Discussion
Mouse MARF1 Cleaves ssRNA in Vitro.
Phylogenetic analysis indicates that MARF1 is highly conserved in mammals (SI Appendix, Fig. S1). To test whether mammalian MARF1 exhibits nuclease activity, we purified residues 158–1,381 of the recombinant Mus musculus MARF1 protein containing all the functional domains (Fig. 1A) and incubated it with various RNA substrates labeled with 32P at the 5′ end. MARF1 efficiently cleaved ssRNAs but had only weak cleavage activity toward dsRNAs of the same size (Fig. 1B). Moreover, this activity of MARF1 was dose- and time-dependent (Fig. 1 B and C), required the presence of specific divalent metal ions, i.e., Mg2+ or Mn2+, and was inhibited by EDTA (Fig. 1D). MARF1 cleaved circular RNA, although with weaker activity than toward the linear RNA substrate (Fig. 1E), indicating that it has exo- and endo-ribonuclease activities.
The RNA-Cleavage Activity of Mouse MARF1 Depends upon the RNA-Binding Activity of Its LOTUS Domains.
The RNA-cleaving activity of MARF1 was severely compromised when it was truncated to delete the C-terminal repeat of LOTUS domains (Fig. 1F). This suggests that the presence of LOTUS domains is critical for MARF1 to elicit the RNase function. Given that the originally defined function of the LOTUS domain was RNA binding, it is tempting to speculate that the LOTUS domain could facilitate the cleavage activity of MARF1 by recognizing and binding the RNA substrates of MARF1. To determine whether, and to what extent, the M. musculus MARF1-LOTUS domain binds RNA, EMSAs were performed on different versions of MARF1 proteins. The C-terminal repeat of LOTUS domains itself binds ssRNA and dsRNA substrates with affinities comparable to MARF1 (residues 158–1,381) (SI Appendix, Fig. S2). However, recent studies in Drosophila suggested that the LOTUS domain of Oskar, a maternal effect gene product essential for germ cell formation, does not bind RNAs. Instead, it functions as a conserved regulator for the DEAD-box RNA helicase VASA in a protein–protein interaction module (40, 41). To determine the structural basis for the RNA-binding ability of M. musculus MARF1-LOTUS domains, we resolved the crystal structure of the first repeat of MARF1’s C-terminal LOTUS domains (LOTUS1) (Fig. 1G and SI Appendix, Table S1). Sequence alignment and structural superimposition of the LOTUS domains from M. musculus MARF1 and other proteins revealed that the M. musculus MARF1-LOTUS domains are different from Oskar and lack the C-terminal extended helix required for protein interaction (SI Appendix, Fig. S3). This result is consistent with the recent observation made by Jeske et al. (40) in Drosophila and provides more structural basis for dividing the LOTUS domain into two subclasses—the extended (e) and minimal (m) LOTUS depending on the presence (eLOTUS) or absence (mLOTUS) of the C-terminal extension. Therefore, the data presented here indicate that LOTUS domains are implicated in substrate recognition and binding by MARF1 and are therefore indispensable for the RNase function of MARF1.
The RNase Activity of Mouse MARF1 Is Determined by the Conserved Aspartic Residues Present in Its Catalytic NYN Domains.
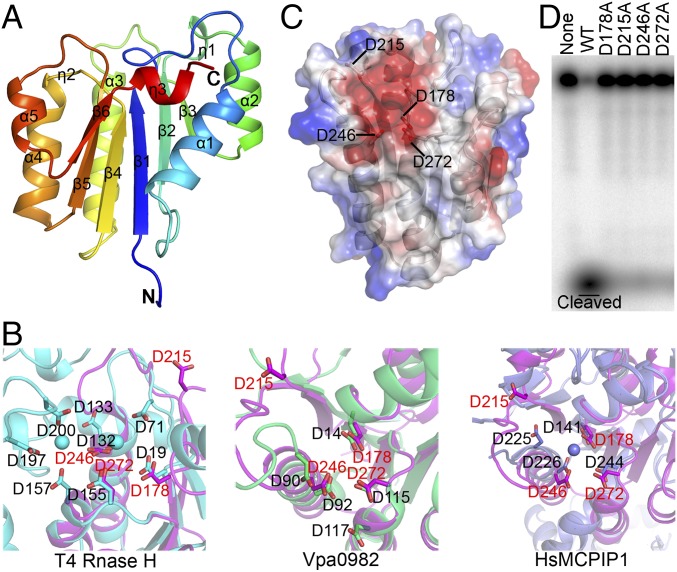
To gain mechanistic insight into the RNase activity of MARF1, we determined the crystal structure of the NYN domain (residues 158–320) at 1.75-Å resolution (SI Appendix, Table S1). The structure adopts an α/β/α sandwich fold, with β-strands 1–6 forming a parallel β-sheet surrounded by sets of α helices on both sides (Fig. 2A). Dali server analysis showed that MARF1-NYN resembled the nuclease domains of T4 RNase H [Protein Data Bank (PDB) ID code ITRF], Vpa0982 (PDB ID code 2QIP), and HsMCPIP1 (PDB ID code 3V32) (SI Appendix, Fig. S4). However, unlike these three proteins that contain an additional helix, MARF1-NYN has a loop formed after α1. This additional loop orthogonally faces the core β-sheet and α1-helix (Fig. 2A). Unlike the other two RNase H and HsMCPIP1 PIN-domain nucleases that have a helix insertion after β2, a structure tending to form a cap over the active site, the MARF1-NYN domain and Vpa0982 lack such a helix insertion after β2 (Fig. 2A and SI Appendix, Fig. S4), making the MARF1-NYN domain a relatively exposed active site.
Fig. 2.
Crystal structure and mutagenic analysis of the NYN domain of MARF1. (A) Cartoon diagram of the overall structure of the NYN domain. The secondary structure elements are labeled. (B) Superimposition of the conserved amino acids of the MARF1-NYN structure (magenta) on the PIN domain of T4 RNase H (cyan) (Left), VPa0982 (green) (Center), and MCPIP1(slate) (Right). (C) Electrostatic surface potential analysis of the NYN structure. Conserved residues D178, D215, D246, and D272 are in the negatively charged pocket. (D) In vitro RNase activity assay of WT and mutant MARF1 in which the conserved aspartate residues (D178, D215, D246, and D272) were mutated individually into alanine.
Sequence alignment and structure superimposition of the typical NYN domains with other PIN proteins revealed that the MARF1-NYN domain contains four conserved aspartates, D178, D215, D246, and D272 (Fig. 2B and SI Appendix, Fig. S5A). Three of them, D178, D215, and D272, are located at the C termini of strands β1, β2, and β4, respectively, and one acidic residue, D246, is at the N terminus of helix α3. Analysis of the electrostatic surface potential of the NYN domain revealed that these four aspartates are in a negatively charged pocket (Fig. 2C). These structural features and their conservation among the key nucleases suggest that these four aspartate residues could be critical for the nuclease activity of MARF1. We therefore tested this possibility by mutating these aspartate residues into alanine and assessing the RNA-cleavage activity of the resultant MARF1-mutant proteins. We first expressed and purified the mutant proteins (SI Appendix, Fig. S5B) and then individually tested their ability to carry out cleavage in vitro. Mutations at the residues of D178, D215, D246, and D272, respectively, caused failure of MARF1 to cleave the ssRNA substrate (Fig. 2C), demonstrating that these four conserved aspartates are indispensable for the catalytic activity of MARF1. In addition to these catalytic aspartates, two other amino acids in the NYN domain are also conserved: One is N181 located at the end of strand β1, and the other is S270 (SI Appendix, Fig. S5A). The N181 residue possibly undergoes a conformational change upon nucleic acid binding, while the hydroxyl group of S270 may form hydrogen bonds with acidic residues in the active site (37).
Mouse MARF1 Exhibits RNase Activity in Oocytes.
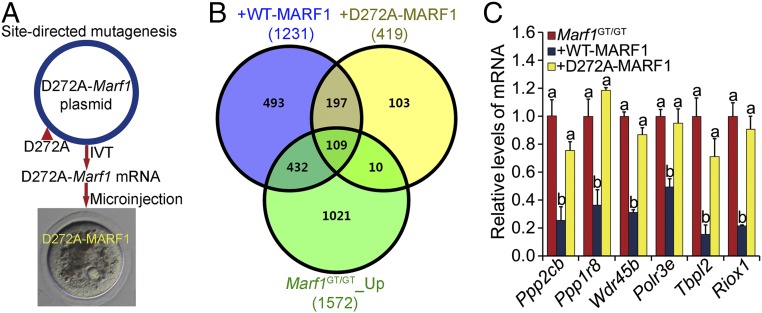
Since MARF1 is highly expressed and functions in oocytes, we tested whether it also exhibits RNase activity in oocytes. We microinjected mRNA encoding the WT and the D272A-mutated MARF1 protein (D272A-MARF1), respectively, into Marf1-genetrap (hereafter referred to as Marf1GT/GT) GV-stage oocytes isolated from large antral follicles and assessed their impact on the oocyte transcriptome during culture (Fig. 3A). Marf1GT/GT oocytes were used because they do not express the endogenous MARF1 protein and possess a cohort of up-regulated transcripts that are potential targets of MARF1 RNase activity. We hypothesized that supplementing the Marf1GT/GT oocytes with exogenous WT, but not mutated, MARF1 would reduce some, or all, of the transcripts that are expressed at higher than WT levels. Indeed, transcriptomic comparison by RNA-sequencing (RNA-seq) analysis demonstrated that supplementation with WT but not D272A-MARF1 caused profound changes in the transcriptome of the Marf1GT/GT oocytes. Steady-state levels of 925 transcripts were uniquely down-regulated by WT MARF1, while only 113 transcripts were uniquely reduced by D272A-MARF1 (Fig. 3B and Datasets S1 and S2). Nearly half (46.7%) of the 925 down-regulated transcripts were among the transcripts identified as up-regulated in the GV-stage oocytes of Marf1GT/GT mice (Fig. 3B). These included Ppp2cb, whose up-regulation causes meiotic arrest in the Marf1GT/GT oocytes (3), and others that were validated by real-time qRT-PCR (Fig. 3C). Since transcription in oocytes isolated from large antral follicles is normally silent (42), the decrease in the levels of these 925 transcripts in Marf1GT/GT oocytes after overexpression of WT MARF1 is most likely attributed to their direct degradation by MARF1.
Fig. 3.
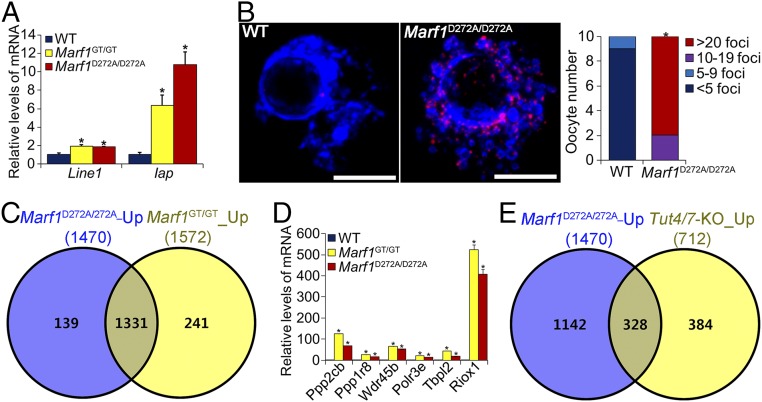
(A) Schematic illustration of the experimental design for the analysis of the RNase activity of MARF1 in oocytes by mutating the conserved D272 residue in the NYN domain to alanine. IVT, in vitro transcription. (B) Venn diagram illustrating the relationship of the transcripts down-regulated by supplementation with either WT MARF1 (+WT-MARF1) or D272A-mutant MARF1 (+D272A-MARF1) in Marf1GT/GT oocytes with those up-regulated in Marf1GT/GT oocytes (Marf1GT/GT_Up) compared with WT mouse oocytes. (C) Real-time RT-PCR validation of representative transcripts down-regulated by supplementation with WT but not D272A-mutant MARF1 in Marf1GT/GT oocytes. Fold changes relative to the Marf1GT/GT group are shown as mean ± SEM (n = 3). Different letters indicate a significant difference, P < 0.05, by one-way ANOVA and Tukey’s HSD test.
Point-Mutation of the Conserved D272 Residue in the NYN Domain of MARF1 Causes a Female-Only Infertile Phenotype in Mice.
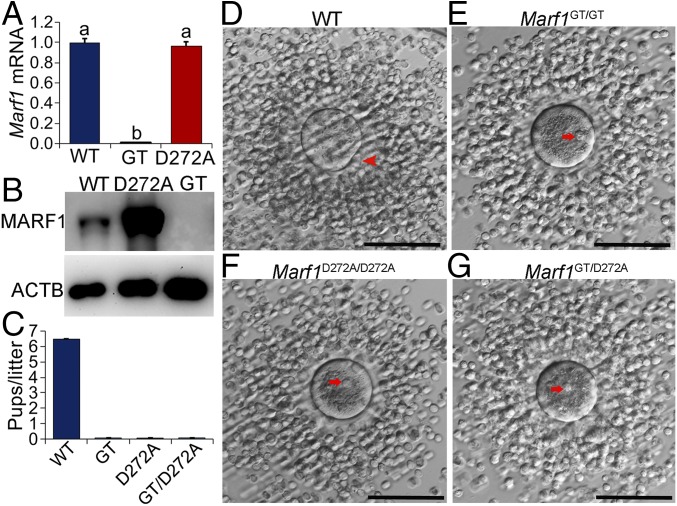
What is the physiological role of the RNase activity of MARF1? To address this question, we generated knockin mice carrying the D272A mutation (hereafter referred to as “Marf1D272A/D272A mice”) by CRISPER/CAS9-mediated genome editing (SI Appendix, Fig. S6 A and B) and assessed the phenotype in the resultant mutant mice. No reduction in the expression of Marf1 mRNA (Fig. 4A) was detected in Marf1D272A/D272A oocytes. However, a dramatic increase in the levels of MARF1 protein was unexpectedly observed in Marf1D272A/D272A oocytes (Fig. 4B). The underlying cause for this increase is not known, but it could be a feedback response of the mutant oocytes to the loss of MARF1-NYN function. Marf1D272A/D272A females, like the Marf1GT/GT females, are completely infertile, while males are not affected (Fig. 4C). This indicates that the RNase activity of MARF1 is indispensable for female fertility. Furthermore, mice carrying both the Marf1-knockin allele and the Marf1GT allele (referred to hereafter as “Marf1GT/D272A mice”) display the same phenotype as Marf1D272A/D272A mice (Fig. 4C), thus indicating that infertility in Marf1D272A/D272A females is not caused by potential off-target effects.
Fig. 4.
Oocyte meiotic arrest and female infertility in Marf1D272A/D272A mice. (A) Real-time RT-PCR analysis of the expression of Marf1 mRNA in WT, Marf1GT/GT (GT), and Marf1D272A/D272A (D272A) GV-stage fully grown oocytes. Fold changes relative to the WT group are shown as mean ± SEM (n = 3). Different letters indicate a significant difference, P < 0.05, by one-way ANOVA and Tukey’s HSD test. (B) Western blot analysis of the expression of MARF1 protein in WT, Marf1D272A/D272A (D272A), and Marf1GT/GT (GT) fully grown GV-stage oocytes. β-Actin (ACTB) was used as internal control (n = 3). (C) Number of pups per litter born by WT, Marf1GT/GT (GT), Marf1D272A/D272A (D272A), and Marf1GT/D272A (GT/D272A) females during the 8-mo period of the fertility test (n = 3 mice per genotype). (D–G) Microphotographs of the ovulated cumulus oocyte complexes by WT (D), Marf1GT/GT (E), Marf1D272A/D272A (F), and Marf1GT/D272A (G) females. The red arrowhead points to the first polar body, and red arrows indicate the GV. (Scale bars: 100 μm.)
Meiotic Arrest and Activation of Retrotransposons in MARF1-D272A Point-Mutant Oocytes.
There was no overt abnormality in folliculogenesis in either Marf1D272A/D272A or Marf1GT/D272A females (SI Appendix, Fig. S7), and ovulation occurred normally as determined by the number of eggs recovered and the morphology of the expanded cumuli oophorus (Fig. 4 D–G and SI Appendix, Fig. S6C). However, the oocytes ovulated by both Marf1D272A/D272A and Marf1GT/D272A females were immature and arrested at the GV stage (Fig. 4 F and G and SI Appendix, Fig. S6C). Therefore the inability of the oocyte to resume the first meiosis is the immediate cause of the infertility in Marf1D272A/D272A females. Moreover, we found that levels of Line1 and Iap retrotransposon mRNAs were elevated in Marf1D272A/D272A fully grown GV-stage oocytes isolated from large antral follicles (Fig. 5A), and the frequency of DNA double-stranded breaks, as demonstrated by γH2AX staining, was also increased in these oocytes (Fig. 5B). These data collectively indicate that the RNase activity of NYN domains mediates the function of MARF1 in controlling oocyte meiosis and retrotransposon silencing.
Fig. 5.
Compromised integrity of the genome and transcriptome in Marf1GT/GT oocytes. (A) Real-time RT-PCR analysis of retrotransposon Line1 and Iap mRNA expression in WT, Marf1GT/GT, and Marf1D272A/D272A GV-stage oocytes. Fold changes relative to the WT group are shown as mean ± SEM (n = 3). *P < 0.05, compared with WT by Student’s t test. (B), Left) Representative confocal micrographs of the γH2AX-stained WT and Marf1D272A/D272A GV-stage oocytes. γH2AX is stained in red, and DNA is in blue. (Scale bars: 5 μm.) (Right) Quantification of this staining. *P < 0.05, compared with WT by χ2 test. (C) Venn diagram illustrating the relationship of the transcripts identified by transcriptomic analysis that are dramatically up-regulated in Marf1D272A/D272A (Marf1D272A/D272A-Up) and Marf1GT/GT (Marf1GT/GT-Up) GV-stage oocytes as compared with WT. (D) Real-time RT-PCR validation of representative transcripts upregulated in Marf1D272A/D272A and Marf1GT/GT GV-stage oocytes. Fold changes relative to the Marf1GT/GT group are shown as mean ± SEM. (n = 3). *P < 0.05, compared with WT by Student’s t test. (E) Venn diagram illustrating the relationship of the upregulated transcripts in Marf1D272A/D272A oocytes (Marf1D272A/D272A-Up) with those up-regulated in Tut4/7-KO oocytes (Tut4/7-KO_Up) (45).
MARF1 Is an Executor of RNA Degradation in Mouse Oocytes.
RNA-seq analysis revealed that the integrity of oocyte transcriptome was also profoundly impaired in Marf1D272A/D272A mice. There were dramatically more transcripts in which the steady-state levels of expression were up-regulated (1,470) than down-regulated (103) in Marf1D272A/D272A fully grown GV-stage oocytes isolated from large antral follicles (Fig. 5C, SI Appendix, Fig. S8B, and Dataset S3). Moreover, there was consistent overlap of the overexpressed transcripts in Marf1D272A/D272A and Marf1GT/GT oocytes; 90.5% were identical (Fig. 5C, SI Appendix, Fig. S8A, and Dataset S4). Ppp2cb is among the commonly changed transcripts (Fig. 5D). Gene-enrichment analysis revealed that the up-regulated transcripts in Marf1D272A/D272A oocytes participate in processes similar to those of transcripts up-regulated in Marf1GT/GT oocytes (SI Appendix, Fig. S8A). Therefore, the RNase activity of MARF1 regulates the homeostasis of oocyte mRNA populations, which is crucial for oocyte development and function. Could the up-regulation of the large number of transcripts in Marf1D272A/D272A and Marf1GT/GT oocytes be attributed to increased transcription? Our previous observation that steady-state levels of mRNA, but not heteronuclear RNA, for a given group of genes were up-regulated in Marf1GT/GT oocytes suggests that this scenario is unlikely (3). However, we cannot rule out the possibility that the RNase activity of MARF1 participates in silencing oocyte global transcriptional activity during the final growth stage of normal WT oocytes. This potential additional function of MARF1 awaits further exploration.
Interestingly, mRNA 3′ terminal uridylation catalyzed by the uridyltransferases TUT4 and -7 mediates the degradation of a cohort transcripts in the oocyte transcriptome, which is crucial for oocyte maturation and female fertility (43). Given the indispensable role of both TUT4/7- and MARF1-dependent pathways in sculpting the maternal transcriptome, we hypothesized that the two pathways may share some of the same mRNA targets in oocytes for degradation. By comparing our transcriptomic data with those published by Morgan et al. (43), we found that the expression levels of nearly half (46.1%) of the transcripts that are up-regulated in Tut4/7-deleted oocytes are also elevated in Marf1D272A/D272A oocytes (Fig. 5E). These data suggest that an interaction exists between these two pathways and that MARF1 could function as a downstream executor of the TUT4/7-mediated mRNA degradation pathway in oocytes. Interestingly, MARF1 and TUT4, along with some other proteins involved in mRNA decay pathways, are components that constitute the proteome of the process (P) bodies of human epithelial cells (44). Thus, MARF1 may coordinate with TUT4/7 in the same functional module in oocytes to specifically shape the maternal transcriptome for supporting oocyte maturation and preimplantation development. LOTUS domains in MARF1 may play an important role in selecting substrate for degradation by binding specific RNA sequences. Nevertheless, although the data presented here demonstrate that MARF1 is a bona fide RNase, they do not imply that MARF1 functions in the same way as PIWI endonucleases to specifically slice the target into smaller fragments (45, 46). Also, whether MARF1 catalyzes the production of small RNAs in oocytes, particularly piRNAs and endo-siRNAs, remains to be determined, as do the direct RNA targets of MARF1.
Both the NYN and LOTUS domains are well conserved across eukaryotes and bacteria. Although the NYN and LOTUS domains in eukaryotic proteins are usually fused with diverse types of RNA-binding or protein–protein interaction domains, the bacterial versions of these two domains almost always combine together in one protein, with an N-terminal NYN domain fused to a single or duplicated LOTUS domain (36). Therefore, MARF1 is an ancient protein that probably originated in bacteria, and the NYN domain fused with the LOTUS domain is probably the first version that entered the eukaryotes and retained the original function of its bacterial cognates (36). Remarkably, despite this fascinating quality of being an ancient protein, the molecular properties of MARF1, as well as its biological function in species other than mammals, remain surprisingly unknown. Here, following up on our initial identification of MARF1 as a key oogenic regulator essential for mouse oocyte meiotic progression and silencing of retrotransposon, we resolved the crystal structure of the NYN and LOTUS domains in mouse MARF1 and demonstrated both in vivo and in vitro that MARF1 is indeed an RNase that controls these essential oogenic processes. These findings not only shed light on the molecular properties of MARF1 but also provide insight into the molecular mechanisms by which MARF1 controls oocyte meiosis and genome integrity.
The quality of oocytes is well recognized as the key to reproductive success and the creation of healthy individuals (47, 48). Oocyte quality is largely determined by the autonomous gene-expression program built within the oocytes (42, 49). Key players in pathways involved in RNA degradation-mediated posttranscriptional control have recently emerged as regulators of oocyte gene expression; these are indispensable for oocyte acquisition of meiotic and developmental competence (42, 43, 50–54). The observations put forward here not only clarify the structural basis for MARF1 function as a mammalian female-specific regulator of germ cell meiosis and retrotransposon surveillance but also provide insight into the posttranscriptional control of oocyte gene expression. By functioning as an executor of RNA-degradation processes in oocytes, MARF1 controls the mRNA homeostasis and genome integrity of mammalian oocytes.
Materials and Methods
Unless otherwise specified, all reagents and chemicals were purchased from Sigma-Aldrich Co.
Mice.
Marf1GT/GT mice were imported from the Jackson Laboratory. D272A-MARF1 knockin mice were generated by CRISPER-Cas9 technology as detailed in SI Appendix, Materials and Methods. All mouse procedures and protocols were approved by the Institutional Animal Care and Use Committee of Nanjing Medical University and were conducted in accordance with the institutional guidelines for the care and use of laboratory animals.
Ovarian Histology and Oocyte Isolation, Microinjection, and Staining.
Ovarian sections were stained with periodic acid–Schiff reagent and Lillie–Mayer hematoxylin. Fully grown GV-stage oocytes were isolated from mice that were primed with equine chorionic gonadotropin (eCG). Microinjection and immunofluorescent staining of the oocytes were carried out as described in SI Appendix, Materials and Methods.
Western Blot and qRT-PCR Analyses.
The same number of WT and Marf1-mutant fully grown GV-stage oocytes were collected and subjected to Western blot and qRT-PCR analyses.
RNA-Seq and Gene-Enrichment Analyses.
Four sets of oocyte samples of each genotype or treatment were collected and subjected to RNA-seq analysis as described in SI Appendix, Materials and Methods. RNA-seq data have been deposited in the Gene Expression Omnibus (accession no. GSE109213). Gene-enrichment analysis of differentially expressed transcripts was carried out using Metascape (metascape.org).
Cloning, Expression, Amino Acid Substitution, and Purification of Recombinant MARF1.
The coding sequence of MmMARF1 (residues 158–320, 158–407, 158–690, 158–1381, and 687–1381) were cloned into the modified pET28a vector with an N-terminal His6-SUMO tag (residues 158-320, 158-407, and 158-690; Merck), the pfastBacHTB vector (residues 158-1381; Thermo Fisher Scientific, Inc.), and the pET28a vector with a C-terminal His6 tag (residues 687-1381; Merck), respectively, and the recombinant protein was then produced and purified as described in SI Appendix, Materials and Methods.
Crystallization, Data Collection, Structure Determination, and Refinement.
Crystals were grown by hanging-drop vapor diffusion at 18 °C and were harvested for diffraction data collection as detailed in SI Appendix, Materials and Methods. Atomic coordinates and structure factors for the reported crystal structures have been deposited in the Protein Data Bank [ID codes 5YAA (NYN domain) and 5YAD (LOTUS1 domain)].
Nucleic Acid Preparation and Nuclease Activity Assay.
The dsRNA and circular ssRNA substrates were prepared using the labeled ssRNA H49 and H49AS that were obtained by in vitro transcription and were 5′ radiolabeled with 32P-γ-ATP (Perkin-Elmer Health Sciences). These 5′ 32P-labeled substrates were then incubated at 37 °C with the recombinant proteins for the nuclease activity assay.
EMSA.
The EMSA was carried out by incubating the recombinant proteins with 5′ 32P-labeled substrates at 25 °C for 30 min.
Statistical Analysis.
Experiments were performed at least three times independently; data are presented as mean ± SEM. Differences between two groups were analyzed by t tests. Experiments having more than two groups were compared by one-way ANOVA followed by Tukey’s Honestly Significant Difference (HSD) test using GraphPad Prism software (GraphPad Software, Inc.). P < 0.05 was defined as significantly different.
Supplementary Material
Acknowledgments
We thank Prof. Donal O’Carroll for providing the data on Tut4/7-KO mouse oocyte-related work; Prof. Shaorong Gao and his group for help with establishing the oocyte microinjection platform; Prof. John Eppig for comments and edits of the manuscript; the staff of the Beamline BL17U1 and BL19U1 at the Shanghai Synchrotron Radiation Facility for assistance with data collection; and the Geekgene group for RNA-seq analyses. This work was supported by National Basic Research (973) Program of China Grants 2014CB943200 and 2013CB945500, National Natural Science Foundation of China Grants 31471351 and 31271538 (to Y.-Q.S.) and 31770792 and 31230041 (to J.M.), and National Natural Science Foundation of Jiangsu Province Grant BK20140061 (to Y.-Q.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA-sequencing data reported in this paper have been deposited in the Gene Expression Omnibus database (accession no. GSE109213). Atomic coordinates and structure factors for the reported crystal structures have been deposited in the Protein Data Bank database [ID codes 5YAA (NYN domain) and 5YAD (LOTUS1 domain)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809744115/-/DCSupplemental.
References
- 1.Smallwood SA, Kelsey G. De novo DNA methylation: A germ cell perspective. Trends Genet. 2012;28:33–42. doi: 10.1016/j.tig.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Su YQ, Sun F, Handel MA, Schimenti JC, Eppig JJ. Meiosis arrest female 1 (MARF1) has nuage-like function in mammalian oocytes. Proc Natl Acad Sci USA. 2012;109:18653–18660. doi: 10.1073/pnas.1216904109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Su YQ, et al. MARF1 regulates essential oogenic processes in mice. Science. 2012;335:1496–1499. doi: 10.1126/science.1214680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: Progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ollinger R, Reichmann J, Adams IR. Meiosis and retrotransposon silencing during germ cell development in mice. Differentiation. 2010;79:147–158. doi: 10.1016/j.diff.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Ku HY, Lin H. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl Sci Rev. 2014;1:205–218. doi: 10.1093/nsr/nwu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vagin VV, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siomi MC, Mannen T, Siomi H. How does the royal family of Tudor rule the PIWI-interacting RNA pathway? Genes Dev. 2010;24:636–646. doi: 10.1101/gad.1899210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C, et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci USA. 2009;106:20336–20341. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Nott TJ, Jin J, Pawson T. Deciphering arginine methylation: Tudor tells the tale. Nat Rev Mol Cell Biol. 2011;12:629–642. doi: 10.1038/nrm3185. [DOI] [PubMed] [Google Scholar]
- 11.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: The vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 12.Vagin VV, et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 13.Das PP, et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 2008;31:79–90. doi: 10.1016/j.molcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houwing S, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 15.Seipel K, Yanze N, Schmid V. The germ line and somatic stem cell gene Cniwi in the jellyfish Podocoryne carnea. Int J Dev Biol. 2004;48:1–7. doi: 10.1387/ijdb.15005568. [DOI] [PubMed] [Google Scholar]
- 16.Wilczynska A, Minshall N, Armisen J, Miska EA, Standart N. Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline. RNA. 2009;15:337–345. doi: 10.1261/rna.1422509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J. 2009;28:2945–2958. doi: 10.1038/emboj.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamudio N, Bourc’his D. Transposable elements in the mammalian germline: A comfortable niche or a deadly trap? Heredity (Edinb) 2010;105:92–104. doi: 10.1038/hdy.2010.53. [DOI] [PubMed] [Google Scholar]
- 19.Gainetdinov I, Skvortsova Y, Kondratieva S, Funikov S, Azhikina T. Two modes of targeting transposable elements by piRNA pathway in human testis. RNA. 2017;23:1614–1625. doi: 10.1261/rna.060939.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng K, et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci USA. 2010;107:11841–11846. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yabuta Y, et al. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J Cell Biol. 2011;192:781–795. doi: 10.1083/jcb.201009043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe T, et al. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell. 2011;20:364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka T, et al. Tudor domain containing 7 (Tdrd7) is essential for dynamic ribonucleoprotein (RNP) remodeling of chromatoid bodies during spermatogenesis. Proc Natl Acad Sci USA. 2011;108:10579–10584. doi: 10.1073/pnas.1015447108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka SS, et al. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- 25.Soper SF, et al. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shoji M, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell. 2009;17:775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Saxe JP, Chen M, Zhao H, Lin H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 2013;32:1869–1885. doi: 10.1038/emboj.2013.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandey RR, et al. Tudor domain containing 12 (TDRD12) is essential for secondary PIWI interacting RNA biogenesis in mice. Proc Natl Acad Sci USA. 2013;110:16492–16497. doi: 10.1073/pnas.1316316110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma L, et al. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet. 2009;5:e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuramochi-Miyagawa S, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 31.Huang H, et al. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell. 2011;20:376–387. doi: 10.1016/j.devcel.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frost RJ, et al. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci USA. 2010;107:11847–11852. doi: 10.1073/pnas.1007158107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 34.Chuma S, et al. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci USA. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carmell MA, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Anantharaman V, Zhang D, Aravind L. OST-HTH: A novel predicted RNA-binding domain. Biol Direct. 2010;5:13. doi: 10.1186/1745-6150-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anantharaman V, Aravind L. The NYN domains: Novel predicted RNAses with a PIN domain-like fold. RNA Biol. 2006;3:18–27. doi: 10.4161/rna.3.1.2548. [DOI] [PubMed] [Google Scholar]
- 38.Matelska D, Steczkiewicz K, Ginalski K. Comprehensive classification of the PIN domain-like superfamily. Nucleic Acids Res. 2017;45:6995–7020. doi: 10.1093/nar/gkx494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Callebaut I, Mornon JP. LOTUS, a new domain associated with small RNA pathways in the germline. Bioinformatics. 2010;26:1140–1144. doi: 10.1093/bioinformatics/btq122. [DOI] [PubMed] [Google Scholar]
- 40.Jeske M, Müller CW, Ephrussi A. The LOTUS domain is a conserved DEAD-box RNA helicase regulator essential for the recruitment of Vasa to the germ plasm and nuage. Genes Dev. 2017;31:939–952. doi: 10.1101/gad.297051.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeske M, et al. The crystal structure of the Drosophila germline inducer Oskar identifies two domains with distinct Vasa helicase- and RNA-binding activities. Cell Rep. 2015;12:587–598. doi: 10.1016/j.celrep.2015.06.055. [DOI] [PubMed] [Google Scholar]
- 42.Svoboda P, Franke V, Schultz RM. Sculpting the transcriptome during the oocyte-to-embryo transition in mouse. Curr Top Dev Biol. 2015;113:305–349. doi: 10.1016/bs.ctdb.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Morgan M, et al. mRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature. 2017;548:347–351. doi: 10.1038/nature23318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hubstenberger A, et al. P-body purification reveals the condensation of repressed mRNA regulons. Mol Cell. 2017;68:144–157.e5. doi: 10.1016/j.molcel.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Reuter M, et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature. 2011;480:264–267. doi: 10.1038/nature10672. [DOI] [PubMed] [Google Scholar]
- 46.De Fazio S, et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480:259–263. doi: 10.1038/nature10547. [DOI] [PubMed] [Google Scholar]
- 47.Gosden R, Lee B. Portrait of an oocyte: Our obscure origin. J Clin Invest. 2010;120:973–983. doi: 10.1172/JCI41294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luke B, et al. Cumulative birth rates with linked assisted reproductive technology cycles. N Engl J Med. 2012;366:2483–2491. doi: 10.1056/NEJMoa1110238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan H, O’brien MJ, Wigglesworth K, Eppig JJ, Schultz RM. Transcript profiling during mouse oocyte development and the effect of gonadotropin priming and development in vitro. Dev Biol. 2005;286:493–506. doi: 10.1016/j.ydbio.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 50.Ivanova I, et al. The RNA m6A reader YTHDF2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol Cell. 2017;67:1059–1067.e4. doi: 10.1016/j.molcel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stein P, et al. Essential role for endogenous siRNAs during meiosis in mouse oocytes. PLoS Genet. 2015;11:e1005013. doi: 10.1371/journal.pgen.1005013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tam OH, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang F, et al. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murchison EP, et al. Critical roles for Dicer in the female germline. Genes Dev. 2007;21:682–693. doi: 10.1101/gad.1521307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.