Significance
The metabolic diversity across the tree of life has become increasingly apparent with the growing availability of sequenced genomes and annotations. The basis for such metabolic diversity in organisms is not understood. Here, we evaluate alternative pathways in amino acid biosynthesis from a standpoint of tradeoff between thermodynamic favorability and cofactor-use efficiency. Using 5,203 sequenced genomes and available metabolic network reconstructions, we found evidence that this tradeoff affects pathway choice, which can be related to organism lifestyle. Given the fundamental importance of metabolism on organism survival and adaptation, such choices help us reveal the impact of selection pressures and may ultimately have fundamental implications for our understanding of the phylogenetic tree.
Keywords: thermodynamics, metabolism, evolution, constraint-based modeling
Abstract
The structure of the metabolic network contains myriad organism-specific variations across the tree of life, but the selection basis for pathway choices in different organisms is not well understood. Here, we examined the metabolic capabilities with respect to cofactor use and pathway thermodynamics of all sequenced organisms in the Kyoto Encyclopedia of Genes and Genomes Database. We found that (i) many biomass precursors have alternate synthesis routes that vary substantially in thermodynamic favorability and energy cost, creating tradeoffs that may be subject to selection pressure; (ii) alternative pathways in amino acid synthesis are characteristically distinguished by the use of biosynthetically unnecessary acyl-CoA cleavage; (iii) distinct choices preferring thermodynamic-favorable or cofactor-use–efficient pathways exist widely among organisms; (iv) cofactor-use–efficient pathways tend to have a greater yield advantage under anaerobic conditions specifically; and (v) lysine biosynthesis in particular exhibits temperature-dependent thermodynamics and corresponding differential pathway choice by thermophiles. These findings present a view on the evolution of metabolic network structure that highlights a key role of pathway thermodynamics and cofactor use in determining organism pathway choices.
Metabolism has historically been viewed as a highly conserved network across all branches of life (1). However, as a greater number of organisms are sequenced and characterized (2), there is an increasing appreciation of the diversity of organism-specific metabolic differences (3–5). Diverse organism living conditions, including nutrient availability, electron acceptors, temperature, pH, pressure, and salt concentrations (6), can create environmental niches that have specific metabolic requirements (7–9). How these environmental conditions impact metabolic diversity remains an important question (6).
Metabolic network reconstructions are highly curated knowledge bases of metabolic function that provide a way to systematically investigate the differences in metabolic capabilities among various organisms (10, 11). Metabolic network models, derived from metabolic reconstructions, are mathematical representations of the metabolic capabilities of an organism that can be used to compute organism phenotypes. Recent efforts reconstructing genome-scale metabolic networks for various organisms have offered a quantitative route to begin to understand the principles underlying metabolic diversity across the tree of life (12, 13).
Metabolic network reconstructions enable a number of powerful computational analyses. First, flux balance analysis (FBA) of metabolic models can calculate the flow of metabolites through the metabolic network by utilizing optimization principles (14). FBA can be used for a number of calculations such as product yields and substrate utilization efficiency at a network level (15, 16). Second, FBA can be integrated with thermodynamic equilibrium constants to calculate additional network properties such as thermodynamically feasible optimal states (17) and thermodynamic bottlenecks (18). These methods thus allow us to evaluate the properties of metabolic pathways in an organism-specific context and provide the basis toward understanding pathway choice among various organisms.
In this study, we utilized metabolic network analysis to evaluate alternative pathway choice in terms of the underlying physicochemical properties of pathways in organisms with diverse lifestyles. We first collected all available information on de novo biosynthesis pathways for biomass precursors and identified organisms containing these pathways. We focused on the biosynthetic pathways for five amino acids with differential use of acyl-CoA cleavage (lysine, arginine, cysteine, isoleucine, and methionine). We examined the basis for preference of Escherichia coli for alternative pathway choice in amino acid biosynthesis using in vivo metabolite and protein concentration measurements. We also identified clusters of organisms with distinct pathway choices related to a tradeoff between thermodynamic favorability and cofactor-use efficiency. Lastly, we focused on two specific cases, isoleucine and lysine biosynthesis, to investigate how organisms’ lifestyles relate to the choice of biosynthetic pathways.
Results
Identifying Biosynthetic Pathway Alternatives Found in Sequenced Genomes.
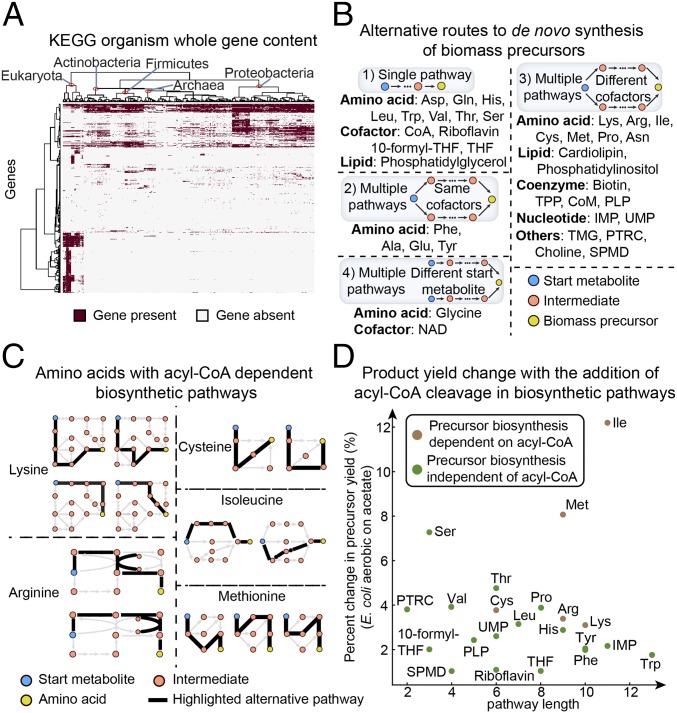
First, we collected the gene content of 5,203 organisms from the Kyoto Encyclopedia of Genes and Genomes (KEGG) Database of genome annotations (19). The organisms spanned three domains of life, with major phyla including Proteobacteria (n = 2,167), Firmicutes (n = 908), Actinobacteria (n = 575), Bacteroidetes (n = 234), Euryarchaeota (n = 179), Tenericutes (n = 134), Chlamydiae (n = 118), Chordata (n = 108), and Cyanobacteria (n = 102). Based on the available genome annotations in KEGG, we obtained a total of 8,247 genes with KEGG orthology identifiers from all organisms. The list of genes corresponds to specific Enzyme Commission numbers and includes both metabolic functions and cellular processes such as assembly of macromolecules, signal transduction, etc. We found that organisms cluster by relationship on the phylogenetic tree based on their gene content (Fig. 1A). For example, organisms in the Archaea and Eukaryota domains each belong to individual clusters, while organisms in the major phyla of the Bacteria domain (Proteobacteria, Actinobacteria, and Firmicutes) fall into separate clusters.
Fig. 1.
Alternative biosynthetic routes of biomass precursors. (A) Hierarchical clustering on 5,203 KEGG organisms, with 8,247 genes having Enzyme Commission annotations. (B) Categories of alternative routes to de novo biosynthesis of biomass precursors. For amino acids, we provided their three letter codes for simplicity. (C) Amino acids with acyl-CoA–dependent alternative biosynthetic pathways. (D) Product yield change due to the addition of acyl-CoA cleavage in biosynthetic pathways of biomass precursors. 10-formyl-THF, 10-formyltetrahydrofolate; CoM, coenzyme M; PLP, pyridoxal phosphate; PTRC, putrescine; SPMD, spermidine; THF, tetrahydrofolate; TMG, trimethylglycine; TPP, thiamine diphosphate.
We then identified alternative pathways for de novo synthesis of biomass precursors using the KEGG PATHWAY and MetaCyc databases (19, 20). The list of biomass precursors examined included amino acids, nucleotides, lipids, and certain small molecules such as vitamins and polyamines (Dataset S1, Table S1 includes all alternative pathway reactions). We classified the precursors based on the types of alternative biosynthetic pathways present (Fig. 1B). Specifically, the pathways either (i) have only one biosynthetic route for the precursor, (ii) start from the same metabolite and use the same cofactors but with different intermediate metabolites, (iii) start from the same metabolite and use different cofactors, or (iv) start from different metabolites and use different cofactors. For precursors with multiple alternative routes, we attempted to trace the pathways back until they intersect at a common starting metabolite. However, for alternative routes that reach central metabolic pathways (e.g., glycolysis and TCA cycle) but have not converged to a common starting metabolite, we considered them as having different starting points.
We found that while some biomass precursors have only a single de novo biosynthetic pathway, a large number display multiple pathways (Fig. 1B). We distinguished between pathways that share common starting metabolites and pathways that start from different metabolites. Pathways that start from the same metabolite but have alternate routes with different cofactor usage include those for a number of amino acids (arginine, asparagine, cysteine, lysine, and methionine), nucleotides (IMP and UMP), and essential small metabolites (biotin, putrescine, spermidine, and thiamine diphosphate). These alternative pathways allowed us to control for any possible factors associated with concentrations or thermodynamics of the starting metabolites themselves when evaluating alternatives. Lastly, pathways starting from alternate metabolites were those for glycine (from 3-phosphoglycerate, glyoxylate, or oxaloacetate via threonine) and NAD (from tryptophan or aspartate).
Alternative Pathways in Amino Acid Biosynthesis Differ by Acyl-CoA Cleavage and Show Distinct Yield Differences.
We examined the thermodynamics (21) (SI Appendix, Fig. S1 and Dataset S1, Table S1) and cofactor use of the alternative biosynthetic pathways for biomass precursors. Pathways with lower standard transformed reaction Gibbs energies () are considered more thermodynamically favorable than those with higher energies. We found that alternative pathways can vary substantially in thermodynamic favorability due to their differences in cofactor use. Examining the common cofactors involved, we found that certain cofactor pairs are prevalent in biosynthetic pathways, including those providing energy (ATP hydrolysis), those serving as the oxidizing/reducing agent (NADH/NAD and NADPH/NADP), and those donating the amino group (glutamate/α-ketoglutarate and glutamine/glutamate).
However, the use of acyl-CoA cleavage to drive biosynthetic pathways is present for only a subset of amino acids, including lysine, arginine, cysteine, isoleucine, and methionine (Fig. 1C and SI Appendix, Supplementary Information Text). Interestingly, these five amino acids have both acyl-CoA–dependent and –independent pathways present. We found the acyl-CoA–dependent pathways of these amino acids to be identical with the other alternatives in cofactor use, except for the additional acyl-CoA cleavage, which for lysine, arginine, and cysteine results in more favorable pathway thermodynamics. On the other hand, the acyl-CoA–independent pathway in isoleucine biosynthesis through threonine has lower energy than the acyl-CoA–dependent route, because it is coupled to a greater energetic cost of hydrolysis of three ATP molecules and oxidation of three equivalent NADH molecules per isoleucine produced.
We then investigated why these five amino acids have alternative biosynthetic pathways that differ by acyl-CoA use while the other biomass precursors have only acyl-CoA–independent pathways. We identified two factors contributing to the presence of acyl-CoA–dependent pathways: the pathway length in terms of reaction number and the change in precursor yield from using pathways with the additional acyl-CoA cleavage. First of all, alternative pathways are unlikely to arise when the production of the precursor takes very few steps (e.g., a single step for alanine, aspartate, glutamate, and glutamine synthesis). Additionally, a large difference in precursor yield due to the additional acyl-CoA cleavage in the pathway may benefit organisms with certain lifestyles, thus motivating the presence of alternative pathways differing in acyl-CoA use.
We obtained biosynthetic pathway length for all biomass precursors (Fig. 1B) from the MetaCyc database and calculated the median length for precursors with multiple alternative routes. We also compared the difference in precursor yield due to the use of acyl-CoA–dependent versus acyl-CoA–independent pathways, through simulations with organism-specific genome-scale metabolic networks (Materials and Methods). For precursors without acyl-CoA–dependent pathways present, we created pseudo pathways similar to their original pathways but with the additional acyl-CoA cleavage. Taking E. coli grown on acetate aerobically, for example (Fig. 1D), we found that the five amino acids (brown dots) with acyl-CoA–dependent pathways present generally have longer pathway length and larger yield change from using the acyl-CoA–dependent pathways, with isoleucine and methionine showing the largest yield change. This trend can also be extended to a number of organisms (Dataset S1, Table S2) (22) under different conditions, for which we found the yield change for the five amino acids to be significantly higher than other precursors in 24 of 43 conditions examined (P < 0.05) (SI Appendix, Fig. S2). Notably, for organisms grown on acetate, we found a significant difference in 11 of 13 conditions examined (SI Appendix, Fig. S2A). In contrast, when examining yield change in pathways differing by the use of ATP hydrolysis, we found only 6 of 43 conditions to have a significantly higher yield change for precursors with ATP-dependent alternative routes compared to those without such alternates (P < 0.05) (SI Appendix, Fig. S3). Therefore, we demonstrate that alternative pathways differing by the use of acyl-CoA show significant yield differences, as is the case for the five amino acids.
E. coli Uses Thermodynamically-Favorable but Cofactor-Use–Inefficient Amino Acid Biosynthetic Pathways.
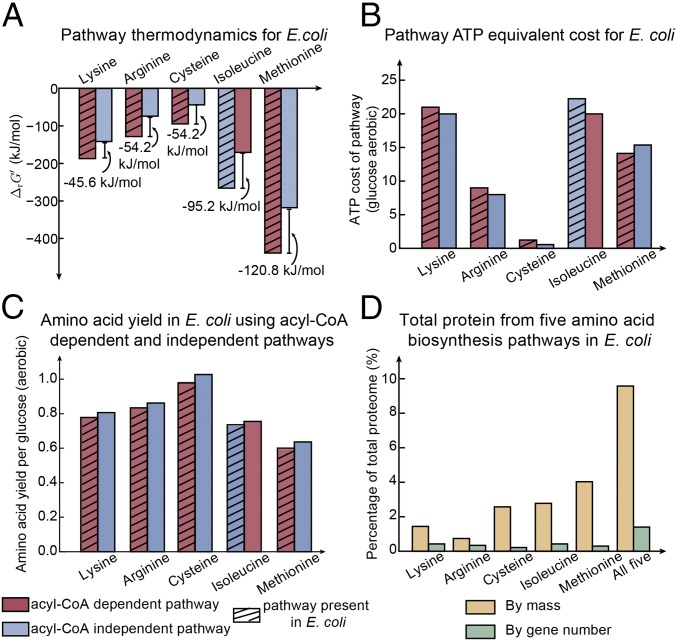
We sought to further compare the acyl-CoA–dependent and –independent alternative pathways for the five amino acids using E. coli, taking advantage of its well-curated metabolic network and abundant quantitative physiological data available. Specifically, we focused on two aspects: pathway thermodynamic favorability and cofactor-use efficiency. E. coli uses acyl-CoA–dependent pathways for biosynthesis of four of the five targeted amino acids, with the exception being isoleucine. Compared with pathways that are not present in E. coli, we found that the pathways used by E. coli are thermodynamically more favorable in each case in terms of intrinsic pathway energy (i.e., lower standard Gibbs energy, , which does not take into account metabolite concentrations). We then calculated transformed Gibbs energy () values for each pathway using measured quantitative metabolomics data of E. coli (23–25) (Dataset S1, Table S3) and verified that these pathways are indeed substantially further from equilibrium (more negative ) (Fig. 2A).
Fig. 2.
Thermodynamics and cofactor-use efficiency of alternative biosynthetic pathways in E. coli. (A) Gibbs energies of reaction () of acyl-CoA–dependent and –independent alternative pathways using E. coli in vivo metabolite concentrations. (B) ATP equivalent cost of acyl-CoA–dependent and –independent pathways calculated from an E. coli metabolic model grown on glucose aerobically. (C) Amino acid yield from acyl-CoA–dependent and –independent pathways simulated using the E. coli metabolic model grown on glucose aerobically. (D) The proteins from five amino acid biosynthesis pathways in terms of fraction in mass and gene number of the total proteome in E. coli.
We next calculated the ATP equivalent cost of the pathways to evaluate the cofactor-use efficiency of the pathways (Materials and Methods). A high ATP cost of the pathway corresponds to a low efficiency in cofactor use. We found that the ATP equivalent costs of the pathways used by E. coli (using glucose aerobically) are greater than those of the alternative pathways in four of five cases (Fig. 2B), indicating that E. coli uses cofactor-use–inefficient pathways. This result was further confirmed by the fact that pathways present in E. coli have lower product yield than those not present in E. coli (Fig. 2C).
Thermodynamically favorable pathways can be beneficial in terms of protein cost, as the enzyme level required to achieve a given flux can increase dramatically for reactions near equilibrium (18). Therefore, it is possible that organisms already with significant resources invested in synthesizing pathway proteins select the thermodynamically favorable routes for efficiency in protein use. We found evidence supporting this hypothesis using E. coli proteomics data (26), wherein the proteins required for biosynthesis of each of the five amino acids occupy a higher fraction of the whole E. coli proteome by mass compared to gene number (Fig. 2D). Together, the proteins from all five amino acid biosynthesis pathways occupy 10% of the proteome by mass, while only 2% by number of genes.
To summarize, we found that tradeoffs between thermodynamic favorability and cofactor-use efficiency exist in pathway alternatives. In the case of E. coli, the use of thermodynamically more favorable pathways may improve the efficiency of pathway protein use.
Distinct Acyl-CoA–Dependent Pathway Choices Exist Among Organisms.
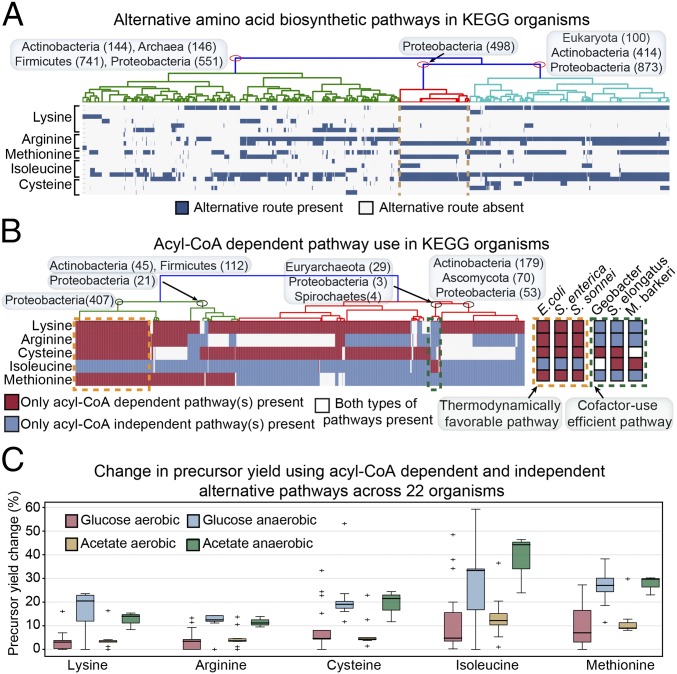
To understand the underlying factors for alternative pathway choice, we first clustered the organisms based on their presence/absence information of alternative pathways for the five amino acids. We found that the organism clusters did not separate cleanly by phylogeny (Fig. 3A), suggesting that factors other than phylogenetics may underlie the choice of alternative pathways. However, we observed interesting patterns when examining the use of acyl-CoA–dependent pathways for the five amino acids among organisms. For each amino acid, we separated the KEGG organisms into three categories: (i) those containing only acyl-CoA–dependent pathway(s); (ii) those containing only acyl-CoA–independent pathway(s); and (iii) those containing both acyl-CoA–dependent and –independent pathways. For methionine biosynthesis, we labeled the pathway using two acyl-CoA molecules as acyl-CoA dependent, and the pathways using only one as acyl-CoA independent.
Fig. 3.
(A) Hierarchical clustering on 5,203 KEGG organisms based on the presence of alternative biosynthetic pathways for the five amino acids. (B) Hierarchical clustering of KEGG organisms based on their use of acyl-CoA–dependent pathways. We show two clusters of organisms: one mostly uses thermodynamically favorable pathways (yellow dashed box), and the other mostly uses cofactor-efficient pathways (green dashed box). (C) Change in precursor yield using the acyl-CoA–dependent and –independent alternative pathways under different conditions for the five amino acids. We predicted the product yield change across 22 organisms, using their available genome-scale metabolic models.
Examining patterns of pathway use within organisms, we did not find any organism choosing acyl-CoA–dependent pathways for all five amino acid biosynthesis pathways, but for only a selection of them. As we clustered the organisms based on the type of acyl-CoA pathways used for the five amino acids, we found that the pathway choice did not break down cleanly by phylogeny. Further analysis on the metabolic genes related to the use of acyl-CoA pathways also shows complex traits in metabolic functions (SI Appendix, Fig. S10), indicating that nonspecific factors, such as lifestyle or organism history, may underlie the acyl-CoA pathway use broadly.
On the other hand, we identified groups of organisms with distinct pathway choices. We found one cluster containing E. coli and other Gammaproteobacteria (Fig. 3B, yellow box) for which the choice of acyl-CoA–dependent pathways is the same as in E. coli. This cluster represents a set of organisms choosing thermodynamically favorable pathways. We also identified a different cluster of organisms that select cofactor-use–efficient pathways instead, including Geobacter metallireducens, the methanogen Methanosarcina barkeri, and the cyanobacterium [Synechococcus elongatus (Fig. 3B, green box)]. These two opposing clusters indicate that tradeoffs between efficiency in product yield and proteome cost in biosynthesis may widely exist in organisms’ pathway choice, perhaps with similar underlying principles to the recent observation that cellular overflow metabolism results from the balance between efficient pathway yield and efficient protein use (27).
A closer look at the lifestyles of these two organism clusters shows that organisms favoring thermodynamic-favorable pathways generally depend on complex carbon sources with aerobic respiration, while organisms favoring cofactor-use–efficient pathways depend on simple carbon sources with anaerobic respiration. To understand how different lifestyles affect the product yield, we attempted to compare the yield change from using the alternative acyl-CoA–dependent pathways under four growth conditions: glucose aerobic, glucose anaerobic, acetate aerobic, and acetate anaerobic. We used the curated genome-scale metabolic models of a total of 22 organisms (Dataset S1, Table S2) for simulations (22, 28, 29). It is worth noting that not all 22 organisms examined here fall into the two organism clusters identified above. We found that anaerobic respiration results in the most significant change in precursor yield for all five amino acids (Fig. 3C), possibly due to the different cofactor cost and availability under different respirations. On the other hand, the carbon sources do not seem to significantly affect the product yield change.
Trade-Off Between Pathway Thermodynamic Favorability and Efficiency of Cofactor Use Underlies Organisms’ Pathway Choice for Isoleucine Biosynthesis.
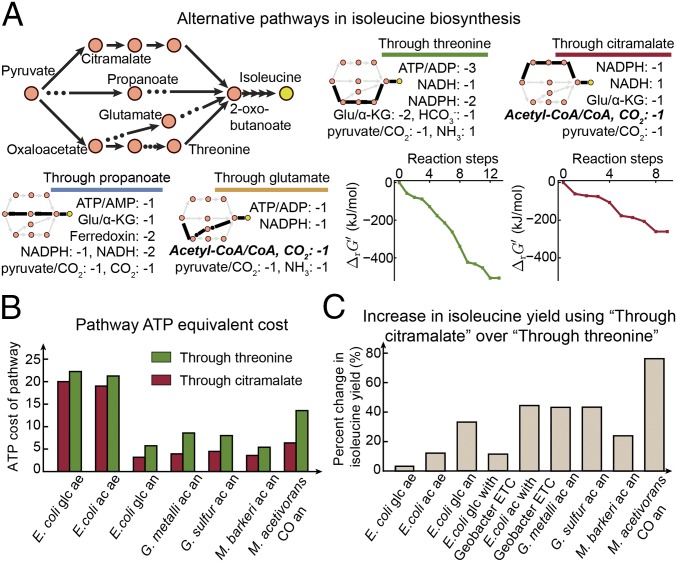
To understand the choice of alternative isoleucine biosynthesis pathways among various organisms, we focused on two alternative pathways and compared their properties in terms of thermodynamic favorability and cofactor-use efficiency. The first pathway uses threonine as the intermediate (Fig. 4A, green) and is present in a large number of organisms from the Bacteria and Eukarya domains. The second pathway uses citramalate as the intermediate (Fig. 4A, red) and is typically present in Archaea but is also found in bacteria from the Spirochaetes phylum (30, 31). A recent study showed that both pathways are present in Geobacter spp., which primarily uses the one through citramalate (32). We selected organisms from each category described above, including E. coli (contains pathway through threonine), M. barkeri and Methanosarcina acetivorans (contain pathway through citramalate), and Geobacter sulfurreducens and G. metallireducens (contain both pathways but mainly use the one through citramalate).
Fig. 4.
Alternative pathways for isoleucine biosynthesis. (A) Sketches of four alternative isoleucine biosynthetic pathways with their cofactor usage. We also included the Gibbs energies (), considering metabolite concentrations for each reaction step in the pathways through threonine and citramalate. (B) Cofactor use in terms of ATP cost for “Through threonine” and “Through citramalate” pathways in different organisms. The organism name, the substrate used, and the respiration type are labeled for each simulated organism condition. (C) Increase in isoleucine yield using the “Through citramalate” pathway compared with “Through threonine” for different organisms. ac, acetate; ae, aerobic; an, anaerobic; CO, carbon monoxide; glc, glucose; Glu/α-KG, glutamate/α-ketoglutarate.
Although the standard energies () of the pathway through threonine are significantly lower than those of the citramalate pathway across different conditions (SI Appendix, Fig. S6B), we further compared the of both pathways by taking metabolite concentrations into account. Using the quantitative metabolomics data of E. coli (23–25) (Dataset S1, Table S3), we calculated the reactionwise energy profile for each pathway (Materials and Methods) and confirmed that the overall of the pathway through threonine is much lower than that through citramalate (Fig. 4A).
Using the available genome-scale metabolic models (28, 29, 33–35), we calculated the ATP equivalent cost of the two pathways for five organisms with their respective carbon sources and types of respiration (Fig. 4B) (Materials and Methods). We used both glucose and acetate as the substrates for E. coli; the latter is a common carbon source for the other four organisms. We also allowed both aerobic and anaerobic respirations for E. coli, although only the latter is possible for the other four organisms. We found that the pathway through threonine is always more costly in cofactor use compared with the one through citramalate (Fig. 4B), while being thermodynamically more favorable.
To examine the possible benefit of using the cofactor-efficient pathway in E. coli, we inserted the citramalate pathway into the E. coli metabolic model. We observed marginal improvement (3.6%) in isoleucine yield per mole of glucose when comparing the two pathways for E. coli (Fig. 4C). On the other hand, we observed a relatively large improvement in isoleucine yield using the citramalate pathway for organisms dependent on simple carbon sources (such as acetate) and grown anaerobically (Geobacter spp., 43.2% and 43.4%; methanogens, 23.9% and 76.2%) (Fig. 4C). To examine whether the carbon source or anaerobic respiration contributes the most to such a large difference in yield change between E. coli and the other four organisms, we performed the same calculations on E. coli grown on acetate aerobically, glucose anaerobically, glucose using the electron transport chain (ETC) from Geobacter spp., and acetate using the ETC from Geobacter. We found the yield change to be most significant under anaerobic respiration (34.7% for E. coli glucose anaerobic). Interestingly, the isoleucine yield change is almost identical for E. coli with the Geobacter ETC and grown on acetate. We obtained similar results for a larger set of organisms shown in Fig. 3C, whereby under anaerobic respiration, the product yield changes the most between acyl-CoA–dependent and –independent pathways.
Thus, the citramalate pathway is more beneficial for organisms under anaerobic respiration and leads to much greater isoleucine yield. In contrast, for E. coli that can utilize aerobic respiration, the threonine pathway with greater thermodynamic favorability is selected over the citramalate pathway, which brings marginal benefit in terms of product yield.
Lysine Biosynthesis in Thermophiles Shows Differential Temperature Dependence of Thermodynamics.
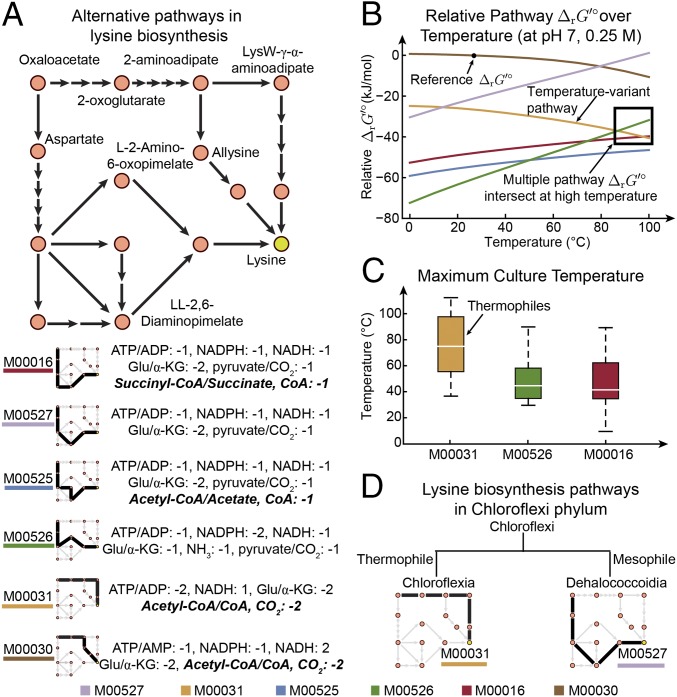
Examining the change of pathway thermodynamics with respect to temperature, we found that values of the diaminopimelate pathways (M00016, M00525, M00526, and M00527) generally increase with temperature, while values of the 2-aminoadipate pathways (M00030 and M00031) decrease with respect to temperature (Fig. 5B). These trends lead to the intersection of values at high temperature between pathway M00031 and pathways M00016 and M00526. Notably, organisms with pathway M00031 have culture temperature around the crossing point (80 to 100 °C) (Fig. 5C), at which the thermodynamics between the three lysine biosynthesis branches are almost equivalent. These organisms living at high temperature are called thermophiles. In contrast, organisms with pathways M00016 and M00526 have much lower culture temperature and are typically termed mesophiles (Fig. 5C). We also note that the organisms containing these three pathways did not show an appreciable difference in culture pH, with medians around pH 7 (SI Appendix, Fig. S8C). Therefore, at high temperature, pathway M00031 might become less disadvantageous in terms of thermodynamics compared with the other pathways and more likely to be selected by thermophiles. On the other hand, mesophiles still favor pathways with greater thermodynamic favorability, such as M00016 and M00526.
Fig. 5.
Alternative pathways for lysine biosynthesis. (A) Sketches of six alternative lysine biosynthetic pathways (with KEGG identifiers), along with their cofactor usage. (B) Thermodynamics of alternative pathways as a function of temperature. The pathway values relative to that of the pathway with the highest at 298.15 K, pH 7, and ionic strength 0.25 M (reference ). (C) Maximum culture temperature of organisms containing pathways M00031, M00526, and M00016. (D) Alternative lysine biosynthetic pathways in Chloroflexi phylum. Glu/α-KG, glutamate/α-ketoglutarate.
Interestingly, we found that organisms in the same phylum select different lysine biosynthesis pathways, and such choice is correlated with organism culture temperature. Specifically, for organisms in the Chloroflexi phylum, those from the Chloroflexia class are thermophiles with the M00031 pathway, while those from the Dehalococcoidia class are mesophiles with the M00527 pathway (Fig. 5D). We found that these two pathways have relatively similar thermodynamics at low temperature, but the M00031 pathway is much more favorable at high temperature than the M00527 pathway (Fig. 5B). This difference in thermodynamic favorability may explain the choice of thermophiles and mesophiles between these two pathways. Additionally, the results demonstrate how factors other than phylogeny can affect pathway choice, whereby organisms that are close in phylogenetic distance select different lysine biosynthesis pathways due to their different environmental conditions.
Discussion
In this work, we examined the basis for the presence of alternative biosynthetic routes for biomass precursors. We showed that acyl-CoA–dependent biosynthetic pathways are only present for five amino acids and investigated the possible factors related to the presence of acyl-CoA–dependent alternative routes. We evaluated the tradeoff between thermodynamic favorability and cofactor-use efficiency of the biosynthetic pathways and identified two clusters of organisms with distinct pathway choices. We found that organism living environment, rather than inherent metabolic capabilities, was the driving factor for alternative pathway choice. Specifically, organisms normally under aerobic respiration benefit from the thermodynamically more favorable routes, while organisms under anaerobic respiration benefit from cofactor-efficient routes, which are usually more advantageous in product yield. Additionally, we showed that organisms living at different temperatures can select alternative lysine biosynthesis pathways.
Examination of the thermodynamics of alternative pathways revealed that many pathways show high- and low-energy routes that are both prevalent among organism genomes. It may be argued that these pathways could have come about through evolution with insufficient selection to distinguish between these energetic alternatives. However, the presence of high- and low-energy alternatives with consistent cofactor structure (e.g., using acyl-CoA cleavage in amino acid biosynthesis), would suggest that these energetic alternatives may exist by selection. Because studies have demonstrated the advantage of thermodynamic favorability in increasing efficiency in protein use (18), a hypothesis emerges for why this might be the case. Presumably, the cell can choose to “waste” additional cofactor for higher thermodynamic favorability, reducing the downstream resources allocated for unnecessary pathway protein synthesis. While the current study examined only E. coli due to the limited organism-specific proteomics data available, more quantitative proteomics data across organisms and conditions may be used to probe this hypothesis.
We have found that organism lifestyle, such as type of respiration and temperature, can affect the alternative pathway choice for amino acid biosynthesis. Organisms in poor/anaerobic environments often choose cofactor-use–efficient pathways, which lead to significant yield improvement under those conditions. While those pathways may or may not depend on acyl-CoA cleavage, it is interesting to note that acyl-CoA is the distinguishing cofactor between the pathway alternatives. The underlying reason might be related to the cost of acyl-CoA under different conditions. Furthermore, while the improvement in growth rate can be small when using the cofactor-use–efficient pathways, such a benefit may not be relevant to organisms living under nutrient-poor and anaerobic conditions. For example, Geobacter spp. may rarely encounter favorable conditions to achieve maximum growth rate and, thus, would not select thermodynamically favorable routes to achieve better efficiency in protein use. In another specific case, thermophiles select pathways that become thermodynamically favorable at higher temperature in lysine biosynthesis. This observation suggests the possibility that certain alternatives may become viable when they become thermodynamically equivalent to other pathways under certain environmental conditions. This work complements known adaptations to high temperature, such as increase in protein stability (36) and alteration in membrane compositions (37).
Together, these results show how alternative pathway choice can be related to organism lifestyle, due to the tradeoff in thermodynamic favorability and cofactor-use efficiency. This study is one of a number of recent efforts aimed at discovering connections between thermodynamics and constraints on metabolic pathways. For example, one study showed that autotrophic amino acid synthesis was exergonic under the conditions in hydrothermal vents, rather than endergonic at surface conditions (38). Another recent effort looked at thermodynamic bottlenecks and proteomic constraints underlying the use of the Entner-Doudoroff pathway (18), indicating that similar tradeoffs in protein efficiency can be observed in central metabolism. As methods for estimating the thermodynamic properties of metabolic networks continue to be refined and as genome annotations continue to improve, these efforts are likely to continue to reveal the physical constraints underlying the adaptation and evolution of metabolic networks to meet organisms’ lifestyles.
Materials and Methods
The specific procedure for collecting the information of KEGG organisms and alternative biosynthetic pathways is described in SI Appendix, Supplementary Information Text. The workflow for calculation of pathway thermodynamics and product yield using metabolic network reconstructions can be found in SI Appendix, Supplementary Information Text.
Supplementary Material
Acknowledgments
This work was supported by the Novo Nordisk Foundation Grant NNF10CC1016517.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805367115/-/DCSupplemental.
References
- 1.Peregrín-Alvarez JM, Sanford C, Parkinson J. The conservation and evolutionary modularity of metabolism. Genome Biol. 2009;10:R63. doi: 10.1186/gb-2009-10-6-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venter JC, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 3.Monk J, Nogales J, Palsson BO. Optimizing genome-scale network reconstructions. Nat Biotechnol. 2014;32:447–452. doi: 10.1038/nbt.2870. [DOI] [PubMed] [Google Scholar]
- 4.Hatzimanikatis V, et al. Exploring the diversity of complex metabolic networks. Bioinformatics. 2005;21:1603–1609. doi: 10.1093/bioinformatics/bti213. [DOI] [PubMed] [Google Scholar]
- 5.Monk JM, et al. Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proc Natl Acad Sci USA. 2013;110:20338–20343. doi: 10.1073/pnas.1307797110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horikoshi K, Antranikian G, Bull AT, Robb FT, Stetter KO, editors. Extremophiles Handbook. Springer; Tokyo: 2010. [Google Scholar]
- 7.Barrie Johnson D, Hallberg KB. Carbon, iron and sulfur metabolism in acidophilic micro-organisms. Adv Microb Physiol. 2009;54:201–255. doi: 10.1016/S0065-2911(08)00003-9. [DOI] [PubMed] [Google Scholar]
- 8.Steunou A-S, et al. In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc Natl Acad Sci USA. 2006;103:2398–2403. doi: 10.1073/pnas.0507513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falb M, et al. Metabolism of halophilic archaea. Extremophiles. 2008;12:177–196. doi: 10.1007/s00792-008-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiele I, Palsson BØ. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santos F, Boele J, Teusink B. A practical guide to genome-scale metabolic models and their analysis. Methods Enzymol. 2011;500:509–532. doi: 10.1016/B978-0-12-385118-5.00024-4. [DOI] [PubMed] [Google Scholar]
- 12.Magnúsdóttir S, et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat Biotechnol. 2017;35:81–89. doi: 10.1038/nbt.3703. [DOI] [PubMed] [Google Scholar]
- 13.Ma H, Zeng A-P. Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms. Bioinformatics. 2003;19:270–277. doi: 10.1093/bioinformatics/19.2.270. [DOI] [PubMed] [Google Scholar]
- 14.Orth JD, Thiele I, Palsson BØ. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blank LM, Ebert BE, Bühler B, Schmid A. Metabolic capacity estimation of Escherichia coli as a platform for redox biocatalysis: Constraint-based modeling and experimental verification. Biotechnol Bioeng. 2008;100:1050–1065. doi: 10.1002/bit.21837. [DOI] [PubMed] [Google Scholar]
- 16.Campodonico MA, Andrews BA, Asenjo JA, Palsson BO, Feist AM. Generation of an atlas for commodity chemical production in Escherichia coli and a novel pathway prediction algorithm, GEM-Path. Metab Eng. 2014;25:140–158. doi: 10.1016/j.ymben.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Henry CS, Broadbelt LJ, Hatzimanikatis V. Thermodynamics-based metabolic flux analysis. Biophys J. 2007;92:1792–1805. doi: 10.1529/biophysj.106.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noor E, et al. Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLOS Comput Biol. 2014;10:e1003483. doi: 10.1371/journal.pcbi.1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caspi R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du B, et al. Temperature-dependent estimation of Gibbs energies using an updated group-contribution method. Biophys J. 2018;114:2691–2702. doi: 10.1016/j.bpj.2018.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.King ZA, et al. BiGG models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016;44:D515–D522. doi: 10.1093/nar/gkv1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kukko E, Heinonen J. The intracellular concentration of pyrophosphate in the batch culture of Escherichia coli. Eur J Biochem. 1982;127:347–349. doi: 10.1111/j.1432-1033.1982.tb06878.x. [DOI] [PubMed] [Google Scholar]
- 24.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asplund-Samuelsson J, Janasch M, Hudson EP. Thermodynamic analysis of computed pathways integrated into the metabolic networks of E. coli and Synechocystis reveals contrasting expansion potential. Metab Eng. 2018;45:223–236. doi: 10.1016/j.ymben.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt A, et al. The quantitative and condition-dependent Escherichia coli proteome. Nat Biotechnol. 2016;34:104–110. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basan M, et al. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature. 2015;528:99–104. doi: 10.1038/nature15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahadevan R, et al. Characterization of metabolism in the Fe(III)-reducing organism Geobacter sulfurreducens by constraint-based modeling. Appl Environ Microbiol. 2006;72:1558–1568. doi: 10.1128/AEM.72.2.1558-1568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benedict MN, Gonnerman MC, Metcalf WW, Price ND. Genome-scale metabolic reconstruction and hypothesis testing in the methanogenic archaeon Methanosarcina acetivorans C2A. J Bacteriol. 2012;194:855–865. doi: 10.1128/JB.06040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ekiel I, Smith ICP, Sprott GD. Biosynthesis of isoleucine in methanogenic bacteria: A carbon-13 NMR study. Biochemistry. 1984;23:1683–1687. [Google Scholar]
- 31.Charon NW, Johnson RC, Peterson D. Amino acid biosynthesis in the spirochete Leptospira: Evidence for a novel pathway of isoleucine biosynthesis. J Bacteriol. 1974;117:203–211. doi: 10.1128/jb.117.1.203-211.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Risso C, Van Dien SJ, Orloff A, Lovley DR, Coppi MV. Elucidation of an alternate isoleucine biosynthesis pathway in Geobacter sulfurreducens. J Bacteriol. 2008;190:2266–2274. doi: 10.1128/JB.01841-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feist AM, Scholten JCM, Palsson BØ, Brockman FJ, Ideker T. Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol Syst Biol. 2006;2:2006.0004. doi: 10.1038/msb4100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feist AM, et al. Constraint-based modeling of carbon fixation and the energetics of electron transfer in Geobacter metallireducens. PLOS Comput Biol. 2014;10:e1003575. doi: 10.1371/journal.pcbi.1003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monk JM, et al. iML1515, a knowledgebase that computes Escherichia coli traits. Nat Biotechnol. 2017;35:904–908. doi: 10.1038/nbt.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Razvi A, Scholtz JM. Lessons in stability from thermophilic proteins. Protein Sci. 2006;15:1569–1578. doi: 10.1110/ps.062130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koga Y. Thermal adaptation of the archaeal and bacterial lipid membranes. Archaea. 2012;2012:789652. doi: 10.1155/2012/789652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amend JP, Shock EL. Energetics of amino acid synthesis in hydrothermal ecosystems. Science. 1998;281:1659–1662. doi: 10.1126/science.281.5383.1659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.