Significance
The Wnt/β-catenin signaling pathway plays prominent roles during embryonic development and adult tissue homeostasis by maintaining somatic stem cell functions. The mammalian target of rapamycin complex 1 (mTORC1) signaling pathway has also been implicated in regulating stem cell functions in multiple tissue types. However, the crosstalk between these two pathways remains largely unclear. Herein, using in vitro cell lines, ex vivo organoids, and an in vivo mouse model, we made striking findings in support of a paradigm that mTORC1 signaling cell autonomously suppresses Wnt/β-catenin signaling through down-regulating the Wnt receptor FZD level to influence stem cell functions, with implications in the aging process.
Keywords: CRISPR screen, mTORC1 signaling, intestinal stem cells, Frizzled, organoids
Abstract
Wnt/β-catenin signaling plays pivotal roles in cell proliferation and tissue homeostasis by maintaining somatic stem cell functions. The mammalian target of rapamycin (mTOR) signaling functions as an integrative rheostat that orchestrates various cellular and metabolic activities that shape tissue homeostasis. Whether these two fundamental signaling pathways couple to exert physiological functions still remains mysterious. Using a genome-wide CRISPR-Cas9 screening, we discover that mTOR complex 1 (mTORC1) signaling suppresses canonical Wnt/β-catenin signaling. Deficiency in tuberous sclerosis complex 1/2 (TSC1/2), core negative regulators of mTORC1 activity, represses Wnt/β-catenin target gene expression, which can be rescued by RAD001. Mechanistically, mTORC1 signaling regulates the cell surface level of Wnt receptor Frizzled (FZD) in a Dishevelled (DVL)-dependent manner by influencing the association of DVL and clathrin AP-2 adaptor. Sustained mTORC1 activation impairs Wnt/β-catenin signaling and causes loss of stemness in intestinal organoids ex vivo and primitive intestinal progenitors in vivo. Wnt/β-catenin–dependent liver metabolic zonation gene expression program is also down-regulated by mTORC1 activation. Our study provides a paradigm that mTORC1 signaling cell autonomously regulates Wnt/β-catenin pathway to influence stem cell maintenance.
Wnt signaling is critical for development and tissue homeostasis, and its deregulation has been implicated in a variety of diseases (1–4). The canonical Wnt/β-catenin signaling pathway is initiated upon binding of Wnt ligand to the seven-pass transmembrane receptor FZD and the single-pass low-density lipoprotein receptor-related protein 5 or 6 (LRP5/6) on the cell surface. This event further triggers phosphorylation of LRP5/6, inactivation of β-catenin destruction complex, and accumulation of active β-catenin and partnering with TCF/LEF transcription factors, leading to the activation of Wnt/β-catenin target gene expression (2, 5). Although the core Wnt/β-catenin pathway components have been extensively investigated over the past decades, additional contributors and the crosstalk with other signaling pathways await further exploration.
As the core receptor of Wnt proteins, the strength of the Wnt/β-catenin signaling pathway is exquisitely sensitive to the level of FZD on the plasma membrane. Regulation of FZD turnover has emerged as a key regulatory node of the Wnt signaling pathway. For instance, FZD is degraded by membrane E3 ligase ZNRF3 and RNF43, and this activity of ZNRF3 and RNF43 is blocked by secreted R-spondin proteins (6, 7). Mutations of RNF43 and ZNRF3 and amplification of R-spondin genes are found in various tumors (8, 9). Later, DVL was identified to act as a negative regulator of FZD turnover, largely through recruitment of ZNRF3/RNF43 to FZD receptors (10). Considering the critical role of FZD in Wnt signaling, it would be expected that other signaling pathways might impact Wnt signaling through regulation of FZD turnover. Nevertheless, whether and how other pathways control FZD turnover is unknown.
The mammalian target of rapamycin complex 1 (mTORC1) signaling coordinates anabolic and catabolic metabolism to support cell growth and tissue homeostasis (11–13). The mTORC1 activity is controlled by various upstream signals, including growth factors, nutrients, and stress. Numerous growth factor and mitogen-dependent pathways converge on Tuberous Sclerosis Complex (TSC) to impact mTORC1 signaling transduction (12). TSC is composed of TSC1, TSC2, and TBC1D7, among which TSC2 is the catalytic subunit with GTPase-activating protein (GAP) activity toward the small GTPase Rheb which binds and directly activates mTORC1 activity through unclear mechanisms (14). The physical interaction of TSC1 and TSC2 is also essential for maintaining the protein stability of TSC2 (15, 16). Activated mTORC1 phosphorylates a variety of downstream effectors, such as S6K, 4EBP1, ULK1, TFEB, and Grb10, and consequently controls a number of cellular processes, including protein synthesis, autophagy, and growth factor signaling, among others (12). Recent studies have highlighted the roles of mTORC1 signaling in stem cell (SC) functions and aging (11–13). For example, Tsc1 deletion in hematopoietic stem cells (HSCs) led to elevated mTORC1 activity and concomitant loss of stemness in HSCs (17). Despite some similarities in regulating diverse fundamental cellular processes, whether Wnt and mTORC1 signaling pathways couple to exert physiological functions is still unclear.
In this study, we discovered a surprising inhibitory effect of mTORC1 signaling on Wnt/β-catenin signaling through an unbiased CRISPR screen. Mechanistic studies revealed that mTORC1 signaling decreases the expression of membrane FZD through enhancing DVL-mediated FZD turnover. Activation of mTORC1 signaling in vivo inhibits Wnt signaling in the intestine and liver. Our findings provide a mechanistic connection of these two fundamental signaling pathways and enhance our understanding of these pathways in stem cell maintenance and tissue homeostasis.
Results
A Genome-Wide CRISPR-Cas9 Screening Reveals that mTORC1 Signaling Negatively Modulates Wnt/β-Catenin Signaling.
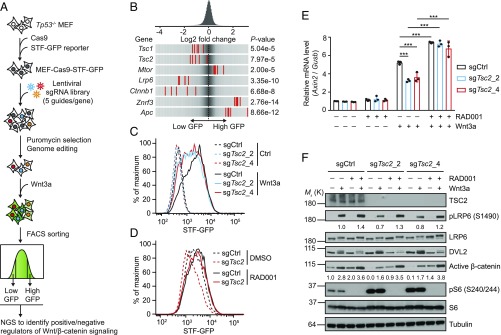
We set out to identify regulators of Wnt/β-catenin signaling by carrying out a genome-wide CRISPR-Cas9 screening (Fig. 1A). First, we engineered Tp53−/− mouse embryonic fibroblast (MEF) cells to express Cas9 and Super-Topflash green fluorescent protein (STF-GFP) reporter (MEF–Cas9–STF-GFP). The MEF–Cas9–STF-GFP cells were then infected with a pooled lentiviral sgRNA library covering 18,360 genes (5 sgRNAs per gene) at a sufficiently low multiplicity of infection to bias for integration of a single lentiviral sgRNA cassette per cell. After puromycin selection and multiple passages to allow gene editing to take place, STF-GFP reporter activity was induced by Wnt3a. Cells were then sorted for populations with the lowest 25% (low GFP) and the highest 25% (high GFP) of the GFP signal and subjected to next-generation sequencing (NGS) and informatics analysis to identify positive and negative regulators of Wnt/β-catenin signaling, respectively (Fig. 1A).
Fig. 1.
A genome-wide CRISPR-Cas9 screening reveals that mTORC1 signaling negatively modulates Wnt/β-catenin signaling. (A) Workflow of the CRISPR-Cas9 screening process. STF-GFP reporter activity was induced by Wnt3a stimulation overnight, allowing an ideal window for identifying both positive and negative regulators of Wnt/β-catenin signaling. (B) Frequency histograms of sgRNAs identified in the low GFP or high GFP population. sgRNAs targeting indicated genes are shown by the red lines. (C) Deletion of TSC2 decreases the STF reporter activity, assessed by flow cytometry analysis of GFP. (D) RAD001 (50 nM) increases the STF reporter activity and rescues TSC2-deletion–induced reduction of STF reporter activity in the presence of Wnt3a, assessed by flow cytometry analysis of GFP. (E) Quantitative RT-PCR analysis of Axin2 mRNA level (relative to Gusb). Data shown as mean ± SD for n = 3 independent experiments; significance tested using ANOVA; ***P < 0.001. (F) Immunoblots of indicated proteins for the same cells and treatments as described in E. Densitometry quantification of pLRP6 and active β-catenin are shown.
Inspection of the list of genes targeted by sgRNAs that are recovered from low GFP and high GFP populations revealed several genes encoding known core positive (Lrp6 and Ctnnb1) and negative regulators (Znrf3 and Apc) of Wnt/β-catenin pathway (Fig. 1B). Interestingly, sgRNAs targeting Tsc1/2 which encode core negative regulators of the mTORC1 signaling and Mtor itself were identified in the low GFP and high GFP populations (Fig. 1B), respectively, suggesting a suppressive role of mTORC1 signaling in the Wnt/β-catenin pathway. To validate the screening results, we deleted Tsc2 in MEF–Cas9–STF-GFP cells using two independent sgRNAs and confirmed that loss of Tsc2 decreased Wnt3a-induced STF-GFP reporter activity (Fig. 1C and SI Appendix, Fig. S1A). Conversely, inhibition of mTORC1 activity by RAD001 treatment increased Wnt3a-induced STF-GFP reporter activity and rescued Tsc2 knockout-induced STF-GFP reduction (Fig. 1D). Consistent with the alteration in reporter activity, expression of Axin2, a classical Wnt/β-catenin target gene, was suppressed by Tsc2 knockout and increased by RAD001 treatment (Fig. 1E). Western blot analysis further demonstrated that Wnt3a-induced accumulation of phosphorylated LRP6 (pLRP6) and active β-catenin were hampered by Tsc2 knockout while enhanced by RAD001 treatment (Fig. 1F). Additionally, RAD001 treatment also increased Wnt3a-induced accumulation of pLRP6 and active β-catenin in Tsc2−/− MEF (SI Appendix, Fig. S1B) and HEK293 (SI Appendix, Fig. S1C) cells. A recent report showed that mTORC1 activation resulted in feedback inactivation of Akt, leading to reduced GSK3β phosphorylation (pGSK3β) at serine 9 and concomitant loss of β-catenin activation in melanoma cell lines (18). However, mTORC1 activation in MEF cells exerted no effect on pGSK3β (SI Appendix, Fig. S1D). Moreover, inhibition of mTORC1 activity by RAD001 in Tsc2−/− MEF cells increased active β-catenin while decreasing the level of pGSK3β (SI Appendix, Fig. S1B), which is in agreement with the previous finding that S6K1 is the predominant GSK3 regulatory kinase under mTORC1-activated condition (19). These results are not consistent with the model that mTORC1 signaling inhibits Wnt signaling through decreasing GSK3 phosphorylation. Importantly, Tsc2 knockout decreased, while RAD001 treatment increased phosphorylation of LRP6 (Fig. 1F). These results together suggest that mTORC1 signaling negatively modulates Wnt/β-catenin signaling, probably through regulation of Wnt receptor activation on the cell surface.
mTORC1 Signaling Regulates the Cell Surface Protein Level of Wnt Receptor Frizzled.
Next we explored the molecular mechanisms through which mTORC1 regulates Wnt signaling. Frizzled (FZD) is the Wnt receptor responsible for Wnt/β-catenin signaling initiation and transduction (5). Since mTORC1 inhibits LRP6 phosphorylation, which is downstream of FZD activation, we examined whether mTORC1 signaling regulates cell surface level of FZD by flow cytometry using pan-FZD antibody 18R5 (20). Indeed, mTORC1 activation by Tsc2 deletion or mTORC1 inhibition by RAD001 treatment resulted in decrease or increase of cell surface FZD level, respectively, in MEF cells (Fig. 2A and SI Appendix, Fig. S2A). We also isolated primary MEF cells (Tp53 wild type, nonimmortalized) from a wild-type mouse and showed that RAD001 treatment increased the cell surface FZD level (SI Appendix, Fig. S2B), ruling out the possibility that FZD regulation by mTORC1 activity is p53 dependent. In addition, RAD001 was also able to increase the cell surface FZD levels in Tsc2−/− MEF (SI Appendix, Fig. S2C), HEK293 (SI Appendix, Fig. S2D), SK-MEL-30 (SI Appendix, Fig. S2F), and TSC2-null SNU-398 (21) (SI Appendix, Fig. S2G) cells. Taken together, these results demonstrate that mTORC1 signaling regulates the membrane levels of Wnt receptor FZD in different cell types.
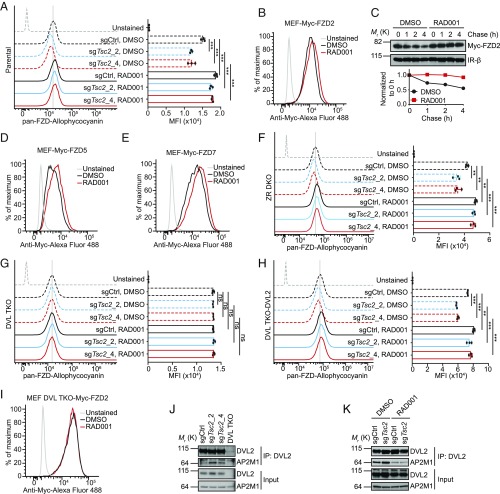
Fig. 2.
mTORC1 signaling regulates the cell surface protein level of Wnt receptor Frizzled in a DVL-dependent manner. (A) Flow cytometry analysis of membrane frizzled levels in control or TSC2-deleted MEF cells treated with DMSO or 50 nM RAD001. MFI, median fluorescence intensity. (B) Flow cytometry analysis of membrane Myc-FZD2 in MEF cells stably expressing ectopic Myc-FZD2 treated with DMSO or 50 nM RAD001. (C) mTORC1 inhibition by RAD001 treatment extends the half-life of Myc-FZD2 in a cell surface protein biotinylation-based pulse-chase assay in MEF–Myc–FZD2 cells. Insulin receptor-β (IR-β) acts as a control. Bottom, densitometry quantification of Myc-FZD2. (D and E) Flow cytometry analysis of membrane Myc-FZD5 (D) or Myc-FZD7 (E) in MEF cells stably expressing ectopic Myc-FZD5 or Myc-FZD7 treated with DMSO or 50 nM RAD001. (F) Flow cytometry analysis of membrane frizzled levels in control or TSC2-deleted MEF ZNRF3/RNF43 double knockout (ZR DKO) cells treated with DMSO or 50 nM RAD001. (G) Flow cytometry analysis of membrane frizzled levels in control or TSC2-deleted MEF DVL1/2/3 triple knockout (DVL TKO) cells treated with DMSO or 50 nM RAD001. (H) Flow cytometry analysis of membrane frizzled levels in control or TSC2-deleted MEF DVL TKO-DVL2 rescued cells treated with DMSO or 50 nM RAD001. (I) Flow cytometry analysis of membrane Myc-FZD2 in MEF DVL TKO cells stably expressing Myc-FZD2 treated with DMSO or 50 nM RAD001. (J) Deletion of TSC2 enhances the interaction between DVL2 and AP2M1, assessed by coimmunoprecipitation assay. (K) RAD001 decreases the interaction between DVL2 and AP2M1, assessed by coimmunoprecipitation assay. Error bars denote SD for n = 3 independent experiments (A, F, G, and H); significance tested using ANOVA; **P < 0.01, ***P < 0.001; ns, not significant.
We then asked whether FZD regulation by mTORC1 signaling is on the transcript or protein level. Assessment of the mRNA levels of all 10 Fzd genes in MEF cells revealed that Fzd2 is the dominant Fzd in MEF cells (SI Appendix, Fig. S3A). In line with this finding, knockout of FZD2 drastically reduced the cell surface FZD level (SI Appendix, Fig. S3B) and significantly reduced Wnt3a-induced STF reporter activity in MEF cells (SI Appendix, Fig. S3C). Importantly, perturbation of mTORC1 activity exhibited little effect on the mRNA levels of Fzd2 and other Fzds (SI Appendix, Fig. S3D), suggesting that mTORC1 signaling regulates the cell surface FZD at the protein level. Consistently, RAD001 treatment increased cell surface Myc-FZD2 level (Fig. 2B) and extended its half-life in a cell surface protein biotinylation-based pulse-chase experiment (Fig. 2C) in MEF cells stably expressing ectopic Myc-FZD2. Interestingly, RAD001 was still able to increase the residual cell surface FZD level in FZD2-deleted cells (SI Appendix, Fig. S3B), implying that mTORC1 signaling regulates several, if not all, FZDs. Indeed, RAD001 treatment increased the cell surface levels of ectopic Myc-FZD5 (Fig. 2D) and Myc-FZD7 (Fig. 2E) in MEF cells. Collectively, these results indicate that mTORC1 signaling regulates cell surface protein levels of multiple FZDs.
Regulation of FZD Protein Level by mTORC1 Signaling Is DVL Dependent.
ZNRF3 and RNF43 E3 ubiquitin ligases have previously been identified as responsible for FZD degradation (6, 7). To investigate whether the regulation of cell surface FZD level by mTORC1 signaling is through ZNRF3/RNF43, we knocked out Znrf3 and Rnf43 in MEF cells (SI Appendix, Fig. S4 A and B). Interestingly, manipulation of the mTORC1 signaling could still cause the alteration of cell surface FZD level in Znrf3/Rnf43 double knockout MEF (MEF ZR DKO) cells (Fig. 2F), in a similar manner to the observations in parental MEF cells (Fig. 2A), implying a ZNRF3/RNF43-independent mechanism by which mTORC1 signaling regulates cell surface FZD level. More recently, Dishevelled (DVL) was shown to be a negative regulator of cell surface FZD level (10). To test whether DVL is required for mTORC1-mediated FZD regulation, we knocked out Dvl1/2/3 in MEF cells (SI Appendix, Fig. S4C). Consistent with Dvl1/2/3 knockout in HEK293 cells (10), Wnt3a-induced STF reporter activity was abrogated in Dvl1/2/3 triple knockout MEF (MEF DVL TKO) cells (SI Appendix, Fig. S4D), whereas the response to GSK3 inhibitor CHIR99021 was retained (SI Appendix, Fig. S4E). Importantly, neither Tsc2 deletion nor RAD001 supplementation affected the cell surface FZD level in MEF DVL TKO cells (Fig. 2G), whereas the regulation of cell surface FZD level by mTORC1 signaling was restored when DVL2 was reintroduced into MEF DVL TKO cells (Fig. 2H and SI Appendix, Fig. S4F). Moreover, RAD001 treatment had no effect on the cell surface level of ectopic Myc-FZD2 in MEF DVL TKO cells (Fig. 2I) or endogenous FZD in HEK293 DVL TKO cells (SI Appendix, Fig. S2E). These findings suggest that mTORC1 signaling regulates cell surface FZD level in a DVL-dependent manner. Given that Dvl1/2/3 knockout abolished Wnt signaling transduction from the cell membrane receptors, it is possible that manipulation of mTORC1 activity failed to alter FZD levels simply due to the loss of Wnt signaling resulting from Dvl1/2/3 knockout. To rule out this possibility, we knocked out Ctnnb1, which encodes β-catenin protein, in MEF cells (SI Appendix, Fig. S5A). As expected, knockout of β-catenin completely abolished Wnt signaling in MEF cells (SI Appendix, Fig. S5B). However, RAD001 treatment still increased the cell surface FZD level in β-catenin–deleted cells (SI Appendix, Fig. S5C). These data further strengthen the idea that regulation of FZD protein level by mTORC1 signaling is DVL dependent, but ZNRF3/RNF43 or β-catenin independent.
Next we sought to gain further insight into how DVL is involved in mTORC1-mediated FZD regulation. Although DVL acts as an adaptor for ZNRF3/RNF43 to recognize and down-regulate FZD (10), DVL TKO cells manifested a higher cell surface FZD level than ZR DKO cells and parental cells in both MEF (SI Appendix, Fig. S4G) and HEK293 (10) contexts, suggesting the existence of an alternative route for DVL to regulate the cell surface FZD level. A previous report has demonstrated that association of DVL with the clathrin AP-2 adaptor is essential for FZD internalization (22). Therefore, DVL likely promotes FZD degradation through promoting ZNRF3/RNF43-mediated FZD ubiquitination and through enhancing clathrin-mediated FZD internalization. Since mTORC1-mediated FZD regulation is DVL dependent and ZNRF3/RNF43 independent, it is possible that mTORC1 signaling regulates FZD turnover through affecting DVL–AP-2 interaction. To test this hypothesis, we assessed the interaction of endogenous DVL2 and AP2M1 proteins by coimmunoprecipitation assay. AP2M1 was coimmunoprecipitated with DVL2 antibody in control cells, but not DVL TKO cells, demonstrating the specificity of the coimmunoprecipitation assay (Fig. 2J). Knockout of Tsc2 using two independent sgRNAs increased DVL2–AP2M1 association (Fig. 2J), while RAD001 treatment decreased DVL2–AP2M1 association in both control cells and Tsc2 knockout cells (Fig. 2K). Notably, effects of Tsc2 deletion or RAD001 treatment on DVL2–AP2M1 interactions correlated perfectly with the effects of these treatments on cell surface FZD level (Fig. 2A). Collectively, these results suggest that mTORC1 signaling regulates cell surface FZD level by influencing the association of DVL with AP-2 adaptor.
Activation of mTORC1 Signaling Impairs Wnt Signaling and Stem Cell Functions in Intestinal Organoids.
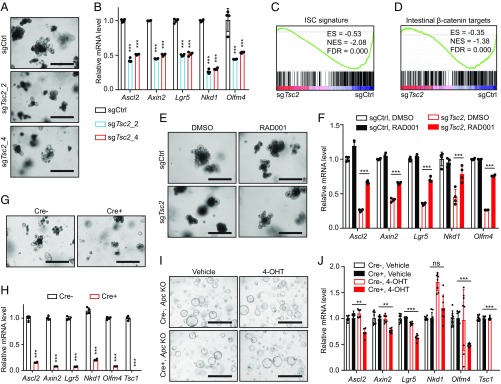
Wnt/β-catenin signaling is essential for intestinal stem cell (ISC) maintenance and organoid functions ex vivo (23, 24). Having established the crosstalk between mTORC1 and Wnt/β-catenin signaling in vitro, we wondered whether sustained mTORC1 activation could destroy Wnt/β-catenin–regulated ISC functions in organoids. To test this idea, we isolated small intestine (SI) crypts from a Cas9-expressing C57BL6/J mouse and cultured them as organoids ex vivo, followed by lentiviral infection of two independent sgRNAs targeting Tsc2 to achieve genetic Tsc2 deletion (SI Appendix, Fig. S6A). Tsc2 deletion markedly impaired crypt formation in organoids (Fig. 3A), indicating a decline in ISC functions typically associated with reduced Wnt/β-catenin signaling (25). Accordingly, gene expression analysis revealed that Tsc2 deletion repressed the expression of Wnt/β-catenin targets and ISC markers (Fig. 3B). Gene set enrichment analysis (GSEA) further demonstrated significant down-regulation of both ISC gene signature (26) (Fig. 3C) and intestinal β-catenin targets (27) (Fig. 3D) in Tsc2-deficient organoids. Conversely, mTORC1 inhibition by RAD001 treatment largely rescued Tsc2-deletion–induced crypt-loss phenotype (Fig. 3E) and gene expression defect (Fig. 3F) in organoids. Collectively, these data suggest that sustained mTORC1 activation suppresses Wnt/β-catenin signaling and impairs ISC functions in an organoid model.
Fig. 3.
Activation of mTORC1 signaling impairs Wnt signaling and stem cell functions in intestinal organoids. (A) Morphology of control or TSC2-deleted intestinal organoids cultured for 4 d. (B) Quantitative RT-PCR analysis of relative mRNA levels of Wnt/β-catenin target genes and intestinal stem cell marker genes in organoids described in A. n = 4. (C and D) GSEA of the ISC gene signature (C) and intestine-specific β-catenin target gene sets (D) in TSC2-deleted versus control organoids. (E) Morphology of control or TSC2-deleted organoids cultured in the presence of DMSO or 50 nM RAD001 for 5 d. (F) Quantitative RT-PCR analysis of relative mRNA levels of Wnt/β-catenin target genes and intestinal stem cell marker genes in organoids described in E. n = 4. (G) Morphology of Tsc1fl/fl (Cre−) or Tsc1fl/fl; Rosa26-CreER (Cre+) organoids cultured in the presence of 1 µM 4-OHT for 4 d. (H) Quantitative RT-PCR analysis of relative mRNA levels of Wnt/β-catenin target genes, intestinal stem cell marker genes, and Tsc1 gene in organoids described in G. n = 4. (I) Morphology of APC-deleted Tsc1fl/fl (Cre−) or Tsc1fl/fl; Rosa26-CreER (Cre+) organoids cultured in the presence of vehicle or 1 µM 4-OHT for 4 d. (J) Quantitative RT-PCR analysis of relative mRNA levels of Wnt/β-catenin target genes, intestinal stem cell marker genes, and Tsc1 gene in APC-deleted Tsc1fl/fl (Cre−) or Tsc1fl/fl; Rosa26-CreER (Cre+) organoids cultured in the presence of vehicle or 1 µM 4-OHT for 4 d. n = 8. Data shown as mean ± SD (B, F, H, and J); significance tested using two-tailed t test (B and H) and ANOVA (F and J); **P < 0.01, ***P < 0.001; ns, not significant. [Scale bars: 200 µm (A and E), 400 µm (G), and 1,000 µm (I).] ES, enrichment score; FDR, false discovery rate; NES, normalized enrichment score.
To further strengthen the above findings, we isolated SI crypts from Tsc1fl/fl (Cre−) littermates or Tsc1fl/fl; Rosa26-creERT2 (Cre+) mice and allow for organoid formation ex vivo. Tsc1 deletion by 4-OH tamoxifen (4-OHT) treatment remarkably inhibited Cre+ organoid formation, whereas Cre− organoids exhibited normal expansion upon 4-OHT treatment (Fig. 3G and SI Appendix, Fig. S6B). In line with the observations in MEF cells in vitro (Fig. 1F), mTORC1 activation in organoids significantly decreased the levels of pLRP6 and active β-catenin (SI Appendix, Fig. S6B). Additionally, Tsc1 deletion resulted in severe repression of Wnt/β-catenin targets and ISC gene signature (26, 27) (Fig. 3H and SI Appendix, Fig. S6 C and D), confirming that persistent mTORC1 activity is detrimental to Wnt/β-catenin–mediated ISC maintenance. Compared with Tsc2 CRISPR knockout, deletion of Tsc1 using Cre-ERT2 showed stronger effects on organoids, likely due to more uniform gene inactivation. To evaluate whether mTORC1 activation-induced down-regulation of Wnt/β-catenin signaling is causative to impaired ISC functions, we activated Wnt/β-catenin signaling by lentiviral coinfection of Cas9 and Apc targeting sgRNA in Cre− or Cre+ organoids to delete Apc, a core suppressive factor of the Wnt/β-catenin pathway. Apc-null organoids were established by R-spondin withdrawal from the organoid culture media. Activation of the Wnt/β-catenin pathway effectively prevented Tsc1 deletion-induced organoid degeneration (Fig. 3I). The gene expression defect resulting from Tsc1 deletion was also markedly ameliorated with Wnt/β-catenin pathway activation (Fig. 3J). The residual reduction in gene expression could be due to the effect of upstream Wnt signaling in APC KO cells (28) or the existence of a minor mechanism that affects Wnt/β-catenin signaling at the level of β-catenin. Taken together, these results demonstrate that persistent mTORC1 activity disrupts ISC functions by suppressing Wnt/β-catenin signaling.
Activation of mTORC1 Signaling Decreases Wnt Signaling in Vivo.
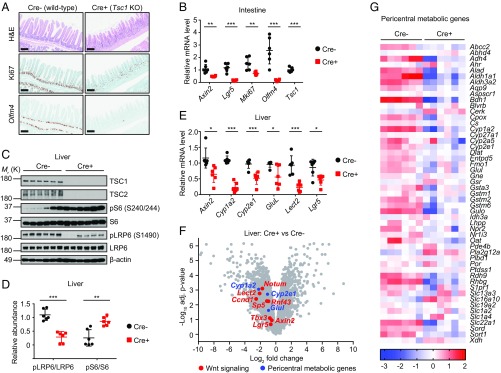
We next determined the effect of mTORC1 activation on Wnt/β-catenin signaling and intestinal epithelium homeostasis in vivo. Histological evaluation of the small intestines of Cre− or Cre+ mice after tamoxifen administration revealed a degenerative phenotype associated with loss of Ki67+ proliferating crypts upon Tsc1 deletion (Fig. 4A). To examine whether Tsc1 deletion reduced ISC numbers, we performed in situ hybridization (ISH) for Olfm4, a marker that is coexpressed by Lgr5+ ISCs (29, 30). Olfm4+ primitive ISCs were remarkably reduced in Tsc1-deficient mice (Fig. 4A). Consistent with the degenerative phenotype, Wnt/β-catenin target gene expression was also significantly decreased in Tsc1-deficient small intestines (Fig. 4B). Together, these results suggest that sustained mTORC1 activation down-regulates Wnt/β-catenin signaling and inhibits the function of ISCs in vivo.
Fig. 4.
Activation of mTORC1 signaling by Tsc1 deletion decreases Wnt signaling in vivo. (A) Hematoxylin and eosin (H&E) staining, Ki67 immunohistochemistry staining, and Olfm4 in situ hybridization analysis in small intestines of Tsc1fl/fl (Cre−) or Tsc1fl/fl; Rosa26-CreER (Cre+) mice dosed with tamoxifen. (B) Quantitative RT-PCR analysis of relative mRNA levels of Wnt/β-catenin target genes, intestinal stem cell marker genes, and Tsc1 gene in small intestines of Tsc1fl/fl (Cre−) or Tsc1fl/fl; Rosa26-CreER (Cre+) mice dosed with tamoxifen. n = 6 mice per group. (C) Immunoblots of indicated proteins in livers of Tsc1fl/fl (Cre−) or Tsc1fl/fl; Rosa26-CreER (Cre+) mice dosed with tamoxifen. n = 6 mice per group. (D) Densitometry quantification of immunoblots in C showing the ratio of pLRP6/total LRP6 and pS6/total S6. (E) Quantitative RT-PCR analysis of relative mRNA levels of Wnt/β-catenin target genes and pericentral metabolic genes in livers of Tsc1fl/fl (Cre−) or Tsc1fl/fl; Rosa26-CreER (Cre+) mice dosed with tamoxifen. n = 6 mice per group. (F) Volcano plot showing selective pericentral genes (in blue) and Wnt/β-catenin target genes (in red) that are differentially expressed in livers of Tsc1fl/fl (Cre−) or Tsc1fl/fl; Rosa26-CreER (Cre+) mice dosed with tamoxifen. n = 6 mice per group. (G) Heatmap showing mRNA expression of pericentral metabolic genes in livers of Tsc1fl/fl (Cre−) or Tsc1fl/fl; Rosa26-CreER (Cre+) mice dosed with tamoxifen. n = 6 mice per group. Error bars denote SD (B, D, and E); significance tested using two-tailed t test; *P < 0.05, **P < 0.01, ***P < 0.001. [Scale bars: 200 µm (A).]
Emerging evidence have highlighted the Wnt/β-catenin pathway as a major regulator of metabolic liver zonation which dictates hepatocyte functions by its position along the portocentral axis of the liver lobule (31–33). Wnt/β-catenin activity is persistently high around the central vein of the liver lobule (34). To explore the impact of mTORC1 activation on Wnt/β-catenin–mediated metabolic zonation regulation, we collected livers from Cre− or Cre+ mice after tamoxifen administration and observed significant down-regulation of pLRP6 upon Tsc1 deletion (Fig. 4 C and D). In keeping with this notion, expression of classical Wnt/β-catenin targets Axin2, Lect2, and Lgr5 was strongly inhibited (Fig. 4E), indicating the down-regulation of Wnt/β-catenin signaling upon Tsc1 deletion in liver. Furthermore, expression of Wnt/β-catenin–dependent metabolic genes Cyp1a2, Cyp2e1, and Glul was also markedly reduced in Tsc1-deficient livers (Fig. 4E). Unbiased high-throughput RNA sequencing (RNA-seq) on total liver RNA samples isolated from Cre− or Cre+ mice affirmed the down-regulation of Wnt/β-catenin target genes (Fig. 4F) and pericentrally expressed metabolic genes (33) (Fig. 4 F and G) in Tsc1-deficient livers. Collectively, these data support the idea that mTORC1 activation down-regulates the Wnt/β-catenin–mediated gene expression program involved in liver metabolic zonation. It should be noted that the in vivo study involved whole-body Tsc1 deletion, so a paracrine effect from other tissues cannot be ruled out. Nevertheless, the effect of mTORC1 activation on Wnt/β-catenin signaling is highly consistent in in vitro cell culture, ex vivo organoid, and in vivo mouse models.
Discussion
In this study, we discover an inhibitory role of mTORC1 activation in the Wnt/β-catenin signaling pathway through an unbiased genome-wide CRISPR screen and demonstrate its importance using ex vivo intestinal organoids and an in vivo mouse model. Although the regulatory outputs of mTORC1 signaling on Wnt/β-catenin targets could still be indirect, we favor a model in which mTORC1 signaling cell autonomously suppresses Wnt/β-catenin signaling through promoting DVL–AP-2 interaction and enhancing DVL-mediated FZD internalization. Thus, DVL-mediated FZD turnover represents a regulatory node of Wnt signaling for pathway crosstalk.
Although DVL–AP-2 interaction is required for FZD internalization (22), the exact binding mode between DVL and the AP-2 adaptor is not completely clear. The Kirchhausen laboratory initially identified that both a tyrosine motif (YHEL) and the DEP domain, particularly the K446 residue, of DVL contribute to AP-2 adaptor binding (22). Subsequently, the same group determined the crystal structure of a single-chain chimera with a variable linker between DEP and YHEL and a second variable linker between YHEL and the C-terminal domain of the µ2 subunit of the AP-2 adaptor (µ2C) (35). In this structure, both the DEP domain and YHEL motif are involved in µ2C interaction, but the K446 residue of the DEP domain is not (35). Although the YHEL motif makes a major contribution to DVL–AP-2 interaction in these two studies, the DVL2–AHEA mutant was fully capable of down-regulating the FZD level in both MEF DVL TKO cells (SI Appendix, Fig. S7) and HEK293 DVL TKO cells (10, 36). In contrast, the DVL2–K446M mutant failed to decrease the FZD level in both MEF DVL TKO cells (SI Appendix, Fig. S7) and HEK293 DVL TKO cells (10), consistent with the model that the K446 residue is directly involved in DVL–FZD interaction (37). Other regions of DVL, and even other proteins, might facilitate the physical interaction between DVL and AP-2.
Our data clearly demonstrated that mTORC1 signaling increases DVL–AP-2 interaction, though the exact mechanism is unclear. Given that DVL is a phosphoprotein, it is possible that mTORC1 regulates phosphorylation of DVL. Two previous mass spectrometry studies systematically identified mTOR-regulated phosphoproteome in immortalized MEF cells (38, 39). Neither DVL nor AP-2 adaptor was among the list of mTORC1 substrates which are sensitive to perturbation of mTORC1 activity. However, focused quantitative phosphoproteomic analysis on DVL is required to further delineate whether DVL phosphorylation is regulated by mTORC1. On the other hand, we noticed that RAD001 treatment appears to decrease the DVL2 protein level in MEF cells (Fig. 1F). Whether this observation is functionally relevant is still not clear. It is possible that a certain form of DVL promotes FZD degradation, and the expression of this form of DVL is decreased upon mTORC1 inhibition. Future studies are clearly needed to map out the mechanistic details of the mTORC1-regulated DVL–AP-2 interaction.
Accumulating studies have suggested an important regulatory role of the TSC/mTORC1 signaling in SC maintenance in multiple tissue types (11, 40). For instance, sustained activation of mTORC1 signaling leads to loss of stemness in hematopoietic SCs (17), hair follicle SCs (41), and tracheal epithelium SCs (42) in mice. Chronic mTORC1 activation also results in a decline of ISC numbers in flies (42), the phenotype of which is now extended to the murine model in the present study (Fig. 4A). Interestingly, reduction of mTORC1 activity by calorie restriction promotes ISC functions and increases the size of intestinal crypts in mice (43). Although a noncell-autonomous mechanism through Paneth cells is proposed (43), it is possible that cell-autonomous regulation of Wnt/β-catenin signaling by mTORC1 signaling in ISCs also contributes to this observation. The cell-autonomous effects of mTORC1 signaling on SC maintenance have been supported by another finding that dietary supplementation with rapamycin is sufficient to promote mouse tracheal SC maintenance and thus preserve regenerative capacity of airway epithelia (42), although whether Wnt/β-catenin signaling is accordingly involved remains to be resolved. Our data demonstrate cell-autonomous down-regulation of Wnt/β-catenin signaling induced by mTORC1 activation, which is associated with the loss of ISCs as well as repression of the liver metabolic zonation-related gene expression program. Given that mTORC1 signaling coordinates various cellular events, including protein translation, autophagy, and metabolism, among others, we can thus not exclude the possibility that such downstream pathways are also involved in the degenerative phenotype resulting from mTORC1 activation.
In normal mouse intestines, mTORC1 signaling is active in normal intestinal crypts where Wnt/β-catenin signaling is also highly active (44, 45). Our data, together with the observation that calorie restriction increases the size of intestinal crypts, appear to be counterintuitive. Nevertheless, these findings suggest a potential self-check mechanism to precisely regulate mTORC1 signaling and Wnt/β-catenin signaling for tissue homeostasis. Interestingly, Inoki et al. (46) reported that Wnt activates mTOR signaling by inhibition of GSK3. Therefore, high mTORC1 signaling in ISCs could represent a negative feedback mechanism to prevent overactivation of Wnt signaling. Hyperactivation of Wnt/β-catenin signaling, typically associated with loss-of-function mutations in the APC tumor suppressor, is the leading cause of development of colorectal cancer (1, 3). Numerous studies have demonstrated that mTORC1 inhibition blocks APC loss-driven colorectal tumorigenesis (45, 47–49), consistent with a critical role of mTORC1 signaling in tumorigenesis. Since mTORC1 inhibits the upstream part of the Wnt/β-catenin signaling pathway, mTORC1 inhibition is not expected to block β-catenin signaling in APC-deficient cells.
Lastly, emerging evidence has demonstrated that aging is accompanied by activation of mTORC1 signaling, and long-term inhibition of mTORC1 activity is capable of extending lifespan in various organisms (13). A recent study reported a decline of ISC functions upon aging due to decreased canonical Wnt/β-catenin signaling, and reactivating Wnt/β-catenin signaling in aged ISCs restores function in organoids (50). Considering that mTORC1 signaling inhibits Wnt/β-catenin signaling, enhanced Wnt/β-catenin signaling in stem cells might contribute to mTORC1 inhibition-associated increased function of stem cells and extension of life span. Our findings provide a mechanistic connection of these two fundamental signaling pathways and may facilitate the deeper understanding of the interplay between mTORC1 and Wnt/β-catenin signaling pathways in stem cell maintenance and aging.
Materials and Methods
All animal work was performed in accordance with protocols approved by the Novartis Institutes for Biomedical Research Institutional Animal Care and Use Committee and the study was compliant with ethical regulations regarding animal research. Detailed procedures for the genome-wide CRISPR screen, generation of knockout cell lines, flow cytometry, immunoblotting, and immunoprecipitation, quantitative RT-PCR and RNA-seq, animal experiments, and intestinal organoid culture are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Melissa Paziuk for mice maintenance; Jacob Lafauce, Alyssa Riley, and Abbie Griggs for assisting in animal experiments; Hua Wu for isolating primary MEF cells; Alicia Lindeman, Scott Clarkson, Qiong Wang, Jian Wang, Qinhui Song, and Debora Bonenfant for technical assistance; and Sue Menon, Hui Wang, Xiaomo Jiang, Yan Feng, and David Glass for comments and advice.
Footnotes
Conflict of interest statement: All authors were employees of Novartis Pharma AG when the research was conducted.
This article is a PNAS Direct Submission.
Data deposition: The raw RNA-seq reads are available in the NCBI Short Read Archive under the accession no. SRP140499 (for liver RNA-seq samples) and no. SRP155719 (for intestinal organoid samples).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808575115/-/DCSupplemental.
References
- 1.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nusse R, Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 4.Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 5.MacDonald BT, He X. Frizzled and LRP5/6 receptors for Wnt/β-catenin signaling. Cold Spring Harb Perspect Biol. 2012;4:a007880. doi: 10.1101/cshperspect.a007880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hao HX, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485:195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- 7.Koo BK, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- 8.Jiang X, et al. Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci USA. 2013;110:12649–12654. doi: 10.1073/pnas.1307218110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giannakis M, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. 2014;46:1264–1266. doi: 10.1038/ng.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang X, Charlat O, Zamponi R, Yang Y, Cong F. Dishevelled promotes Wnt receptor degradation through recruitment of ZNRF3/RNF43 E3 ubiquitin ligases. Mol Cell. 2015;58:522–533. doi: 10.1016/j.molcel.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 11.Russell RC, Fang C, Guan KL. An emerging role for TOR signaling in mammalian tissue and stem cell physiology. Development. 2011;138:3343–3356. doi: 10.1242/dev.058230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zoncu R, Efeyan A, Sabatini DM. mTOR: From growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, Manning BD. The TSC1-TSC2 complex: A molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benvenuto G, et al. The tuberous sclerosis-1 (TSC1) gene product hamartin suppresses cell growth and augments the expression of the TSC2 product tuberin by inhibiting its ubiquitination. Oncogene. 2000;19:6306–6316. doi: 10.1038/sj.onc.1204009. [DOI] [PubMed] [Google Scholar]
- 16.Chong-Kopera H, et al. TSC1 stabilizes TSC2 by inhibiting the interaction between TSC2 and the HERC1 ubiquitin ligase. J Biol Chem. 2006;281:8313–8316. doi: 10.1074/jbc.C500451200. [DOI] [PubMed] [Google Scholar]
- 17.Chen C, et al. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao J, et al. Tuberous sclerosis complex inactivation disrupts melanogenesis via mTORC1 activation. J Clin Invest. 2017;127:349–364. doi: 10.1172/JCI84262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang HH, Lipovsky AI, Dibble CC, Sahin M, Manning BD. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol Cell. 2006;24:185–197. doi: 10.1016/j.molcel.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gurney AL, et al. 2017. Frizzled-binding agents and uses thereof (US Patent 20170234854A1)
- 21.Huynh H, et al. Loss of tuberous sclerosis complex 2 (TSC2) is frequent in hepatocellular carcinoma and predicts response to mTORC1 inhibitor everolimus. Mol Cancer Ther. 2015;14:1224–1235. doi: 10.1158/1535-7163.MCT-14-0768. [DOI] [PubMed] [Google Scholar]
- 22.Yu A, et al. Association of Dishevelled with the clathrin AP-2 adaptor is required for Frizzled endocytosis and planar cell polarity signaling. Dev Cell. 2007;12:129–141. doi: 10.1016/j.devcel.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barker N. Adult intestinal stem cells: Critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 24.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 25.Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muñoz J, et al. The Lgr5 intestinal stem cell signature: Robust expression of proposed quiescent '+4′ cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fevr T, Robine S, Louvard D, Huelsken J. Wnt/beta-catenin is essential for intestinal homeostasis and maintenance of intestinal stem cells. Mol Cell Biol. 2007;27:7551–7559. doi: 10.1128/MCB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voloshanenko O, et al. Wnt secretion is required to maintain high levels of Wnt activity in colon cancer cells. Nat Commun. 2013;4:2610. doi: 10.1038/ncomms3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 30.van der Flier LG, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 31.Yang J, et al. β-catenin signaling in murine liver zonation and regeneration: A Wnt-Wnt situation! Hepatology. 2014;60:964–976. doi: 10.1002/hep.27082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monga SP. Role and regulation of β-catenin signaling during physiological liver growth. Gene Expr. 2014;16:51–62. doi: 10.3727/105221614X13919976902138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Planas-Paz L, et al. The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat Cell Biol. 2016;18:467–479. doi: 10.1038/ncb3337. [DOI] [PubMed] [Google Scholar]
- 34.Gebhardt R, Hovhannisyan A. Organ patterning in the adult stage: The role of Wnt/beta-catenin signaling in liver zonation and beyond. Dev Dyn. 2010;239:45–55. doi: 10.1002/dvdy.22041. [DOI] [PubMed] [Google Scholar]
- 35.Yu A, Xing Y, Harrison SC, Kirchhausen T. Structural analysis of the interaction between Dishevelled2 and clathrin AP-2 adaptor, a critical step in noncanonical Wnt signaling. Structure. 2010;18:1311–1320. doi: 10.1016/j.str.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gammons MV, Rutherford TJ, Steinhart Z, Angers S, Bienz M. Essential role of the Dishevelled DEP domain in a Wnt-dependent human-cell-based complementation assay. J Cell Sci. 2016;129:3892–3902. doi: 10.1242/jcs.195685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gammons MV, Renko M, Johnson CM, Rutherford TJ, Bienz M. Wnt signalosome assembly by DEP domain swapping of dishevelled. Mol Cell. 2016;64:92–104. doi: 10.1016/j.molcel.2016.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu PP, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu Y, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meng D, Frank AR, Jewell JL. mTOR signaling in stem and progenitor cells. Development. 2018;145:dev152595. doi: 10.1242/dev.152595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castilho RM, Squarize CH, Chodosh LA, Williams BO, Gutkind JS. mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell. 2009;5:279–289. doi: 10.1016/j.stem.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haller S, et al. mTORC1 activation during repeated regeneration impairs somatic stem cell maintenance. Cell Stem Cell. 2017;21:806–818 e805. doi: 10.1016/j.stem.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yilmaz OH, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Metcalfe C, et al. Dvl2 promotes intestinal length and neoplasia in the ApcMin mouse model for colorectal cancer. Cancer Res. 2010;70:6629–6638. doi: 10.1158/0008-5472.CAN-10-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujishita T, Aoki K, Lane HA, Aoki M, Taketo MM. Inhibition of the mTORC1 pathway suppresses intestinal polyp formation and reduces mortality in ApcDelta716 mice. Proc Natl Acad Sci USA. 2008;105:13544–13549. doi: 10.1073/pnas.0800041105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inoki K, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 47.Brandt M, et al. mTORC1 inactivation promotes colitis-induced colorectal cancer but protects from APC loss-dependent Tumorigenesis. Cell Metab. 2018;27:118–135 e118. doi: 10.1016/j.cmet.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 48.Faller WJ, et al. mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature. 2015;517:497–500. doi: 10.1038/nature13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gulhati P, et al. Targeted inhibition of mammalian target of rapamycin signaling inhibits tumorigenesis of colorectal cancer. Clin Cancer Res. 2009;15:7207–7216. doi: 10.1158/1078-0432.CCR-09-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nalapareddy K, et al. Canonical Wnt signaling ameliorates aging of intestinal stem cells. Cell Rep. 2017;18:2608–2621. doi: 10.1016/j.celrep.2017.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.