Significance
UBQLN2, a ubiquitin-linked quality-control protein, accumulates in common neurodegenerative diseases and, when mutated, directly causes neurodegeneration. Employing a range of model systems, we show that UBQLN2 is intrinsically prone to self-assemble, leading to the formation of liquid-like droplets and amyloid aggregates. A disease-causing mutation in UBQLN2 impairs droplet dynamics and favors amyloid-like aggregation associated with neurotoxicity. Self-assembly is regulated by ubiquitin-linked domains in UBQLN2, implying a functional relationship between oligomerization and ubiquitin-dependent protein quality control. Our results emphasize a critical link between UBQLN2’s role in ubiquitin-dependent pathways and its propensity to self-assemble and aggregate in neurodegenerative diseases.
Keywords: UBQLN2, ALS, FTD, liquid–liquid phase separation, membraneless organelle
Abstract
UBQLN2 is one of a family of proteins implicated in ubiquitin-dependent protein quality control and integrally tied to human neurodegenerative disease. Whereas wild-type UBQLN2 accumulates in intraneuronal deposits in several common age-related neurodegenerative diseases, mutations in the gene encoding this protein result in X-linked amyotrophic lateral sclerosis/frontotemporal dementia associated with TDP43 accumulation. Using in vitro protein analysis, longitudinal fluorescence imaging and cellular, neuronal, and transgenic mouse models, we establish that UBQLN2 is intrinsically prone to self-assemble into higher-order complexes, including liquid-like droplets and amyloid aggregates. UBQLN2 self-assembly and solubility are reciprocally modulated by the protein’s ubiquitin-like and ubiquitin-associated domains. Moreover, a pathogenic UBQLN2 missense mutation impairs droplet dynamics and favors amyloid-like aggregation associated with neurotoxicity. These data emphasize the critical link between UBQLN2’s role in ubiquitin-dependent pathways and its propensity to self-assemble and aggregate in neurodegenerative diseases.
The accumulation of insoluble proteins in extracellular or intracellular deposits is a central theme among neurodegenerative diseases, including the amyotrophic lateral sclerosis/frontotemporal dementia (ALS/FTD) spectrum. The proteins comprising these deposits accumulate spontaneously in common, sporadic forms of disease and, less commonly when mutated, these same proteins—including VCP, P62, TDP43, and FUS—also underlie rare inherited forms of disease (1–3). A recent addition to this class of proteins is UBQLN2, a ubiquitin-binding protein implicated in neuronal protein quality control. Neuronal UBQLN2+ deposits are found in many common neurodegenerative disorders, including ALS/FTD, Lewy body dementia and polyglutamine expansion diseases (4–6), and mutations in the UBQLN2 gene directly cause a rare form of X-linked ALS/FTD (7) (Fig. 1A). Understanding conserved features of WT UBQLN2 accumulation in sporadic disease and mutant UBQLN2 in hereditary disease should shed light on basic mechanisms underlying neurodegeneration.
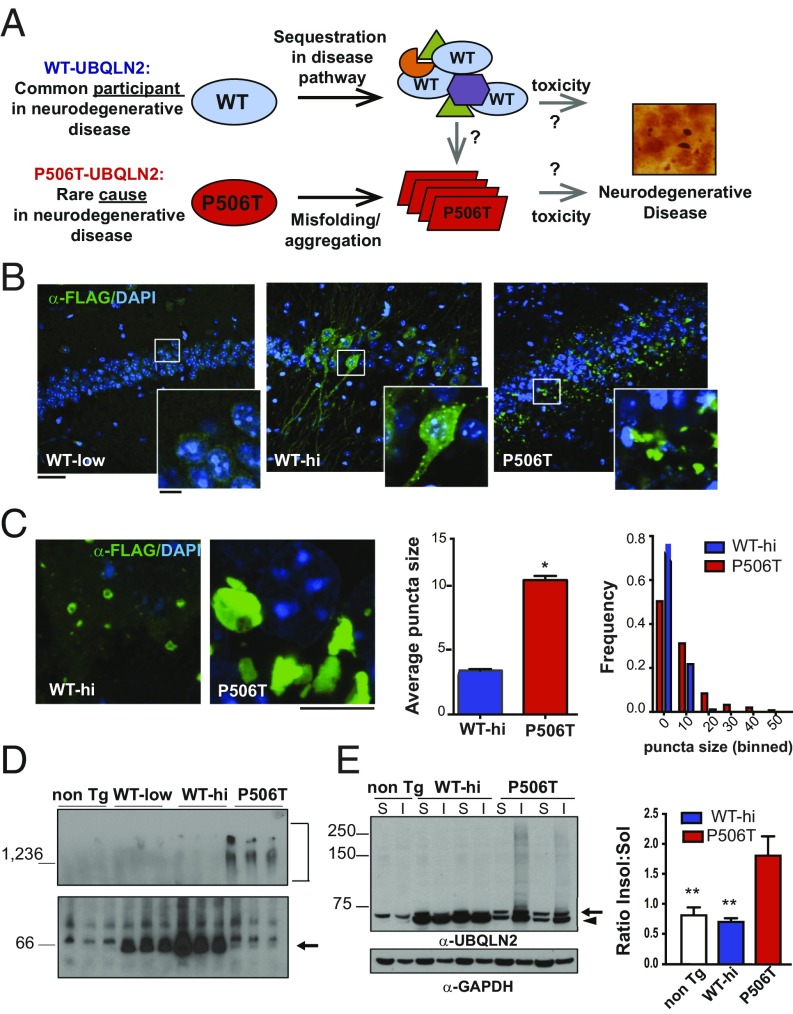
Fig. 1.
Transgenic mice show that the intrinsic propensity of UBQLN2 to aggregate is enhanced by a pathogenic mutation (P506T). (A) Reminiscent of other disease-linked proteins, UBQLN2 commonly accumulates in neurodegenerative diseases and, when mutated, directly causes neurodegeneration along the ALS/FTD spectrum. Studies reported here seek to explain this behavior. (B) Immunofluorescence of the hippocampus in UBQLN2 transgenic mouse lines expressing FLAG-tagged WT-UBQLN2 or P506T-UBQLN2 under control of the mouse prion promoter (3-mo old). Tissue sections were stained for FLAG-UBQLN2 (green) and DAPI (blue). Higher-magnification Insets highlight differences in puncta/aggregate formation by WT- and P506T-UBQLN2. (Scale bars, 50 μM; 10 μM Insets.) (C) Neuronal puncta/aggregates in WT-hi and P506T mouse lines differ in size and appearance. (Scale bars, 5 μM.) (Right) Quantification of puncta/aggregate size in WT-UBQLN2-hi and P506T-UBQLN2 hippocampal neurons, plotted as mean size compared by unpaired t test, (error bars = SE, *P < 0.05), and as relative distribution of puncta size (n = 5 WT-hi mice, 776 total puncta: n = 6 P506T mice, 3,745 total puncta). The WT and P506T puncta size distributions differ significantly based on the Kolmogorov–Smirnov test (D = 0.26402, P < 2.2e-16). (D) Native-PAGE of brain lysates from nontransgenic or transgenic mice probed with anti-UBQLN2 antibody. Bracket indicates high molecular-weight species present only in P506T-UBQLN2 mice, and arrow indicates UBQLN2 monomer. (E) Western blot of soluble (S) and insoluble (I) brain fractions from transgenic and nontransgenic mice probed with anti-UBQLN2 antibody (arrowhead, FLAG-UBLQN2; arrow, murine UBLQN2). (Right) Average insoluble/soluble ratio for UBQLN2 monomer in each line (**P < 0.005).
Many neurodegenerative disease proteins naturally form dynamic, or liquid-like, assemblies as part of their normal function (8–12). While beneficial under most physiological circumstances (13), this propensity to self-assemble may promote the formation of energetically stable complexes or even amyloid-like fibrils, with deleterious consequences for the cell. This phenomenon is observed for many RNA-binding proteins underlying multisystem proteinopathies and ALS/FTD (14). Several proteins in this group have been linked to UBQLN2: hnRNPA1 (15) and TDP43 (16) have been reported to interact with UBQLN2, and ubiquilins form a complex with VCP, an AAA+ ATPase that assists in the clearance of RNA granules (12, 15, 17). Together, these observations raise the possibility that UBQLN2 helps regulate RNA granule dynamics. Indeed, like many proteins implicated in ALS/FTD, UBQLN2 undergoes liquid–liquid phase separation (LLPS) in vitro (18). Even so, it remains unknown whether UBQLN2 undergoes LLPS as part of its normal participation in protein quality control pathways, and whether pathogenic UBQLN2 mutations alter this behavior.
UBQLN2 is involved in a wide range of activities in protein quality control. UBQLN2 acts principally as a proteasomal adaptor protein that, by virtue of its ubiquitin-like (UBL) and ubiquitin-associated (UBA) domains, binds to and shuttles ubiquitinated substrates to the proteasome for degradation (19). UBQLN2 also interacts with molecular chaperones, possibly through its STI1 motifs, and may function in autophagy and other quality-control pathways (20, 21). At least a dozen UBQLN2 missense mutations have been identified in X-linked ALS and FTD, with most of the mutations occurring in or near a proline-rich repeat (PXX) domain. Moreover, disease-associated mutant UBQLN2 forms intraneuronal aggregates in human disease and in rodent models of the disease (7, 14, 22–24), suggesting that missense mutations promote aggregation of the disease protein.
Despite the prevalence of aggregated proteins in most neurodegenerative disorders, a direct role for protein aggregation in disease pathogenesis remains unsettled. The extent to which aggregation elicits a toxic gain-of-function versus a deleterious loss-of-function likely differs in various diseases and remains an open question in UBQLN2-mediated disease. Here we use a broad range of experimental approaches, including in vitro protein analysis, cellular, and transgenic mouse models, to explore the assembly formation and aggregation properties of UBQLN2, assess how a pathogenic UBQLN2 mutation affects these properties, and determine the impact of mutant UBQLN2 expression on protein aggregation and neuronal toxicity. Our results establish that UBQLN2 readily self-assembles into dynamic structures with liquid-like properties similar to those formed by RNA-binding proteins in related neurodegenerative diseases and observed in vitro (18). The UBA domain is a key driver of oligomerization and a pathogenic missense mutation stabilizes UBQLN2-self assemblies, promoting amyloid-like aggregation associated with neurotoxicity. These data suggest that protein aggregation linked to disease mutations contributes to UBQLN2-mediated neurodegeneration.
Results
UBQLN2 Expression in Transgenic Mice Reveals an Intrinsic Aggregation Propensity Accelerated by a Pathogenic Mutation.
Evidence suggests that UBQLN2, like many other neurodegenerative disease proteins, is prone to oligomerize and aggregate (4, 7, 25). To study UBQLN2 behavior in vivo, we generated transgenic mice expressing WT UBQLN2 or UBQLN2 harboring the pathogenic P506T mutation. We chose this particular mutation in the PXX domain because it causes a broad neurodegenerative phenotype in humans (7) and animal models (14). WT- and P506T-UBQLN2 cDNA were expressed from the mouse prion promoter, which drives broad CNS expression (26) (SI Appendix, Fig. S1A). After screening founders, we retained two WT-expressing lines, one each with a high and low level of expression (WT-hi, WT-low), and a single P506T line expressing UBQLN2 at an intermediate level between that of the two WT lines (SI Appendix, Fig. S1B). All animal procedures were approved by the University of Michigan Institutional Animal Care and Use Committee and conducted in accordance with the U.S. Public Health Service’s Policy on Humane Care and Use of Laboratory Animals.
UBQLN2 immunofluorescence in the brains of transgenic mice demonstrates marked differences in the aggregation behavior of WT-UBQLN2 vs. P506T-UBQLN2 (Fig. 1B). When expressed at low levels, WT-UBQLN2 diffusely resides in the neuronal cytoplasm, but at high levels WT-UBQLN2 begins to accumulate in spherical, cytoplasmic puncta, with much of the UBQLN2 signal still remaining diffuse. In contrast, P506T-UBQLN2 accumulates in larger, irregular cytoplasmic inclusions with no detectable diffuse protein. Higher-resolution imaging of WT-hi and P506T mice (Fig. 1C) confirmed that the puncta formed by WT-UBQLN2 are smaller and spherical, whereas the puncta formed by P506T-UBQLN2 are larger and highly irregular in shape.
In transgenic mice, WT- and P506T-UBQLN2 also differ in solubility and self-assembly. On native-PAGE of soluble brain lysates, endogenous and transgenic WT-UBQLN2 migrate as monomers, whereas P506T-UBQLN2 primarily migrates as high molecular-weight complexes (Fig. 1D). When analyzed by SDS/PAGE, WT-UBQLN2 partitions in both the soluble and insoluble fractions of brain lysate, similar to endogenous murine UBQLN2 in nontransgenic mice. In contrast, P506T-UBQLN2 tends to accumulate solely in the insoluble fraction (blot and graph in Fig. 1E). These results suggest that mutant UBQLN2 forms highly stable, detergent-resistant complexes in the CNS, whereas WT-UBQLN2 does not, even when expressed at much higher levels. The detergent-resistant nature of mutant UBQLN2 complexes suggests that the large irregular puncta observed by immunofluorescence in P506T-UBQLN2 mice represent aggregates, unlike the smaller, spherical puncta seen in WT-UBQLN2. We speculate that these smaller puncta are dynamic complexes still capable of disassembly.
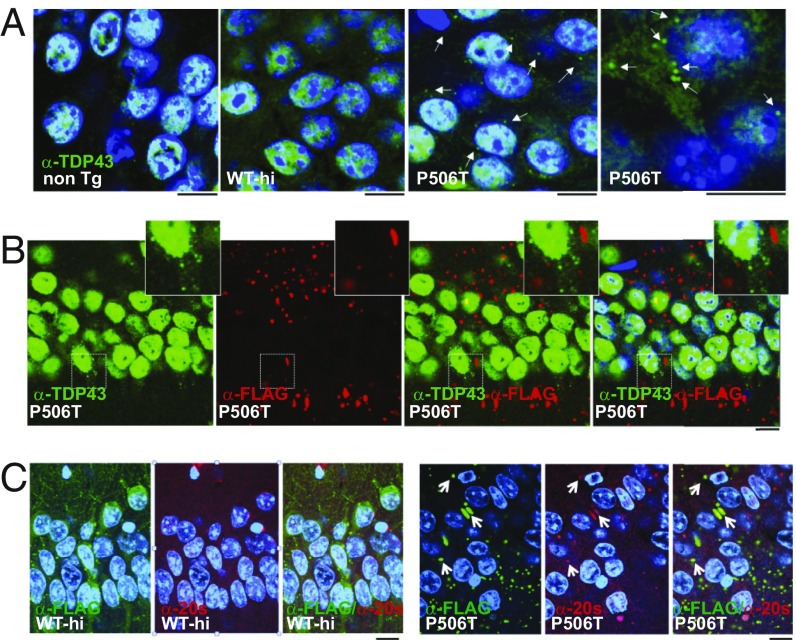
Subcellular redistribution and cytoplasmic aggregation of the RNA-binding protein TDP43 is a hallmark of UBQLN2-mediated neurodegeneration (7, 27–29). Reminiscent of human disease, TDP43 accumulates in discrete cytoplasmic foci in neurons of P506T-UBQLN2 mice, but not in neurons of WT-UBQLN2 mice (Fig. 2A). These TDP43 puncta are distinct from UBQLN2 inclusions (Fig. 2B). We do not, however, observe a marked redistribution of TDP43 from the nucleus to the cytoplasm in P506T mice.
Fig. 2.
P506T-UBQLN2 transgenic mice demonstrate mislocalization of TDP-43 and sequestration of proteasome subunits. (A) Immunostaining with anti-TDP43 antibody (green) demonstrates TDP43-rich cytoplasmic puncta (arrows) in neurons of P506T-UBQLN2 mice, but not WT-UBQLN2 mice. (Scale bars, 10 μm.) DAPI: blue. (B) Immunostaining with anti-FLAG (red) and anti-TDP43 (green) antibodies in P506T-UBQLN2 mice show that the TDP43-rich puncta do not overlap with UBQLN2 puncta. (Scale bar, 10 μm; Insets are magnified 2×.) DAPI: blue. (C) Immunostaining with anti-20s core proteasome subunits (red) and anti-FLAG (green) antibodies shows that proteasome subunits are sequestered in some P506T-UBQLN2 puncta (arrows), but not in WT-UBQLN2 puncta. (Scale bars, 10 μm.) DAPI: blue.
Intraneuronal aggregates found in several neurodegenerative diseases include components of the proteasome (30, 31). Transgenic mice expressing UBQLN2-P497H were previously found to form hippocampal inclusions that costained for proteasome subunits (32). Similarly, a subset of the puncta in P506T-UBQLN2 mice, but not WT- UBQLN2 mice, immunostain for the 20s proteasome core (Fig. 2C), raising the possibility that sequestration of proteasomal subunits may impair the ubiquitin proteasome system (UPS) and contribute to neurotoxocity. This result also underscores the heterogeneity of mutant UBQLN2 complexes in neurons and confirms that WT- and P506T-UBQLN2 puncta differ in their composition.
Formation of Dynamic Assemblies by UBQLN2 Is Altered by a Pathogenic Mutation.
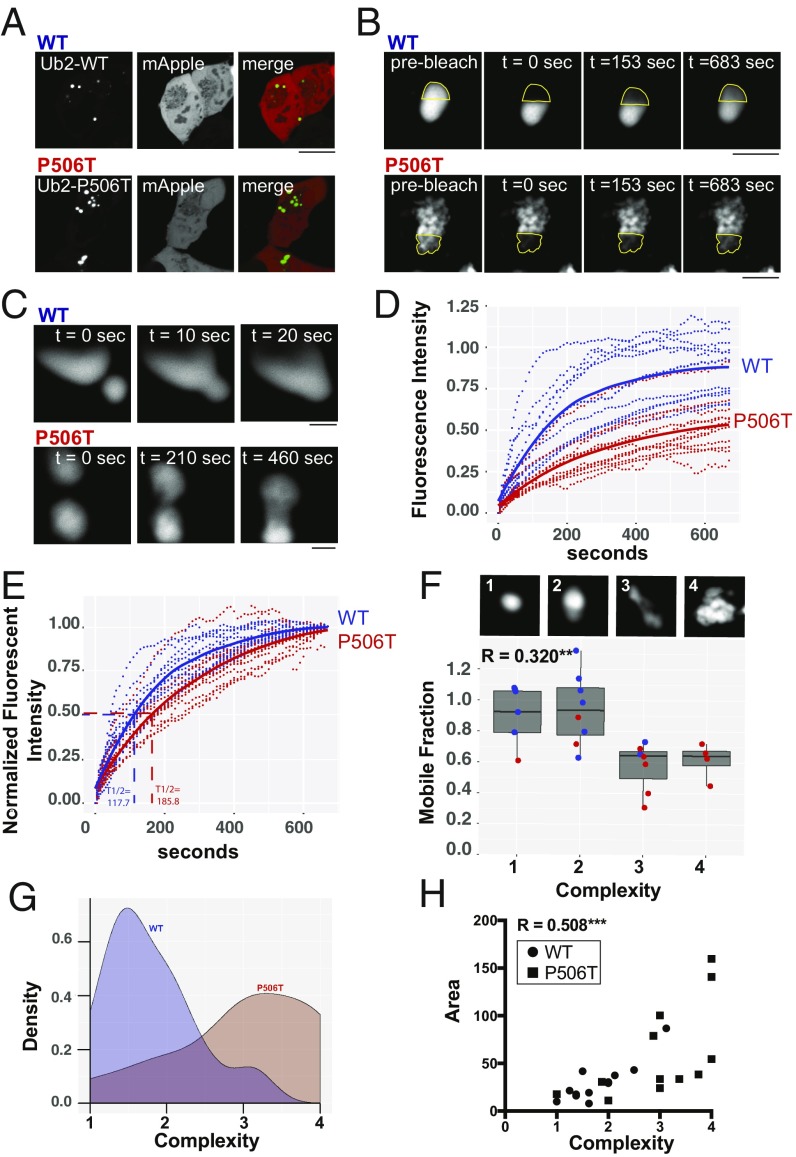
The above results suggest UBQLN2 is naturally prone to form molecular assemblies and that a pathogenic mutation alters this behavior in a manner that promotes formation of larger aggregates. In vitro, UBQLN2 undergoes LLPS that is reversed by ubiquitin binding (18), suggesting that liquid-like properties may be crucial to UBQLN2’s function as a ubiquitin-dependent proteasomal shuttle protein. To explore the possibility that the spherical puncta observed in neurons of WT-UBQLN2 transgenic mice are liquid droplets, we expressed GFP-tagged WT- or P506T-UBQLN2 in HEK293 cells and assessed fluorescence recovery after photobleaching (FRAP) of UBQLN2 puncta. We specifically tested whether UBQLN2 puncta are indeed dynamic and, if so, whether a pathogenic mutation alters this property (Fig. 3A). Puncta in cells expressing GFP-tagged WT- or P506T-UBQLN2 were visualized by confocal laser-scanning microscopy, after which a region-of-interest (ROI) comprising approximately half of a punctum was photobleached and recovery of fluorescence was then monitored for 10 min. Fig. 3B shows representative images of WT or P506T puncta before photobleaching, immediately after bleaching, at midrecovery and at the end of recovery. Movies S1 and S2 show the full imaging time course for additional representative WT and P506T puncta. In these studies, WT-UBQLN2 puncta were more dynamic than P506T-UBQLN2 puncta: the former recovered faster more completely from photobleaching (Fig. 3 C–E). Nevertheless, both WT- and P506T-UBQLN2 puncta demonstrated properties more suggestive of liquid-like droplets than solid aggregates. Consistent with this, we observed fusion events for both WT and P506T-UBQLN2 puncta (Fig. 3C and Movies S1–S4).
Fig. 3.
In vivo FRAP demonstrates that WT-UBQLN2 puncta are more liquid-like and structurally less complex that P506T-UBQLN2 puncta. (A) Representative images of HEK293 cells transfected to express WT (Upper) or P506T (Lower) UBQLN2 together with mApple. Intracellular puncta were selected for FRAP analysis as in B and C and Movies S1 and S2. (Scale bar, 10 μM.) (B) Representative high-magnification images of UBQLN2 puncta from two FRAP experiments. Time points are shown before photobleach, immediately after photobleach, at time to half-maximal recovery (t 1/2 max), and at the end of the 10-min recovery period. Dotted outline shows the photobleached area. (Scale bars, 2 μM.) (C) Images showing fusion of two WT (Upper) or P506T (Lower) UBQLN2 puncta. (Scale bars, 2 μM.) (D) Time course and maximum recovery for individual puncta following photobleaching (dotted lines; WT, blue and P506T, red). Mean recovery curves for WT and P506T are shown with solid lines. (E) As in D, except that the extent of recovery (mobile fraction) has been normalized to highlight differences in the rate of recovery between WT- and P506T-UBQLN2. Average t 1/2 max is shown for both. (F) Box plot depicting strong correlation between mobile fraction and puncta complexity (R2 = 0.320; **P ≤ 0.01). Images (Upper) show representative puncta for each complexity score 1–4 (imaged at 100× with 10× digital zoom). Dots show each individual values for WT (blue) or P506T (red) puncta. An average complexity score for each punctum was calculated by combining ratings from a panel of eight scorers blinded to UBQLN2 genotype. (G) Density plot showing the distribution of complexity scores for WT and P506T UBQLN2 puncta. The WT and P506T population distributions differ significantly from each other based on the Kolmogorov–Smirnov test (D = 0.66667, ***P value < 0.01). (H) Graph showing a significant correlation between puncta area and complexity (R2 = 0.508, ***P ≤ 0.001). Circles and squares depict values for WT- and P506T-UBQLN2 puncta, respectively.
Although WT- and P506T-UBQLN2 puncta vary in appearance, P506T-UBQLN2 puncta are on average more irregularly shaped, reminiscent of the irregular inclusions noted in transgenic mice (Fig. 1C). Using criteria illustrated in Fig. 3F, P506T-UBQLN2 puncta proved to be more complex than WT-UBQLN2 puncta (Fig. 3 F and G). Moreover, this greater complexity correlates with size, with larger puncta displaying greater complexity (Fig. 3H). Additionally, complexity correlates inversely with the extent of fluorescence recovery. Because the morphologies and dynamic behavior of WT- and P506T-UBQLN2 puncta overlap, we analyzed all puncta jointly as a single population. In doing so, we noted that less-complex puncta recovered more completely (i.e., had a higher mobile fraction) than highly complex puncta (Fig. 3F). Movie analysis (Movie S2) suggests that more complex puncta possess internal heterogeneity, including regions of less mobile UBQLN2. This internal heterogeneity, coupled with the decrease in UBQLN2 mobility, suggests that more complex, slowly recovering puncta are analogous to the gel-like state observed in in vitro FRAP experiments (12, 33). Within complex puncta, moreover, microdomains lacking robust fluorescence recovery may correspond to highly immobilized aggregated or fibrillar protein.
The UBA Domain Promotes Amyloid-Like Aggregation of UBQLN2.
The observation that both WT- and P506T-UBQLN2 form puncta or aggregates in vivo and form liquid-like droplets in cells suggests that UBQLN2 assembles into higher-order structures, such as amyloid-like aggregates. Disease-causing mutations like P506T may further increase this propensity and lead to amyloid-mediated neuronal toxicity. To predict regions of UBQLN2 likely to mediate aggregation, we analyzed the protein sequence using four separate bioinformatics algorithms. Predictions by CamSol and TANGO are empirical, based on parameters experimentally determined to correlate with protein aggregation (34–36). Waltz and ZipperDB apply structural information to predict regions likely to form β-sheets (37, 38). In Fig. 4A, colored boxes corresponding to each algorithm highlight regions of UBQLN2 predicted to aggregate or form β-sheets. All algorithms identified three overlapping segments of the protein, highlighted in yellow, as potential domains responsible for driving aggregation. The UBA, UBL, and middle domains of UBQLN2 each possess one of these segments. In contrast, no algorithm identified the PXX domain—which harbors most disease-causing mutations—and none of the pathogenic UBQLN2 mutations identified to date reside within the segments predicted to be important for aggregation.
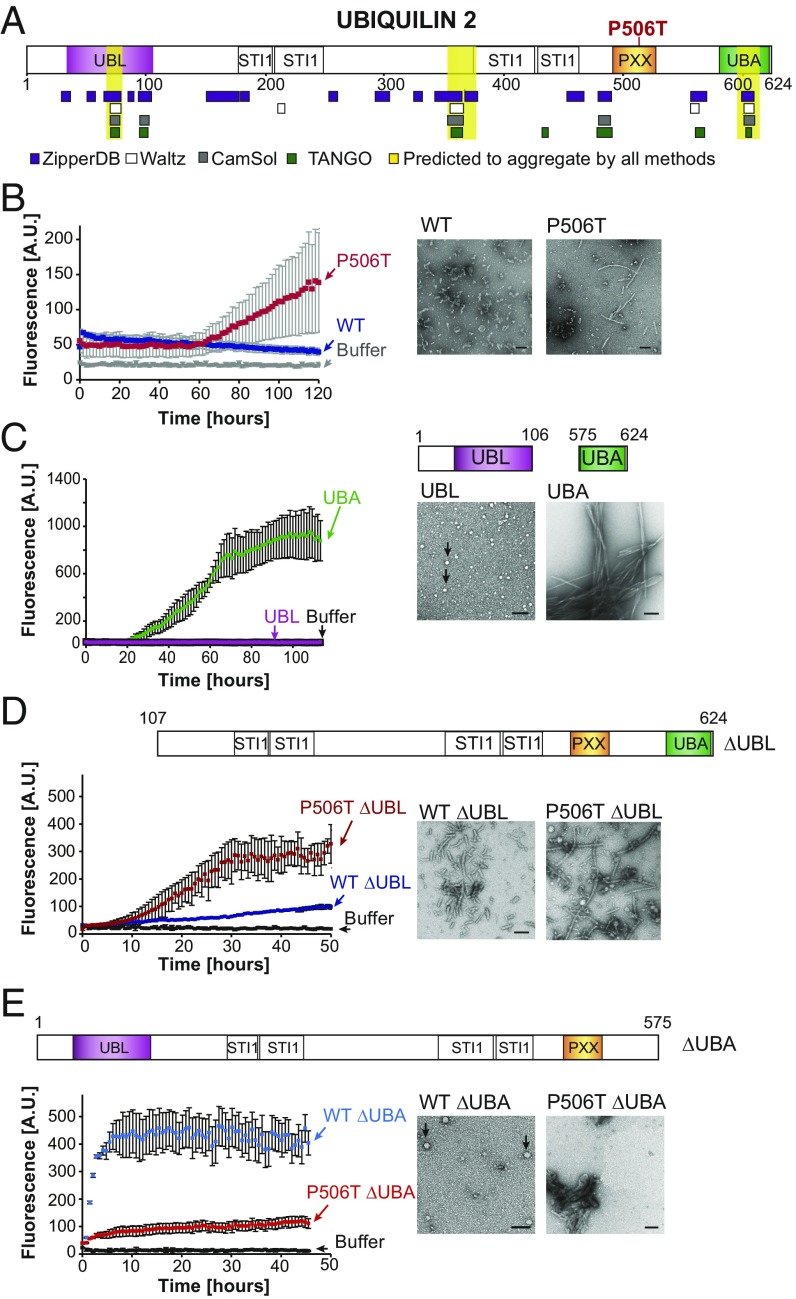
Fig. 4.
UBQLN2 is intrinsically prone to aggregate and the UBA domain is a key determinant of aggregation. (A) Schematic of UBQLN2, highlighting functional domains. The UBL and UBA domains reside at the N and C termini, flanking a central domain that contains multiple STI1 motifs predicted to bind chaperones and the PXX domain of unknown function. Four protein-folding algorithms were applied to the UBQLN2 sequence; three regions predicted to be aggregate-prone by all four algorithms are highlighted in yellow. (B) Recombinant WT-UBQLN2 and P506T-UBQLN2 differ in the kinetics and extent of aggregation in vitro, assessed by ThT reactivity (Left). Mean fluorescence was calculated from at least four independent experiments, with error bars representing SD. By 5 d, P506T-UBQLN2 forms abundant fibrils visualized by TEM, whereas WT-UBQLN2 accumulates in fusiform structures (Right). (C) The UBA domain readily forms ThT+ fibrils, whereas the UBL domain does not, instead forming ThT−, circular species (black arrows) reminiscent of liquid droplets. Schematic of UBQLN2 domain deletion constructs are depicted above each TEM image. Mean fluorescence of UBA and UBL was calculated from four and three independent experiments, respectively. (D) Deletion of the UBL domain promotes fibril formation. While both WT- and P506T-ΔUBL bind ThT, P506T-ΔUBL aggregates earlier and produces greater ThT fluorescence. Mean fluorescence was calculated from two independent experiments. After 1 wk of incubation, both WT- and P506T-ΔUBL form predominantly fibrillar species, with P506T-ΔUBL fibrils being longer. (E) Deletion of the UBA domain suppresses fibril formation. While both WT- and P506T-ΔUBA fluoresce in the presence of ThT, fibrillar species are absent with WT-ΔUBA and rarely observed with P506T-ΔUBA. Mean fluorescence was calculated from four independent experiments. (Scale bars, 100 nm.)
To explore intrinsic properties of UBQLN2, we assessed in vitro the aggregation propensity of recombinant WT- and P506T-UBQLN2. Thioflavin-T (ThT), an amyloid-binding dye, was used to monitor the aggregation of UBQLN2 and assess the kinetics of amyloid fibril formation. P506T-UBQLN2 displayed three- to fivefold greater ThT fluorescence than WT-UBQLN2 (Fig. 4B), suggesting that the pathogenic P506T mutation promotes the formation of aggregates with amyloid-like properties. Some variability was observed in the aggregation kinetics for P506T-UBQLN2, but the comparatively greater fluorescence of P506T-UBQLN2 remained consistent in all experiments (e.g., SI Appendix, Fig. S2).
Corresponding to its enhanced ThT fluorescence, P506T-UBQLN2 consistently formed short fibrils visualized by transmission electron microscopy (TEM) (Fig. 4B). In contrast, fibrillar species were absent or only rarely observed with WT-UBQLN2. Both WT- and P506T-UBQLN2 also formed round and fusiform species of variable size (Fig. 4B). From these results, we conclude that both WT- and P506T-UBQLN2 form heterogeneous species in solution and those formed by mutant UBQLN2 exhibit increased amyloid-like properties.
Guided by the above algorithms predicting aggregation domains, we investigated aggregation of the UBA and UBL domains in isolation. The UBA domain robustly formed amyloid fibrils based on ThT fluorescence and TEM analysis (Fig. 4C). In contrast, the UBL domain failed to form ThT+ aggregates under physiological conditions, even after 2 wk of incubation. These data suggest that the UBA domain, but not the UBL domain, contributes to amyloid-like aggregation.
These findings imply that removing the UBL domain might increase fibril formation, whereas removing the aggregate-prone UBA domain should decrease fibril formation. Indeed, UBQLN2 lacking the UBL domain (ΔUBL) consistently formed fibrils visible by TEM. WT-ΔUBL fibrils were uniformly short, whereas fibrils formed by P506T-ΔUBL varied in length and displayed higher affinity for ThT (Fig. 4D). Conversely, removing the UBA domain markedly suppressed UBQLN2 fibril formation (Fig. 4E): only very small, round species of WT-ΔUBA were observed by TEM, reminiscent of species formed by the UBL domain itself. P506T-ΔUBA formed amorphous structures and relatively rare bundles of fibrils. Reinforcing our observation that the P506T mutation changes the aggregation properties of UBQLN2, WT- and P506T-ΔUBA also differed in affinity for ThT; intriguingly, WT-ΔUBA, which did not form fibrils, actually showed higher ThT binding (Fig. 4E). Complexes formed by WT-ΔUBA appeared to bind ThT despite lacking fibrillar morphology. Typically, ThT interacts with the extended β-sheets of amyloid fibrils (39, 40), but structural motifs and chemical features of β-sheets also contribute to ThT binding (41, 42). Although less commonly observed, nonfibrillar oligomeric species can likewise bind ThT (43, 44). We suspect this is the case for the nonfibrillar aggregates of ΔUBA.
Collectively, our in vitro results suggest that the UBA domain promotes—whereas the UBL domain delays—UBQLN2 aggregation, and that introduction of a pathogenic mutation, P506T, increases the aggregation propensity of the protein.
The P506T Mutation Promotes Puncta Formation by UBQLN2 in Neurons.
Next, we asked if differences in WT-UBQLN2 and P506T-UBQLN2 self-assembly—observed in mice, HEK cells, and in vitro—are recapitulated in mature neurons. Moreover, to determine if UBQLN2 self-assembly is pathogenic, epiphenomenal, or potentially protective, we sought to identify the factors governing UBQLN2 self-assembly and investigate their respective contributions to neurodegeneration. To pursue these questions, we employed longitudinal automated fluorescence microscopy, a platform with the capacity to relate factors, such as protein expression and aggregation to neuronal survival on the single-cell level in an unbiased and high-throughput manner (Fig. 5A) (45–47).
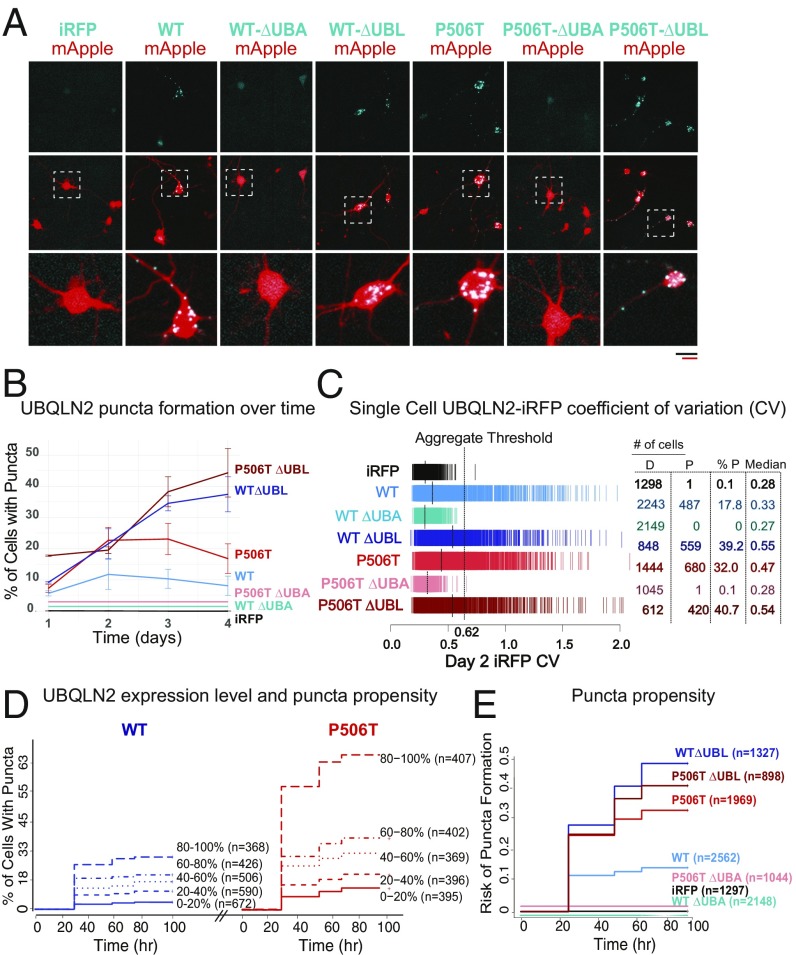
Fig. 5.
Serial imaging of neurons by automated fluorescence microscopy demonstrates increased puncta formation by the P506T mutation in neurons. (A) Neurons were cotransfected at day in vitro 4 with the indicated UBQLN2-iRFP plasmids (blue) together with mApple (red) as a morphology marker, then imaged 48 h later. Both WT-UBQLN2 and P506T-UBQLN2 form puncta in neurons, whereas UBQLN2 lacking the UBA domain remains diffuse. (Scale bars, black is for Upper two rows and red scale bar for Lower Inset, both 50 µm.) Neurons within checkered boxes are enlarged in Bottom row. (B) Plot representing percentage of neurons containing UBQLN2 puncta over time. A neuron was classified as containing puncta if it had an iRFP CV great than the aggregation threshold of 0.62. Only cells that lived for the duration of the 4 d of analysis were included. Error bars represent the SEM from three replicate experiments. Lines for iRFP, WT ∆UBA, and P506T ∆UBA are offset for visualization purposes but all equal 0. (C) Single-cell iRFP CV, a proxy for the extent of aggregation, measured 2 d after transfection. Each hash mark represents a single cell, and the dashed lines indicate the median CV value for a population. A CV of 0.62 was established as an effective aggregate threshold to differentiate neurons with visible aggregates from those with diffuse UBQLN2 (SI Appendix, Fig. S4). The number of cells with a CV value below (diffuse: D) or above (punctate: P) this threshold, as well as the median CV value for each population, are listed for each variant. (D) Percentage of neurons containing puncta with increasing UBQLN2 expression levels. Neurons within the groups WT-UBQLN2 (blue) and P506T-UBQLN2 (red) were divided into quintiles based on day 1 expression level (iRFP intensity) and plotted on a cumulative hazard plot with the y axis representing the percentage of neurons containing aggregates. Hazard ratios are reported in SI Appendix, Table S1. Neurons are pooled from three replicate experiments. Both plots share the same y axis. (E) The relative risk of puncta formation was compared between groups. Cells from three replicate experiments were rank-ordered by day 1 iRFP intensity, then divided into quintiles. For Cox proportional hazards analysis, cells were stratified by day 1 expression quintile to minimize the effects of differential expression between genotypes. Only cells that lived the duration of analysis (4 d) were included. Hazard ratios are reported in SI Appendix, Table S2.
Primary rodent cortical neurons were transfected with WT-UBQLN2, P506T-UBQLN2, or the corresponding ∆UBA and ∆UBL mutants. Each variant was tagged at the amino terminus with a near-infrared fluorescent protein (48) and neurons were cotransfected with a second construct (mApple) to provide a morphology marker. By comparing the UBQLN2 immunoreactivity of transfected and untransfected cells, we determined that transfection resulted, on average, in a 6.1-fold increase in single-cell UBQLN2 levels (SI Appendix, Fig. S3). Loss of mApple signal and compromised cell integrity, which represent sensitive and accurate criteria for determining cell death (49), were used to identify time of death for each cell (Fig. 6A)
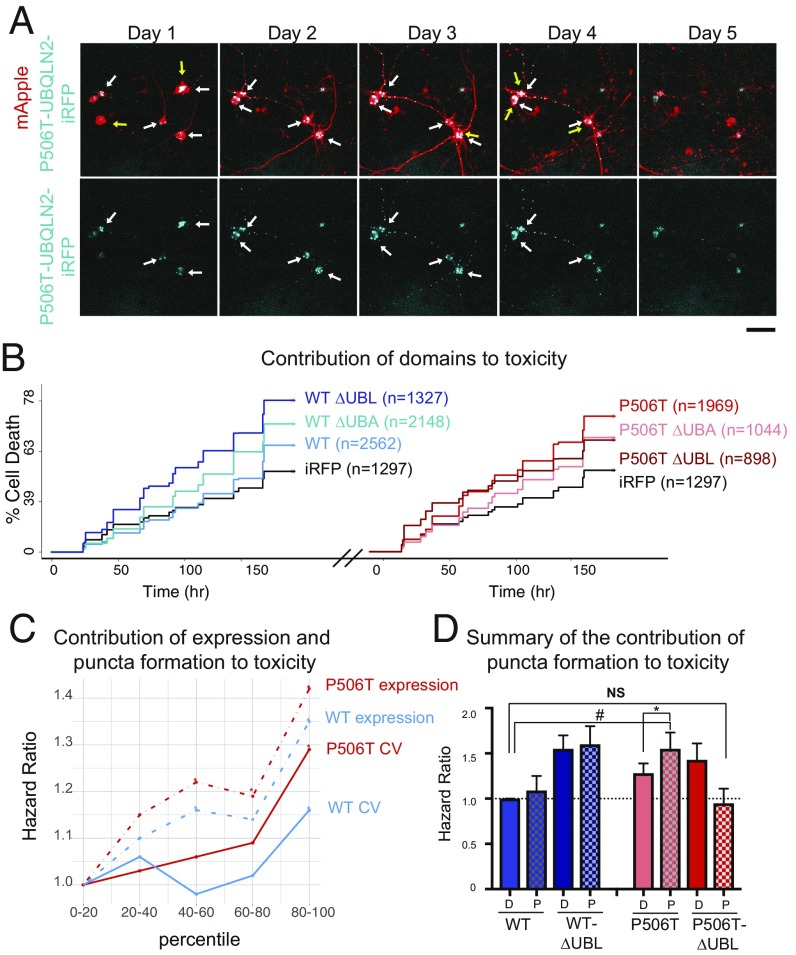
Fig. 6.
Increased propensity of UBQLN2 to form puncta correlates with neuronal toxicity. (A) To assess survival, transfected neurons expressing iRFP-tagged P506T-UBQLN2 (blue) and mApple (red) were imaged daily and the last time a cell was seen alive (yellow arrows) was recorded as time of death. Neurons expressing iRFP-P506T-UBQLN2 form aggregates (white arrows). (Upper) Merged images; (lower) only iRFP-P506T-UBQLN2. (Scale bar, 50 μm.) (B) Cumulative risk of death over time for neurons expressing WT-UBQLN2 or P506T-UBQLN2 as well as the corresponding UBA and UBL deletion mutants. Neurons in each group were pooled from three replicates, stratified by experiment date. Only neurons that lived to day 2 with diffuse UBQLN2 on day one were used for analysis, to keep the data consistent with subsequent analyses. Both plots share the same y axis. Hazard ratios are reported in SI Appendix, Table S3. (C) Dose-dependent toxicity is observed for both UBQLN2 expression and puncta formation. In neurons expressing WT- or P506T-UBQLN2, cells were rank-ordered by day 1 expression and degree of punctateness (CV), then divided into quintiles. For each genotype the relative toxicity (hazard ratio) for each quintile was compared with its lowest quintile. Solid lines = CV, dashed lines = expression, *P < 0.05 by Cox proportional hazards analysis. Hazard ratios and significance values for all groups are reported in SI Appendix, Table S4. (D) Puncta formation by UBQLN2-P506T enhances toxicity. For neurons expressing WT- or P506T-UBQLN2 and their UBL domain deletion mutants, cells were stratified into two groups: diffuse or punctate (D or P) UBQLN2 on day 2 after transfection. Cells that formed inclusions by day 1 were excluded to ensure that only cells in which the inclusion formation was observed were included. Cells also were rank-ordered by day 1 expression levels, then separated into quintiles. Cox proportional hazards analysis was performed stratifying by expression quintile to ensure that cells with comparable expression were compared (hazard ratios and significance values are reported in SI Appendix, Table S7) (*P < 0.01, #P < 0.0001, NS, not significant).
As in transgenic mice, we observed abundant WT-UBQLN2 and P506T-UBQLN2 puncta throughout the soma and neuritic processes of transfected neurons. For simplicity, we refer to visible UBQLN2+ structures within the transfected neurons as “puncta.” Based on our analysis of UBQLN2 behavior in transgenic mice, FRAP experiments, and in vitro aggregation assays, we expect that the inclusions in transfected neurons represent a mixture of liquid-like droplets, gel-like structures, and aggregated protein. The negative control (iRFP alone) was diffusely distributed, indicating that aggregation of iRFP-UBQLN2 fusions is driven by UBQLN2. Mirroring our in vitro results, deletion of the UBA domain, but not the UBL domain, prevented puncta formation of both WT-UBQLN2 and P506T-UBQLN2 (Fig. 5A). These data are consistent with the recent finding that the UBA domain is essential for UBQLN2 LLPS in vitro and in HeLa cells (18).
To measure the formation of UBQLN2 puncta in living neurons, we developed a reliable single-cell metric that can be employed in an automated fashion. Because puncta represent peaks of florescence intensity surrounded by a low diffuse signal, we hypothesized that cells containing puncta should have a higher SD of florescence intensity relative to cells without puncta. To account for varying intensities between cells, we chose the coefficient of variation (CV), which takes into account mean florescence, as the most appropriate metric for punctate UBQLN2 distribution. Indeed, we found that the CV of a cell’s iRFP fluorescence intensity represents a sensitive and specific predictor of punctate UBQLN2 (Fig. 5B). As used here, the CV is an unbiased measure of UBQLN2 distribution, agnostic to UBQLN2 secondary structure. We systematically assessed the sensitivity and specificity of the CV for detecting visible UBQLN2 puncta in transfected neurons by constructing a receiver operating characteristic curve (SI Appendix, Fig. S4). From these data, we selected a CV value of 0.62 as the optimal “aggregation threshold” for subsequent experiments, corresponding to a sensitivity of 83% and a false-positive rate of less than 1%. Applying this threshold, we can quickly and accurately measure the percentage of cells containing UBQLN2 puncta over time (Fig. 5B).
In agreement with manual analysis, the CV threshold technique demonstrated that P506T-UBQLN2 is more prone to inclusion formation than WT-UBQLN2, while deleting the UBA domain prevents it (Fig. 5B). In addition to representing a dichotomous state (puncta or no puncta), CV can also be used as a continuous variable, wherein higher CV values indicate a progressively more punctate UBQLN2 distribution. Neurons expressing P506T-UBQLN2 displayed a significantly higher median CV than neurons expressing WT-UBQLN2, corresponding to a higher percentage of neurons exceeding the visible puncta threshold (Fig. 5C). Deleting the UBL domain raised the median CV value, while deleting the UBA domain reduced the median CV value to that observed in iRFP-expressing neurons, essentially the absence of puncta (Fig. 5C).
The expression level of aggregation-prone proteins is a strong driver of aggregate formation (46). To determine if UBQLN2 puncta formation is similarly affected by protein levels, we took advantage of the strong linear relationship between the intensity of a fluorescently tagged protein and its concentration (49) to estimate levels of UBQLN2 in transfected neurons. First, we rank-ordered cells based on their day 1 fluorescence intensity and stratified the population into five quintiles (SI Appendix, Fig. S5A). The relative risk of puncta formation in each quintile was then calculated using Cox proportional hazards analysis (Fig. 5D). Often used to compare the relative risk of death in clinical trials, Cox analysis can also estimate the risk of other time-dependent outcomes, including puncta formation (49). This analysis demonstrated a direct relationship between UBQLN2 expression and risk of puncta formation for both WT- and P506T-UBQLN2. In each case, the risk of puncta formation was six- to sevenfold higher for neurons in the top expression quintile, compared with those in the lowest expression quintile (Fig. 5D and SI Appendix, Table S1).
Because expression level impacts puncta formation, and UBQLN2 expression levels varied between groups (SI Appendix, Fig. S5B), we wanted to rule out the possibility that the observed differences in UBQLN2 puncta formation were driven by expression level. To do so, we compared the risk of UBQLN2 puncta formation across all quintiles by Cox analysis (Fig. 5E). We noted the same trends in puncta propensity as earlier (Fig. 5 B and C), indicating that the differences in UBQLN2 aggregation between groups are driven by intrinsic differences in protein composition and not simply due to changes in expression. In fact, when comparing neurons of comparable expression, the risk of puncta formation for P506T-UBQLN2 was approximately twice that of WT-UBQLN2 (Fig. 5E and SI Appendix, Table S2).
UBQLN2 Puncta Formation in Neurons Correlates with Increased Neurotoxicity.
We next explored the impact of UBLQN2 expression and puncta formation on neuronal survival. Relative to neurons transfected with iRFP alone, overexpression of WT-UBQLN2 was mildly toxic, increasing the risk of death by 20% (P = 6.84 × 10−5, Cox hazards analysis). Neurons expressing P506T-UBQLN2, however, exhibited a much greater (60%) increase in the risk of death (P = 2 × 10−16, Cox hazards analysis) (Fig. 6B and SI Appendix, Table S3). Deleting WT-UBQLN2’s UBL domain, which enhances puncta formation (Fig. 5 A–C and E), similarly increased WT-UBQLN2 toxicity. However, so did deleting the UBA domain, which completely prevents WT-UBQLN2 puncta formation (Fig. 5 A–C and E). As these deletion constructs are expected to impact UBQLN2’s interaction with the proteasome and ubiquitinated proteins (50–53), their overexpression may exert dominant-negative toxicity on WT-UBQLN2 distinct from effects of UBQLN2 aggregation itself or the P506T mutation.
In light of the proportional relationship between UBLQN2 expression and puncta formation (Fig. 5D) and the toxicity of overexpressed UBLQN2 variants in neurons (Fig. 6B), we sought to examine the individual and combined contribution of UBQLN2 expression and aggregation to neuronal survival. Upon stratifying each population into quintiles based on UBLQN2 expression and extent of puncta formation, or “punctateness” (i.e., CV), we noted a strong dose-dependent relationship between each factor and the risk of death for WT- and P506T-UBQLN2 (Fig. 6C). In other words, both UBQLN2 expression and extent of puncta formation are associated with neurotoxicity. Confirming this observation, univariate Cox hazards models incorporating expression level or CV as continuous variables indicated that both factors are significantly and positively associated with risk of cell death (SI Appendix, Fig. S6 A and B). Because UBQLN2 expression can influence puncta formation (Fig. 5D), we also established a multivariate Cox hazards model to determine if the toxicity observed for UBQLN2 aggregation was dependent on expression level. In this model, both expression and punctateness remained significantly associated with risk of death, suggesting that they contribute independently to neuronal toxicity (SI Appendix, Table S5). To account for the effects of each factor, stratification by both expression and punctateness were incorporated into the Cox hazards model (SI Appendix, Fig. S6C and Table S6). In this analysis, P506T-UBQLN2 significantly elevated neuronal risk of death relative to both WT-UBQLN2 and iRFP (P = 4.2E-15, Cox hazards analysis), suggesting that intrinsic properties of mutant UBQLN2 drive toxicity independent of expression level and extent of puncta formation.
In some studies, the formation of protein inclusions has correlated with neuronal survival, presumably reflecting protective sequestration of toxic oligomers (45). To explore whether UBLQN2 aggregation itself influences neuronal survival, we tracked neuronal survival and UBQLN2 puncta formation in cells longitudinally (Fig. 6D). In this analysis we excluded all cells exhibiting UBLQN2 puncta on day 1, instead following only neurons with comparable levels of diffuse UBQLN2 at day 1. Subsequent puncta formation had no clear effect on the survival of neurons expressing WT-UBQLN2, but significantly increased the risk of death in P506T-UBQLN2–expressing neurons, suggesting that aggregation of the mutant protein is toxic. Furthermore, while deleting the UBL domain had no observable effect on the survival of neurons displaying diffuse WT- or P506T-UBQLN2, this domain appeared to be necessary for the toxicity of aggregated P506T-UBQLN2 (Fig. 6D and SI Appendix, Fig. S5). These data raise the intriguing possibility that P506T-UBQLN2 aggregates exert toxicity through UBL-dependent interactions with the proteasome, although further experiments are required to determine whether alterations in proteasome function contribute to the neurotoxicity we observe.
UBQLN2 Puncta Formation and Toxicity Are Regulated by Ubiquitin Binding.
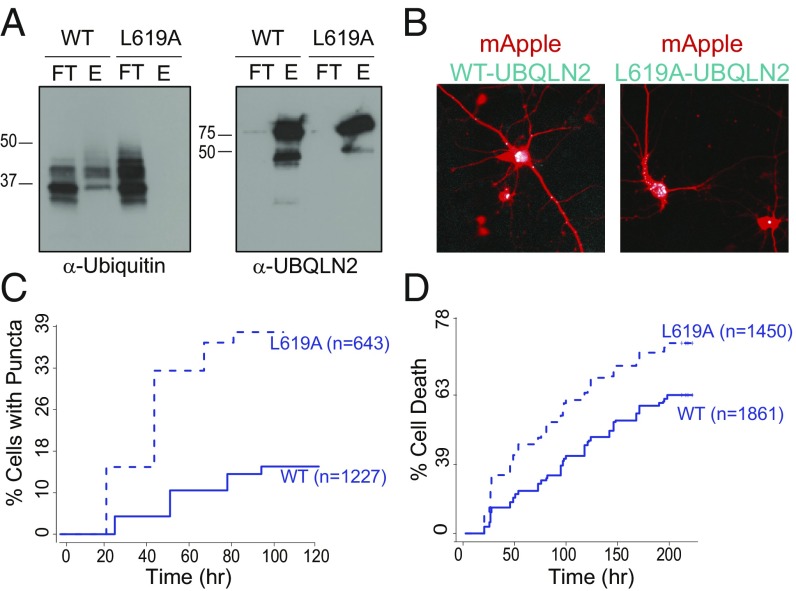
Recent studies on LLPS of UBQLN2 showed that ubiquitin binding by UBQLN2 results in dissolution of liquid droplets in vitro (18). To determine if ubiquitin binding changed the aggregation propensity of UBQLN2, we examined the behavior of the ubiquitin-binding–deficient form of UBQLN2 (L619A) in neurons. We first confirmed that the L619A mutant can no longer bind polyubiquitin chains in pull-down assays (Fig. 7A). Consistent with the hypothesis that ubiquitin binding inhibits LLPS of UBQLN2 (18), L619A-UBQLN2 is threefold more prone to form puncta than WT-UBQLN2 in neurons (P = 2 × 10−16, Cox hazards analysis) (Fig. 7 B and C). Neurons expressing L619A-UBQLN2 also exhibit a 50% increase in the risk of death compared with neurons expressing WT-UBQLN2 (P = 2 × 10−16, Cox hazards analysis) (Fig. 7D). These observations suggest that the unoccupied UBA domain promotes UBQLN2 self-assembly, leading to an increase in UBQLN2 aggregation with deleterious consequences for the neuron.
Fig. 7.
The ubiquitin-binding deficient mutation, L619A, increases both puncta formation and toxicity in neurons. (A) Western blots showing the results of a pull-down experiment to assess L619A-UBQLN2 ubiquitin binding. Recombinant His-tagged WT or L619A-UBQLN2 was preincubated with poly-ubiquitin/Ub3-Ub7 WT chains (K48 linked) followed by a subsequent incubation with Ni-NTA agarose beads. The flow-through (FT) and the elution (E) were visualized on Western blots probed with α-UBQLN2 or α-ubiqutin antibodies. (B) Representative images of primary neurons cotransfected with either iRFP or iRFP-tagged L619A-UBQLN2 (blue) and mApple (red) on day 2 following transfection (magnification, 20×). (C) The percent of neurons that form puncta is significantly increased in L619A-UBQLN2 transfected neurons compared with WT. (hazard ratio = 3.09, P = 2E-16) Neurons were rank-ordered by expression level and stratified by expression quintile. (D) The cumulative risk of death is increased in L619A-UBQLN2–transfected neurons compared with WT (hazard ratio = 1.54, P = 2E-16). Neurons were rank-ordered by expression level and stratified by expression quintile.
Discussion
We employed a range of model systems in an effort to understand why UBQLN2 accumulates in common age-related neurodegenerative diseases and, when mutated, directly causes neurodegeneration. Collectively, our results suggest that the behavior of UBQLN2, both in sporadic disease and in UBQLN2-linked neurodegeneration, reflects an intrinsic property of the protein to self-assemble and aggregate. Even in the absence of known interactors, such as ubiquitin and chaperones, UBQLN2 forms oligomers in vitro and displays amyloid-like features, especially when harboring a disease-associated missense mutation. UBQLN2 thus belongs to a growing group of neurodegenerative disease proteins that naturally form multiprotein assemblies that not only regulate native protein function but also promote aggregation under conditions of proteotoxic stress.
Studies of UBQLN2 to date have provided important snapshots of UBQLN2 behavior: LLPS of UBQLN2 has been observed in vitro (18) and published studies of rodent models of UBQLN2-mediated disease have shown that mutant forms of UBQLN2 accumulate in puncta consistent with aggregate-like structures (14, 20, 32). Using a broad repertoire of methods, we sought to provide a link between the in vitro properties of UBQLN2 and its behavior in vivo. The UBQLN2 puncta observed across our model systems are not homogenous, but rather represent structures along a continuum, ranging from liquid-like droplets to detergent-resistant aggregates. This continuum is influenced by several factors, including pathogenic mutations, intact functional domains, and the ability to bind ubiquitin.
The mutant UBQLN2 transgenic mice reported here recapitulate key aspects of human disease associated with UBQLN2 mutations, including the formation of large UBQLN2 inclusions in neurons throughout the CNS, broad deposition of detergent-resistant, high molecular-weight UBQLN2 species that are consistent with aggregates, and the formation of discrete cytoplasmic TDP43 puncta (7, 22, 27, 54–57). While these features are not observed in WT-UBQLN2 mice, we do note that even WT-UBQLN2 accumulates in smaller, spherical puncta when overexpressed. These WT-UBQLN2 puncta may be functionally relevant to UBQLN2 accumulation in various neurodegenerative diseases. Our results mirror those seen in other rodent and Drosophila UBQLN2 models, confirming that mutant UBQLN2 variants harboring missense mutations are increasingly prone to aggregate (20, 22, 23, 58) and in some cases elicit neurodegeneration (20, 24, 59).
UBQLN2 oligomerization and LLPS likely underlie its aggregation propensity, which is regulated by the very domains critical for UBQLN2 function in ubiquitin-dependent protein quality control (SI Appendix, Table S8). The carboxyl-terminal UBA domain, through which UBQLN2 binds ubiquitinated substrates, appears to drive aggregation, while the amino-terminal UBL domain, which facilitates the delivery of bound substrates to the proteasome, inhibits aggregation. Our in vitro studies establish that UBQLN2 is intrinsically capable of forming amyloid-like fibrillar aggregates, even in the absence of known interactors, such as ubiquitin and chaperones. Nevertheless, the process is likely regulated by such interactions in vivo. Indeed, the fact that the domain responsible for binding ubiquitinated substrates also promotes aggregation suggests that the propensity of UBQLN2 to self-assemble and aggregate is modulated by ubiquitin binding in cells. For example, ubiquitin chain binding by the UBA domain could prevent UBQLN2 self-assembly and favor the disassembly of oligomerized UBQLN2. Increased aggregation by the ubiquitin-binding mutant L619A-UBQLN2 in neurons supports this view. It is intriguing to speculate that oligomerization of UBQLN2 functions as a sensor of ubiquitin status in the cell, determining the availability of UBQLN2 to shuttle substrates to the proteasome.
The relationship of protein aggregation or inclusion formation to neurotoxicity remains unsettled in neurodegenerative diseases. Automated microscopy allowed us to explore this relationship in neurons expressing UBQLN2. In general, the propensity of each UBQLN2 variant to form puncta correlated directly with the observed risk of neuronal death. Studies in neurons also confirmed the importance of the UBA domain for puncta formation in vivo, and showed that a pathogenic mutation (P506T) enhances the propensity of UBQLN2 to form puncta in neurons. Together, these results suggest that UBQLN2 accumulation is deleterious rather than protective to neurons. We should add, however, that deleting the UBA domain reduced puncta formation while actually enhancing toxicity compared with the full-length protein, implying a complex relationship between UBQLN2 function, puncta formation (aggregation), and neuronal fate. Sorting out the precise relationship between UBQLN2 aggregation and neuronal toxicity will require the study of additional disease-associated UBQLN2 mutants, ideally at physiological concentration rather than as overexpressed protein, which itself can represent a confounding variable.
Accumulating evidence suggests that proteins containing low-complexity domains naturally form dynamic assemblies (8–12). The coalescence of these proteins— including many directly implicated in FTD/ALS, such as tau, TDP43, and FUS—into liquid-like droplets is reversible and can be modified by temperature, ionic strength, binding partners, protein concentration, and posttranslational modifications. Under conditions of chronic stress, such as impaired protein homeostasis or in the setting of pathogenic mutations, this propensity to form complexes leads to energetically stable hydrogels, oligomers, or fibrils with limited return to the liquid-droplet or soluble state (60). Recent work and our study suggest that UBQLN2 also belongs to this class of proteins. UBQLN2 undergoes LLPS in vitro that is regulated by ubiquitin binding to UBQLN2’s UBA domain (18). Collectively, our results support the view that LLPS may be an important physiological function of UBQLN2 in vivo. Specifically, our FRAP studies on UBQLN2 puncta demonstrate dynamic, liquid-like behavior entirely consistent with that of membraneless organelles (Fig. 3) (9, 10, 12, 18, 61, 62). Furthermore, the internal substructure evident within puncta implies they are structurally—and potentially functionally—heterogeneous. Although our results were obtained in overexpression systems, we speculate that endogenous UBQLN2 undergoes similar transitions under specific cellular stress conditions, analogous to the formation of stress granules by RNA-binding proteins. These UBQLN2-containing membraneless organelles may serve to bring components of the UPS into close proximity, and their formation and dissolution may depend on the proteostatic needs and ubiquitin status of the cell.
The differences we observed in complexity and dynamism of WT- vs. P506T-UBQLN2 liquid droplets are consistent with a model in which a pathological mutation stabilizes the UBQLN2-containing membraneless organelle, leading to the formation of hydrogel complexes and, eventually, amyloid fibrils. Based on this, we favor the interpretation that the smaller puncta observed in neurons of WT-UBQLN2 mice represent membraneless organelles linked to the protein’s function in ubiquitin-dependent protein quality control, while the much larger structures in P506T-UBQLN2 mice correspond to amyloid-like aggregates formed by the mutant protein. This aggregation, in turn, may impair protein quality control pathways and, over time, lead to neuronal dysfunction and neurodegeneration. Future studies aimed at determining the composition, identity, and behavior of UBQLN2 puncta, and the impact of other disease-associated UBQLN2 mutations on puncta dynamics, will be critical for assessing their contributions to neurodegeneration.
In light of these results and evidence from related studies (14, 18, 20, 24, 58, 59), we propose a model (Fig. 8) of UBQLN2 function/dysfunction in which oligomerization and aggregation are central elements and the dynamic qualities and morphology of UBQLN2 puncta exist along a continuum. Disease-associated mutations and proteotoxic stress shift the equilibrium toward the formation of more complex, stable amyloid fibrils. The distribution across this equilibrium likely is influenced by the state of protein homeostasis and ubiquitin availability. We favor the view that amyloid-like species can be generated even by WT UBQLN2, particularly under times of proteotoxic stress, and that the missense mutations underlying UBQLN2-mediated neurodegeneration shift the equilibrium toward amyloid-like fibril formation.
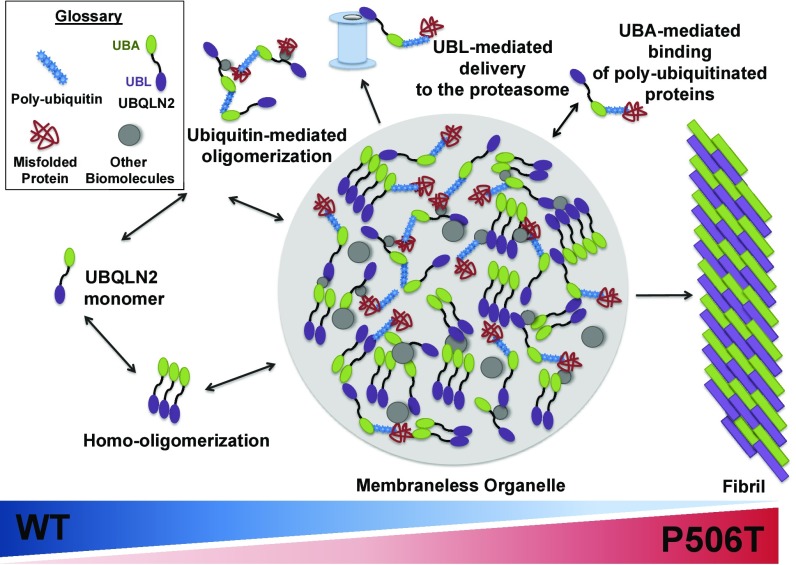
Fig. 8.
Schematic of dynamic continuum between oligomerization, liquid–liquid phase transition and fibril formation for WT and mutant UBQLN2. In this speculative model, UBQLN2 forms oligomers with itself and associates with ubiquitinated substrates and other biomolecules, including chaperones. While the UBA and UBL domains of UBQLN2 may interact intermolecularly, this model is not meant to imply particular structures for oligomeric states of UBQLN2. Under physiological conditions, UBQLN2 oligomers assemble into membraneless organelles that function as reservoirs for UPS components, thereby facilitating chaperone-mediated folding or proteasomal degradation of UBQLN2-bound substrates. The gradients (Bottom) reflect the potential shift in equilibrium from dynamic molecular assemblies (Left) to more stable hydrogels or amyloid fibrils (Right) in the presence of disease-causing mutations (such as P506T) or proteotoxic stress.
The inherent ability of UBQLN2 to oligomerize is probably crucial for self-assembly into membraneless organelles. The potential downside of this property, however, is the ease with which various stressors may tip the balance toward the formation of irreversible amyloid aggregates. A growing number of pathogenic mutations responsible for ALS/FTD affect the formation or resolution of membraneless organelles (63, 64). Our data suggest that UBQLN2 mutations have analogous and equally disruptive effects.
Collectively, our results suggest that the toxicity caused by mutant UBQLN2 arises from a combination of dominant-negative effects and gain-of-function toxicity associated with protein aggregation. Disease is unlikely to be caused by a simple loss-of-function because UBQLN2-null animals show few disease-relevant phenotypes (24). Instead, aggregation of mutant UBQLN2 may not only impair its ability to function in protein quality control, but also create dominant-negative effects by sequestering proteasome subunits and related brain-expressed UBQLNs 1 and 4 into mutant UBQLN2 aggregates. In addition, our data show that the aggregates themselves elicit a gain-of-function toxicity. There is no reason to believe these mechanisms are mutually exclusive, and instead likely operate together.
Methods
For more detailed methods, see SI Appendix.
Plasmids.
The pCMV4-FLAG-UBQLN2 plasmid (p4455 FLAG-hPLIC-2; Addgene plasmid # 8661) was a gift from Peter Howley, Harvard Medical School, Boston. The P506T mutation was introduced using site directed mutagenesis (QuikChange II; Agilent Genomics). For bacterial expression and purification, constructs were cloned into pETite C-His Kan DNA vector (Lucigen) or the pET28 vector. For expression in primary neurons the constructs were subcloned into the expression vector, pGW1.
Generation of Transgenic Mice.
Both WT and P506T were subcloned into the MoPrP vector (courtesy of David Borchelt, University of Florida, Gainesville, FL). Transgenic mouse founders were generated through the UM Transgenic Animal Core in a (C57BL/6 X SJL)F2 background.
Immunofluorescence.
For immunofluorescence, 40-μm free-floating, fixed brain sections and fixed primary neurons were labeled with primary and secondary antibodies at at 4 °C. Primary antibodies used in these studies included the following: antiubiquitin (MAB1510; 1:500; Millipore Sigma), guinea pig anti-p62 C-terminal (1:200; Progen), rabbit polyclonal anti-TDP43 antibodies (G. Yu and J. Herz, University of Texas Southwestern Medical Center, Dallas, TX; 1:5,000 and Proteintech 10782–2-AP, 1:1,000), antiproteasome 20S core subunits (Enzo BML-PW8155-0025, 1:1,000), and rabbit anti-UBQLN2 (1:50; NBP1-85639; Novus Biologicals). Secondary antibodies were Alexa Fluor 488 or 568 antibodies (1:1,000; Invitrogen).
Western Blotting.
Lysates from brain tissue were prepared in RIPA buffer (cat #R0278; Sigma) or PBS with a protease inhibitor mixture for detection of transgenic UBQLN2 in mouse brains. For Native blots, lysates were prepared in 4× Native Page Sample Buffer (Novus) as per the manufacturer’s instructions. Primary antibodies included, rabbit α-FLAG (cat#F7425; Sigma) 1:1,000, rabbit anti-UBQLN2 1:250 (NBP1-85639; Novus Biologicals), rabbit anti–α-Tubulin 1:10000 (cat# 2144; Cell Signaling), and mouse anti-GAPDH (1:5,000, MAB374; Millipore) poly-ubiquitinated substrate antibody, FK1 (1:1,000; Enzo). Secondary antibodies were peroxidase-conjugated goat anti-rabbit or goat anti-mouse (1:10,000; Jackson Immunoresearch). Blots were quantified using ImageJ analysis software (NIH).
FRAP.
HEK 293 cells were transfected with 0.5 μg of UBQLN2-GFP and then imaged 24 h after with a Nikon A-1 confocal microscope FRAP analysis was performed in ImageJ. Following thresholding one ROI was generated that corresponded to the prebleach signal and another to the postbleach signal. A mask was created from the prebleach ROI, inverted, and the postbleach ROI was subtracted from this image. This created a region of bleached signal deemed the FRAP ROI. At each time point, the integrated density of the FRAP ROI was divided by the integrated density of the prebleach ROI. These values were then normalized such that the mean of the five prebleach values was set to 1 and the first postbleach value was set to 0. Values from each granule were then fitted to the equation I(t) = A(1− e−t/τ), where I is normalized fluorescent intensity, A is mobile fraction, t is time, and τ is the time constant (65). To quantify complexity, eight observers were asked to score the complexity of UBQLN2 granules on a scale from 1 to 4 based on representative images corresponding to a granule matching each number.
Protein Expression.
UBQLN2 constructs were transformed and expressed in Rosetta (DE3) Escherichia coli bacteria.
ThT Binding Assay.
ThT assays were carried out with 10 μM protein in 0.1 M NaCl, 25 mM Na phosphate pH 7.5. ThT was added to a final concentration of 10 μM. Plates were incubated at 37 °C in a FLUOstar Omega (BMG Labtech) by shaking at 200 rpm. Fluorescence was measured with gain set at 90%, an excitation wavelength of 440 nm, and emission wavelength of 490 nm. Data shown in Fig. 3 and SI Appendix, Fig. S2 are averaged over two to four independent experiments done with different protein preparations.
TEM.
Negatively stained specimens for TEM were prepared by applying 5 μL of protein sample to hydrophilic 400-mesh carbon-coated Formvar support films mounted on copper grids (01702-F; Ted Pella).
Primary Neuron Culture, Transfection, Imaging, and Analysis.
Primary mixed cortical neurons were dissected from embryonic day 20 rat pups and transfected using lipofectamine 2000 (Invitrogen) on day in vitro 4. Each well was cotransfected with 100 ng of either pGW1-iRFP or a pGW1-UBQLN2-iRFP plasmid and 100 ng of pGW1-mApple. Neurons were tracked longitudinally using an automated fluorescence microscopy platform, as previously described (47, 66). Statistical analyses were performed using the survival package and custom scripts in R. To calculate the CV on a single-cell basis, the following equation was used: CV = SDCy5/µCy5, where SD is the SD of fluorescence intensity in the Cy5 channel (representing iRFP fluorescence) for each cell, and µCy5 is the mean fluorescence intensity (integrated density/area) in the Cy5 channel for each cell.
WT-UBQLN2 and L619A-UBQLN2 + Ubiquitin Pulldown.
Full length His-tagged UBQLN2 WT and L619A were expressed as described in the SI Appendix. Next, 0.1 mg/mL WT or L619A UBQLN2 and 0.01 mg/mL poly-Ubiquitin/Ub3-Ub7 WT Chains (K48 linked) (R&D Systems) were incubated on a Labquake tube rotator (Fisher Scientific) at room temperature for 1 h. The solution was added to Ni-NTA agarose, washed with 1× TBS and eluted by 1× TBS with 300 mM imidazole. The flow-through and elution samples were assayed by Western blot. Membranes were probed with rabbit polyclonal Ubiquilin-2 antibody (Novus Biologicals) and rabbit polyclonal Ubiquitin antibody (#3933; Cell Signal Technology).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810522115/-/DCSupplemental.
References
- 1.Hutton M, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 2.Polymeropoulos MH, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 3.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori F, et al. Ubiquilin immunoreactivity in cytoplasmic and nuclear inclusions in synucleinopathies, polyglutamine diseases and intranuclear inclusion body disease. Acta Neuropathol. 2012;124:149–151. doi: 10.1007/s00401-012-0999-z. [DOI] [PubMed] [Google Scholar]
- 5.Rutherford NJ, et al. Unbiased screen reveals ubiquilin-1 and -2 highly associated with huntingtin inclusions. Brain Res. 2013;1524:62–73. doi: 10.1016/j.brainres.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng L, et al. Differential recruitment of UBQLN2 to nuclear inclusions in the polyglutamine diseases HD and SCA3. Neurobiol Dis. 2015;82:281–288. doi: 10.1016/j.nbd.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng HX, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernández-Vega A, et al. Local nucleation of microtubule bundles through tubulin concentration into a condensed tau phase. Cell Rep. 2017;20:2304–2312. doi: 10.1016/j.celrep.2017.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambadipudi S, Biernat J, Riedel D, Mandelkow E, Zweckstetter M. Liquid-liquid phase separation of the microtubule-binding repeats of the Alzheimer-related protein tau. Nat Commun. 2017;8:275. doi: 10.1038/s41467-017-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel A, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 11.Lin Y, Protter DS, Rosen MK, Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molliex A, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riback JA, et al. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell. 2017;168:1028–1040.e19. doi: 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le NT, et al. Motor neuron disease, TDP-43 pathology, and memory deficits in mice expressing ALS-FTD-linked UBQLN2 mutations. Proc Natl Acad Sci USA. 2016;113:E7580–E7589. doi: 10.1073/pnas.1608432113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilpin KM, Chang L, Monteiro MJ. ALS-linked mutations in ubiquilin-2 or hnRNPA1 reduce interaction between ubiquilin-2 and hnRNPA1. Hum Mol Genet. 2015;24:2565–2577. doi: 10.1093/hmg/ddv020. [DOI] [PubMed] [Google Scholar]
- 16.Cassel JA, Reitz AB. Ubiquilin-2 (UBQLN2) binds with high affinity to the C-terminal region of TDP-43 and modulates TDP-43 levels in H4 cells: Characterization of inhibition by nucleic acids and 4-aminoquinolines. Biochim Biophys Acta. 2013;1834:964–971. doi: 10.1016/j.bbapap.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dao TP, et al. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol Cell. 2018;69:965–978.e6. doi: 10.1016/j.molcel.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko HS, Uehara T, Tsuruma K, Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004;566:110–114. doi: 10.1016/j.febslet.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 20.Hjerpe R, et al. UBQLN2 mediates autophagy-independent protein aggregate clearance by the proteasome. Cell. 2016;166:935–949. doi: 10.1016/j.cell.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chuang KH, Liang F, Higgins R, Wang Y. Ubiquilin/Dsk2 promotes inclusion body formation and vacuole (lysosome)-mediated disposal of mutated huntingtin. Mol Biol Cell. 2016;27:2025–2036. doi: 10.1091/mbc.E16-01-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ceballos-Diaz C, et al. Viral expression of ALS-linked ubiquilin-2 mutants causes inclusion pathology and behavioral deficits in mice. Mol Neurodegener. 2015;10:25. doi: 10.1186/s13024-015-0026-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gorrie GH, et al. Dendritic spinopathy in transgenic mice expressing ALS/dementia-linked mutant UBQLN2. Proc Natl Acad Sci USA. 2014;111:14524–14529. doi: 10.1073/pnas.1405741111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Q, et al. Pathogenic Ubqln2 gains toxic properties to induce neuron death. Acta Neuropathol. 2015;129:417–428. doi: 10.1007/s00401-014-1367-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams KL, et al. UBQLN2/ubiquilin 2 mutation and pathology in familial amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:2527.e3–2527.e10. doi: 10.1016/j.neurobiolaging.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 26.Borchelt DR, et al. A vector for expressing foreign genes in the brains and hearts of transgenic mice. Genet Anal. 1996;13:159–163. doi: 10.1016/s1050-3862(96)00167-2. [DOI] [PubMed] [Google Scholar]
- 27.Barmada SJ, et al. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimura AL, et al. Allele-specific knockdown of ALS-associated mutant TDP-43 in neural stem cells derived from induced pluripotent stem cells. PLoS One. 2014;9:e91269. doi: 10.1371/journal.pone.0091269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee EB, Lee VM, Trojanowski JQ. Gains or losses: Molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci. 2011;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cummings CJ, et al. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 31.Kwak S, Masaki T, Ishiura S, Sugita H. Multicatalytic proteinase is present in Lewy bodies and neurofibrillary tangles in diffuse Lewy body disease brains. Neurosci Lett. 1991;128:21–24. doi: 10.1016/0304-3940(91)90751-e. [DOI] [PubMed] [Google Scholar]
- 32.Gorrie GH, et al. Dendritic spinopathy in transgenic mice expressing ALS/dementia-linked mutant UBQLN2. Proc Natl Acad Sci USA. 2014;111:14524–14529. doi: 10.1073/pnas.1405741111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murakami T, et al. ALS/FTD mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron. 2015;88:678–690. doi: 10.1016/j.neuron.2015.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sormanni P, Aprile FA, Vendruscolo M. The CamSol method of rational design of protein mutants with enhanced solubility. J Mol Biol. 2015;427:478–490. doi: 10.1016/j.jmb.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 35.Linding R, Schymkowitz J, Rousseau F, Diella F, Serrano L. A comparative study of the relationship between protein structure and beta-aggregation in globular and intrinsically disordered proteins. J Mol Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- 36.Rousseau F, Schymkowitz J, Serrano L. Protein aggregation and amyloidosis: Confusion of the kinds? Curr Opin Struct Biol. 2006;16:118–126. doi: 10.1016/j.sbi.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 37.Thompson MJ, et al. The 3D profile method for identifying fibril-forming segments of proteins. Proc Natl Acad Sci USA. 2006;103:4074–4078. doi: 10.1073/pnas.0511295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maurer-Stroh S, et al. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat Methods. 2010;7:237–242. doi: 10.1038/nmeth.1432. [DOI] [PubMed] [Google Scholar]
- 39.Biancalana M, Koide S. Molecular mechanism of thioflavin-T binding to amyloid fibrils. Biochim Biophys Acta. 2010;1804:1405–1412. doi: 10.1016/j.bbapap.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Groenning M. Binding mode of thioflavin T and other molecular probes in the context of amyloid fibrils-current status. J Chem Biol. 2010;3:1–18. doi: 10.1007/s12154-009-0027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biancalana M, Makabe K, Koide A, Koide S. Molecular mechanism of thioflavin-T binding to the surface of beta-rich peptide self-assemblies. J Mol Biol. 2009;385:1052–1063. doi: 10.1016/j.jmb.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfe LS, et al. Protein-induced photophysical changes to the amyloid indicator dye thioflavin T. Proc Natl Acad Sci USA. 2010;107:16863–16868. doi: 10.1073/pnas.1002867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kardos J, et al. Structural studies reveal that the diverse morphology of beta(2)-microglobulin aggregates is a reflection of different molecular architectures. Biochim Biophys Acta. 2005;1753:108–120. doi: 10.1016/j.bbapap.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 44.Lindgren M, Sörgjerd K, Hammarström P. Detection and characterization of aggregates, prefibrillar amyloidogenic oligomers, and protofibrils using fluorescence spectroscopy. Biophys J. 2005;88:4200–4212. doi: 10.1529/biophysj.104.049700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 46.Miller J, et al. Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation, and neuronal death provide novel insight into Huntington’s disease molecular pathogenesis. J Neurosci. 2010;30:10541–10550. doi: 10.1523/JNEUROSCI.0146-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barmada SJ, et al. Amelioration of toxicity in neuronal models of amyotrophic lateral sclerosis by hUPF1. Proc Natl Acad Sci USA. 2015;112:7821–7826. doi: 10.1073/pnas.1509744112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Filonov GS, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arrasate M, Finkbeiner S. Automated microscope system for determining factors that predict neuronal fate. Proc Natl Acad Sci USA. 2005;102:3840–3845. doi: 10.1073/pnas.0409777102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kleijnen MF, et al. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 51.Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41:1767–1777. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- 52.Zhang D, Raasi S, Fushman D. Affinity makes the difference: Nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2008;377:162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang KY, Yang S, Warraich ST, Blair IP. Ubiquilin 2: A component of the ubiquitin-proteasome system with an emerging role in neurodegeneration. Int J Biochem Cell Biol. 2014;50:123–126. doi: 10.1016/j.biocel.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 54.Al-Sarraj S, et al. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- 55.Barmada SJ, Finkbeiner S. Pathogenic TARDBP mutations in amyotrophic lateral sclerosis and frontotemporal dementia: Disease-associated pathways. Rev Neurosci. 2010;21:251–272. doi: 10.1515/revneuro.2010.21.4.251. [DOI] [PubMed] [Google Scholar]
- 56.Blokhuis AM, Groen EJ, Koppers M, van den Berg LH, Pasterkamp RJ. Protein aggregation in amyotrophic lateral sclerosis. Acta Neuropathol. 2013;125:777–794. doi: 10.1007/s00401-013-1125-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brettschneider J, et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann Neurol. 2013;74:20–38. doi: 10.1002/ana.23937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim SH, et al. Mutation-dependent aggregation and toxicity in a Drosophila model for UBQLN2-associated ALS. Hum Mol Genet. 2018;27:322–337. doi: 10.1093/hmg/ddx403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang B, Wu Q, Zhou H, Huang C, Xia XG. Increased Ubqln2 expression causes neuron death in transgenic rats. J Neurochem. 2016;139:285–293. doi: 10.1111/jnc.13748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murray DT, et al. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171:615–627.e16. doi: 10.1016/j.cell.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell Commun Signal. 2016;14:1. doi: 10.1186/s12964-015-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boeynaems S, et al. Protein phase separation: A new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Purice MD, Taylor JP. Linking hnRNP function to ALS and FTD pathology. Front Neurosci. 2018;12:326. doi: 10.3389/fnins.2018.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.St George-Hyslop P, et al. The physiological and pathological biophysics of phase separation and gelation of RNA binding proteins in amyotrophic lateral sclerosis and fronto-temporal lobar degeneration. Brain Res. 2018;1693:11–23. doi: 10.1016/j.brainres.2018.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barmada SJ, et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10:677–685. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.