Significance
Accurate gene expression is essential in all organisms. During protein synthesis, transfer RNAs (tRNAs) decode the genetic information contained in messenger RNA (mRNA) on the ribosome into amino acids using a defined 3-nt code. A fundamental question in biology is how the ribosome maintains this 3-nt code, or mRNA frame, during the dynamic processes that move the mRNA-tRNA pairs through the different tRNA-binding sites. We solved structures of a frameshift-prone tRNA bound to the bacterial ribosome after mRNA decoding. We find that the tRNA undergoes conformational rearrangements in the peptidyl (P) and exit (E) sites that cause the ribosome to lose its grip on the mRNA and allow the tRNA to shift into a new reading frame.
Keywords: recoding, tRNA, mRNA, ribosome, frameshift
Abstract
Accurate translation of the genetic code is critical to ensure expression of proteins with correct amino acid sequences. Certain tRNAs can cause a shift out of frame (i.e., frameshifting) due to imbalances in tRNA concentrations, lack of tRNA modifications or insertions or deletions in tRNAs (called frameshift suppressors). Here, we determined the structural basis for how frameshift-suppressor tRNASufA6 (a derivative of tRNAPro) reprograms the mRNA frame to translate a 4-nt codon when bound to the bacterial ribosome. After decoding at the aminoacyl (A) site, the crystal structure of the anticodon stem-loop of tRNASufA6 bound in the peptidyl (P) site reveals ASL conformational changes that allow for recoding into the +1 mRNA frame. Furthermore, a crystal structure of full-length tRNASufA6 programmed in the P site shows extensive conformational rearrangements of the 30S head and body domains similar to what is observed in a translocation intermediate state containing elongation factor G (EF-G). The 30S movement positions tRNASufA6 toward the 30S exit (E) site disrupting key 16S rRNA–mRNA interactions that typically define the mRNA frame. In summary, this tRNA-induced 30S domain change in the absence of EF-G causes the ribosome to lose its grip on the mRNA and uncouples the canonical forward movement of the tRNAs during elongation.
The ribosome coordinates the concerted movement of mRNA and tRNA pairs during translation while accurately maintaining the mRNA reading frame for the correct sequential addition of amino acids (1). Three-nucleotide mRNA sequences, or codons, encode for amino acids, and the selection of correct tRNAs helps maintain the precise mRNA frame. In the absence of translational fidelity, nonsense protein products are expressed or else premature termination occurs. Nevertheless, programmed recoding of the mRNA coding sequence can occur and is well established in viruses, prokaryotes, and eukaryotes (2–4). Certain tRNAs can cause a shift out of frame (i.e., frameshifting) due to imbalances in tRNA concentrations (5–8), lack of RNA modifications (9–14), or insertions or deletions in tRNAs known as frameshift suppressors (15–21). Synthetic biology has attempted to reprogram the genetic code by using frameshift-suppressor tRNAs for the incorporation of unnatural amino acids into proteins to probe questions of cellular protein localization, protein–protein interactions, and posttranslational modifications (22). Currently, however, unnatural amino acid incorporation suffers from low efficiency, leading to the inability to add multiple and different chemical moieties at defined locations in proteins. Such issues stem from a poor understanding of how the ribosome controls the normal mRNA frame and how this can be repurposed for recoding.
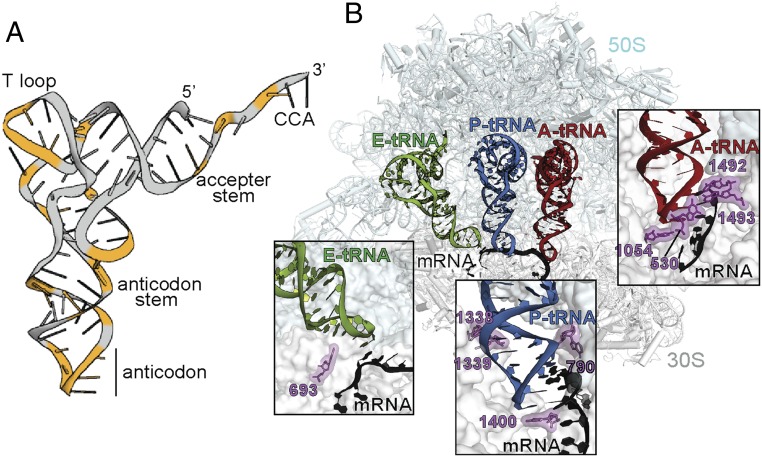
To determine whether the genetic code can be altered to nontriplet codons, experiments were conducted to identify suppressors of nucleotide insertions or deletions in essential genes (23–25). The ability to restore the mRNA reading frame by decoding a noncanonical mRNA codon was mediated predominately by mutated tRNAs, called frameshift suppressor tRNAs (reviewed in ref. 21). These tRNAs contained insertions or deletions along their entire body, but the majority were located in their anticodon stem-loop (ASL) region (Fig. 1A). Recoding events induced by these frameshift-prone tRNAs could occur in either direction on the mRNA (5′ or 3′), resulting in positive or negative mRNA frameshifting, that is, a −1 event (a codon containing two nucleotides) or a +1 event (a codon containing four nucleotides). Although the structure and function of the ribosome has been extensively studied, it remains unclear at what point during elongation these tRNAs cause ribosome dysregulation and mRNA frameshifting.
Fig. 1.
Interaction of tRNAs with the ribosome. (A) tRNA tertiary structure with nucleotides in which mutations cause mRNA frameshifts, highlighted in orange. (B) tRNA interactions with the A, P, and E sites on the ribosome (PDB ID code 4V51). (Insets) The extensive interactions of the 16S rRNA with the mRNA-tRNA pair at the A site, which are absent in the P and the E sites.
The structures of +1 frameshift-suppressor or frameshift-prone tRNAs bound to the 70S ribosome provide important insights into how an ASL containing an extra nucleotide interacted with the aminoacyl (A) site during decoding of the mRNA codon (26–28). These studies established that the frameshift-prone tRNA decoded the mRNA in the unshifted or zero frame, thus indicating that the move into the frameshift must be in a postdecoding step. However, the point at which the shift occurs after the tRNA-mRNA pair leaves the A site is not clear. Indeed, the interactions between the ribosome and the mRNA-tRNA pairs bound at the A, peptidyl (P), and exit (E) sites change substantially, providing additional support for the idea that the move into the +1 frame occurs after the frameshift-prone tRNA has moved from the A site (Fig. 1B). For example, in the A site, the mRNA codon and the tRNA anticodon are closely monitored by 16S rRNA nucleotides G530, C1054, A1492, and A1493 to ensure cognate or correct tRNA selection (29) (Fig. 1B). These interactions likely enforce a 3-nt codon-anticodon pairing between frameshift-prone tRNA and a 4-nt codon, thus preventing a shift into the new +1 frame (26–28). Once the tRNA-mRNA pair is moved or translocated to the P site by elongation factor G (EF-G), the ribosome minimally inspects the codon-anticodon pairing; P-site 16S rRNA nucleotides A790, A1338, and A1339 engage the anticodon stem, while nucleotide C1400 packs beneath the third codon-anticodon pair (30) (Fig. 1B). At the opposite end of the tRNA on the 50S, 23S rRNA nucleotides form extensive interactions to stabilize the tRNA for peptidyl transfer with the incoming aminoacyl-tRNA (aa-tRNA). On translocation of the tRNA to the E site, the lack of interactions between the ribosome and the codon-anticodon helix persists, and only 16S rRNA nucleotide G693 flanks the 5′ of the mRNA path (Fig. 1B). The absence of interactions of the ribosome with the codon-anticodon helix in the P and the E sites provides opportunities for disruption of the mRNA-tRNA pairing.
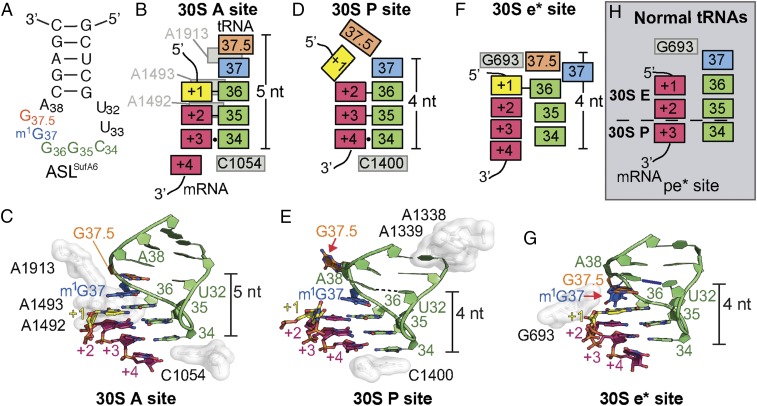
Several models have been proposed to explain the mechanism of frameshift-suppressor tRNA dysregulation of the mRNA frame (31); however, how these tRNAs alone induce the ribosome out-of-frame remains unknown. Our previous structures of frameshift-suppressor tRNASufA6 bound to the ribosomal A site provided important insights into how this tRNA pairs with a 4-nt mRNA codon (28). Frameshift suppressor tRNASufA6 is derived from proline tRNA (tRNAPro) and contains an insertion between ASL nucleotides 37 and 38 (referred to as G37.5) that enables high levels of +1 frameshifting within mRNAs containing 5′-CCC-U/G/C-3′ (the proline codon is underlined, and the U, G, or C nucleotides can be decoded as the 4-nt codon) (15, 32) (Fig. 2A and SI Appendix, Fig. S1). Likewise, parental tRNAPro also undergoes recoding in the absence of a posttranslational chemical modification at nucleotide position 37 in its ASL (17). Despite models suggesting that the ribosome decodes the +1 codon as four bases instead of the normal three in the A site (i.e., “quadruplet” decoding), our results establish that tRNASufA6 decodes the mRNA as a standard 3-nt codon-anticodon in the zero frame (28) (Fig. 2 B and C). Although we demonstrated that this recoding does not occur in the A site, there is significant disruption of evolutionarily conserved base-pairing interactions between anticodon stem nucleotides 32 and 38 of the tRNA (Fig. 2B). The nucleotide identity of the 32–38 interaction is a fundamental feature of all tRNAs and is directly correlated to the strength of the codon–anticodon interaction (high vs. low GC base pairs) (33). Disrupting this relationship leads to the inability of the ribosome to distinguish correct from incorrect tRNAs (34–36). These structures suggest that disruption of the tertiary structure of the tRNA is likely one consequence of the inserted nucleotide in the ASL that causes a +1 frameshift. Here we present X-ray crystal structures of the Thermus thermophilus 70S ribosome undergoing a +1 recoding event induced by tRNASufA6 after decoding that provide molecular insights into mRNA frame dysregulation.
Fig. 2.
Interaction of frameshift-prone tRNASufA6 with the tRNA-binding sites on the ribosome. (A) Secondary structure of ASLSufA6 with the anticodon nucleotides (green), the inserted nucleotide G37.5 (orange), and the modified G37 (blue). (B and C) Schematic (B) and structure (C) of the codon–anticodon interaction in the A site. The anticodon and mRNA are surrounded by 16S rRNA nucleotides A1492/93, A1913, and C1054 (white), which aid correct tRNA selection. ASLSufA6 forms a 5-nt stack and is decoded in the zero frame despite containing an additional anticodon loop nucleotide, G37.5 (orange) (PDB ID code 4L47). Watson–Crick base pairs between the first two interactions of the anticodon and the codon (+1–36 and +2–35) are shown with a black bar, and the +3–34 wobble interaction is shown with a circle (B). There is no interaction between U32 and A38. (D and E) In the P site, the anticodon and codon are minimally monitored, and instead 16S rRNA nucleotides A1338/9 (white) recognize the anticodon stem and C1400 packs beneath the +4–34 base pair. In this relaxed state, ASLSufA6 forms a 4-nt stack by base-flipping G37.5 (red arrow in E) and engages the mRNA codon nucleotides +2, +3, and +4 in the new +1 frame. This movement also reestablishes the U32-A38 pairing (shown with a dotted line). Watson–Crick base pairs between the first two interactions of the anticodon and the codon (+2–36 and +3–35) are shown with a black line, and the +4–34 wobble interaction is shown with a circle (D). (F and G) Frameshift-prone tRNASufA6 is biased toward the E site, adopting a new position (e*) on the 30S that pulls the mRNA codon by one nucleotide into the E site. Additional remodeling of the ASL ejects m1G37 (red arrow in G) to maintain the 4-nt stack and the U32-A38 pairing (show with a blue line). The single interaction between the codon and the anticodon is denoted as a black bar (+1–36) in F. (H) Comparison of a 70S·EF-G translocation intermediate state containing a pe/E tRNA (PDB ID code 4V9L) reveals that during canonical translocation, there is space between 16S rRNA nucleotide G693 and the +1 mRNA of the codon. This allows for the full accommodation of the E site codon on completion of translocation. The anticodon is pulled away from the mRNA, as shown by the lack of Watson–Crick base pair interactions. The dotted line signifies the barrier between the 30S E and P sites.
Results and Discussion
To precisely define how an 8-nt anticodon loop interacts with a +1 codon after decoding, we solved the 3.3-Å X-ray structure of the ASL of tRNASufA6 programmed to a 4-nt codon in the P site on the ribosome (Fig. 2 D and E and SI Appendix, Fig. S2 and Table S1). The anticodon loop of ASLSufA6 is fully methylated at position 37, a critical feature of this tRNA family for selection by the ribosome (37). Our structure reveals that P site-bound ASLSufA6 interacts with the mRNA in the new +1 frame, engaging the +2 to +4 nucleotides of the codon (Fig. 2 D and E). This result demonstrates unequivocally that the frameshift-suppressor tRNA alone promotes movement into the +1 frame, consistent with primer extension studies of tRNAs containing expanded ASLs programmed in the P site (38, 39). Moreover, the +1 nucleotide of the codon is positioned toward the 30S E site, compacting the mRNA on the ribosome to accommodate seven nucleotides, instead of the usual six, of the E and P site mRNA codons (SI Appendix, Fig. S3).
Although the anticodon of ASLSufA6 forms a canonical 3-nt interaction with the mRNA codon in the P site, albeit in the +1 frame, its ASL undergoes conformational remodeling in two distinct ways compared with normal tRNAs (Fig. 2 D and E) First, the extra nucleotide G37.5 is ejected from the anticodon loop, allowing the important base-pairing interactions between nucleotides U32 and A38 to reform. Previously, we found that the 32–38 interaction was disrupted when tRNASufA6 decoded the mRNA codon in the A site (28). One likely reason for the restoration of the 32–38 base pair is the lack of interactions of the ribosome with the codon-anticodon pair in the P site. This relaxed environment allows the 32–38 pair to form and consequently causes the ejection of G37.5. A second difference is that the G37.5 ejection causes a reduction in the base-stacking interactions of the 3′ end of the anticodon loop from five to four bases (compare Fig. 2 B and C with Fig. 2 D and E). Almost all tRNAs contain a 7-nt anticodon loop, and these nucleotides form a 4-nt stack (residues 34–37), a stabilizing feature important for interactions with mRNA and the ribosome. Modification at ASL position 37 enhances the stability of the codon–anticodon interaction as well as this stacking interaction (37, 40) (SI Appendix, Fig. S4). tRNASufA6 forms a 5-nt stack in the A site during decoding (28), further supporting the hypothesis that the restrictive environment of the A site influences the tRNA structure and likely prevents the ejection of G37.5 (Fig. 2 B and C and SI Appendix, Fig. S4). Our structure demonstrates that once the ASL of tRNASufA6 transits to the more relaxed environment of the ribosomal P site, the important U32–A38 interaction reforms at the expense of extruding G37.5, allowing the mRNA to shift into the +1 frame. These data suggest that the intrinsic geometric plasticity of frameshift-prone tRNAs likely drives unusual tRNA conformations that promote frameshifting.
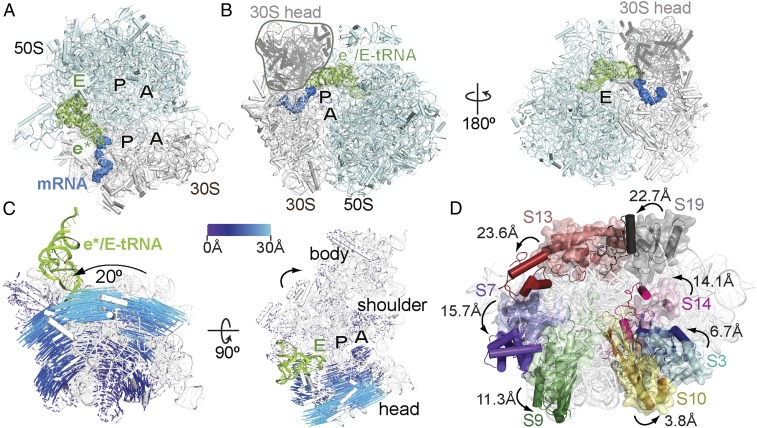
The structure of ASLSufA6 in the P site addresses the fundamental question of the timing of recoding by frameshift-suppressor tRNAs. We next sought to understand how such a change would propagate along the entire tRNA by solving a 3.9-Å resolution X-ray structure of the tRNASufA6 programmed to the 70S ribosomal P site (SI Appendix, Fig. S2 D–F and Table S1). Although tRNASufA6 is deacylated, we assume that the tRNA would adopt a canonical position in the P site (e.g., a P/P position denoting its location on the 30S/50S) similar to other deacylated tRNAs bound to the 70S ribosome (30, 41). Surprisingly, however, tRNASufA6 adopted a previously unidentified tRNA position on the ribosome whereby the ASL is biased toward the 30S E site (Fig. 3 A and B). We refer to this position as the e* site, with the lowercase letter denoting a chimeric position between the P and E sites on the 30S. The CCA 3′−end of the tRNA is positioned in the 50S E site; however, its position is likely influenced by its deacylated state (SI Appendix, Fig. S5). We note that if the P-tRNA contained a peptidyl moiety, the CCA 3′-end would be located in the 50S P site. In the e* position on the 30S, the ASL of tRNASufA6 undergoes additional and distinct remodeling compared with when it is bound in the P or A site (28). This remodeling of the tRNA is likely due to the absence of interactions of the tRNA with the interior of the ribosome. Nucleotide G37.5, which was ejected from the anticodon loop in the P site, swaps positions with m1G37; m1G37 now extends toward 16S rRNA helix 24 nucleotide A790 (Fig. 2 F and G and SI Appendix, Fig. S2F). Since methylation of nucleotide 37 stabilizes the frame during translocation events in normal tRNAs (37, 42), we propose that base-flipping of modified m1G37 contributes to the frameshifting capability of tRNASufA6.
Fig. 3.
Frameshift-prone tRNASufA6 is biased toward the E site concurrent with 30S head domain swiveling. (A) Overview of the 70S bound to the e*/E tRNASufA6 with the A, P, and E tRNA-binding sites indicated. (B) The 70S shown from the A site (Left) and, rotated 180° around the vertical axis, from the E site (Right), with the 30S head domain indicated. (C) Conformational changes of the 30S on tRNASufA6 binding. Shifts in Cα atoms of the ribosomal proteins in the unrotated 70S (PDB ID code 1VY4) and the 70S- tRNASufA6 are indicated by the color-coded scale mapped onto the 30S subunit of the 70S- tRNASufA6 complex. (Left) The same view as in A. (Right) A 90° horizontal rotation. (D) Movement of the 30S head domain proteins S3, S7, S9, S10, S13, S14, and S19 (∼4–24 Å) on binding of tRNASufA6 in the e* site (same view as A). The original locations of 30S proteins and 16S rRNA are shown as semitransparent molecular surfaces, and the locations occupied in the context of bound tRNASufA6 are shown as cartoons (and darker shades of the same colors).
Translocation of the tRNAs through the ribosome requires the disruption and reestablishment of interactions of ribosomal proteins, rRNA, and intersubunit molecular bridges, and the flexing of the tRNAs (43–46). Ribosomes actively undergoing translocation of tRNAs by EF-G reveal the disruptive nature of this movement (44, 47, 48) (SI Appendix, Fig. S6). In the context of a chimeric pe*/E tRNA trapped during translocation between the 30S P and E sites, EF-G causes large-scale conformational changes, including counterclockwise intrasubunit rotation of the 30S body domain (1–3°) and counterclockwise intersubunit swiveling of the 30S head domain (18–21°) (44, 47–49). In addition, these states were shown to be functionally relevant in the translocation mechanism in the context of biochemical experiments (50). These conformational changes result in the significant displacement (3–24 Å) of 30S ribosomal proteins S3, S7, S9, S10, S13, S14, and S19 and a 14-Å inward movement of the L1 stalk to contact the tRNA elbow. The location of tRNASufA6 resembles a tRNA trapped in the pe*/E translocation intermediate state (47), with some notable differences. For example, tRNASufA6 induces an ∼20° swiveling of the 30S head domain and a∼0.3–2.5° rotation of the body domain relative to the 50S (Fig. 3 B–D and SI Appendix, Fig. S7 and Table S2). In addition, tRNASufA6 is also positioned ∼3.5 Å toward the 30S E site as compared with pe*/E tRNA (47) (SI Appendix, Fig. S5B). Finally, these large conformational changes caused by tRNASufA6 occur in the absence of EF-G. This remarkable translocation-like movement induced by frameshift-prone tRNASufA6 alone in the absence of EF-G strongly suggests that the process of translocation is disrupted by tRNAs able to recode.
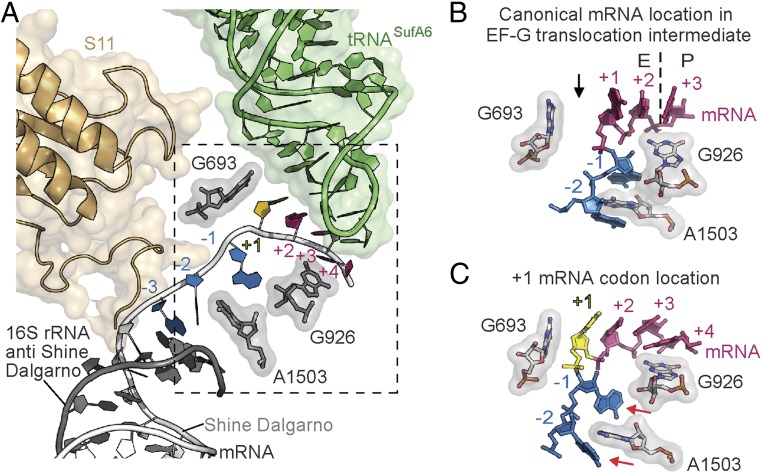
As EF-G translocates the tRNA-mRNA pairs through the ribosome, the large conformational changes cause the ablation of interactions between the codon and the anticodon even in the cognate case (47) (SI Appendix, Fig. S6). Consistent with these findings, the near-cognate codon–anticodon interaction in the 70S-e*/E tRNASufA6 complex is also disrupted (Fig. 4 A and B and SI Appendix, Fig. S8B). Therefore, although the anticodon loop of tRNASufA6 moves toward the E site on the 30S in the absence of EF-G, the disengagement of the tRNA from the codon is likely not influenced by this lack of interaction, as this occurs normally even with a cognate codon–anticodon interaction during translocation (47).
Fig. 4.
Interactions of e*/E tRNASufA6 with 16S rRNA. (A) tRNASufA6 binds in a newly defined e*/E site where the CCA end is located on the 50S E site and the ASL is closer to the 30S E site. The mRNA is numbered with +1 (yellow) corresponding to the 5′ nucleotide of the 4-nt frameshifting codon. The boxed region is shown in more detail in C. (B) Structure of 70·EF-G translocation intermediates state showing the tRNA located between the 30S P and the E sites, termed pe*/E (PDB ID code 4V9L). In this state, the +1 and +2 mRNA codon nucleotides (magenta) are in the E site, the +3 nucleotide is in the P site, and 16S rRNA nucleotide G926 is packed beneath. The dotted line signifies the boundary between the E and the P sites, and the black arrow denotes the 1-nt gap between G693 and the +1 nucleotide of the codon. 16S rRNA nucleotide A1503 is stably intercalated between the −1 and −2 nucleotides of the mRNA. (C) In the e*/E tRNASufA6 structure, the +1 mRNA nucleotide (yellow) is pulled into the E site and packs directly against G693. 16S rRNA nucleotide G926 also flanks the +3/+4 mRNA codon, but A1503 no longer stacks with mRNA nucleotides −1 and −2 (denoted by red arrows).
The ribosome has been proposed to play an active role in maintaining the mRNA frame via base-stacking interactions of 16S rRNA residues A1503 and C1397 with mRNA 5′ and 3′ nucleotides, respectively (47). One role for these stabilizing interactions is to prevent reverse movement of the mRNA during translocation, thereby preserving the correct mRNA frame (47). In the case of the mRNA-tRNASufA6 pair in the e*/E site, base-stacking interactions between A1503 with the mRNA nucleotides 5′ of the E-site codon are disrupted (Fig. 4 A and C and SI Appendix, Fig. S8). A possible reason for this disruption could be the additional nucleotide in the codon that structurally constrains the mRNA to accommodate seven instead of six nucleotides in the E and the P sites. Then, as the mRNA progresses through the path after the E site, the ribosome loses its grip on the mRNA due to the absence of base-stacking interactions between the 16S rRNA and the mRNA.
Although the inability of the ribosome to monitor the mRNA frame is likely influenced by the scrunching of the mRNA codon in the E site, one important question regarding the mechanism of frameshifting is how does the extra mRNA nucleotide of the expanded codon fit into this restricted space? In 70S-EF-G translocation intermediate states where tRNAs are trapped between sites (e.g., pe*/E and ap/ap chimeric positions), only two mRNA nucleotides of the E-site codon are translocated, allowing space for the additional nucleotide of the E-site mRNA codon on the completion of translocation (44, 47, 48) (SI Appendix, Fig. S7D). In contrast, when tRNASufA6 adopts an e* position on the 30S in the absence of EF-G, the +1 nucleotide of the 4-nt codon occupies this typically vacant space in the E site and directly stacks with 16S rRNA G693 (Figs. 2F and 4 A and C). We predict that once tRNASufA6 is fully translocated to the E site by EF-G, the first nucleotide of the codon (+1) will be required to exit the E site, ensuring and sustaining the +1 frame of the mRNA.
These studies provide critical insight into how tRNA structural plasticity can result in dysregulation of the reading frame by inducing large-scale conformational changes of the 30S head domain in the absence of EF-G. In addition, our present results may have important implications in understanding other recoding events caused by mRNAs, including that of viral mRNAs that are required for gene expression (2, 51). Frameshift-suppressor tRNAs are a potentially powerful tool in chemical biology for genetic reprogramming to incorporate unnatural amino acids (52). Our mechanistic insights into recoding induced by these tRNAs suggests that tRNAs can be rationally designed to promote higher efficiencies to facilitate innovative and new protein-based technologies.
Experimental Procedures
mRNA, ASL, and Ribosome Purification.
ASLSufA6 containing an m1G37-modified ASL (18 nucleotides) and mRNA (19 nucleotides) were purchased from GE Healthcare Dharmacon and dissolved in 10 mM Tris⋅HCl pH 7.0 and 5 mM MgCl2 (SI Appendix, Table S2). We used a chemically synthesized 18-nt ASL to ensure complete 1-methyl modification at position 37 (m1G37), as previously used to examine interactions in the A site (28). Purification of T. thermophilus 70S ribosomes was performed as described previously (53).
tRNASufA6 Ligation and Purification.
To ensure that tRNASufA6 was methylated at tRNA nucleotide 37 (m1G), the 5′ half of the tRNA was chemically synthesized (GE Healthcare Dharmacon) to position 39 and enzymatically ligated to the chemically synthesized 3′ half (SI Appendix, Table S2). To anneal the 5′- and 3′-ends of the tRNA, the following protocol was followed (54, 55). In brief, after 5′-end phosphorylation of the 3′ half by T4 polynucleotide kinase (New England BioLabs), the enzyme was heat-inactivated, and the two 5′ and 3′ halves of each tRNA were mixed and annealed in the T4 RNA ligase buffer at 80 °C for 5 min and then slow-cooled on the heat block to room temperature. T4 RNA ligase (New England BioLabs) was added (final concentration of 500 U/mL), followed by incubation at 37 °C for 18 h. The ligation reaction was run on a 12% denaturing 8 M urea-polyacrylamide gel, and the ligated fragment was excised and purified using a modified crush-and-soak method (55). The RNA was ethanol-precipitated, and the pellet was thoroughly air-dried and resuspended in 10 mM Tris⋅HCl pH 7.0 and 5 mM MgCl2, followed by annealing at 70 °C for 2 min and then slow-cooling to room temperature on the benchtop. The RNA was aliquoted and stored at −20 °C.
Complex Formation and Structural Studies of 70S-ASLSufA6 and 70S-tRNASufA6.
The 70S ribosomes (4.4 µM) were programmed with mRNA (8.8 µM) for 6 min at 37 °C. A 2.5-molar excess of ASLSufA6 (11 µM) and a 2-molar excess of tRNASufA6 (8.8 µM) were individually incubated for 30 min at 37 °C. Each tRNA was programmed in the ribosomal P site by designing the Shine–Dalgarno sequence eight nucleotides from the first nucleotide of the P-site codon. tRNASufA6 was deacylated and should bind optimally in the P site, as numerous 70S-deacylated P site-bound tRNAs structures have been solved (30, 41). Deoxy BigCHAP (2.8 μM; Hampton Research) was added just before crystallization. Crystals were grown by sitting-drop vapor diffusion in 4–5% polyethylene glycol (PEG) 20K, 4–5% PEG 550 MME, 0.1 M Tris-acetate pH 7.0, 0.2 M KSCN, and 10 mM MgCl2 and cryoprotected by increasing PEG 550 MME to a final concentration of 30%. Crystals were flash-frozen in liquid nitrogen for data collection.
X-ray diffraction data were collected at either the Southeast Regional Collaborative Access Team beamline 22-ID or the Northeastern Collaborative Access Team beamline ID24-C at the Advanced Photon Source. Data were integrated and scaled using the XDS program (56). In the 70S-ASLSufA6–containing mRNA, the structure was solved by molecular replacement in PHENIX using coordinates from a 70S structure containing mRNA and tRNAs (SI Appendix, Table S1) (PDB ID code 4V6G) (57, 58). For the 70S complex containing an e*/E tRNA and the mRNA, the strategy was modified to include the 30S subunit from 70S translocation intermediate containing EF-G and neomycin (PDB ID code 4W29) (48). In this structure, the 30S head domain is swiveled 18° relative to the 50S. The structure was solved by molecular replacement in PHENIX (58) followed by iterative rounds of manual building in Coot (59). All figures were prepared in PyMOL (60).
Structural Comparisons.
30S head swivel and body rotation were calculated relative to a nonrotated 70S complex (PDB ID code 4V51) using the structural alignment of 50S large subunit (23S rRNA residues 200–800) (SI Appendix, Table S2). The PyMOL plugin for Euler–Rodriguez transformations was used (49) in which the 30S head is defined as 16S rRNA residues 921–1396 and the body is defined as 16S rRNA 1–920 and 1397–1542.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant R01 GM093278. C.M.D. is a Burroughs Wellcome Fund Pathogenesis of Infectious Diseases Fellow. The X-ray crystallography datasets were collected at the Northeastern Collaborative Access Team beamlines, which are funded by National Institute of General Medical Sciences Grant P41 GM103403, and at the Southeast Regional Collaborative Access Team beamlines. The Pilatus 6M detector on beamline 24-IDC is funded by National Institutes of Health Office of Research Infrastructure Programs High-End Instrumentation Grant S10 RR029205. This research used resources of the Advanced Photon Source, a US Department of Energy Office of Science User Facility operated by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.D.D. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB codes 5VPO and 5VPP).
See Commentary on page 11121.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809319115/-/DCSupplemental.
References
- 1.Zaher HS, Green R. Fidelity at the molecular level: Lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farabaugh PJ. Programmed translational frameshifting. Annu Rev Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- 3.Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016;44:7007–7078. doi: 10.1093/nar/gkw530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinman JD. Control of gene expression by translational recoding. Adv Protein Chem Struct Biol. 2012;86:129–149. doi: 10.1016/B978-0-12-386497-0.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkins JF, Gesteland RF, Reid BR, Anderson CW. Normal tRNAs promote ribosomal frameshifting. Cell. 1979;18:1119–1131. doi: 10.1016/0092-8674(79)90225-3. [DOI] [PubMed] [Google Scholar]
- 6.Vimaladithan A, Farabaugh PJ. Special peptidyl-tRNA molecules can promote translational frameshifting without slippage. Mol Cell Biol. 1994;14:8107–8116. doi: 10.1128/mcb.14.12.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sundararajan A, Michaud WA, Qian Q, Stahl G, Farabaugh PJ. Near-cognate peptidyl-tRNAs promote +1 programmed translational frameshifting in yeast. Mol Cell. 1999;4:1005–1015. doi: 10.1016/s1097-2765(00)80229-4. [DOI] [PubMed] [Google Scholar]
- 8.O’Connor M. Imbalance of tRNA(Pro) isoacceptors induces +1 frameshifting at near-cognate codons. Nucleic Acids Res. 2002;30:759–765. doi: 10.1093/nar/30.3.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Björk GR, Wikström PM, Byström AS. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science. 1989;244:986–989. doi: 10.1126/science.2471265. [DOI] [PubMed] [Google Scholar]
- 10.Hagervall TG, Tuohy TM, Atkins JF, Björk GR. Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J Mol Biol. 1993;232:756–765. doi: 10.1006/jmbi.1993.1429. [DOI] [PubMed] [Google Scholar]
- 11.Lecointe F, et al. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J Biol Chem. 2002;277:30445–30453. doi: 10.1074/jbc.M203456200. [DOI] [PubMed] [Google Scholar]
- 12.Licznar P, et al. Programmed translational −1 frameshifting on hexanucleotide motifs and the wobble properties of tRNAs. EMBO J. 2003;22:4770–4778. doi: 10.1093/emboj/cdg465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Björk GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waas WF, Druzina Z, Hanan M, Schimmel P. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J Biol Chem. 2007;282:26026–26034. doi: 10.1074/jbc.M703391200. [DOI] [PubMed] [Google Scholar]
- 15.Qian Q, et al. A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol Cell. 1998;1:471–482. doi: 10.1016/s1097-2765(00)80048-9. [DOI] [PubMed] [Google Scholar]
- 16.Qian Q, Björk GR. Structural alterations far from the anticodon of the tRNAProGGG of Salmonella typhimurium induce +1 frameshifting at the peptidyl-site. J Mol Biol. 1997;273:978–992. doi: 10.1006/jmbi.1997.1363. [DOI] [PubMed] [Google Scholar]
- 17.Sroga GE, Nemoto F, Kuchino Y, Björk GR. Insertion (sufB) in the anticodon loop or base substitution (sufC) in the anticodon stem of tRNA(Pro)2 from Salmonella typhimurium induces suppression of frameshift mutations. Nucleic Acids Res. 1992;20:3463–3469. doi: 10.1093/nar/20.13.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herr AJ, Nelson CC, Wills NM, Gesteland RF, Atkins JF. Analysis of the roles of tRNA structure, ribosomal protein L9, and the bacteriophage T4 gene 60 bypassing signals during ribosome slippage on mRNA. J Mol Biol. 2001;309:1029–1048. doi: 10.1006/jmbi.2001.4717. [DOI] [PubMed] [Google Scholar]
- 19.Bossi L, Roth JR. Four-base codons ACCA, ACCU, and ACCC are recognized by frameshift suppressor sufJ. Cell. 1981;25:489–496. doi: 10.1016/0092-8674(81)90067-2. [DOI] [PubMed] [Google Scholar]
- 20.Bossi L, Smith DM. Suppressor sufJ: A novel type of tRNA mutant that induces translational frameshifting. Proc Natl Acad Sci USA. 1984;81:6105–6109. doi: 10.1073/pnas.81.19.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Atkins JF, Björk GR. A gripping tale of ribosomal frameshifting: Extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev. 2009;73:178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis L, Chin JW. Designer proteins: Applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 23.Riddle DL, Roth JR. Suppressors of frameshift mutations in Salmonella typhimurium. J Mol Biol. 1970;54:131–144. doi: 10.1016/0022-2836(70)90451-1. [DOI] [PubMed] [Google Scholar]
- 24.Riyasaty S, Atkins JF. External suppression of a frameshift mutant in Salmonella. J Mol Biol. 1968;34:541–557. doi: 10.1016/0022-2836(68)90179-4. [DOI] [PubMed] [Google Scholar]
- 25.Yourno J, Tanemura S. Restoration of in-phase translation by an unlinked suppressor of a frameshift mutation in Salmonella typhimurium. Nature. 1970;225:422–426. doi: 10.1038/225422a0. [DOI] [PubMed] [Google Scholar]
- 26.Dunham CM, et al. Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit. RNA. 2007;13:817–823. doi: 10.1261/rna.367307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fagan CE, Maehigashi T, Dunkle JA, Miles SJ, Dunham CM. Structural insights into translational recoding by frameshift suppressor tRNASufJ. RNA. 2014;20:1944–1954. doi: 10.1261/rna.046953.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maehigashi T, Dunkle JA, Miles SJ, Dunham CM. Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem-loops. Proc Natl Acad Sci USA. 2014;111:12740–12745. doi: 10.1073/pnas.1409436111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 30.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 31.Curran JF, Yarus M. Reading frame selection and transfer RNA anticodon loop stacking. Science. 1987;238:1545–1550. doi: 10.1126/science.3685992. [DOI] [PubMed] [Google Scholar]
- 32.Roth JR. Frameshift mutations. Annu Rev Genet. 1974;8:319–346. doi: 10.1146/annurev.ge.08.120174.001535. [DOI] [PubMed] [Google Scholar]
- 33.Olejniczak M, Uhlenbeck OC. tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie. 2006;88:943–950. doi: 10.1016/j.biochi.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Olejniczak M, Dale T, Fahlman RP, Uhlenbeck OC. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat Struct Mol Biol. 2005;12:788–793. doi: 10.1038/nsmb978. [DOI] [PubMed] [Google Scholar]
- 35.Ledoux S, Uhlenbeck OC. Different aa-tRNAs are selected uniformly on the ribosome. Mol Cell. 2008;31:114–123. doi: 10.1016/j.molcel.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ledoux S, Olejniczak M, Uhlenbeck OC. A sequence element that tunes Escherichia coli tRNA(Ala)(GGC) to ensure accurate decoding. Nat Struct Mol Biol. 2009;16:359–364. doi: 10.1038/nsmb.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gustilo EM, Vendeix FA, Agris PF. tRNA’s modifications bring order to gene expression. Curr Opin Microbiol. 2008;11:134–140. doi: 10.1016/j.mib.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker SE, Fredrick K. Recognition and positioning of mRNA in the ribosome by tRNAs with expanded anticodons. J Mol Biol. 2006;360:599–609. doi: 10.1016/j.jmb.2006.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phelps SS, et al. Translocation of a tRNA with an extended anticodon through the ribosome. J Mol Biol. 2006;360:610–622. doi: 10.1016/j.jmb.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 40.Grosjean H, Söll DG, Crothers DM. Studies of the complex between transfer RNAs with complementary anticodons, I: Origins of enhanced affinity between complementary triplets. J Mol Biol. 1976;103:499–519. doi: 10.1016/0022-2836(76)90214-x. [DOI] [PubMed] [Google Scholar]
- 41.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 42.Gamper HB, Masuda I, Frenkel-Morgenstern M, Hou YM. Maintenance of protein synthesis reading frame by EF-P and m(1)G37-tRNA. Nat Commun. 2015;6:7226. doi: 10.1038/ncomms8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340:1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tourigny DS, Fernández IS, Kelley AC, Ramakrishnan V. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou J, Lancaster L, Donohue JP, Noller HF. How the ribosome hands the A-site tRNA to the P site during EF-G–catalyzed translocation. Science. 2014;345:1188–1191. doi: 10.1126/science.1255030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohan S, Donohue JP, Noller HF. Molecular mechanics of 30S subunit head rotation. Proc Natl Acad Sci USA. 2014;111:13325–13330. doi: 10.1073/pnas.1413731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wasserman MR, Alejo JL, Altman RB, Blanchard SC. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat Struct Mol Biol. 2016;23:333–341. doi: 10.1038/nsmb.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dinman JD. Mechanisms and implications of programmed translational frameshifting. Wiley Interdiscip Rev RNA. 2012;3:661–673. doi: 10.1002/wrna.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang K, Schmied WH, Chin JW. Reprogramming the genetic code: From triplet to quadruplet codes. Angew Chem Int Ed Engl. 2012;51:2288–2297. doi: 10.1002/anie.201105016. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, Hong S, Ruangprasert A, Skiniotis G, Dunham CM. Alternative mode of E-site tRNA binding in the presence of a downstream mRNA stem loop at the entrance channel. Structure. 2018;26:437–445.e3. doi: 10.1016/j.str.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherlin LD, et al. Chemical and enzymatic synthesis of tRNAs for high-throughput crystallization. RNA. 2001;7:1671–1678. [PMC free article] [PubMed] [Google Scholar]
- 55.Stark MR, Rader SD. Efficient splinted ligation of synthetic RNA using RNA ligase. Methods Mol Biol. 2014;1126:137–149. doi: 10.1007/978-1-62703-980-2_10. [DOI] [PubMed] [Google Scholar]
- 56.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol. 2010;17:555–560. doi: 10.1038/nsmb.1790. [DOI] [PubMed] [Google Scholar]
- 58.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schrödinger LLC 2010. The PyMOL Molecular Graphics System, version 1.3r1 (Schrödinger LLC, New York)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.