Significance
Alphaviruses are mosquito-borne RNA viruses that cause encephalitis, rash, and arthritis. Alphavirus nonstructural protein 3 (nsP3) has a highly conserved macrodomain that can bind and remove ADP-ribose (ADPr) residues from ADP-ribosylated proteins, but its role in virus replication was not known. We show that alphavirus replication in neural cells depends on protein ADP ribosylation and that nsP3s with mutations that eliminate ADPr binding cannot form a functional replicase. Mutations that decrease ADPr binding result in fewer infected cells, while mutations that increase binding but decrease hydrolase activity infect cells normally but amplify replication complexes less well. Therefore, alphavirus replication requires nsP3 macrodomain interaction with one or more ADP-ribosylated proteins, as is consistent with the observed high conservation of this region.
Keywords: alphavirus, macrodomain, ADP ribosylation, replication complexes, PARP
Abstract
Alphaviruses are plus-strand RNA viruses that cause encephalitis, rash, and arthritis. The nonstructural protein (nsP) precursor polyprotein is translated from genomic RNA and processed into four nsPs. nsP3 has a highly conserved macrodomain (MD) that binds ADP-ribose (ADPr), which can be conjugated to protein as a posttranslational modification involving transfer of ADPr from NAD+ by poly ADPr polymerases (PARPs). The nsP3MD also removes ADPr from mono ADP-ribosylated (MARylated) substrates. To determine which aspects of alphavirus replication require nsP3MD ADPr-binding and/or hydrolysis function, we studied NSC34 neuronal cells infected with chikungunya virus (CHIKV). Infection induced ADP-ribosylation of cellular proteins without increasing PARP expression, and inhibition of MARylation decreased virus replication. CHIKV with a G32S mutation that reduced ADPr-binding and hydrolase activities was less efficient than WT CHIKV in establishing infection and in producing nsPs, dsRNA, viral RNA, and infectious virus. CHIKV with a Y114A mutation that increased ADPr binding but reduced hydrolase activity, established infection like WT CHIKV, rapidly induced nsP translation, and shut off host protein synthesis with reduced amplification of dsRNA. To assess replicase function independent of virus infection, a transreplicase system was used. Mutant nsP3MDs D10A, G32E, and G112E with no binding or hydrolase activity had no replicase activity, G32S had little, and Y114A was intermediate to WT. Therefore, ADP ribosylation of proteins and nsP3MD ADPr binding are necessary for initiation of alphavirus replication, while hydrolase activity facilitates amplification of replication complexes. These observations are consistent with observed nsP3MD conservation and limited tolerance for mutation.
Alphaviruses are icosahedral, enveloped, plus-strand RNA viruses in the Togaviridae family that are mostly transmitted by mosquitoes and cause a range of diseases in humans (1–3). New World alphaviruses often cause encephalitis, while the Old World viruses primarily cause rash and arthritis. However, in recent epidemics chikungunya virus (CHIKV), an Old World alphavirus, has also been associated with neurological disease (4).
The alphavirus genome is capped, polyadenylated, and organized in two ORFs (5). The 5′ ORF encodes the four nonstructural replicase proteins (nsPs), and the 3′ ORF encodes the structural proteins (6, 7). The nsP precursors, polyproteins P123 and P1234, are translated from the genomic RNA. P123 together with nsP4, released through proteolytic processing by a papain-like protease in nsP2, synthesizes a minus-strand RNA copy of the genome. Sequential polyprotein processing switches RNA synthesis from minus strand to full-length plus strand genomic and subgenomic (sg) RNAs (8–13). Formation of replication complexes and synthesis of dsRNA is accompanied by shutoff of host protein synthesis. Because synthesis of P123 and P1234 from amplified full-length plus-strand RNAs has requirements for translation that are similar to most host cell mRNAs (14), synthesis of new P1234 required for minus-strand viral RNA synthesis also ceases. RNA synthesis therefore shifts to synthesis of only plus-strand genomic and sgRNAs by the processed individual nsPs (14–16). sgRNA is translated to produce the structural proteins required for virion assembly (8–13, 17).
Functions of most of the alphavirus nsPs in replication are known (11). nsP1 has methyl transferase and guanylyl transferase activities, is palmitoylated, and binds the replication complex to membranes (18, 19). nsP2 has helicase, ATPase, GTPase, and RNA 5′-triphosphatase as well as protease activities. nsP4 is the RNA-dependent RNA polymerase with a conserved C-terminal GDD RNA polymerase motif and adenylyl transferase (20, 21). Identification of the function(s) of nsP3, a phosphoprotein found both in replication complexes and cytoplasmic aggregates (22, 23), has been more difficult.
nsP3, a determinant of mosquito vector specificity (24, 25) and mouse neurovirulence (24, 26–31), is an essential part of the viral replicase, induces membrane remodeling necessary for the formation of spherules, and regulates RNA synthesis in a cell type-dependent manner (32–35). nsP3 has three domains: a highly conserved N-terminal macrodomain (MD), a zinc-binding oligomerization domain, and a poorly conserved, unstructured, acidic, and highly Ser/Thr phosphorylated C-terminal hypervariable domain (HVD). nsP3 functions initially as part of the polyprotein P123 in conjunction with nsP4 for negative-strand synthesis and later as P23 and nsP3 for plus-strand RNA synthesis. The cellular proteins bound to nsP3 and the locations of nsP3–host protein complexes change during replication from early plasma membrane spherules to late cytoplasmic granules (32). Phosphorylation of the HVD occurs early in replication and is required for optimal RNA synthesis (22, 34, 36). The HVD interacts with multiple host proteins and serves as a hub for the assembly of factors required for replication complex formation and initiation of RNA synthesis (26, 37–41). The MD affects phosphorylation of nsP3, P23 processing, minus-strand synthesis, shutoff of host protein synthesis, and virulence (27, 31, 34, 42) by poorly understood mechanisms.
MDs are conserved 130- to 190-aa structural modules characterized by the ability to bind ADP-ribose (ADPr) and its derivatives (43–45). ADP ribosylation is a posttranslational modification accomplished by transfer of ADPr moieties, either singly as mono ADPr (MAR) or multiply in branched or linear polymeric chains as poly ADPr (PAR), from NAD+ to target proteins (46). Intracellular ADP ribosylation regulates a range of cellular functions and is catalyzed by enzymes of the ADP-ribosyltransferases including a subfamily commonly known as “poly (ADPr) polymerases” (PARPs) (47, 48).
The nsPs of a subset of plus-strand RNA viruses (i.e., alphaviruses, rubella virus, coronaviruses, hepatitis E virus) have MDs, originally identified as a highly conserved “X” domain of unknown function (43, 44) and classified as macroD-type by recent informatics analyses (45). We and others recently demonstrated that the alphaviral nsP3MD, like other viral and macroD-type MDs, possesses the ability not only to bind ADPr but also to hydrolyze the bond between ADPr and the acidic side chains of mono ADP-ribosylated (MARylated) protein residues (27, 49–53). Viruses with nsP3 mutations that substantially decrease or eliminate ADPr binding or hydrolase activity are not viable (27). Viruses with mutations that reduce but do not eliminate ADPr binding or hydrolase activity replicate less well in neuronal cells, with reduced RNA synthesis and attenuated virulence in mice (27, 31, 54–58).
However, the requirement for interaction with MARylated cellular proteins and the step(s) in replication affected by the loss of ADPr binding or hydrolase activity are not known. Because several PARPs can be induced by IFN, are under diversifying selection, and have antiviral activity, it has been postulated that the nsP3MD is important for countering the antiviral activities of host proteins ADP-ribosylated by antiviral PARPs (59–65). In the present study, we determined that nsP3MD-mediated ADPr binding is critical for establishing replication complexes to initiate infection and that hydrolase activity facilitates replication-complex amplification in infected neuronal cells; these observations are consistent with the high conservation of this region and its importance for neurovirulence.
Results
nsP3MD Mutations That Affect ADPr-Binding and Hydrolase Activities Impair CHIKV Replication.
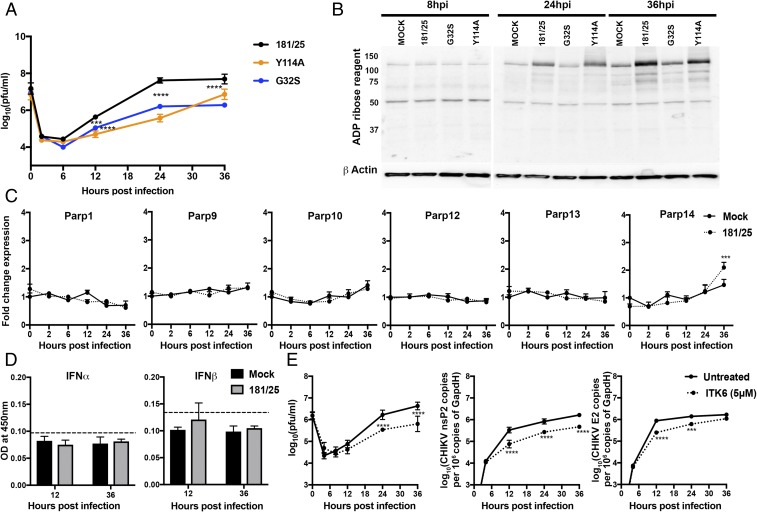
To assess the effects of nsP3MD mutations that alter ADPr binding or hydrolase activity on replication in neuronal cells, NSC34 cells were infected with the nsP3MD mutants G32S (decreased binding and hydrolase activities) and Y114A (increased binding and decreased hydrolase activities) (Table 1), and virus production was compared with WT CHIKV-infected cells (Fig. 1A). As previously described (27), G32S replicates with similar kinetics but lower virus production at all times than the WT virus, while Y114A replicates with distinct kinetics characterized by delayed production of virus but with similar induction of cell death by 48 h.
Table 1.
Characteristics of nsP3 MDs relative to WT (++++) and viability of viruses studied
| nsP3MD mutant | ADPr binding | ADPr hydrolase | Viable virus |
| WT | ++++ | ++++ | Yes |
| D10A | — | + | No |
| G32E | — | — | No |
| G32S | ++ | ++ | Yes |
| G112E | — | — | No |
| Y114A | +++++ | ++ | Yes |
Data from ref. 27.
Fig. 1.
CHIKV infection facilitates virus replication in NSC34 cells by increasing ADP ribosylation of cellular proteins without inducing PARP gene transcription. (A) NSC34 cells were infected with CHIKV 181/25 (WT) and nsP3MD mutants G32S and Y114A at an MOI of 10. Virus production was measured by plaque formation in Vero cells. Each value represents the average from three independent experiments; error bars indicate SDs. ***P < 0.001, ****P < 0.0001 (181/25 vs. nsP3MD mutants G32S and Y114A). (Adapted with permission from ref. 27.) (B) NSC34 cells were infected with CHIKV 181/25 and nsP3MD mutants G32S and Y114A at an MOI of 5, and cell lysates were immunoblotted for ADPr. Antibody against β-actin was used for loading controls. A representative image from three independent experiments is shown. (C) NSC34 cells were infected with CHIKV 181/25 at an MOI of 5, and PARP mRNA expression was measured by qRT-PCR. Mock-infected cells (solid line) were compared with CHIKV-infected cells (dashed line). CT values were normalized to Gapdh, and fold change was calculated relative to uninfected 0-h (ΔΔCT) data. Each value represents the average ± SD from three independent experiments. ***P < 0.001 (mock vs. infected). (D) NSC34 cells were infected with CHIKV 181/25 (MOI = 5), and supernatant fluids were assayed for IFNα and IFNβ by enzyme immunoassay. Dashed lines indicate the lower limit of detection for the assays. (E) NSC34 cells were infected with CHIKV 181/25 (MOI = 1) and were treated or not treated with the pan mono-ADPr inhibitor ITK6 (5 μM) at the time of infection. Supernatant fluids were assayed for virus production by plaque formation in Vero cells (Left), and intracellular RNAs were assayed by qRT-PCR for viral genomic (nsP2; Middle) and genomic+sg E2 RNA and were compared with standard curves of CHIKV RNAs (Right). Each value represents the average ± SD from three independent experiments. ***P < 0.001, ****P < 0.0001.
CHIKV Infection of NSC34 Cells Increases ADP Ribosylation of Cellular Proteins Without Inducing IFN or PARP Gene Transcription.
To determine the response of neuronal cells to alphavirus infection, NSC34 cells infected with CHIKV were assessed for induction of ADP-ribosylated proteins by immunoblot using an ADPr-binding reagent that detects both MARs and PARs (Fig. 1B) (66). Infection with all viruses induced detectable ADP ribosylation of cellular proteins from the host or virus that increased through 36 h. Y114A induced similar, if not higher, amounts of protein ADP ribosylation than the WT virus at 24 h but slightly less at 36 h, while the G32S mutant induced substantially less ADP ribosylation than either the WT virus or Y114A at all times (Fig. 1B). To determine whether increased ADP ribosylation was due to increased expression of one or more of the enzymatically active PARPs or was associated with IFN production, infected cells were assessed from 2 to 36 h after infection for expression of PARP 1, 9, 10, 12, 13, and 14 mRNAs by qRT-PCR (Fig. 1C), and supernatant fluids were assayed for IFNα and IFNβ protein by enzyme immunoassay (Fig. 1D). Except for Parp14 at 36 h, none of the PARP mRNAs examined was increased, and type I IFN was not detectable at any time after infection. Therefore, the increased ADP ribosylation of NSC34 cellular proteins induced by CHIKV infection is likely due to increased PARP activity without evidence of IFN induction or increased PARP gene expression.
Optimal CHIKV Replication Requires MARylation.
To determine whether MARylation is important for virus replication, NSC34 cells were treated with noncytotoxic levels (89% viability at 24 h) of ITK6, an inhibitor of PARPs 10, 11, and 14 (67), at the time of infection with WT CHIKV (Fig. 1E). Virus production and plus-strand RNA syntheses were significantly decreased when MARylation was broadly inhibited (P < 0.0001), indicating that MARylation of viral or host proteins promotes CHIKV replication in neural cells. However, the role of nsP3MD ADPr-binding and hydrolase activity for replication is not clear.
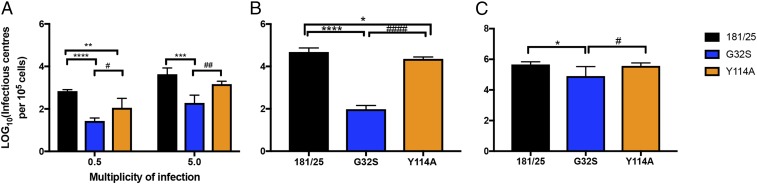
nsP3MD Mutations Affecting ADPr-Binding and Hydrolase Activities Differentially Affect Initiation of Infection.
Initiation of infection is dependent on the likelihood that an individual genome can successfully establish an early replication complex and amplify incoming genomic viral RNA. To determine whether mutations affecting nsP3MD ADPr-binding and/or hydrolase activities affect the likelihood of establishing infection, infectious center assays were performed in NSC34 cells infected at Vero cell multiplicities of infection (MOIs) of 0.5 and 5. Percentages of productively infected NSC34 cells were determined by plaque formation after cocultivation on Vero cells (Fig. 2A). The G32S mutant virus infected cells less well than WT virus (30 G32S vs. 691 WT infectious centers per 105 cells at an MOI of 0.5, P < 0.0001; 240 G32S vs. 5,035 WT infectious centers per 105 cells at an MOI of 5; P < 0.001). The Y114A mutant virus was less impaired than the G32S mutant (30 vs. 150 infectious centers per 105 cells at MOI 0.5, P < 0.05; 240 vs. 1,515 infectious centers per 105 cells at MOI 5, P < 0.01) but established fewer infectious centers than WT CHIKV (150 vs. 691 infectious centers per 105 cells at MOI 0.5, P < 0.01). In vitro-transcribed RNA was also transfected directly into BHK21 cells as well as into the less susceptible NSC34 cells for a more accurate comparison of efficiency of initiation of infection (Fig. 2B). Transfection of RNA into NSC34 cells yielded results similar to infection at an MOI of 5 (Fig. 2A), while BHK21 cells were better able to support initiation of infection with similar infectious centers for WT and Y114A and a 10-fold reduction for G32S (Fig. 2C). Because G32S and Y114A have similar deficiencies in hydrolase activity (Table 1), but Y114A binds ADPr better than either the WT virus or G32S, these data indicate that hydrolase activity is important for successful infection but that improved binding can partially compensate for decreased hydrolase activity and that the relative importance of each is cell type dependent.
Fig. 2.
nsP3MD mutations affecting ADP ribosyl binding and hydrolase activities differentially affect the initiation of infection. (A) Infectious center assays for NSC34 cells infected at MOIs of 0.5 and 5 with CHIKV 181/25 (WT) and nsP3MD mutants G32S or Y114A. Infected cells were trypsinized, viable cells were counted, and serially diluted cells were plated on Vero cells to identify virus-producing cells by plaque assay. (B and C) Infectious center assays for NSC34 (B) and BHK21 (C) cells after electroporation of 10 μg of viral RNA transcribed in vitro from full-length clones of 181/25 and nsP3MD G32S and Y114A mutants into 105 cells that were plated onto subconfluent monolayers of BHK21 cells to identify virus-producing cells by plaque formation. The data are presented as log10 infectious centers per 105 cells. Each value represents the average ± SD from three independent experiments; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 (WT vs. G32S/Y114A); #P < 0.05; ##P < 0.01, ####P < 0.0001 (Y114A vs. G32S).
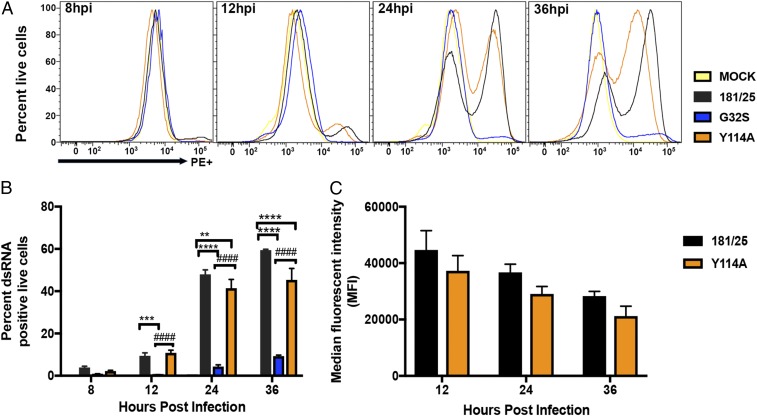
Formation of Replication Complexes Containing dsRNA.
After entry, the first steps in replication are translation of the nsPs and establishment of spherules at the plasma membrane for synthesis of minus-strand template RNA and formation of dsRNA-containing replication complexes that amplify genomic RNA and produce sgRNA. To determine whether nsP3MD ADPr-binding and hydrolase activities are critical for establishing replication complexes or affect a subsequent step in infection, NSC34 cells infected with CHIKV WT, G32S, and Y114A were stained with antibody to dsRNA and were analyzed by flow cytometry (Fig. 3). Fewer cells infected with G32S were positive for dsRNA than cells infected with WT virus or Y114A at 12, 24, and 36 h after infection, while the numbers of cells staining positively for dsRNA were similar in WT and Y114A-infected cells at 12 h [0.48 (G32S) vs. 9.4 (WT), P < 0.001, and 10.8 (Y114A), P < 0.0001] and were only slightly less at 24 and 36 h, reflecting the ability of the viruses to spread through the culture (Fig. 3 A and B). Individual cells infected with WT virus amplified replication complexes with dsRNA better than cells infected with Y114A virus, as is evident from a comparison of the fluorescent intensities (Fig. 3 A and C).
Fig. 3.
Formation of replication complexes containing dsRNA. NSC34 cells infected at an MOI of 5 with CHIKV 181/25 (WT) virus and nsP3MD mutants G32S or Y114A were gated for live cells, fixed, permeabilized, stained for dsRNA, and analyzed by flow cytometry. (A) Representative histograms from three independent experiments. (B) Quantification of the percent of live cells positive for dsRNA. (C) The median fluorescent intensities for WT and Y114A-infected cells. The data are presented as the means ± SD from three independent experiments; **P < 0.01; ***P < 0.001; ****P < 0.0001 (WT vs. G32S/Y114A); ####P < 0.0001 (Y114A vs. G32S). hpi, hours postinfection.
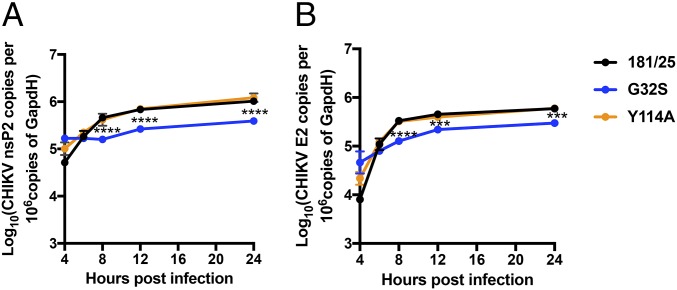
Synthesis of Viral RNAs and Proteins.
To determine whether differences in replication complex formation and amplification resulted in differences in the synthesis of viral RNAs, levels of genomic and sgRNAs were determined by RT-qPCR (Fig. 4). The amounts of both genomic RNA (Fig. 4A) and genomic plus sgRNA (Fig. 4B) synthesized by the G32S mutant virus were significantly less than the amounts synthesized by either the WT or Y114A viruses. Amounts of RNAs synthesized by Y114A were not detectably different from those synthesized by the WT virus.
Fig. 4.
Synthesis of viral RNAs. Levels of genomic (A) and genomic plus sg (B) RNA from NSC34 cells infected with CHIKV 181/25 (WT) and nsP3MD mutants G32S or Y114A at an MOI of 5 were measured by qRT-PCR. Total cellular RNA was collected, and cDNA was produced and was quantified by qPCR compared with a standard curve of CHIKV cDNA fragments corresponding to genomic or sgRNA. The data represent the means ± SD from three independent experiments. ***P < 0.001; ****P < 0.0001 (WT/Y114A vs. G32S).
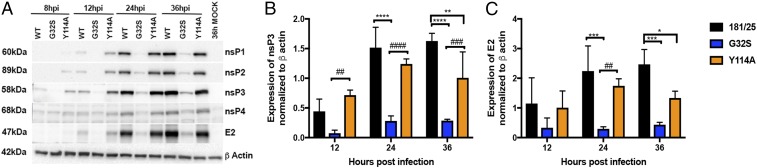
To determine whether synthesis of viral RNA reflected translation of nsPs from genomic RNA, lysates from NSC34 cells infected with WT, G32S, or Y114A viruses were assessed for nsP synthesis by immunoblot (Fig. 5A). The G32S mutant virus was deficient in viral protein synthesis, with only low levels of nsPs even at 36 h after infection compared with Y114A and WT viruses, likely reflecting the inadequate amplification of viral RNA and production of virus for subsequent rounds of infection. Interestingly, Y114A produced more viral nsPs at 8 h and 12 h than the WT virus, suggesting that better ADPr binding facilitated the translation of genomic RNA early after infection (Fig. 5B). To begin to assess the synthesis of structural proteins from sgRNA, lysates were also probed for the E2 glycoprotein. As anticipated, G32S produced little E2, while amounts of E2 were initially similar for Y114A and the WT virus, but at 36 h Y114A produced less E2 than WT (Fig. 5C), indicating that hydrolase activity is required at later phases of virus production.
Fig. 5.
Effect of nsP3MD mutation on viral protein synthesis. NSC34 cells were infected with CHIKV 181/25 (WT) or nsP3MD mutants G32S or Y114A at an MOI of 5, and cell lysates were probed by immunoblot for production of viral proteins using rabbit polyclonal antibodies to nsP1, nsP2, nsP3, nsP4, and E2. Antibody against β-actin was used for loading controls. (A) Representative image. (B and C) Quantification of expression of nsP3 (B) and E2 (C) relative to actin by densitometry on blots from three independent experiments. Data indicate the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (WT vs. mutant); ##P < 0.01, ###P < 0.001, ####P < 0.0001 (Y114A vs. G32S). hpi, hours postinfection.
Therefore, the CHIKV with the nsP3MD mutation G32S that decreased both ADPr-binding and hydrolase activity initiated infection in neuronal cells less efficiently, resulting in decreased production of viral RNA and protein compared with the WT virus. In contrast, virus with the Y114A mutation that decreased hydrolase activity but increased binding was only slightly impaired for infection initiation, with efficient translation of nsPs (Fig. 5 A and B), but was less able to amplify replication complexes than the WT virus (Fig. 3) and produced less structural protein (Fig. 5C) and less infectious virus (Fig. 1A) even though the amounts of sgRNA were not identifiably different (Fig. 4B).
Phosphorylation of eIF2α and Shutoff of Host Protein Synthesis.
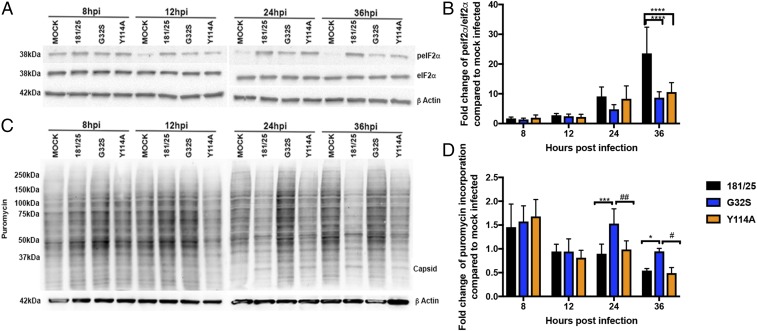
Because alphavirus replication induces host translational shutoff that also affects polyprotein translation from full-length genomic RNA, we assessed the effects of WT and mutant virus infection on the phosphorylation of the translation initiation factor eIF2α by immunoblot (Fig. 6 A and B) and on host protein synthesis by incorporation of puromycin, a structural analog of aminoacyl tRNA, into newly synthesized proteins (Fig. 6 C and D) (68). Phosphorylated eIF2α was evident by 8–12 h for all viruses, was increased at 24 h for WT and Y114A, and was maintained at increased levels relative to total eIF2α for WT virus-infected cells at 36 h (Fig. 6B). Host translational shutoff was evident earliest (at 12 h) in cells infected with Y114A (Fig. 6C) and was more pronounced at 24 h and 36 h for cells infected with WT and Y114A viruses (Fig. 6D). For G32S-infected cells, there was little evidence of shutoff of host cell protein synthesis, with levels of newly synthesized protein similar to those of uninfected cells even at late times (Fig. 6 C and D).
Fig. 6.
Effect of nsP3MD mutation on phosphorylation of eIf2α and host protein translation. NSC34 cells were infected with CHIKV 181/25 (WT) or nsP3MD mutants G32S or Y114A at an MOI of 5. (A and B) Cell lysates were probed by immunoblot for levels of total and phosphorylated eIF2α. Antibody against β-actin was used for loading controls. (A) Representative image. (B) Blots from three independent experiments were quantitated by densitometry, and the ratios of phosphorylated eIF2α to total eIF2α were calculated. Data are expressed as the mean fold change over mock-infected cells ± SD. (C and D) Host protein translation was analyzed by the incorporation of puromycin. At the time points indicated, medium was replaced with medium containing puromycin (5 μg/mL) and was incubated for 10 min. Cell lysates were probed by immunoblot with antibody to puromycin to detect newly synthesized protein. Antibody against β-actin was used for loading controls. (C) Representative image. (D) Blots from three independent experiments were quantitated by densitometry, and puromycin incorporation was compared with mock-infected cells. Data are presented as the mean fold change over mock ± SD. *P < 0.05, ***P < 0.001, ****P < 0.0001 (WT vs. mutant); #P < 0.05, ##P < 0.01 (Y114A vs. G32S).
Analysis of nsP Function in the Absence of Virus Infection Using a Transreplicase System.
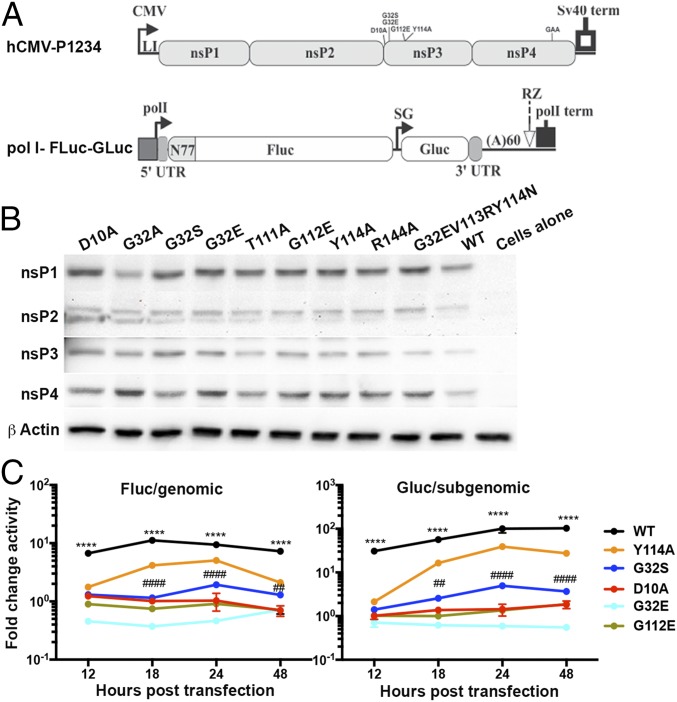
To allow functional analysis of nsPs for nonviable as well as viable CHIKV nsP3MD mutants with similar production of nsPs independent of virus replication, a modified CHIKV transreplication system was used to generate host-mediated synthesis of nsPs and template RNAs for amplification of viral RNAs (Fig. 7A) (8, 69, 70). For this system, one plasmid encodes the CHIKV P1234 polyprotein behind a human cytomegalovirus (hCMV)-driven polymerase II (pol II) promoter, and a second plasmid encodes a pol I promoter-driven viral template RNA for synthesis of luciferase reporters from both the genomic 5′ UTR (firefly luciferase, Fluc) and subgenomic 5′ UTR (Gaussia luciferase, Gluc) by the viral proteins encoded by the P1234 plasmid. The pol II promoter/terminator was substituted for the previously described hCMV-Fluc-Gluc reporter to reduce the background reporter expression (69, 71) and improve assay sensitivity. All previously analyzed nsP3 mutations (27) were introduced into viral polymerase-competent hCMV-P1234 and polymerase-mutated control CMV-P1234GAA plasmids by site-directed mutagenesis and were transfected into NSC34 cells along with the pol I-Fluc-Gluc template plasmid (Fig. 7). Because nsP mRNA synthesis is accomplished by RNA pol II in the nucleus rather than in the cytoplasm, similar levels of nsPs will be produced independent of viral replicase function, and reporter expression will reflect the levels of genomic and sgRNAs produced by nsPs that include WT and mutant nsP3.
Fig. 7.
Analysis of nsP function in the absence of virus infection using a transreplicase system. (A) The transreplicase system includes an hCMV-P1234 plasmid with and without nsP3MD mutations, an identical inactivated nsP4 polymerase hCMV-P1234GAA plasmid, and a pol I 5′ UTR–Fluc–sg promoter–Gluc–3′ UTR plasmid. (B) NSC34 cells were transfected with 1 μg of each plasmid and were incubated at 37 °C for 24 h. Viral protein production was analyzed by immunoblotting with antibodies to CHIKV nsP1, nsP2, nsP3, nsP4, and E2. Antibody against β-actin was used for loading controls. The image is representative of three independent experiments. (C) Fluc reporter activity for genomic RNA synthesis and Gluc reporter activity for subgenomic RNA synthesis. Luciferase activities were read on a luminometer and were normalized to the total protein content in the lysate. The reporter activities generated by the inactive replicase P1234GAA were subtracted from reporter activities generated by the active replicase P1234 and are reported as fold change. The data are presented as the means ± SD from three independent experiments. ****P < 0.0001, WT vs. G32S; ##P < 0.01, ####P < 0.0001, Y114A vs. G32S.
Levels of nsPs expressed from plasmids with the protein-coding regions of viable (G32A, G32S, T111A, and Y114A) and nonviable (D10A, G32E, G112E, R144A, and G32EV113RY114N) mutant viruses were similar (Fig. 7B). However, for the nsPs representing viable viruses, most notably WT, less protein was present at 24 h with expression of the WT replicase, suggesting shutoff of host transcription (72, 73). These protein levels suggest that transcriptional shutoff is impaired by mutations in nsP3, as it is for mutations in nsP2 (Fig. 7B) (69). Although lower levels of WT nsPs were detected on immunoblot, reporter expression from synthesized RNAs was robust, indicating fully functional replicase activity (Fig. 7C). Peak genomic RNA synthesis for the WT virus was 18 h after transfection (P < 0.0001), while sgRNA synthesis increased gradually and was maximal at 24 h and 48 h (P < 0.0001) after transfection. Reporter expression from genomic and sgRNAs for representative nonviable nsP3 mutants D10A, G32E, and G112E (no viruses rescued) (Table1) (27) did not exceed background levels of reporter protein expression, indicating that without nsP3MD binding and/or hydrolase activities the replicase was not functional (Fig. 7C). Levels of reporter protein expression for the representative viable mutants G32S and Y114A (rescued viruses with impaired replication) were intermediate between the WT and the nonviable mutants (Fig. 7C). G32S produced much lower genomic (WT vs. G32S, P < 0.0001; Y114A vs. G32S, P < 0.01) and sgRNA compared with WT (P < 0.0001). Levels of genomic and sgRNAs were better for Y114A than for G32S (Y114A vs. G32S, P < 0.01) but were impaired compared with WT (P < 0.0001), although this was not evident from the RT-qPCR data obtained from infected cells, possibly because of differences in the sensitivity of the assays (Fig. 4). Therefore, viruses with nsP3MD mutations D10A, G32E, and G112E, which are deficient in ADPr binding, are not viable because viral RNA could not be synthesized.
Discussion
Alphavirus nsP3 is an essential component of the viral RNA replicase and has an N-terminal MD highly conserved among togaviruses, coronaviruses, and hepevirus, but specific functions important for virus replication have not been identified (32, 35, 74). Viral nsP3MDs have mono ADPr hydrolase as well as binding activity (27, 49, 50, 75), and mutations that affect these functions of alphavirus nsP3MDs attenuate virus replication and reduce neurovirulence in mice (27, 31). Because several enzymatically active PARPs have demonstrated antiviral activity, are induced by IFN, and are under diversifying selective pressure (59–63, 65), the MD and particularly its hydrolase function have been postulated to function primarily by countering the antiviral effects of ADP-ribosylated cellular proteins (64, 65, 75). Here, we show that ADP ribosylation is induced by alphavirus infection of neural cells independent of IFN production, that MARylation of cellular proteins promotes virus replication, and that nsP3 ADPr-binding and hydrolase activities are important for different aspects of virus replication. If both functions are absent, the replicase is inactive, viral RNA is not synthesized, and the virus is not viable. If both functions are present but diminished, virus can be rescued, but replicase function is severely compromised, initiation of infection is inefficient, and little virus is produced. If binding is increased but hydrolase decreased, initial nsP synthesis is facilitated, and replication complexes are efficiently established, but host and nsP protein synthesis are shut off more rapidly, replicase function is reduced, amplification of replication complexes is less efficient, and virus production is delayed. Therefore, nsP3MD binding to one or more ADP-ribosylated host cell or viral proteins is critical for initiating infection, while ADPr hydrolase activity facilitates later stages in the virus replication cycle.
The induction of ADP ribosylation of cellular proteins by infection of neural cells and the requirement for ADPr binding for initiation of replication indicate that this posttranslational modification of cellular proteins is important for efficient alphavirus replication. Previous studies that focused on the poly ADP-ribosylating (PARylating) enzyme PARP1 showed activation by Sindbis virus (SINV) infection of neural cells and association with nsP3 through the HVD, but neither PARP1 deficiency nor inhibition of its activity affected SINV replication (41, 76, 77). However, most enzymatically active PARPs are ADPr transferases with MARylating activity rather than polymerases (65, 78). MARylation was induced in NSC34 neuronal cells during CHIKV infection without increased PARP expression or evidence of IFN induction, suggesting that virus replication per se induced PARP activation or reduced the rate of ADPr removal. Although several PARPs can inhibit alphavirus replication and are induced in vivo (61, 79–82), reduced CHIKV replication with the pan-MARylation inhibitor ITK6 indicates that the virus uses one or more ADP-ribosylated cellular proteins to its advantage for replication and benefits from the availability of at least some ADP-ribosylated proteins during initiation of viral infection (although others may work to restrict replication). This fact may explain why the MD is so highly conserved in alphaviruses and perhaps why it is conserved in other MD-containing plus-strand RNA viruses.
The identification of the nsP3-mediated replication defect early in infection was confirmed using a transreplicase system that allows evaluation of replicase function without requiring virus infection. In this system nsPs are produced through pol II-dependent cellular transcription, and reporter protein expression from a replication-competent template is dependent on the ability of the nsPs to form a functional replicase to amplify genomic RNA and synthesize sgRNA. Interestingly, nsP synthesis was suppressed by 24 h in cells transfected with the plasmid encoding WT nsPs but not nonviable mutant nsPs, with viable mutants showing an intermediate phenotype. For SINV, nsP2 in the nucleus induces ubiquitin-mediated proteasomal degradation of Rpb1, the catalytic subunit of RNA pol II (72), a function that is impaired with mutations in either the helicase or C domain of nsP2 (73, 83). Our data (Fig. 7B) suggest that mutations in the MD of nsP3 also impair inhibition of pol II transcription and/or translation of pol II-generated mRNAs, a finding that merits further investigation. For nsP3 mutants D10A, G32E, and G112E that lacked ADPr-binding or hydrolase activity, reporter expression did not exceed background levels even though nsPs were stably expressed, indicating that nsP3MD binding and/or hydrolase activities were crucial for replicase function. Therefore, viruses with nsP3MD mutations D10A, G32E, and G112E were not viable because viral RNA could not be synthesized. Levels of reporter protein expression for the viable mutants G32S and Y114A with similarly diminished hydrolase activity allowed separate assessment of the relative importance of ADPr binding. G32S with low ADPr binding produced much lower reporter expression than Y114A with binding that was enhanced compared with WT. Our previous studies with SINV nsP3MD mutants also showed decreased RNA synthesis, and the synthesis and processing of viral proteins were also affected (31).
ADPr binding is a titratable function because viruses with increased binding initiate replication faster, while viruses with decreased binding show a cell type-specific defect in initiation of replication. Initiation of infection is more dependent on ADPr binding in neural cells than in BHK cells. Cell type specificity has been previously noted for SINV nsP3MD mutants in which impaired replication was greater for differentiated neurons than for undifferentiated neurons and replication was not impaired in BHK cells (31). These differences could reflect variable induction of ADP ribosylation or the interaction of nsP3 with an ADP-ribosylated cellular protein with variable availability that is important for the formation of replication complexes. The determinant(s) of cell type-dependent effects of nsP3MD function deserve further investigation.
Alphavirus nsP3 interacts with many cellular proteins, some of which can be ADP-ribosylated, but to date all interactions have been mapped to the nsP3 HVD, not the MD (32, 41, 84–87). Host cell proteins localized to early replication complexes and important for RNA synthesis include sphingosine kinase 2 (88), amphiphysins (26), and RasGAP SH3-domain–binding proteins (G3BPs) (37). G3BP1 and 2 are RNA-binding proteins that interact with the FGDF motif in the nsP3 HVD through a NTF2-like motif in G3BP (89, 90), that are essential for early steps in replication of Old World alphaviruses (91), and that are consistently associated with nsP3 both early and late in the replication cycle (85, 86, 92). Reduced nsP3 HVD interaction with G3BP impairs CHIKV replication (90). While the importance of G3BP in stress granule assembly is clearly established (93, 94), the role(s) of G3BP and the identification of the relevant posttranslational modifications in early and late phases of alphavirus replication remain unclear. It has been suggested that G3BP has both antiviral and proviral activities by activating PKR (95, 96) and suppressing P1234 translation (97), that it escorts the incoming genomic RNA to the plasma membrane (37), and that it regulates the switch from translation to transcription of genomic RNA (91). G3BP1 is the target for several posttranslational modifications (e.g., phosphorylation, ADP ribosylation, acetylation, methylation, NEDDylation, and ubiquitylation) that regulate its endonuclease activity and its participation in the formation of stress granules and other protein complexes (94, 98–107). The presence or absence and the location of these modifications likely also play a regulatory role in interaction with nsP3 and genomic RNA for initiation of alphavirus replication. Although the documented interaction is with the nsP3 HVD, the MD is exposed in the P23 polyprotein (108) and possibly in the P123 polyprotein and may provide an additional regulatory binding site that could evolve during replication with activation of the nsP3MD hydrolase function. Additional interaction with the MD through an ADP-ribosylated site may be functionally important. Many other associated host proteins may also play a role, but their ADP-ribosylation status is not known (20).
Removal of ADPr is also important. For G32S, a deficiency in MD hydrolase function as well as a binding defect resulted in the impairment of initiation of replication and replicase activity that was substantially greater than observed with the better-binding Y114A. In addition, Y114A was not able to amplify replication complexes as well as the WT virus, potentially because, in the context of virus infection, more rapid synthesis of nsPs and dsRNA led to earlier shutdown of cap-dependent translation of both cellular mRNAs and genomic viral RNA. Comparison of Y114A replication with the replication of WT virus (Fig. 1A) suggests that hydrolase deficiency may also adversely affect later stages of replication when nsP3 is found mainly in cytoplasmic granules formed after the disruption of stress granules.
The MD is highly conserved among the coronavirus, alphavirus, and hepevirus families of plus-strand RNA viruses (53) and is critical for replication in mammalian as well as mosquito cells. Thus, the identification of an important role in alphavirus replication opens approaches to the identification of cellular MD functions. A better understanding of the replication of this group of plus-strand RNA viruses that include several important human pathogens may also provide opportunities for new therapeutic interventions.
Materials and Methods
Cell Culture and Inhibitor Treatment.
The murine neuronal NSC34 cell line, generated by fusion of mouse N18TB2 neuroblastoma cells with mouse motor neurons (a kind gift from Neil Cashman, University of British Columbia, Victoria, BC, Canada) (109, 110), baby hamster kidney 21 (BHK21) and African green monkey epithelial (Vero) cells, were grown in DMEM supplemented with 10% heat-inactivated FBS, l-glutamine (2 mM), penicillin (100 U/mL), and streptomycin (100 μg/mL) (Gibco, Life Technologies), at 37 °C in a 5% CO2 incubator. To assess the effect of inhibition of MARylation on virus replication, cells were treated at the time of infection with the pan mono PARP inhibitor ITK6 (5 μM) (67) in DMEM plus 1% FBS and were assessed for virus production by plaque assay in Vero cells. Toxicity of ITK6 for uninfected NSC34 cells after 24 h was assessed using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) assay (ATCC) and showed 91% viability at 2 μM, 87% viability at 5 μM, and 73% viability at 10 μM.
Viruses, Mutagenesis, and Sequencing.
The WT CHIKV vaccine strain 181/25 was grown in BHK21 cells from transfected RNA that had been transcribed from a full-length cDNA clone of the virus (gift from Naomi Forrester, University of Texas Medical Branch, Galveston, TX) (111). Mutations were introduced into the nsP3 gene and sequenced, viral RNAs were transcribed and transfected into BHK21 cells, and mutant viruses were rescued as previously described (Table 1) (27). Viruses that could be recovered (G32S and Y114A) were passaged once in BHK21 cells, and RNAs from virions were sequenced to confirm the presence of the mutations. Viral stocks were grown in BHK21 cells and assayed by plaque formation in Vero cells.
qRT PCR for PARP Gene Expression.
NSC34 cells were mock infected or were infected with 5 pfu WT CHIKV, as measured in Vero cells, for an hour. Cells were washed with PBS, and fresh medium (DMEM and 1% FBS) was replaced. Infected cells were harvested in RLT buffer (Qiagen), and RNA was isolated using the RNeasy Mini kit (Qiagen). Briefly, cDNA was synthesized with random primers using the High Capacity cDNA reverse transcription kit (Life Technologies), and qRT-PCR for Parp1, 9, 10, 12, 13, and 14 mRNAs was performed using TaqMan gene-expression arrays (Integrated DNA Technologies) and Universal PCR Master Mix (Applied Biosystems). Relative gene expression was determined by the ΔΔCT method using 0-h control samples and rodent Gapdh for normalization. All reactions were run on the Applied Biosystems 7500 Real-time PCR machine under the following conditions: 50 °C for 2 min, 95 °C for 10 min, 95 °C for 15 s, and 60 °C for 1 min for 40 cycles and were analyzed with Sequence Detector Software version 1.4 (Applied Biosystems).
IFNα and β Enzyme Immunoassays.
IFNα and β levels in supernatant fluids from mock-infected and CHIKV-infected (MOI 5) NSC34 cells were measured at intervals from 2 to 36 h after infection using VeriKine ELISA kits (PBL Assay Science) according to the manufacturer’s instructions. Supernatant fluids from three independent experiments were tested. Assay ranges were 12.5–400 pg/mL for IFNα and 15.6–1000 pg/mL for IFNβ.
Infectious Center Assay.
NSC34 cells were infected with WT and nsP3MD G32S and Y114A mutant viruses at MOIs of 0.5 and 5 for 1 h at 4 °C and were shifted to 37 °C for 4 h or were transfected with 10 μg RNA transcribed from a full-length cDNA clone of WT CHIKV or nsP3MD G32S or Y114A mutant viruses using Amaxa-Nucleofector II. Infected and transfected cells were trypsinized and counted, and 10-fold dilutions were plated on Vero cells (infected cells) or BHK cells (transfected cells) and were overlaid with 0.6% Bacto Agar (BD) in Modified Eagle Medium (Gibco) and incubated at 37 °C for 48 h. Cells were fixed with 10% formaldehyde in PBS and stained with 0.02% crystal violet, and plaques were counted. Data are reported as number of infectious centers per 105 NSC34 cells.
dsRNA Flow Cytometry.
NSC34 cells, mock infected or infected at an MOI of 5 with WT and nsP3MD G32S and Y114A mutant viruses, were incubated at 37 °C. After live/dead staining (Invitrogen) on ice for 30 min in the dark, cells were fixed with 2% paraformaldehyde and were permeabilized with 0.2% Triton in FACS buffer (PBS with 0.4% 0.5 M EDTA and 0.5% BSA). Cells were stained for dsRNA with J2 mouse monoclonal antibody (1:1,000; Scicons) for 1 h on ice followed by PE-conjugated goat anti-mouse IgG and were analyzed on a FACSCanto flow cytometer. Histograms were plotted to determine median fluorescence intensity and the percent of live cells positive for dsRNA.
Viral RNA Synthesis.
Total RNA from WT (ITK6-treated or untreated) and nsP3MD G32S and Y114A mutant virus-infected NSC34 cells was isolated using the RNeasy Plus mini kit (Qiagen). RNA was quantified with a nanodrop spectrophotometer, and cDNA was synthesized using the High Capacity cDNA Synthesis Kit (Applied Biosystems) and random primers. qRT-PCR was performed using TaqMan primers and probes specific for the genomic (nsP2) and genomic plus sg (E2) regions of the CHIKV genome: for the CHIKV E2 gene: E2 922F 5′-GAAGAGTGGGTGACGCATAAG-3′; E2 1011R 5′-TGGATAACTGCGGCCAATAC-3′); for the TaqMan probe: CHK E2 949 5′-6-carboxyfluorescein [FAM]-ATCAGGTTAACCGTGCCGACTGAA-Minor groove binder (MGB) nonfluorescent quencher (NFQ)-3′ (Applied Biosystems); for the CHIKV nsP2 gene: nsP2 1247F 5′-GTACGGAAGGTAAACTGGTATGG-3′; nsP2 1359R 5′-TCCACCTCCCACTCCTTAAT-3′); and for the TaqMan probe: CHK nsP2 1304 5′-(FAM)-TGCAGAACCCACCGAAAGGAAACT-(MGB NFQ)-3′ (Applied Biosystems). Copies of viral RNA were quantified using a standard curve constructed of 10-fold dilutions of a pCRII-TOPO plasmid containing the CHIKV E2 or nsP2 region cDNA and normalized to endogenous rodent Gapdh. Data are plotted as mean CHIKV RNA copies per 106 copies of Gapdh.
Transreplicase System and Mutagenesis.
To measure the replicase activity of WT, viable (G32S and Y114A), and nonviable (D10A, G32E, and G112E) nsP3MD mutants without requiring virus infection, a CHIKV transreplicase system was used (69). Two plasmids were transfected: one encoding P1234 controlled by the immediate early promoter of hCMV and the second encoding a pol I-synthesized viral RNA template that includes (i) the CHIKV 5′ UTR, a fusion ORF consisting of the 77 N-terminal residues of nsP1 (including the 51-nt conserved sequence element), and Fluc; (ii) the CHIKV subgenomic promoter, intergenic region and the gene for Gluc; and (iii) the CHIKV 3′ UTR, 60 adenine residues, the hepatitis delta virus antisense strand ribozyme, and the pol I termination signal (Fig. 7A). To control for reporter baseline expression, an hCMV-driven P1234 plasmid with a GDD→GAA mutation in the catalytic domain of nsP4 was included (69).
P1234 and P1234GAA plasmids with point mutations in the nsP3MD were generated using the QuickChange Site-Directed Mutagenesis Kit (Agilent Technologies). Mutant clones prepared were D10A, G32S or G32E, G112E, and Y114A (Table 1). Clones were sequenced to confirm the mutations with TrNSP3MDR primer 5′-CTTCTCCCACTCTTTGTCGCGGCAGTAGAT-3′. One microgram each of CMV-P1234 or CMV P1234GAA and pol I-Fluc-Gluc were cotransfected into NSC34 cells using lipofectamine LTX (Invitrogen). Cells were lysed with Renilla luciferase assay lysis buffer (Promega) after 12, 18, 24, and 48 h, and Fluc and Gluc activities were analyzed in a luminometer (IVIS Spectrum Imager) using the Dual-Luciferase reporter assay (Promega) and Living Image software (PerkinElmer). Protein concentration was measured using the DC assay (Bio-Rad). Luciferase activity per microgram of protein for P1234GAA was subtracted from luciferase activity for P1234, and data are expressed as the fold change over background.
Immunoblot Analysis of Protein Expression and ADP Ribosylation.
Virus-infected NSC34 cells or transreplicase-transfected NSC34 cells were lysed using radioimmune precipitation assay (RIPA) buffer [1% Nonidet P-40, 0.1% SDS, 0.5% sodium orthovanadate, 50 mM Tris (pH 8), 150 mM NaCl, 1 mM EDTA] containing protease and phosphatase inhibitors (Sigma). To assess ADP ribosylation of cellular proteins, RIPA buffer also contained PARP inhibitor 3-aminobenzamide (Sigma) and poly(ADPr) glycohydrolase inhibitor ADP-HPD (Millipore). Cell lysates were centrifuged at 15,700 × g for 10 min, total protein was estimated using the DC assay (Bio-Rad), and 10 μg was loaded onto 10% polyacrylamide gels, electrophoresed, and transferred to nitrocellulose membranes. Membranes were incubated overnight at 4 °C with polyclonal rabbit antibodies to CHIKV nsP1, nsP2, nsP3 (1:10,000), and nsP4 (1:200) (69), monoclonal mouse antibodies to E2 (CHK-187 11A4. F1.F4), p-eIF2α, and eIF2α (Cell Signaling) or β actin (Millipore) or pan ADPr-binding reagent (MABE1016; Millipore) diluted in 5% BSA. Secondary antibodies were HRP-conjugated anti-rabbit or anti-mouse IgG (Cell Signaling) diluted in 2% milk (1:10,000) and incubated for 1 h at RT. Membranes were developed using the Amersham ECL Plus Western Blot Developing kit (GE Healthcare). ImageJ software (NIH) was used for densitometric analysis of blots from three independent experiments.
Puromycin Assay for Cellular Protein Synthesis.
NSC34 cells were infected with WT and nsP3MD mutant viruses at an MOI of 5 for 1 h and were washed once with PBS; then fresh DMEM with 1% FBS was added. At 12, 24, 36, and 48 h, medium containing 5 μg/mL puromycin dihydrochloride (Sigma) was substituted, incubated for 10 min, and cells were collected in RIPA buffer for immunoblot analysis. After electrophoresis and transfer, membranes were probed with mouse anti-puromycin monoclonal antibody clone 12D10 (Millipore) (68) and HRP-conjugated anti-mouse IgG (1:10,000) and were developed using the Amersham ECL Plus Western Blot Developing Kit. ImageJ software was used for densitometric analysis of blots from three or four independent experiments.
Statistical Analysis.
Differences between groups during infection were determined using two-way ANOVA and Bonferroni post tests. Differences between groups at a single time point were determined using an unpaired, two-tailed Student t test with a 95% confidence interval. All statistical analysis was done using Prism 7 (GraphPad); results are expressed as means ± SD.
Acknowledgments
We thank Naomi Forrester (University of Texas Medical Branch) for the cDNA clone for CHIKV strain 181/25; Michael Diamond (Washington University in St. Louis) for CHIKV E2 antibody; and Easwaran Sreekumar (Rajiv Gandhi Centre for Biotechnology) for initiating these studies. This work was supported by a Johns Hopkins Catalyst Award (to A.K.L.L.); Pilot Research Grants from the Johns Hopkins University School of Medicine Sherrilyn and Ken Fisher Center for Environmental Infectious Disease (to D.E.G. and A.K.L.L.); the Pew Charitable Trust (M.S.C.); NIH Grants R01GM104135S1 and T32CA009110 (to R.L.M.); and Estonian Research Council Institutional Research Funding Grant IUT20-27 (to A.M.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Gubler DJ. The global emergence/resurgence of arboviral diseases as public health problems. Arch Med Res. 2002;33:330–342. doi: 10.1016/s0188-4409(02)00378-8. [DOI] [PubMed] [Google Scholar]
- 2.Weaver SC, Lecuit M. Chikungunya virus and the global spread of a mosquito-borne disease. N Engl J Med. 2015;372:1231–1239. doi: 10.1056/NEJMra1406035. [DOI] [PubMed] [Google Scholar]
- 3.Ronca SE, Dineley KT, Paessler S. Neurological sequelae resulting from encephalitic alphavirus infection. Front Microbiol. 2016;7:959. doi: 10.3389/fmicb.2016.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandak NH, et al. Neurological complications of chikungunya virus infection. Neurol India. 2009;57:177–180. doi: 10.4103/0028-3886.51289. [DOI] [PubMed] [Google Scholar]
- 5.Strauss JH, Strauss EG. The alphaviruses: Gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voss JE, et al. Glycoprotein organization of chikungunya virus particles revealed by X-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 7.Leung JY, Ng MM, Chu JJ. Replication of alphaviruses: A review on the entry process of alphaviruses into cells. Adv Virol. 2011;2011:249640. doi: 10.1155/2011/249640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kallio K, Hellström K, Jokitalo E, Ahola T. RNA replication and membrane modification require the same functions of alphavirus nonstructural proteins. J Virol. 2015;90:1687–1692. doi: 10.1128/JVI.02484-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lemm JA, Rümenapf T, Strauss EG, Strauss JH, Rice CM. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: A model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Utt A, et al. Mutations conferring a noncytotoxic phenotype on chikungunya virus replicons compromise enzymatic properties of nonstructural protein 2. J Virol. 2015;89:3145–3162. doi: 10.1128/JVI.03213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. Alphavirus RNA synthesis and non-structural protein functions. J Gen Virol. 2015;96:2483–2500. doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy WR, Strauss JH. Processing the nonstructural polyproteins of Sindbis virus: Nonstructural proteinase is in the C-terminal half of nsP2 and functions both in cis and in trans. J Virol. 1989;63:4653–4664. doi: 10.1128/jvi.63.11.4653-4664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirako Y, Strauss JH. Regulation of Sindbis virus RNA replication: Uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrasco L, Sanz MA, González-Almela E. The regulation of translation in alphavirus-infected cells. Viruses. 2018;10:E70. doi: 10.3390/v10020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawicki DL, Silverman RH, Williams BR, Sawicki SG. Alphavirus minus-strand synthesis and persistence in mouse embryo fibroblasts derived from mice lacking RNase L and protein kinase R. J Virol. 2003;77:1801–1811. doi: 10.1128/JVI.77.3.1801-1811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawicki DL, Sawicki SG. Short-lived minus-strand polymerase for Semliki Forest virus. J Virol. 1980;34:108–118. doi: 10.1128/jvi.34.1.108-118.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Froshauer S, Kartenbeck J, Helenius A. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spuul P, et al. Role of the amphipathic peptide of Semliki Forest virus replicase protein nsP1 in membrane association and virus replication. J Virol. 2007;81:872–883. doi: 10.1128/JVI.01785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zusinaite E, et al. Mutations at the palmitoylation site of non-structural protein nsP1 of Semliki Forest virus attenuate virus replication and cause accumulation of compensatory mutations. J Gen Virol. 2007;88:1977–1985. doi: 10.1099/vir.0.82865-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pietilä MK, Hellström K, Ahola T. Alphavirus polymerase and RNA replication. Virus Res. 2017;234:44–57. doi: 10.1016/j.virusres.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Coffey LL, Beeharry Y, Bordería AV, Blanc H, Vignuzzi M. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc Natl Acad Sci USA. 2011;108:16038–16043. doi: 10.1073/pnas.1111650108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li GP, La Starza MW, Hardy WR, Strauss JH, Rice CM. Phosphorylation of Sindbis virus nsP3 in vivo and in vitro. Virology. 1990;179:416–427. doi: 10.1016/0042-6822(90)90310-n. [DOI] [PubMed] [Google Scholar]
- 23.Peränen J, Takkinen K, Kalkkinen N, Kääriäinen L. Semliki Forest virus-specific non-structural protein nsP3 is a phosphoprotein. J Gen Virol. 1988;69:2165–2178. doi: 10.1099/0022-1317-69-9-2165. [DOI] [PubMed] [Google Scholar]
- 24.Fros JJ, et al. Mosquito Rasputin interacts with chikungunya virus nsP3 and determines the infection rate in Aedes albopictus. Parasit Vectors. 2015;8:464. doi: 10.1186/s13071-015-1070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxton-Shaw KD, et al. O’nyong nyong virus molecular determinants of unique vector specificity reside in non-structural protein 3. PLoS Negl Trop Dis. 2013;7:e1931. doi: 10.1371/journal.pntd.0001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuvonen M, et al. SH3 domain-mediated recruitment of host cell amphiphysins by alphavirus nsP3 promotes viral RNA replication. PLoS Pathog. 2011;7:e1002383. doi: 10.1371/journal.ppat.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McPherson RL, et al. ADP-ribosylhydrolase activity of chikungunya virus macrodomain is critical for virus replication and virulence. Proc Natl Acad Sci USA. 2017;114:1666–1671. doi: 10.1073/pnas.1621485114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saul S, et al. Differences in processing determinants of nonstructural polyprotein and in the sequence of nonstructural protein 3 affect neurovirulence of Semliki Forest virus. J Virol. 2015;89:11030–11045. doi: 10.1128/JVI.01186-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tuittila M, Hinkkanen AE. Amino acid mutations in the replicase protein nsP3 of Semliki Forest virus cumulatively affect neurovirulence. J Gen Virol. 2003;84:1525–1533. doi: 10.1099/vir.0.18936-0. [DOI] [PubMed] [Google Scholar]
- 30.Tuittila MT, Santagati MG, Röyttä M, Määttä JA, Hinkkanen AE. Replicase complex genes of Semliki Forest virus confer lethal neurovirulence. J Virol. 2000;74:4579–4589. doi: 10.1128/jvi.74.10.4579-4589.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park E, Griffin DE. The nsP3 macro domain is important for Sindbis virus replication in neurons and neurovirulence in mice. Virology. 2009;388:305–314. doi: 10.1016/j.virol.2009.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Götte B, Liu L, McInerney GM. The enigmatic alphavirus non-structural protein 3 (nsP3) revealing its secrets at last. Viruses. 2018;10:E105. doi: 10.3390/v10030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foy NJ, Akhrymuk M, Shustov AV, Frolova EI, Frolov I. Hypervariable domain of nonstructural protein nsP3 of Venezuelan equine encephalitis virus determines cell-specific mode of virus replication. J Virol. 2013;87:7569–7584. doi: 10.1128/JVI.00720-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dé I, Fata-Hartley C, Sawicki SG, Sawicki DL. Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus. J Virol. 2003;77:13106–13116. doi: 10.1128/JVI.77.24.13106-13116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.LaStarza MW, Lemm JA, Rice CM. Genetic analysis of the nsP3 region of Sindbis virus: Evidence for roles in minus-strand and subgenomic RNA synthesis. J Virol. 1994;68:5781–5791. doi: 10.1128/jvi.68.9.5781-5791.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vihinen H, Ahola T, Tuittila M, Merits A, Kääriäinen L. Elimination of phosphorylation sites of Semliki Forest virus replicase protein nsP3. J Biol Chem. 2001;276:5745–5752. doi: 10.1074/jbc.M006077200. [DOI] [PubMed] [Google Scholar]
- 37.Kim DY, et al. New World and Old World alphaviruses have evolved to exploit different components of stress granules, FXR and G3BP proteins, for assembly of viral replication complexes. PLoS Pathog. 2016;12:e1005810. doi: 10.1371/journal.ppat.1005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fros JJ, et al. Chikungunya virus nsP3 blocks stress granule assembly by recruitment of G3BP into cytoplasmic foci. J Virol. 2012;86:10873–10879. doi: 10.1128/JVI.01506-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thaa B, et al. Differential phosphatidylinositol-3-kinase-Akt-mTOR activation by Semliki Forest and chikungunya viruses is dependent on nsP3 and connected to replication complex internalization. J Virol. 2015;89:11420–11437. doi: 10.1128/JVI.01579-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fros JJ, et al. Chikungunya virus nonstructural protein 2 inhibits type I/II interferon-stimulated JAK-STAT signaling. J Virol. 2010;84:10877–10887. doi: 10.1128/JVI.00949-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park E, Griffin DE. Interaction of Sindbis virus non-structural protein 3 with poly(ADP-ribose) polymerase 1 in neuronal cells. J Gen Virol. 2009;90:2073–2080. doi: 10.1099/vir.0.012682-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lulla A, Lulla V, Merits A. Macromolecular assembly-driven processing of the 2/3 cleavage site in the alphavirus replicase polyprotein. J Virol. 2012;86:553–565. doi: 10.1128/JVI.05195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorbalenya AE, Koonin EV, Lai MM. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koonin EV, et al. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: Delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci USA. 1992;89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rack JG, Perina D, Ahel I. Macrodomains: Structure, function, evolution, and catalytic activities. Annu Rev Biochem. 2016;85:431–454. doi: 10.1146/annurev-biochem-060815-014935. [DOI] [PubMed] [Google Scholar]
- 46.Ribet D, Cossart P. Pathogen-mediated posttranslational modifications: A re-emerging field. Cell. 2010;143:694–702. doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koch-Nolte F, Kernstock S, Mueller-Dieckmann C, Weiss MS, Haag F. Mammalian ADP-ribosyltransferases and ADP-ribosylhydrolases. Front Biosci. 2008;13:6716–6729. doi: 10.2741/3184. [DOI] [PubMed] [Google Scholar]
- 48.Ueda K, Hayaishi O. ADP-ribosylation. Annu Rev Biochem. 1985;54:73–100. doi: 10.1146/annurev.bi.54.070185.000445. [DOI] [PubMed] [Google Scholar]
- 49.Eckei L, et al. The conserved macrodomains of the non-structural proteins of chikungunya virus and other pathogenic positive strand RNA viruses function as mono-ADP-ribosylhydrolases. Sci Rep. 2017;7:41746. doi: 10.1038/srep41746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li C, et al. Viral macro domains reverse protein ADP-ribosylation. J Virol. 2016;90:8478–8486. doi: 10.1128/JVI.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosenthal F, et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat Struct Mol Biol. 2013;20:502–507. doi: 10.1038/nsmb.2521. [DOI] [PubMed] [Google Scholar]
- 52.Jankevicius G, et al. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat Struct Mol Biol. 2013;20:508–514. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malet H, et al. The crystal structures of chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J Virol. 2009;83:6534–6545. doi: 10.1128/JVI.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fehr AR, et al. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J Virol. 2015;89:1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fehr AR, et al. The conserved coronavirus macrodomain promotes virulence and suppresses the innate immune response during severe acute respiratory syndrome coronavirus infection. MBio. 2016;7:e01721-16. doi: 10.1128/mBio.01721-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eriksson KK, Cervantes-Barragán L, Ludewig B, Thiel V. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1′′-phosphatase, a viral function conserved in the alpha-like supergroup. J Virol. 2008;82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Putics A, Filipowicz W, Hall J, Gorbalenya AE, Ziebuhr J. ADP-ribose-1"-monophosphatase: A conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J Virol. 2005;79:12721–12731. doi: 10.1128/JVI.79.20.12721-12731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuri T, et al. The ADP-ribose-1′′-monophosphatase domains of severe acute respiratory syndrome coronavirus and human coronavirus 229E mediate resistance to antiviral interferon responses. J Gen Virol. 2011;92:1899–1905. doi: 10.1099/vir.0.031856-0. [DOI] [PubMed] [Google Scholar]
- 59.Shaw AE, et al. Fundamental properties of the mammalian innate immune system revealed by multispecies comparison of type I interferon responses. PLoS Biol. 2017;15:e2004086. doi: 10.1371/journal.pbio.2004086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 61.Atasheva S, Akhrymuk M, Frolova EI, Frolov I. New PARP gene with an anti-alphavirus function. J Virol. 2012;86:8147–8160. doi: 10.1128/JVI.00733-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Atasheva S, Frolova EI, Frolov I. Interferon-stimulated poly(ADP-ribose) polymerases are potent inhibitors of cellular translation and virus replication. J Virol. 2014;88:2116–2130. doi: 10.1128/JVI.03443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuny CV, Sullivan CS. Virus-host interactions and the ARTD/PARP family of enzymes. PLoS Pathog. 2016;12:e1005453. doi: 10.1371/journal.ppat.1005453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leung AKL, McPherson RL, Griffin DE. Macrodomain ADP-ribosylhydrolase and the pathogenesis of infectious diseases. PLoS Pathog. 2018;14:e1006864. doi: 10.1371/journal.ppat.1006864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Daugherty MD, Young JM, Kerns JA, Malik HS. Rapid evolution of PARP genes suggests a broad role for ADP-ribosylation in host-virus conflicts. PLoS Genet. 2014;10:e1004403. doi: 10.1371/journal.pgen.1004403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gibson BA, Conrad LB, Huang D, Kraus WL. Generation and characterization of recombinant antibody-like ADP-ribose binding proteins. Biochemistry. 2017;56:6305–6316. doi: 10.1021/acs.biochem.7b00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirby IT, et al. A potent and selective PARP11 inhibitor suggests coupling between cellular localization and catalytic activity. Cell Chem Biol. 2018 doi: 10.1016/j.chembiol.2018.09.011. in press. [DOI] [PubMed] [Google Scholar]
- 68.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 69.Utt A, et al. Versatile trans-replication systems for chikungunya virus allow functional analysis and tagging of every replicase protein. PLoS One. 2016;11:e0151616. doi: 10.1371/journal.pone.0151616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spuul P, et al. Assembly of alphavirus replication complexes from RNA and protein components in a novel trans-replication system in mammalian cells. J Virol. 2011;85:4739–4751. doi: 10.1128/JVI.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bartholomeeusen K, et al. A chikungunya virus trans-replicase system reveals the importance of delayed nonstructural polyprotein processing for efficient replication complex formation in mosquito cells. J Virol. 2018;92:e00152-18. doi: 10.1128/JVI.00152-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Akhrymuk I, Kulemzin SV, Frolova EI. Evasion of the innate immune response: The Old World alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II. J Virol. 2012;86:7180–7191. doi: 10.1128/JVI.00541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gorchakov R, Frolova E, Frolov I. Inhibition of transcription and translation in Sindbis virus-infected cells. J Virol. 2005;79:9397–9409. doi: 10.1128/JVI.79.15.9397-9409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang YF, Sawicki SG, Sawicki DL. Alphavirus nsP3 functions to form replication complexes transcribing negative-strand RNA. J Virol. 1994;68:6466–6475. doi: 10.1128/jvi.68.10.6466-6475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fehr AR, Jankevicius G, Ahel I, Perlman S. Viral macrodomains: Unique mediators of viral replication and pathogenesis. Trends Microbiol. 2018;26:598–610. doi: 10.1016/j.tim.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ubol S, et al. Temporal changes in chromatin, intracellular calcium, and poly(ADP-ribose) polymerase during Sindbis virus-induced apoptosis of neuroblastoma cells. J Virol. 1996;70:2215–2220. doi: 10.1128/jvi.70.4.2215-2220.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nargi-Aizenman JL, Simbulan-Rosenthal CM, Kelly TA, Smulson ME, Griffin DE. Rapid activation of poly(ADP-ribose) polymerase contributes to Sindbis virus and staurosporine-induced apoptotic cell death. Virology. 2002;293:164–171. doi: 10.1006/viro.2001.1253. [DOI] [PubMed] [Google Scholar]
- 78.Kleine H, et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 79.Bick MJ, et al. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol. 2003;77:11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kozaki T, et al. Role of zinc-finger anti-viral protein in host defense against Sindbis virus. Int Immunol. 2015;27:357–364. doi: 10.1093/intimm/dxv010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y, Burke CW, Ryman KD, Klimstra WB. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J Virol. 2007;81:11246–11255. doi: 10.1128/JVI.01282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nair SR, Abraham R, Sundaram S, Sreekumar E. Interferon regulated gene (IRG) expression-signature in a mouse model of chikungunya virus neurovirulence. J Neurovirol. 2017;23:886–902. doi: 10.1007/s13365-017-0583-3. [DOI] [PubMed] [Google Scholar]
- 83.Garmashova N, Gorchakov R, Frolova E, Frolov I. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J Virol. 2006;80:5686–5696. doi: 10.1128/JVI.02739-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lark T, Keck F, Narayanan A. Interactions of alphavirus nsP3 protein with host proteins. Front Microbiol. 2018;8:2652. doi: 10.3389/fmicb.2017.02652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Frolova E, et al. Formation of nsP3-specific protein complexes during Sindbis virus replication. J Virol. 2006;80:4122–4134. doi: 10.1128/JVI.80.8.4122-4134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cristea IM, et al. Tracking and elucidating alphavirus-host protein interactions. J Biol Chem. 2006;281:30269–30278. doi: 10.1074/jbc.M603980200. [DOI] [PubMed] [Google Scholar]
- 87.Meshram CD, et al. Multiple host factors interact with hypervariable domain of chikungunya virus nsP3 and determine viral replication in cell-specific mode. J Virol. 2018;92:JVI.00838-18. doi: 10.1128/JVI.00838-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Reid SP, et al. Sphingosine kinase 2 is a chikungunya virus host factor co-localized with the viral replication complex. Emerg Microbes Infect. 2015;4:e61. doi: 10.1038/emi.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kristensen O. Crystal structure of the G3BP2 NTF2-like domain in complex with a canonical FGDF motif peptide. Biochem Biophys Res Commun. 2015;467:53–57. doi: 10.1016/j.bbrc.2015.09.123. [DOI] [PubMed] [Google Scholar]
- 90.Schulte T, et al. Combined structural, biochemical and cellular evidence demonstrates that both FGDF motifs in alphavirus nsP3 are required for efficient replication. Open Biol. 2016;6:160078. doi: 10.1098/rsob.160078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scholte FE, et al. Stress granule components G3BP1 and G3BP2 play a proviral role early in chikungunya virus replication. J Virol. 2015;89:4457–4469. doi: 10.1128/JVI.03612-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gorchakov R, Garmashova N, Frolova E, Frolov I. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J Virol. 2008;82:10088–10101. doi: 10.1128/JVI.01011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kedersha N, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tourrière H, et al. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160:823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 95.Reineke LC, Lloyd RE. The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses. J Virol. 2015;89:2575–2589. doi: 10.1128/JVI.02791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Reineke LC, Dougherty JD, Pierre P, Lloyd RE. Large G3BP-induced granules trigger eIF2α phosphorylation. Mol Biol Cell. 2012;23:3499–3510. doi: 10.1091/mbc.E12-05-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cristea IM, et al. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: Role for G3BP1 and G3BP2 in virus replication. J Virol. 2010;84:6720–6732. doi: 10.1128/JVI.01983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jayabalan AK, et al. NEDDylation promotes stress granule assembly. Nat Commun. 2016;7:12125. doi: 10.1038/ncomms12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leung AK, et al. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes Dev. 2007;21:3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reineke LC, et al. Casein kinase 2 is linked to stress granule dynamics through phosphorylation of the stress granule nucleating protein G3BP1. Mol Cell Biol. 2017;37:e00596-16. doi: 10.1128/MCB.00596-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ohn T, Anderson P. The role of posttranslational modifications in the assembly of stress granules. Wiley Interdiscip Rev RNA. 2010;1:486–493. doi: 10.1002/wrna.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tsai WC, Reineke LC, Jain A, Jung SY, Lloyd RE. Histone arginine demethylase JMJD6 is linked to stress granule assembly through demethylation of the stress granule-nucleating protein G3BP1. J Biol Chem. 2017;292:18886–18896. doi: 10.1074/jbc.M117.800706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tsai WC, et al. Arginine demethylation of G3BP1 promotes stress granule assembly. J Biol Chem. 2016;291:22671–22685. doi: 10.1074/jbc.M116.739573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tourrière H, et al. RasGAP-associated endoribonuclease G3Bp: Selective RNA degradation and phosphorylation-dependent localization. Mol Cell Biol. 2001;21:7747–7760. doi: 10.1128/MCB.21.22.7747-7760.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Li WM, Barnes T, Lee CH. Endoribonucleases–enzymes gaining spotlight in mRNA metabolism. FEBS J. 2010;277:627–641. doi: 10.1111/j.1742-4658.2009.07488.x. [DOI] [PubMed] [Google Scholar]
- 108.Shin G, et al. Structural and functional insights into alphavirus polyprotein processing and pathogenesis. Proc Natl Acad Sci USA. 2012;109:16534–16539. doi: 10.1073/pnas.1210418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cashman NR, et al. Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn. 1992;194:209–221. doi: 10.1002/aja.1001940306. [DOI] [PubMed] [Google Scholar]
- 110.Durham HD, Dahrouge S, Cashman NR. Evaluation of the spinal cord neuron X neuroblastoma hybrid cell line NSC-34 as a model for neurotoxicity testing. Neurotoxicology. 1993;14:387–395. [PubMed] [Google Scholar]
- 111.Gorchakov R, et al. Attenuation of chikungunya virus vaccine strain 181/clone 25 is determined by two amino acid substitutions in the E2 envelope glycoprotein. J Virol. 2012;86:6084–6096. doi: 10.1128/JVI.06449-11. [DOI] [PMC free article] [PubMed] [Google Scholar]