Significance
Nonalcoholic fatty liver disease (NAFLD) is the fastest rising cause of hepatocellular carcinoma (HCC) in Western countries; however, the molecular mechanisms driving NAFLD-HCC remain elusive. Using Sleeping Beauty transposon mutagenesis in two mouse models of NAFLD-HCC, we identified hundreds of NAFLD-HCC candidate cancer genes that were enriched in pathways often associated with NAFLD and HCC. We also showed that Sav1, which functions in the Hippo signaling pathway and was the most frequently mutated gene identified by SB in both screens, prevents progression of steatohepatitis and subsequent HCC development in coordination with PI3K signaling via suppression of Yap, a downstream effector of the Hippo pathway. Our forward genetic screens have thus identified pathways and genes driving the development of NAFLD-HCC.
Keywords: Sleeping Beauty, liver cancer, Hippo, Sav1, NAFLD
Abstract
Nonalcoholic fatty liver disease (NAFLD) is the fastest rising cause of hepatocellular carcinoma (HCC) in Western countries; however, the molecular mechanisms that cause NAFLD-HCC remain elusive. To identify molecular drivers of NAFLD-HCC, we performed Sleeping Beauty (SB) transposon mutagenesis screens in liver-specific Pten knockout and in high-fat diet-fed mice, which are murine models of NAFLD-HCC. SB mutagenesis accelerated liver tumor formation in both models and identified 588 and 376 candidate cancer genes (CCGs), respectively; 257 CCGs were common to both screens and were enriched in signaling pathways known to be important for human HCC. Comparison of these CCGs with those identified in a previous SB screen of hepatitis B virus-induced HCC identified a core set of 141 CCGs that were mutated in all screens. Forty-one CCGs appeared specific for NAFLD-HCC, including Sav1, a component of the Hippo signaling pathway and the most frequently mutated gene identified in both NAFLD-HCC screens. Liver-specific deletion of Sav1 was found to promote hepatic lipid accumulation, apoptosis, and fibrogenesis, leading to the acceleration of hepatocarcinogenesis in liver-specific Pten mutant mice. Sav1/Pten double-mutant livers also showed a striking up-regulation of markers of liver progenitor cells (LPCs), along with synergistic activation of Yap, which is a major downstream effector of Hippo signaling. Lastly, Yap activation, in combination with Pten inactivation, was found to accelerate cell growth and sphere formation of LPCs in vitro and induce their malignant transformation in allografts. Our forward genetic screens in mice have thus identified pathways and genes driving the development of NAFLD-HCC.
Hepatocellular carcinoma (HCC) is a deadly cancer, with a mortality rate ranking second among all cancers worldwide (1). In addition, the incidence of HCC in the United States and worldwide has continuously increased for the past 20 to 30 y (1, 2). Until recently, the major risk factors for HCC were hepatitis B virus (HBV) and hepatitis C virus (HCV) infection. However, recent advances in antiviral therapy have made it possible to eradicate HCV at extremely high rates and efficiently suppress HBV replication, contributing to the reduction of HCC incidence in patients with viral hepatitis (3). In contrast, the incidence of HCC in patients with nonalcoholic fatty liver disease (NAFLD), characterized by excessive accumulation of lipid in the liver, is rising. Surveillance, Epidemiology, and End Results program registry data show that 14.1% of HCC that developed in patients in the United States between 2004 and 2009 was associated with NAFLD (2). Therefore, to discover effective therapies to prevent or treat HCC, it is important to comprehensively understand the molecular mechanism of hepatocarcinogenesis in NAFLD patients.
The International Cancer Genome Consortium (4) and The Cancer Genome Atlas (TCGA) (5) projects have successfully profiled the cancer genomes of over 500 HCC patients. They revealed extensive intra- and intertumor heterogeneity and numerous passenger mutations in more than 10,000 coding genes (5). On average, each cancer genome contained 40 to 80 mutations in protein-coding genes, with a different combination of mutations present in each cancer genome, suggesting that complex combinations of mutations contribute to the development and progression of HCC. Such extreme heterogeneity has made it difficult to generate a comprehensive catalog of HCC drivers due to the lack of statistical power. In addition, the etiologies of most sequenced HCCs are chronic HBV or HCV infection. Therefore, even with substantial international collaborative efforts, the mutational landscape linking NAFLD to HCC remains elusive.
Insertional mutagenesis using a Sleeping Beauty (SB) or PiggyBac transposon is a versatile tool for identifying cancer genes in a variety of cancer types (6–8). Genome-wide random mutagenesis using high-copy transposons, combined with high-throughput sequencing of transposon insertion sites, enabled us to discover HCC driver genes in a comprehensive manner, including those mutated infrequently in human HCC or that were missed because of the lack of statistical power. In the current study, we aimed to discover drivers of NAFLD-HCC by performing whole-genome, unbiased SB mutagenesis screens in liver-specific Pten-knockout (PtenKO) mice (9) and in high-fat diet (HFD)-fed mice, both of which are murine models of NAFLD-HCC (10). Both screens successfully identified hundreds of candidate cancer genes (CCGs), and the comparison with genes identified in other SB screens of HCC further refined the list of CCGs important for NAFLD-HCC. In addition, our validation studies using liver-specific Sav1 and Pten double-knockout mice underscore the important role of Hippo signaling in NAFLD-associated HCC.
Results
SB Mutagenesis Significantly Accelerates NAFLD-HCC in PtenKO Mice.
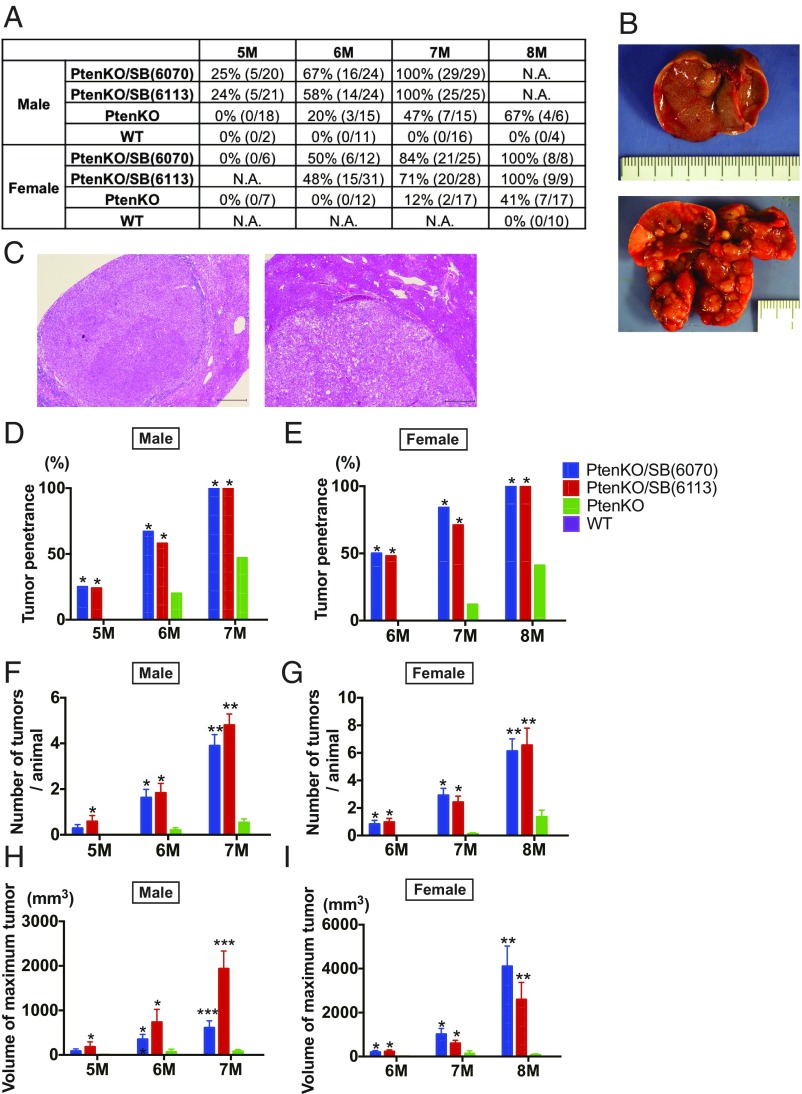
To comprehensively profile the genes and evolutionary forces driving NAFLD-HCC, we first performed an SB mutagenesis screen in PtenKO mice, a genetically perturbed NAFLD-HCC model (9). PtenKO mice are known to spontaneously develop steatohepatitis and liver fibrosis, leading to HCC development with a 1-y latency (9), recapitulating human NAFLD-HCC well. To achieve genome-wide coverage, we used two high-copy SB transposon transgenic lines (T2Onc2;6113 and T2Onc2;6070), in which the transposons were mobilized from transposon concatamers located on chromosomes 1 and 4, respectively. T2Onc2 contains a transcriptional stop cassette to inactivate tumor suppressor genes (11), as well as a murine stem cell virus promoter and downstream splice donor to activate oncogenes. We first confirmed that mice with liver-specific deletion of Pten developed steatohepatitis, regardless of whether they underwent active SB transposition (SI Appendix, Fig. S1 A–C). PtenKO mice with SB transposition (PtenKO/SB) started to develop liver tumors at 5 mo of age, and all of the male and female mice developed tumors by 7 and 8 mo of age, respectively (Fig. 1 A–C). Tumor penetrance was significantly higher in both male and female PtenKO/SB mice compared with PtenKO mice (Fig. 1 D and E). The number of tumors and maximum tumor volume per animal also increased over time and was significantly higher in PtenKO/SB mice compared with PtenKO mice (Fig. 1 F–I). These data clearly indicate that SB mutagenesis accelerated liver tumor development in a genetically perturbed NAFLD model. Tumor penetrance and latency were similar for both transposon lines, and the data were thus combined for subsequent analysis.
Fig. 1.
SB mutagenesis significantly accelerates NAFLD-HCC in PtenKO mice. (A) Tumor penetrance of WT, PtenKO, PtenKO/SB(6113) and PtenKO/SB(6070) mice at 5, 6, 7, 8 mo of age. The numbers of mice that have tumors, compared with the number of mice analyzed, are listed in brackets. N.A., not assessed. (B) Gross pictures of the livers from a PtenKO (Top) and a PtenKO/SB(6113) (Bottom) mouse at 7 mo of age. (C) H&E staining of liver tumors from an PtenKO/SB(6113) mouse. (Scale bar: 500 μm.) (D and E) Tumor penetrance of male (D) and female (E) WT, PtenKO, PtenKO/SB(6113), and PtenKO/SB(6070) mice at 5, 6, and 7 and at 6, 7, and 8 mo of age, respectively (*P < 0.05 vs. PtenKO). (F and H) Number of tumors (F) and maximum tumor volume (H) per animal, observed in male WT, PtenKO, PtenKO/SB(6113) and PtenKO/SB(6070) mice at 5, 6, and 7 mo of age (*P < 0.05 vs. PtenKO, **P < 0.05 vs. PtenKO and WT, ***P < 0.05 vs. all; data are provided as mean ± SEM). (G and I) Number of tumors (G) and maximum tumor volume (I) per animal observed in female WT, PtenKO, PtenKO/SB(6113) and PtenKO/SB(6070) mice at 6, 7, and 8 mo of age (*P < 0.05 vs. PtenKO, **P < 0.05 vs. PtenKO and WT; data are provided as mean ± SEM).
To comprehensively identify genes mutated by SB that drive liver tumor development in PtenKO mice, we PCR-amplified and sequenced the transposon insertion sites from 225 liver tumors [118 tumors from 58 PtenKO/SB(6113) mice and 107 tumors from 52 PtenKO/SB(6070) mice]. To obtain the maximum number of sequencing reads and avoid PCR bias (12), we used splink HiSeq methods (6, 13) and identified 2,623,466 nonredundant transposon insertion sites (an average of 11,660 insertion sites per tumor) (Dataset S1). Using the gene-centric common insertion site (gCIS)-calling method (12) that looks for a higher density of transposon insertions within the coding regions of all RefSeq genes than is predicted by chance, we identified 588 statistically significant CCGs (P < 0.05, calculated by χ2 test followed by Bonferroni correction) (SI Appendix, Table S1).
SB Mutagenesis Significantly Accelerates NAFLD-HCC in HFD-Fed Mice.
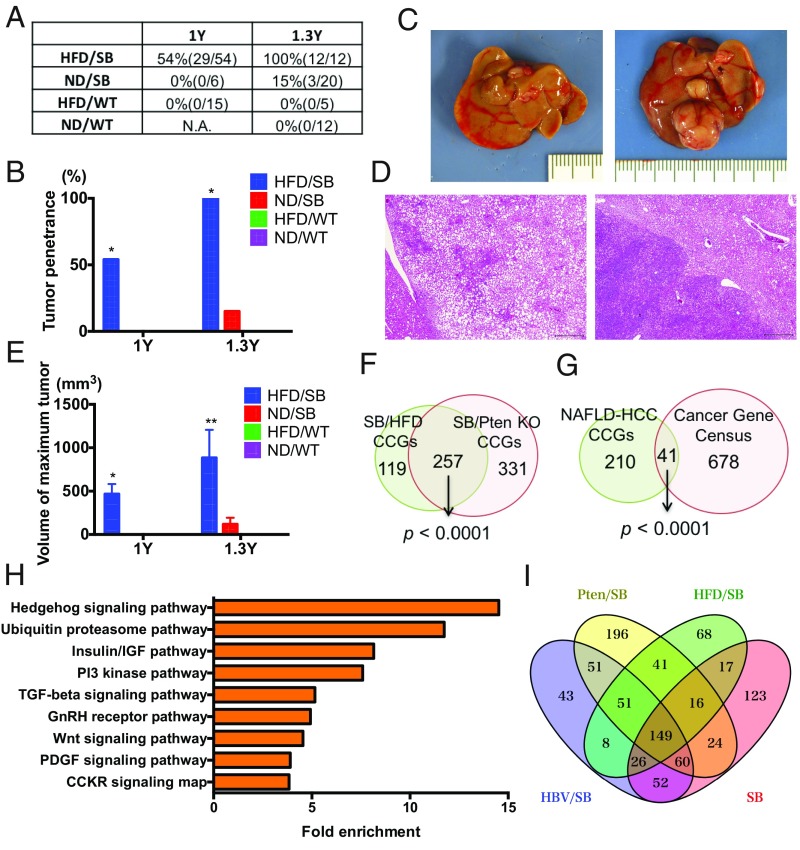
Next, to avoid the screening bias possibly caused by genetic perturbation of the PI3K pathway in PtenKO mice, we performed another SB screen using a diet-based NAFLD model. By combining the data from these two screens, we hoped to identify HCC drivers important for human NAFLD-HCC in an unbiased manner. HFD feeding induced lipid deposition in the liver, regardless of SB transposon status (T2Onc2;6113) (SI Appendix, Fig. S2A). HFD feeding also induced severe glucose intolerance (SI Appendix, Fig. S2B), recapitulating the human NAFLD disease (14). Male mice with or without SB transposition were then aged and monitored for tumor formation. Tumor penetrance in HFD-fed mice with SB transposition (HFD/SB) was 54% at 1 y of age but reached 100% by 1.3 y of age (Fig. 2 A–D). In contrast to PtenKO mice, there was no tumor development in the HFD-fed mice in the absence of active SB transposition, while normal diet-fed mice rarely developed tumors, even in the presence of active SB transposition (Fig. 2 A and B). Tumor penetrance was thus significantly higher in HFD/SB mice compared with others (Fig. 2 A and B). The maximum tumor volume per animal also increased over time, and was significantly higher in HFD/SB mice than in other mice (Fig. 2E). These data demonstrate that SB mutagenesis also significantly accelerates liver tumor formation in a diet-based NAFLD model. To identify CCGs driving liver tumors in HFD-fed mice, we sequenced the transposon insertion sites from 66 tumors and identified 721,684 nonredundant transposon insertion sites (average of 10,934 insertions per tumor) (Dataset S2). gCIS analysis of these insertion sites subsequently identified 376 statistically significant CCGs (P < 0.05 calculated by χ2 test followed by Bonferroni correction) (SI Appendix, Table S2).
Fig. 2.
SB mutagenesis significantly accelerates NAFLD-HCC in HFD-fed mice. (A and B) Tumor penetrance of normal diet (ND)-fed WT, HFD-fed WT, ND-fed SB, and HFD-fed SB mice at 1 and 1.3 y of age. The numbers of mice that have tumors compared with the numbers of mice analyzed, are listed in brackets. *P < 0.05 vs. all. (C) Gross pictures of the livers from a HFD-fed WT mouse (Left) and a HFD-fed SB mouse (Right) at 1 y of age. (D) H&E staining of liver tumors from a HFD-fed SB mouse. (Scale bar: 500 μm.) (E) Maximum tumor volume per animal observed in male ND-fed WT, HFD-fed WT, ND-fed SB, and HFD-fed SB mice at 1 and 1.3 y of age (*P < 0.05 vs. HFD/WT, **P < 0.05 vs. all; data are provided as mean ± SEM). (F) Venn diagram comparing the list of CCGs obtained from SB mutagenesis screens of PtenKO and HFD-fed mice. (G) Venn diagram comparing the list of human orthologs of common CCGs obtained from NAFLD-HCC SB mutagenesis screens and known human cancer genes found in the Cancer Gene Census database (15). (H) Pathway analysis performed with PANTHER. Enriched pathways among CCGs are shown along with the fold enrichment value. (I) Venn diagram comparing lists of CCGs obtained from four SB mutagenesis screens [PtenKO and HFD-fed mice in the current study, and HBV surface antigen transgenic (HBV/SB) and ND-fed mice (SB) in previous studies] (21, 22).
Comparison of CCGs Identified in Different SB Screens.
To identify the drivers of NAFLD-HCC, we compared the lists of CCGs identified in PtenKO and HFD-fed mice. We found that 257 CCGs were common in both models (Fig. 2F), and this overlap is highly significant (P < 0.0001 by Fisher’s exact test), proving the similarity of CCGs identified in both NAFLD-HCC screens. In addition, we found that these common CCGs were significantly enriched in the 719 known human cancer genes found in the Cancer Gene Census database (15) (Fig. 2G). We then identified the pathways perturbed by transposon insertions in SB-derived NAFLD-HCC, including hedgehog (Hh) signaling, insulin/insulin-like growth factor (IGF), PI3K, TGF-β, and Wnt pathways (Fig. 2H). Activation of Hh signaling is known to be correlated with the severity of human NAFLD; it also accelerates murine NAFLD and hepatocarcinogenesis (16). In addition to the well-known role of insulin resistance in NAFLD (14), IGF-1 levels are reported to be correlated with the severity of human NAFLD (17). Moreover, a recent report identified aberrant epigenetic activation of Wnt/β-catenin in the development of NAFLD-associated HCC (18). Together, these results suggest that our screens identified cancer genes involved in human NAFLD-HCC in a high-throughput manner.
Human cancer genome sequence efforts have revealed that the most frequent HCC drivers, such as TERT, TP53, and CTNNB1, are mutated at similar rates, regardless of background etiologies, indicating the existence of common HCC drivers (19). On the other hand, human HBV-HCC has unique mutational features caused by HBV genomic integration (19, 20), suggestive of etiology-dependent unique HCC drivers. We have also previously performed SB mutagenesis screens in the presence or absence of an HBV surface antigen transgene, reporting the candidate drivers that are important for HBV-HCC (21, 22). Here, taking advantage of analyzing different sets of SB mutagenesis screens (HBV-HCC, non–HBV-HCC, and the current two NAFLD-HCC cohorts), we aimed to identify both common and unique HCC drivers of NAFLD-HCC. The transposon insertion sites from the earlier screens were analyzed using SB splinkerette PCR, coupled with 454 sequencing, which is different from the splink HiSeq methods used here. However, we previously showed that high-read depth sites are reproducibly detected across the two different methods (6). We thus reanalyzed the 454 sequencing data and performed gCIS analysis using insertions with read counts ≥3, and identified 440 and 467 CCGs, respectively, among the HBV-HCC and non–HBV-HCC cohorts. A total of 149 CCGs were identified in all four cohorts and were considered to be the common drivers of HCC (Fig. 2I), while 41 CCGs were detected only in the NAFLD cohorts (Table 1), suggesting they are especially important for NAFLD-HCC.
Table 1.
Forty-one candidate cancer genes specific for NAFLD-HCC
| PtenKO cohort | HFD cohort | ||||||
| Gene name | Locus | gCIS P value | Insertion frequency, % | Mean sequence read counts | gCIS P value | Insertion frequency, % | Mean sequence read counts |
| Sav1 | chr12 | 0 | 25.8 | 1948.2 | 0 | 21.2 | 597.0 |
| Qrich1 | chr9 | 3.50E-65 | 8.4 | 599.0 | 1.66E-18 | 6.1 | 81.0 |
| Son | chr16 | 7.41E-40 | 6.2 | 1048.8 | 5.60E-34 | 7.6 | 245.0 |
| Slc25a17 | chr15 | 1.03E-39 | 6.7 | 390.1 | 3.87E-21 | 7.6 | 369.5 |
| Acox2 | chr14 | 2.32E-37 | 5.3 | 1021.1 | 1.69E-27 | 6.1 | 158.2 |
| Mob1b | chr5 | 2.37E-37 | 6.7 | 1192.5 | 1.38E-17 | 6.1 | 523.8 |
| Ccdc50 | chr16 | 3.24E-33 | 8.0 | 335.5 | 7.19E-11 | 6.1 | 869.4 |
| Fbxo11 | chr17 | 3.66E-31 | 8.9 | 348.7 | 3.30E-08 | 6.1 | 153.4 |
| Rtn4 | chr11 | 1.67E-30 | 7.6 | 389.6 | 2.83E-11 | 6.1 | 528.4 |
| Myo10 | chr15 | 5.19E-28 | 11.6 | 166.1 | 3.20E-14 | 10.6 | 534.2 |
| Btaf1 | chr19 | 3.22E-27 | 9.3 | 376.4 | 1.64E-06 | 6.1 | 181.0 |
| Kcmf1 | chr6 | 1.08E-26 | 6.7 | 641.1 | 4.99E-54 | 12.1 | 223.7 |
| Nup153 | chr13 | 5.01E-25 | 6.7 | 307.9 | 2.84E-19 | 7.6 | 68.7 |
| Eif3h | chr15 | 1.75E-23 | 8.0 | 165.2 | 7.35E-08 | 6.1 | 204.4 |
| Ptpn2 | chr18 | 1.37E-20 | 6.2 | 169.4 | 4.48E-27 | 9.1 | 121.3 |
| Kpna3 | chr14 | 1.93E-20 | 7.6 | 119.9 | 8.63E-08 | 6.1 | 64.6 |
| Pnisr | chr4 | 4.61E-20 | 5.1 | 107.7 | 2.98E-36 | 7.6 | 63.7 |
| Nfatc3 | chr8 | 3.43E-18 | 6.7 | 159.8 | 1.79E-14 | 7.6 | 321.4 |
| Psmd14 | chr2 | 4.04E-18 | 8.4 | 362.8 | 1.66E-05 | 6.1 | 153.3 |
| Nup98 | chr7 | 7.21E-18 | 8.0 | 234.4 | 1.47E-14 | 9.1 | 199.4 |
| Otud7b | chr3 | 2.16E-17 | 5.8 | 999.1 | 5.80E-27 | 9.1 | 142.0 |
| Spopl | chr2 | 2.18E-17 | 6.2 | 400.1 | 5.30E-10 | 6.1 | 85.2 |
| Abhd2 | chr7 | 1.36E-16 | 5.8 | 263.6 | 3.98E-11 | 6.1 | 506.2 |
| Tbl1xr1 | chr3 | 7.91E-15 | 8.9 | 103.1 | 5.25E-14 | 10.6 | 198.2 |
| Alcam | chr16 | 1.26E-14 | 11.6 | 188.4 | 4.58E-08 | 10.6 | 639.9 |
| St5 | chr7 | 1.81E-14 | 6.2 | 277.4 | 1.74E-08 | 6.1 | 176.6 |
| Bptf | chr11 | 4.86E-14 | 6.2 | 178.3 | 2.91E-08 | 6.1 | 103.6 |
| Smad3 | chr9 | 5.90E-14 | 5.8 | 309.5 | 2.61E-22 | 9.1 | 332.2 |
| Hipk2 | chr6 | 1.13E-13 | 9.3 | 85.2 | 7.01E-12 | 10.6 | 875.6 |
| Asap1 | chr15 | 1.33E-13 | 11.6 | 214.7 | 2.92E-10 | 12.1 | 370.2 |
| Abcb4 | chr5 | 2.13E-13 | 5.3 | 84.8 | 3.46E-17 | 7.6 | 266.9 |
| Map2k4 | chr11 | 8.95E-13 | 7.1 | 235.5 | 3.35E-19 | 10.6 | 363.7 |
| Mbnl2 | chr14 | 1.52E-12 | 8.0 | 210.5 | 8.47E-11 | 9.1 | 72.1 |
| Diap1 | chr18 | 1.73E-11 | 6.2 | 132.7 | 1.92E-16 | 9.1 | 148.3 |
| Rbm26 | chr14 | 2.70E-10 | 5.3 | 349.5 | 1.36E-08 | 6.1 | 195.3 |
| Met | chr6 | 2.79E-09 | 6.2 | 159.1 | 2.30E-09 | 7.6 | 331.3 |
| Cers6 | chr2 | 6.10E-08 | 9.3 | 87.1 | 1.94E-10 | 12.1 | 247.3 |
| Mob3b | chr4 | 1.12E-06 | 7.6 | 288.9 | 1.88E-08 | 9.1 | 136.0 |
| Ints6 | chr14 | 1.60E-06 | 5.3 | 140.3 | 2.09E-41 | 15.2 | 123.0 |
| D930015E06Rik | chr3 | 1.09E-05 | 5.3 | 165.8 | 2.83E-05 | 6.1 | 215.4 |
| Osbpl1a | chr18 | 1.41E-05 | 6.7 | 73.6 | 1.85E-05 | 7.6 | 221.6 |
Identification of NAFLD-HCC Trunk Driver Genes.
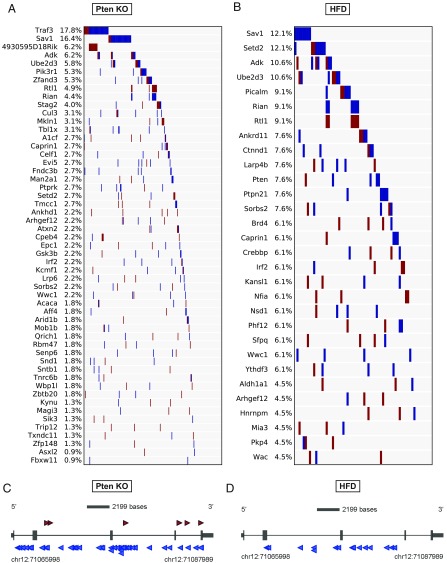
Previously (21), we showed that most CCGs identified by SB are branch drivers, which likely function during late stages of tumor progression. We also showed that trunk drivers, which function early in tumor development, could be identified by focusing on transposon insertion sites with high sequencing reads (21). To identify trunk drivers in NAFLD-HCC, we focused on insertion sites with read counts of ≥2% of the total reads on each tumor basis. This analysis identified 53 and 30 trunk drivers in the PtenKO/SB and HFD/SB cohorts, respectively (Fig. 3 A and B). Ten genes (Adk, Arhgef12, Caprin1, Irf2, Rtl1/Rian, Sav1, Setd2, Sorbs2, Ube2d3, and Wwc1) were identified as trunk drivers in both PtenKO/SB and HFD/SB tumors. Among them, Sav1 was the only gene that was identified as a CCG unique to NAFLD-HCC. Most of the SB insertions in Sav1 are located throughout the coding region in antisense orientations (Fig. 3 C and D), suggesting that Sav1 is functioning as a tumor suppressor gene in NAFLD-HCC.
Fig. 3.
Identification of trunk driver genes. (A and B) Fifty-three and 30 trunk driver genes identified in the PtenKO SB screen (A) and the HFD-fed SB screen (B), respectively. Transposon insertion sites with read counts of ≥2% of the total reads on each tumor basis were used for gCIS analysis. Each column indicates a single tumor and the frequency of each trunk driver gene is presented as the percentage of total tumor number. Red and blue bars indicate transposons inserted in the sense or antisense orientation, respectively. (C and D) Insertion maps showing the location of transposon insertions in Sav1 in PtenKO/SB mice (C) and HFD/SB mice (D). Insertions represented by the highest sequencing reads (≥100) are shown. Each arrow indicates a single transposon insertion event. Transcription orientation is from 5′ to 3′. Red and blue arrows indicate transposons inserted in the sense or antisense orientation, respectively.
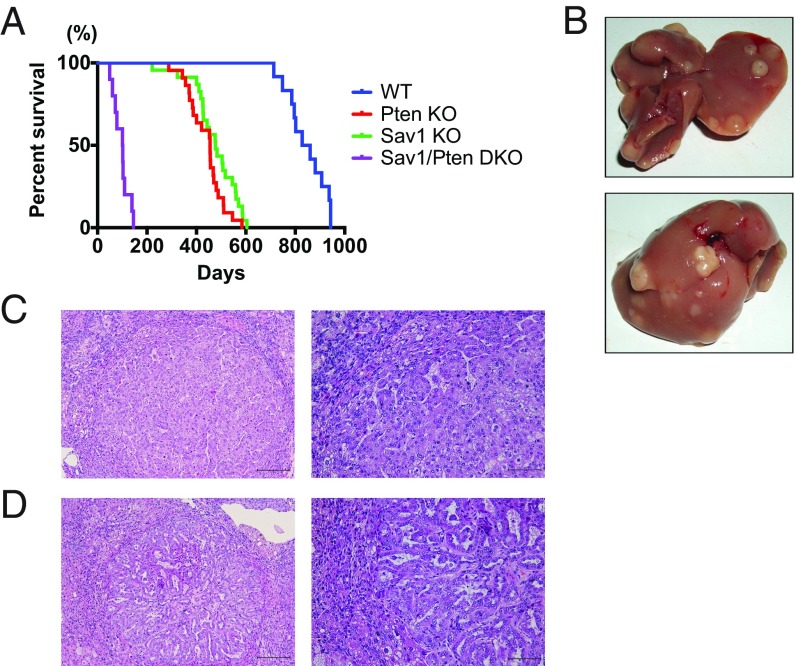
Sav1 Loss Greatly Accelerates HCC in Pten-Deficient NAFLD Liver.
To identify a link between Sav1 and NAFLD tumorigenesis, we investigated the effect of Sav1 deletion on the NAFLD liver caused by Pten loss. While mice with liver-specific deletion of either Sav1 or Pten survived longer than a year, liver-specific Sav1/Pten double-knockout mice showed reduced survival, starting around 3 mo of age (Fig. 4A). Their life span was significantly shorter than Sav1 or Pten single-knockout mice or littermate control mice (Fig. 4A). Importantly, all double-knockout mice developed multiple liver tumors when moribund (Fig. 4B), clearly indicating that Sav1 functions as a strong tumor suppressor in Pten-deficient NAFLD liver. Histological analysis showed that most liver tumors consisted of well- to moderately differentiated HCCs (Fig. 4C), but some contained ductal-like structures, consistent with intrahepatic cholangiocarcinoma (Fig. 4D).
Fig. 4.
Sav1 loss greatly accelerates HCC in Pten-deficient NAFLD liver. (A) Kaplan-Meier survival curves for WT, liver-specific Sav1 or Pten knockout, or liver-specific Sav1/Pten double-knockout (DKO) mice. The median survival for DKO mice was 101.5 d vs. 455 d for PtenKO mice, 477 d for Sav1KO mice, and 844 d for WT mice (log-rank test P < 0.0001). (B) Gross picture of the livers from a DKO mouse at 3 mo of age. (C and D) H&E staining of liver tumors from DKO mice. (Scale bar: Left, 200 μm; Right, 100 μm.)
Sav1 Prevents Hepatic Lipid Accumulation Caused by Pten Loss.
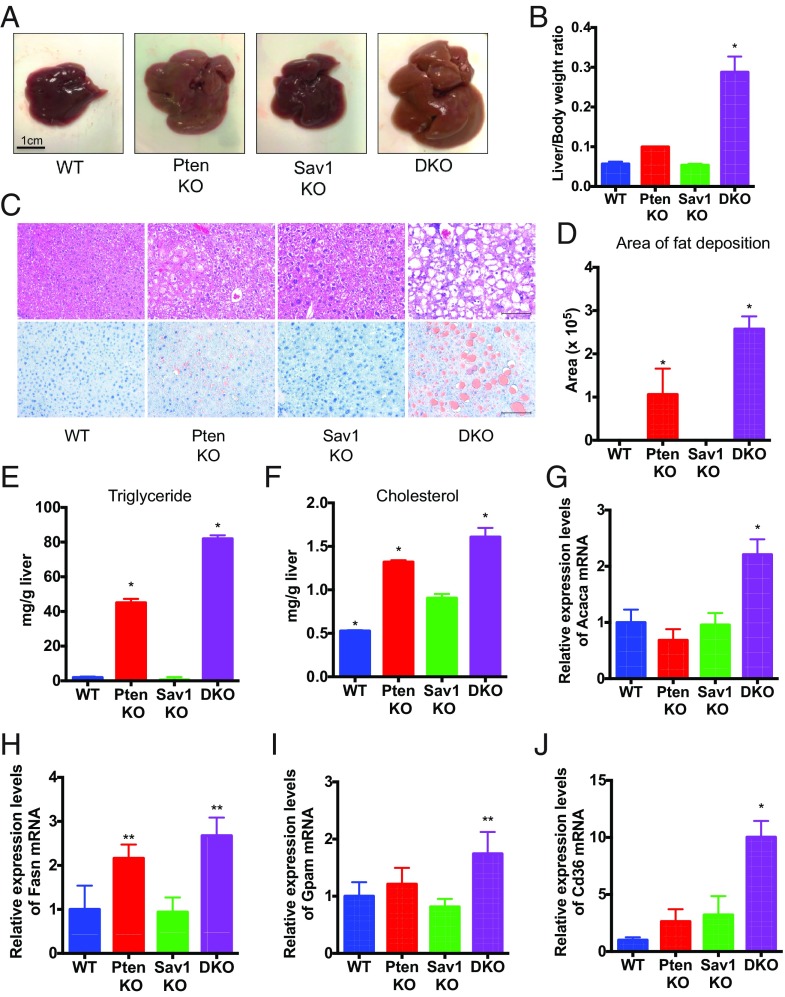
Next, we sought to determine how deletion of Sav1 accelerates NAFLD-HCC in PtenKO mutant liver. At 2 mo of age, Sav1/Pten double-mutant mice, which were confirmed to have reduced expression of Sav1 and Pten in their liver (SI Appendix, Fig. S3), showed marked hepatomegaly and significantly higher liver/body weight ratios than single-mutant mice (Fig. 5 A and B). Hepatic lipid accumulation observed in Pten-deficient liver was significantly augmented by the deletion of Sav1, as assessed by H&E and Oil Red O staining (Fig. 5 C and D). Triglyceride and cholesterol levels were also significantly increased in the liver of double-knockout mice compared with other mice (Fig. 5 E and F). These data show that Sav1 suppresses liver fat accumulation induced by Pten loss.
Fig. 5.
Sav1 prevents hepatic lipid accumulation caused by Pten loss. (A) Gross pictures of the livers from WT, PtenKO or Sav1KO, or Sav1/Pten double-knockout (DKO), mice at 2 mo of age. (B) Ratio of liver weight to body weight in WT, PtenKO, Sav1KO, and DKO mice (n = 3 to 4 for each; *P < 0.05 vs. all). (C) H&E (Top) and Oil Red O (Bottom) staining of liver tissue from WT, PtenKO, Sav1KO, and DKO mice. (Scale bar: 100 μm.) (D) Area of fat deposition in the liver was quantified by ImageJ in WT, PtenKO, Sav1KO, and DKO mice (n = 3 to 4 for each; *P < 0.05 vs. all). (E and F) Triglyceride (E) and cholesterol (F) levels in the livers of WT, PtenKO, Sav1KO, and DKO mice (n = 3 for each; *P < 0.05 vs. all). (G–J) Acaca (G), Fasn (H), Gpam (I), and Cd36 (J) mRNA abundance in liver tissue from WT, PtenKO, Sav1KO, and DKO mice (n = 3 from each; *P < 0.05 vs. all, **P < 0.05 vs. WT and Sav1KO).
Hepatic lipid accumulation, one of the hallmarks of NAFLD, arises from the dysregulation of lipid metabolism, including de novo lipogenesis, fatty acid uptake, mitochondrial fatty acid oxidation, and very low density lipoprotein cholesterol export (23). We thus examined the expression levels of genes involved in lipid metabolism in the double-knockout mouse livers, to elucidate the underlying mechanisms of lipid accumulation. We found elevated expression of Acaca, Fasn, and Gpam (Fig. 5 G–I), which are all core components of the de novo lipogenesis pathway, in addition to the Cd36 receptor for fatty acid (Fig. 5J). The expression of both Cpt1, important for mitochondrial uptake of fatty acid for β-oxidation, and Pparα, a transcriptional regulator of fatty acid oxidation, were unchanged (SI Appendix, Fig. S4). Together, our data suggest that Sav1 prevents hepatic lipid accumulation in Pten-deficient liver through suppression of de novo lipogenesis and fatty acid uptake into hepatocytes.
Sav1 Deletion Promotes Liver Injury with Macrophage Activation and Fibrogenesis, Leading to NAFLD Progression in Pten-Deficient Liver.
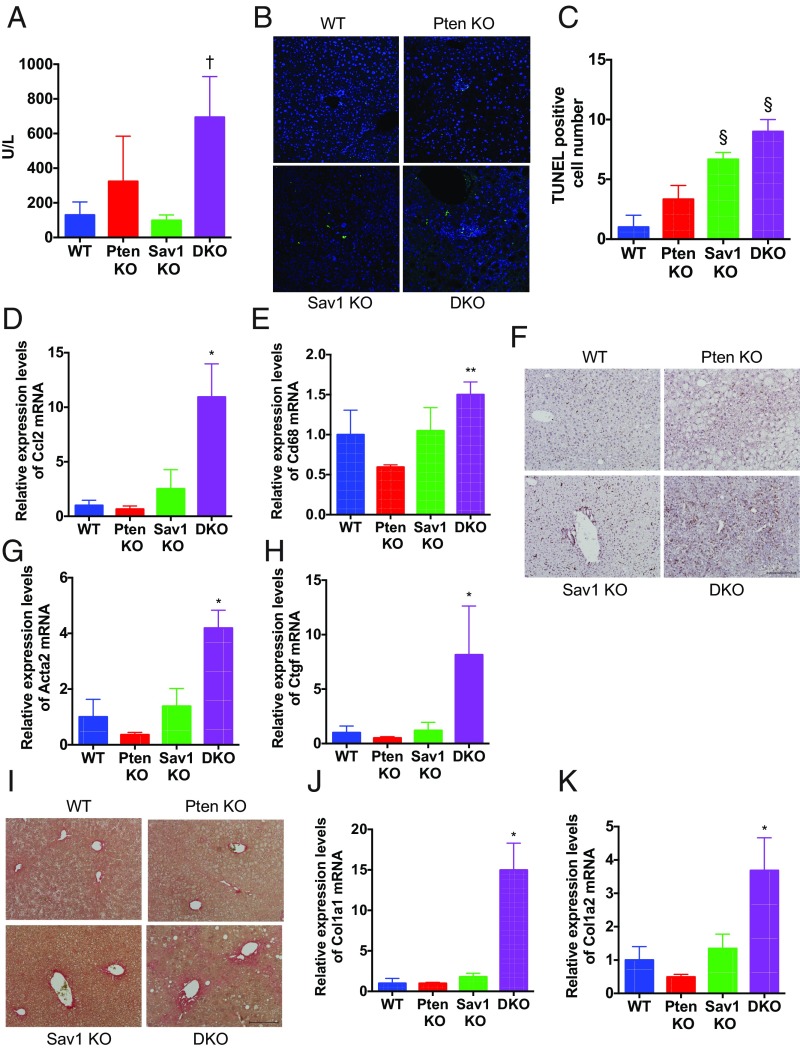
Excess lipid accumulation is known to cause lipotoxicity, leading to hepatocyte damage and apoptosis (24). Consistent with this, Sav1/Pten double-knockout mice exhibited elevated levels of serum alanine aminotransferase, a marker of hepatocyte damage (Fig. 6A), and a significant increase in the number of TUNEL-positive cells (Fig. 6 B and C) at 2 mo of age. Double-knockout mice also showed an increase in the expression of inflammatory chemokines, including Mcp1/Ccl2, in addition to CD68, a protein highly expressed in monocyte lineage cells (Fig. 6 D and E). F4/80 antibody staining, which recognizes a 160-kDa glycoprotein expressed by murine macrophages, also showed an increase in the number of hepatic macrophages in these mice (Fig. 6F). In addition, we found high Nos2 and low Arg1 levels, which are M1 and M2 subtype markers, respectively (25), in the double-knockout liver (SI Appendix, Fig. S5). These findings suggest that M1 macrophage activation by hepatocyte lipoapoptosis may also contribute to liver injury in these mice.
Fig. 6.
Sav1 deletion promotes liver injury and fibrogenesis, leading to NASH progression in Pten-deficient liver. (A) Serum aminotransferase levels in WT, PtenKO, Sav1KO, and Sav1/Pten double-knockout (DKO) mice at 2 mo of age (n = 3 to 4 for each; †P < 0.05 vs. WT and Sav1KO). (B and C) TUNEL staining of liver tissue (B) and the number of TUNEL-positive cells (C) in WT, PtenKO, Sav1KO, and DKO mice (n = 3 to 4 for each; §P < 0.05 vs. WT and PtenKO). (D and E) Ccl2 (D) and Cd68 (E) mRNA abundance in liver tissue of WT, PtenKO, Sav1KO, and DKO mice (n = 3 for each; *P < 0.05 vs. all, **P < 0.05 vs. PtenKO). (F) F4/80 staining of liver tissue in WT, PtenKO, Sav1KO, and DKO mice. (Scale bar: 200 μm.) (G and H) Acta2 (G) and Ctgf (H) mRNA abundance in liver tissue of WT, PtenKO, Sav1KO, and DKO mice (n = 3 for each; *P < 0.05 vs. all). (I) Picro-Sirius Red staining of liver tissue in WT, PtenKO, Sav1KO, and DKO mice. (Scale bar: 200 μm.) (J and K) Col1a1 (J) and Col1a2 (K) mRNA abundance in liver tissue of WT, PtenKO, Sav1KO, and DKO mice (n = 3 for each; *P < 0.05 vs. all).
Hepatic fibrosis is one of the most important phenotypes observed in NAFLD patients, whose fibrosis levels are closely related to the risk of developing HCC (26). We thus assessed the levels of liver fibrosis in double-knockout mice. Expression levels of smooth muscle actin (Acta2), a marker of hepatic stellate cell activation, were significantly increased in double-knockout liver (Fig. 6G). We and others have also demonstrated a profibrogenic role for connective tissue growth factor (Ctgf) (27). Ctgf expression was similarly significantly increased in double-knockout liver (Fig. 6H). In addition, we observed marked collagen deposition by Picro-Sirius Red staining in double-knockout liver (Fig. 6I), along with a significant elevation of type I collagen synthesis (Fig. 6 J and K), indicative of the exacerbation of hepatic fibrosis. Together, our findings suggest that hepatocyte Sav1 loss promotes NAFLD progression by inducing hepatocyte injury and fibrosis in Pten-deficient liver.
Yap Activation Cooperates with Pten Loss to Induce the Malignant Transformation of Liver Progenitor Cells.
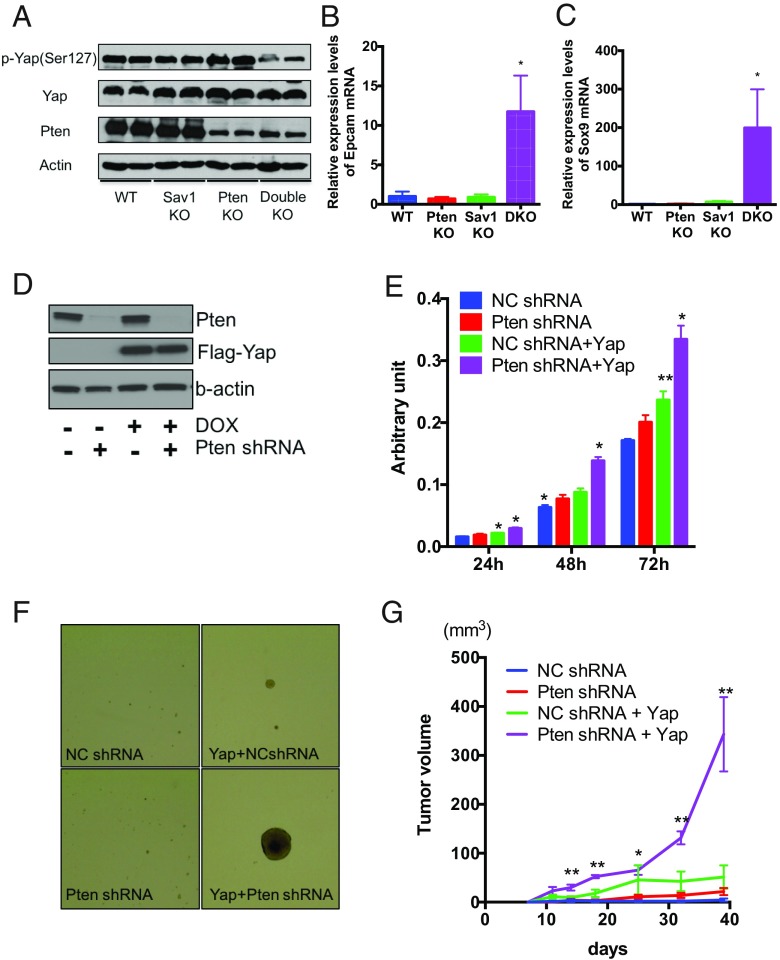
Sav1 is a known suppressor of Yap, which is a major downstream effector of Hippo signaling. Consistent with this, in Sav1/Pten double-knockout liver, we found the down-regulation of phospho-Yap levels, the inactive form of Yap, as well as the nuclear accumulation of Yap (Fig. 7A and SI Appendix, Fig. S6A). In addition, expression levels of multiple Yap target genes, including Ctgf (28, 29), increased in these mice (Fig. 6H and SI Appendix, Fig. S6 B–D), indicating strong Yap activation. Hippo signaling is also known to regulate the differentiation of progenitor cells during organ development (30), raising the possibility that activation of Yap may also affect the differentiation of liver progenitor cells (LPCs). In support of this hypothesis, we found that expression of Epcam and Sox9, two markers of LPCs (31, 32), was significantly higher in double-mutant liver (Fig. 7 B and C).
Fig. 7.
Yap activation and Pten loss cooperatively induce malignant transformation of LPCs. (A) Western blot analysis of protein expression levels of phospho-Yap, Yap, Pten, and β-actin in liver tissue of WT, PtenKO, Sav1KO, and Sav1/Pten double-knockout (Double KO) mice. (B and C) Epcam (B) and Sox9 (C) mRNA abundance in liver tissue of WT, PtenKO, Sav1KO, and Sav1/Pten double-knockout (DKO) mice (n = 3 for each; *P < 0.05 vs. all). (D–G) LPCs were transduced with lentivirus containing Pten shRNA or/and doxycycline (DOX)-inducible Flag-Yap-cDNA. Transduced LPCs were incubated with 100 ng/mL DOX for 24 h, and protein expression levels of Yap, Pten, and β-actin were determined by Western blot analysis (D). Cell proliferation was measured by WST-1 assays in transduced LPCs (E) (n = 4 for each; *P < 0.05 vs. all, **P < 0.05 vs. WT). (F) Tumorsphere assays of transduced LPCs. Representative images of tumorspheres are shown. (Magnification: 4×.) (G) LPCs were s.c. injected into NSG mice. Tumor volumes were measured at various different times after injection of 2.0 × 106 cells (n = 6 for each; *P < 0.05 vs. NC shRNA, **P < 0.05 vs. all). NC, negative control.
To determine whether LPCs contribute to hepatocarcinogenesis in double-knockout mice, we isolated LPCs from wild-type fetal liver and assessed their tumorigenic potential upon lentiviral introduction of Pten shRNA and Yap cDNA (Fig. 7D). Knockdown of Pten, combined with Yap overexpression, significantly increased LPC proliferation (Fig. 7E) and resulted in the formation of large tumorspheres (Fig. 7F). Importantly, LPCs with Pten shRNA and Yap cDNA formed tumors that continuously grew in vivo (Fig. 7G), showing that Yap activation along with Pten inhibition is sufficient to induce the malignant transformation of LPCs.
SAV1 and PTEN Down-Regulation Is Observed in Human Nonviral-Caused HCCs and Correlated with Poor Prognosis.
Lastly, to examine the clinical significance of Sav1 and Pten in human NAFLD-HCC, we looked at the RNA-sequencing data present in the publically available TCGA database. Interestingly, mRNA expression levels of SAV1 and PTEN are significantly down-regulated in non–viral-caused HCCs [NAFLD, alcoholic liver disease, or no specific etiology, which presumably contains end-stage burned-out nonalcoholic steatohepatitis (NASH) that loses lipid deposition] compared with the surrounding nontumor liver tissues (SI Appendix, Fig. S7 A and B). Expression levels of multiple Yap target genes are also inversely correlated with Sav1 mRNA levels (SI Appendix, Fig. S7C), suggesting that Sav1 inactivation may lead to Yap activation in non–viral-caused HCCs. Moreover, among those nonviral HCCs, patients with lower levels of SAV1 and PTEN mRNA manifested significantly poorer survival rates than those with higher levels of both mRNAs (SI Appendix, Fig. S7D). Together, these data support an important role for Sav1 and Pten as a tumor suppressor in NAFLD-related HCC.
Discussion
In this study, both SB transposon screens in diet-induced and genetically perturbed NAFLD mouse models identified Sav1 as the most frequently mutated NAFLD-HCC trunk driver gene. Sav1 is an essential component of Hippo signaling and is known to be involved in the regulation of organ size, including the liver (33). Sav1 is a scaffolding protein required for maximal Hippo kinase (Mst1/2) activation. Activated Mst1/2, in turn, activates Lats1/2 kinases, resulting in phosphorylation of the effector proteins Yap and Taz (34) and their cytoplasmic retention and subsequent degradation (34). We have previously reported that liver-specific Sav1 deletion induces hepatomegaly and the development of liver tumors with a 1-y latency (35), showing the importance of the Hippo pathway dysregulation as a driver of HCC. Our current study further reveals that Sav1 prevents NAFLD progression and subsequent liver tumor development in Pten-deficient liver, identifying the important link between the Hippo pathway and NAFLD-HCC. There are a few other reports of a connection similar to ours. Wang et al. (36) reported that hepatocyte Taz promotes NAFLD progression via secretion of Indian Hh, acting on stellate cells, to promote fibrosis. Aylon et al. (37) showed that Lats2 down-regulation activates sterol regulatory element binding proteins, resulting in hepatic cholesterol accumulation. Ye et al. (38) showed that junctional protein associated with coronary artery disease inhibits Lats2 activity, resulting in Yap-associated up-regulation of Gli2 and hepatoma proliferation. They also found an increase in Yap1 protein levels and a decrease in the phospho-Yap/Yap ratio in human NAFLD-HCC. Together, our studies and those of others have reinforced the important role of Hippo pathway dysregulation as a driver of NAFLD-HCC.
Very recently, Jeong et al. (39) reported bilateral crosstalk between the Hippo and PI3K pathways in NAFLD-HCC. They showed that deletion of Sav1 and Pten synergistically activated Yap/Taz, downstream effectors of Hippo signaling, and IRS2/Akt, downstream molecules of PI3K signaling, leading to the acceleration of hepatic steatosis and liver cancer development. IRS2/Akt signaling is known to play a major role in hepatic lipogenesis, including up-regulation of Fasn, a key enzyme of de novo fatty acid synthesis, through the activation of Srebp1c (40). Consistent with their findings, in the current study, we found strong activation of de novo fatty acid synthesis and hepatic lipid accumulation in Sav1/Pten double-mutant mice, leading to NAFLD progression. In addition, we found marked up-regulation of hepatic Epcam and Sox9 expression, both of which are markers of LPCs (31, 32), suggesting the dysregulation of liver differentiation in these mice. Interestingly, we showed that Yap activation, in conjunction with Pten inhibition, synergistically increased cell proliferation and sphere formation of LPCs and, most importantly, induced their malignant transformation. These results suggest the synergism of the Hippo and PI3K pathways promoting hepatocarcinogenesis from progenitor cells. Human liver cancers with the feature of stem/progenitor cells are known to be associated with a poor prognosis (41, 42). In this study, we also found that non–viral-caused HCC patients with lower levels of both PTEN and SAV1 manifested poor survival rate, possibly suggesting the clinical significance of this oncogenic synergism between the Hippo and PI3K signaling in human NAFLD-HCC.
While Sav1 was the most frequent trunk driver in the HFD cohort, we identified Traf3 as the most frequent trunk driver in the Pten cohort. The genes Traf3 and 4930595D18Rik, the third most frequent trunk driver in this cohort, are located adjacent to each other on opposite strands, and have some sequence overlap (SI Appendix, Fig. S8A). While most insertions in 4930595D18Rik are located in the 5′ region in the sense orientation (SI Appendix, Fig. S8B), insertions in Traf3 are located throughout the coding region in the antisense orientation (SI Appendix, Fig. S8C). It is therefore difficult to determine whether either or both genes are important for HCC, based on the insertion patterns alone. The function of 4930595D18Rik is not well studied and may not have a human ortholog. On the other hand, Traf3 is a member of the TNF receptor-associated factor protein family, and is a positive regulator of type I IFN signaling and a negative regulator of alternative nuclear factor κB signaling (43). Traf3 has been reported to function as a tumor suppressor in nasopharyngeal carcinoma as well as in macrophages and myeloid cells (44). Interestingly, Li and coworkers (45) also recently reported that Traf3 promotes liver steatosis, suggesting the potential link between Traf3 and NAFLD-HCC. In addition, Traf3 and 4930595D18Rik were detected as trunk divers only in the Pten cohort, suggesting that these genes and the PI3K pathway might have a crosstalk in NAFLD-HCC.
Tschida et al. (46) reported an SB mutagenesis screen performed in the context of the alcohol-induced steatotic liver. In these studies, Tschida et al. identified 203 candidate steatosis-associated CCGs and a number of cancer driver pathways, including Wnt/β-catenin and PKA/cAMP. They also validated the role of Nat10 and Prkaca in HCC. Surprisingly, Sav1 was not identified in this screen, although the reason for this is not clear. Meanwhile, the overlaps between the CCGs identified by Tschida et al. and the two NAFLD-HCC screens reported here are all highly statistically significant, and 54 CCGs were common to all three cohorts (SI Appendix, Fig. S9A and Table S3). Therefore, these genes might be important drivers of steatohepatitis-based HCC. Very recently, Riordan et al. (47) also reported a SB mutagenesis screen performed in the context of chronic liver injury induced by chronic exposure to carbon tetrachloride. Fifty-eight CCGs were identified in injury-associated tumors (≥2% of tumors) and proved the oncogenic role of Gli2, a transcription factor activated downstream of Hh signaling, in injury-associated HCC. Importantly, their CCGs also highly significantly overlapped with the CCGs identified in our two NAFLD-HCC screens, and 24 genes were common to all three cohorts (SI Appendix, Fig. S9B and Table S4). Considering that our models also induce chronic liver injury, these genes might be important HCC drivers in a chronic liver damage environment.
In summary, by comparing the list of CCGs identified by SB mutagenesis in two NAFLD-HCC mouse models, we have been able to generate an unbiased catalog of CCGs driving NAFLD-HCC. We have also validated a tumor suppressive role for Sav1 in NAFLD-HCC, in addition to identifying a role for Hippo signaling in conjunction with PI3K signaling in NASH progression and hepatocarcinogenesis.
Materials and Methods
We used Alb-Cre/+; T2Onc2/+; Rosa26-lsl-SB11/+; Pten fl/fl mice and HFD-fed Alb-Cre/+; T2Onc2/+; Rosa26-lsl-SB11/+ mice for the transposon mutagenesis screens and Alb-Cre/+; Pten fl/fl; Sav1 fl/fl mice for the validation study. Generation of these knockout mice and all other information regarding material and methods can be found in SI Appendix, Material and Methods. All mouse procedures were approved by the Institutional Animal Care and Use Committee of the Osaka University and Houston Methodist Research Institute.
Supplementary Material
Acknowledgments
We thank E. Freiter and H. Lee (Houston Methodist Research Institute) for monitoring mice and for animal technical assistance. This work was supported by the Cancer Prevention Research Institute of Texas (CPRIT) (N.G.C. and N.A.J.); the National Institutes of Health (R.L.J.); the Japan Society for the Promotion of Science KAKENHI Grant JP17K09422 (to T.K.); the Japan Agency for Medical Research and Development under Grants JP18fk0210026, JP18fk0210041, and JP18cm0106533 (to T.K.); the Astellas Foundation for Research on Metabolic Disorders (T.K.); and The YASUDA Medical Foundation (T.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808968115/-/DCSupplemental.
References
- 1.Ferlay J, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Said A, Ghufran A. Epidemic of non-alcoholic fatty liver disease and hepatocellular carcinoma. World J Clin Oncol. 2017;8:429–436. doi: 10.5306/wjco.v8.i6.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ioannou GN, Green PK, Berry K. HCV eradication induced by direct-acting antiviral agents reduces the risk of hepatocellular carcinoma. J Hepatol. September 5, 2017 doi: 10.1016/j.jhep.2017.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudson TJ, et al. International Cancer Genome Consortium International network of cancer genome projects. Nature. 2010;464:993–998, and erratum (2010) 465:966. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169:1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mann KM, et al. Analyzing tumor heterogeneity and driver genes in single myeloid leukemia cells with SBCapSeq. Nat Biotechnol. 2016;34:962–972. doi: 10.1038/nbt.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rad R, et al. A conditional piggyBac transposition system for genetic screening in mice identifies oncogenic networks in pancreatic cancer. Nat Genet. 2015;47:47–56. doi: 10.1038/ng.3164. [DOI] [PubMed] [Google Scholar]
- 8.Takeda H, et al. Transposon mutagenesis identifies genes and evolutionary forces driving gastrointestinal tract tumor progression. Nat Genet. 2015;47:142–150. doi: 10.1038/ng.3175. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe S, et al. Non-alcoholic steatohepatitis and hepatocellular carcinoma: Lessons from hepatocyte-specific phosphatase and tensin homolog (PTEN)-deficient mice. J Gastroenterol Hepatol. 2007;22(Suppl 1):S96–S100. doi: 10.1111/j.1440-1746.2006.04665.x. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa H. Recent advances in mouse models of obesity- and nonalcoholic steatohepatitis-associated hepatocarcinogenesis. World J Hepatol. 2015;7:2110–2118. doi: 10.4254/wjh.v7.i17.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupuy AJ, et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69:8150–8156. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brett BT, et al. Novel molecular and computational methods improve the accuracy of insertion site analysis in Sleeping Beauty-induced tumors. PLoS One. 2011;6:e24668. doi: 10.1371/journal.pone.0024668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kodama T, et al. Transposon mutagenesis identifies genes and cellular processes driving epithelial-mesenchymal transition in hepatocellular carcinoma. Proc Natl Acad Sci USA. 2016;113:E3384–E3393. doi: 10.1073/pnas.1606876113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitade H, Chen G, Ni Y, Ota T. Nonalcoholic fatty liver disease and insulin resistance: New insights and potential new treatments. Nutrients. 2017;9:E387. doi: 10.3390/nu9040387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Futreal PA, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verdelho Machado M, Diehl AM. Role of hedgehog signaling pathway in NASH. Int J Mol Sci. 2016;17:E857. doi: 10.3390/ijms17060857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi Y. The role of growth hormone and insulin-like growth factor-I in the liver. Int J Mol Sci. 2017;18:E1447. doi: 10.3390/ijms18071447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian Y, Mok MT, Yang P, Cheng AS. Epigenetic activation of Wnt/β-catenin signaling in NAFLD-associated hepatocarcinogenesis. Cancers (Basel) 2016;8:E76. doi: 10.3390/cancers8080076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Totoki Y, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46:1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 20.Tu T, Budzinska MA, Shackel NA, Urban S. HBV DNA integration: Molecular mechanisms and clinical implications. Viruses. 2017;9:E75. doi: 10.3390/v9040075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kodama T, et al. Two-step forward genetic screen in mice identifies Ral GTPase-activating proteins as suppressors of hepatocellular carcinoma. Gastroenterology. 2016;151:324–337.e12. doi: 10.1053/j.gastro.2016.04.040. [DOI] [PubMed] [Google Scholar]
- 22.Bard-Chapeau EA, et al. Transposon mutagenesis identifies genes driving hepatocellular carcinoma in a chronic hepatitis B mouse model. Nat Genet. 2014;46:24–32. doi: 10.1038/ng.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawano Y, Cohen DE. Mechanisms of hepatic triglyceride accumulation in non-alcoholic fatty liver disease. J Gastroenterol. 2013;48:434–441. doi: 10.1007/s00535-013-0758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marra F, Svegliati-Baroni G. Lipotoxicity and the gut-liver axis in NASH pathogenesis. J Hepatol. 2018;68:280–295. doi: 10.1016/j.jhep.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 25.Alisi A, et al. The role of tissue macrophage-mediated inflammation on NAFLD pathogenesis and its clinical implications. Mediators Inflamm. 2017;2017:8162421. doi: 10.1155/2017/8162421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angulo P, et al. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015;149:389–397.e10. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kodama T, et al. Increases in p53 expression induce CTGF synthesis by mouse and human hepatocytes and result in liver fibrosis in mice. J Clin Invest. 2011;121:3343–3356. doi: 10.1172/JCI44957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venkataramani V, et al. CD31 expression determines redox status and chemoresistance in human angiosarcomas. Clin Cancer Res. 2018;24:460–473. doi: 10.1158/1078-0432.CCR-17-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zanconato F, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Jiang D, Chi F, Zhao B. The Hippo pathway regulates stem cell proliferation, self-renewal, and differentiation. Protein Cell. 2012;3:291–304. doi: 10.1007/s13238-012-2919-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okabe M, et al. Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development. 2009;136:1951–1960. doi: 10.1242/dev.031369. [DOI] [PubMed] [Google Scholar]
- 32.Furuyama K, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- 33.Patel SH, Camargo FD, Yimlamai D. Hippo signaling in the liver regulates organ size, cell fate, and carcinogenesis. Gastroenterology. 2017;152:533–545. doi: 10.1053/j.gastro.2016.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu L, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, et al. Hepatocyte TAZ/WWTR1 promotes inflammation and fibrosis in nonalcoholic steatohepatitis. Cell Metab. 2016;24:848–862. doi: 10.1016/j.cmet.2016.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aylon Y, et al. The LATS2 tumor suppressor inhibits SREBP and suppresses hepatic cholesterol accumulation. Genes Dev. 2016;30:786–797. doi: 10.1101/gad.274167.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye J, et al. JCAD promotes progression of nonalcoholic steatohepatitis to liver cancer by inhibiting LATS2 kinase activity. Cancer Res. 2017;77:5287–5300. doi: 10.1158/0008-5472.CAN-17-0229. [DOI] [PubMed] [Google Scholar]
- 39.Jeong SH, et al. Hippo-mediated suppression of IRS2/AKT signaling prevents hepatic steatosis and liver cancer. J Clin Invest. 2018;128:1010–1025. doi: 10.1172/JCI95802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Viscarra J, Kim SJ, Sul HS. Transcriptional regulation of hepatic lipogenesis. Nat Rev Mol Cell Biol. 2015;16:678–689. doi: 10.1038/nrm4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee JS, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 42.Yong KJ, et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med. 2013;368:2266–2276. doi: 10.1056/NEJMoa1300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Häcker H, Tseng PH, Karin M. Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat Rev Immunol. 2011;11:457–468. doi: 10.1038/nri2998. [DOI] [PubMed] [Google Scholar]
- 44.Lalani AI, Luo C, Han Y, Xie P. TRAF3: A novel tumor suppressor gene in macrophages. Macrophage (Houst) 2015;2:e1009. doi: 10.14800/macrophage.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang PX, et al. Hepatocyte TRAF3 promotes liver steatosis and systemic insulin resistance through targeting TAK1-dependent signalling. Nat Commun. 2016;7:10592. doi: 10.1038/ncomms10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tschida BR, et al. Sleeping Beauty insertional mutagenesis in mice identifies drivers of steatosis-associated hepatic tumors. Cancer Res. 2017;77:6576–6588. doi: 10.1158/0008-5472.CAN-17-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riordan JD, et al. Chronic liver injury alters driver mutation profiles in hepatocellular carcinoma in mice. Hepatology. 2018;67:924–939. doi: 10.1002/hep.29565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.