Significance
Our findings shift the paradigm of NRF2 as a transcriptional activator to one in which NRF2 can also act as a transcriptional repressor, which we believe will stimulate new research areas and interests among scientists from other fields. While the majority of the data provided in this paper center on suppression of MYLK expression and the resulting pathological significance, the more far-reaching findings are the in silico and RNA-seq datasets indicating that the NRF2-replication protein A1 (RPA1)-ARE-NRE complex transcriptionally represses other genes as well, again highlighting the broad scope and significance of NRF2 repression of target genes.
Keywords: NRF2, MYLK/MLCK, RPA1, acute lung injury, transcriptional regulation
Abstract
NRF2 regulates cellular redox homeostasis, metabolic balance, and proteostasis by forming a dimer with small musculoaponeurotic fibrosarcoma proteins (sMAFs) and binding to antioxidant response elements (AREs) to activate target gene transcription. In contrast, NRF2-ARE–dependent transcriptional repression is unreported. Here, we describe NRF2-mediated gene repression via a specific seven-nucleotide sequence flanking the ARE, which we term the NRF2-replication protein A1 (RPA1) element (NRE). Mechanistically, RPA1 competes with sMAF for NRF2 binding, followed by interaction of NRF2-RPA1 with the ARE-NRE and eduction of promoter activity. Genome-wide in silico and RNA-seq analyses revealed this NRF2-RPA1-ARE-NRE complex mediates negative regulation of many genes with diverse functions, indicating that this mechanism is a fundamental cellular process. Notably, repression of MYLK, which encodes the nonmuscle myosin light chain kinase, by the NRF2-RPA1-ARE-NRE complex disrupts vascular integrity in preclinical inflammatory lung injury models, illustrating the translational significance of NRF2-mediated transcriptional repression. Our findings reveal a gene-suppressive function of NRF2 and a subset of negatively regulated NRF2 target genes, underscoring the broad impact of NRF2 in physiological and pathological settings.
The important role of maintaining redox homeostasis to preserve lung architecture and function in response to inflammatory challenges has been attributed to the activation of NRF2, a transcription factor and master regulator of the cellular antioxidant response (1, 2). NRF2 induces gene expression via binding to antioxidant response elements (AREs) in the regulatory region of target genes that encode proteins involved in redox homeostasis, xenobiotic metabolism, anabolic metabolism, DNA damage, proliferation, and survival responses (3–7). NRF2 binds AREs as heterodimers with small musculoaponeurotic fibrosarcoma proteins (sMAFs) (8), thereby recruiting chromatin remodeling complexes, coactivators, and mediator proteins to up-regulate gene expression (9). Similar to NRF2, sMAFs are leucine zipper proteins; however, unlike NRF2, sMAFs lack transcription activation domains (10). While sMAF homodimers repress associated transcription units by shielding spurious activation by neighboring regulatory regions, sMAFs also mark NRF2-responsive genes, maintaining accessibility for rapid NRF2-mediated gene activation (11, 12).
The nonmuscle isoform of myosin light chain kinase (nmMLCK; 210 kDa) is encoded by the MYLK gene and is a key actin cytoskeletal regulatory protein (13, 14). The contractile activity elicited by nmMLCK-mediated phosphorylation of myosin light chains (MLCs) is involved in multiple pleiotropic biological and pathobiological processes, including cellular proliferation and apoptosis (15), leukocyte recruitment to tissues (16), regulation of vascular barrier integrity (17), and generation of reactive oxygen species (ROS) (18). The translational impact of nmMLCK activities was validated by studies identifying MYLK polymorphisms that alter nmMLCK expression and enzymatic function, increasing the inflammatory burden and mortality associated with acute respiratory distress syndrome (ARDS) and severe asthma (19, 20). Clearly, nmMLCK and ROS generation are each critical to acute and chronic inflammatory pathobiologies, including lipopolysaccharide (LPS)-induced acute lung injury, severe ARDS in humans (21, 22), and ventilator-induced lung injury (VILI) (1, 23). Exposure of nmMLCK null mice to LPS, mechanical ventilation, or hyperoxia results in reduced ROS production and lung leukocyte recruitment, attenuation of pulmonary vascular permeability, and reduced expression of genes involved in biological pathways, such as the NRF2-mediated antioxidant response, coagulation, leukocyte extravasation, and IL-6 signaling (18, 24). Thus, nmMLCK is an attractive and proven target for ameliorating the adverse effects associated with lung inflammation.
NRF2-dependent regulation of MYLK has not been described, however. As nmMLCK null mice exhibit reduced NRF2 target gene expression when exposed to acute inflammatory stimuli (24), the potential for cross-talk between nmMLCK and NRF2 exists. Although this down-regulation of oxidative responses could conceivably reflect reduced inflammatory burden and ROS generation, MYLK contains an ARE-like sequence within the nmMYLK promoter. Here we report the identification of a pathophysiologically important mechanism of NRF2-ARE–dependent gene suppression. NRF2 negatively regulates MYLK transcription via direct interaction with replication protein A1 (RPA1) and binding of the NRF2-RPA1 (NRE) complex to the ARE and an adjacent 7-nt negative regulatory sequence. This ARE-NRE–dependent transcriptional mechanism of gene repression is in stark contrast to the well-recognized ARE-dependent transcriptional activation evoked by NRF2. Genome-wide and RNA-seq analyses revealed NRF2-mediated negative transcriptional regulation as a fundamental regulatory mechanism controlling expression of a subset of ARE-containing genes. Furthermore, the translational impact of NRF2-dependent negative MYLK regulation was confirmed using in vitro models of vascular permeability, as well as in vivo models of inflammatory lung injury in genetically engineered mice (Nrf2−/−, Mylk−/−, and Nrf2−/−;Mylk−/−). These results constitute an important paradigm shift in the understanding of NRF2-mediated transcriptional regulation of genes harboring AREs.
Results
NRF2 Negatively Regulates MYLK Expression.
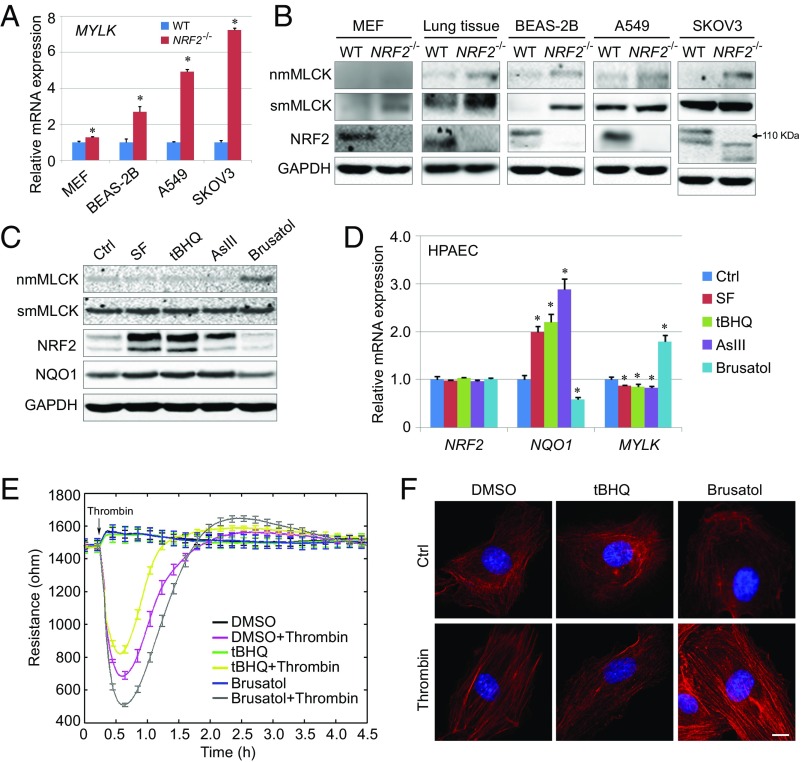
To gain a deeper mechanistic understanding of cross-talk between MYLK and NRF2, potential MYLK transcriptional regulation by NRF2 was explored in murine embryonic fibroblasts (MEFs), in lung tissues isolated from wild-type (WT) mice or Nrf2 knockout mice (Nrf2−/−), and in human isogenic cell lines in which NRF2 is either present (WT) or genetically deleted (NRF2−/−). MYLK mRNA expression was increased in all NRF2−/− cells compared with their WT counterparts (Fig. 1A and SI Appendix, Fig. S1A). Consistent with the increase in MYLK mRNA expression, higher protein levels of nmMLCK and smooth muscle myosin light chain kinase (smMLCK) were observed in each human NRF2−/− cell line tested (Fig. 1B and SI Appendix, Fig. S1B), with protein levels of smMLCK also elevated in Nrf2−/− MEFs, whereas nmMLCK was not detected (Fig. 1B and SI Appendix, Fig. S1B). This apparent negative regulation of MYLK expression by NRF2 was further explored in WT and Nrf2−/− mouse lung tissues, revealing significantly increased nmMLCK and smMLCK protein levels in Nrf2−/− lung, compared with WT (Fig. 1B and SI Appendix, Fig. S1B). Moreover, this NRF2-mediated negative regulation of MLCK was also confirmed in Keap1−/− MEFs and Keap1−/− H1299 cells, as decreased expression of MLCK at both the mRNA (SI Appendix, Fig. S1C) and protein (SI Appendix, Fig. S1D) levels were observed.
Fig. 1.
NRF2 negatively regulates MYLK expression. (A) qRT-PCR analysis of MYLK expression in WT and NRF2−/− cell lines. n = 3. Each gene was normalized to its control (Ctrl); unnormalized results are shown in SI Appendix, Fig. S1A. Data are presented as mean ± SD. *P < 0.05. (B) Immunoblot analysis of WT and NRF2−/− cell lines and lung tissue lysates from C57BL/6J mice. Quantification is shown in SI Appendix, Fig. S1B. (C) Immunoblot analysis of HPAECs treated (16 h) with NRF2 activators SF (5 μM), tBHQ (20 μM), and AsIII (1 μM) and with the NRF2 inhibitor brusatol (40 nM). Quantification is shown in SI Appendix, Fig. S1C. (D) qRT-PCR analysis of gene expression in HPAECs pretreated with NRF2 activators and brusatol as in C. Each gene is normalized to its control (Ctrl). Unnormalized results are shown in SI Appendix, Fig. S1F. (E) TEER of HPAECs pretreated (16 h) with DMSO, tBHQ (20 μM), or brusatol (40 nM), with or without thrombin (1 U/mL) challenge. Data were collected continuously every 30 s during the entire 4.5-h period. n = 4. Data are presented as mean ± SD. (F) Representative immunofluorescence images of polymerized F-actin stained with phalloidin (red) in HPAECs pretreated for 16 h with SF (5 μM) or brusatol (40 nM), followed by a 5-min challenge with thrombin (1 U/mL). Nuclei were labeled with DAPI (blue). (Scale bar: 50 μm.)
We next explored MYLK-NRF2 transcriptional dynamics using well-defined pharmacologic NRF2 modulators in human pulmonary artery endothelial cells (HPAECs). As expected, the NRF2 activators sulforaphane (SF), tert-butylhydroquinone (tBHQ), and arsenic (AsIII), each enhanced protein levels of both NRF2 and NAD(P)H quinone dehydrogenase 1 (NQO1), a representative NRF2 target gene (Fig. 1C and SI Appendix, Fig. S1E). The mRNA levels of NQO1, but not of NRF2, were also increased (Fig. 1D and SI Appendix, Fig. S1F). In contrast, the NRF2 inhibitor brusatol decreased NRF2 and NQO1 protein levels and NQO1 mRNA expression (Fig. 1 C and D and SI Appendix, Fig. S1 E and F). However, in stark contrast to NQO1, nmMLCK protein expression (Fig. 1C and SI Appendix, Fig. S1E) and nmMYLK mRNA expression (Fig. 1D and SI Appendix, Fig. S1F) were reduced by NRF2 activation and enhanced by NRF2 inhibition in brusatol-challenged HPAECs.
The functional effect of NRF2-mediated nmMYLK suppression was next determined by assessing human lung endothelial cell (EC) barrier integrity, a critical nmMLCK homeostatic function, using measurements of transendothelial electrical resistance (TEER) and phalloidin staining of F-actin. Vascular barrier-regulatory responses to thrombin, a potent EC barrier-disrupting agonist (13), were exacerbated in ECs pretreated with the NRF2 inhibitor brusatol compared with DMSO controls (Fig. 1E). This was further highlighted by the presence of increased contractile actin stress fibers (Fig. 1F) indicative of increased nmMLCK enzymatic activity and activation of the contractile apparatus. These functional analyses indicate a potential role for NRF2 in suppressing nmMLCK-regulated cytoskeletal rearrangement and mediating vascular barrier integrity.
An NRE Element Exists Adjacent to the MYLK ARE.
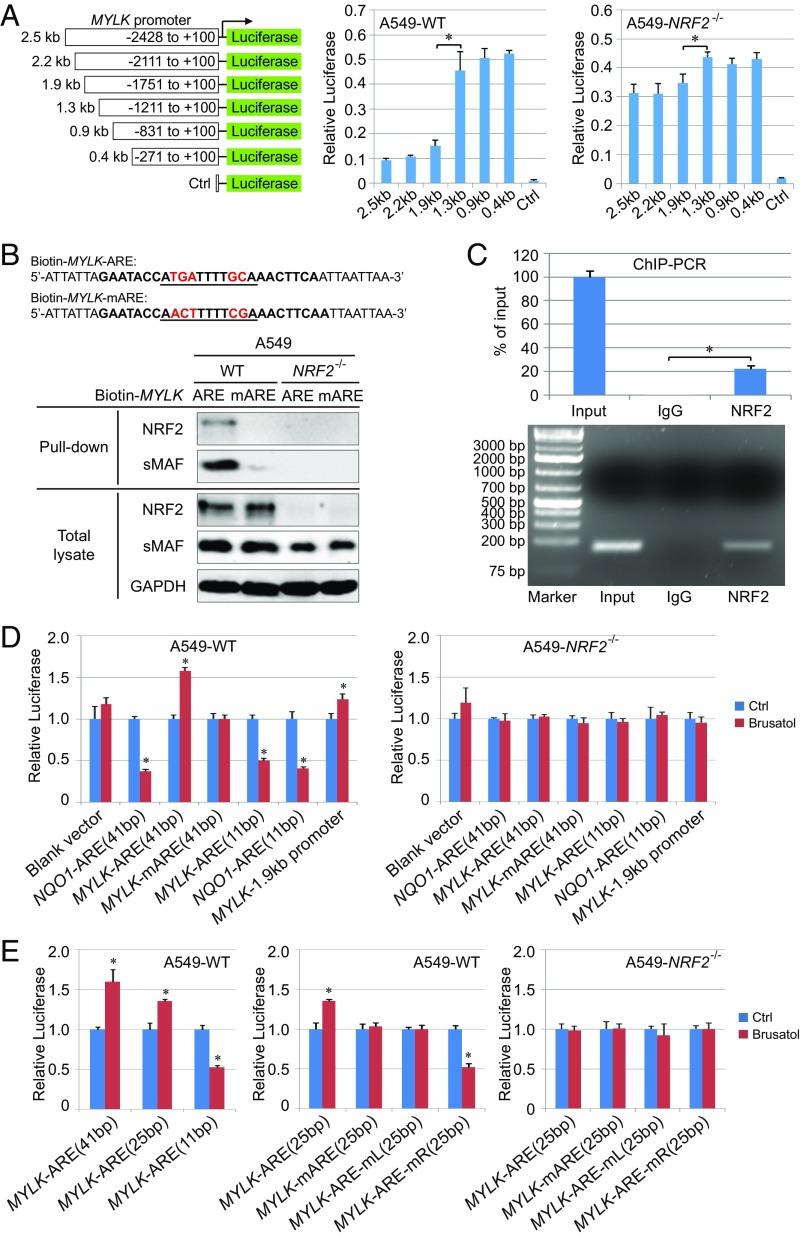
We next sought to identify the exact site of NRF2-mediated negative regulation of MYLK, including the potential involvement of the ARE in this repressive mechanism. Luciferase activity was measured in A549-WT and A549-NRF2−/− cells transfected with luciferase reporters containing a deletion series of the nmMYLK promoter (Fig. 2A). These studies identified the −1.3 to −1.9-kb promoter region is critical for NRF2-dependent negative regulation of MYLK, which is further supported by the minimal change in promoter activity in A549-NRF2−/− cells (Fig. 2A). In silico analysis identified an ARE-like sequence within this −1.3 to −1.9-kb region (MYLK-ARE). Using biotinylated 41-bp DNA probes of either the WT MYLK-ARE promoter sequence (MYLK-ARE) or a mutated MYLK-ARE promoter sequence (MYLK-mARE), NRF2 was confirmed to specifically bind to the MYLK-ARE but not to the MYLK-mARE (Fig. 2B), which was further verified by ChIP-PCR (Fig. 2C). These studies demonstrate that the MYLK promoter contains a functional ARE that is regulated by NRF2.
Fig. 2.
An NRE exists adjacent to the MYLK ARE. (A) Relative basal luciferase activity of A549 WT and NRF2−/− cells transfected with the pGL3-Luc vector containing portions of the human MYLK promoter cloned upstream of the firefly luciferase gene. Cells were cotransfected with Renilla luciferase to normalize firefly luciferase activity to this transfection control. n = 3. Data are presented as mean ± SD. *P < 0.05. (B) Pull-down assay of A549 WT and NRF2−/− cells. The biotinylated DNA probe containing the MYLK ARE WT or mutant (mARE) was incubated with lysates from either A549 WT or NRF2−/− cells. DNA-binding proteins were pulled down using streptavidin beads and detected by immunoblot analysis. (C) ChIP-PCR of A549 WT cells. Rabbit IgG served as a negative control. The expected size of the PCR product was 133 bp. (D and E) Relative luciferase activity of A549 WT and NRF2−/− cells transfected with the pGL4.22-Luc vector containing different AREs. WT and NRF2−/− cells were untreated or pretreated with brusatol (40 nM, 16 h). The sequences of the different AREs are shown in SI Appendix, Fig. S2 A and B. Cells were cotransfected with Renilla luciferase to normalize firefly luciferase activity to this transfection control. Results were further normalized to each untreated control (Ctrl). Unnormalized results are shown in SI Appendix, Fig. S2 C and D. n = 3. Data are presented as mean ± SD. *P < 0.05.
NRF2 positively regulates ARE-containing genes via formation of NRF2-sMAF heterodimers that bind AREs to up-regulate transcription. To interrogate the mechanistic basis for negative MYLK regulation, luciferase activities of the 11-bp core nmMYLK-ARE and an extended 41-bp nmMYLK-ARE (sequences shown in SI Appendix, Fig. S2A) were measured in A549-WT and A549-NRF2−/− cells, with the 41-bp and 11-bp NQO1-ARE reporters serving as controls. In A549-WT cells, NRF2 inhibition by brusatol decreased luciferase activities of both the 41-bp and 11-bp NQO1-ARE reporters (Fig. 2D and SI Appendix, Fig. S2C). In contrast, brusatol enhanced the activity of both the 1.9-kb MYLK promoter and the 41-bp MYLK-ARE reporter, while decreasing the core 11-bp MYLK-ARE reporter. Luciferase activity of the mutated MYLK-mARE was refractory to brusatol (Fig. 2D and SI Appendix, Fig. S2C). The effects of brusatol were abolished in NRF2−/− cells (Fig. 2D and SI Appendix, Fig. S2C), strongly supporting the notion that MYLK-ARE promoter activity is modulated by NRF2 in a manner distinct from NQO1 and suggesting the presence of a repressive element (i.e., NRE) flanking the 11-bp core ARE but residing within the 41-bp ARE sequence of the MYLK promoter.
To more clearly define the NRE, the following MYLK-ARE luciferase vectors (25 bp long, with 7 bp flanking the 11-bp core ARE) were generated: MYLK-ARE (WT), MYLK-mARE (mutations in the core ARE), MYLK-ARE-mL (5′ or “left” flanking mutations), and MYLK-ARE-mR (3′ or “right” flanking mutations) (SI Appendix, Fig. S2B). Brusatol enhanced the activity of both the 41-bp and 25-bp MYLK-ARE constructs but decreased the activity of the 11-bp MYLK-ARE (Fig. 2E and SI Appendix, Fig. S2D). These results indicate that the NRE resides in a sequence flanking the core. Consistent with this speculation, mutation of three nucleotides flanking the 3′ end of MYLK-ARE in MYLK-ARE-mR, but not in MYLK-ARE-mL, reversed NRF2-mediated transcriptional repression of MYLK (Fig. 2E and SI Appendix, Fig. S2D). Predictably, brusatol did not alter MYLK-ARE luciferase activity in NRF2−/− cells (Fig. 2E and SI Appendix, Fig. S2D). These results demonstrate that the 7-bp 3′ flanking sequence (AACTTCA) of the core MYLK-ARE represents the NRE required for NRF2-dependent MYLK-ARE repression.
NRE-Mediated Attenuation of MYLK Transcription Is ARE-Specific.
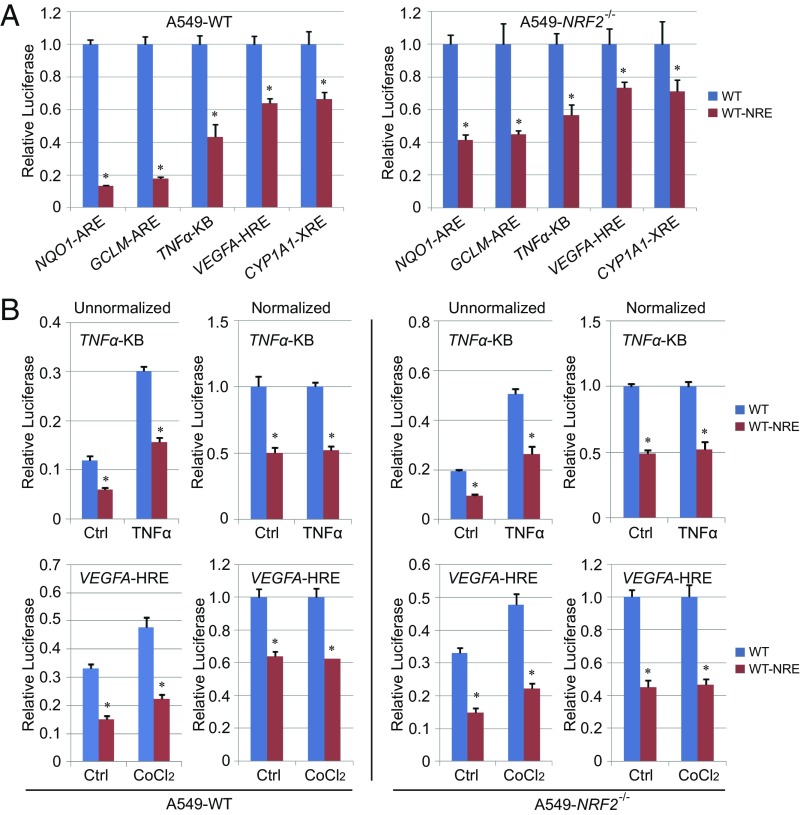
We next investigated whether the inhibitory MYLK NRE sequence could alter the expression of other genes harboring an ARE, such as NQO1 and GCLM, or influence the transcription of other response elements, including the hypoxia response element (HRE) present in VEGFA, the NF-κB response element (κB) in TNFA, and the xenobiotic response element (XRE) in CYP1A1. Compared with the endogenous sequence (SI Appendix, Fig. S3A), direct insertion of the NRE into the ARE of either NQO1 or GCLM reduced luciferase reporter activities by sixfold in A549-WT cells but by only approximately twofold in A549-NRF2−/− cells (Fig. 3A and SI Appendix, Fig. S3B). In contrast, NRE insertion into the HRE, κB, or XRE reduced luciferase activities only modestly (approximately twofold) in both A549-WT and A549-NRF2−/− cells (Fig. 3A) under either basal or induced conditions (Fig. 3), suggesting NRF2-independent effects. These results indicate that NRE repression is specific and highly dependent on the presence of an ARE enhancer sequence.
Fig. 3.
NRE-mediated attenuation of MYLK transcription is ARE-specific. (A) Relative basal luciferase activity of A549 WT and NRF2−/− cells transfected with the pGL4.22-Luc vector containing different response elements with and or without the NRE sequence. The promoter sequences are shown in SI Appendix, Fig. S3A. Cells were cotransfected with Renilla luciferase to normalize firefly luciferase activity to this transfection control. Results were further normalized to the cells transfected with response elements without NRE (WT) for each pair. Unnormalized results are shown in SI Appendix, Fig. S3B. n = 3. Data are presented as mean ± SD. *P < 0.05. (B) Relative luciferase activity of A549 WT and NRF2−/− cells transfected with different response elements with or without the NRE in uninduced (Ctrl) or induced condition [TNFα (20 ng/mL) for 4 h, CoCl2 (0.2 mM) for 16 h]. n = 3. Data are presented as mean ± SD. *P < 0.05.
Involvement of RPA1-NRE Binding in Repression of MYLK Transcription.
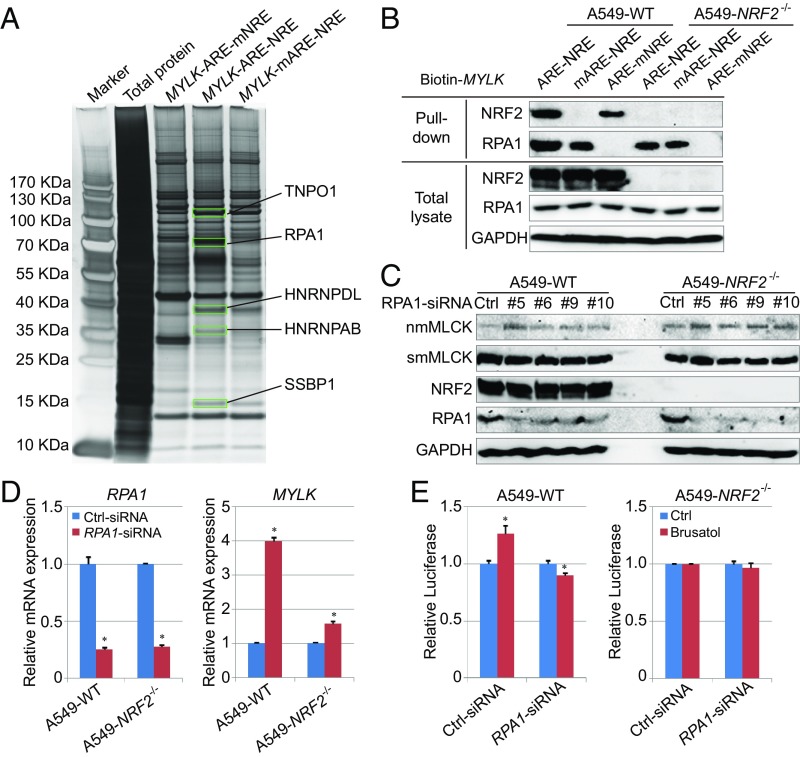
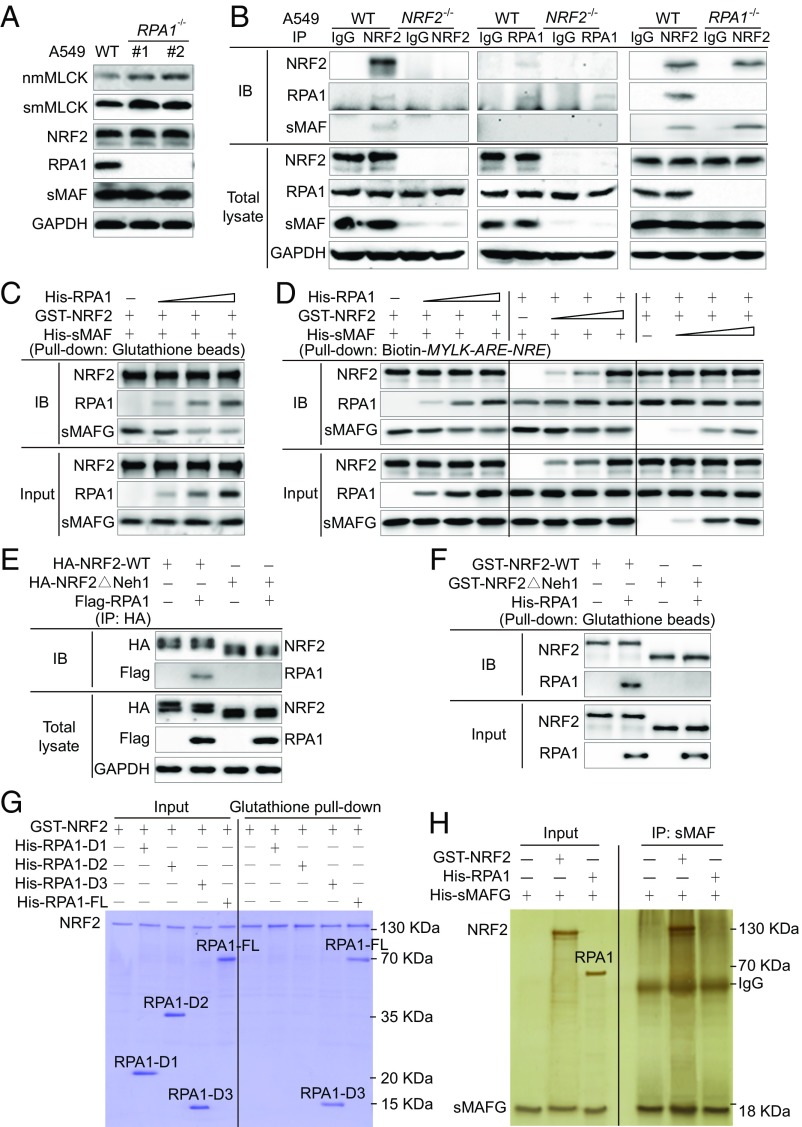
To identify transcription cofactors potentially mediating NRF2-ARE repression via NRE binding, biotinylated 41-bp double-stranded probes of MYLK-ARE-NRE (WT), MYLK-mARE-NRE (mutation in ARE), and MYLK-ARE-mNRE (mutation in NRE) were generated (sequences in SI Appendix, Fig. S4A). Each probe was incubated with A549-WT cell lysates to assess protein–DNA interactions, and DNA-binding proteins were identified by mass spectrometry. Unique proteins binding only to MYLK-ARE-NRE, and not to MYLK-ARE-mNRE, were identified (Fig. 4A). Of the potential candidates identified (SI Appendix, Fig. S4B), RPA1, the sole gene repressor, was selected for further analysis. Immunoblot analysis demonstrated that the MYLK-ARE (ARE) could pull down both NRF2 and RPA1, whereas mutation of the ARE (mARE) disrupted NRF2 binding completely and reduced RPA1 binding, and mutation of the NRE (mNRE) prevented RPA1 binding (Fig. 4B). RPA1 is known to be an ssDNA-binding protein that forms a heterotrimeric complex with RPA2 and RPA3 during DNA replication or repair (SI Appendix, Fig. S4C) (25). However, RPA2 and RPA3 were not detected in the MYLK-ARE-RPA1 complex (SI Appendix, Fig. S4D), suggesting a new function of RPA1 in sequence-specific binding to dsDNA.
Fig. 4.
Involvement of RPA1-NRE binding in repression of MYLK transcription. (A) Pull-down assay using a biotinylated dsDNA probe in A549-WT cell lysates. The probes used include WT ARE and WT NRE (MYLK-ARE-NRE), mutated ARE and WT NRE of MYLK (MYLK-mARE-NRE), and WT ARE and mutated NRE (MYLK-ARE-mNRE). Proteins identified by SDS/PAGE and silver staining as differentially pulled down (green rectangles, present in MYLK-ARE-NRE only) were identified by mass spectrometry. The probe sequences are shown in SI Appendix, Fig. S4A, and the peptides identified in MYLK-ARE-NRE pull-down are shown in SI Appendix, Fig. S4B. (B) Immunoblotting of biotinylated dsDNA probe pull-down of A549 WT and NRF2−/− cell lysates. (C) Immunoblot analysis of A549 WT and NRF2−/− cells transfected with control siRNA (Ctrl) or four different siRNAs targeting RPA1. Quantification is shown in SI Appendix, Fig. S4E. (D) qRT-PCR analysis of RPA1 and MYLK levels in A549 WT and NRF2−/− cells transfected with control siRNA or RPA1 siRNA. Unnormalized results are shown in SI Appendix, Fig. S4F. n = 3. Data are presented as mean ± SD. *P < 0.05. (E) Relative luciferase activity of A549 WT and NRF2−/− cells transfected with Ctrl siRNA or RPA1 siRNA. Cells were cotransfected with Renilla luciferase to normalize firefly luciferase activity to this transfection control. Results were further normalized to each untreated control (Ctrl). Unnormalized results are shown in SI Appendix, Fig. S4G. n = 3. Data are presented as mean ± SD. *P < 0.05.
Next, the contribution of RPA1 to the negative regulation of MYLK was explored. RPA1 silencing using multiple siRNA constructs reduced RPA1 protein levels without affecting NRF2 or smMLCK, but significantly increased nmMLCK protein levels (Fig. 4C and SI Appendix, Fig. S4E). Consistently, RPA1 siRNA reduced its mRNA expression by ∼75% in both A549-WT and A549-NRF2−/− cells and increased the mRNA expression of MYLK by fourfold in A549-WT and by ∼1.5-fold in A549-NRF2−/− cells (Fig. 4D and SI Appendix, Fig. S4F). Furthermore, RPA1 silencing reversed the brusatol-mediated increase in MYLK-ARE activity in A549-WT cells but failed to affect MYLK-ARE activity in A549-NRF2−/− cells (Fig. 4E and SI Appendix, Fig. S4G). These data indicate synergism between RPA1 and NRF2 in repressing MYLK transcription.
RPA1 Competes with sMAF to Directly Bind NRF2.
The mechanism by which RPA1 represses ARE-driven gene expression was explored further. Consistent with the RPA1 siRNA results (Fig. 4C), RPA1−/− cells exhibited increased smMLCK and nmMLCK expression without altering NRF2 or sMAF expression (Fig. 5A and SI Appendix, Fig. S5A). Protein–protein interactions involving NRF2, sMAF, and RPA1 were next investigated using WT, NRF2−/−, and RPA1−/− A549 cell lines (Fig. 5B). Both RPA1 and sMAF coimmunoprecipitated with NRF2 in A549 WT cells, whereas only sMAF immunoprecipitated with NRF2 in A549-RPA1−/− cells, confirming the direct interaction of NRF2 with sMAF and RPA1 (Fig. 5B). Since RPA1 immunoprecipitated with NRF2 but not with sMAF directly (Fig. 5B), these results indicate a potential competition between RPA1 and sMAF for NRF2 binding. This competitive RPA1- and sMAF-NRF2–binding model was verified using purified proteins, with increasing amounts of RPA1 decreasing the level of sMAF in the sMAF-NRF2 complexes (Fig. 5C). Furthermore, this competitive binding between sMAF and RPA1 was also confirmed by biotin-MYLK-ARE-NRE pulldown after the DNA was incubated with increasing concentrations of all three purified proteins (Fig. 5D).
Fig. 5.
RPA1 competes with sMAFs to directly bind NRF2. (A) Immunoblot analysis of A549 WT and RPA1−/− cells. Quantification is shown in SI Appendix, Fig. S5A. (B) Immunoprecipitation assay using different antibodies with A549 WT, NRF2−/−, and RPA1−/− cell lysates. Rabbit and mouse IgG served as negative controls for NRF2 and RPA1 antibodies, respectively. (C) In vitro pull-down assay of purified GST-NRF2 (1 μg for each group) incubated with His-RPA1 (0, 0.5, 1, and 2 μg, respectively) and His-sMAF (1 μg for each group). (D) Immunoblotting of biotinylated dsDNA probe pull-down of purified GST-NRF2 (10 μg for each group), His-RPA1 (10 μg for each group), and His-sMAF (10 μg for each group); 0, 2.5, 5, and 10 μg of protein was used in the different dose-dependent groups. (E) Immunoprecipitation assay of HEK293 cells transfected with HA-tagged WT NRF2 (HA-NRF2-WT) or Neh1 domain deletion mutant (HA-NRF2ΔNeh1) alone or in combination with Flag-RPA1. (F) In vitro pull-down assay of purified His-RPA1 incubated with GST-NRF2-WT or GST-NRF2ΔNeh1. (G) In vitro pull-down assay and Coomassie blue staining of purified GST-NRF2-WT and His-tagged FL RPA1 (His-RPA1-FL) or RPA1 deletion mutants (His-RPA1-D1, His-RPA1-D2, or His-RPA1-D3). (H) In vitro immunoprecipitation assay and silver staining of purified His-sMAFG alone or in combination with GST-NRF2-WT and His-RPA1.
Since sMAF proteins form heterodimers with NRF2 via binding to the Neh1 domain (8), we next assessed the requirement of Neh1 domain binding by RPA1. RPA1 coimmunoprecipitated with NRF2-WT but not with an NRF2 mutant with the Neh1 domain deleted (NRF2ΔNeh1) (Fig. 5E). Pulldown analysis using purified NRF2 and NRF2ΔNeh1 proteins further confirmed direct binding of NRF2 with RPA1 via the Neh1 domain (Fig. 5F). The Neh1 domain, containing the CNC basic leucine zipper, is highly conserved within the NRF family. Therefore, to determine whether other NRF family members could bind to RPA1, the binding of NRF1 and NRF3 to RPA1 was also tested. As expected, RPA1 also immunoprecipitated with NRF1 and NRF3, confirming that it binds to the Neh1 domain of NRF transcription factors (SI Appendix, Fig. S5B). To determine which domain in RPA1 binds to NRF2, pull-down experiments were performed using full-length (FL) RPA1 and three RPA1 deletion mutants: RPA1-D1, RPA1-D2, and RPA1-D3 (Fig. 5G and SI Appendix, Fig. S5C). NRF2 interaction was observed only with RPA1-D3 and RPA1-FL, demonstrating that the D3 domain is required for NRF2 interaction (Fig. 5G). Examination of sMAF interactions with either NRF2 or RPA1 confirmed an sMAF–NRF2 interaction, but no direct sMAF–RPA1 binding (Fig. 5H). These studies support a previously unappreciated mode of NRF2-mediated negative transcriptional regulation of ARE-NRE–containing genes via RPA1 competing with sMAF for NRF2 binding.
NRF2-Mediated Negative Transcriptional Regulation Is a Fundamental Mechanism Controlling the Expression of Other Genes.
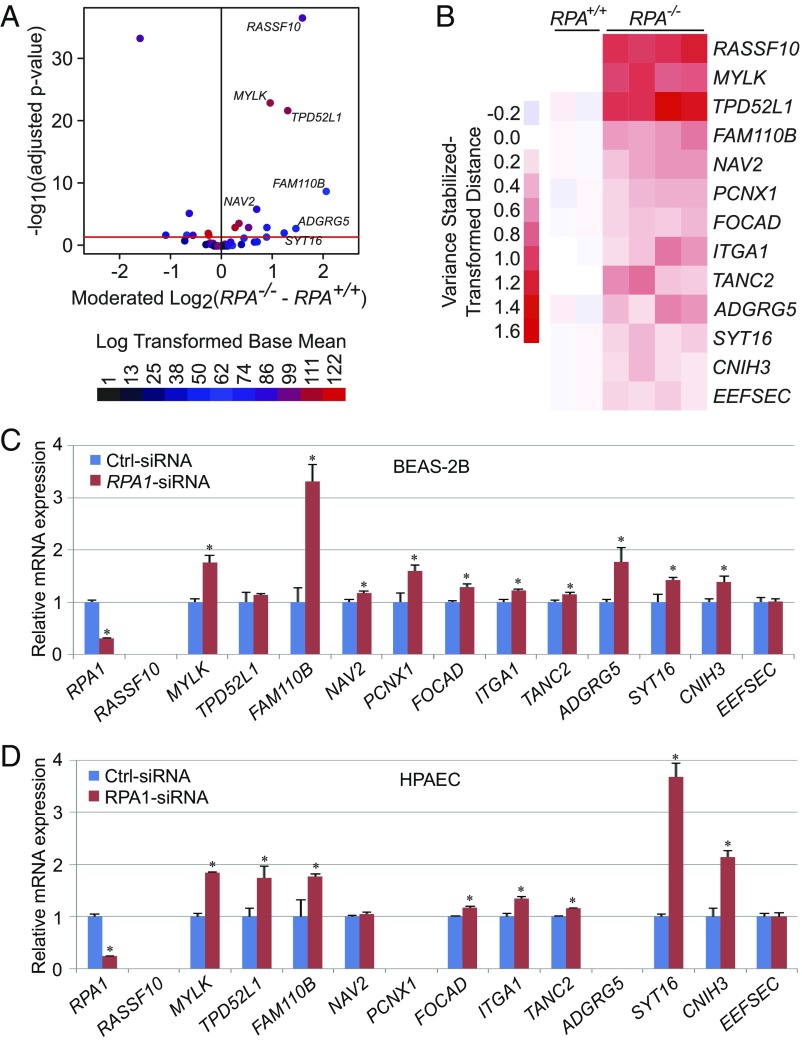
We next sought to investigate whether the NRF2-RPA1 complex transcriptionally represses ARE-containing genes in addition to MYLK. A genome-wide in silico analysis identified 428 unique genomic loci containing the exact ARE-NRE consensus sequence derived from the nmMYLK promoter (TGABNNNGCAAACTTCA) (SI Appendix, Table S1), which were further filtered by location within the promoter region (≤5 kb upstream of the transcription start site) or residing within the first intron. Of the identified loci, 10.3% were within the first intron and only 4.8% resided within promoter regions (SI Appendix, Fig. S6A) yielding a total of 55 genes potentially regulated by the NRF2-RPA1 complex (SI Appendix, Table S2). To validate these in silico findings, RNA sequencing was performed to compare genome-wide mRNA expression in RPA1 knockout (RPA1−/−) and control (RPA1+/+) A549 cells. Of the 55 genes identified by in silico approaches, 13 genes, including MYLK, exhibited significant increases in transcript levels in RPA1−/− cells compared with control cells (Fig. 6 A and B). These results were further validated by qRT-PCR in RPA1-silenced BEAS-2B cells and HPAECs, which exhibited increased transcript levels in both cell lines (11 of 13 genes) (Fig. 6 C and D). Furthermore, 12 of these 13 genes were transcriptionally up-regulated in NRF2−/− A549 cells (SI Appendix, Fig. S6B). Thus, NRF2-RPA1 modulation of ARE-NRE sites, similar to MYLK, is a previously unappreciated and fundamental mechanism of negative regulation of gene expression.
Fig. 6.
The NRF2-mediated negative transcriptional regulation is a fundamental mechanism controlling the expression of other genes. (A) Volcano plot showing differentially expressed genes harboring ARE-NRE sites for RNA-seq data comparing A549-RPA1+/+ and A549-RPA1−/− cells. The red line indicates −log10 (adjusted P value) = 1.30, which corresponds to an adjusted P value of 0.05. Points are colored according to their log-transformed base mean value. (B) Heatmap representing variance stabilized-transformed distance data from RNA sequencing of A549-RPA+/+ and A549-RPA−/− cells. The presented genes harbor ARE-NRE sites within their promoter regions or the first intron and follow the same expression pattern as MYLK following RPA1 knockout. (C and D) qRT-PCR analysis of candidate gene mRNA expression in Ctrl-siRNA– and RPA1-siRNA–transfected BEAS-2B cells (C) and HPAECs (D). n = 3. Data are presented as mean ± SD. *P < 0.05.
NRF2-Driven Repression of MYLK Expression Attenuates Inflammatory Lung Injury.
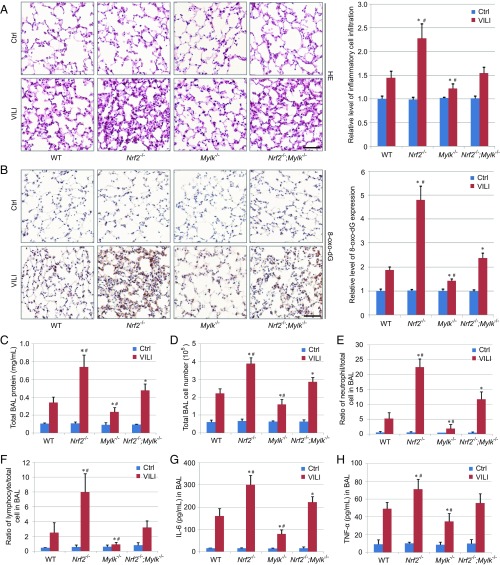
NRF2-mediated protection in models of acute inflammatory lung injury has been attributed largely to the induction of antioxidant genes (1). To explore the importance of negative MYLK regulation by NRF2, we exposed WT, Nrf2 knockout (Nrf2−/−), nmMLCK isoform-specific Mylk knockout (Mylk−/−), and double- knockout (Nrf2−/−;Mylk−/−) mice to a well-established model of inflammatory lung injury produced by exposure to high-tidal volume mechanical ventilation (i.e., VILI). Immunohistochemistry (IHC) analysis for NRF2 and nmMLCK (SI Appendix, Fig. S7 A and B) demonstrated VILI-induced lung tissue expression of both proteins, which was confirmed by immunoblot analysis (SI Appendix, Fig. S7C). Both smMLCK and nmMLCK protein levels were increased in Nrf2−/− mice compared with WT mice, whereas KEAP1 or GAPDH protein levels were similar across all groups (SI Appendix, Fig. S7C). For each index of VILI-induced inflammatory injury, Nrf2−/− mice exhibited the greatest degree of injury, followed by Nrf2−/−;Mylk−/− mice and WT mice, with Mylk−/− mice exhibiting the least degree of inflammatory injury as assessed by lung morphology alterations and leukocyte infiltration (Fig. 7A), levels of 8-deoxyguanosine (8-oxo-dG; an indicator of oxidative DNA damage) (Fig. 7B), levels of bronchoalveolar lavage (BAL) fluid protein (vascular leakage), inflammatory cell infiltration (Fig. 7 C–F), and levels of BAL proinflammatory cytokines IL-6 and TNF-α (Fig. 7 G and H). These results confirm that NRF2 reduces acute inflammatory lung injury via both induction of antioxidant responses and enhancement of lung vascular barrier integrity by repression of MYLK expression.
Fig. 7.
NRF2-driven repression of MYLK expression attenuates inflammatory lung injury. (A, Left) Hematoxylin and eosin staining of lung tissue sections from control (Ctrl; n = 3) and VILI (n = 6) mice. (Scale bar: 50 μm.) (A, Right) Quantification of inflammatory cell infiltration is shown on the right. (B, Left) IHC staining of 8-oxo-dG in lung tissue sections from Ctrl (n = 3) and VILI (n = 6) mice. (Scale bar: 50 μm.) (B, Right) Relative intensity of 8-oxo-dG staining. (C) Total protein concentration in BAL fluid from Ctrl (n = 3) and VILI (n = 6) mice. Data are presented as mean ± SD. *P < 0.05 compared with WT mice; # P < 0.05 compared with Nrf2−/−;Mylk−/− mice. (D) Total BAL cell numbers from Ctrl (n = 3) and VILI (n = 6) mice. Data are presented as mean ± SD. *P < 0.05 compared with WT mice; #P < 0.05 compared with Nrf2−/−;Mylk−/− mice. (E) Percentage of BAL neutrophils in Ctrl (n = 3) and VILI (n = 6) mice. Data are presented as mean ± SD. *P < 0.05 compared with WT mice; #P < 0.05 compared with Nrf2−/−;Mylk−/− mice. (F) Percentage of BAL lymphocytes in Ctrl (n = 3) and VILI (n = 6) mice. Data are presented as mean ± SD. *P < 0.05 compared with WT mice; # P < 0.05 compared with Nrf2−/−;Mylk−/− mice. (G) BAL IL-6 quantification in Ctrl (n = 3) and VILI (n = 6) mice. Data are presented as mean ± SD. *P < 0.05 compared with WT mice; #P < 0.05 compared with Nrf2−/−;Mylk−/− mice. (H) BAL TNFα quantification in Ctrl (n = 3) and VILI (n = 6) mice. Data are presented as mean ± SD. *P < 0.05 compared with WT mice; #P < 0.05 compared with Nrf2−/−;Mylk−/− mice.
Discussion
The role of nmMLCK in EC barrier regulation and inflammatory lung injury has been extensively characterized, with nmMYLK deletion being protective in LPS-induced lung injury and VILI and nmMLCK overexpression in ECs profoundly increasing lung vascular permeability, which can be reversed by nmMLCK enzymatic inhibition (13, 24). The contributions of NRF2 in reducing acute inflammatory lung injury have previously been attributed to enhanced antioxidant and anti-inflammatory responses (1). In this report, we demonstrate that NRF2-mediated lung protective effects extend beyond redox regulation and include repression of MYLK transcription, which in turn improves vascular barrier integrity and reduces lung inflammation in clinically relevant inflammatory mouse models.
NRF2 transcriptionally up-regulates more than 300 target genes by dimerizing with sMAFs and triggering the recruitment of coactivator complexes and other transcription factors to activate gene expression (9). In contrast, MYLK is the first NRF2 target gene shown to be directly repressed by NRF2 in an ARE-dependent manner. This novel mechanism of transcriptional MYLK repression is dependent on the formation of an NRF2-RPA1-ARE-NRE complex, with RPA1 as an NRF2-binding partner that competes with sMAF for NRF2 binding. We hypothesize that an NRF2-sMAF–containing transcriptional activator complex is replaced by an NRF2-RPA1–containing repressor complex that results in NRF2-ARE–dependent gene repression. Genome-wide in silico and RNA-seq analyses revealed NRF2-RPA1-ARE-NRE–mediated repression of transcription as a fundamental gene-regulatory mechanism, with a new subset of NRF2 target genes identified as negatively regulated by NRF2. These results deepen our understanding of the cellular responses mediated by NRF2, as exemplified by the functional importance of the controlled negative regulation of NRF2 target gene, MYLK, in maintaining EC barrier integrity.
ARE transcriptional activity has been reported to be repressed when occupied by NRF3-sMAF, sMAF-sMAF, or BACH1/2-sMAF dimers (26–29). However, these mechanisms fail to explain NRF2-ARE–dependent transcriptional repression, since greater levels of NRF2 would successfully replace sMAF homodimers with NRF2-sMAF heterodimers to activate, rather than repress, the expression of ARE-containing genes. Transcription factors such as ATF3 and RXRα directly bind to NRF2 and repress gene expression (30–32), suggesting the formation of a complex with NRF2 and its sequestration from AREs. This mechanism, however, would indiscriminately repress the entire NRF2 transcriptional program, and it does not address specific ARE-containing gene repression.
Genome-wide profiling of macrophages, astrocytes, liver, and small intestine from mice of diverse genetic backgrounds (Nrf2−/−, Keap1f/f, and Nrf2−/−;Keap1−/−), either under basal conditions or following challenge with NRF2 inducers, implicated NRF2 in the down-regulation of fatty acid and cholesterol synthesis enzymes (ACLY, FABP1, FASN, SCD1, and HMGCS1), transcription factors (PPARA and SREBF1), growth factors (FGF21), proinflammatory cytokines (IL1B and IL6), cell receptors (ERR1 and RON), and DNA nucleases (33–42). However, these genes lack functional AREs and thus likely involve indirect mechanisms of NRF2-mediated repression, a speculation strongly supported by the failure to identify any of these genes by RNA-seq analysis of RPA1−/− and RPA1+/+ A549 cells. It is worth mentioning that no previous study has identified MYLK as a gene repressed by NRF2. While most of the data supplied in this study center on suppression of MYLK expression and the resulting pathological significance, the more far-reaching findings are the in silico and RNA-seq datasets indicating that the NRF2-RPA1-ARE-NRE complex transcriptionally represses other genes as well. Among these are genes implicated in selenoprotein translation (EEFSEC), tumor suppression (RASSF10 and FOCAD), cell growth and proliferation (FAM110B and NAV2), calcium signaling (TPD52L1), cell–cell adhesion (ITGA1 and TANC2), membrane channel regulation (CNIH3), vesicle transport (SYT16), Notch signaling (PCNX1), and the immune response (ADGRG5), again highlighting the broad scope and significance of NRF2 repression of target genes.
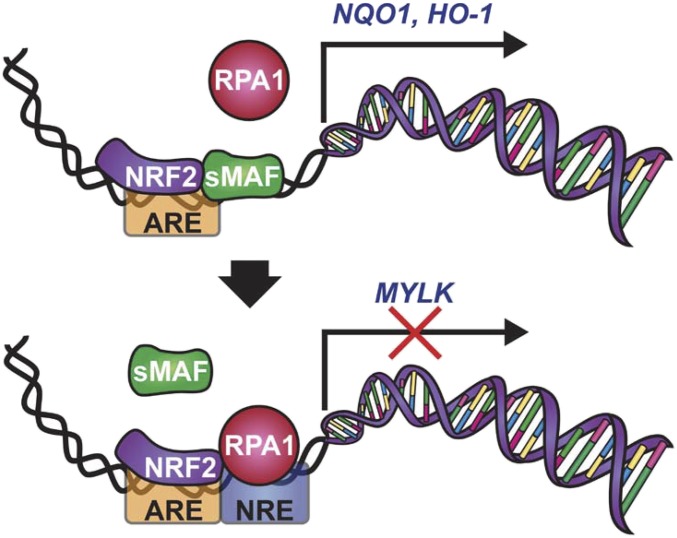
The mechanism of locus-specific NRF2-dependent gene repression identified in this study requires an RPA1-binding NRE adjacent to the 3′ end of the ARE. Flanking site NRE insertion into non-ARE response elements modestly altered gene expression in an NRF2-independent manner, whereas insertion of the NRE adjacent to AREs significantly reduced gene expression, suggesting synergism with NRF2. RPA1 was originally characterized as the largest subunit of the heterotrimeric RPA complex (43, 44). RPA1 is the initial protein recruited to ssDNA during replication, recombination, or DNA damage repair and provides nuclease protection, prevents hairpin formation, and recruits numerous DNA processing proteins via protein–protein interactions (45–47). RPA1 has been suggested to be important for the transcriptional activation of heat shock factor protein and BRCA1 (48, 49). In contrast, RPA1 involvement in transcriptional repression of metallothionein IIA and endothelial nitric oxide synthase has also been reported (50, 51). It has also been suggested that specific binding of RPA1 to dsDNA may depend on its interaction with other DNA-binding proteins independent of the DNA sequence (45, 46). As shown in the model (Fig. 8), our results emphatically support the requirement for the NRE in NRF2-RPA1–dependent gene repression. We believe that both RPA1–NRE (protein–DNA) interaction and RPA1–NRF2 (protein–protein) interaction result in synergistic repression of ARE-NRE–containing genes. This notion is supported by the results shown in Fig. 3 demonstrating much greater RPA1-dependent repression of ARE-NRE promoter activity than non–ARE-NRE promoter activity. On the other hand, the absence of the NRE adjacent to the ARE of NRF2 target genes results in formation of NRF2-sMAF heterodimers that up-regulate expression of ARE-containing genes (Fig. 8). Future studies will be aimed at determining the specific repressor complexes recruited by the NRF2-RPA1 complex, the biological functions of this class of NRF2-repressed genes, and the evolutionary advantage evoked by positively vs. negatively regulated NRF2 target genes.
Fig. 8.
Model of NRF2-RPA1-ARE-NRE–dependent transcriptional repression.
Materials and Methods
All mice were handled according to the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals (52), and the study protocols were approved by the University of Arizona’s Institutional Animal Care and Use Committee.
Detailed descriptions of the study materials and methods are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01 ES026845, R01 DK109555, and P42 ES004940 (to D.D.Z.) and R01 HL91889, P01 HL126609, R01 HL125615, and P01 HL134610 (to J.G.N.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.P.W. is a guest editor invited by the Editorial Board.
Data deposition: RNA-seq data have been deposited in the National Center for Biotechnology Information’s BioProject database, http://www.ncbi.nlm.nih.gov/bioproject487650 (BioProject ID PRJNA487650).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812125115/-/DCSupplemental.
References
- 1.Tao S, et al. Bixin protects mice against ventilation-induced lung injury in an NRF2-dependent manner. Sci Rep. 2016;6:18760. doi: 10.1038/srep18760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mills EL, et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Rojo de la Vega M, Chapman E, Zhang DD. NRF2 and the hallmarks of cancer. Cancer Cell. 2018;34:21–43. doi: 10.1016/j.ccell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitsuishi Y, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 6.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 8.Itoh K, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 9.Tonelli C, Chio IIC, Tuveson DA. 2017 doi: 10.1089/ars.2017.7342. Transcriptional regulation by Nrf2. Antioxid Redox Signal. [DOI]
- 10.Fujiwara KT, Kataoka K, Nishizawa M. Two new members of the maf oncogene family, mafK and mafF, encode nuclear b-Zip proteins lacking putative trans-activator domain. Oncogene. 1993;8:2371–2380. [PubMed] [Google Scholar]
- 11.Kataoka K, et al. Small Maf proteins heterodimerize with Fos and may act as competitive repressors of the NF-E2 transcription factor. Mol Cell Biol. 1995;15:2180–2190. doi: 10.1128/mcb.15.4.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Motohashi H, Katsuoka F, Shavit JA, Engel JD, Yamamoto M. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell. 2000;103:865–875. doi: 10.1016/s0092-8674(00)00190-2. [DOI] [PubMed] [Google Scholar]
- 13.Garcia JG, Davis HW, Patterson CE. Regulation of endothelial cell gap formation and barrier dysfunction: Role of myosin light chain phosphorylation. J Cell Physiol. 1995;163:510–522. doi: 10.1002/jcp.1041630311. [DOI] [PubMed] [Google Scholar]
- 14.Lazar V, Garcia JG. A single human myosin light chain kinase gene (MLCK; MYLK) Genomics. 1999;57:256–267. doi: 10.1006/geno.1999.5774. [DOI] [PubMed] [Google Scholar]
- 15.Petrache I, et al. Caspase-dependent cleavage of myosin light chain kinase (MLCK) is involved in TNF-alpha-mediated bovine pulmonary endothelial cell apoptosis. FASEB J. 2003;17:407–416. doi: 10.1096/fj.02-0672com. [DOI] [PubMed] [Google Scholar]
- 16.Garcia JG, Verin AD, Herenyiova M, English D. Adherent neutrophils activate endothelial myosin light chain kinase: Role in transendothelial migration. J Appl Physiol (1985) 1998;84:1817–1821. doi: 10.1152/jappl.1998.84.5.1817. [DOI] [PubMed] [Google Scholar]
- 17.Dudek SM, Garcia JG. Cytoskeletal regulation of pulmonary vascular permeability. J Appl Physiol (1985) 2001;91:1487–1500. doi: 10.1152/jappl.2001.91.4.1487. [DOI] [PubMed] [Google Scholar]
- 18.Usatyuk PV, et al. Novel role for non-muscle myosin light chain kinase (MLCK) in hyperoxia-induced recruitment of cytoskeletal proteins, NADPH oxidase activation, and reactive oxygen species generation in lung endothelium. J Biol Chem. 2012;287:9360–9375. doi: 10.1074/jbc.M111.294546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao L, et al. Polymorphisms in the myosin light chain kinase gene that confer risk of severe sepsis are associated with a lower risk of asthma. J Allergy Clin Immunol. 2007;119:1111–1118. doi: 10.1016/j.jaci.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 20.Wang T, Zhou T, Saadat L, Garcia JG. A MYLK variant regulates asthmatic inflammation via alterations in mRNA secondary structure. Eur J Hum Genet. 2015;23:874–876. doi: 10.1038/ejhg.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ware LB, Matthay MA. Clinical practice: Acute pulmonary edema. N Engl J Med. 2005;353:2788–2796. doi: 10.1056/NEJMcp052699. [DOI] [PubMed] [Google Scholar]
- 22.Rojo de la Vega M, et al. Role of Nrf2 and autophagy in acute lung injury. Curr Pharmacol Rep. 2016;2:91–101. doi: 10.1007/s40495-016-0053-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papaiahgari S, et al. Genetic and pharmacologic evidence links oxidative stress to ventilator-induced lung injury in mice. Am J Respir Crit Care Med. 2007;176:1222–1235. doi: 10.1164/rccm.200701-060OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirzapoiazova T, et al. Non-muscle myosin light chain kinase isoform is a viable molecular target in acute inflammatory lung injury. Am J Respir Cell Mol Biol. 2011;44:40–52. doi: 10.1165/rcmb.2009-0197OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Byrne BM, Oakley GG. Replication protein A, the laxative that keeps DNA regular: The importance of RPA phosphorylation in maintaining genome stability. Semin Cell Dev Biol. April 20, 2018 doi: 10.1016/j.semcdb.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Sankaranarayanan K, Jaiswal AK. Nrf3 negatively regulates antioxidant-response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. J Biol Chem. 2004;279:50810–50817. doi: 10.1074/jbc.M404984200. [DOI] [PubMed] [Google Scholar]
- 27.Motohashi H, O’Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 28.Oyake T, et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoshino H, et al. Oxidative stress abolishes leptomycin B-sensitive nuclear export of transcription repressor Bach2 that counteracts activation of Maf recognition element. J Biol Chem. 2000;275:15370–15376. doi: 10.1074/jbc.275.20.15370. [DOI] [PubMed] [Google Scholar]
- 30.Brown SL, Sekhar KR, Rachakonda G, Sasi S, Freeman ML. Activating transcription factor 3 is a novel repressor of the nuclear factor erythroid-derived 2-related factor 2 (Nrf2)-regulated stress pathway. Cancer Res. 2008;68:364–368. doi: 10.1158/0008-5472.CAN-07-2170. [DOI] [PubMed] [Google Scholar]
- 31.Wang XJ, Hayes JD, Henderson CJ, Wolf CR. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proc Natl Acad Sci USA. 2007;104:19589–19594. doi: 10.1073/pnas.0709483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, et al. RXRα inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res. 2013;73:3097–3108. doi: 10.1158/0008-5472.CAN-12-3386. [DOI] [PubMed] [Google Scholar]
- 33.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2–dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J Biol Chem. 2003;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 34.Thimmulappa RK, et al. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- 35.Kwak MK, et al. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway: Identification of novel gene clusters for cell survival. J Biol Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- 36.Wu KC, Cui JY, Klaassen CD. Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol Sci. 2011;123:590–600. doi: 10.1093/toxsci/kfr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yates MS, et al. Genetic versus chemoprotective activation of Nrf2 signaling: Overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis. 2009;30:1024–1031. doi: 10.1093/carcin/bgp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitteringham NR, et al. Proteomic analysis of Nrf2 deficient transgenic mice reveals cellular defence and lipid metabolism as primary Nrf2-dependent pathways in the liver. J Proteomics. 2010;73:1612–1631. doi: 10.1016/j.jprot.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi EH, et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat Commun. 2016;7:11624. doi: 10.1038/ncomms11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang C, et al. NRF2 promotes breast cancer cell proliferation and metastasis by increasing RhoA/ROCK pathway signal transduction. Oncotarget. 2016;7:73593–73606. doi: 10.18632/oncotarget.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thangasamy A, Rogge J, Krishnegowda NK, Freeman JW, Ammanamanchi S. Novel function of transcription factor Nrf2 as an inhibitor of RON tyrosine kinase receptor-mediated cancer cell invasion. J Biol Chem. 2011;286:32115–32122. doi: 10.1074/jbc.M111.245746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen B, et al. Curcumin inhibits proliferation of breast cancer cells through Nrf2-mediated down-regulation of Fen1 expression. J Steroid Biochem Mol Biol. 2014;143:11–18. doi: 10.1016/j.jsbmb.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 43.Lin YL, Chen C, Keshav KF, Winchester E, Dutta A. Dissection of functional domains of the human DNA replication protein complex replication protein A. J Biol Chem. 1996;271:17190–17198. doi: 10.1074/jbc.271.29.17190. [DOI] [PubMed] [Google Scholar]
- 44.Bastin-Shanower SA, Brill SJ. Functional analysis of the four DNA binding domains of replication protein A: The role of RPA2 in ssDNA binding. J Biol Chem. 2001;276:36446–36453. doi: 10.1074/jbc.M104386200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen R, Wold MS. Replication protein A: Single-stranded DNA’s first responder. Dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. BioEssays. 2014;36:1156–1161. doi: 10.1002/bies.201400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh KK, Samson L. Replication protein A binds to regulatory elements in yeast DNA repair and DNA metabolism genes. Proc Natl Acad Sci USA. 1995;92:4907–4911. doi: 10.1073/pnas.92.11.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujimoto M, et al. RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Mol Cell. 2012;48:182–194. doi: 10.1016/j.molcel.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 49.Thakur S, et al. Regulation of BRCA1 transcription by specific single-stranded DNA binding factors. Mol Cell Biol. 2003;23:3774–3787. doi: 10.1128/MCB.23.11.3774-3787.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang CM, Tomkinson AE, Lane WS, Wold MS, Seto E. Replication protein A is a component of a complex that binds the human metallothionein IIA gene transcription start site. J Biol Chem. 1996;271:21637–21644. doi: 10.1074/jbc.271.35.21637. [DOI] [PubMed] [Google Scholar]
- 51.Miyamoto Y, et al. Replication protein A1 reduces transcription of the endothelial nitric oxide synthase gene containing a −786T→C mutation associated with coronary spastic angina. Hum Mol Genet. 2000;9:2629–2637. doi: 10.1093/hmg/9.18.2629. [DOI] [PubMed] [Google Scholar]
- 52.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.