Significance
We report optical mapping of DNA from a single cell. Notably, we demonstrate isolation of single cells, DNA extraction, and optical mapping, all within a single integrated micro-/nanofluidic device. Single-cell optical mapping is less complex than sequencing, which we performed after whole-genome amplification of DNA extracted from a single cell isolated on-chip. In some cases, optical mapping was more efficient than sequencing at detecting structural variation. As single-cell analysis can address genomic heterogeneity within a tumor, it may prove useful for the selection of cancer therapies. Thus, optical mapping of the long-range features of single-cell genomes and sequencing of the short-range features may become complementary tools for the analysis of tumors.
Keywords: nanofluidics, single cell, DNA, sequencing, optical mapping
Abstract
To elucidate cellular diversity and clonal evolution in tissues and tumors, one must resolve genomic heterogeneity in single cells. To this end, we have developed low-cost, mass-producible micro-/nanofluidic chips for DNA extraction from individual cells. These chips have modules that collect genomic DNA for sequencing or map genomic structure directly, on-chip, with denaturation–renaturation (D-R) optical mapping [Marie R, et al. (2013) Proc Natl Acad Sci USA 110:4893–4898]. Processing of single cells from the LS174T colorectal cancer cell line showed that D-R mapping of single molecules can reveal structural variation (SV) in the genome of single cells. In one experiment, we processed 17 fragments covering 19.8 Mb of the cell’s genome. One megabase-large fragment aligned well to chromosome 19 with half its length, while the other half showed variable alignment. Paired-end single-cell sequencing supported this finding, revealing a region of complexity and a 50-kb deletion. Sequencing struggled, however, to detect a 20-kb gap that D-R mapping showed clearly in a megabase fragment that otherwise mapped well to the reference at the pericentromeric region of chromosome 4. Pericentromeric regions are complex and show substantial sequence homology between different chromosomes, making mapping of sequence reads ambiguous. Thus, D-R mapping directly, from a single molecule, revealed characteristics of the single-cell genome that were challenging for short-read sequencing.
Sequencing of the genomes of individual isolated cells or nuclei has become well established (1, 2) and is used to resolve intercellular variations in a heterogeneous population of cells from tissues to tumours (3–5). However, it is difficult to accurately delineate the long-range structure of the genome by sequencing alone because of short read lengths and variant detection based on alignment to a reference, which discards a substantial proportion of reads (6). By contrast, single-molecule optical mapping of DNA (7–9) provides a sweeping view of megabase lengths of the genome (10–13). Several schemes exist to locate sequence information along the length of stretched DNA, and some have been combined with superresolution microscopy (14, 15). In early realizations of optical mapping, DNA molecules were stretched on a surface (7, 16). More recently, nanoconfinement has emerged as a powerful technique to stretch DNA molecules uniformly and in a controlled manner without attaching them to a surface (17). Das et al. (18) were able to image sequence-specific fluorescent optical maps in nanochannels with a cross-section below the persistence length of DNA. The combination of nanoconfinenent with additional mechanisms such as entropic recoil (19–21), hydrodynamic drag (22, 23), mechanical confinement (24), or thermophoresis (25) can enhance the degree of stretching.
Although genomic DNA extracted from individual cells has been stretched in micro- (26) and nanofluidic (27) devices, single-molecule optical mapping of DNA from individual cells has not previously been demonstrated. The critical challenge is to couple single-cell genome extraction with a means of handling the DNA at the appropriate length scales and performing the mapping reaction in an integrated device. Stained single genomic DNA molecules can become patterned according to their AT/GC content by partial denaturation and renaturation (D-R) (11). As the DNA double helix opens up, the intercalating dye leaves the AT-rich regions, thus creating dark spots along the DNA strand (see SI Appendix and Fig. 1D). A high degree of stretching, typically above 90%, provided by a flow-stretch device allows imaging of D-R patterns at diffraction-limited resolution. D-R mapping identifies structural variations (SVs) relative to a human reference genome in single copies of the genome (11), and this makes it a unique candidate for optical mapping of single cells.
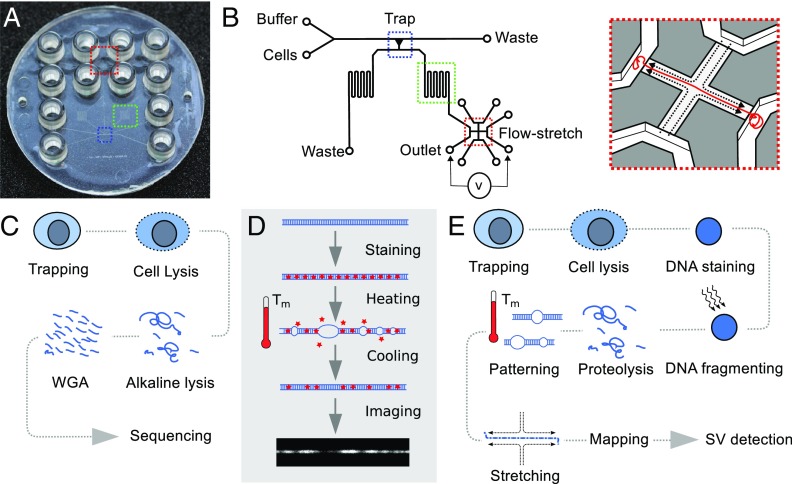
Fig. 1.
Architecture of single-cell processing device and workflow for single-cell D-R mapping and whole-genome sequencing. (A) An all-polymer lab-on-a-chip device with 12 connectors comprises a cell trap (blue), a meandering channel (green), and a flow-stretch device (red). (B) A single cell is captured by hydrodynamic trapping, DNA is extracted and patterned according to AT/GC composition by a heating–cooling cycle, and genomic DNA is stretched and visualized. (C) Workflow using device shown in SI Appendix, Fig. S1 for single-cell trapping and extracting and amplifying DNA before sequencing. (D) Principle of D-R pattern generation: The genomic DNA is homogeneously stained with YOYO-1. During partial denaturation at a temperature between the melting temperatures of AT and GC bonds, , the double helix opens up in AT-rich regions and dye leaves these regions. After renaturation, dark and light segments along the DNA constitute the D-R pattern used for mapping. The D-R map is compared with an in silico map of the reference genome (see SI Appendix). (E) Workflow using device in A and B to prepare genomic DNA for D-R mapping for SV detection.
Results
Using valve-less polymer devices that can be easily mass-produced by injection moulding (e.g., Fig. 1A), we have implemented a microfluidic architecture for trapping and isolating single cells from a population (Fig. 1B and SI Appendix, Fig. S1) and extracting their genomic DNA. We show that this trap design can be aligned within the device to an architecture for harvesting the genomic DNA from a single cell for sequencing outside of the device (Fig. 1C) or, alternatively, to an architecture for D-R mapping of the genomic DNA from a single cell within the device (Fig. 1D). Sequencing of DNA from single cells was enabled by the design shown in SI Appendix, Fig. S1. We fed a population of cells from the LS174T colorectal cancer cell line (SI Appendix, Fig. S2) into the polymer device with a flow design that steered cells toward the side of a microchannel having a series of funnel-shape traps. We stopped the feeding when each individual cell occupied a separate trap. The DNA was then extracted by alkaline lysis and collected in the outlet port for multiple displacement amplification (MDA) and Illumina sequencing. The sequence coverage from single cells was comparable to that obtained from a population of cells (SI Appendix, Fig. S3). As with commercial single-cell processing devices (e.g., Fluidigm C1), using our device for isolating single cells, DNA extraction and DNA collection in the outlets gave superior whole-genome single-cell sequencing results compared with processing single cells without the use of the microfluidic environment: Library preparation was successful for 97% of single-cell samples; there was less than 2% contamination from nonhuman DNA; 87% of bases were represented at a sequencing depth as low as 10×; allelic dropout in the single cells across all high-quality heterozygous variants found in bulk was as low as 30% at 15× to 20× coverage (28).
D-R mapping of DNA from single cells was enabled by the design shown in Fig. 1B and SI Appendix, Fig. S4. The operation of this device is depicted in Fig. 2. We fed cells into the polymer device wherein a single cell was captured in a trap. The DNA in the nucleus was then stained uniformly by adding the fluorescent intercalating dye YOYO-1 to the lysis buffer (SI Appendix, Figs. S5 and S6). A dose of light was used to nick the chromosomal DNA within the nucleus by YOYO-1–induced oxidative damage. Proteolysis was then used to extricate the chromosomal DNA from the nucleus and remove histone proteins (Fig. 3A). This resulted in a significant number of chromosome subfragments approximating megabase lengths. These fragments are easier to manipulate in the microfluidic structure than chromosomal DNA that remains close to its native length. The relationship between dose of light used for photonicking and fragment length is reciprocal because the relationship between dose and nicking probability per kilobase is linear (SI Appendix, Figs. S7–S11). The nuclear lysate was passed through a meandering microchannel (Fig. 3B) in which the DNA was D-R patterned using a cycle of heating and cooling (Fig. 1D). The patterned molecules were then individually threaded into the cross-flow nanoslit using electrophoretic forces, stretched, and imaged at 85% to 100% stretching (Fig. 3C), which was sufficient for them to be mapped to the reference genome GRCh37 (Fig. 3 D and E). The cross-flow structure (Fig. 1B, Inset) is particularly appropriate for the purpose because a useable map can readily be obtained by imaging just one patterned molecule (11). This is important because single-cell mapping requires that the map is derived from a unitary molecule, as there are only two homologous but distinct copies of the genome in a normal diploid cell before replication.
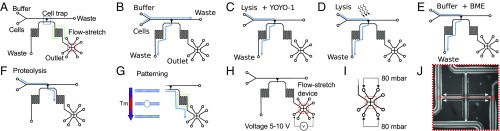
Fig. 2.
Device operation to realize the workflow shown in Fig. 1D. (A) The device comprises a main channel with inlets for cells and buffer, a cell trap connecting the main channel to the outlets. On the outlet side, a flow stretch device is placed. (B) Cells are introduced, and a single cell is captured in the trap. The flow is stopped when a cell occupies the trap. (C) Lysis buffer is introduced to remove the cytoplasm. The lysis solution contains YOYO-1 to stain the DNA of the nucleus. (D) The stained nucleus is illuminated with blue light (480 nm) to induce photonicking. (E) Buffer with BME is introduced to prevent further photonicking. (F) Same buffer to which protease K is added is introduced to release the genomic DNA by proteolysis. (G) When the DNA reaches the meandering channel, temperature is raised to partially denaturate the DNA before the device is cooled again. (H) DNA molecules are introduced to the flow-stretch device by electrophoresis and (I) stretched by a flow of buffer at a pressure of 80 mbar. (J) Brightfield image of the flow-stretch device indicating the buffer flow (dotted line) and the position of the DNA (red line). The nanoslit is 450 m across.
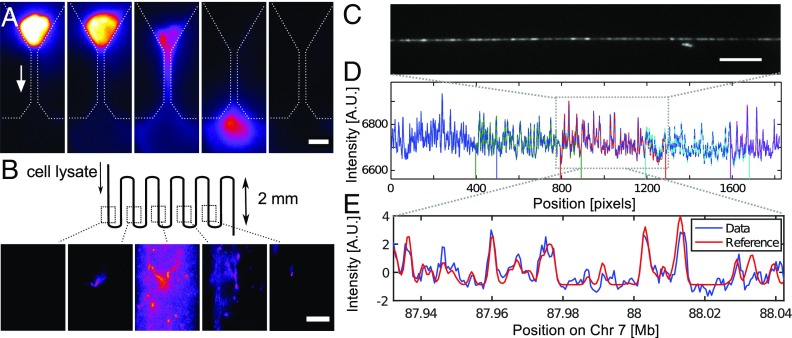
Fig. 3.
Extraction and mapping of DNA from a trapped cell. (A) Proteolysis liberates the genomic DNA from the trapped nucleus under flow (arrow). (Scale bar, 10 m.) (B) DNA is collected as a 10-nL plug of solution in a section of the device where D-R is performed. (Scale bar, 10 m.) (C) Single field-of-view of a segment of a stretched DNA molecule. (Scale bar, 10 m.) (D) Full D-R map of a molecule is stitched together from five fields-of-view. (E) Match between the D-R map (pixel 900 to 1100) and a position on Chr7 of computer-simulated whole-genome melting map.
We sampled 21 molecules of 1 Mb length from one cell and found that in 17 out of 21 cases, the molecule could be mapped to unique coordinates in the GRCh37 reference genome (see SI Appendix, Table S1). Mapping is achieved by comparing the experimental D-R map and the in silico map generated for the entire reference genome by calculating the sum of squared differences (11). The certainty of the match arises from multiple independent D-R map segments mapping on the genome at predictable distances from each other (i.e., they fall on a diagonal in the plots displayed in SI Appendix, Figs. S12–S20). When mapped against the mouse genome (GRCm38/mm10), we found no such match for the experimental D-R maps. Fig. 3 D and E shows the D-R map derived from the image of one of the molecules extracted from the single cell and its matching to the in silico-generated melting map of a reference genome. Two of the molecules that did not match anywhere along the reference genome lacked a pattern that was rich enough in features to map back to the genome (see SI Appendix, Fig. S21). This complication might arise because all molecules were denatured at a single temperature. This temperature had been found to yield informative D-R maps for most molecules. But a uniformly GC-rich region can appear featureless and bright at this temperature, while a uniformly AT-rich region can appear featureless and dark (see SI Appendix, section 2E). The two other molecules that did not find a match in the reference genome did contain sufficient patterning for mapping to the reference; our inability to find a match for these molecules, or even parts of the molecules, suggests that they came from the fraction of the genome that remains uncharted or is subject to extreme structural change (29), which renders it unmappable and is therefore not represented in the reference genome (SI Appendix, section 2C).
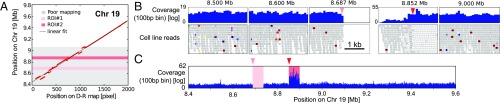
A number of the molecules that mapped to the genome had peaks in their values that coincide with line shifts larger than a few kilobases in their plots of the genome versus D-R map position (SI Appendix, Figs. S12–S20). These peaks indicate poor alignment to the reference; however, if no line shift is detected, we cannot pinpoint the nature of the SV (11). Insertions and deletions down to 5 kb can be detected with high confidence (for the SV detection sensitivity of D-R mapping, see SI Appendix, section 3). Significant SV was detected on molecule 17 by D-R mapping (SI Appendix, Table S1); half of the 1 Mb-long molecule mapped poorly to Chr19:8.5 to 9.1 Mb (SI Appendix, Fig. S22 and Fig. 4A). This is consistent with the LS174T whole-genome sequencing data. The read-depth analysis suggests a 50-kb deletion at 8.687 to 8.737 Mb, which is likely homozygous, as well as a 50-kb complex region around 8.87 Mb characterized by a higher read depth but with a poor mapping of the reads (Fig. 4 B and C). In this case, single-cell D-R mapping data and bulk sequencing concur and point to a complex region. However, the poor agreement of the D-R map with the reference genome leading to a high -value background does not allow the SV detection by D-R mapping alone (SI Appendix, section 2B).
Fig. 4.
Optical D-R map and paired-end sequencing on Chr 19. (A) Plot of sliding window analysis of experimental versus in silico D-R maps, revealing a region where the experimental D-R map aligns poorly to the reference. (B) The coverage and the visual analysis of the reads in the Integrative Genomics Viewer (IGV) show three normal regions and two ROIs in this region, one is a homozygous deletion and one is a complex region. (C) The coverage of reads over the whole fragment.
We found two segments clearly missing in molecule 12 (8 and 12 kb) at 51.012 and 51.513 Mb. Their absence could not be detected in the bulk or the single-cell data. However, we found a 20-kb region limited by two regions of reads with an incorrect insert size centered at the position of interest, 51.012 Mb. At the other position of interest, in one of the single-cell sequencings, we see two deletions smaller than a kilobase positioned at 23 kb on each side of the position 51.510 Mb. Both regions are shown in SI Appendix, Figs. S23 and S24. In this case, the D-R mapping’s SV detection points to SVs detected by sequencing but without a one-to-one correspondence. This lack of one-to-one correspondence may be due to the fact that it is not the very same cell that we D-R map and sequence.
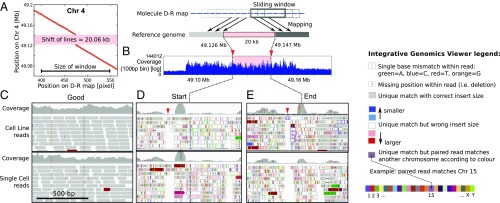
A molecule from another single cell was found to map to the pericentromeric region of Chr4, but a 20-kb segment was absent between Chr4:49.127 and 49.147 Mb on GRCh37 (region of interest, ROI) (Fig. 5A). The sliding-window analysis showed a clear signature for the absence of the segment (11). Whole-genome sequencing indicates that the ROI is situated in a 66-kb region with high but uneven coverage in both the bulk and single-cell sequencing data (Fig. 5B). Single-nucleotide variants, indels, and gaps were pervasive around the ROI and varied between individual cells (SI Appendix, Fig. S25); this intercell variation would be impossible to resolve by bulk sequencing.
Fig. 5.
Optical D-R map and paired-end sequencing on Chr 4. (A) Plot of sliding window analysis, revealing a 20-kb discontinuity in the experimental D-R map compared with reference: This is the ROI. (B) Coverage plot of the cell line bulk-sequencing data displayed with a 100-bp bin around the molecule, including the ROI. (C–E) IGV plot of read-pair data for three genomic regions in the bulk sample and a single cell comprising (C) an arbitrary 1-kb region 1 Mb before the ROI, (D) 1 kb at the start of the ROI, and (E) 1 kb at the end of the ROI. For each case, the coverage is shown in gray in the top row. A stack of reads is shown in the rows below (arrows indicating orientation); gray indicates that both reads of the pair are consistent with the reference, including orientation and insert size; blue indicates a decrease and red color an increase in insert size between read pairs on the same chromosome; the color palette indicates that one read of the pair is on another chromosome according to the color key provided; vertical colored lines within reads indicate the identity of a single-nucleotide polymorphism (with color code: A, green; C, blue; T, red; G, orange); the block dots or short line within reads indicate insertions. The red downward arrowheads point to a gap in reads that is present in all single-cell and bulk data in the vicinity of the start and end of the ROI.
To confirm the verity of our single-cell sequencing data, we looked at an arbitrary position in the genome (Fig. 5C) and found high coverage, low sequence variation, and alignment of both reads to the expected locations in the reference (see also ref. 28). This contrasted to the substantial discordance between read pairs we found in the ROI, with one read mapping to the ROI while the other did not (Fig. 5 D and E and SI Appendix, Fig. S26). This paints a picture of local genome instability. This is unsurprising given the region’s pericentromeric location where poorly delineated tracts of satellite and nonsatellite sequences are known to span hundreds of kilobases (30). Segmental duplications are enriched in these regions and may seed copy-number changes through errors in homologous recombination.
Not unexpectedly, pericentromeric regions are poorly represented in reference genomes, and sequence reads are difficult to align and would typically be discarded. Although we found that some reads in our data could be mapped to this region, the same reads found matches to several other locations in pericentromeric regions of the genome (SI Appendix, Table S3). Clearly, the mapping of the single-cell sequencing data for this region could not be easily interpreted. This makes the clear detection of the missing segment by D-R mapping highly significant and shows that our approach can provide answers in regions where sequencing does not.
Discussion
A substantial proportion of somatic SVs in cancer genomes are complex events and are rich in interchromosomal rearrangements (29, 31). Short-read sequencing does not easily reveal mid- to long-range complex structural changes without a substantial bioinformatics effort (32). By contrast, the megabase-range views of the genome provided by single-cell D-R optical mapping detect such changes.
We have shown that D-R mapping can determine that a region of the genome (on Chr19) in a LS174T cell has a 0.6-Mb region that matches poorly to the reference and that our single-cell sequencing data are consistent with this. However, D-R mapping of a megabase fragment from another single cell shows the absence of a 20-kb segment on Chr4, which is not picked up by single-cell read-depth or read pair sequence analysis. However, single-cell sequencing does suggest that this is a complex, unstable region of the genome that sequencing has not been able to chart accurately. Computational reconstruction of the region by paired reads is unproductive because the region is not well mapped in the reference and has a high degree of similarity with other parts of the genome. By contrast, with D-R mapping, we are able to directly visualize the structure of the fragment and unequivocally assign its genome location by comparing its D-R melting map with an in silico-generated melting map of the human reference genome (SI Appendix, section 2); most of the megabase fragments match with high confidence to Chr4, but if the reference genome is an accurate representation of this region, a 20-kb segment is clearly deleted in this fragment or translocated to another location. Because of the visual nature of the D-R experimental data, there is little doubt that what we are looking at is real and not an artifact. The LS174T cell line has a 45,X karyotype, and spectral karyotyping does not reveal any large-scale rearrangements (33, 34). Moreover, bulk SNP analysis of Chr19 does not find microdeletions (SI Appendix, Table S4). The single-cell D-R mapping might therefore be providing information on an uncommon variant. Single-cell D-R mapping thus expands the tools available to detect genomic heterogeneity, and such a capability might help provide deeper insight into the structure and dynamics of a genomic region and may aid the mapping of disease genes.
The modest throughput of our current device design is balanced by the fact that we obtain a map from just one molecule. While other nanofluidic platforms may currently be able to process more molecules, the optical maps generated rely intrinsically on the data from several overlapping molecules and thus do not have the true single-molecule sensitivity required to map genomic DNA from single cells. Moreover, analysis based on individual megabase molecules means that we can resolve the long-range structure of haplotypes in a diploid genome. With the application of our full automation method (35), it will be possible to apply D-R optical mapping to whole single-cell genomes. Single-cell D-R mapping will then have the potential to simply and inexpensively screen whole-genome SV between cells as a complement or alternative to sequencing.
Materials and Methods
Optical Mapping Device Design.
The device architecture for optical mapping is summarized in Fig. 1B. Microchannels are designed to accommodate cells from a human cancer cell line with a typical diameter of 15 m (SI Appendix, Fig. S2), so the microfluidic network has minimal depths and widths of 33 m. The device comprises an inlet for cells and an inlet for buffer (Fig. 1B) that merge into a single channel to feed the single-cell trap (blue box in Fig. 1B). At the intersection between the cell and buffer inlets, cells get aligned along the side wall of the feeding channel where the trap is located. The trap is a simple constriction dimensioned to capture cells from a human cancer cell line. The constriction for cell trapping has a trapezoidal cross-section: It is 4.3 m wide at the bottom, 6 m at middle depth, and 8 m at the top with a depth of 33 m. The cell trap connects the feeding channel with the outlet channels, which consist of two meandering channels: one for waste and the other (green box in Fig. 1B) leading to the outlet and the flow-stretch device (red box in Fig. 1B). The flow-stretch device consists of a 20 m-wide, 450 m-long, 100 nm-deep cross-shaped nanoslit that connects the outlet channel to three other microchannels (11).
Optical Mapping Device Fabrication.
The device is fabricated by replicating a nickel shim using injection molding of TOPAS 5013 (TOPAS) as previously reported (36, 37). Briefly, a silicon master is produced by UV lithography and reactive ion etching. A 100-nm NiV seeding layer is deposited and nickel is electroplated to a final thickness of 330 m. The Si master is chemically etched away in KOH. Injection molding is performed using a melt temperature of C, a mould temperature of C, a maximum holding pressure of 1,500 bar for 2 s, and an injection rate varying between 20 /s and 45 /s. Finally, a 150 m TOPAS foil is used to seal the device by a combined UV and thermal treatment under a maximum pressure of 0.51 MPa. The surface roughness of the foil is reduced by pressing the foil at C and 5.1 MPa for 20 min between two flat nickel plates electroplated from silicon wafers before sealing the device. This ensures that the lid of the device is optically flat, allowing for high-NA optical microscopy. The device is mounted on an inverted fluorescence microscope (Nikon T2000) equipped with an air objective (50×/0.8), an oil objective (60×/1.40), and an EMCCD camera (Photometrics cascade II 512). Fluids are driven through the device using a pressure controller (MFCS, Fluigent) at pressures in the 0 to 10 mbar range. The device is primed with ethanol, and then degassed FACSFlow Sheath Fluid (BD Biosciences) is loaded in all microchannels but the microchannel connecting the flow-stretch device. Instead, a buffer suitable for single-molecule imaging and electrophoresis (0.5× TBE + 0.5% v/v Triton-X100 + 1% v/v -mercaptoethanol, BME) is loaded in the channels of the flow-stretch device. This buffer prevents DNA sticking in the nanoslit of the flow-stretch device and suppresses electroosmotic flow that can counteract the introduction of DNA to the nanoslit later in the experiment.
Cells.
LS174T colorectal cancer cells are cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) with 10% fetal bovine serum (FBS; Autogen-Bioclear UK Ltd.) and 1% penicillin/streptomycin (Lonza) before freezing at a concentration of 1.7 cells per milliliter in 10% DMSO in FBS. After thawing, cell suspension is mixed 1:1 with FACSFlow buffer, centrifuged at 28.8 × g (A-4-44, Eppendorf) for 5 min, and resuspended in FACSFlow. Finally, the cells are stained with 1 M Calcein AM (Invitrogen) and loaded in the chip at 0.35 cells per milliliter. So out of the 7,000 cells loaded, the first cell trapped is analyzed.
Optical Mapping Device Operation.
The protocol for operating the device is sketched in Figs. 1D and 2 A–I. First, cells and buffer are introduced simultaneously, aligning the cells along the side wall of the microchannel where the trap is located. A cell is captured and kept in the trap for a buffer flow through the trap up to 30 nL/min. Cell doublets are present in the cell culture and may be captured in the cell trap. In this case, the experiment is aborted. The lysis buffer composed of 0.5× TBE + 0.5% v/v Triton-X100 + 0.1 M YOYO-1 (Invitrogen) is loaded in one of the inlets and injected at 10 nL/min through the trap for 10 min (see SI Appendix). Then, the solution is exchanged to a buffer without YOYO-1 in all wells to stop the staining. Next, the cell nucleus is exposed to blue excitation light at a dose of 1 nW/(m)2 for up to 300 s, causing a partial photonicking of the DNA (see SI Appendix). Then, the buffer is changed to a solution containing BME (0.5× TBE + 0.5% v/v triton-X100 + 1% v/v BME), and the excitation of the fluorescence lamp is lowered to the minimum intensity that still allows fluorescence imaging. Finally, the temperature is raised to C, and the proteolysis solution (0.5× TBE + 0.5% v/v Triton-X100 + 1% v/v BME + 200 g/mL) is introduced, pushing the lysate through the trap. In experiments where a denaturation pattern is created on DNA extracted from a single cell, the DNA staining is modified. Thus, the lysis buffer contains 0.04 M YOYO-1 instead of 0.1 M. In addition, the device is heated further after the proteolysis for 10 min at C before rapid cooling to room temperature by moving the oil immersion objective in place. The temperature at which the D-R pattern is created is highly dependent on the dye loading and the ionic strength of the buffer and should be adjusted by steps of C if necessary. DNA travels through the meandering channel as the temperature is lowered, and an oil immersion objective is moved into place for single-molecule imaging (60×/1.4, with a 1.5× lens giving a 180-nm pixel size). DNA fragments are introduced from the microchannel to the nanoslit of the flow-stretch device using electrophoresis by applying a voltage of 5 to 10 V across the nanoslit. When a DNA fragment has both ends in opposite microchannels, voltage is turned off, and the final stretching of the DNA is obtained by the flow stretch at a pressure drop of 80 mbar (11).
Imaging and Mapping.
Imaging and mapping are performed as previously reported, and the match on the human reference genome is validated as shown in detail in the supplementary information to ref. 11. Briefly, the 450-m portion of the molecule stretched in the nanoslit is imaged using 50-ms exposures in five overlapping fields-of-view. Since the DNA is at a predictable position in the nanoslit, this can be done using simple automation of the motorized stage and image acquisition (Metamorph, Molecular Devices) (11). In each field-of-view, the D-R pattern along the DNA (intensity averaged over 3 to 5 pixel width) was extracted from the average of 20 frames. Consecutive fields-of-view are stitched together using a chi-square fit (Matlab) to produce a continuous D-R pattern covering the entire length of the nanoslit. Mapping of the experimental D-R map with the theoretical melting profile for the whole human genome (GRCh37) produced with the software Bubblyhelix (www.bubblyhelix.org) (38) is according to the supplementary information in ref. 8.
Device Design for DNA Extraction.
In experiments, where DNA is prepared for sequencing, LS174T cells are trapped in a separate device fabricated as described above and comprising only cell traps (and not the flow-stretch part) so that it allows processing several cells in parallel. Detailed description of the following can be found in ref. 28. The device consists of a microfluidic network at a single depth (31 m), and cell traps have a geometry similar to the trap in the device for optical mapping (i.e., they are 4.5 m-wide constrictions). The device architecture is shown in SI Appendix, Fig. S1. On this device, eight single-cell traps are fed through a common channel and lead to separate outlets. Cells introduced to the common channel are aligned along the side wall of the channel by a flow of buffer incoming from inlet B1 and B2.
Device Operation and DNA Amplification.
The device is wetted with degassed 0.1% Triton-X100 in BD FACSFlow buffer followed by degassed 0.1 mg/mL BSA in BD FACSFlow buffer and left in the fridge overnight. Cells are loaded in the cell inlet, and BD FACSFlow is loaded in all other wells. Cells are trapped by applying 4 mbar to the cell and 5 mbar to the B1 and B2 inlets until single cells occupy the traps. The trapped cells are lysed by loading the alkaline lysis solution (Repli-g kit, Qiagen) in inlets B1 and B2. Lysis occurs at room temperature by applying 5 to 10 mbar to B1 and B2, and DNA is pushed in separate outlets of the device. After adding neutralization solution, MDA is performed in the outlet wells using the Repli-g Mini kit. MasterMix with Phi29 polymerase was added to the chip outlets containing gDNA, and the device is placed in a thermal cycler for 8 h at C. Finally, polymerase is inactivated 5 min at C. The amplified DNA is collected and quantified using Qubit dsDNA HS kit (Thermo Fischer). DNA from single cells, one of them presented in Fig. 5 C–E, was extracted on different devices, on different days.
DNA Sequencing.
To confirm the quality of whole genome amplification (WGA) products, multiplex PCR is performed on the amplified DNA. Five primers targeting five chromosomes (2, 4, 12, 13, 22) are used. PCR products (295 bp Chr13, 235 bp Chr12, 196 bp Chr2, 150 bp Chr22, and 132 bp Chr4) are visualized on a 4% agarose gel. The DNA material that passes quality controls—that is, five bands on gel after multiplex PCR—was then prepared for sequencing. Library preparation for next-generation sequencing was done using Nextera DNA Library Prep kit (Illumina) with 10 ng of amplified DNA as input. Illumina paired-end sequencing (2 100 bp or 2 125 bp) was performed on HiSeq 2000/2500 instruments using HiSeq SBS and Cluster kits (v3 or v4 chemistry). For comparison, we also prepared a library from DNA extracted off-chip from the LS174T cell line. For this sample (cell line in Figs. 4 B and C and 5 B–E), the library was prepared using the Illumina TruSeq Genomic kit (this is because, with a larger amount of DNA, this is the best preparation method and represents the gold standard of Illumina sequencing) but was run on four of the Illumina HiSeq high output lanes, with 2 100 bp paired-end reads, using the v3 chemistry. Read alignment is performed on the GRCh37 genome and visualized in the IGV (39).
Supplementary Material
Acknowledgments
We thank Charles Cantor and Wilhelm Ansorge for advice. This work was supported by European Union’s Seventh Framework Programme FP7/2007-2013 Grant 278204 (CellOMatic) and Danish Council for Strategic Research Grant 10-092322 (PolyNano).
Footnotes
Conflict of interest statement: R.M., J.N.P., A.K., and K.U.M. filed patents. K.U.M. declares that XGenomes is developing nucleic acid sequencing technologies.
This article is a PNAS Direct Submission.
Data deposition: The single-cell sequencing data are available at the European Nucleotide Archive (accession nos. ERS2168085–ERS2168123), https://www.ebi.ac.uk/ena/browse.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804194115/-/DCSupplemental.
References
- 1.Navin NE. Delineating cancer evolution with single-cell sequencing. Sci Transl Med. 2015;7:296fs29. doi: 10.1126/scitranslmed.aac8319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: Current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 3.McConnell MJ, et al. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheow LF, et al. Single-cell multimodal profiling reveals cellular epigenetic heterogeneity. Nat Methods. 2016;13:833–836. doi: 10.1038/nmeth.3961. [DOI] [PubMed] [Google Scholar]
- 5.Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501:355–364. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwartz DC, et al. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science. 1993;262:110–114. doi: 10.1126/science.8211116. [DOI] [PubMed] [Google Scholar]
- 8.Reisner W, et al. Single-molecule denaturation mapping of DNA in nanofluidic channels. Proc Natl Acad Sci USA. 2010;107:13294–13299. doi: 10.1073/pnas.1007081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neely RK, et al. DNA fluorocode: A single molecule, optical map of DNA with nanometre resolution. Chem Sci. 2010;1:453–460. [Google Scholar]
- 10.Teague B, et al. High-resolution human genome structure by single-molecule analysis. Proc Natl Acad Sci USA. 2010;107:10848–10853. doi: 10.1073/pnas.0914638107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marie R, et al. Integrated view of genome structure and sequence of a single DNA molecule in a nanofluidic device. Proc Natl Acad Sci USA. 2013;110:4893–4898. doi: 10.1073/pnas.1214570110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hastie AR, et al. Rapid genome mapping in nanochannel arrays for highly complete and accurate de novo sequence assembly of the complex aegilops tauschii genome. PLoS One. 2013;8:e55864. doi: 10.1371/journal.pone.0055864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freitag C, et al. Visualizing the entire DNA from a chromosome in a single frame. Biomicrofluidics. 2015;9:044114. doi: 10.1063/1.4923262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baday M, et al. Multicolor super-resolution DNA imaging for genetic analysis. Nano Lett. 2012;12:3861–3866. doi: 10.1021/nl302069q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vranken C, et al. Super-resolution optical DNA mapping via DNA methyltransferase-directed click chemistry. Nucleic Acids Res. 2014;42:e50. doi: 10.1093/nar/gkt1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bensimon A, et al. Alignment and sensitive detection of DNA by a moving interface. Science. 1994;265:2096–2098. doi: 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- 17.Tegenfeldt JO, et al. The dynamics of genomic-length DNA molecules in 100-nm channels. Proc Natl Acad Sci USA. 2004;101:10979–10983. doi: 10.1073/pnas.0403849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das SK, et al. Single molecule linear analysis of DNA in nano-channel labeled with sequence specific fluorescent probes. Nucleic Acids Res. 2010;38:e177. doi: 10.1093/nar/gkq673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeh JW, Taoni A, Chen YL, Chou CF. Entropy-driven single molecule tug-of-war of DNA at micro-nanofluidic interfaces. Nano Lett. 2012;12:1597–1602. doi: 10.1021/nl2045292. [DOI] [PubMed] [Google Scholar]
- 20.Jo K, et al. A single-molecule barcoding system using nanoslits for DNA analysis. Proc Natl Acad Sci USA. 2007;104:2673–2678. doi: 10.1073/pnas.0611151104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kounovsky-Shafer KL, et al. Presentation of large DNA molecules for analysis as nanoconfined dumbbells. Macromolecules. 2013;46:8356–8368. doi: 10.1021/ma400926h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perkins T, Smith D, Chu S. Single polymer dynamics in an elongational flow. Science. 1997;276:2016–2021. doi: 10.1126/science.276.5321.2016. [DOI] [PubMed] [Google Scholar]
- 23.Cipriany BR, et al. Real-time analysis and selection of methylated DNA by fluorescence-activated single molecule sorting in a nanofluidic channel. Proc Natl Acad Sci USA. 2012;109:8477–8482. doi: 10.1073/pnas.1117549109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berard DJ, et al. Convex lens-induced nanoscale templating. Proc Natl Acad Sci USA. 2014;111:13295–13300. doi: 10.1073/pnas.1321089111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedersen JN, et al. Thermophoretic forces on DNA measured with a single-molecule spring balance. Phys Rev Lett. 2014;113:268301. doi: 10.1103/PhysRevLett.113.268301. [DOI] [PubMed] [Google Scholar]
- 26.Benitez JJ, et al. Microfluidic extraction, stretching and analysis of human chromosomal DNA from single cells. Lab Chip. 2012;12:4848–4854. doi: 10.1039/c2lc40955k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahshid S, et al. Development of a platform for single cell genomics using convex lens-induced confinement. Lab Chip. 2015;15:3013–3020. doi: 10.1039/c5lc00492f. [DOI] [PubMed] [Google Scholar]
- 28.Marie R, et al. Sequencing of human genomes extracted from single cancer cells isolated in a valveless microfluidic device. Lab Chip. 2018;18:1891–1902. doi: 10.1039/c8lc00169c. [DOI] [PubMed] [Google Scholar]
- 29.Stephens PJ, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horvath J, Schwartz S, Eichler E. The mosaic structure of human pericentromeric dna: A strategy for characterizing complex regions of the human genome. Genome Res. 2000;10:839–852. doi: 10.1101/gr.10.6.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang L, et al. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell. 2013;153:919–929. doi: 10.1016/j.cell.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baslan T, et al. Genome-wide copy number analysis of single cells. Nat Protoc. 2012;7:1024–1041. doi: 10.1038/nprot.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rutzky L, Kaye C, Siciliano M, Chao M, Kahan B. Longitudinal karyotype and genetic signature analysis of cultured human-colon adenocarcinoma cell-lines LS180 and LS174T. Cancer Res. 1980;40:1443–1448. [PubMed] [Google Scholar]
- 34.Abdel-Rahman WM, et al. Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proc Natl Acad Sci USA. 2001;98:2538–2543. doi: 10.1073/pnas.041603298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sørensen KT, et al. Automation of a single-DNA molecule stretching device. Rev Sci Instrum. 2015;86:063702. doi: 10.1063/1.4922068. [DOI] [PubMed] [Google Scholar]
- 36.Utko P, Persson F, Kristensen A, Larsen NB. Injection molded nanofluidic chips: Fabrication method and functional tests using single-molecule DNA experiments. Lab Chip. 2011;11:303–308. doi: 10.1039/c0lc00260g. [DOI] [PubMed] [Google Scholar]
- 37.Østergaard PF, et al. Optical mapping of single-molecule human DNA in disposable, mass-produced all-polymer. J Micromech Microeng. 2015;25:105002. [Google Scholar]
- 38.Tøstesen E, Liu F, Jenssen TK, Hovig E. Speed-up of DNA melting algorithm with complete nearest neighbor properties. Biopolymers. 2003;70:364–376. doi: 10.1002/bip.10495. [DOI] [PubMed] [Google Scholar]
- 39.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.