Abstract
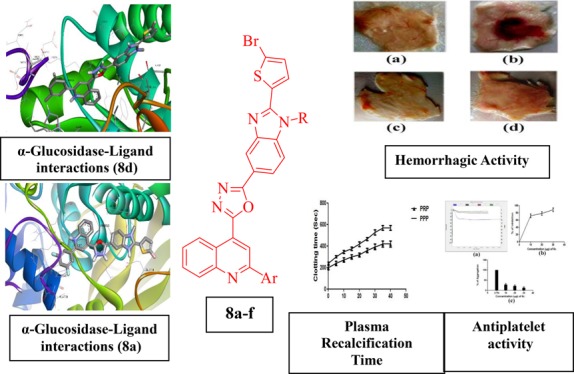
The current study evaluates antidiabetic, anticoagulant, and antiplatelet activity of novel benzimidazole-containing quinolinyl oxadiazoles. These derivatives are synthesized and characterized using spectroscopy (FT-IR, 1H NMR, and mass spectroscopy) and single-crystal X-ray diffraction methods. The inhibitory effects of these compounds were evaluated by the α-glucosidase inhibitory assay and shows the activity in the range of IC50 = 0.66 ± 0.05 to 3.79 ± 0.46 μg/mL. In addition, molecular docking studies revealed that benzimidazole-containing quinolinyl oxadiazoles can correctly dock into the target receptor protein of the human intestinal α-glucosidase, while their bioavailability/drug-likeness was predicted to be acceptable but requires further optimization. On the other hand, compound 8a and 8d showed anticoagulant activity as they enhanced the clotting time from control 180–410 and 180–390 s, respectively, in platelet rich plasma and 230–460 and 230–545 s in platelet poor plasma. Furthermore, only 8a showed antiplatelet activity by inhibiting epinephrine-induced platelet aggregation, and the observed aggregation inhibition was found to be 93.4%. Compounds 8a–f show nontoxic properties because of the non-hydrolyzing properties in the RBC cells. In addition, 8a and 8d show anti-edema and anti-hemorrhagic properties in the experimental mice. These findings reveal that benzimidazole-containing quinolinyl oxadiazoles act as α-glucosidase inhibitors to develop novel therapeutics for treating type-II diabetes mellitus and can act as lead molecules in drug discovery as potential antidiabetic and antithrombotic agents.
Introduction
As one of the chronic diseases, diabetes mellitus is expanding worldwide rapidly. It is characterized by high blood sugar levels for a longer period.1 This uncontrolled hyperglycemia can cause serious damage to many vital organs in the body, including the kidneys, heart, and nervous tissues.2,3 Postprandial hyperglycemia has emerged as a prominent and early defect in type-2 diabetes and as a predictor of cardiovascular or even all-cause mortality, as well as an independent risk factor for atherosclerosis.4
The membrane-bound enzyme, α glucosidase, is found in the epithelium of the small intestine. It cleaves α-1,4 glycosidic linkage at the nonreducing end of starch and disaccharides to release glucose units. Diabetes is caused with increasing glucose levels in the blood stream and relatively increases postprandial blood glucose levels.5−7 Inhibition of α-glucosidase is involved in the reduction of glucose absorption (rate) in the intestine and further decreasing plasma glucose levels. With these properties, α-glucosidase (therapeutic target) is used for the modulation of postprandial hyperglycemia in type-2 diabetes.8 The inhibitors, acarbose, miglitol, and voglibose, used to inhibit α-glucosidase are used to control postprandial blood glucose levels in type-2 diabetic patients.9,10
Over the past decade, α-glucosidase inhibitors, such as acarbose, miglitol, and voglibose, have been used to decrease the postprandial blood glucose levels in type-2 diabetic patients.9,10 Furthermore, various studies show that the molecules which exhibit the α-glucosidase inhibitor property are also used in treating cancer, HIV, virus, and tumors.11−13 Therefore, the present scenario needs a proper designing and synthesizing of better α-glucosidase inhibitors to enrich the discipline of medicinal chemistry.
Blood plays a pivotal role in supplying micronutrients and macronutrients to different parts of the body.14 Meanwhile, blood oozing out during a vascular injury leads to many consequences. Thus, prevention of blood loss plays a major role in physiological condition and the phenomenon is termed as hemostasis, which is a highly regulated pathway.15 Some environmental and genetic factors may alter the hemostatic pathway, which leads to thrombosis.16 Thrombosis is nothing but the generation of unusual clots in arteries and veins, which is a major cause for death around the world.17 Whereas, antiplatelet and anticoagulant agents play a major role in treating thrombotic disorders.18 The present study finds the beneficial role of benzimidazole-containing quinolinyl oxadiazoles on thrombotic disorders.
Several quinoline derivatives show various pharmacological activities such as antifungal,19,20 antimalarial,21 antibacterial,22 anthelmintic,23,24 anticancer,25,26 anticonvulsant,27 anti-inflammatory,28 analgesic,29 and antihyperglycemic activities.30−32 The quinoline derivatives isolated from natural systems have been reported for potent α-glucosidase inhibition.33−36
On the other hand, benzimidazoles have potency for the anti-inflammatory, antisplasmodic, analgesic, antihistaminic, antidiabetic, antimicrobial, antitubercular, antiproliferative, antitumor, anti-HIV-RT, anticancer, antiulcer, and cyclooxygenase inhibitor activities.37−46 The benzimidazole derivatives also exhibited α-glucosidase47−49 as well as antidiabetic50 activity; hence, it is important to obtain novel benzimidazole derivatives as antidiabetic compounds. There are only limited studies of benzimidazole derivatives used on α-glucosidase inhibitory (AGI) activity and cytotoxicity.51
It was reported that oxadiazoles act as a potential class of α-glucosidase inhibitors34,52,53 and are found to have pharmacological activities such as anticancer,54,55 antimicrobial,56−58 anti-inflammatory,59 anticonvulsant,60 antioxidant,61 and antidiabetic.34,62 The recent report on 1,3,4-oxadiazole scaffolds discusses potential antiproliferative agents.63−65
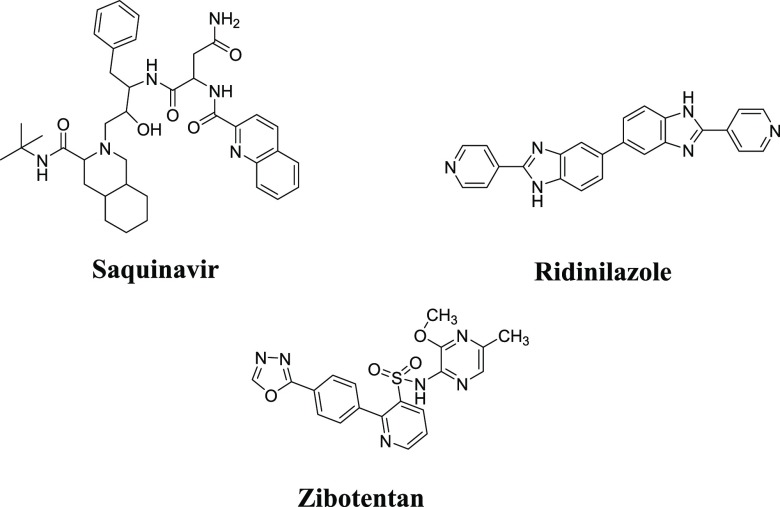
Drugs in market containing quinoline, benzimidazole, and 1,3,4-oxadiazole as a heterocycle shown in the Figure 1.
Figure 1.
Drugs in market containing quinoline, benzimidazole, and 1,3,4-oxadiazole moieties.
In view of the above observations in the quest to design better drugs for diabetes and thrombotic disorders in our current work, we have synthesized benzimidazole-containing quinolinyl oxadiazoles and subjected to anticoagulant, antiplatelet, and α-glucosidase inhibition activities.
Results and Discussion
Chemistry
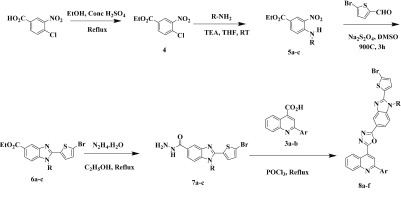
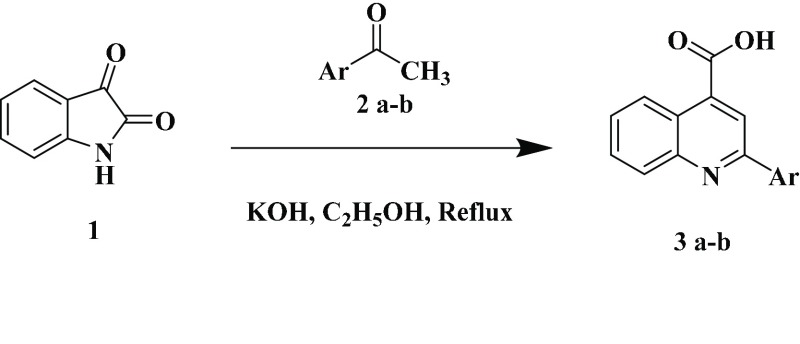
The synthetic methods for benzimidazole-containing quinolinyl oxadiazoles 8a–f are illustrated in Scheme 1. The synthesis of precursors substituted quinoline-4-carboxylic acids 3a–b with various acetophenones, as shown in Scheme 2. Furthermore, key intermediates 7a–c were obtained initially from the core nucleus benzimidazole-5-carboxylates 6a–c that were efficiently synthesized by the “one pot” nitro reductive cyclization reaction between ethyl 3-nitro-4-(substituted amino) benzoates 5a–c and 5-bromothiophene-2-carbaldehyde using sodium dithionite in dimethylsulfoxide. Interestingly, the “one-pot” reaction was proceeded very smoothly, in short reaction time with an excellent yield. The benzimidazole esters 6a–c were converted into the corresponding hydrazides 7a–c and then to highly functionalized quinolinyloxadiazoles 8a–f. All the studied compounds were synthesized by using the same procedure and characterized by spectroscopic analysis.66,67 The solid-state properties of the target compounds are listed in Supporting Information Table S1.
Scheme 1. Synthetic Route for the Preparation of 8a–f. Ar = 3,5-F2C6H3, 2,4-Cl2C6H3. R = CH3, C3H7, C4H9.
Scheme 2. Synthetic Route for the Preparation of 3a–b. Ar = 3,5-F2C6H3, 2,4-Cl2C6H3.
X-ray Diffraction Analysis and Hirshfeld Surfaces Analysis
Figure 2 describes the ORTEP of the 6c. The dihedral angle between rings Cg1: S1/C11–C14 and Cg4: N1/C4–C10/N2 is 27.4(2)0. Crystal structure is stabilized through C1···H1B···O2 intermolecular hydrogen bond in the form of 1D-infinite chain and C16···H16A···Cg3 (C4–C9) intermolecular interactions (Figure 3 and Table 1). The molecular arrangement within the crystal structure is shown along a, b, and c-axis (Figure 3). The crystal data and refinement parameters are given in Table 2.
Figure 2.
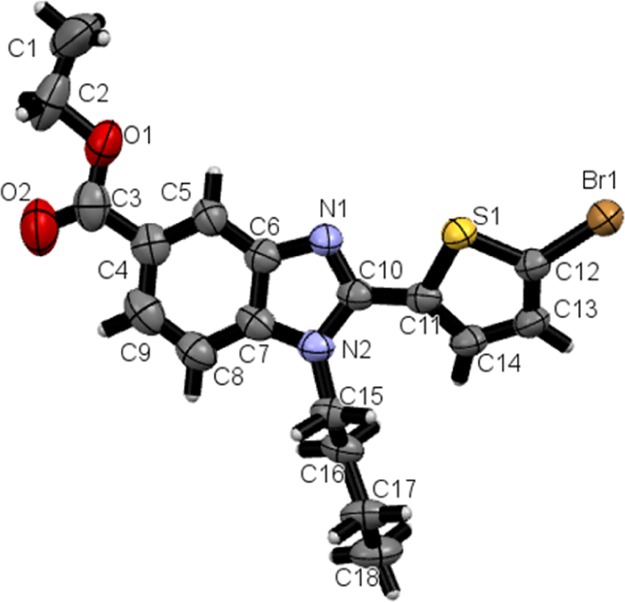
ORTEP diagram of the molecule 6c with thermal ellipsoids drawn at 50% probability.
Figure 3.
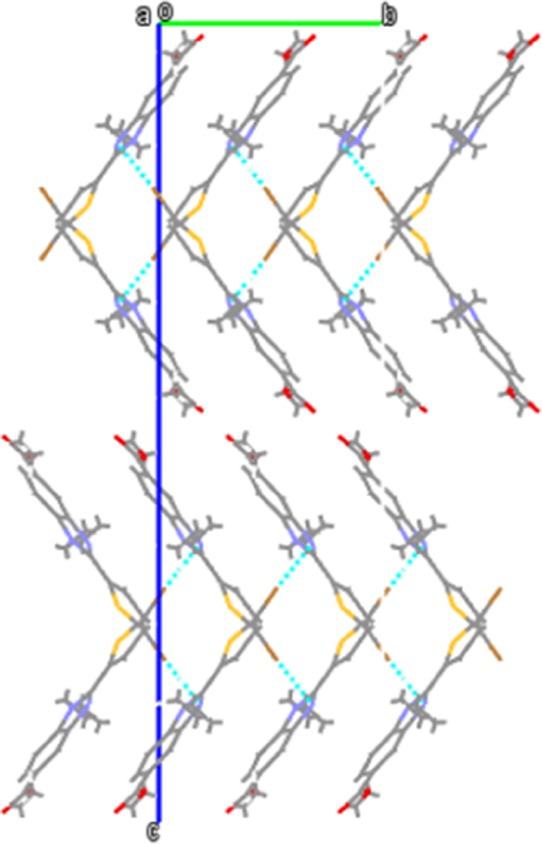
Molecular arrangement of the molecules viewed along the b-axis within the crystal structure. Dotted lines represent intermolecular hydrogen bonds.
Table 1. Intramolecular and Intermolecular Hydrogen Bonds of Compound 6c.
| D···H/X···A/Cg | D–H | H/X···A/Cg | D···A/Cg | D···H/X···A/Cg |
|---|---|---|---|---|
| C1···H1B···O2a | 0.96 | 2.54 | 3.412(13) | 152 |
| C16···H16A···Cg3b | 2.90 | 3.698(6) | 141 |
–1/2 + x, 5/2 – y, 1 – z.
3/2 – x, −1/2 + y, z.
Table 2. Crystal Data and Refinement Statistics of Compound 6c.
| formula | C18H19BrN2O2S |
| formula weight | 407.32 |
| crystal system | orthorhombic |
| space group | Pbca (no. 61) |
| a, b, c (Å) | 11.1173(7), 9.6158(7), 34.457(2) |
| V (Å3) | 3683.5(4) |
| Z | 8 |
| D(calc) (g/cm3) | 1.469 |
| μ(Mo Kα) (/mm) | 2.358 |
| F(000) | 1664 |
| crystal size (mm) | 0.21 × 0.23 × 0.26 |
| Data Collection | |
| temperature (K) | 293 |
| radiation (Å) | Mo Kα 0.71073 |
| θ min–max (deg) | 2.2, 26.4 |
| dataset | –13: 13; −10: 12; −43: 43 |
| tot., uniq. data, R(int) | 370 38, 3761, 0.187 |
| observed data [I > 0.0σ(I)] | 2377 |
| Refinement | |
| Nref, Npar | 3761, 219 |
| R, wR2, S | 0.0747, 0.2225, 1.05 |
| max. and av. shift/error | 0.03, 0.00 |
| min. and max. resd. dens. (e/Å3) | –0.63, 0.83 |
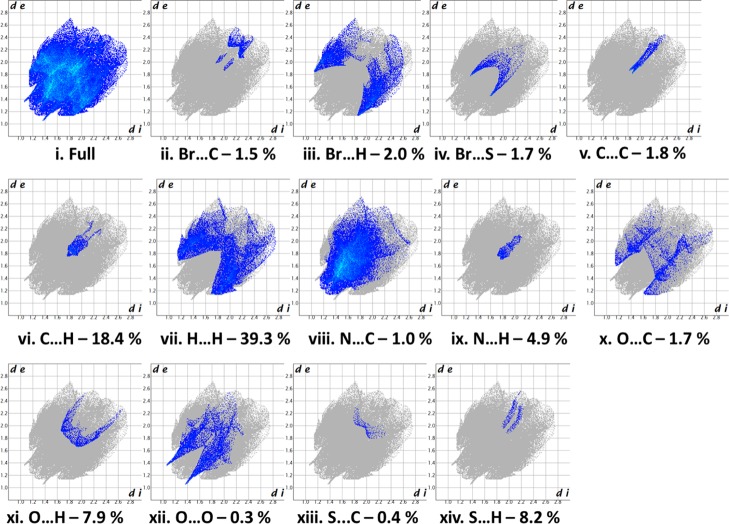
The intercontacts in the crystal structure are analyzed and visualized using Hirshfeld surfaces and 2D fingerprint plots (Figures 4 and 5). The intercontacts found for 6c are Br···C—1.5%, Br···H—11.1%, Br···N—2.0%, Br···S—1.7%, C···C—1.8%, C···H—18.4%, H···H—39.3%, N···C—1.0%, N···H—4.9%, O···C—1.7%, O···H—7.9%, O···O—0.3%, S···C—0.4%, and S···H—8.2%. The 2D finger print plots of these intercontacts are shown in Figure 5. The major contributions are from H···H, C···H, O···H, and N···H when compared to other intercontacts.
Figure 4.
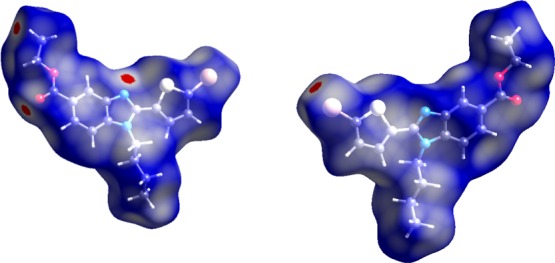
dnorm mapped on Hirshfeld surface for visualizing the intercontact in different orientations. Color scale is between −0.18 au (blue) to 1.4 au (red). The ball and stick model represents the molecule orientation.
Figure 5.
Fingerprint of the title compound 6c.
Computational Screening (Structure–Activity Relationship)
The docking study was implemented using AutoDock 4.2.6 software for benzimidazole-containing quinolinyl oxadiazole derivatives and acarbose against N-terminal domain of the human intestinal α-glucosidase. The parameters of binding energy and binding mode of each compound were carefully analyzed. Using these parameters, hydrogen bond interactions and intermolecular interactions (π–π) with the enzymes were identified. Docking studies corroborate the significant interactions of all the in vitro active benzimidazole-containing quinolinyloxadiazole derivatives with the active site (acarbose inhibition site) of α-glucosidase enzyme. The docking scores also show positive correlation with the experimental results, in general. The docking pose of compounds 8a–f was shown in (Figure 6 and Table 3). The docking study predicted that the quinoline ring of compound 8d binds to enzyme forming π–π stacking with the amino group of Lys776 and benzimidazole ring forms π–π interaction with the amino group of Lys513 amino acid residue with the least binding energy of −8.45 kcal/mol. The second most active in this series is compound 8a with the least binding energy of −8.08 kcal/mol, in which quinoline ring forms π–π interaction with the amino group of Lys776 while benzimidazole ring forms π–π interaction with the amino group of Lys513. Furthermore, the benzimidazole ring of compound 8b forms π–π stacking with the amino group of Lys513 with the least binding energy of −7.77 kcal/mol, and in compound 8f with the least binding energy of −7.5 kcal/mol, the benzimidazole ring forms π–π interaction with amino group of Lys513. At the binding site of a-glucosidase, the amino groups of Lys776 and Lys513 residues construct the π–π stacking interaction with quinoline ring and benzimidazole ring of the ligand 8c, respectively. However, in the ligand 8e with binding energy −7.95 kcal/mol, no interaction has been seen with amino acid residues.
Figure 6.
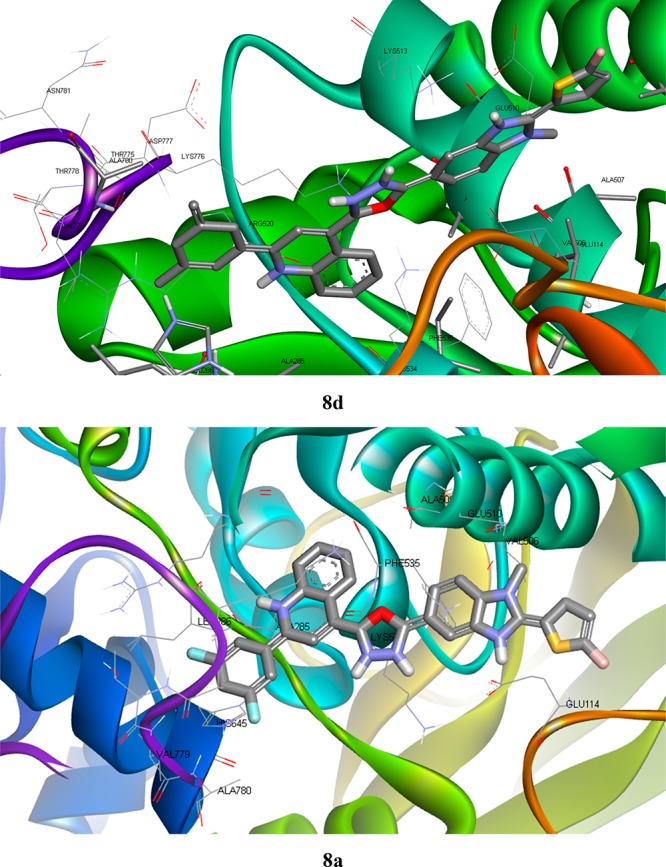
Docked poses of α-glucosidase–ligand interactions of 8d and 8a showing best binding affinity.
Table 3. Comparison of Predicted Binding Affinities of Active Benzimidazole-Containing Quinolinyl Oxadiazoles and Quinoline Schiff Bases and Acarbose against AGI Activity.
| ligand | binding energy | intermol. or internal energy | internal energy | torsional energy | unbound energy | H bonds/AA | π–π bonds/AA |
|---|---|---|---|---|---|---|---|
| 8d | –8.45 | –8.75 | 0 | 0.3 | 0 | NA | 2/Lys776, Lys513 |
| 8a | –8.08 | –8.38 | 0.05 | 0.3 | 0.05 | NA | 2/Lys776, Lys513 |
| 8e | –7.95 | –8.84 | –0.46 | 0.89 | –0.46 | NA | NA |
| 8b | –7.77 | –8.66 | –0.65 | 0.89 | –0.65 | NA | 1/Lys513 |
| 8f | –7.5 | –8.4 | –0.51 | 0.89 | –0.51 | NA | 1/Lys513 |
| 8c | –7.08 | –7.98 | –0.53 | 0.89 | –0.53 | NA, 3/Arg520, Lys534 | 2/Lys776, Lys513 |
| acarbose | –14.38 | –18.25 | –5.52 | 3.88 | –5.52 | Val779 | NA |
In general, all the ligand molecules except 8e showed more promising results against α-glucosidase compared to standard compounds. Hence, they may act as potential specific inhibitors for this target enzyme.
Biological Evaluation
Antidiabetic Activity: AGI Effect
α-Glucosidase inhibition assay was carried out for the newly synthesized compounds 8a–f in vitro. All the compounds exhibited significant α-glucosidase-inhibiting potential compared to standard acarbose (IC50 = 1460. 28 ± 244.365 μg/mL). It was demonstrated that compounds 8a and 8d had the best AGI activity with IC50 values 0.66 ± 0.05 and 0.68 ± 0.02 μg/mL as represented in Table 4.
Table 4. IC50 Values of Benzimidazole-Containing Quinolinyl Oxadiazoles and Quinoline Schiff Bases for AGI Activity.
| sample | IC50 value (μg/mL) (n = 3 ± standard deviation) |
|---|---|
| 8a | 0.66 ± 0.05 |
| 8b | 2.76 ± 0.37 |
| 8c | 3.79 ± 0.46 |
| 8d | 0.68 ± 0.02 |
| 8e | 1.40 ± 0.23 |
| 8f | 2.81 ± 0.24 |
| acarbose | 1460.28 ± 244.365 |
Plasma Recalcification Time
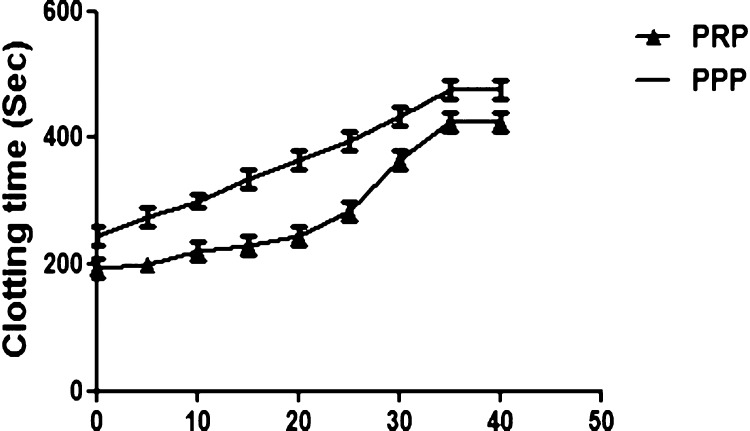
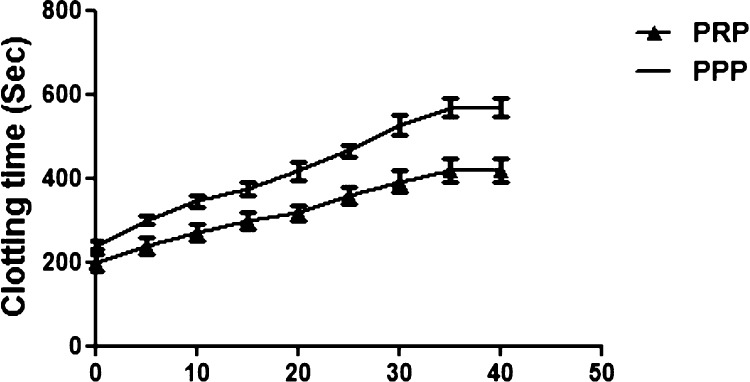
To pinpoint the possible role of 8a and 8d in the blood coagulation cascade, plasma recalcification time was accomplished using both human platelet rich plasma (PRP) and platelet poor plasma (PPP). Astonishingly, 8a and 8d displayed anticoagulant effect by enhancing the clotting time of both PRP and PPP from control 180–410, 180–390 and 230–460, 230–545 s, respectively. The supreme concentration consumed in both the cases was found to be 35 μg and remain unchanged upon increased dose to 40 μg, Figures 7 and 8. Coagulation factors are responsible for blood clot which is a physiological phenomenon, and the blood coagulation cascade could be activated just to arrest the bleeding in the case of injury. It involves several factors such as factor VIIa to factor XIIIa, which mainly encompasses three pathways which are intrinsic, extrinsic, and common pathways. Some genetic imbalance and some environmental factors alter the normal coagulation system, which further leads to thrombosis, a pathological phenomenon. Plentiful of anticoagulants were reported from the biological sources and nanoparticles,14,68−71 but reports available on the anticoagulants from benzimidazole-containing quinolinyl oxadiazoles are very rare.
Figure 7.
Plasma recalcification time. 8a (0–40 μg) was pre-incubated with 0.2 mL of citrated human plasma PRP/PPP in the presence of 20 μL of 10 mM Tris-HCl buffer (pH 7.4) for 1 min at 37 °C. CaCl2 (20 μL; 0.25 M) was added to the pre-incubated mixture and clotting time was recorded.
Figure 8.
Plasma recalcification time. 8d (0–40 μg) was pre-incubated with 0.2 mL of citrated human plasma PRP/PPP in the presence of 20 μL of 10 mM Tris-HCl buffer (pH 7.4) for 1 min at 37 °C. CaCl2 (20 μL; 0.25 M) was added to the pre-incubated mixture and clotting time was recorded.
Antiplatelet and Nontoxic Properties of 8a–f
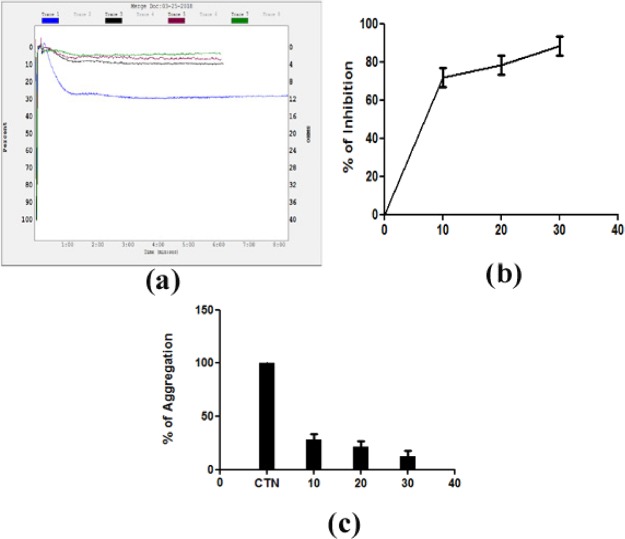
Furthermore, to ascertain the role of 8a and 8d on platelets, these were analyzed for platelet aggregation assay using PRP with agonist epinephrine. Curiously, only 8a is able to inhibit the epinephrine-induced platelet aggregation but not the 8d sample. The 8a sample inhibits epinephrine-induced platelet aggregation of about 93% at the concentration of 30 μg as shown in Figure 9. However, platelets are very much essential to stop bleeding along with coagulation factors. Aggregation of platelets leads to the formation of platelet plugs when blood vessels are injured. Many physiological agents act as agonists such as ADP, epinephrine, collagen, thrombin, arachidonic acid, and thromboxane to activate platelets in order to arrest the blood by forming platelet plugs at the site injury. No control over the activation of platelets leads to pathophysiology of thrombotic disorders. Several antiplatelet agents have been identified from the biological sources,72,73 but very few are studied from synthesized compound sources.
Figure 9.
Platelet aggregation was initiated by adding epinephrine as an agonist of 8a. (a) Traces of platelet aggregation: trace 1 (epinephrine 5 μM); trace 2 (epinephrine 5 μM + 10 μg of 8a); trace 3 (epinephrine 5 μM + 20 μg of 8a); and trace 4 (epinephrine 5 μM + 30 μg of 8a). The values represent of three independent experiments. (b) Dose-dependent platelet aggregation inhibition %. (c) Dose-dependent platelet aggregation %.
Moreover, all the synthesized compounds of benzimidazole-containing quinolinyl oxadiazoles 8a–f went unhydrolyzed with RBC cells. Molecules 8a and 8d were active in preventing hemorrhage and edema in experimental mice up to the concentration level of 100 μg. The positive control, Daboia russelli venom, encouraged hemorrhage and edema in experimental mice, suggesting its nontoxic property as shown in Figure 10.
Figure 10.
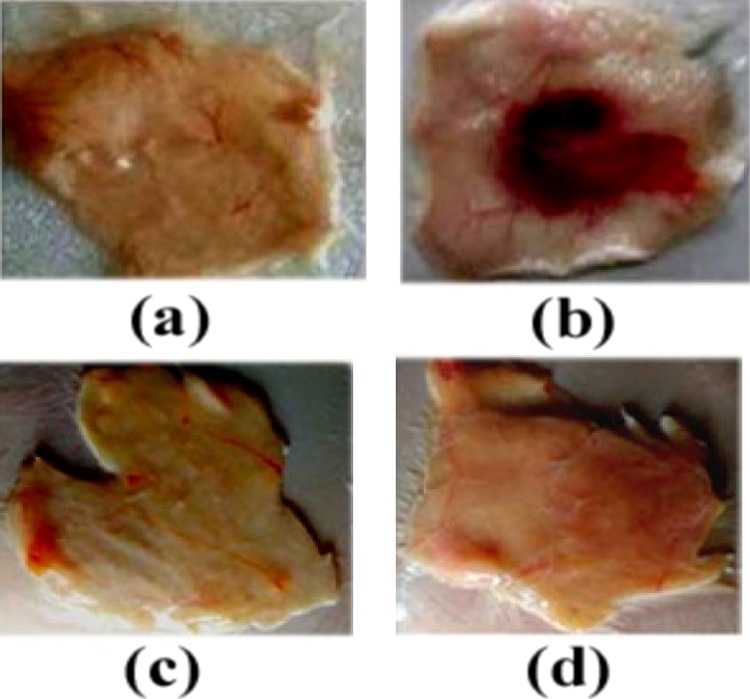
Dose-dependent hemorrhagic activity of 8a and 8d: (a) saline, (b) positive control 2 MDH venom, (c) 100 μg of 8a, and (d) 100 μg of 8d were injected independently into mice in a total volume of 50 μL intradermal.
Conclusions
Benzimidazole-containing quinolinyloxadiazoles 8a–f were synthesized. The novel synthesized compounds were characterized by spectral and analytical data and were screened for antidiabetic and antithrombotic activity. The results are correlated with docking studies. The molecular docking data provided positive correlation with in vitro antidiabetic activity in comparison with the standards revealed that these compounds can act as potential inhibitors. The core nucleus benzimidazole-5-carboxylates 6a–c were efficiently synthesized by a “one pot” nitro reductive cyclization reaction between ethyl 3-nitro-4-(substituted amino) benzoates 5a–c and 5-bromothiophene-2-carbaldehyde using sodium dithionite in dimethylsulfoxide. This “one-pot” reaction was proceeded very smoothly, in short reaction time with an excellent yield. Compounds benzimidazole-containing quinolinyloxadiazoles 8a–f showed good binding interactions with the target enzyme with least binding energies. Therefore, these compounds may be further evaluated and useful for diabetic treatment; on the other hand, this study for the first time demonstrates that benzimidazole-containing quinolinyl oxadiazoles (8a–f) are responsible for anticoagulant (8a and 8d) and antiplatelet (8a) properties. Furthermore, they exhibit nontoxic property on RBC cells. Thus, they could play a major role in the treatment of thrombotic disorders, thus providing a new scope in drug discovery.
Experimental Section
Materials and Methods
All reagents were purchased from Sigma-Aldrich/Spectrochem, India, and used without further purification. Melting points were determined in an open capillary tube and were uncorrected. The progress of each reaction was monitored by ascending thin layer chromatography on silica gel G (Merck 1.05570.0001), visualized by UV light. The IR spectra for samples were recorded using a Shimadzu IRPrestige-21 FT-IR spectrophotometer, and the wave numbers were given in cm–1. The 1H NMR spectra were recorded (CDCl3/DMSO-d6 mixture) on an Agilent 400 MHz with an ATB probe and operated by VnmrJ software with TMS as the internal standard and Bruker; model AV 400. The X-ray intensity data were collected on a Rigaku Saturn70 diffractometer (Mo Kα radiation) at 293 K, and data were collected and processed using Crystal Clear. Mass spectra were recorded in a WATERS model SynaptG2 LC-mass spectrometer.
Source for Anticoagulant and Antiplatelet Activity
Fresh human blood was collected from healthy donors for the PRP and PPP.
Preparation of 2-Arylquinoline-4-carboxylic Acids (3a–b)
2-arylquinoline-4-carboxylic acid (Figure 1) was prepared according to the literature method.74,75
2-(3,5-Difluorophenyl)quinoline-4-carboxylic acid (3a): mp 230.0–232 °C.
2-(3,5-Dichlorophenyl)quinoline-4-carboxylic acid (3b): mp 198–204 °C.
Preparation of Ethyl 4-(Alkylamino)-3-nitrobenzoates (5a–c)
Ethyl 4-chloro-3-nitro benzoate (0.01 mol) was taken in 10 mL of tetrahydrofuran. To this solution, primary amine (0.015 mol) and triethylamine (0.03 mol) were added. The reaction mixture was kept for stirring overnight at room temperature. After completion of the reaction, the reaction mixture was poured onto crushed ice. The product obtained was filtered, dried, and purified by recrystallization. Different ethyl 4-(alkylamino)-3-nitrobenzoates prepared using this method are as follows.
Ethyl 4-(Methylamino)-3-nitrobenzoate (5a): mp 98–100 °C (Lit. mp 101 °C).76
Ethyl 4-(Propylamino)-3-nitrobenzoate (5b): mp 77–78 °C (Lit. mp 78 °C).77
Ethyl 4-(Butylamino)-3-nitrobenzoate (5c): mp 52–54 °C (Lit. mp 50–52 °C).78
General Procedure for the Preparation of Ethyl 2-(5-Bromothiophen-2-yl)-1-alkyl-1H-benzo[d]imidazole-5-carboxylates (6a–c)
Sodium dithionite (3.0 equiv) was added to a stirred solution of ethyl-4-(4-alkylamino)-3-nitrobenzoate (0.01 mol) and 5-bromothiophene-2-carbaldehyde (0.01 mol) in DMSO (20 mL). The reaction mixture was stirred at 90 °C for 3 h. After the completion of reaction, it was poured onto crushed ice. The solid separated was filtered off, washed with water, and dried. The crude product 6a–c was recrystallized using dimethylformamide.
Ethyl 2-(5-Bromothiophen-2-yl)-1-methyl-1H-benzo[d]imidazole-5-carboxylate (6a)
Yield 98%; mp 160–165 °C; FT IR (ATR, ν cm–1): 3040 (Ar-H), 2943 (C–H), 1713 (C=O), 1616 (C=N), 1538 (C=C); 1H NMR (DMSO-d6 δ ppm): 1.43 (t, 3H, CH3), 4.04 (s, 3H, CH3), 4.52 (q, 2H, CH2), 7.05 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 7.28 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 7.48 (d, 1H, benzimidazolyl-C6-H, J = 8.4 Hz), 7.66 (d, 1H, benzimidazolyl-C7-H, J = 8.4 Hz), 8.03 (s, 1H, benzimidazolyl-C4-H); 13C NMR (100 MHz, CDCl3 δ ppm): 14.4 (CH3), 30.1 (CH3), 61.5 (CH2), 110.2, 110.5, 113.0, 129.5, 129.8, 130.9, 134.2, 135.1, 139.4, 141.5, 153.9, 167.2 (C=O); mass m/z: 364.54 (M + H)+, 366.79 (M + H + 2)+; Anal. found (calcd) for C15H13BrN2O2S (%): C, 49.17 (49.33); H, 3.45 (3.59); 7.86 (7.67).
Ethyl 2-(5-Bromothiophen-2-yl)-1-propyl-1H-benzo[d]imidazole-5-carboxylate (6b)
Yield 98%; mp 120–124 °C; FT IR (ATR, ν cm–1): 3060 (Ar-H), 2970 (C–H), 1728 (C=O), 1629 (C=N), 1462 (C=C); 1H NMR (DMSO-d6 δ ppm): 1.08 (t, 3H, CH3), 1.39 (t, 3H, CH3), 2.08–2.19 (m, 2H, CH2), 4.35 (q, 2H, CH2), 4.42 (t, 2H, CH2), 7.13 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 7.35 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 7.55 (d, 1H, benzimidazolyl-C6-H, J = 8.4 Hz), 7.71 (d, 1H, benzimidazolyl-C7-H, J = 8.4 Hz), 8.09 (s, 1H, benzimidazolyl-C4-H); 13C (100 MHz, CDCl3 δ ppm): 11.5 (CH3), 14.1 (CH3), 20.9 (CH2), 49.6 (CH2), 61.2 (CH2), 111.2, 11.5, 114.8, 128.3, 129.2, 131.2, 135.4, 137.9, 139.2, 143.2, 154.2, 168.1 (C=O); mass m/z: 393.29 (M + H)+, 395.72 (M + H + 2)+; Anal. found (calcd) for C17H17BrN2O2S (%): C, 51.75 (51.92); H, 4.17 (4.36); 7.30 (7.12).
Ethyl 2-(5-Bromothiophen-2-yl)-1-butyl-1H-benzo[d]imidazole-5-carboxylate (6c)
Yield 98%; mp 95–98 °C; FT IR (ATR, ν cm–1): 3090 (Ar-H), 2985 (C–H), 1735 (C=O), 1612 (C=N), 1563 (C=C); 1H NMR (DMSO-d6 δ ppm): 0.96 (t, 3H, CH3), 1.31 (t, 3H, CH3), 1.39–1.51 (m, 2H, CH2), 1.81–1.95 (m, 2H, CH2), 4.22 (q, 2H, CH2), 4.39 (t, 2H, CH2), 7.19 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 7.45 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 7.56 (d, 1H, benzimidazolyl-C6-H, J = 8.4 Hz), 7.79 (d, 1H, benzimidazolyl-C7-H, J = 8.4 Hz), 8.12 (s, 1H, benzimidazolyl-C4-H); 13C NMR (100 MHz, CDCl3 δ ppm): 13.9 (CH3), 14.1 (CH3), 21.9 (CH2), 30.1 (CH2), 49.8 (CH2), 62.1 (CH2), 110.1, 112.2, 113.9, 127.9, 132.5, 136.4, 137.3, 138.4, 140.8, 145.2, 155.3, 168.0 (C=O); mass m/z: 407.21 (M + H)+, 409.53 (M + H + 2)+; Anal. found (calcd) for C18H19BrN2O2S (%): C, 53.25 (53.08); H, 4.55 (4.70); 6.71 (6.88).
General Procedure for the Preparation of 2-(5-Bromothiophen-2-yl)-1-alkyl-1H-benzo[d]imidazole-5-carbohydrazide (7a–c)
A mixture of ethyl 2-(5-bromothiophen-2-yl)-1-alkyl-1H-benzo[d]imidazole-5-carboxylate (0.1 mol), hydrazine hydrate (0.15 mol), and 20 mL of ethanol was refluxed on an oil bath for 10 h. The excess solvent was then distilled off under reduced pressure, and the concentrated solution was quenched with ice cold water. The solid separated was filtered, washed, and dried. The crude product was purified by recrystallization from ethanol.
2-(5-Bromothiophen-2-yl)-1-methyl-1H-benzo[d]imidazole-5-carbohydrazide (7a)
Yield 98%; mp >230 °C; FT IR (ATR, ν cm–1): 3435, 3277 (NH2), 3599 (NH), 3146 (Ar-H), 2947 (C–H), 1679 (C=O), 1663 (C=N), 1454 (C=C), 633 (C–S), 544 (C–Br); 1H NMR (DMSO-d6) δ (ppm): 4.01 (s, 3H, CH3), 4.79 (s, 2H, NH2), 7.17 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 7.40 (dd, 1H, benzimidazolyl-C6-H, J = 8.4 Hz), 7.58 (dd, 1H, benzimidazolyl-C7-H, J = 8.4 Hz), 7.67 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 8.08 (s, 1H, benzimidazolyl-C4-H), 9.96 (s, 1H, NH); 13C NMR (100 MHz, CDCl3 δ ppm): 30.8 (CH3), 108.8, 113.0, 120.4, 128.7, 129.2, 129.8, 134.6, 134.8, 135.7, 141.0, 152.9, 165.4; mass m/z: 350.23 (M + H)+, 352.57 (M + H + 2)+; Anal. found (calcd) for C13H11BrN4OS (%): C, 44.58 (44.46); H, 3.30 (3.16); 15.78 (15.95).
2-(5-Bromothiophen-2-yl)-1-propyl-1H-benzo[d]imidazole-5-carbohydrazide (7b)
Yield 98%; mp 140–144 °C; FT IR (ATR, ν cm–1): 3325, 3284 (NH2), 3511 (NH), 3188 (Ar-H), 2968 (C–H), 1636 (C=O), 1533 (C=N), 1472 (C=C), 697 (C–S), 561 (C–Br); 1H NMR (DMSO-d6 δ ppm): 1.06 (t, 3H, CH3), 2.06–2.18 (m, 2H, CH2), 4.31 (t, 2H, CH2), 4.81 (s, 2H, NH2), 7.14 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 7.44 (dd, 1H, benzimidazolyl-C6-H, J = 8.4 Hz), 7.55 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 7.77 (dd, 1H, benzimidazolyl-C7-H, J = 8.4 Hz), 8.08 (s, 1H, benzimidazolyl-C4-H), 10.02 (s, 1H, NH); 13C NMR (100 MHz, CDCl3 δ ppm): 12.5 (CH3), 21.9 (CH2), 49.6 (CH2), 110.8, 113.0, 122.4, 125.7, 127.8, 132.8, 134.8, 136.5, 141.0, 152.9, 155.3, 165.4; mass m/z: 379.37 (M + H)+, 381.71 (M + H + 2)+; Anal. found (calcd) for C15H15BrN4OS (%): C, 47.67 (47.50); H, 3.81 (3.99); 14.65 (14.77).
2-(5-Bromothiophen-2-yl)-1-butyl-1H-benzo[d]imidazole-5-carbohydrazide (7c)
Yield 98%; mp >230 °C; FT IR (ATR, ν cm–1): 3345, 3298 (NH2), 3523 (NH), 3159 (Ar-H), 3009 (C–H), 1675 (C=O), 1621 (C=N), 1459 (C=C), 679 (C–S), 578 (C–Br); 1H NMR (DMSO-d6 δ ppm): 0.91 (t, 3H, CH3), 1.29–1.42 (m, 2H, CH2), 1.85–1.98 (m, 2H, CH2), 4.37 (t, 2H, CH2), 4.87 (s, 2H, NH2), 7.25 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 7.52 (dd, 1H, benzimidazolyl-C6-H, J = 8.4 Hz), 7.68 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 7.72 (dd, 1H, benzimidazolyl-C7-H, J = 8.4 Hz), 8.16 (s, 1H, benzimidazolyl-C4-H), 10.14 (s, 1H, NH); 13C NMR (100 MHz, CDCl3 δ ppm): 13.9 (CH3), 20.1 (CH2), 30.1 (CH2), 49.6 (CH2), 109.3, 111.5, 115.8, 121.6, 129.1, 132.5, 133.3, 137.5, 139.2, 144.1, 148.2, 165.1; mass m/z: 393.29 (M + H)+, 395.68 (M + H + 2)+; Anal. found (calcd) for C16H17BrN4OS (%): C, 48.71 (48.86); H, 4.20 (4.36); 14.44 (14.25).
General Procedure for the Preparation of 2-(2-(5-Bromothiophen-2-yl)-1-alkyl-1H-benzo[d]imidazol-5-yl)-5-(2-(3,5-difluorophenyl)quinolin-4-yl)-1,3,4-oxadiazole (8a–f)66
A mixture of 2-(5-bromothiophen-2-yl)-1-alkyl-1H-benzo[d]imidazole-5-carbohydrazide (1 mmol), substituted quinoline-4-carboxylic acid (1 mmol) and phosphorus oxychloride (2.5 mmol) was refluxed (100–110 °C for 6 h) and the residue was quenched with ice cold water. The solid separated was processed (filtered, washed, dried, and recrystallized).
2-(2-(5-Bromothiophen-2-yl)-1-methyl-1H-benzo[d]imidazol-5-yl)-5-(2-(3,5-difluorophenyl)quinolin-4-yl)-1,3,4-oxadiazole (8a)
Yield 82%; mp 214–218 °C; FT IR (ATR, ν cm–1): 3080 (Ar-H), 2969 (C–H), 1604 (C=N), 1478 (C=C), 1101 (C–F), 692 (C–S); 1H NMR (DMSO-d6 δ ppm): 3.97 (s, 3H, CH3), 7.38 (overlapped d, 1H, difluorophenyl-C4-H), 7.49 (d, 1H, Br-thienyl-H, J = 8.0 Hz), 7.64 (d, 2H, difluorophenyl, J = 7.2 Hz), 7.75 (d, 1H, benzimidazolyl-C6-H, J = 8.4 Hz), 7.82 (d, 1H, Br-thienyl-H, J = 8.0 Hz), 7.91–8.01 (m, 2H, quinolinyl-H), 8.17 (d, 1H, benzimidazolyl-C7-H, J = 8.4 Hz), 8.25 (s, 1H, benzimidazolyl-C4-H), 8.37 (d, 1H, quinolinyl-H, J = 8.4 Hz), 8.56 (s, 1H, quinolinyl-H), 8.77 (d, 1H, quinolinyl-H, J = 8.0 Hz); 13C NMR (100 MHz, CDCl3 δ ppm): 30.5 (CH3), 109.0, 111.8, 114.0, 116.0, 116.2, 117.4, 120.2, 121.3, 126.5, 127.0, 127.7, 128.5, 129.1, 130.2, 130.8, 131.0, 132.3, 135.8, 136.7, 137.7, 142.0, 146.5, 153.9, 159.4, 161.7, 161.9, 164.7, 166.2; mass m/z: 600.41 (M + H)+, 602.29 (M + H + 2)+; Anal. found (calcd) for C29H16BrF2N5OS (%): C, 58.19 (58.01); H, 2.81 (2.69); 11.51 (11.66).
2-(2-(5-Bromothiophen-2-yl)-1-propyl-1H-benzo[d]imidazol-5-yl)-5-(2-(3,5-difluorophenyl)quinolin-4-yl)-1,3,4-oxadiazole (8b)
Yield 81%; mp 189–193 °C; FT IR (ATR, ν cm–1): 3077 (Ar-H), 2962 (C–H), 1606 (C=N), 1477 (C=C), 1089 (C–F), 606 (C–S); 1H NMR (DMSO-d6 δ ppm): 1.10 (t, 3H, CH3), 1.99–2.05 (m, 2H, −CH2), 4.48 (t, 2H, −CH2), 6.92 (overlapped d, 1H, difluorophenyl-C4-H), 7.22 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 7.55 (d, 1H, Br-thienyl-H, J = 7.6 Hz), 7.65–7.85 (m, 2H, quinolinyl-H), 8.15 (d, 1H, benzimidazolyl-C7-H, J = 8.4 Hz), 8.22 (d, 1H, benzimidazolyl-C6-H, J = 8.4 Hz), 8.33 (d, 2H, difluorophenyl-H, J = 8.0 Hz), 8.44 (s, 1H, quinolinyl-H), 8.65 (s, 1H, benzimidazolyl-C4-H), 8.73 (d, 1H, quinolinyl-H, J = 8.4 Hz), 9.17 (d, 1H, quinolinyl-H, J = 8.4 Hz); 13C NMR (100 MHz, CDCl3 δ ppm): 12.5 (CH3), 21.9 (CH2), 49.6 (CH2), 108.0, 110.5, 113.0, 117.9, 123.0, 125.5, 125.9, 128.4, 129.0, 130.4, 130.8, 131.7, 133.4, 137.8, 139.1, 140.2, 141.3, 144.5, 145.9, 148.3, 151.1, 153.5, 156.5, 158.6, 159.3, 160.3, 162.8, 163.1; mass m/z: 628.10 (M + H)+, 630.25 (M + H + 2)+; Anal. found (calcd) for C31H20BrF2N5OS (%): C, 59.10 (59.24); H, 3.07 (3.21); 11.33 (11.14). HPLC purity at 254 nm, 95.2%.
2-(2-(5-Bromothiophen-2-yl)-1-butyl-1H-benzo[d]imidazol-5-yl)-5-(2-(3,5-difluorophenyl)quinolin-4-yl)-1,3,4-oxadiazole (8c)
Yield 80%; mp 192–194 °C; FT IR (ATR, ν cm–1): 3051 (Ar-H), 2943 (C–H), 1623 (C=N), 1491 (C=C), 1123 (C–F), 623 (C–S); 1H NMR (DMSO-d6 δ ppm): 0.96 (t, 3H, CH3), 1.27–1.35 (m, 2H, −CH2), 2.18–2.25 (m, 2H, −CH2), 4.58 (t, 2H, −CH2), 7.34 (overlapped d, 1H, difluorophenyl-C4-H), 7.41 (d, 1H, Br-thienyl-H, J = 8.0 Hz), 7.69 (d, 2H, difluorophenyl, J = 7.2 Hz), 7.77 (d, 1H, benzimidazolyl-C6-H, J = 8.4 Hz), 7.84 (d, 1H, Br-thienyl-H, J = 8.0 Hz), 7.94–8.09 (m, 2H, quinolinyl-H), 8.20 (d, 1H, benzimidazolyl-C7-H, J = 8.4 Hz), 8.29 (s, 1H, benzimidazolyl-C4-H), 8.40 (d, 1H, quinolinyl-H, J = 8.4 Hz), 8.51 (s, 1H, quinolinyl-H), 8.80 (d, 1H, quinolinyl-H, J = 8.0 Hz); 13C NMR (100 MHz, CDCl3 δ ppm): 13.9 (CH3), 20.1 (CH2), 30.1 (CH2), 49.6 (CH2), 109.0, 111.1, 118.5, 124.6, 126.1, 126.5, 129.0, 130.1, 131.0, 131.4, 132.3, 134.0, 138.4, 140.7, 141.8, 142.9, 145.1, 146.5, 149.9, 152.7, 154.1, 157.1, 159.2, 160.9, 161.9, 163.4, 165.5, 165.9; Mass m/z: 642.21 (M + H)+, 644.39 (M + H + 2)+; Anal. found (calcd) for C32H22BrF2N5O2S (%): C, 59.97 (58.82); H, 3.33 (3.45); 10.78 (10.90).
2-(2-(5-Bromothiophen-2-yl)-1-methyl-1H-benzo[d]imidazol-5-yl)-5-(2-(2,4-dichlorophenyl)quinolin-4-yl)-1,3,4-oxadiazole (8d)
Yield 84%; mp >225 °C; FT IR (ATR, ν cm–1): 3092 (Ar-H), 2951 (C–H), 1655 (C=N), 1455 (C=C), 777 (C–Cl), 649 (C–S); 1H NMR (DMSO-d6 δ ppm): 4.08 (s, 3H, CH3), 7.23 (d, 1H, Br-thienyl-H, J = 8.0 Hz), 7.29 (d, 1H, Br-thienyl-H, J = 8.0 Hz), 7.45 (overlapped d, 1H, dichlorophenyl-C3-H), 7.55 (s, 1H, benzimidazolyl-C4-H), 7.63 (dd, 2H, benzimidazolyl-C6-H and C7-H, J = 8.4 Hz), 7.77–7.92 (m, 4H, dichlorophenyl-H and quinolinyl-H), 8.18 (s, 1H, quinolinyl-H), 8.54 (d, 1H, quinolinyl-H, J = 8.4 Hz), 8.75 (d, 1H, quinolinyl-H, J = 8.4 Hz); 13C NMR (100 MHz, CDCl3 δ ppm): 30.8 (CH3), 105.0, 108.2, 111.6, 114.4, 115.5, 120.7, 121.2, 122.0, 122.7, 123.3, 123.7, 124.4, 124.6, 125.2, 126.0, 127.7, 128.4, 129.5, 130.5, 130.9, 136.2, 140.7, 147.1, 148.4, 149.3, 153.1, 159.3, 160.4; mass m/z: 631.53 (M + H)+, 633.18 (M + H + 2)+; Anal. found (calcd) for C29H16BrCl2N5OS (%): C, 55.13 (55.00); H, 2.41 (2.55); 11.22 (11.06). HPLC purity at 254 nm, 95.5%.
2-(2-(5-Bromothiophen-2-yl)-1-propyl-1H-benzo[d]imidazol-5-yl)-5-(2-(2,4-dichlorophenyl)quinolin-4-yl)-1,3,4-oxadiazole (8e)
Yield 83%; mp 172–176 °C; FT IR (ATR, ν cm–1): 3061 (Ar-H), 2981 (C–H), 1673 (C=N), 1463 (C=C), 759 (C–Cl), 635 (C–S); 1H NMR (DMSO-d6 δ ppm): 1.08 (s, 3H, CH3), 2.29–2.38 (m, 2H, −CH2), 4.75–4.82 (t, 2H, −CH2), 7.32 (d, 1H, Br-thienyl-H, J = 8.0 Hz), 7.40 (overlapped d, 1H, dichlorophenyl-C3-H), 7.47–7.53 (m, 2H, dichlorophenyl-H), 7.65–7.79 (m, 2H, quinolinyl-H), 7.91 (d, 1H, Br-thienyl-H, J = 8.0 Hz), 8.19 (dd, 2H, benzimidazolyl-C6-H and C7-H, J = 8.4 Hz), 8.28 (s, 1H, benzimidazolyl-C4-H), 8.37 (s, 1H, quinolinyl-H), 8.42 (d, 1H, quinolinyl-H, J = 8.4 Hz), 8.78 (d, 1H, quinolinyl-H, J = 8.4 Hz); 13C NMR (100 MHz, CDCl3 δ ppm): 11.5 (CH3), 20.9 (CH2), 48.6 (CH2), 109.8, 112.0, 115.4, 118.2, 119.3, 124.5, 125.0, 125.7, 126.5, 126.8, 127.1, 127.5, 128.2, 128.6, 128.8, 129.0, 129.7, 132.2, 133.3, 134.3, 134.7, 140.0, 142.5, 144.5, 151.9, 157.1, 162.2, 162.7; mass m/z: 659.75 (M + H)+, 661.29 (M + H + 2)+; Anal. found (calcd) for C31H20BrCl2N5OS (%): C, 56.17 (56.30); H, 3.21 (3.05); 10.44 (10.59).
2-(2-(5-Bromothiophen-2-yl)-1-butyl-1H-benzo[d]imidazol-5-yl)-5-(2-(2,4-dichlorophenyl)quinolin-4-yl)-1,3,4-oxadiazole (8f)
Yield 85%; mp >225 °C; FT IR (ATR, ν cm–1): 3049 (Ar-H), 2933 (C–H), 1644 (C=N), 1444 (C=C), 741 (C–Cl), 658 (C–S); 1H NMR (DMSO-d6 δ ppm): 0.90 (s, 3H, CH3), 1.33–1.40 (m, 2H, −CH2), 2.81–2.89 (m, 2H, −CH2), 4.95 (t, 2H, −CH2), 7.28 (d, 1H, Br-thienyl-H, J = 8.0 Hz), 7.35 (overlapped d, 1H, dichlorophenyl-C3-H), 7.49–7.58 (m, 2H, dichlorophenyl-H), 7.65–7.76 (m, 2H, quinolinyl-H), 7.88 (dd, 2H, benzimidazolyl-C6-H and C7-H, J = 8.4 Hz), 7.97 (d, 1H, Br-thienyl-H, J = 8.0 Hz), 8.24 (s, 1H, benzimidazolyl-C4-H), 8.33 (s, 1H, quinolinyl-H), 8.45 (d, 1H, quinolinyl-H, J = 8.4 Hz), 8.81 (d, 1H, quinolinyl-H, J = 8.4 Hz); 13C NMR (100 MHz, CDCl3 δ ppm): 14.5 (CH3), 22.1 (CH2), 33.1 (CH2), 48.6 (CH2), 112.2, 115.2, 118.6, 120.4, 121.5, 127.7, 128.2, 129.7, 130.0, 131.4, 131.6, 132.2, 133.0, 134.7, 135.5, 136.4, 137.5, 137.9, 144.2, 147.7, 154.1, 155.4, 156.3, 160.1, 165.2, 166.3, 167.4; mass m/z: 674.29 (M + H)+, 676.54 (M + H + 2)+; Anal. found (calcd) for C32H20BrCl2N5OS (%): C, 56.79 (56.91); H, 3.17 (3.28); 10.49 (10.37).
Single Crystal X-ray Diffraction Studies
The single-crystal X-ray diffraction intensities collection and data process were carried out using the Rigaku Saturn 724 diffractometer (Mo Kα radiation; 296 K) and CrystalClear software, respectively.79 Direct methods were used for solving the structure (SHELXS), and refinement was made by the full-matrix least squares method on F2 using SHELX.80 All the non-hydrogen atoms were exhibited in the first difference Fourier map itself, in which all the hydrogen bonds were geometrically positioned (C–H = 0.93 Å, O–H = 0.82 Å) and a riding model with Uiso(H) = 1.2Ueq and 1.5Ueq(O) was used for the refinement. The geometry of the 6c is calculated using PLATON program.81 The ORTEP and packing diagrams were generated using MERCURY.82Table 2 represents the details of crystal structure and data refinement. Figure 2 represents the ORTEP82 of the molecule with thermal ellipsoids drawn at 50% probability.
Hirshfeld Surfaces Analysis
The Hirshfeld surfaces computational analysis was accomplished to determine the intercontacts in the crystal structure of compound 6c as shown in Figure 4, in which the intercontacts of the intermolecular interaction were indicated by the red colored spots over the Hirshfeld surface. The short interatomic contacts emerged by the strong hydrogen bonds were indicated by the dark red spots on the dnorm surface, while the light red spots indicate the other intermolecular interactions. The intercontacts with respect to di and de were plotted using 2D-fingerprint plot as shown in the Figure 5.74,83−86
Computational Screening by Molecular Docking Studies
Protein and Ligand Preparation
The N-terminal domain of α-glucosidase protein in the human intestine (3CTT) was obtained from PDB (http://www.rcsb.org/pdb). The bound ligands, heteroatoms, and water molecules were removed from each of the protein molecules. Polar hydrogens and Gasteiger charges were assigned after merging of nonpolar hydrogen atoms. After preprocessing of the crystal structure, it was saved in the PDBQT file format using ADT. Ligands were optimized and saved in SDF format using Maestro 2D Draw (version 9.7). Using Marvin Sketch (ver 17.1.9.0; http://www.chemaxon.com), all the SDF formatted ligands were converted to the PDB format and subsequently to the PDBQT format in AutoDockTools (ADT)-1.5.6.
Molecular Docking Simulations
AutoDock 4.2.6 was used to carry out the docking simulations. A standard protocol for molecular docking was performed. To validate drug–target association, automated docking was performed with AutoDock 4.2.6.87 The grid map was set on the whole protein by AutoGrid 4 with grid dimensions to their maximum values. Ten independent genetic algorithm runs were performed for each ligand. The resulted values differing by <2.0 Å in positional root-mean-square deviation show the most favorable free energy of binding. All torsions were allowed to rotate during docking. Docked poses were visualized considering a maximum cutoff distance of 3.0 Å for hydrogen bonds. Analysis and visualizations were carried out using MacPymol,88 ADT, and PoseView.89
Antidiabetic activity: AGI Effect
AGI assay was performed by the hydrolysis of chromogenic substrate p-nitrophenyl-α-d-glucopyranoside (PNPG) using a previously explained method elsewhere.90 Newly synthesized compounds 8a–f each were dissolved in DMSO to obtain 1 mg/mL stock solution. From 1 mg/mL stock solution, further dilution was made into 0.1, 0.005, and 0.001 mg/mL using 0.1 M phosphate buffer (pH 7) to find their IC50 values. Briefly, 100 μL of reaction mixture in a 96-well microplate containing 40 μL (0.0015 units) of α glucosidase enzyme, 50 μL of compounds, and 10 μL of phosphate buffer (0.1 M) pH 7 was incubated at 37 °C for 20 min. After that, 50 μL of substrate (20 mM, PNPG) was added. The reaction mixture was further incubated at 37 °C for 30 min, and immediately, 100 μL of sodium carbonate (200 mM) solution was added for terminating the reaction. The reaction blank was prepared by replacing compounds and enzyme with buffer. Similarly, the control was prepared using buffer in the place of compound. Also, sample blanks were prepared for each sample replacing enzyme with buffer. The acarbose (50 mg) tablets were dissolved in buffer at 10 mg/mL concentration and used as a standard. The absorbance of p-nitrophenol released was quantified by reading at 405 nm using a Thermoscientific Multiscan plate reader. The percent inhibition of α glucosidase enzyme was calculated using the following formula.
The amount of sample required for the 50% inhibition of glucosidase enzyme was represented by IC50 values.
Plasma Recalcification Time
Plasma recalcification time was measured as described by Quick et al.91 Different concentrations of compounds 8a and 8d (0–40 μg) were pre-incubated with 0.2 mL citrated human plasma at 37 °C for 1 min. Clotting was measured after the addition of 20 μL CaCl2 (0.25 M).
Preparation of PRP and PPP
PRP and PPP were prepared using the method described by Ardlie and Han.92 The concentration of platelets in PRP was conserved as 3.1 × 108 platelets/mL with PPP at 37 °C.
Platelet Aggregation93
Platelet aggregation was performed in a Chronology dual channel whole blood/optical lumi aggregation system (model-700) as described by Born et al.93 PRP was pre-incubated with various concentrations of 6c (0–15 μg) in 0.25 mL reaction volume mixture. The platelet aggregation process was initiated by addition of agonists independently such as ADP and epinephrine.
Direct Hemolytic Activity
Direct hemolytic activity was performed as described previously.18 Concisely, packed human erythrocytes and phosphate-buffered saline (PBS) were mixed in the ratio 1:9. Different concentrations of compounds 8a–f (100 μg) were pre-incubated with 1 mL of this above solution for 1 h at 37 °C. Then, the reaction was arrested by adding 9 mL of ice cold PBS and centrifuged at 1000g for 10 min at 37 °C. Finally, the amount of hemoglobin released was measured at 540 nm.
Edema Inducing Activity94
Edema inducing activity was performed as described by Sannanaik Vishwanath et al.94 Briefly, compounds 8a and 8b were injected at different doses (10–100 μg) for the selected group of five mice at the right foot pads. After 1 h, mice were anaesthetized using diethyl ether, and then, the hind-limbs were removed carefully at the ankle joint and weighed. Saline (20 μL) was injected alone into the left foot serves as control. Minimum edema dose was defined as the amount of protein required to cause an edema ratio of 120%.
Hemorrhagic Activity95
Hemorrhagic activity was performed as described by Kondo et al.95 Briefly, higher concentrations of compounds 8a and 8d were injected with saline into the selected group of mice. Mice that received only saline act as the negative control whereas a group of mice that received snake venom serves as the positive control. After 3 h, all groups of mice were anesthetized, and then, hemorrhage was noticed by removing the dorsal patch of skin surface carefully. The minimum hemorrhagic dose was defined as the amount of the compound producing 10 mm of hemorrhage in diameter.
Statistical Analysis
The data are presented as mean ± SD. Statistical analyses were performed by Student’s t-test. A significant difference between the groups was considered if P < 0.01.
Acknowledgments
The authors are thankful to the Institute of Excellence, University of Mysore, Saif NMR Research Centre, and IISC Bangalore for providing NMR facilities and DST-PURSE Mangalore University for analytical data. The authors also thankful to Prof. Usha, Department of Biochemistry, Alva’s College, Moodbidri, for her help in studying biological activity.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b01476.
Physical properties and structural features of target compounds; IR spectra, 1H NMR spectra, RP-HPLC-spectrum, and LC-MS spectrum of specific compounds (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- DeFronzo R. A. Pathogenesis of Type 2 Diabetes Mellitus. Med. Clin. North Am. 2004, 88, 787–835. 10.1016/j.mcna.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Deshpande A.; Hayes M. H.; Schootman M. Diabetes-Related Complications. Phys. Ther. 2018, 88, 1254–1264. 10.2522/ptj.20080020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A. D.; Sherf M.; Vidavsky L.; Vardy D. A.; Shapiro J.; Meyerovitch J. Association between Psoriasis and the Metabolic Syndrome. Dermatology 2008, 216, 152–155. 10.1159/000111512. [DOI] [PubMed] [Google Scholar]
- Boutati E. I.; Raptis S. A. Postprandial Hyperglycaemia in Type 2 Diabetes: Pathophysiological Aspects, Teleological Notions and Flags for Clinical Practice. Diabetes. Metab. Res. Rev. 2004, 20, S13–S23. 10.1002/dmrr.528. [DOI] [PubMed] [Google Scholar]
- Alberti K. G. M. M.; Taylor R.. Informed Written Consent Was Obtained from Each Subject, and the Protocol Was Approved by the Newcastle Health Authority Ethical Committee. 337. 1988, 11, 337–344. [Google Scholar]
- Puls W.; Keup U.; Krause H. P.; Thomas G.; Hoffmeister F. Glucosidase inhibition. Naturwissenschaften 1977, 64, 536–537. 10.1007/bf00483562. [DOI] [PubMed] [Google Scholar]
- Shim Y.-J.; Doo H.-K.; Ahn S.-Y.; Kim Y.-S.; Seong J.-K.; Park I.-S.; Min B.-H. Inhibitory Effect of Aqueous Extract from the Gall of Rhus Chinensis on Alpha-Glucosidase Activity and Postprandial Blood Glucose. J. Ethnopharmacol. 2003, 85, 283–287. 10.1016/s0378-8741(02)00370-7. [DOI] [PubMed] [Google Scholar]
- Joshi S. R.; Standl E.; Tong N.; Shah P.; Kalra S.; Rathod R. Therapeutic potential of α-glucosidase inhibitors in type 2 diabetes mellitus: an evidence-based review. Expert Opin. Pharmacother. 2015, 16, 1959–1981. 10.1517/14656566.2015.1070827. [DOI] [PubMed] [Google Scholar]
- Ranilla L. G.; Kwon Y.-I.; Apostolidis E.; Shetty K. Phenolic Compounds, Antioxidant Activity and in Vitro Inhibitory Potential against Key Enzymes Relevant for Hyperglycemia and Hypertension of Commonly Used Medicinal Plants, Herbs and Spices in Latin America. Bioresour. Technol. 2010, 101, 4676–4689. 10.1016/j.biortech.2010.01.093. [DOI] [PubMed] [Google Scholar]
- Toeller M. Α-Glucosidase Inhibitors in Diabetes: Efficacy in NIDDM Subjects. Eur. J. Clin. Invest. 1994, 24, 31–35. 10.1111/j.1365-2362.1994.tb02253.x. [DOI] [PubMed] [Google Scholar]
- Atsumi S.; Nosaka C.; Ochi Y.; Iinuma H.; Umezawa K. Inhibition of Experimental Metastasis by an Alpha-Glucosidase Inhibitor, 1,6-Epi-Cyclophellitol. Cancer Res. 1993, 53, 4896–4899. [PubMed] [Google Scholar]
- Gamblin D. P.; Scanlan E. M.; Davis B. G. Glycoprotein Synthesis: An Update. Chem. Rev. 2009, 109, 131–163. 10.1021/cr078291i. [DOI] [PubMed] [Google Scholar]
- Rawlings A. J.; Lomas H.; Pilling A. W.; Lee M. J.-R.; Alonzi D. S.; Rountree J. S. S.; Jenkinson S. F.; Fleet G. W. J.; Dwek R. A.; Jones J. H.; et al. Synthesis and Biological Characterisation of NovelN-Alkyl-Deoxynojirimycin α-Glucosidase Inhibitors. ChemBioChem 2009, 10, 1101–1105. 10.1002/cbic.200900025. [DOI] [PubMed] [Google Scholar]
- Zanetti V. C.; da Silveira R. B.; Dreyfuss J. L.; Haoach J.; Mangili O. C.; Veiga S. S.; Gremski W. Morphological and Biochemical Evidence of Blood Vessel Damage and Fibrinogenolysis Triggered by Brown Spider Venom. Blood Coagul. Fibrinolysis 2002, 13, 135–148. 10.1097/00001721-200203000-00009. [DOI] [PubMed] [Google Scholar]
- Manjappa B.; Gangaraju S.; Girish K. S.; Kemparaju K.; Gonchigar S. J.; Shankar R. L.; Shinde M.; Sannaningaiah D. Momordica Charantia Seed Extract Exhibits Strong Anticoagulant Effect by Specifically Interfering in Intrinsic Pathway of Blood Coagulation and Dissolves Fibrin Clot. Blood Coagul. Fibrinolysis 2015, 26, 191–199. 10.1097/mbc.0000000000000191. [DOI] [PubMed] [Google Scholar]
- Devaraja S.; Girish K. S.; Devaraj V. R.; Kemparaju K. Factor Xa-like and fibrin(ogen)olytic activities of a serine protease from Hippasa agelenoides spider venom gland extract. J. Thromb. Thrombolysis 2010, 29, 119–126. 10.1007/s11239-009-0341-3. [DOI] [PubMed] [Google Scholar]
- Devaraja S.; Girish K. S.; Gowtham Y. N. J.; Kemparaju K. The Hag-protease-II is a fibrin(ogen)ase from Hippasa agelenoides spider venom gland extract: Purification, characterization and its role in hemostasis. Toxicon 2011, 57, 248–258. 10.1016/j.toxicon.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Nandish S. K.; Kengaiah J.; Ramachandraiah C.; Shivaiah A.; Chandramma; Girish K. S.; Kemparaju K.; Sannaningaiah D.. Anticoagulant, antiplatelet and fibrin clot hydrolysing activities of flax seed buffer extract. Phcog. Mag. 2018. [Google Scholar]
- Musiol R.; Jampilek J.; Buchta V.; Silva L.; Niedbala H.; Podeszwa B.; Palka A.; Majerz-Maniecka K.; Oleksyn B.; Polanski J. Antifungal Properties of New Series of Quinoline Derivatives. Bioorg. Med. Chem. 2006, 14, 3592–3598. 10.1016/j.bmc.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Musiol R.; Serda M.; Hensel-Bielowka S.; Polanski J. Quinoline-Based Antifungals. Curr. Med. Chem. 2010, 17, 1960–1973. 10.2174/092986710791163966. [DOI] [PubMed] [Google Scholar]
- Foley M. Quinoline Antimalarials Mechanisms of Action and Resistance and Prospects for New Agents. Pharmacol. Ther. 1998, 79, 55–87. 10.1016/s0163-7258(98)00012-6. [DOI] [PubMed] [Google Scholar]
- Sanchez J. P.; Domagala J. M.; Hagen S. E.; Heifetz C. L.; Hutt M. P.; Nichols J. B.; Trehan A. K. Quinolone Antibacterial Agents. Synthesis and Structure-Activity Relationships of 8-Substituted Quinoline-3-Carboxylic Acids and 1,8-Naphthyridine-3-Carboxylic Acids. J. Med. Chem. 1988, 31, 983–991. 10.1021/jm00400a016. [DOI] [PubMed] [Google Scholar]
- Alaimo R. J.; Spencer C. F.; Sheffer J. B.; Storrin R. J.; Hatton C. J.; Kohls R. E. Imidazo[4,5-f]quinolines. 4. Synthesis and anthelmintic activity of a series of imidazo[4,5-f]quinolin-9-ols. J. Med. Chem. 1978, 21, 298–300. 10.1021/jm00201a014. [DOI] [PubMed] [Google Scholar]
- Rossiter S.; Péron J.-M.; Whitfield P. J.; Jones K. Synthesis and Anthelmintic Properties of Arylquinolines with Activity against Drug-Resistant Nematodes. Bioorg. Med. Chem. Lett. 2005, 15, 4806–4808. 10.1016/j.bmcl.2005.07.044. [DOI] [PubMed] [Google Scholar]
- Chen Y.-L.; Hung H.-M.; Lu C.-M.; Li K.-C.; Tzeng C.-C. Synthesis and Anticancer Evaluation of Certain Indolo[2,3-b]Quinoline Derivatives. Bioorg. Med. Chem. 2004, 12, 6539–6546. 10.1016/j.bmc.2004.09.025. [DOI] [PubMed] [Google Scholar]
- Via L. D.; Gia O.; Gasparotto V.; Ferlin M. G. Discovery of a New Anilino-3H-Pyrrolo[3,2-f]Quinoline Derivative as Potential Anti-Cancer Agent. Eur. J. Med. Chem. 2008, 43, 429–434. 10.1016/j.ejmech.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Xie Z.-F.; Chai K.-Y.; Piao H.-R.; Kwak K.-C.; Quan Z.-S. Synthesis and anticonvulsant activity of 7-alkoxyl-4,5-dihydro-[1,2,4]triazolo[4,3-a]quinolines. Bioorg. Med. Chem. Lett. 2005, 15, 4803–4805. 10.1016/j.bmcl.2005.07.051. [DOI] [PubMed] [Google Scholar]
- Bekhit A. A.; El-Sayed O. A.; Aboulmagd E.; Park J. Y. Tetrazolo[1,5-a]Quinoline as a Potential Promising New Scaffold for the Synthesis of Novel Anti-Inflammatory and Antibacterial Agents. Eur. J. Med. Chem. 2004, 39, 249–255. 10.1016/j.ejmech.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Boini K. M.; Graf D.; Kuhl D.; Häussinger D.; Lang F. SGK1 Dependence of Insulin Induced Hypokalemia. Pflugers Arch. Eur. J. Physiol. 2009, 457, 955–961. 10.1007/s00424-008-0559-5. [DOI] [PubMed] [Google Scholar]
- Desai N. C.; Kotadiya G. M.; Trivedi A. R. Studies on Molecular Properties Prediction, Antitubercular and Antimicrobial Activities of Novel Quinoline Based Pyrimidine Motifs. Bioorg. Med. Chem. Lett. 2014, 24, 3126–3130. 10.1016/j.bmcl.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Sangani C. B.; Makawana J. A.; Zhang X.; Teraiya S. B.; Lin L.; Zhu H.-L. Design, Synthesis and Molecular Modeling of Pyrazole-Quinoline-Pyridine Hybrids as a New Class of Antimicrobial and Anticancer Agents. Eur. J. Med. Chem. 2014, 76, 549–557. 10.1016/j.ejmech.2014.01.018. [DOI] [PubMed] [Google Scholar]
- Taha M.; Ismail N. H.; Imran S.; Wadood A.; Rahim F.; Ali M.; Rehman A. U. Novel quinoline derivatives as potent in vitro α-glucosidase inhibitors: in silico studies and SAR predictions. Medchemcomm 2015, 6, 1826–1836. 10.1039/c5md00280j. [DOI] [Google Scholar]
- Taha M.; Ismail N. H.; Baharudin M. S.; Lalani S.; Mehboob S.; Khan K. M.; yousuf S.; Siddiqui S.; Rahim F.; Choudhary M. I. Synthesis crystal structure of 2-methoxybenzoylhydrazones and evaluation of their α-glucosidase and urease inhibition potential. Med. Chem. Res. 2015, 24, 1310–1324. 10.1007/s00044-014-1213-8. [DOI] [Google Scholar]
- Taha M.; Ismail N. H.; Imran S.; Rokei M. Q. B.; Saad S. M.; Khan K. M. Synthesis of new oxadiazole derivatives as α-glucosidase inhibitors. Bioorg. Med. Chem. 2015, 23, 4155–4162. 10.1016/j.bmc.2015.06.060. [DOI] [PubMed] [Google Scholar]
- Taha M.; Naz H.; Rasheed S.; Ismail N.; Rahman A.; Yousuf S.; Choudhary M. Synthesis of 4-Methoxybenzoylhydrazones and Evaluation of Their Antiglycation Activity. Molecules 2014, 19, 1286–1301. 10.3390/molecules19011286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-W.; Yang J.-Y.; Lee H.-S. Quinoline-2-carboxylic acid isolated from Ephedra pachyclada and its structural derivatives show inhibitory effects against α-glucosidase and α-amylase. J. Korean Soc. Appl. Biol. Chem. 2014, 57, 441–444. 10.1007/s13765-014-4156-3. [DOI] [Google Scholar]
- Merglen A.; Theander S.; Rubi B.; Chaffard G.; Wollheim C. B.; Maechler P. Glucose Sensitivity and Metabolism-Secretion Coupling Studied during Two-Year Continuous Culture in INS-1E Insulinoma Cells. Endocrinology 2004, 145, 667–678. 10.1210/en.2003-1099. [DOI] [PubMed] [Google Scholar]
- Terzioglu N.; Gürsoy A. Synthesis and Anticancer Evaluation of Some New Hydrazone Derivatives of 2,6-Dimethylimidazo[2,1-b][1,3,4]Thiadiazole-5-Carbohydrazide. Eur. J. Med. Chem. 2003, 38, 781–786. 10.1016/s0223-5234(03)00138-7. [DOI] [PubMed] [Google Scholar]
- Gadad A. K.; Noolvi M. N.; Karpoormath R. V. Synthesis and Anti-Tubercular Activity of a Series of 2-Sulfonamido/Trifluoromethyl-6-Substituted Imidazo[2,1-b]-1,3,4-Thiadiazole Derivatives. Bioorg. Med. Chem. 2004, 12, 5651–5659. 10.1016/j.bmc.2004.07.060. [DOI] [PubMed] [Google Scholar]
- Achar K. C. S.; Hosamani K. M.; Seetharamareddy H. R. In-Vivo Analgesic and Anti-Inflammatory Activities of Newly Synthesized Benzimidazole Derivatives. Eur. J. Med. Chem. 2010, 45, 2048–2054. 10.1016/j.ejmech.2010.01.029. [DOI] [PubMed] [Google Scholar]
- Labanauskas L. K.; Brukštus A. B.; Gaidelis P. G.; Buchinskaite V. A.; Udrenaite É. B.; Daukšas V. K. Synthesis and Antiinflammatory Activity of Some New 1-Acyl Derivatives of 2-Methylthio-5,6-Diethoxybenzimidazole. Pharm. Chem. J. 2000, 34, 353–355. 10.1023/a:1005213306544. [DOI] [Google Scholar]
- Sondhi S.; Bhattacharjee G.; Jameel R.; Shukla R.; Raghubir R.; Lozach O.; Meijer L. Antiinflammatory, analgesic and kinase inhibition activities of some acridine derivatives. Open Chem. 2004, 2, 1–15. 10.2478/bf02476181. [DOI] [Google Scholar]
- Garuti L.; Roberti M.; Pizzirani D.; Pession A.; Leoncini E.; Cenci V.; Hrelia S. Differential Antiproliferative Activity of New Benzimidazole-4,7-Diones. Il Farmaco 2004, 59, 663–668. 10.1016/j.farmac.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Kumar D.; Jacob M. R.; Reynolds M. B.; Kerwin S. M. Synthesis and Evaluation of Anticancer Benzoxazoles and Benzimidazoles Related to UK-1. Bioorg. Med. Chem. 2002, 10, 3997–4004. 10.1016/s0968-0896(02)00327-9. [DOI] [PubMed] [Google Scholar]
- Paramashivappa R.; Phani Kumar P.; Subba Rao P. V.; Srinivasa Rao A. Design, Synthesis and Biological Evaluation of Benzimidazole/Benzothiazole and Benzoxazole Derivatives as Cyclooxygenase Inhibitors. Bioorg. Med. Chem. Lett. 2003, 13, 657–660. 10.1016/s0960-894x(02)01006-5. [DOI] [PubMed] [Google Scholar]
- Özkay Y.; Tunalı Y.; Karaca H.; Işıkdağ I. Antimicrobial Activity and a SAR Study of Some Novel Benzimidazole Derivatives Bearing Hydrazone Moiety. Eur. J. Med. Chem. 2010, 45, 3293–3298. 10.1016/j.ejmech.2010.04.012. [DOI] [PubMed] [Google Scholar]
- Taha M.; Ismail N. H.; Imran S.; Mohamad M. H.; Wadood A.; Rahim F.; Saad S. M.; Rehman A.; Khan K. M. Synthesis, α-glucosidase inhibitory, cytotoxicity and docking studies of 2-aryl-7-methylbenzimidazoles. Bioorg. Chem. 2016, 65, 100–109. 10.1016/j.bioorg.2016.02.004. [DOI] [PubMed] [Google Scholar]
- Zawawi N. K. N. A.; Taha M.; Ahmat N.; Wadood A.; Ismail N. H.; Rahim F.; Azam S. S.; Abdullah N. Benzimidazole derivatives as new α-glucosidase inhibitors and in silico studies. Bioorg. Chem. 2016, 64, 29–36. 10.1016/j.bioorg.2015.11.006. [DOI] [PubMed] [Google Scholar]
- Dinparast L.; Valizadeh H.; Bahadori M. B.; Soltani S.; Asghari B.; Rashidi M.-R. Design, synthesis, α-glucosidase inhibitory activity, molecular docking and QSAR studies of benzimidazole derivatives. J. Mol. Struct. 2016, 1114, 84–94. 10.1016/j.molstruc.2016.02.005. [DOI] [Google Scholar]
- Kwak H. J.; Pyun Y. M.; Kim J. Y.; Pagire H. S.; Kim K. Y.; Kim K. R.; Rhee S. D.; Jung W. H.; Song J. S.; Bae M. A.; et al. Synthesis and Biological Evaluation of Aminobenzimidazole Derivatives with a Phenylcyclohexyl Acetic Acid Group as Anti-Obesity and Anti-Diabetic Agents. Bioorg. Med. Chem. Lett. 2013, 23, 4713–4718. 10.1016/j.bmcl.2013.05.081. [DOI] [PubMed] [Google Scholar]
- Kumar J. A.; Tiwari A. K.; Ali A. Z.; Madhusudhana K.; Reddy B. S.; Ramakrishna S.; China Raju B. New antihyperglycemic, α-glucosidase inhibitory, and cytotoxic derivatives of benzimidazoles. J. Enzyme Inhib. Med. Chem. 2010, 25, 80–86. 10.3109/14756360903017122. [DOI] [PubMed] [Google Scholar]
- Taha M.; Ismail N. H.; Imran S.; Wadood A.; Ali M.; Rahim F.; Khan A. A.; Riaz M. Novel thiosemicarbazide-oxadiazole hybrids as unprecedented inhibitors of yeast α-glucosidase and in silico binding analysis. RSC Adv. 2016, 6, 33733–33742. 10.1039/c5ra28012e. [DOI] [Google Scholar]
- Taha M.; Ismail N. H.; Imran S.; Wadood A.; Rahim F.; Saad S. M.; Khan K. M.; Nasir A. Synthesis, molecular docking and α-glucosidase inhibition of 5-aryl-2-(6′-nitrobenzofuran-2′-yl)-1,3,4-oxadiazoles. Bioorg. Chem. 2016, 66, 117–123. 10.1016/j.bioorg.2016.04.006. [DOI] [PubMed] [Google Scholar]
- Bondock S.; Adel S.; Etman H. A.; Badria F. A. Synthesis and Antitumor Evaluation of Some New 1,3,4-Oxadiazole-Based Heterocycles. Eur. J. Med. Chem. 2012, 48, 192–199. 10.1016/j.ejmech.2011.12.013. [DOI] [PubMed] [Google Scholar]
- Salahuddin; Shaharyar M.; Mazumder A.; Ahsan M. J. Synthesis, Characterization and Anticancer Evaluation of 2-(Naphthalen-1-Ylmethyl/Naphthalen-2-Yloxymethyl)-1-[5-(Substituted Phenyl)-[1,3,4]Oxadiazol-2-Ylmethyl]-1H-Benzimidazole. Arab. J. Chem. 2014, 7, 418–424. 10.1016/j.arabjc.2013.02.001. [DOI] [Google Scholar]
- Aziz-ur-Rehman; Siddiqa A.; Abbasi M. A.; Rasool S.; Siddiqui S. Z.; Ahmad I.; Afzal S. Synthesis of Some New 5-Substituted-2-((6-Chloro-3,4-Methylenedioxyphenyl)Methylthio)-1,3,4-Oxadiazole Derivatives as Suitable Antibacterial Inhibitors. Bull. Fac. Pharm. (Cairo Univ.) 2015, 53, 37–43. 10.1016/j.bfopcu.2014.10.001. [DOI] [Google Scholar]
- Desai N. C.; Dodiya A. M.; Rajpara K. M.; Rupala Y. M. Synthesis and Antimicrobial Screening of 1,3,4-Oxadiazole and Clubbed Thiophene Derivatives. J. Saudi Chem. Soc. 2014, 18, 255–261. 10.1016/j.jscs.2011.06.020. [DOI] [Google Scholar]
- Malladi S.; Isloor A. M.; Peethambar S. K.; Fun H. K. Synthesis and Biological Evaluation of Newer Analogues of 2,5-Disubstituted 1,3,4-Oxadiazole Containing Pyrazole Moiety as Antimicrobial Agents. Arab. J. Chem. 2014, 7, 1185–1191. 10.1016/j.arabjc.2013.12.020. [DOI] [Google Scholar]
- Gadegoni H.; Manda S. Synthesis and Screening of Some Novel Substituted Indoles Contained 1,3,4-Oxadiazole and 1,2,4-Triazole Moiety. Chin. Chem. Lett. 2013, 24, 127–130. 10.1016/j.cclet.2013.01.001. [DOI] [Google Scholar]
- Harish K. P.; Mohana K. N.; Mallesha L.; Prasanna kumar B. N. Synthesis of novel 1-[5-(4-methoxy-phenyl)-[1,3,4]oxadiazol-2-yl]-piperazine derivatives and evaluation of their in vivo anticonvulsant activity. Eur. J. Med. Chem. 2013, 65, 276–283. 10.1016/j.ejmech.2013.04.054. [DOI] [PubMed] [Google Scholar]
- Musad E. A.; Mohamed R.; Ali Saeed B.; Vishwanath B. S.; Lokanatha Rai K. M. Synthesis and Evaluation of Antioxidant and Antibacterial Activities of New Substituted Bis(1,3,4-Oxadiazoles), 3,5-Bis(Substituted) Pyrazoles and Isoxazoles. Bioorg. Med. Chem. Lett. 2011, 21, 3536–3540. 10.1016/j.bmcl.2011.04.142. [DOI] [PubMed] [Google Scholar]
- Zawawi N. K. N. A.; Taha M.; Ahmat N.; Wadood A.; Ismail N. H.; Rahim F.; Ali M.; Abdullah N.; Khan K. M. Novel 2,5-disubtituted-1,3,4-oxadiazoles with benzimidazole backbone: A new class of β-glucuronidase inhibitors and in silico studies. Bioorg. Med. Chem. 2015, 23, 3119–3125. 10.1016/j.bmc.2015.04.081. [DOI] [PubMed] [Google Scholar]
- Rashid M.; Husain A.; Mishra R. Synthesis of Benzimidazoles Bearing Oxadiazole Nucleus as Anticancer Agents. Eur. J. Med. Chem. 2012, 54, 855–866. 10.1016/j.ejmech.2012.04.027. [DOI] [PubMed] [Google Scholar]
- Husain A.; Rashid M.; Mishra R.; Parveen S.; Shin D.-S.; Kumar D. Benzimidazole Bearing Oxadiazole and Triazolo-Thiadiazoles Nucleus: Design and Synthesis as Anticancer Agents. Bioorg. Med. Chem. Lett. 2012, 22, 5438–5444. 10.1016/j.bmcl.2012.07.038. [DOI] [PubMed] [Google Scholar]
- Puthiyapurayil P.; Poojary B.; Chikkanna C.; Buridipad S. K. Design, Synthesis and Biological Evaluation of a Novel Series of 1,3,4-Oxadiazole Bearing N-Methyl-4-(Trifluoromethyl)Phenyl Pyrazole Moiety as Cytotoxic Agents. Eur. J. Med. Chem. 2012, 53, 203–210. 10.1016/j.ejmech.2012.03.056. [DOI] [PubMed] [Google Scholar]
- Bharadwaj S. S.; Poojary B.; Madan Kumar S.; Byrappa K.; Nagananda G. S.; Chaitanya A. K.; Zaveri K.; Yarla N. S.; Shiralgi Y.; Kudva A. K.; et al. Design, Synthesis and Pharmacological Studies of Some New Quinoline Schiff Bases and 2,5-(Disubstituted-[1,3,4])-Oxadiazoles. New J. Chem. 2017, 41 (). 10.1039/c6nj03913h [DOI] [Google Scholar]
- Vasantha K.; Basavarajaswamy G.; Vaishali Rai M.; Boja P.; Pai V. R.; Shruthi N.; Bhat M. Rapid ’one-pot’ synthesis of a novel benzimidazole-5-carboxylate and its hydrazone derivatives as potential anti-inflammatory and antimicrobial agents. Bioorg. Med. Chem. Lett. 2015, 25, 1420–1426. 10.1016/j.bmcl.2015.02.043. [DOI] [PubMed] [Google Scholar]
- Devaraja S.; Nagaraju S.; Mahadeswaraswamy Y. H.; Girish K. S.; Kemparaju K. A Low Molecular Weight Serine Protease: Purification and Characterization from Hippasa Agelenoides (Funnel Web) Spider Venom Gland Extract. Toxicon 2008, 52, 130–138. 10.1016/j.toxicon.2008.04.168. [DOI] [PubMed] [Google Scholar]
- Rachidi S.; Aldin E. S.; Greenberg C.; Sachs B.; Streiff M.; Zeidan A. M. The Use of Novel Oral Anticoagulants for Thromboprophylaxis after Elective Major Orthopedic Surgery. Expert Rev. Hematol. 2013, 6, 677–695. 10.1586/17474086.2013.853430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenawy H. I.; Boral I.; Bevington A. Complement-Coagulation Cross-Talk: A Potential Mediator of the Physiological Activation of Complement by Low PH. Front. Immunol. 2015, 6, 1–10. 10.3389/fimmu.2015.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchen S.; Gray E.; Mackie I.; Baglin T.; Makris M. Measurement of Non-Coumarin Anticoagulants and Their Effects on Tests of Haemostasis: Guidance from the British Committee for Standards in Haematology. Br. J. Haematol. 2014, 166, 830–841. 10.1111/bjh.12975. [DOI] [PubMed] [Google Scholar]
- Sim P. J.; Wiedmers T.; Esmonsliqb C. T.; Weissll H. J.; Shattil S. J. Assembly of the Platelet Prothrombinase Complex Is Linked to Vesiculation of the Platelet Plasma Membrane. Studies in Scott syndrome: an isolated defect in platelet procoagulant activity.. J. Biol. Chem. 1989, 264, 17049. [PubMed] [Google Scholar]
- Poon M.-C.; dʼOiron R. Recombinant activated factor VII (NovoSeven) treatment of platelet-related bleeding disorders. Blood Coagulation Fibrinolysis 2000, 11, S55–S68. 10.1097/00001721-200004001-00013. [DOI] [PubMed] [Google Scholar]
- Bharadwaj S. S.; Poojary B.; Kumar S. M.; Byrappa K. Synthesis, characterization, thermal analysis, crystal structure and Hirshfeld surface analysis of 2-(3, 5-difluorophenyl)quinoline-4-carboxylic acid. Chem. Data Collect. 2017, 9–10, 134–142. 10.1016/j.cdc.2017.05.003. [DOI] [Google Scholar]
- Bharadwaj S. S.; Kumara K.; Poojary B.; Yathirajan H. S.; Byrappa K.; Lokanath N. K.; Kumar S. M. Ethyl 2-(3,5-Difluorophenyl)Quinoline-4-Carboxylate: A Second Triclinic Polymorph. IUCrData 2016, 1, x160739. [Google Scholar]
- Alkhader M. A.; Perera R. C.; Sinha R. P.; Smalley R. K. Synthesis of polynuclear heterocycles. Part 4. Imidazo[4,5-g][3,1]benzoxazinones, imidazo[4,5-g]quinazolinones, imidazo[4,5-g]quinazolinediones, and imidazo[4,5-f]indazolinones. J. Chem. Soc., Perkin Trans. 1 1979, 1056–1062. 10.1039/p19790001056. [DOI] [Google Scholar]
- Özden S.; Atabey D.; Yıldız S.; Göker H. Synthesis and Potent Antimicrobial Activity of Some Novel Methyl or Ethyl 1H-Benzimidazole-5-Carboxylates Derivatives Carrying Amide or Amidine Groups. Bioorg. Med. Chem. 2005, 13, 1587–1597. 10.1016/j.bmc.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Kumar V.; Poojary B.; Prathibha A.; Shruthi N. Synthesis of Some Novel 1,2-Disubstituted Benzimidazole-5-carboxylates via One-Pot Method Using Sodium Dithionite and Its Effect onN-Debenzylation. Synth. Commun. 2014, 44, 3414–3425. 10.1080/00397911.2014.943347. [DOI] [Google Scholar]
- Ogi A.; Inaba K. X-ray thin film measurement techniques. The Rigaku J 2011, 27, 1–26. [Google Scholar]
- Sheldrick G. M. A short history ofSHELX. Acta Crystallogr., Sect. A: Found. Crystallogr. 2008, 64, 112–122. 10.1107/s0108767307043930. [DOI] [PubMed] [Google Scholar]
- Spek A. L. PLATON, An Integrated Tool for the Analysis of the Results of a Single Crystal Structure Determination. Acta Crystallogr., Sect. A: Found. Crystallogr. 1990, 46, 34. 10.1107/S0108767390099780. [DOI] [Google Scholar]
- Macrae C. F.; Bruno I. J.; Chisholm J. A.; Edgington P. R.; McCabe P.; Pidcock E.; Rodriguez-Monge L.; Taylor R.; van de Streek J.; Wood P. A. Mercury CSD 2.0- new features for the visualization and investigation of crystal structures. J. Appl. Crystallogr. 2008, 41, 466–470. 10.1107/s0021889807067908. [DOI] [Google Scholar]
- McKinnon J. J.; Spackman M. A.; Mitchell A. S. Novel Tools for Visualizing and Exploring Intermolecular Interactions in Molecular Crystals. J. Acta. Crystallogr. 2004, 60, 627–668. 10.1107/s0108768104020300. [DOI] [PubMed] [Google Scholar]
- Spackman M. A.; Jayatilaka D. Hirshfeld Surface Analysis. CrystEngComm 2009, 11, 19–32. 10.1039/b818330a. [DOI] [Google Scholar]
- Wood P. A.; McKinnon J. J.; Parsons S.; Pidcock E.; Spackman M. A. Analysis of the Compression of Molecular Crystal Structures Using Hirshfeld Surfaces. CrystEngComm 2008, 10, 368–376. 10.1039/b801571f. [DOI] [Google Scholar]
- Kumar S. M.; Manjunath B. C.; Lingaraju G. S.; Abdoh M. M. M.; Sadashiva M. P.; Lokanath N. K. A Hirshfeld Surface Analysis and Crystal Structure of 2’-[1-(2-Fluoro-Phenyl)-1H-tetrazol-5-Yl]-4-Methoxy-Biphenyl-2-Carbaldehyde. Cryst. Struct. Theory Appl. 2013, 2, 124–131. 10.4236/csta.2013.23017. [DOI] [Google Scholar]
- Buzko O. V.; Bishop A. C.; Shokat K. M. Modified AutoDock for Accurate Docking of Protein Kinase Inhibitors. J. Comput. Aided. Mol. Des. 2002, 16, 113–127. 10.1023/a:1016366013656. [DOI] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkóczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L.-W.; Kapral G. J.; Grosse-Kunstleve R. W.; et al. PHENIX: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 213–221. 10.1107/s0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierand K.; Rarey M. From Modeling to Medicinal Chemistry: Automatic Generation of Two-Dimensional Complex Diagrams. ChemMedChem 2007, 2, 853–860. 10.1002/cmdc.200700010. [DOI] [PubMed] [Google Scholar]
- Tao Y.; Zhang Y.; Cheng Y.; Wang Y. Rapid screening and identification ofα-glucosidase inhibitors from mulberry leaves using enzyme-immobilized magnetic beads coupled with HPLC/MS and NMR. Biomed. Chromatogr. 2013, 27, 148–155. 10.1002/bmc.2761. [DOI] [PubMed] [Google Scholar]
- Quick A. J.; Stanley-Brown M.; Bancroft F. W. A Study of the Coagulation Defect in Hemophilia and in Jaundice. Am. J. Med. Sci. 1935, 190, 501–510. 10.1097/00000441-193510000-00009. [DOI] [Google Scholar]
- Ardlie N. G.; Han P. Enzymatic Basis for Platelet Aggregation and Release: The Significance of the ’Platelet Atmosphere’ and the Relationship between Platelet Function and Blood Coagulation. Br. J. Haematol. 1974, 26, 331–356. 10.1111/j.1365-2141.1974.tb00477.x. [DOI] [PubMed] [Google Scholar]
- Born G. V. R.; Bergquist D.; Arfors K.-E. Evidence for inhibition of platelet activation in blood by a drug effect on erythrocytes. Nature 1976, 259, 233–235. 10.1038/259233a0. [DOI] [PubMed] [Google Scholar]
- Sannanaik Vishwanath B.; Manjunatha Kini R.; Veerabasappa Gowda T. Characterization of three edema-inducing phospholipase A2 enzymes from habu (Trimeresurus flavoviridis) venom and their interaction with the alkaloid aristolochic acid. Toxicon 1987, 25, 501–515. 10.1016/0041-0101(87)90286-8. [DOI] [PubMed] [Google Scholar]
- Kondo H.; Kondo S.; Irezawa H.; Murata R.; Ohsaka A. Studies on the Quantitative Method for Determination of Hemorrhagic Activity of Habu Snake Venom. Jpn. J. Med. Sci. Biol. 1960, 13, 43–51. 10.7883/yoken1952.13.43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.