Abstract
Hexose transporter-deficient yeast strains are valuable testbeds for the study of sugar transport by native and heterologous transporters. In the popular Saccharomyces cerevisiae strain EBY.VW4000, deletion of 21 transporters completely abolished hexose transport. However, repeated use of the LoxP/Cre system in successive deletion rounds also resulted in major chromosomal rearrangements, gene loss and phenotypic changes. In the present study, CRISPR/SpCas9 was used to delete the 21 hexose transporters in an S. cerevisiae strain from the CEN.PK family in only three deletion rounds, using 11 unique guide RNAs. Even upon prolonged cultivation, the resulting strain IMX1812 (CRISPR-Hxt0) was unable to consume glucose, while its growth rate on maltose was the same as that of a strain equipped with a full set of hexose transporters. Karyotyping and whole-genome sequencing of the CRISPR-Hxt0 strain with Illumina and Oxford Nanopore technologies did not reveal chromosomal rearrangements or other unintended mutations besides a few SNPs. This study provides a new, ‘genetically unaltered’ hexose transporter-deficient strain and supplies a CRISPR toolkit for removing all hexose transporter genes from most S. cerevisiae laboratory strains in only three transformation rounds.
Keywords: Saccharomyces cerevisiae, hexose transport, CRISPR/SpCas9, genome editing, multiplexing, genome rearrangements
This study supplies a CRISPR-based toolkit enabling the simple, fast and precise deletion of 21 hexose-transporter genes in Saccharomyces cerevisiae laboratory strains and thereby the construction of hexose-transport deficient strains.
INTRODUCTION
Redundancy in hexose transporters is a common feature in living cells, which is best exemplified by the multiplicity of transmembrane proteins that are able to transport glucose, with a broad range of rates and affinities, found in Saccharomyces cerevisiae (Kruckeberg 1996; Boles and Hollenberg 1997; Özcan and Johnston 1999; Maier et al.2002). This complexity has turned S. cerevisiae into an attractive and powerful model to study the mechanisms connecting extracellular signals to transcriptional responses in eukaryotes (Rolland, Winderickx and Thevelein 2001). While its redundancy in hexose transporter genes initially complicated the use of S. cerevisiae as a testbed for heterologous transporters, the construction of an S. cerevisiae hexose-transport deficient strain (named EBY.VW4000 or Hxt0) (Wieczorke et al.1999) provided a unique and intensively used platform for functional analysis of native hexose transporters and studies on hexose transport (see for instance Wieczorke et al.2003; Schussler et al.2006; Price et al.2010; Young et al.2011; Xuan et al.2013; Boles and Oreb 2018) and of transport of other biotechnologically relevant sugars (Young, Lee and Alper 2010; Thomik et al.2017).
In EBY.VW4000, which was derived from the CEN.PK strain lineage (Entian and Kötter 2007), complete abolishment of hexose uptake required the deletion of 21 genes: HXT1 to HXT16, GAL2, STL1, AGT1 (also known as MAL11), MPH2 and MPH3 (HXT17 is absent in the CEN.PK family; Wieczorke et al.1999) in 17 deletion rounds. With the exception of GAL2, whose deletion was performed using URA3 as selectable marker and 5-FOA for marker recycling, all genes were deleted using the marker module KanMX (Wach et al.1994) flanked by LoxP sites. Each deletion was followed by excision of the marker module by the Cre recombinase targeting the LoxP sites (Güldener et al.1996). While the deletion of 21 genes in 17 sequential rounds of selective cultivation resulted in only a small number of single-nucleotide mutations, the repetitive use of the LoxP/Cre system for gene deletion and marker recycling has caused large genome alterations (Solis-Escalante et al.2015). During the strain construction process, four different chromosomal translocations occurred between six distinct LoxP scars, resulting in four neo-chromosomes and two truncated versions of existing chromosomes, IV-t and XV-t. These translocations also resulted in the loss of two subtelomeric regions harboring eight genes. While the physiology of EBY.VW4000 has not been extensively explored, its sporulation and spore-germination deficiencies upon crossing with strains from the parental lineage indicated that drastic chromosomal rearrangements and gene loss influenced its physiology (Solis-Escalante et al.2015).
Since the construction of EBY.VW4000, the advent of CRISPR-based genome editing has revolutionized strain construction. Extensive genetic engineering programs that would cost years of intensive effort in the pre-CRISPR era are now attainable within months (Mans et al.2017). The RNA-guided endonucleases, such as Streptococcus pyogenes Cas9 (SpCas9), can efficiently and precisely edit multiple genomic loci to simultaneously perform gene deletion or integration, or to introduce single nucleotide mutations. While several CRISPR-based approaches have been developed to enable multiplex genome editing (Bao et al.2015; Świat et al.2017; Ferreira et al.2018), so far the record in S. cerevisiae is of six simultaneously edited loci with six single guide RNAs using SpCas9 (Mans et al.2017). Furthermore, while genome editing can be specifically designed to cause chromosomal rearrangements, intensive utilization of SpCas9 for strain construction has not been reported to induce unwanted and untargeted recombinations between chromosomes. CRISPR/SpCas9 therefore appears to be a very promising methodology to rapidly and accurately construct novel hexose transport-deficient S. cerevisiae strains.
The goal of the present study was to develop a simple and fast CRISPR-based strategy to construct hexose-transport deficient S. cerevisiae strains with minimum alterations in genome sequence and structure. To this end, a series of six plasmids carrying combinations of single guide RNAs (sgRNAs) targeting the 21 hexose transporters was constructed. Exploiting the strong homology of hexose transporters, the number of single guide RNAs could be minimized to 11, enabling the complete deletion of all hexose transporters in three transformations rounds only. The genome sequence and copy number variation of the resulting CRISPR-Hxt0 strain IMX1812 were characterized by Illumina sequencing and its chromosomal architecture was explored by a combination of Nanopore sequencing, karyotyping and flow cytometry-based ploidy analysis. Finally, the ability of the CRISPR-Hxt0 strain to grow on various hexoses was investigated.
MATERIALS AND METHODS
Strains, media and storage
The Saccharomyces cerevisiae strains used in this work belong to the CEN.PK family (Entian and Kötter 2007; Salazar et al.2017) and are listed in Table 1. The strains were grown at 30°C in 500 mL flasks containing 100 mL chemically defined medium (synthetic medium, SM) (Verduyn et al.1992) or yeast extract peptone (YP) medium supplemented with 20 g L−1 glucose (YPD) or 6.8 g L−1 maltose (YPM) in an Innova incubator shaker (New Brunswick Scientific, Edison, NJ) set at 200 rpm. SM contains 3 g L−1 KH2PO4, 0.5 g L−1 MgSO4·7H2O, 5 g L−1 (NH4)2SO4, 1 ml L−1 of trace element solution and 1 ml L−1 of vitamin solution as described in (Verduyn et al.1992). YP medium contains 10 g L−1 yeast extract and 20 g L−1 peptone. When required, SM was supplemented with 150 mg L−1 uracil, 125 mg L−1 histidine, 500 mg L−1 leucine and/or 75 mg L−1 tryptophan (Pronk 2002).
Table 1.
Strains used in this study.
| Name | Relevant genotype | Parental strain | Origin |
|---|---|---|---|
| CEN.PK113-7D | MATa URA3 TRP1 LEU2 HIS3 | (Entian and Kötter 2007) | |
| CEN.PK2-1C | MATa ura3-52 trp1-1 his3Δ | (Entian and Kötter 2007) | |
| CEN.PK122 | MATa/MATα | (Entian and Kötter 2007) | |
| EBY.VW4000 | MATa ura3-52 trp1-1 leu2-3,112 his3Δ hxt13Δ::loxP hxt15Δ::loxP hxt16Δ::loxP hxt14Δ::loxP hxt12Δ::loxP hxt9Δ::loxP hxt11Δ::loxP hxt10Δ::loxP hxt8Δ::loxP hxt4-1-5Δ::loxP hxt2Δ::loxP hxt3-6-7Δ::loxP gal2Δ stl1Δ::loxP agt1Δ::loxP mph2(ydl247w)Δ::loxP mph3(yjr160c)Δ::loxP | CEN.PK2-1C | (Wieczorke et al.1999) |
| IMX672 | MATa ura3-52 trp1-1 leu2-3,112 his3Δ can1Δ::Spcas9-natNT2 | CEN.PK2-1C | (Mans et al.2015) |
| IMX1521 | MATa ura3-52 trp1-1 leu2-3,112 his3Δ can1Δ::Spcas9-natNT2 gal2Δ hxt4-1-5Δ hxt3-6-7Δ::ars4 hxt8Δ hxt14Δ | IMX672 | This study |
| IMX1541 | MATa ura3-52 trp1-1 leu2-3,112 his3Δ can1Δ::Spcas9-natNT2 gal2Δ hxt4-1-5Δ hxt3-6-7Δ::ars4 hxt8Δ hxt14Δ hxt2Δ hxt9Δ hxt10Δ hxt12Δ hxt13Δ hxt15Δ hxt16Δ | IMX1521 | This study |
| IMX1812 | MATa ura3-52 trp1-1 leu2-3,112 his3Δ can1Δ::Spcas9-natNT2 gal2Δ hxt4-1-5Δ hxt3-6-7Δ::ars4 hxt8Δ hxt14Δ hxt2Δ hxt9Δ hxt10Δ hxt11Δ hxt12Δ hxt13Δ hxt15Δ hxt16Δ mph2(ydl247w)Δ mph3(yjr160c)Δ mal11Δ stl1Δ | IMX1541 | This study |
To obtain solid media, 2% (w/v) agar was added. Yeast strains and Escherichia coli cultures were stored by adding glycerol to the cultures to a final concentration of 30% (v/v) and stored at –80°C.
Molecular biology techniques
Plasmids were isolated from E. coli with the GenElute Plasmid Miniprep Kit (Sigma Aldrich, St. Louis, MI) according to the supplier's instruction. DNA amplification was performed by PCR using Phusion® High-Fidelity DNA polymerase (Thermo Fisher Scientific, Waltham, MA) as previously described (Mans et al.2015). Separation of DNA fragments was done in 1% (w/v) agarose gel (Thermo Fisher Scientific) with SERVA DNA Stain Clear G in 1x TAE buffer (Thermo Fischer Scientific). DNA fragments were isolated from gel using the Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA). DNA concentrations were measured with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific). Plasmids were constructed in vitro with NEBuilder® HiFi DNA Assembly Master Mix (New England Biolabs, Beverly, MA) according to manufacturer's protocol, but with reaction volumes down-scaled by 4-fold. Colony PCR was performed using DreamTaq PCR Master Mix (2x) (Thermo Fisher Scientific). Following the manufacturer's instruction. Yeast transformation was performed with the LiAc/ssDNA method (Gietz and Woods 2002). Yeast genomic DNA was extracted as previously described (Lõoke, Kristjuhan and Kristjuhan 2011) prior to colony PCR.
Plasmid construction
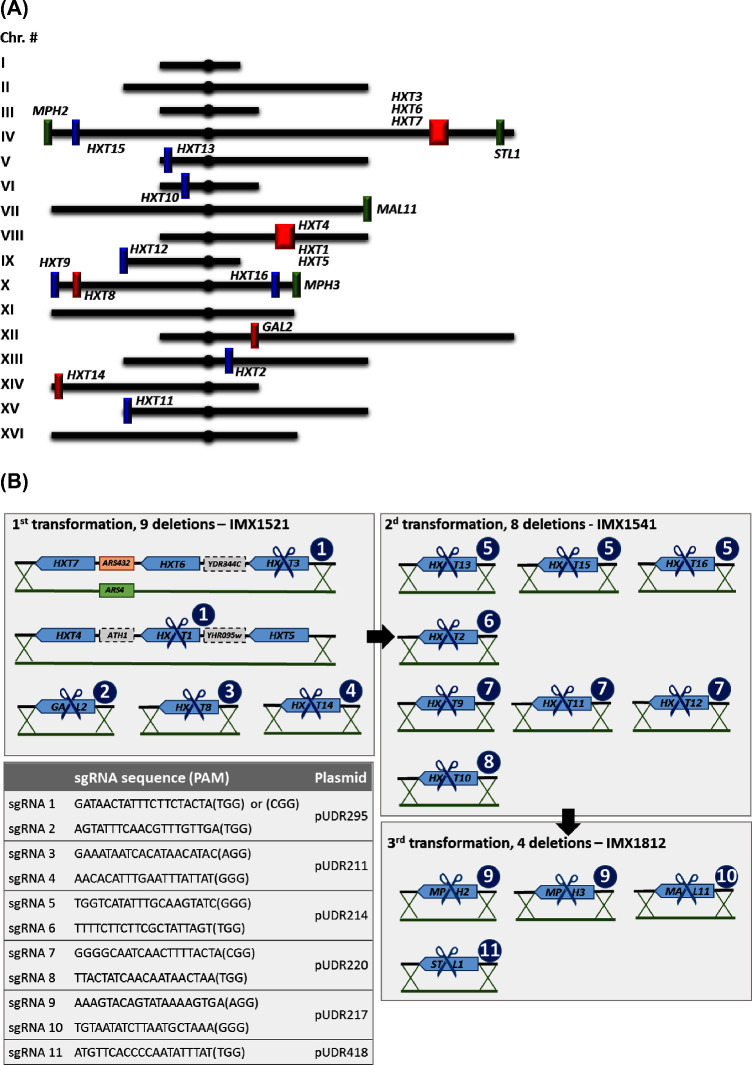
The double sgRNA method for CRISPR/SpCas9-mediated deletions based on pROS plasmid series (Mans et al.2015, 2018) was used to construct several sgRNA plasmids. The guide RNA sequences were selected to perform the 21 hexose transporter deletions in a minimal number of transformation rounds. Based on alignments of HXT gene sequences, considering sequences with AT content above 65% and the absence of strong secondary structures, 11 sgRNAs were designed (sgRNA labelled 1 to 11, Fig. 1B) to yield a kit of six sgRNA-carrying plasmids.
Figure 1.
(A) Overview of the chromosomal localization of the deleted hexose transporters. Genes indicated with the same color were removed in the same deletion round. Red, first round; blue, second round; green, third round. (B) Deletion strategy. The scissors indicate the gene targeted by SpCas9 editing. The circled numbers indicate the sgRNA used to guide SpCas9 for editing.
The sgRNA plasmids were assembled in vitro from two DNA parts (using NEBuilder HiFi DNA Assembly Master Mix, New England Biolabs). The first part was the plasmid backbone obtained by PCR with Phusion polymerase, using a single primer (6005, Supplemental Material 1) binding at each of the two SNR52 promoters of either pROS10, pROS14 or pROS16 (Table 2; Mans et al.2018). The second part was a 2 μm fragment surrounded by two sgRNA sequences. This fragment was obtained by PCR amplification (DreamTaq polymerase) of the 2 μm of pROS10 using primers containing the specific 20 bp sgRNA recognition (target) sequence and a 50 bp sequence, homologous to the linearized plasmid backbone. Plasmid assembly was followed by chemical transformation to chemically competent E. coli XL1-blue cells according to the supplier's instructions (Agilent Technologies, Santa Clara, CA) for storage and plasmid propagation, in Lysogeny Broth (LB) medium with 100 mg L−1 ampicillin.
Table 2.
Plasmids used in this study.
| Name | Relevant characteristics | Addgene # | Origin |
|---|---|---|---|
| pRS416 | CEN6/ARS4 ampR URA3 | (Sikorski and Hieter 1989) | |
| pROS10 | 2 μm ampR URA3 sgRNA-CAN1.Y sgRNA-ADE2.Y | #107924 | (Mans et al.2015) |
| pROS14 | 2 μm ampR KlLEU2 sgRNA-CAN1.Y sgRNA-ADE2.Y | #107928 | (Mans et al.2015) |
| pROS16 | 2 μm ampR HIS3 sgRNA-CAN1.Y sgRNA-ADE2.Y | #107930 | (Mans et al.2015) |
| pUDR211 | 2 μm ampR KlLEU2 sgRNA3-HXT8 sgRNA4-HXT14 | #113870 | This study |
| pUDR214 | 2 μm ampR URA3 sgRNA5- HXT13-15-16 sgRNA6-HXT2 | #113871 | This study |
| pUDR217 | 2 μm ampR HIS3 sgRNA9-MPH2-3 sgRNA10-MAL11 | #113872 | This study |
| pUDR220 | 2 μm ampR KlLEU2 sgRNA8-HXT10 sgRNA7-HXT9-11-12 | #113873 | This study |
| pUDR295 | 2 μm ampR HIS3 sgRNA2-GAL2 sgRNA1-HXT4-1-5;HXT3-6-7 | #113874 | This study |
| pUDR418 | 2 μm ampR URA3 sgRNA11-STL1 sgRNA11-STL1 | #113875 | This study |
The E. coli colonies were picked and mixed directly in the DreamTaq PCR mix, containing primers binding to the specific 20 bp recognition sequence (primers 11662-11670, 11756) together with two primers binding in the plasmid backbone (primers 4034 and 5941), confirming the presence of one or two target sgRNAs. This PCR will result in four products, since the sgRNA primer can bind on either side of the 2 μM fragment. The orientation of the 2 μm fragment was confirmed by restriction with FastDigest enzymes (Thermo Fisher Scientific) following the supplier's manual. All primers for plasmid construction, diagnostic PCR and sequencing are listed in Supplemental Material 1.
The plasmids pUDR211 and pUDR220 were constructed by in vitro assembly with the backbone of pROS14 (KlLEU2, Addgene plasmid #107928; Mans et al.2018) and insert fragment amplified with primers containing the HXT8 (primer 9575) and HXT14 (9577) recognition sequences and primers containing the HXT10 (9576) and HXT9, HXT11, HXT12 (9572) recognition sequences, respectively. Similarly, the plasmids pUDR214 and pUDR418 were constructed with the pROS10 backbone (URA3) (Addgene plasmid #107924; Mans et al.2018) and insert fragment amplified with primers containing spacer sgRNA targeting HXT2 (9574) and HXT13, HXT15, HXT16 (9573) recognition sequences and insert fragment amplified with primers containing the spacer sgRNA targeting STL1 (primer 13616) respectively (Fig. 1). The plasmids pUDR217 and pUDR295 were constructed with the pROS16 backbone (HIS3, Addgene plasmid #107930; Mans et al.2018) and insert fragments amplified with primers containing spacers sgRNA targeting MPH2, MPH3 (primer 9579) and MAL11 (primer 9585) recognition sequences and insert fragments amplified with primers containing spacers sgRNA targeting GAL2 and HXT1,3-7 respectively. Plasmid confirmation was performed by Sanger sequencing (using primers 2758, 6197, 8556 and 2918) (BaseClear B.V., Leiden, The Netherlands).
Strain construction
The CRISPR/SpCas9 system with double sgRNA-containing plasmids was used to delete 21 genes in the quadruple auxotrophic strain IMX672 in three transformation rounds (Mans et al.2015). As described above, six sgRNA expression plasmids were constructed, pUDR211, pUDR214, pUDR217, pUDR220, pUDR295 and pUDR418 (Table 2). Strains were transformed with combinations of two plasmids together with double-stranded DNA fragments (repair DNA) designed to repair the double strand break created by SpCas9. With one exception, repair DNA was a 120 bp oligonucleotide sequence composed of two adjacent 60 bp sequences homologous to sequences located up- and downstream of the DNA break. To prepare the repair DNA fragments, two 120 bp complementary single-stranded oligonucleotides (Supplemental Material 1) were heated for 5 min at 95°C and then cooled to room temperature. To confirm correct annealing, the dsDNA concentration was measured with Qubit fluorometer 2.0 and Qubit dsDNA BR Assay kit (Thermo Fisher Scientific). Deletion of the adjacent HXT3, HXT6 and HXT7 resulted in the loss of ARS432. To prevent potential replication problems, the repair DNA carried ARS4, amplified by PCR from pRS416 with primers 9525 and 9526. Transformation of S. cerevisiae was performed as previously described (Gietz and Woods 2002).
The first transformation was accomplished with 1 μg of each plasmid pUDR211 and pUDR295 (Table 2), together with 2 μg of each corresponding repair oligonucleotide (Supplemental Material 1). The gene deletions were confirmed by PCR and plasmids were subsequently removed by growing one positive clone in 100 mL YPM medium at 30°C with aeration until the end of the exponential growth on glucose. This procedure was repeated by inoculating 1 μL of the culture to a new 100 mL YPM flask. At the end of the exponential growth phase, the culture was plated on YPM agar to obtain single colonies. Plasmid loss was confirmed by re-streaking single colonies on plates with selective and non-selective media. A single colony, unable to grow on selective medium, was inoculated in liquid YPM and grown overnight in an incubator at 30°C. The strain was stocked as IMX1521 and used for the second round of transformation, in which 2 μg of each plasmid, pUDR214 and pUDR220, and 2 μg of each corresponding repair oligonucleotide were transformed. The genotype was confirmed by PCR and the plasmids were recycled as described above, resulting in strain IMX1541. This strain was transformed with 2 μg of each plasmid pUDR217 and pUDR418, and with 2 μg of each corresponding repair oligonucleotide. The genotype was confirmed by PCR and the plasmids were removed, resulting in strain IMX1812. For the second and third transformation rounds, liquid and solid cultures were supplied with maltose as carbon source.
CHEF electrophoresis
For chromosome separation, contour-clamped homogeneous electric field (CHEF) electrophoresis was used. The CHEF Yeast Genomic DNA Plug Kit (Bio-Rad, Richmond, CA) was used to prepare 1% agarose plugs as recommended by the supplier. The agarose plugs were placed in a 1% megabase agarose gel in 0.5x TBE buffer (Thermo Fisher Scientific), together with the Lambda PFG Ladder (New England Biolabs). The CHEF-DRIII Pulsed Field Electrophoresis system (Bio-Rad) chilled to 14°C was used with a voltage of 5 V/cm, a pulse angle of 120° and pulse time of 60 s during 28 h followed by a pulse time of 90 s for another 16 h. The CHEF gel was stained in 200 mL 0.5x TBE with 3 μg mL−1 ethidium bromide followed by de-staining in 200 mL 0.5× TBE. Images were taken using a UV transilluminator.
Flow cytometric measurement of DNA content
A sample of an exponentially growing aerobic shake-flasks culture was washed with demineralized water, fixed in 70% ethanol and stored at 4°C. Approximately 1 × 107 ethanol-fixed cells were washed with 50 mM Tris-HCl buffer at pH 7.5. The pellet was suspended in the same buffer supplemented with 1 mg mL−1 RNase A and incubated at 37°C for 2 h. Trypsin was added to a final concentration of 3.3 mg mL−1, and the cell suspension was incubated at 37°C for 2 h. Cells were subsequently washed with 50 mM Tris-HCl pH 7.5 buffer, suspended to a final concentration of 2 × 107 cells per mL in 50 mM Tris-HCl pH 7.5 buffer and stored on ice. A total of 2 × 106 cells were mixed in 50 mM Tris-HCl pH 7.5 buffer with or without 1μM Sytox Green Nucleic Acid stain (Sigma), sonicated at 6 μm peak-to-peak amplitude (MSE Soniprep 150, Fisher Scientific, Loughborough, United Kingdom) for 15 s and stored on ice. Analysis was done on a BD-AccuriTM C6 flow cytometer equipped with a 488 nm excitation laser (Becton Dickinson, Franklin lakes, NJ). A minimum of 1000 events were analyzed in FlowJo v10.4.1 (FlowJo LLC, Ashland, OR) to determine the DNA content of the constructed strains. Exponentially growing aerobic shake-flask cultures of the haploid S. cerevisiae strain CEN.PK113-7D and the diploid strain CEN.PK122 were used to identify the fluorescence intensity corresponding to 1 and 2 N peaks, respectively.
Whole genome sequencing
Illumina sequencing
Genomic DNA of IMX1812, IMX672, IMX1541 and CEN.PK2-1C was isolated using the Qiagen 100/G Kit (Qiagen, Hilden, Germany) following the manufacturer's recommendations. DNA concentration was quantified using a Qubit® Fluorometer 2.0 with Qubit dsDNA BR Assay kit. The genomic DNA of IMX1812 was used to obtain a 300 cycle paired-end library with insert-size of 550 bp and sequenced in-house on a MiSeq sequencer (Illumina, San Diego, CA) using TruSeq DNA PCR-free library preparation. Sequence data are available at NCBI under Bioproject accession number PRJNA478763.
MinION sequencing
For Nanopore sequencing, a 1D sequencing library (SQK-LSK108) was prepared according to the manufacturer's recommendation, shearing DNA with g-TUBE (Covaris Ltd, Brighton), and loaded onto an FLO-MIN106 (R9.4) flow cell, connected to a MinION Mk1B unit (Oxford Nanopore Technology, Oxford, UK). MinKNOW software (version 1.11.5; Oxford Nanopore Technology) was used for quality control of active pores and for sequencing. Raw files generated by MinKNOW were base called, on a local compute server (HP ProLiant DL360 G9, 2x XEON E5-2695v3 14 Cores and 256 GB RAM), using Albacore (version 1.2.5; Oxford Nanopore). Reads, in fastq format, with minimum length of 1000 bp were extracted, yielding 7.15 Gigabase of sequence with an average read length of 7.3 kb. Sequencing data are available at NCBI under Bioproject accession number PRJNA478763.
De novo assembly
De novo assembly was performed using Canu (v1.4, settings: genome size = 12 m) (Koren et al.2017) producing a 12.16 Megabase genome. Paired-end Illumina library was aligned, using BWA (Li and Durbin 2010), to the assembly and the resulting BAM file (Binary Alignment Map file) was processed by Pilon (Walker et al.2014) for polishing the assembly (for correcting assembly errors), using correction of only SNPs and short indels (–fix bases parameter). Gene annotations were performed using the MAKER2 annotation pipeline (version 2.31.9) (Holt and Yandell 2011) using SNAP (version 2013–11-29) (Korf 2004) and Augustus (version 3.2.3) (Stanke et al.2006) as ab initio gene predictors. S288C EST and protein sequences were obtained from SGD (Saccharomyces Genome Database, http://www.yeastgenome.org/) and were aligned using BLASTX (BLAST version 2.2.28+) (Camacho et al.2009). Translated protein sequences of the final gene model were aligned using BLASTP to S288C protein Swiss-Prot database. Custom made Perl scripts were used to map systematic names to the annotated gene names.
Analysis of copy number variation
Illumina reads from IMX1812 and CEN.PK2-1C were co-assembled to detect copy number variation between the two strains by applying the Magnolya algorithm (Nijkamp et al.2012).
Growth tests on solid media
Cells from a frozen stock were inoculated in liquid YPM or SM medium supplemented with maltose, uracil, histidine, leucine and tryptophan and grown aerobically at 30°C, 200 rpm. After 8 h, the optical density at 660 nm was measured with a JENWAY 7200 spectrophotometer (Cole-Parmer, Stone, UK). An appropriate volume of cell suspension was spun down (3000 g, 2 min at 4°C) and washed with sterile water, then transferred to a fresh 500 mL shake flask with an initial OD660 of 0.5 and cultivated for 4 hours at 30°C. After measuring OD660 in triplicate, cells were collected by centrifugation (3000 g, 2 min at 4°C), washed with sterile water and resuspended to a concentration of 107 cells mL−1. From this cell suspension, a 10x serial dilution was prepared in water with final concentration of 103 cells mL−1. Then, 10 μL of each concentration were spotted on selective SM agar plates supplemented with uracil, tryptophan, leucine and histidine with 20 g L−1 glucose, 6.8 g L−1 maltose, 20 g L−1 mannose, 20 g L−1 galactose, or 20 g L−1 fructose as carbon source and grown for 2 days at 30°C.
Growth test in shake flask
All liquid cultures with SM were supplemented with uracil, histidine, leucine and tryptophan and grown aerobically in 500 mL shake-flasks at 30°C, 200 rpm. For growth rate determination, cells were inoculated from a frozen stock culture in 100 mL SM medium with 6.8 g L−1 maltose as carbon source. After 8 h of incubation, optical density at 660 nm was measured with a JENWAY 7200 spectrophotometer. Cells were transferred to an initial OD660 of 0.01 and grown overnight in the same medium. The next morning, exponentially growing cultures were transferred to fresh 100 mL SM maltose medium (initial OD660 ∼0.1) and the OD was monitored over time. For growth tests with glucose as carbon source, cells were washed once in water and inoculated to an OD660 of 0.2 in SM with 20 g L−1 glucose. OD660 was monitored until stationary phase was reached. Specific growth rates were calculated from at least six data points evenly distributed during the exponential growth phase.
Samples were taken and spun down (3 min at 13,000 g) for substrate and extracellular metabolite concentration determination by high-performance liquid chromatography (HPLC) analysis. HPLC analysis was performed with an Agilent 1100 HPLC (Agilent Technologies) equipped with an Aminex HPX-87H ion-exchange column (Bio-Rad, Veenendaal, The Netherlands) kept at 60°C, eluted with sulfuric acid (5 mM, 0.6 mL min−1). Detection of glucose, ethanol and glycerol was performed by a refractive-index detector (Agilent G1362A).
RESULTS AND DISCUSSION
Experimental design for CRISPR-SpCas9-mediated deletion of 21 hexose transporter genes
To construct a new hexose null S. cerevisiae strain, the complete set of 21 transporter genes that were shown to promote growth on hexoses in the CEN.PK strain background (Wieczorke et al.1999) were deleted (Fig. 1A). The starting strain, IMX672, was constructed by integration of a copy of the constitutively expressed SpCas9-encoding gene in the CAN1 locus of CEN.PK2-1C, a quadruple auxotrophic strain belonging to the CEN.PK family (trp1-1 leu2-3,112 his3Δ ura3-52, Table 1; Mans et al.2015). Integration of a single copy of SpCas9-encoding gene under the control of a strong promoter in S. cerevisiae has been shown to enable high genome editing efficiency, while preventing toxicity caused by excessive SpCas9 expression levels (Mans et al.2015).
The sequences of S. cerevisiae hexose transporter genes are highly homologous. A single, carefully designed sgRNA can therefore be used to simultaneously target multiple homologous HXT genes (Marques et al.2017). sgRNA 1 was used to target simultaneously HXT1 and HXT3, although the PAM sequence was different for the two loci (Fig. 1B). The PAM sequence is not present on the delivered sgRNA but is essential to guide SpCas9 to the correct target (Mojica et al.2009), which enables a single sgRNA to target the same sequence even when it is framed by different PAM sequences. sgRNA 5 was designed to edit HXT13, HXT15 and HXT16 simultaneously, sgRNA 7 targeted HXT9, HXT11 and HXT12 and sgRNA 9 targeted MPH2 and MPH3. Conversely sgRNAs 2, 3, 4, 6, 8, 10 and 11 targeted unique loci. In total, 11 sgRNAs were designed to edit the 21 hexose transporter genes. Following the method described by Mans et al. (2015), plasmids carrying two identical or different sgRNAs (called sgRNA plasmids) were constructed and transformed in combinations of two to achieve multiplexing. Three transformation rounds were performed to delete all 21 hexose transporter genes. During transformation, double-stranded DNA fragments of 120 nucleotides (repair DNA) carrying 60 bp homology with sequences upstream and downstream the SpCas9-mediated editing sites were provided to delete the targeted genes. The only exception was the DNA fragment used to delete HXT3, HXT6 and HXT7 which was longer and carried ARS4 to replace the autonomously replicating sequence region located between HXT6 and HXT7 (Fig. 1B). After each transformation round, these sgRNA plasmids were removed by cultivation in rich, non-selective medium.
In the first transformation round, nine hexose transporter genes were deleted simultaneously using two CRISPR plasmids, pUDR211 and pUDR295. HXT1, HXT4 and HXT5 are clustered on chromosome VIII and HXT3, HXT6 and HXT7 on chromosome IV (Fig. 1A). As mentioned above, sgRNA 1 was designed to edit simultaneously HXT1 and HXT3. By supplying repair DNA fragments with homology up- and downstream of the entire clusters, all six hexose transporter genes could be simultaneously deleted. pUDR295 carried sgRNA 1 together with sgRNA 2 targeting GAL2. pUDR211 also contained two sgRNAs, sgRNA 3 targeting HXT8 and sgRNA 4 targeting HXT14. IMX672 was transformed with 1 μg of each plasmid (pUDR211, pUDR295) and 2 μg of each of the five repair DNA fragments. Only two colonies appeared on the transformation plates; however, diagnostic PCR revealed that one of these colonies harbored the expected nine deletions (Supplemental Material 2). Mans et al. (2015) observed a negative correlation between the number of targeted loci and the number of transformants counted on plates, with as few as 14 ± 2 colonies when simultaneously targeting six loci. The low number of transformants obtained with five independent editing events is therefore in line with these earlier observations. After removal of the sgRNA plasmids, one strain was stocked as IMX1521.
In the second transformation, eight hexose transporter genes were simultaneously targeted: HXT2, HXT9, HXT10, HXT11, HXT12, HXT13, HXT15 and HXT16. pUDR214 carried sgRNA 5 targeting HXT13, HXT15 and HXT16 and sgRNA 6 targeting HXT2. pUDR220 carried sgRNA 7 targeting HXT9, HXT11 and HXT12 and sgRNA 8 targeting HXT10. IMX1521 was transformed with pUDR214, pUDR220 and six repair fragments. In an attempt to increase the number of clones obtained after transformation, the quantity of each plasmid (pUDR214 and pUDR220) was increased from 1 to 2 μg in the second transformation, which resulted in 23 transformants. Out of seven colonies checked by diagnostic PCR, one displayed the expected pattern (Supplemental Material 2). The sgRNA plasmids were again removed by cultivation on rich medium with maltose as carbon source, and one strain was stocked as IMX1541.
The final transformation round targeted four genes: MPH2, MPH3, MAL11 (also known as AGT1) and STL1 with three sgRNAs. pUDR217 carried sgRNA 9 targeting the homologs MPH2 and MPH3 and sgRNA 10 targeting MAL11. Harbored by pUDR418, sgRNA 11 was designed to target STL1. IMX1541 was transformed with pUDR217 and pUDR418 (2 μg each) and three repair DNA fragments, resulting in 433 transformants. Four out of eleven transformants tested by diagnostic PCR displayed the expected four deletions (Supplemental Material 2). One of the transformants was grown on non-selective medium to recycle the sgRNA plasmids and stocked as IMX1812.
Physiological characterization of the CRISPR-Hxt0 strain IMX1812
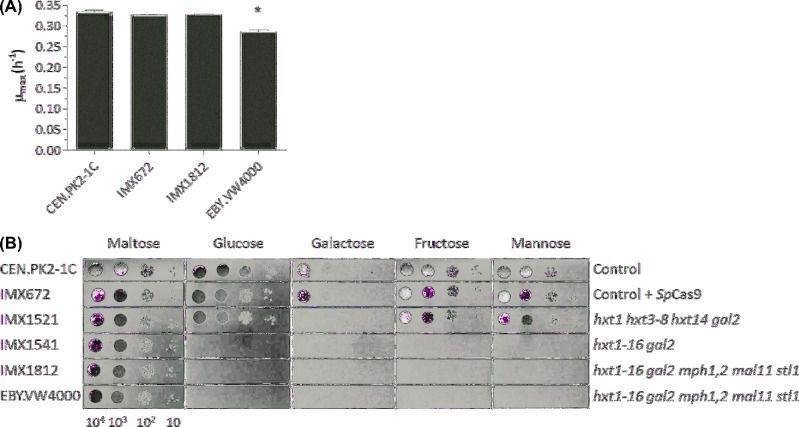
IMX1812 was grown in shake-flask with chemically defined medium using 2% glucose as sole carbon source. As observed for EBY.VW4000, no growth, glucose consumption or ethanol production could be detected after 4 weeks of culture (Supplemental Material 3), confirming that IMX1812 was unable to utilize glucose. While the specific growth rate of EBY.VW4000 on maltose was 15% lower than that of the control strain CEN.PK2-1C (t-test, two-tailed, homoscedastic P < 0.05), IMX1812 grew as fast as the control strains (Fig. 2A). Interestingly, deletion of most major hexose transporters (hxt1 hxt3-8 hxt14) in IMX1521 did not visibly affect its ability to grow on solid medium with 2% glucose. As IMX1521 still contained HXT2, this result was in line with earlier observations that S. cerevisiae carrying only HXT2 grew as rapidly on media with 1% glucose as a control strain equipped with a full set of hexose transporters (Reifenberger, Freidel and Ciriacy 1995). Conversely, IMX1541 did not grow on 2% glucose on solid medium, confirming that HXT2, potentially assisted by some or all of the genes removed in the second deletion round (HXT9, HXT10, HXT12, HXT13, HXT15 and HXT16) contributed to the fast growth of IMX1521 on high glucose concentrations.
Figure 2.
Physiological characterization of the CRISPR-Hxt0 strain IMX1812 and its ancestors. (A) Growth in aerobic shake-flasks with chemically defined medium using maltose as sole carbon source. CEN.PK2-1C: control strain without SpCas9, IMX672: control strain constitutively expressing SpCas9 from its genomic DNA, IMX1812: CRISPR-Hxt0 strain and EBY.VW4000: Hxt0 strain from Wieczorke et al. (1999). All strains are quadruple auxotrophs and were supplied with uracil, leucine, histidine and tryptophan. Data represent the average and mean deviation of biological duplicates for each strain and growth condition. The asterisk indicates a P value below 0.05 (Student t-test) as compared to the control strain CEN.PK2-1C. (B) Growth on chemically defined medium with various carbon sources. IMX1521 and IMX1541 were obtained after the first and second transformation round, respectively. The pictures were taken after 2 days of incubation at 30°C.
As previously described for EBY.VW4000, deletion of 21 hexose transporter genes abolished growth on fructose, galactose and mannose in IMX1812 (Wieczorke et al.1999; Fig. 2B). Growth on galactose was already fully abolished after the first transformation round. During this transformation, GAL2, encoding the major galactose transporter (Tschopp et al.1986) was deleted, as well as HXT14 which encodes a transporter that favors the transport of galactose over other hexoses (Wieczorke et al.1999). While Wieczorke and co-workers showed that galactose uptake was detected when either HXT9, HXT10 or HXT11 were overexpressed in an hxt1-17 gal2 deletion strain, the present results demonstrated that their presence in the genome of IMX1512 in a single copy was not sufficient to promote growth on galactose (Fig. 2B). IMX1521 grew as well as the control strain with fructose or mannose as sole carbon source. Most hexose transporters can transport fructose and mannose, albeit with low affinity (Reifenberger, Boles and Ciriacy 1997; Wieczorke et al.1999). The presence of HXT2, HXT9-13, HXT15 and HXT16 in IMX1521 was therefore sufficient to sustain near-wildtype growth rates on glucose, fructose and mannose. IMX1541 was unable to grow on 2% glucose, fructose and mannose, indicating that, in agreement with Wieczorke et al. (1999), MPH2, MPH3, MAL11 and STL1 were not sufficient to sustain growth on these carbon sources.
The CRISPR-Hxt0 strain IMX1812 is devoid of chromosomal alterations
In the HXT0 EBY.VW4000, a sequential deletion campaign that involved repeated use of the LoxP/Cre system has resulted in extensive chromosomal rearrangements (Fig. 3A and B; Solis-Escalante et al.2015). While SpCas9 editing has not been reported to lead to similar chromosomal rearrangements in haploid S. cerevisiae strains, our deletion strategy involved simultaneous editing of highly homologous genes located on different chromosomes, using the same target sequence. Upon editing, the homologous recombination machinery has two types of homologous sequences at its disposal that can be used for repair of double-strand breaks induced by SpCas9. As an alternative to the repair DNA fragments supplied during transformation, freshly edited genomic DNA with high sequence homology might be used. The possibility of chromosomal rearrangements could therefore not be excluded, more particularly for genes located in subtelomeric regions, which are known to be recombinogenic and a source of genetic variability (Horowitz, Thorburn and Haber 1984; Dujon and Louis 2017). In particular HXT9, HXT11 and HXT12 are located close to telomeres on chromosomes X, XV and IX respectively, and were deleted in the second transformation round with a single sgRNA (Fig. 1). Similarly, MPH2 and MPH3, which are highly homologous and located near the telomeres of chromosomes IV and X respectively, were deleted simultaneously with a single sgRNA in the third transformation round (Fig. 1).
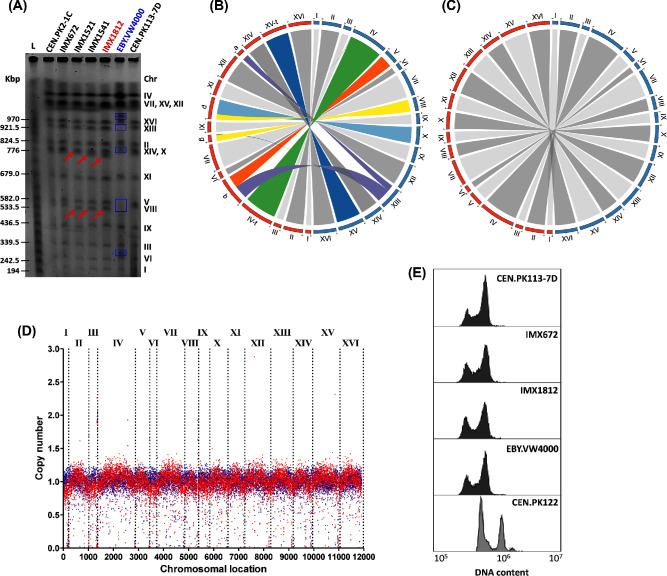
Figure 3.
Genomic characterization of the CRISPR-Hxt0 strain IMX1812, its ancestors and EBY.VW4000. (A) Karyotyping using pulse-field electrophoresis. Red arrows indicate size decrease of chromosomes as consequence of CRISPR-based hexose transporter genes deletion (Supplemental Material 4). Blue boxes indicate polymorphisms in EBY.VW4000 as compared to the control strains CEN.PK113-7D. (B) Circos plot comparing the chromosomes of EBY.VW4000 (left side of the circle, indicated in red) and of the control strain CEN.PK113-7D (right side of the circle, indicated in blue). (C) Circos plot comparing the chromosomes of IMX1812 (left side of the circle, indicated in red) and of the control strain CEN.PK113-7D (right side of the circle, indicated in blue). (D) Chromosome copy number variation of IMX1812 based on Nanopore sequencing data. Blue dots represent the parental strain CEN.PK2-1C and red dots the Hxt0 strain IMX1812. (E) Cellular DNA content measurement by flow cytometry. CEN.PK113-7D and CEN.PK122 are the haploid and diploid controls, respectively.
Karyotyping by CHEF electrophoresis of IMX1812 revealed a chromosomal pattern that was near-identical to that of its parental strain IMX672 (Fig. 3A). The small shift towards smaller size of chromosomes VIII and X was readily explained by the deletion of multiple genes on these chromosomes in IMX1812 (HXT1, HXT4 and HXT5 deletion on chromosome VIII with expected size reduction of 9253 bp; HXT8, HXT9, HXT16 and MPH3 on chromosome X with expected size reduction of 9319 bp; see Supplemental Material 4). In other cases, deletions were either too small or the chromosomes in which they were introduced too large to detect deletions by CHEF electrophoresis (Supplemental Material 4). The similarity between IMX1812 and the control strains CEN.PK113-7D, CEN.PK2-1C and IMX672 suggested the absence of chromosomal rearrangements in IMX1812.
Whole genome sequence of the CRISPR-Hxt0 strain IMX1812
Although Illumina sequencing technology was able to detect chromosomal translocations in the EBY.VW4000 genome (Solis-Escalante et al.2015), the short size of the sequencing reads prevented resolution of the sequence of repeated regions. Especially in subtelomeric regions, this limitation resulted in a fragmented assembly. The advent of third generation sequencing technology, such as nanopore sequencing, has considerably improved the completeness of sequenced genomes, yielding near complete draft chromosome sequences with telomere-to-telomere coverage. The only chromosomal region still resisting accurate sequencing is the region of chromosome XII carrying rDNA (Venema and Tollervey 1999). To check for the occurrence of chromosomal rearrangements, the CRISPR-Hxt0 strain IMX1812 was sequenced with Oxford Nanopore Technology MinION platform.
The MinION assembly of the IMX1812 genome resulted in only 19 contigs, which represents a near 6-fold reduction in the number of contigs as compared to the EBYWV4000 genome assembly that was derived from multiple (mate and paired-end) libraries (Solis-Escalante et al.2015). All nuclear chromosomes, with the exception of chromosome XII, were assembled in single contigs with telomere-to-telomere coverage. Chromosome XII was split in three contigs; the first included sequences ranging from the left telomere to the left end of the ribosomal DNA (rDNA) locus, the second included sequences ranging from the right end of the rDNA locus to the right telomere and the third contig, smaller in size (∼90 kb), representing an embryo of the rDNA locus. The latter contig was also exhibiting a higher coverage than the rest of the assembly, in line with the repetitive nature of rDNA (Venema and Tollervey 1999). Although assembled separately, the three chromosome XII contigs shared homology at their end, enabling their homology-based concatenation into a chromosome XII scaffold. The mitochondrial genome was also reassembled into a single contig. Considering the error rate of this third-generation sequencing method, IMX1812 was also sequenced by Illumina technology and the resulting sequence was used to polish the Oxford Nanopore assembly. In comparison to the previous EBYVW4000 assembly, the present IMX1812 genome captures an additional 700 kb (Table 3; Solis-Escalante et al.2015).
Table 3.
Quality of genome assembly of the Hxt0 strains IMX1812 and EBY.VW4000.
| EBY.VW4000a | IMX1812 | |
|---|---|---|
| Technology | Illumina paired-end | Nanopore |
| and mate-pair | ||
| Scaffolds/contigs (>500 bp) | 104 | 19 |
| Largest scaffold/contig (Mbp) | 1.045 | 1.479 |
| Average scaffold/contig (Mbp) | 0.116 | 0.640 |
| N50 (Mbp) | 0.423 | 0.804 |
| Total assembly size (Mbp) | 11.5 | 12.2 |
aSolis-Escalante et al. (2015).
Inspection of IMX1812 genome sequence confirmed the complete absence of chromosomal rearrangements (Fig. 3C and D). Ploidy check by flow cytometry was in line with the sequencing data, confirming that the constructed strains remained haploid throughout the successive transformation rounds (Fig. 3E). Further scrutiny of IMX1812 genome sequence confirmed the deletion of the 21 hexose transporter genes. For all genes, SpCas9-mediated genome editing was repaired precisely as expected based on the sgRNA target and the supplied repair DNA fragments. Combining Illumina and nanopore sequencing revealed the absence of gene loss in IMX1812 as compared to its parent CEN.PK2-1C (Fig. 3D), but identified nine SNPs and a single indel located on six ORFs (TMN3, SPT16, MUC1, MNN5, LAP4 and ALT1; Table 4). These ORFs were located at least 26 kb from the deletion sites and shared no homology with any of the 11 sgRNAs, suggesting that they most likely did not result from SpCas9 activity but rather from random events caused by the three consecutive transformation rounds. TMN3, SPT16, MNN5, LAP4 and ALT1 carried a single SNP, leading to an amino acid change. Of these genes only SPT16, encoding a subunit of the FACT complex, is essential, and depletion of Spt16 leads to chromosome rearrangements and loss (Schlesinger and Formosa 2000). The fast growth on maltose and absence of chromosomal rearrangements in IMX1812 suggested that Spt16 function was not affected in the Hxt0 strain. Impairment of Tmn3, Mnn5 and Lap4 (also known as Ape1) functions are not expected to substantially affect S. cerevisiae physiology (Cueva, Garcia-Alvarez and Suarez-Rendueles 1989; Rayner and Munro 1998), in agreement with the absence of phenotypic difference between IMX1812 and its ancestor during growth on maltose (Fig. 2A). Impairment of the activity of the alanine transaminase Alt1 results in growth defects in media supplied with ammonium as sole nitrogen source (Peñalosa-Ruiz et al.2012). Such a phenotype was however not observed for IMX1812 (Fig. 2). Surprisingly, MUC1, also known as FLO11 and involved in flocculation and pseudohyphal growth, was mutated by three SNPs (two of them leading to an amino acid change) and a 269 bp deletion, which most probably led to a loss of function (Table 4). As in many common laboratory S. cerevisiae strains, strains from the CEN.PK family are deficient in flocculation and pseudohyphal growth (Liu, Styles and Fink 1996), and mutations in FLO11 are therefore not likely to affect IMX1812 phenotype. Additionally, Illumina sequencing of the genome of CEN.PK2-1C and of the strains derived from CEN.PK2-1C (IMX672, IMX1541 and IMX1812; Table 1) revealed that, conversely to current records, the TRP1 mutation in this lineage did not correspond to the trp1-289 alteration (Botstein et al.1979; Entian and Kötter 2007) but matched the trp1-1 characterized by a C to T change at position +403, resulting in the introduction of an amber stop codon (Cag-Tag) (https://wiki.yeastgenome.org). Accordingly, EBY.VW4000, based on CEN.PK2-1C, also harbored the trp1-1 mutation.
Table 4.
List of SNPs identified in IMX1812 genome sequence as compared to its parental strain CEN.PK2-1C.
| Chr. | ORF | Syst. name | Position | Mutation typea | Amino acid change |
|---|---|---|---|---|---|
| V | TMN3 | YER113C | 390.575 | NSY | Asp-61-Tyr (G→T) |
| VII | SPT16 | YGL207W | 111.686 | NSY | Ile-165-Val (A→G) |
| IX | MUC1 | YIR019C | 384.267-384.535 | 269 bp deletion | |
| MUC1 | YIR019C | 384.543 | NSY | Gly-1108-Val (G→T) | |
| MUC1 | YIR019C | 384.548 | SYN | Thr-1106-Thr (C→A) | |
| MUC1 | YIR019C | 384.565 | NSY | Ser-1101-Ala (T→G) | |
| X | MNN5 | YJL186W | 69.736 | NSY | Asn-264-Lys (C→A) |
| XI | LAP4 | YKL103C | 253.049 | NSY | Lys-320-Glu (A→G) |
| XII | ALT1 | YLR089C | 298.577 | NSY | Ile-120-Thr (T→C) |
| XVI | FHL1 | YPR104C | 742.545 | NSY | Asn-669-Lys (T→A) |
aNSY: Non-Synonymous mutation, SYN: Synonymous mutation.
CONCLUSION
The present work provides an Hxt0 strain that is genetically near-identical to its hexose transport proficient ancestor. In-depth analysis of IMX1812 revealed the absence of undesired recombination, duplication or excision events during the three transformation rounds performed to delete the 21 hexose transporter genes. Despite the few SNPs caused by these transformations, IMX1812 is genetically and phenotypically closer to its hexose transport proficient ancestor than EBY.VW4000. The extremely high fidelity of genome editing, despite the simultaneous targeting of multiple (subtelomeric) loci with high homology, highlights the power of CRISPR-SpCas9 editing for extensive strain construction programs.
The present study also provides an ‘Hxt0 CRISPR toolkit’ composed of a set of plasmids, efficient sgRNAs and repair DNA that enables the abolishment of glucose transport in strains from the CEN.PK family. Thanks to the high conservation of hexose transporter genes (Bisson, Fan and Walker 2016), the application of this toolkit is not restricted to the CEN.PK strain family. For instance, based on published genome sequences, the Hxt0 CRISPR toolkit can be directly used to turn S288C and its BY4741 derivatives into Hxt0 strains. Although, contrary to CEN.PK strains, S288C and its derivatives contain a genomic copy of HXT17, the strong homology between HXT17, HXT15 and HXT16 enables sgRNA 5, which targets HXT15 and HXT16, to also edit HXT17 during the second transformation round. Not only the plasmids, but also the repair DNA fragments designed for CEN.PK, can be used for these popular laboratory strains. Of course, an additional DNA fragment would have to be designed and supplied to repair the double strand break upon HXT17 editing.
IMX1812 is available upon request and the sgRNA plasmids have been deposited to Addgene (see Table 2 for accession numbers).
Supplementary Material
Acknowledgements
We thank Isaac Molenaar and Mark Bisschops for experimental support during strain construction and flow cytometric analysis, respectively.
FUNDING
This project was funded by the AdLibYeast ERC consolidator 648141 grant awarded to PDL.
REFERENCES
- Bao Z, Xiao H, Liang J et al. . Homology-Integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth Biol 2015;4:585–94. [DOI] [PubMed] [Google Scholar]
- Bisson LF, Fan Q, Walker GA. Sugar and glycerol transport in Saccharomyces cerevisiae. Adv Exp Med Biol 2016;892:125–68. [DOI] [PubMed] [Google Scholar]
- Boles E, Hollenberg CP. The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev 1997;21:85–111. [DOI] [PubMed] [Google Scholar]
- Boles E, Oreb M. A growth-based screening system for hexose transporters in yeast. Methods Mol Biol 2018;1713:123–35. [DOI] [PubMed] [Google Scholar]
- Botstein D, Falco SC, Stewart SE et al. . Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene 1979;8:17–24. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V et al. . BLAST+: architecture and applications. BMC Bioinformatics 2009;10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cueva R, Garcia-Alvarez N, Suarez-Rendueles P. Yeast vacuolar aminopeptidase yscI. Isolation and regulation of the APE1 (LAP4) structural gene. FEBS Lett 1989;259:125–9. [DOI] [PubMed] [Google Scholar]
- Dujon BA, Louis EJ. Genome diversity and evolution in the budding yeasts (Saccharomycotina). Genetics 2017;206:717–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian KD, Kötter P. Yeast genetic strain and plasmid collections. In: Stansfield I, Stark MJR (eds) Yeast Gene Analysis vol. 36, 2nd edn Amsterdam: Academic Press, Elsevier, 2007, 629–66. [Google Scholar]
- Ferreira R, Skrekas C, Nielsen J et al. . Multiplexed CRISPR/Cas9 genome editing and gene regulation using Csy4 in Saccharomyces cerevisiae. ACS Synth Biol 2018;7:10–15. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Meth Enzymol 2002;350:87–96. [DOI] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fielder T et al. . A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 1996;24:2519–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 2011;12:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz H, Thorburn P, Haber JE. Rearrangements of highly polymorphic regions near telomeres of Saccharomyces cerevisiae. Mol Cell Biol 1984;4:2509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K et al. . Canu: Scalable and accurate long-read assembly via adaptive k -mer weighting and repeat separation. Genome Res 2017;27:722–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinformatics 2004;5:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruckeberg AL. The hexose transporter family of Saccharomyces cerevisiae. Arch Microbiol 1996;166:283–92. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 1996;144:967–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lõoke M, Kristjuhan K, Kristjuhan A. Extraction of genomic DNA from yeasts for PCR-based applications. BioTechniques 2011;50:325–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier A, Volker B, Boles E et al. . Characterisation of glucose transport in Saccharomyces cerevisiae with plasma membrane vesicles (countertransport) and intact cells (initial uptake) with single Hxt1, Hxt2, Hxt3, Hxt4, Hxt6, Hxt7 or Gal2 transporters. FEMS Yeast Res 2002;2:539–50. [DOI] [PubMed] [Google Scholar]
- Mans R, Hassing EJ, Wijsman M et al. . A CRISPR/Cas9-based exploration into the elusive mechanism for lactate export in Saccharomyces cerevisiae. FEMS Yeast Res 2017;17, DOI: 10.1093/femsyr/fox085. [DOI] [PubMed] [Google Scholar]
- Mans R, van Rossum HM, Wijsman M et al. . CRISPR/Cas9: A molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res 2015;15, DOI: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans R, Wijsman M, Daran-Lapujade P et al. . A protocol for introduction of multiple genetic modifications in Saccharomyces cerevisiae using CRISPR/Cas9. FEMS Yeast Res 2018;18, DOI:10.1093/femsyr/foy063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques WL, Mans R, Marella ER et al. . Elimination of sucrose transport and hydrolysis in Saccharomyces cerevisiae: a platform strain for engineering sucrose metabolism. FEMS Yeast Res 2017;17, DOI:10.1093/femsyr/fox006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J et al. . Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009;155:733–40. [DOI] [PubMed] [Google Scholar]
- Nijkamp JF, van den Broek M, Datema E et al. . De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb Cell Fact 2012;11:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev 1999;63:554–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalosa-Ruiz G, Aranda C, Ongay-Larios L et al. . Paralogous ALT1 and ALT2 retention and diversification have generated catalytically active and inactive aminotransferases in Saccharomyces cerevisiae. PLoS One 2012;7:e45702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DR, Tibbles K, Shigenobu S et al. . Sugar transporters of the major facilitator superfamily in aphids; from gene prediction to functional characterization. Insect Mol Biol 2010;19:97–112. [DOI] [PubMed] [Google Scholar]
- Pronk JT. Auxotrophic yeast strains in fundamental and applied research. Appl Environ Microbiol 2002;68:2095–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner JC, Munro S. Identification of the MNN2 and MNN5 mannosyltransferases required for forming and extending the mannose branches of the outer chain mannans of Saccharomyces cerevisiae. J Biol Chem 1998;273:26836–43. [DOI] [PubMed] [Google Scholar]
- Reifenberger E, Boles E, Ciriacy M. Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae and their relation to the triggering mechanisms of glucose repression. Eur J Biochem 1997;245:324–33. [DOI] [PubMed] [Google Scholar]
- Reifenberger E, Freidel K, Ciriacy M. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol Microbiol 1995;16:157–67. [DOI] [PubMed] [Google Scholar]
- Rolland F, Winderickx J, Thevelein JM. Glucose-sensing mechanisms in eukaryotic cells. Trends Biochem Sci 2001;26:310–7. [DOI] [PubMed] [Google Scholar]
- Salazar AN, Gorter de Vries AR, van den Broek M et al. . Nanopore sequencing enables near-complete de novo assembly of Saccharomyces cerevisiae reference strain CEN.PK113-7D. FEMS Yeast Res 2017;17, DOI: 10.1093/femsyr/fox074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger MB, Formosa T. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics 2000;155:1593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schussler A, Martin H, Cohen D et al. . Characterization of a carbohydrate transporter from symbiotic glomeromycotan fungi. Nature 2006;444:933–6. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989;122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis-Escalante D, van den Broek M, Kuijpers NG et al. . The genome sequence of the popular hexose-transport-deficient Saccharomyces cerevisiae strain EBY.VW4000 reveals LoxP/Cre-induced translocations and gene loss. FEMS Yeast Res 2015;15, DOI: 10.1093/femsyr/fou004. [DOI] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I et al. . AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res 2006;34:W435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Świat MA, Dashko S, den Ridder M. et al. FnCpf1: a novel and efficient genome editing tool for Saccharomyces cerevisiae. Nucleic Acids Res 2017;45:12585–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomik T, Wittig I, Choe JY et al. . An artificial transport metabolon facilitates improved substrate utilization in yeast. Nat Chem Biol 2017;13:1158–63. [DOI] [PubMed] [Google Scholar]
- Tschopp JF, Emr SD, Field C. et al. GAL2 codes for a membrane-bound subunit of the galactose permease in Saccharomyces cerevisiae. J Bacteriol 1986;166:313–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Broek M, Bolat I, Nijkamp JF et al. . Chromosomal copy number variation in Saccharomyces pastorianus is evidence for extensive genome dynamics in industrial lager brewing strains. Appl Environ Microbiol 2015;81:6253–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet 1999;33:261–311. [DOI] [PubMed] [Google Scholar]
- Verduyn C, Postma E, Scheffers WA et al. . Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 1992;8:501–17. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R et al. . New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 1994;10:1793–808. [DOI] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T et al. . Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 2014;9:e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorke R, Dlugai S, Krampe S et al. . Characterisation of mammalian GLUT glucose transporters in a heterologous yeast expression system. Cell Physiol Biochem 2003;13:123–34. [DOI] [PubMed] [Google Scholar]
- Wieczorke R, Krampe S, Weierstall T et al. . Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett 1999;464:123–8. [DOI] [PubMed] [Google Scholar]
- Xuan YH, Hu YB, Chen LQ et al. . Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc Natl Acad Sci USA 2013;110:E3685–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E, Lee SM, Alper H. Optimizing pentose utilization in yeast: The need for novel tools and approaches. Biotechnol Biofuels 2010;3:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E, Poucher A, Comer A et al. . Functional survey for heterologous sugar transport proteins, using Saccharomyces cerevisiae as a host. Appl Environ Microbiol 2011;77:3311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.