HLA and killer immunoglobulin-like receptor alleles were assessed for association with cervical neoplasia. Our findings suggest HLA-C1 group alleles protect against human papillomavirus type 16–related cervical neoplasia, mainly through a KIR-mediated mechanism.
Keywords: Cervical neoplasia, human leukocyte antigens (HLA), killer immunoglobulin-like receptors (KIRs), HPV16-related cervical neoplasia
Abstract
Background
Cervical cancer is the fourth most common cancer in women, and we recently reported human leukocyte antigen (HLA) alleles showing strong associations with cervical neoplasia risk and protection. HLA ligands are recognized by killer immunoglobulin-like receptors (KIRs) expressed on a range of immune cell subsets, governing their proinflammatory activity. We hypothesized that the inheritance of particular HLA-KIR combinations would increase cervical neoplasia risk.
Methods
Here, we used HLA and KIR dosages imputed from single-nucleotide polymorphism genotype data from 2143 cervical neoplasia cases and 13858 healthy controls of European decent.
Results
The following 4 novel HLA alleles were identified in association with cervical neoplasia, owing to their linkage disequilibrium with known cervical neoplasia–associated HLA-DRB1 alleles: HLA-DRB3*9901 (odds ratio [OR], 1.24; P = 2.49 × 10−9), HLA-DRB5*0101 (OR, 1.29; P = 2.26 × 10−8), HLA-DRB5*9901 (OR, 0.77; P = 1.90 × 10−9), and HLA-DRB3*0301 (OR, 0.63; P = 4.06 × 10−5). We also found that homozygosity of HLA-C1 group alleles is a protective factor for human papillomavirus type 16 (HPV16)–related cervical neoplasia (C1/C1; OR, 0.79; P = .005). This protective association was restricted to carriers of either KIR2DL2 (OR, 0.67; P = .00045) or KIR2DS2 (OR, 0.69; P = .0006).
Conclusions
Our findings suggest that HLA-C1 group alleles play a role in protecting against HPV16-related cervical neoplasia, mainly through a KIR-mediated mechanism.
Cervical cancer is the fourth most common cancer in women, with >500000 new cases presenting worldwide in 2012, and accounts for 7.5% of cancer deaths among female individuals [1]. Its impact is particularly high among young women, ranking as the second commonest cancer affecting women aged 20–39 years [2]. Cervical cancer results from chronic infection with human papillomavirus (HPV), with the HPV genome detected in nearly all cases of cervical cancer. Of the different HPV types, HPV16 and HPV18 are most frequently involved, and together they account for approximately 70% of cervical cancers worldwide [3]. Whereas infection with HPV is essentially universal, most cervical HPV infections are cleared by the immune system [4, 5], and only approximately 1% of women with cervical HPV infection develop cervical cancer [6].
Genetic factors strongly influence the persistence of HPV infection and the risk of cervical cancer. HPV persistence is associated with the monogenic disorders epidermodysplasia verruciformis and WHIM syndrome, from mutations in EVER1/2 and CXCR4, respectively [7, 8]. The only robust common variant genetic associations with cervical cancer are with genes of the major histocompatibility complex (MHC), in particular HLAs. We demonstrated that the haplotypes HLA-DRB1*1501/HLA-DQB1*0602/HLA-DQA1*0102 and HLA-DQA1*0301/HLA-DRB1*0401 increase the risk of HPV-associated cervical neoplasia and that the allele HLA-B*15 and haplotype HLA-DRB1*1301/HLA-DQB1*0603 are protective. Of note, HLA-DRB1*1301/HLA-DQA1*0103/HLA-DQB1*0603 is associated with protection from oral and pharyngeal cancer, particularly among HPV-positive cases [9]. We showed that the HLA risks of cervical neoplasia were determined by amino acids at positions 13 and 71 in pocket 4 of HLA-DRB1 and position 156 in HLA-B [10]. Common genetic variant contribution to cervical neoplasia susceptibility is substantial (36%) [10], although a large component of the heritability has yet to be elucidated.
HLA proteins are critical for antigen presentation to effector cells of the adaptive immune system [11]. HLA class I complexes are expressed on all nucleated cells and present endogenous, intracellular-derived antigens, as well as pathogen-derived peptides (as with viral infection), with a residue length of 8–10 amino acids. These are recognized by CD8+ T cells that can engage foreign peptides through their T-cell receptor. Conversely, natural killer (NK) cells, a component of the innate immune system, are able to respond to downregulated surface HLA, a consequence of the immune evasion strategy of some viruses to avoid CD8+ T-cell recognition. Once activated, these lymphocytes can kill the antigen-presenting cell via release of cytotoxic granules. HLA class II complexes typically present extracellular-derived antigens, such as bacterial pathogens. After endocytosis, these proteins are processed and presented on the cell surface bound to MHC class II to initiate an immune response from CD4+ cells.
Interaction between HLA, viral epitopes, and killer immunoglobulin-like receptors (KIR) expressed on NK cells lead either to activation or inhibition of NK cell cytotoxic activity. KIRs are expressed on all NK cells and a minority of T cells (including some CD4+, CD8+, and γδ cells). Seventeen KIR genes have been identified, encoded within the leukocyte receptor complex on chromosome 19q13.4, all of which share significant homology (85%–99% DNA sequence similarity) [12, 13]. They are encoded in variable gene content haplotypes with activating and inhibitory counterparts. Different inhibitory and activating KIRs demonstrate specificity for different HLA subgroups, providing fine-tuning of NK and KIR-bearing T-cell responses [14].
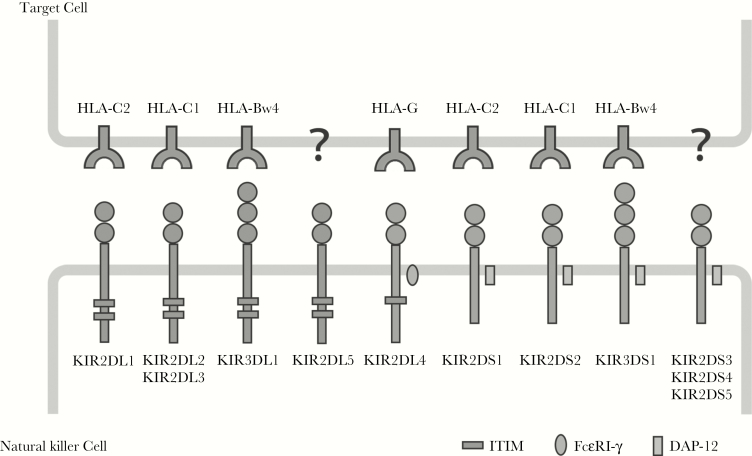
Variability at the KIR locus includes allelic, gene combination (haplotypic), and expression level differences, with the latter under significant epigenetic control [15]. KIR genes are inherited in haplotypes of vastly diverse content, ranging from 4 to 14 receptor-encoding loci, with >50 distinct haplotypes based on gene content alone [13, 16]; approximately 700 allelic variants have been reported [16, 17]. Group 1 HLA-C (HLA-C1) allotypes have an asparagine at residue 80 and are ligands for the inhibitory receptors encoded by KIR2DL2 and KIR2DL3, which segregate as alleles of a single locus, and KIR2DS2 [18]. The remaining HLA-C allotypes (group 2; HLA-C2) have a lysine at position 80 and are ligands for KIR2DL1 (an inhibitory receptor) and KIR2DS1 (the homologous activating receptor). HLA-B Bw4 allotypes serve as ligands for KIR3DL1 and KIR3DS1 (Figure 1).
Figure 1.
Killer immunoglobulin-like receptor (KIR) proteins are classified by the number of extracellular immunoglobulin domains (2D or 3D) and by whether they have a long (L) or short (S) cytoplasmic domain. Inhibitory KIRs and KIR2DL4 have immunoreceptor tyrosine-based inhibitory motifs (ITIMs) in their cytoplasmic domains. Activating KIRs possess a basic amino acid in the transmembrane domain, which allows interactions with the accessory molecule DAP-12, delivering activating signals through its immunoreceptor tyrosine-based activating (ITAM) motif. The ligands for several KIR proteins are subsets of HLA class I proteins. KIR2DL4 has a charged amino acid and ITIM motifs, and it interacts with the accessory protein FcεRI-γ, which sends an activating signal via its ITAM similar to DAP-12. Note that HLA-Bw6 alleles are not known to be KIR ligands.
The immunological mechanisms involved in clearance of HPV are poorly understood. NK cells are crucial for clearing viral infection and for antitumor immunity and are thought to be important in HPV control [19]. Regulation of NK cell responses depend on KIR genotype, HLA genotype, heterozygosity versus homozygosity for each of these, interaction of HLA and KIR, and changes in KIR and HLA expression. The development of imputation-based methods for HLA typing and, more recently, for KIR typing has enabled the use of single-nucleotide polymorphism (SNP)–typed data sets to investigate genetic associations with these loci in large cohorts. Here, HLA and KIR gene imputation and association tests were performed using genotype data from 2143 cervical neoplasia cases and 13858 healthy controls of European descent to test whether HLA-KIR combinations are associated with cervical neoplasia risk.
METHODS AND MATERIALS
Study Population
Phenotypic information, including cervical histologic findings and HPV genotype, appears in Supplementary Table 1. As described by Leo et al [10], all cases needed to have cervical intraepithelial neoplasia types 2 and 3 (if type 2, an age of >30 years was required, to limit analysis to women who did not clear HPV) or cervical cancer (HPV type and histologic findings were not always known). HPV DNA types from tumor tissues were categorized into 6 groups: (1) HPV16 but not HPV18 positive, (2) HPV18 but not HPV16 positive, (3) neither HPV16 nor HPV18 positive, (4) both HPV16 and HPV18 positive, (5) negative for any HPV type (noting that usually samples are only tested for HPV16 and HPV18), and (6) those not tested for HPV genotype [10].
Genotyping
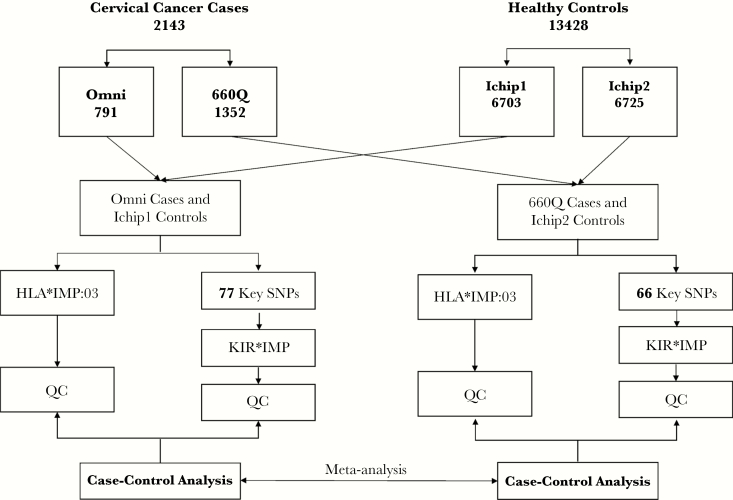
Case samples were SNP microarray genotyped in house. A total 791 cases were genotyped using Illumina OmniExpress BeadChips (Omni), and 1352 cases were genotyped using Illumina Human660-Quad BeadChips (660Q). Controls were genotyped using Illumina Immunochip BeadChips (Ichip; Figure 2). Bead intensity data were processed and normalized for each sample, and genotypes were called within participating studies using GenomeStudio and verified manually and corrected where necessary. Standard quality control measures were performed [10], with particular care taken in comparison of the performance of different chip types.
Figure 2.
HLA and KIR gene imputation and association tests were performed using genotype data from 2143 cervical neoplasia cases and 13858 healthy controls of European descent. A total of 791 individuals were genotyped using the Illumina OmniExpress BeadChip tool (Omni), and 1352 individuals were genotyped using the Illumina Human660-Quad BeadChip tool (660Q). All 13428 controls were genotyped using the Illumina Immunochip BeadChip tool (Ichip). To avoid imputation bias caused by different numbers of key single-nucleotide polymorphisms (SNPs) between Omni and 660Q, these 2 groups of cases were imputed separately and compared to 2 control groups—Ichip control group 1 (Ichip1; n = 6703) and control group 2 (Ichip2; n = 6725)—obtained by randomly dividing the controls. A total of 77 key SNPs for Omni and Ichip1 and 66 key SNPs for 660Q and Ichip2 were reclustered and used for imputation. Case-control analyses were conducted separately, followed by meta-analysis. QC, quality control.
HLA Imputation
HLA alleles were inferred using HLA*IMP:03 (available at: http://www.biorxiv.org/content/early/2016/12/09/091009). Individuals with posterior probability of HLA alleles of <0.6 were excluded from downstream association testing (Supplementary Figure 1). HLA groups (C1, C2, Bw4, and Bw6) were inferred from known allele classifications. HLA amino acids were inferred by the SNP2HLA tool [20], using a reference panel from the Type 1 Diabetes Genetics Consortium (n = 5225). Amino acids imputed by SNP2HLA with an r2 of <0.5 were excluded, and samples where the allele dosage at any HLA type was >2.5 were removed.
KIR Imputation
KIR genes and haplotypes were imputed with the KIR*IMP method [21], using SNP genotypes across the KIR locus. This requires certain informative key SNPs for accurate imputation, which vary according to the SNP chip used (Supplementary Table 2). To avoid imputation bias caused by different numbers of key SNPs genotyped by Omni and 660Q, the 2 groups of cases were imputed separately, with a separate comparator group for each obtained by randomly dividing the control group into 2 groups: Ichip control group 1 (Ichip1; n = 6703) and group 2 (Ichip2; n = 6725; Figure 2). Imputed data for KIR and HLA loci were compared between the 2 control groups. There were no significant differences between the 2 control groups for 271 KIR SNPs (because most of these were only available on Ichip [ie, not on chips used for case genotyping], there was a smaller number available for case-control analyses). Of 256 HLA alleles, 10 (3.9%) were significantly different (P < .05) between the 2 control groups. Most (8 of 10) were rare (≤0.05); the remaining 2 had a frequency of 0.08. None of these SNPs were significant in subsequent analyses. The 77 key SNPs for Omni cases and Ichip1 and the 66 key SNPs for 660Q cases and Ichip2 were reclustered manually (converting array intensity data for each allele of the SNP concerned to genotype calls) before imputation to improve genotyping accuracy (Supplementary Table 3). Individuals with a posterior probability of accurate KIR imputation for each KIR allele <0.6 were excluded in the downstream association testing.
Statistical Methods
Population stratification was assessed via principal component analysis of genome-wide genotypes, using Shellfish (available at: http://www.stats.ox.ac.uk/~davison/software/shellfish/shellfish.php). Association analyses were performed using custom R scripts. Three models were used to test HLA-C1, HLA-C2, HLA-Bw4, HLA-Bw6, and HLA alleles and their combination with KIR genes: (1) a dosage model, which treated genotypes as 0, 1, or 2 copies; (2) a dominant model, which treated 1 or 2 copy genotypes as present and 0 as absent; and (3) a recessive model to study the difference of homozygotes and heterozygotes of HLA-B and HLA-C, which treated 2 copy genotypes as present and 0 or 1 copy as absent. Meta-analysis of combination between Omni-Ichip1 and 660Q-Ichip2 was performed using METAL software (available at: http://csg.sph.umich.edu/abecasis/metal/). For tests over all HLA alleles, the multiple testing correction method was used [22, 23], using a correlation matrix derived from SNP2HLA imputation with all 1027 HLA alleles and amino acids. This analysis estimated 206 independent loci, implying a Bonferroni-corrected P value of 2.4 × 10−4 for a type I error rate of 5%. For the HLA-KIR interaction using 12 KIR alleles, we used the most conservative Bonferroni-corrected P value (ie, P = [0.05/(206 × 12)] = 2.0 × 10−5) for statistical significance.
RESULTS
Quality Control
A total of 2143 cases and 13428 controls passed quality control. Among cases, 736 were squamous cell carcinoma, 542 were adenocarcinoma, and 865 had unspecified histologic findings. Four principal components were used as covariates to control for population stratification, calculated using 11980 common SNPs shared by the Omni, 660Q, and Ichip microarrays. To assess population stratification, we used a subset of 333 null SNPs outside the MHC region included on each chip type and avoided SNPs included on the Ichip, because of their potential immunogenetic significance (ie, they are associated with reading and learning disability, schizophrenia, and psychosis). From this, the genomic inflation factor overall was calculated as 1.018 (Supplementary Figure 2). No divergences were observed between cases and controls or between different genotype platforms (Supplementary Figure 3). All HLA loci were imputed with an accuracy of >95% (Supplementary Table 4), and all KIR loci were imputed with an accuracy of >80% (Supplementary Table 3).
MHC Findings
HLA Association Analysis
Consistent with findings from our recent study [10], we found increased and decreased risks of cervical neoplasia associated with HLA haplotypes (Supplementary Table 5) and determined these associations were carried by amino acids at positions 13 and 71 in pocket 4 of HLA-DRB1 and at position 156 in HLA-B. Using these in-depth imputation methods across HLA and KIR loci, we detected further novel associations from HLA-DRB3 and HLA-DRB5.
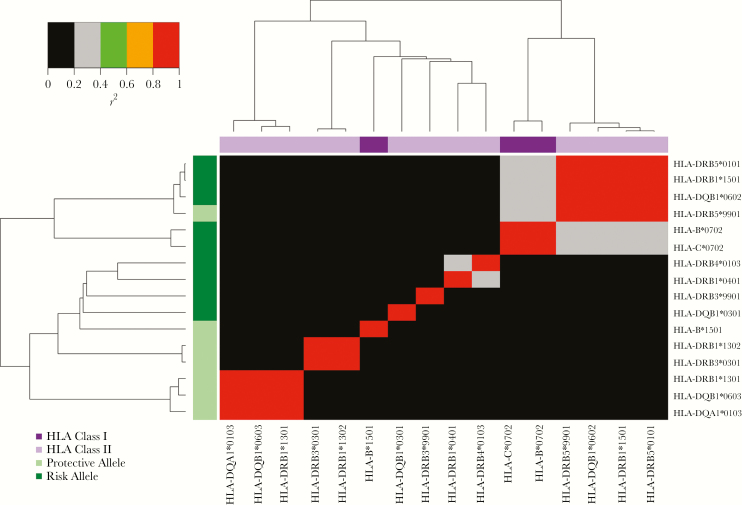
Novel risk associations with cervical neoplasia were identified with HLA-DRB3*9901 (odds ratio [OR], 1.24; P = 2.49 × 10−9; Table 1) and HLA-DRB5*0101 (OR, 1.29; P = 2.26 × 10−8). Although HLA-DRB3*9901 was not in linkage disequilibrium with any individual cervical neoplasia HLA risk allele (P < .05), adjustment for the association with HLA-DRB1 amino acid positions 37, 71, and 96 attenuated the association with HLA-DRB3*9901 (P > .01). HLA-DRB5*0101 was in positive linkage disequilibrium with the HLA class II risk allele HLA-DRB1*1501 (r2 = 0.99) and the protective allele HLA-DRB5*9901 (r2 = 0.93; Table 1 and Figure 3). No residual association for HLA-DRB5*0101 was observed after control for the association of HLA-DRB1*1501, HLA-DRB5*9901, or HLA-DRB1 amino acid positions 13 and 71 (P < .01).
Table 1.
Analysis of Novel Cervical Neoplasia–Associated HLA Alleles, Conditioned on HLA Alleles and HLA Amino Acids
| HLA Alleles | DRB3*9901 | DRB5*0101 | DRB5*9901 | DRB3*0301 |
|---|---|---|---|---|
| Unconditioned P | 2.49 × 10−9 | 2.26 × 10−8 | 1.90 × 10−8 | 4.06 × 10−6 |
| Odds ratio | 1.24 | 1.29 | 0.77 | 0.63 |
| Conditioned P | ||||
| DRB3*9901 | NA | 4.19 × 10−5 | 3.66 × 10−5 | .001033 |
| DRB5*0101 | 3.07 × 10−5 | NA | .45 | 4.43 × 10−5 |
| DRB5*9901 | 3.21 × 10−5 | .66 | NA | 4.50 × 10−5 |
| DRB3*0301 | 1.24 × 10−6 | 8.7 × 10−8 | 7.41 × 10−8 | NA |
| DRB1*1501 | 1.99 × 10−5 | .21 | .37 | 5.28 × 10−5 |
| B*0702 | 9.64 × 10−6 | .0040 | .0040 | 6.49 × 10−5 |
| DQB1*0602 | 6.94 × 10−6 | .0059 | .0043 | 1.81 × 10−5 |
| DRB1*1302 | 3.18 × 10−6 | 4.33 × 10−8 | 4.45 × 10−8 | .14 |
| DRB1_11 | .0018 | .055 | .056 | .0063 |
| DRB1_13 | 4.73 × 10−6 | .01 | .0096 | .013 |
| DRB1_37 | .11 | .01 | .0092 | .0063 |
| DRB1_71 | .013 | .04 | .037 | .84 |
| DRB1_96 | .49 | .00058 | .0006 | .001 |
| DRB1_13 and 71 | 2.26 × 10−5 | .37 | .94 | .54 |
| DRB1_71 and 96 | .84 | .046 | .086 | .7 |
Abbreviation: NA, not applicable.
Figure 3.
Pair-wise linkage disequilibrium (r2) plot of HLA alleles associated with cervical neoplasia. HLA alleles are clustered according to their pair-wise linkage disequilibrium on both the x-axis and y-axis. On the y-axis, alleles are labeled with regard to whether they are risk or protective alleles in the overall cervical neoplasia data set, and on the x-axis, they are labeled according to whether they are HLA class I or II alleles.
Novel inverse associations with cervical neoplasia were observed with HLA-DRB5*9901 (OR, 0.77; P = 1.90 × 10−9) and HLA-DRB3*0301 (OR, 0.63; P = 4.06 × 10−5). HLA-DRB5*9901 is in positive linkage disequilibrium with the HLA class II risk alleles HLA-DRB1*1501 (r2 = 0.93) and HLA-DRB5*0101 (r2 = 0.93; Table 1 and Figure 3). No residual associations at these 2 HLA loci were observed after control for the association of HLA-DRB1 amino acid 71 (P < .01; Table 1).
HLA alleles were associated with disease when assessing HPV genotype without regard to histologic classification. Comparison of HPV16-related cases (n = 667) to all controls (n = 13428) revealed risk associations with HLA-DRB3*9901 (OR, 1.43; P = 1.27 × 10−8; Supplementary Table 6). Control for the association with HLA-DRB1 amino acid positions 13 and 37 controlled for the association of HLA-DRB3*9901 (P > .01). For cases of HPV18-related cervical neoplasia (n = 166), the strongest associated HLA allele was HLA-DPA1*0103 (OR, 1.89; P = .00039; Supplementary Table 7). With regard to histopathologic findings, a reduced risk was seen between HLA-DRB5*9901 and squamous cell carcinoma (OR, 0.72; P = 6.57 × 10−5). No residual association was observed after control for the association of HLA-DRB1 amino acids 13, 17, and 96 (P < .01). An increased risk was seen between HLA-DRB3*9901 and adenocarcinoma (OR, 1.31; P = 7.38 × 10−5; Supplementary Table 8). No residual association was observed after control for the association of HLA-DRB1 amino acids 13, 71, and 96 (P < .01).
HLA-Bw4/Bw6 and HLA-Cw1/Cw2 Association Analysis
Dominant and dosage models showed nominal risk associations between HLA-Bw4 (OR, 1.24; P = .014) and HLA-Bw6 (OR, 1.16; P = .011) and HPV16-related cervical neoplasia (Table 1 and Supplementary Table 9). The dosage model suggests that HLA-Bw4 has a weak inverse association with HPV18-related cervical neoplasia (OR, 0.78; P = .04). No associations were found between HLA-Bw4 or HLA-Bw6 and cervical cancer overall, squamous cell carcinoma, or adenocarcinoma.
All 3 models suggested that HLA-C1/2 alleles are associated with HPV16-related cervical neoplasia (Supplementary Table 10). HLA-C1 and HLA-C2 are mutually exclusive: an individual can have 2 copies of either or 1 copy of each. Both HLA-C1 and HLA-C2 can interact with specific KIRs and lead to inhibitory or activating signaling. To investigate the role of HLA-C1 or HLA-C2 and their combination with KIR, we used a recessive model to distinguish HLA-C homozygotes and heterozygotes. The frequency of individuals with 2 copies of HLA-C1 alleles was lower in HPV16-related cervical neoplasia cases (35.7%) than controls (41.3%; OR, 0.79; P = .005; Table 2). The frequency of 2 copies of HLA-C2 alleles did not differ between the groups. No association was found between HLA-C1 or HLA-C2 and cervical cancer overall, HPV18-related cervical cancer, squamous cell carcinoma, or adenocarcinoma.
Table 2.
KIR-HLA-Bw4/Bw6 and KIR-HLA-C1/C2 Type Combinations Associated With Human Papillomavirus Type 16 (HPV16)–Related Cervical Neoplasia
| Genetic Factor | HPV16-Positive Cases, Proportion (%) | Controls, Proportion (%) | ORa (95% CI) | P b |
|---|---|---|---|---|
| HLA-C1/C1 | 232/649 (35.7) | 5434/13 148 (41.3) | 0.79 (.67–.93) | .005 |
| HLA-C1/C2 | 326/649 (50.2) | 6054/13 148 (46) | 1.18 (1.01–1.38) | .039 |
| HLA-C2/C2 | 91/649 (14.0) | 1660/13 148 (12.6) | 1.13 (.90–1.42) | .29 |
| 2DL2 | 295/633 (46.6) | 6658/13 073 (50.9) | 0.84 (.72–.99) | .04 |
| 2DL3 | 596/647 (92.1) | 11942/13 188 (90.6) | 1.22 (.91–1.63) | .25 |
| 2DS2 | 310/648 (47.8) | 6751/13 167 (51.3) | 0.87 (.74–1.02) | .08 |
| 2DL2-C1/C1 | 96/624 (15.4) | 2754/12 932 (21.3) | 0.67 (.54–.84) | .00045 |
| C1/C1 in 2DL2+ | 96/291 (33.0) | 2754/6587 (41.8) | 0.69 (.51–.88) | .0029 |
| C1/C1 in 2DL2- | 131/333 (39.3) | 2599/6345 (41.0) | 0.93 (.75–1.17) | .56 |
| 2DL3-C1/C1 | 216/638 (33.9) | 4890/13 044 (37.5) | 0.85 (.72–1.01) | .09 |
| 2DL2-C1/C2 | 158/624 (25.3) | 3027/12 932 (23.4) | 1.11 (.92–1.33) | .29 |
| 2DL3-C1/C2 | 291/638 (45.6) | 5432/13 044 (41.6) | 1.18 (1.00–1.38) | .053 |
| 2DS2-C1/C1 | 101/639 (15.8) | 2791/13 023 (21.4) | 0.69 (.55–.85) | .0006 |
| C1/C1 in 2DS2+ | 101/306 (33.0) | 2791/6677 (41.8) | 0.68 (.54–.87) | .0024 |
| C1/C1 in 2DS2- | 131/333 (39.3) | 2599/6346 (41.0) | 0.93 (.75–1.17) | 0.56 |
| 2DS2-C1/C2 | 165/639 (25.8) | 3068/13 023 (23.6) | 1.13 (.94–1.35) | .23 |
| 2DS1-C2/C2 | 22/652 (3.4) | 573/13 199 (4.3) | 0.77 (.50–1.19) | .15 |
| HLA-Bw4 | 406/623 (65.2) | 7459/12 375 (60.3) | 1.24 (1.04–1.46) | .014 |
| 3DL1-HLA-Bw4 | 388/623 (62.3) | 7111/12 375 (57.5) | 1.22 (1.03–1.44) | .0085 |
| HLA-Bw4 in 3DL1+ | 388/601 (64.5) | 7111/11 805 (60.2) | 1.20 (1.02–1.43) | .034 |
| HLA-Bw4 in 3DL1- | 18/22 (81.8) | 348/570 (61.1) | 2.8 (.96–8.6) | .059 |
| 3DS1+HLA-Bw4 | 156/623 (25.0) | 2773/12 371 (22.4) | 1.16 (.96–1.39) | .09 |
Frequencies of HLA-B, HLA-C, and KIR-HLA-B, C combinations among HPV16-infected cervical cancer cases and healthy controls are shown. HLA-C1C1 indicates 2 group 1 HLA-C alleles, HLA-C2C2 indicates 2 group 2 HLA-C alleles, and HLA-C1C2 indicates 1 of each. HLA-Bw4 indicates 1 or 2 group Bw4 alleles.
Abbreviations: CI, confidence interval; HLA-C1, HLA-C01, 03, 07, 08, 12, 13, 14, and 16 and HLA-B4601 and 7301; HLA-C2, HLA-C02, 04, 05, 06, 15, 17, and 18; HLA-Bw4, HLA-B05, 5102, 5103, 13, 17, 27, 37, 38, 44, 47, 49, 51, 52, 53, 57, 58, 59, 63, and 77; - negative; +, positive.
aA positive odds ratio (OR) indicates a protective association with HPV16 infection.
b P values were calculated by using the R code glm model with principle components 1–4; combination P values were calculated from a meta-analysis between Omni-Ichip1 and 660Q-Ichip2 data sets
KIR Findings
KIR imputation concordance was explored by comparing KIR haplotype frequencies among 4 data sets (Supplementary Figure 4), comparing published population prevalences (Supplementary Figure 5), and a control-control association test (Ichip1 vs Ichip2, using the randomly divided Ichip data set; Supplementary Table 11). For the control-control association test of 16 imputed KIR loci, 4 (KIR2DS3, KIR2DL1, KIR2DP1, and KIR2DL5) were inconsistent because of the different key SNP numbers in each group and were not analyzed further. Prevalences of the other 12 KIR genes were concordant between groups. No association was found between KIR genes and cervical cancer overall or HPV18-related cervical cancer. A weak protective association was seen between KIR2DL2 and HPV16-related cervical cancer (ORomni, 0.87; OR660Q, 0.83; Pmeta = .04).
KIR-HLA-Bw4/Bw6 and KIR-HLA-C1/2 Combinations
No KIR-HLA-Bw4/Bw6 or KIR-HLA-C1/2 combination was associated with cervical neoplasia, HPV18-related cervical neoplasia, squamous cell carcinoma, or adenocarcinoma. HLA-Bw4 alleles were associated with an increased risk of HPV16-associated cervical neoplasia; this association was restricted to KIR3DL1 carriers (OR, 1.22; Pmeta = .0085). No association was seen in individuals presenting with Bw6 HLA-B alleles.
KIR2DL3 and KIR2DL2 bind HLA-C1 allotypes, with KIR2DL2 binding with greater affinity [18]. The protective association for HPV16-related cervical neoplasia of HLA-C1/C1 was restricted to individuals carrying KIR2DL2 (Pmeta = 0.00045; OR, 0.67), or KIR2DS2 (Pmeta = 0.0006; OR, 0.69), these KIR alleles often being found together on KIR haplotypes. In our dataset, KIR2DL2 and KIR2DS2 are in near complete linkage disequilibrium (r2 = 0.99). KIR2DL2 and KIR2DL3 were not associated with cervical neoplasia in individuals who were lacking HLA-C1/C1. HLA-C2 can interact with either KIR2DS1 or KIR2DL1. No association with HPV16-related cervical neoplasia was seen in individuals with KIR2DS1 and HLA-C2. KIR2DL1 could not be investigated in this study due to the low KIR2DL1 imputation accuracy.
KIR and HLA Allele Combinations
The strongest associations of any combination of KIR and HLA alleles with cervical neoplasia involved HLA-B*5501 and KIR2DS2 (ORomni, 0.61; OR660Q, 0.19; Pmeta = 5.97 × 10−5) and HLA-B*5501 and KIR2DL2 (ORomni, 0.6; OR660Q, 0.2; Pmeta = .00013; Table 3 and Supplementary Figure 6). HLA-B*5501 was not associated with cervical neoplasia independent of this interactive association with KIR genes.
Table 3.
KIR-HLA Combinations Are Associated With Cervical Neoplasia
| Combination | OR | Sample Size | HLA Frequency | KIR Frequency | KIR+HLA+ | KIR-HLA- | KIR+HLA- | KIR-HLA+ | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HLA Allele | KIR Gene | P a | Omni- Ichip1 | 660Q- Ichip2 | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls |
| HLA-B*5501 | KIR2DS2 | 5.97 × 10−5 | 0.61 | 0.19 | 2019 | 12650 | 0.034 | 0.034 | 0.52 | 0.51 | 21 | 240 | 923 | 5994 | 1028 | 6230 | 47 | 186 |
| HLA-B*5501 | KIR2DL2 | .00013 | 0.6 | 0.21 | 1983 | 12563 | 0.034 | 0.033 | 0.511 | 0.508 | 21 | 233 | 922 | 5993 | 993 | 6151 | 47 | 186 |
| HLA- DQB1*0601 | KIR2DL3 | .0032 | 0.14 | 0.17 | 2062 | 12975 | 0.012 | 0.0097 | 0.905 | 0.905 | 17 | 118 | 188 | 1215 | 1850 | 11634 | 7 | 8 |
| HLA- DRB1*1502 | KIR2DL3 | .0042 | 0.2 | 0.18 | 1920 | 12349 | 0.012 | 0.0104 | 0.908 | 0.908 | 16 | 119 | 170 | 1133 | 1727 | 11088 | 7 | 9 |
| HLA-B*3801 | KIR2DS4TOTAL | .0047 | 0.16 | 0.19 | 2056 | 12800 | 0.025 | 0.023 | 0.956 | 0.955 | 46 | 287 | 85 | 575 | 1919 | 11931 | 6 | 7 |
| HLA-B*3801 | KIR3DL1ex4 | .0047 | 0.16 | 0.16 | 2055 | 12807 | 0.025 | 0.023 | 0.956 | 0.954 | 46 | 287 | 85 | 576 | 1918 | 11937 | 6 | 7 |
| HLA-B*3801 | KIR3DL1ex9 | .0048 | 0.16 | 0.19 | 2053 | 12805 | 0.026 | 0.023 | 0.956 | 0.954 | 47 | 290 | 85 | 575 | 1915 | 11933 | 6 | 7 |
| HLA- DQA1*0103 | KIR2DL3 | .0078 | 0.57 | 0.5 | 2078 | 13106 | 0.084 | 0.120 | 0.905 | 0.906 | 147 | 1422 | 170 | 1086 | 1733 | 10453 | 28 | 145 |
Data are no. of cervical neoplasia cases and healthy controls with the specified KIR-HLA combinations, unless otherwise indicated.
Abbreviations: OR, odds ratio; -, negative; +, positive.
aCombination P values were calculated from a meta-analysis between Omni-Ichip1 and 660Q-Ichip2 data sets; P values of each data set were calculated by using the R code glm model with principal components 1–4.
DISCUSSION
Here, we report novel associations between cervical neoplasia and combinations of HLA and KIR alleles. We also extend our previous findings of protective and risk haplotypes associated with cervical neoplasia, demonstrating associations of HLA-DRB3 and HLA-DRB5 alleles with the disease, and we confirm that these are due to linkage disequilibrium with HLA-DRB1 amino acids [10]. In our previous genome-wide associated study, no significant association was noted at the leukocyte receptor complex on chromosome 19q13, but many SNPs at this locus failed quality control because the complex genetic structure of the locus leads to reduced accuracy of genotype calling by automated algorithms. In the current study, careful manual checking of genotype calling was performed, and KIR genotypes were imputed and analyzed in combination with HLA alleles.
We used imputation methods to infer HLA and KIR genotypes from SNP microarray data to perform one of the largest HLA-KIR association studies reported to date. By comparing our imputed KIR haplotype and KIR gene prevalences with published data generated using direct genotyping approaches, we confirmed that concordance between genotyped and imputed data is high. We further compared our imputation findings with direct HLA and KIR genotype data in 86 1000 genome study samples. The concordance for HLA genes in 2-digit and 4-digit resolution was 99.77% and 99.42%, respectively (data not shown). While we were able to impute 16 KIR genes, the imputation of 4 genes was inconsistent between our 2 cohorts and therefore was not considered. Of the remaining 12, 2 (KIR3DP1 and KIR2DL4) are framework KIR genes present in all individuals, and thus association with disease cannot be calculated for these genes. For the remaining 10 KIR genes, we did not find differences between cases and controls in isolation but demonstrated suggestive associations in combination with specific HLA alleles.
We identified 3 new HLA allelic associations with cervical neoplasia, HLA-DRB5*0101 and HLA-DRB3*9901 as risk factors, and HLA-DRB3*301 as a protective factor. HLA-DRB3, HLA-DRB4, and HLA-DRB5 are paralogs of HLA-DRB1, with which they are in linkage disequilibrium, and their expression level is one fifth that of HLA-DRB1 (RefSeq, July 2008). Consistent with the strong linkage disequilibrium across this locus, the associations between the HLA-DRB3 and HLA-DRB5 alleles and cervical neoplasia in this study were due to linkage disequilibrium with HLA-DRB1 alleles and amino acid variants.
Homozygosity of the HLA-C1 genotype group (C1/C1) overall was associated with protection from HPV16-related cervical neoplasia. This protective association was restricted to carriers of either KIR2DL2 or KIR2DS2, suggesting that it operates through a KIR-mediated mechanism. A role for HLA-C1 genotypes in cervical neoplasia is supported by previous studies, although the direction of association has not been consistent, and studies have generally had small sample sizes. A study of cervical cancer–affected parent-case trios showed that HLA-C1 was overtransmitted among women with invasive cervical cancer (P = .04), particularly in the subgroup infected with HPV16 or HPV18(P = .008) [24]. A study using unrelated cases and controls of European ancestry showed that the frequencies of HLA-C1/KIR2DL2 and HLA-C1/KIR2DL3 pairs were decreased in patients with HPV-positive cervical lesions and increased in those infected with high-risk HPV types, compared with findings for individuals infected with low-risk HPV types [25]. While these studies are not definitive, they suggest involvement of HLA-C1 group alleles in modulating the risk of cervical neoplasia, perhaps through their function as KIR ligands.
Several small candidate gene studies of the KIR locus and cervical cancer have been performed. Consistent with our findings, a Western Australian cohort study of 147 cases demonstrated weak associations between KIR2DL2 and KIR2DS2 and high-grade cervical intraepithelial neoplasia types 2 and 3 overall (P = .046 and P = .049, respectively) [26]. A study of Eastern US and Costa Rican individuals (196 cases and 330 controls) indicated that the frequency of KIR3DS1 increased and that of HLA-C2 alleles decreased the risk of cervical cancer [27]. Our data do not support these findings. Other studies from Sweden (65 cases) [28], Brazil (79 cases) [29], and Korea (132 cases) [30] have not reported positive associations, although their samples sizes were too small to identify anything other than essentially monogenic risk associations.
The strongest HLA-KIR combination association with cervical neoplasia observed in this study was between HLA-B*5501 and KIR2DL2/DS2 (Pmeta = 5.97 × 10−5), although this did not meet the conservative Bonferroni-corrected significance threshold of a P value of 2.0 × 10−5 for HLA-KIR interactions. While the frequency of HLA-B*5501 did not differ between cases (3.5% [71 of 2015]) and controls (3.4% [426 of 12458]), suggestive associations involving the combination of HLA-B*5501 and either KIR2DS2 or KIRSDL2 were observed. The signal was mainly attributable to the 660Q-Ichip2 data. No imputation bias was found in KIR2DS2, KIR2DL2, and HLA-B*5501 between case-case and control-control tests, suggesting that this was not an artifact of imputation, but in the absence of clear replication, further studies are required to determine its significance. HLA-B55 has been reported to be associated with cervical neoplasia, but this has not been replicated in larger studies [31]. KIR2DS2 and KIR2DL2 bind a range of HLA-C1 group allotypes, which include HLA-B46 and HLA-B73 but not HLA-B55. HLA-B55 is encoded by an HLA-Bw6 allele, which does not bind KIR. It is possible that the association we observed here is due to linkage disequilibrium with other HLA types, or it may be a noninformative suggestive association, particularly given that it was only found in comparison with one of the 2 imputation sets.
Cervical neoplasia may be associated with HLA-C1 group alleles that interact with KIR in controlling NK cell activation. Further HLA associations were demonstrated and shown to be driven by linkage disequilibrium with known amino acid components of HLA-DRB1 allelic associations of disease, further supporting that these variants are key to HLA-associations of cervical neoplasia. No definitive KIR associations were noted with cervical neoplasia, although we only examined KIR gene carriage and not allelic variation within KIR genes, for which larger and more densely sequenced reference data sets for imputation will be required. Further studies of KIR allelic variation are warranted, particularly given the role of KIR-bearing cells in immunity to viral infection and the suggestive association of HLA-C1 variants observed with disease.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgments
Acknowledgments. We thank Jessica Darlington-Brown, for coordinating specimen collection for the New South Wales, Australia, component; and Dr David Pennisi, for proofreading and copyediting the manuscript.
Financial support. This work was supported by the National Health and Medical Research Council, Australia (senior principal research fellowship to M. A. B. and grant 387701); the Australian Cancer Research Foundation; the National Cancer Institute, National Institutes of Health (grants P01CA042792 and R01CA112512); the Cancer Council New South Wales, Australia; the Canadian Institutes of Health Research (grant MOP-42532); Réseau sida et maladies infectieuses du Fonds de recherche du Québec – Santé; the Swedish Research Council; the Swedish Foundation for Strategic Research; the LUA-ALF (research grants via the University of Gothenburg and the University of Umeå); the Lundberg Foundation; the Torsten and Ragnar Söderberg’s Foundation; the Novo Nordisk Foundation; the European Commission (grant HEALTH-F2-2008-201865-GEFOS, BBMRI.se); the Swedish Society of Medicine; the Kempe-Foundation (grant JCK-1021); the Medical Faculty of Umeå University; the County Council of Västerbotten (grant Spjutspetsanslag VLL:159:33–2007); the Canada Research Chairs Program (tier 1 Canada research chair in human genome epidemiology to J. L.); and the Royal Women’s Hospital Clinical Research Foundation.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Ferlay JSI, Ervik M, Dikshit R, et al. . GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase no. 11. http://globocan.iarc.fr. Accessed 8 September 2017. [Google Scholar]
- 2. Fidler MM, Gupta S, Soerjomataram I, Ferlay J, Steliarova-Foucher E, Bray F. Cancer incidence and mortality among young adults aged 20–39 years worldwide in 2012: a population-based study. Lancet Oncol 2017; 18:1579–89. [DOI] [PubMed] [Google Scholar]
- 3. Li N, Franceschi S, Howell-Jones R, Snijders PJ, Clifford GM. Human papillomavirus type distribution in 30848 invasive cervical cancers worldwide: Variation by geographical region, histological type and year of publication. Int J Cancer 2011; 128:927–35. [DOI] [PubMed] [Google Scholar]
- 4. Dunne EF, Park IU. HPV and HPV-associated diseases. Infect Dis Clin North Am 2013; 27:765–78. [DOI] [PubMed] [Google Scholar]
- 5. Stewart BW, Wild C, International Agency for Research on Cancer, World Health Organization World cancer report 2014. Lyon, France Geneva, Switzerland: International Agency for Research on Cancer WHO Press, 2014. [Google Scholar]
- 6. Schiffman M, Glass AG, Wentzensen N, et al. . A long-term prospective study of type-specific human papillomavirus infection and risk of cervical neoplasia among 20000 women in the Portland Kaiser Cohort Study. Cancer Epidemiol Biomarkers Prev 2011; 20:1398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ramoz N, Rueda LA, Bouadjar B, Montoya LS, Orth G, Favre M. Mutations in two adjacent novel genes are associated with epidermodysplasia verruciformis. Nat Genet 2002; 32:579–81. [DOI] [PubMed] [Google Scholar]
- 8. Hernandez PA, Gorlin RJ, Lukens JN, et al. . Mutations in the chemokine receptor gene CXCR4 are associated with WHIM syndrome, a combined immunodeficiency disease. Nat Genet 2003; 34:70–4. [DOI] [PubMed] [Google Scholar]
- 9. Lesseur C, Diergaarde B, Olshan AF, et al. . Genome-wide association analyses identify new susceptibility loci for oral cavity and pharyngeal cancer. Nat Genet 2016; 48:1544–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leo PJ, Madeleine MM, Wang S, et al. . Defining the genetic susceptibility to cervical neoplasia-A genome-wide association study. PLoS Genet 2017; 13:e1006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janeway CA Jr, Travers P, Walport M, et al. Immunobiology: The Immune System in Health and Disease, 5th edition. New York: Garland Science; 2001. [Google Scholar]
- 12. Martin AM, Freitas EM, Witt CS, Christiansen FT. The genomic organization and evolution of the natural killer immunoglobulin-like receptor (KIR) gene cluster. Immunogenetics 2000; 51:268–80. [DOI] [PubMed] [Google Scholar]
- 13. Pyo CW, Wang R, Vu Q, et al. . Recombinant structures expand and contract inter and intragenic diversification at the KIR locus. BMC Genomics 2013; 14:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fauriat C, Ivarsson MA, Ljunggren HG, Malmberg KJ, Michaëlsson J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood 2010; 115:1166–74. [DOI] [PubMed] [Google Scholar]
- 15. Schenk A, Bloch W, Zimmer P. Natural killer cells–an epigenetic perspective of development and regulation. Int J Mol Sci 2016; 17:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Norman PJ, Hollenbach JA, Nemat-Gorgani N, et al. . Defining KIR and HLA class I genotypes at highest resolution via high-throughput sequencing. Am J Hum Genet 2016; 99:375–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 2015; 43:D423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol 1998; 161:571–7. [PubMed] [Google Scholar]
- 19. Villa LL. Human papillomaviruses and cervical cancer. Adv Cancer Res 1997; 71:321–41. [DOI] [PubMed] [Google Scholar]
- 20. Jia X, Han B, Onengut-Gumuscu S, et al. . Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 2013; 8:e64683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vukcevic D, Traherne JA, Næss S, et al. . Imputation of KIR types from SNP variation data. Am J Hum Genet 2015; 97:593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 2005; 95:221–7. [DOI] [PubMed] [Google Scholar]
- 23. Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 2004; 74:765–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Martin MP, Borecki IB, Zhang Z, et al. . HLA-Cw group 1 ligands for KIR increase susceptibility to invasive cervical cancer. Immunogenetics 2010; 62:761–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rizzo R, Gentili V, Rotola A, Bortolotti D, Cassai E, Di Luca D. Implication of HLA-C and KIR alleles in human papillomavirus infection and associated cervical lesions. Viral Immunol 2014; 27:468–70. [DOI] [PubMed] [Google Scholar]
- 26. Brestovac B, Wong ME, Tjendera R, Costantino PJ, Mamotte C, Witt CS. Human papillomavirus, high-grade intraepithelial neoplasia and killer immunoglogulin-like receptors: a Western Australian cohort study. Infect Agent Cancer 2013; 8:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carrington M, Wang S, Martin MP, et al. . Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med 2005; 201:1069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arnheim L, Dillner J, Sanjeevi CB. A population-based cohort study of KIR genes and genotypes in relation to cervical intraepithelial neoplasia. Tissue Antigens 2005; 65:252–9. [DOI] [PubMed] [Google Scholar]
- 29. Marangon AV, Guelsin GA, Visentainer JE, et al. . The association of the immune response genes to human papillomavirus-related cervical disease in a Brazilian population. Biomed Res Int 2013; 2013:146079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Song MJ, Lee CW, Kim JH, et al. . Association of KIR genes and HLA-C alleles with HPV-related uterine cervical disease in Korean women. Tissue Antigens 2013; 81:164–70. [DOI] [PubMed] [Google Scholar]
- 31. Krul EJ, Schipper RF, Schreuder GM, Fleuren GJ, Kenter GG, Melief CJ. HLA and susceptibility to cervical neoplasia. Hum Immunol 1999; 60:337–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.