Combining zebrafish and mouse genetics, Cantù et al. identified a conserved β-catenin-associated function for BCL9 and Pygo proteins during vertebrate heart development.
Keywords: CRISPR–Cas9, cardiovascular development, congenital heart disease, heart, transcription, Wnt signaling
Abstract
Bcl9 and Pygopus (Pygo) are obligate Wnt/β-catenin cofactors in Drosophila, yet their contribution to Wnt signaling during vertebrate development remains unresolved. Combining zebrafish and mouse genetics, we document a conserved, β-catenin-associated function for BCL9 and Pygo proteins during vertebrate heart development. Disrupting the β-catenin–BCL9–Pygo complex results in a broadly maintained canonical Wnt response yet perturbs heart development and proper expression of key cardiac regulators. Our work highlights BCL9 and Pygo as selective β-catenin cofactors in a subset of canonical Wnt responses during vertebrate development. Moreover, our results implicate alterations in BCL9 and BCL9L in human congenital heart defects.
The secreted factors of the Wnt family provide key morphogen activities during vertebrate development and tissue homeostasis (Nusse and Clevers 2017). Binding of the Wnt ligand to the transmembrane receptors of the Frizzled family and their LRP5/6 coreceptors triggers cytoplasmic events that culminate in stabilizing β-catenin that then moves into the nucleus to act as a transcriptional regulator (Nusse and Clevers 2017). Nuclear β-catenin orchestrates canonical Wnt target gene expression by recruiting a host of cofactors to Wnt-responsive elements (WREs) occupied by TCF/LEF transcription factors (Mosimann et al. 2009). In Drosophila, the coupling of the histone code reader and plant homology domain (PHD) finger protein Pygopus (Pygo) via the adaptor protein Legless (Lgs) to the β-catenin N-terminal Armadillo repeats is necessary for virtually all canonical Wnt signaling-dependent responses (Kramps et al. 2002; Townsley et al. 2004a; van Tienen et al. 2017). Lgs was shown to simultaneously bind Pygo and β-catenin via two evolutionarily conserved domains: the homology domain 1 (HD1) and HD2 (Kramps et al. 2002; Townsley et al. 2004b). In Drosophila, the protein complex TCF/LEF > β-catenin > Lgs > Pygo, termed “chain of adaptors,” is required to efficiently activate Wnt target gene expression (Städeli and Basler 2005). In support of their role in Drosophila, recent biochemical evidence further suggested that Pygo–BCL9 act as connecting factors of chromatin-engaged complexes for the assembly of a Wnt-specific enhanceosome (van Tienen et al. 2017).
Vertebrate genomes commonly harbor two paralogs of lgs (Bcl9 and Bcl9l [Bcl9/9l]), and two of pygo (Pygo1 and Pygo2 [Pygo1/2]). Morpholino antisense experiments and overexpression studies have suggested functions in β-catenin-associated axis formation during gastrulation in Xenopus and zebrafish (Brembeck et al. 2004; Kennedy et al. 2010). Nonetheless, genetic loss of function in mice has questioned their relevance for canonical Wnt signaling in mammals: The deletion of Bcl9/9l or Pygo1/2 did not cause a general abrogation of canonical Wnt signaling in mouse development (Li et al. 2007; Song et al. 2007; Cantù et al. 2013, 2014). Mouse mutant analysis has further emphasized that BCL9 and Pygo proteins have evolved β-catenin-independent functions in mammals, including the genetic interaction with Pax6 during eye lens formation, a role in chromatin compaction during spermatogenesis, and cytoplasmic activity in enamel formation during tooth development (Song et al. 2007; Cantù et al. 2013, 2014, 2017). Whether and to what extent BCL9/9L and Pygo1/2 contribute at all to β-catenin-dependent processes during mammalian development remain to be resolved.
Here, we provide genetic and molecular evidence that vertebrate BCL9 and Pygo proteins contribute as tissue-specific mediators of β-catenin in the development of specific structures and organs, in particular during heart formation. In zebrafish mutants for the bcl9 and pygo genes or upon selective chemical inhibition of the BCL9–β-catenin interaction, we uncovered that disrupting the β-catenin–BCL9–Pygo complex causes limited developmental phenotypes, including heart defects. In mice, both constitutive and heart-specific conditional loss of Bcl9/9l or Pygo1/2 or the simultaneous impairment of the BCL9/9L–β-catenin and BCL9/9L–PYGO2 interactions leads to heart malformations, which include defects in chamber septation and outflow tract (OFT) and valve formation. These data reveal that, in vertebrates, the Wnt-dependent function of the BCL9–Pygo module is restricted to select processes. Transcriptome analyses established that, in the developing heart and pharyngeal structures, the β-catenin–BCL9–Pygo complex regulates the expression of tissue-specific groups of genes. In addition, genome-wide chromatin-binding profiling revealed that β-catenin and PYGO co-occupy putative cis-regulatory regions of cardiac regulatory factors. Collectively, our results provide functional and molecular evidence for a conserved tissue-specific contribution of the BCL9 and Pygo factors in canonical Wnt signaling in two distant vertebrate species. In addition, our findings suggest a causative link between human congenital heart disease (CHD) and the reported copy number changes in BCL9 at 1q21.1 and mutations in BCL9L (Christiansen et al. 2004; Brunet et al. 2009; Tomita-Mitchell et al. 2012; Dolcetti et al. 2013).
Results
BCL9 and Pygo perturbations cause developmental heart defects in zebrafish and mice
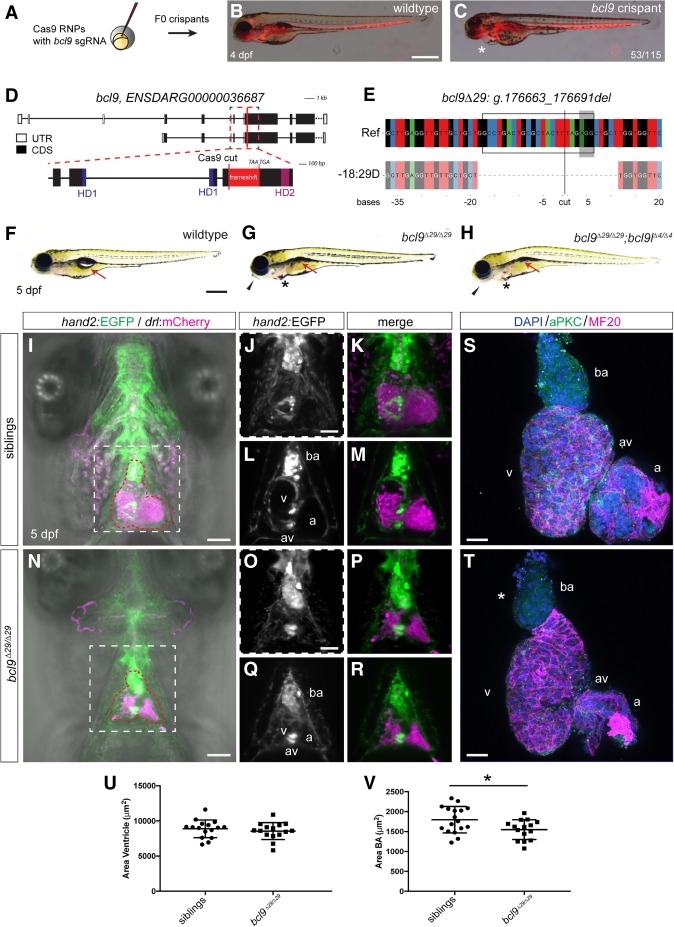
To investigate the contribution of BCL9/9L proteins to vertebrate heart development based on their repeated association with CHD, we applied maximized CRISPR–Cas9-mediated mutagenesis in zebrafish embryos to generate bcl9 crispants (Fig. 1A–C; Burger et al. 2016): We targeted both BCL9 family genes bcl9 and bcl9l with individual single-guide RNAs (sgRNAs) by injection of Cas9 ribonucleoprotein complexes into one-cell stage zebrafish embryos and observed highly penetrant cardiac phenotypes following somatic mutagenesis of bcl9 (Fig. 1B,C). We established mutant alleles for both bcl9 and bcl9l, featuring frameshift deletions in-between the coding sequences for HD1 and HD2 (Fig. 1D), the domains of BCL9 proteins that convey functional interaction with Pygo and β-catenin, respectively (Kramps et al. 2002; Cantù et al. 2014). We refer to these new alleles as bcl9Δ29 and bcl9lΔ4; if not null, their BCL9/9L protein products lost their HD2 and cannot bind β-catenin but still interact with Pygo through the HD1 (Fig. 1D,E; Supplemental Fig. S1; Kramps et al. 2002; Townsley et al. 2004b; Hoffmans et al. 2005; Mosimann et al. 2009). Heterozygous bcl9Δ29 as well as homozygous bcl9lΔ4 zebrafish and their maternal-zygotic mutant offspring (MZbcl9lΔ4) were viable and fertile with no obvious phenotypes (observed for more than four generations) (Supplemental Fig. S1). Zygotic mutant embryos homozygous for bcl9Δ29 displayed unaltered expression of early cardiac markers (nkx2.5, myh6/amhc, and gata4) at 24 h post-fertilization (hpf) (Supplemental Fig. S2) and developed seemingly normally until 48 hpf.
Figure 1.
Mutations in bcl9 lead to cardiac defects in zebrafish. (A) A CRISPR–Cas9-mediated mutagenesis identified bcl9 as a potential regulator of heart morphogenesis. (B,C) bcl9 crispants have heart-looping defects, as visible in drl:mCherry transgenics, leading to cardiac edema (asterisk) (lateral view; anterior is to the left). (D) Schematic representation of the bcl9 gene locus and generation of the bcl9Δ29 germline allele. A sgRNA was designed to target the coding exon 6 between HD1 and HD2 of the zebrafish bcl9 gene. The locus is represented as per annotation Zv10, with two isoforms that differ in the first coding exon and the untranslated regions (UTRs). The dotted box represents a zoomed region of the locus, and the red line marks the location of the sgRNA to introduce the Cas9-mediated cut, which resulted in the indicated frameshift (red box) followed by two stop codons in the isolated bcl9Δ29 allele. In the isolated allele, black boxes mark coding exons (CDS), white boxes mark UTRs, blue boxes represent the CDSs that contribute to HD1, and purple boxes represent the CDSs that contribute to HD2. (E) CrispRVariants panel plot depiction of the bcl9Δ29 germline allele with a 29-base-pair (bp) deletion. The top shows genomic reference bcl9Δ29 (bcl9:g.176663_176691), shown below. bcl9Δ29 features an out-of-frame deletion introducing a frameshift followed by 157 novel amino acids terminated by two consecutive stop codons, thus disconnecting HD1 from HD2. The black box indicates the exact position of the sgRNA sequence, the gray-shaded box indicates the 5′-NGG-3′ PAM sequence, and the black line indicates the predicted Cas9-induced double-strand break position. (F–H) Bright-field images of 5-d post-fertilization (dpf) homozygous bcl9Δ29 and bcl9Δ29;bcl9lΔ4 embryos and their wild-type-looking siblings (lateral views; anterior is to the left). Mutant embryos showed heart-looping defects and cardiac edema (asterisks). Moreover, mutant embryos did not inflate their swim bladders (arrows), presumably due to a failure in gasping air because of craniofacial malformations (black arrowheads). (I–R) Single-plane illumination microscopy (SPIM) images of hand2:EGFP;drl:mCherry-expressing wild-type siblings and homozygous bcl9Δ29 embryos (ventral views; anterior is to the top; imaged after viable heart-stopping BDM treatment). I and N depict maximum-intensity projections, and J, K, O, and P show close-ups of the dotted square in I and N, while L, M, Q, and R depict optical sections at the atrio–ventricular canal level. Compared with siblings that form correctly looped hearts with atrio–ventricular canal valves and a bulbus arteriosus (BA; heart outlined with red dotted line; n = 4; J–M), bcl9Δ29 embryos show heart-looping defects (N–R) and craniofacial malformations in both cartilage and head vascularization (n = 8; N). (S–V) Six-micrometer Z-confocal projections of sibling control (S) and homozygous bcl9Δ29 (T) hearts at 5 dpf. Quantification of the ventricular (U) and BA area (V) in sibling hearts (n = 16) compared with homozygous bcl9Δ29 hearts (n = 15) shows that the BA area is significantly smaller in the bcl9Δ29 hearts. (*) P < 0.0221, unpaired t-test with Welch correction. Each data point represents the averaged ventricular or BA area from one heart; quantification was derived from three independent experiments. N = 3. (ba) BA; (a) atrium; (v) ventricle; (av) atrio–ventricular canal. Bars: B,C,F–H, 500 µm; I–R, 100 µm; S,T, 20 µm.
Between 56 and 72 hpf, we observed in homozygous bcl9Δ29 mutants a variable incidence and expressivity of pericardial edema (38% ± 12%) and misexpression of the cardiac valve marker vcana (Supplemental Fig. S2). By 5 d post-fertilization (dpf), homozygous bcl9Δ29 embryos showed highly penetrant craniofacial and cardiac defects (n = 181 out of 879; 20.6% compared with Mendelian 25%; N = 6) (Fig. 1F–I,N; Supplemental Movies 1, 2). Upon detailed inspection, mutants displayed perturbed heart looping with misaligned atrium and ventricle (Fig. 1I–R; Supplemental Fig. S3) with more variable expressivity as well as an underdeveloped cardiopharyngeal vasculature (Fig. 1I–R; Supplemental Fig. S3). Detailed comparison of dissected sibling hearts versus homozygous bcl9Δ29 hearts at 5 dpf revealed slightly smaller ventricles and a significantly smaller smooth muscle-based bulbus arteriosus (BA) (Fig. 1S–V; Supplemental Fig. S3). Zygotic bcl9Δ29 mutants further developed a deformed pharyngeal skeleton, with abnormal Meckel's and palatoquadrate cartilage development and fusion defects of the ceratohyal and ceratobranchial 1 cartilage (Supplemental Fig. S4). Homozygous bcl9Δ29 mutant embryos failed to properly inflate their swim bladders (Fig. 1F–H; Supplemental Fig. S3) and invariantly died at 11–12 dpf. Notably, double-mutant zebrafish embryos homozygous for both bcl9Δ29 and bcl9lΔ4 looked indistinguishable from homozygous bcl9Δ29 mutants (Fig. 1F–H), suggesting a nonessential function of bcl9l and that no significant genetic compensation (Rossi et al. 2015) by the paralog bcl9l occurs in bcl9Δ29 mutants.
To test for a potential role of the BCL9-interacting partner Pygo1/2 during zebrafish development, we also generated mutant pygo1 and pygo2 alleles. We retained strains that harbor alleles with frameshift mutations within the essential N-terminal homology domain (NHD) and result in a premature stop codon before the BCL9-binding C-terminal PHD domain (Fig. 2A,B); we refer to these alleles as pygo1Δ5 and pygo2Δ1 (Fig. 2B–D). Embryos homozygous for pygo2Δ1 and concomitantly heterozygous or homozygous for pygo1Δ5 displayed cardiac defects and a spectrum of craniofacial cartilage defects that, depending on expressivity, mimicked those observed in homozygous bcl9Δ29 zebrafish mutants (Fig. 2E,F, Supplemental Fig. S4). In contrast, zebrafish embryos homozygous for pygo1Δ5, homozygous for pygo2Δ, or homozygous for pygo1Δ5 with one allele of pygo2Δ were indistinguishable from wild-type siblings throughout development and developed into fertile adults, including maternal-zygotic mutant pygo1Δ5 animals (Supplemental Fig. S4). Taken together, these data reveal select post-gastrulation defects and sensitivity of cardiac development to functional BCL9 and Pygo levels in zebrafish.
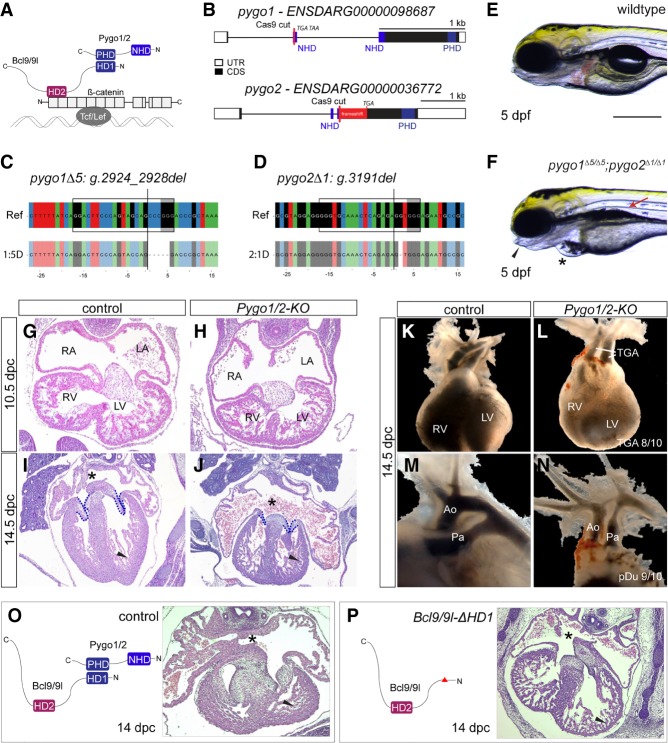
Figure 2.
Pygo1/2 mutant zebrafish and mouse embryos develop cardiac malformations reminiscent of CHD. (A) Schematic of the tripartite complex comprised of β-catenin, BCL9, and Pygo with their individual interaction domains together tethered to a WRE by TCF/LEF. (B) Schematic representation of the zebrafish pygo1 and pygo2 genes with annotated NHDs and PHDs and the Cas9 cutting site to generate mutants. The gene locus is represented as per genome annotation Zv10, with the main isoforms of both genes shown. See also the legend for Figure 1B. (C,D) CrispRVariants panel plot depictions of the germline alleles with a 5-bp or 1-bp deletion in pygo1 and pygo2, respectively. The top shows the genomic reference with the pygo1Δ5:g.2924_2928del and pygo2Δ1:g.3191del alleles shown below. Both alleles result in an out-of-frame deletion introducing a frameshift in the CDS. See also the legend for Figure 1C. (E,F) Bright-field images of live pygo1Δ5;pygo2Δ1 double mutants reveal cardiac edema (asterisks), craniofacial defects (arrowheads), and aberrant swim bladder inflation (arrows) as detected in bcl9Δ29 mutants (lateral views; anterior to the left). (G–J) Hematoxylin/eosin-stained transverse sections of a murine heart at 10.5 d post-coitum (dpc) (G,H) and 14.5 dpc (I,J). At 10.5 dpc, the development of the heart and heart cushion was still largely normal in the mutants. At 14.5 dpc, the mutants displayed markedly smaller and thinner valves (dashed reduced compact outline), reduced compact ventricular myocardium (arrowhead), highly dilated atria, and defective or absent atrial septum (asterisks). (RA) Right atrium; (LA) left atrium; (RV) right ventricle; (LV) left ventricle. (K–N) Gross anatomical view of the heart and great vessels at 13.5/14.5 dpc as revealed by India ink injection. While normally the aorta (Ao) arises from the left ventricle (LV), and the pulmonary artery (Pa) arises from the right ventricle (RV), mutants showed a classic transposition of the great arteries (TGA; arrows) and hypoplastic aorta and pulmonary artery. (O,P) Schematic representation of the Bcl9/9l–Pygo1/2 interaction (O), the molecular configuration of this interaction when the HD1 domain in Bcl9/9l is deleted in Bcl9/9l-ΔHD1 mice (P), and heart sections at 14 dpc, stained with hematoxylin/eosin. The abrogation of this interaction leads to a delayed chamber septation (asterisks), hypoplastic myocardium (arrowhead), and valve deficiency. Bars: E,F, 250 µm.
We next sought to delineate the cardiac phenotypes caused by Pygo loss of function in mice. Combined loss of function of Pygo1 and Pygo2 (Pygo1/2) leads to well-established embryonic lethality at 13.5–14.5 d post-coitum (dpc) (Li et al. 2007; Cantù et al. 2013). Our reanalysis of these mutants revealed that Pygo1/2 mutant mouse embryos develop severe heart defects between 10.5 and 14.5 dpc. Histology and three-dimensional reconstruction of the heart documented hypoplastic ventricular myocardium, dilated atria, shorter and thinner atrio–ventricular valve leaflets, and aberrant chamber septation (Fig. 2G–J; Supplemental Movies 3, 4; Supplemental Fig. S5). In addition, Pygo1/2 mutant embryos displayed prominent OFT anomalies such as transposition of the great arteries (TGA; penetrance of 80%; n = 10) (Fig. 2K,L) and hypoplastic aorta and pulmonary artery (penetrance of 90%; n = 10) (Fig. 2M,N).
To test the requirement of PYGO1/2 binding to BCL9/9L in heart formation, we used Bcl9/9l alleles carrying a deletion in the region encoding the PYGO1/2-binding HD1 (Bcl9/9l-ΔHD1) (Cantù et al. 2014). Bcl9/9l-ΔHD1-homozygous mouse embryos died at the same stage as Pygo1/2 mutants without apparent developmental delay (13.5–14.5 dpc), and histological sections revealed underdeveloped ventricle myocardium, dilated atria, defects in the atrio–ventricular valve leaflets, and perturbed septation (Fig. 2O,P; Supplemental Fig. S6). These results indicate that perturbing the cooperative action of BCL9/9L and PYGO1/2 is sufficient to trigger cardiac defects in mice. Taken together, analyses in mice and zebrafish establish that perturbation of BCL9 and Pygo function causes a range of heart defects in two evolutionarily distant vertebrate species.
The BCL9–Pygo complex drives Wnt/β-catenin signaling in developing hearts
Due to the β-catenin-independent roles of BCL9 and Pygo proteins (Song et al. 2007; Cantù et al. 2013, 2014, 2017), we cannot formally link the phenotypes resulting from our mutants to defective canonical Wnt signaling. Since BCL9/9L can act as linker proteins connecting β-catenin and Pygo (Bienz 2006), we aimed at specifically testing the concurrent requirement of connecting these interaction partners.
To do so, we generated a trans-heterozygous allelic configuration for both Bcl9 and Bcl9l, in which one allele carries a deletion of the HD1 (essential for PYGO binding), while the other lacks the HD2 (essential for β-catenin binding) (Fig. 3A,B; Cantù et al. 2014). Note that mice double-heterozygous for deletion of the HD1 (Bcl9ΔHD1/+;Bcl9lΔHD1/+; referred to here as Bcl9/9l-ΔHD1/+) or the HD2 (Bcl9ΔHD2/+;Bcl9lΔHD2/+; referred to here as Bcl9/9l-ΔHD2/+) are viable and fertile. Crossing Bcl9/9l-ΔHD1/+ and Bcl9/9l-ΔHD2/+ mice should lead to trans-heterozygous progeny (in one out of 16 embryos) in which both domain deletions are present (Bcl9ΔHD1/ΔHD2;Bcl9lΔHD1/ΔHD2; referred to here as Bcl9/9l-ΔHD1/ΔHD2). The resulting protein products can form either BCL9–β-catenin or BCL9–PYGO interactions but not the full tripartite transcriptional module (Fig. 3B), allowing us to test the developmental requirement of the β-catenin–BCL9–Pygo complex.
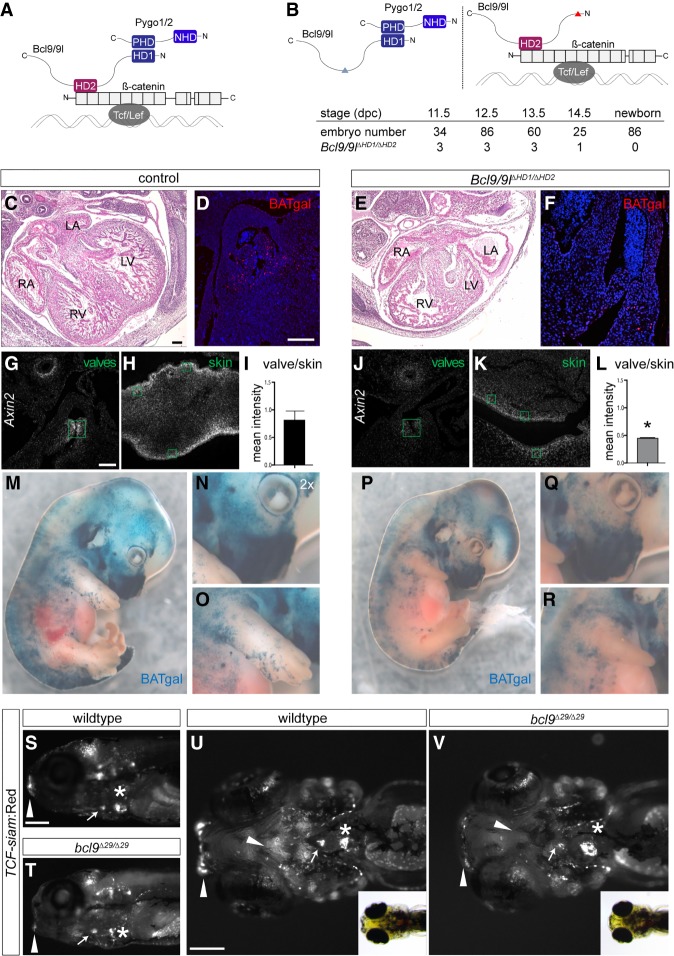
Figure 3.
The BCL9–Pygo complex drives Wnt/β-catenin transcription during heart development. (A) Schematic representation of the β-catenin transcriptional complex to outline the experimental strategy. BCL9/9l simultaneously interacts with Pygo1/2 and β-catenin via the HD1 and HD2, respectively. (B) In Bcl9/9l-Δ1/Δ2 mice, no Bcl9/9l molecule can simultaneously bind Pygo1/2 and β-catenin; the deletion of the HD2 or HD1 abrogates Bcl9/9l binding to β-catenin (left panel) or Pygo1/2 (right panel), respectively. Bcl9/9l-Δ1/Δ2 embryos are never found after 14.5 dpc (table). (C–F) Hematoxylin/eosin staining on heart sections of control (C) and Bcl9/9l-Δ1/Δ2 (E) embryos at 13.5 dpc. Bcl9/9l-Δ1/Δ2 embryos have pronounced heart defects, with thinned myocardium of the ventricular walls and malformations of the forming septum, the atrio–ventricular valves, and OFT. The expression of in vivo BATgal Wnt reporter in the OFT region is markedly reduced in Bcl9/9l-Δ1/Δ2 mutant (F) compared with control (D) hearts. (RA) Right atrium; (LA) left atrium; (RV) right ventricle; (LV) left ventricle. (G–L) Axin2 expression (detected by single-molecule mRNA ISH) in wild-type and Bcl9/9l-Δ1/Δ2 OFT valves (G,J) and skin (H,K) at 13.5 dpc. Green boxes are drawn around transversal sections of individual valves of the OFT (G,J) and in skin sections from the limbs (H,K) for quantification of fluorescence intensity levels (I,L). Axin2 expression is strongly reduced in the valves of Bcl9/9l-Δ1/Δ2 embryos (J), while reduction in the skin is mild (K). (I,L) The quantification of the ratio between Axin2 expression in the valves and the skin in the different genotypes reveals a significant reduction of Axin2 expression in Bcl9/9l-Δ1/Δ2 valves. (*) P-value < 0.05. (M–R) In vivo BATgal reporter expression in 13.5-dpc wild-type (M–O) and Bcl9/9l-Δ1/Δ2 mutant (P–R) embryos. Mutant embryos have a slightly decreased reporter expression, but BATgal reporter expression is generally retained in most tissues. Reduced BATgal activity is observed in the craniofacial region (N,Q) and the forelimbs (O,R). Loss of canonical Wnt signaling transcription in the forelimbs is accompanied by severe developmental limb defects in Bcl9/9l-Δ1/Δ2 embryos (cf. O and R). (S–V) Fluorescent and bright-field images of TCF-siam:Red bcl9Δ29 and wild-type zebrafish embryo lateral (S,T) and ventral (U,V) views; anterior is to the left. TCF reporter activity is specifically reduced in the cardiac OFT (arrow) and craniofacial apparatus (arrowheads) and altered in the atrio–ventricular valve (asterisks). Bars: C–F,G,H,J,K, 100 µm; S–V, 200 µm.
From these crosses, we never recovered Bcl9/9l-ΔHD1/ΔHD2 pups (Fig. 3B, bottom table), indicating embryonic lethality. Even though at lower numbers than the expected Mendelian ratio, Bcl9/9l-ΔHD1/ΔHD2 embryos did reach the 13.5–14.5 dpc stage (Fig. 3B) and displayed heart defects, including ventricular myocardium hypoplasia and smaller and thinner valve leaflets (Fig. 3C–F; Supplemental Fig. S6). This indicated that, for adequate development of these cardiac structures, BCL9/9L are required to physically connect PYGO proteins to β-catenin. Further relating these defects to perturbed canonical Wnt signaling was the reduced expression of the in vivo BATgal reporter (sensing nuclear β-catenin activity) in Bcl9/9l-ΔHD1/ΔHD2 mutants in cardiac valve progenitors, a region with active canonical Wnt signaling (Fig. 3D,F; Bosada et al. 2016). Moreover, single-molecule mRNA in situ hybridization (ISH) of Axin2, a prototypical pan-canonical Wnt target gene, confirmed that, at the time of analysis, canonical Wnt signaling activity was decreased in valve progenitors but not in other tissues with active Wnt signaling, such as the skin (Fig. 3G–L; Zhu et al. 2014). In addition to cardiac defects, we observed underdeveloped limbs (Fig. 3M–R), and mild skeletal malformations including shortened radius and ulna bones, incorrect specification of digit number and bifid ribs (Supplemental Fig. S7). Notably, while BATgal expression remained broadly unaffected in Bcl9/9l-ΔHD1/ΔHD2 embryos, its reduction occurred as for the cardiac valve progenitors mainly in the malformed tissues, including the developing forelimbs (Fig. 3M–R). Correspondingly, in bcl9Δ29 mutant zebrafish at 5 dpf, the pattern and strength of the canonical Wnt signaling reporter Tg(7xTCF-Xla.Siam: nlsmCherry)ia5 (referred to here as TCF-siam:Red) (Moro et al. 2012) remained globally unaffected yet showed mild reduction or pattern changes in the atrio–ventricular valve region, the OFT, and craniofacial structures (Fig. 3S–V). Taken together, these data indicate that disrupting the β-catenin–BCL9–Pygo complex in vivo has a minor impact on overall Wnt signaling but selectively perturbs individual domains of active canonical Wnt signaling.
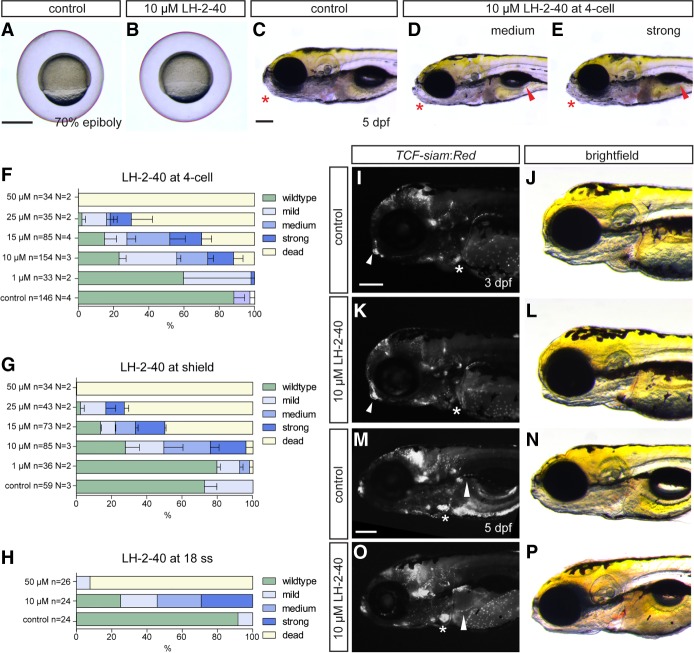
To independently disrupt the protein–protein interaction between BCL9 and β-catenin in zebrafish, we used the chemical compound LH-2-40 (compound 22) (Hoggard et al. 2015) that acts as a uniquely selective inhibitor of the BCL9-HD2 interaction with the β-catenin arm repeats 1 and 2 (Hoggard et al. 2015; Wisniewski et al. 2016). We treated wild-type zebrafish embryos with LH-2-40 at concentrations ranging from 1 to 50 µM at distinct developmental time points: four-cell stage, shield stage, and 18-somite stage, respectively. Pregastrulation and gastrulation stage treatment with LH-2-40 did not induce any observable gastrulation defects (Fig. 4A,B); in contrast, at 5 dpf, treated embryos recapitulated the bcl9Δ29 mutant phenotypes in a dose-dependent manner (Fig. 4C–H). Consistent with our genetic observations, LH-2-40-treated embryos displayed altered TCF-siam:Red expression in the atrio–ventricular valve region and in craniofacial structures at 3 and 5 dpf (Fig. 4I–P) as well as phenotypes resembling those observed in bcl9Δ29 homozygous embryos (Supplemental Fig. S8).
Figure 4.
Chemical manipulation of zebrafish heart development by Bcl9–β-catenin inhibitor LH-2-40. (A,B) Treatment with 10 µM LH-2-40 from the two- to four-cell stages does not result in gastrulation defects (lateral views). (C–E) Bright-field images of 5-dpf DMSO controls (C) and representative embryos treated with 10 µM LH-2-40 from the four-cell stage on (D,E), as assessed for phenotype classes (quantified in F–H) (lateral views; anterior is to the left). Phenotypes in LH-2-40-treated embryos become visible at 5 dpf, comparable with phenotypes observed in bcl9Δ29 mutants. LH-2-40-treated embryos show a variable phenotype expressivity, with mild to strong swim bladder inflation defects (arrowheads; D,E) linked with variable craniofacial defects (asterisks; D,E, cf. control in C). (F–J) Similar dose-dependent phenotype penetrance and expressivity are observed after Bcl9 inhibition at the four-cell, shield, and 18-somite (18 ss) stages, suggesting that the observed phenotypes result from perturbed Bcl9 function in craniofacial and heart development after somitogenesis. Mild phenotypes are characterized by mild craniofacial and swim bladder inflation defects. (D) Both phenotypes are more pronounced in the medium phenotype class. (E) Strong phenotypes are characterized by strong craniofacial defects and a complete failure to inflate the swim bladder. (I–P) Fluorescent and bright-field images of TCF:siamRed Bcl9-inhibited (K,L,O,P) and DMSO-treated (I,J,M,N) wild-type siblings at 3 and 5 dpf (lateral views; anterior is to the left). TCF reporter activity is severely reduced in the atrio–ventricular valve at 3 dpf (asterisks; I,K) and altered in the craniofacial cartilage (arrowheads; I,K). At 5 dpf, the TCF reporter is aberrantly patterned in the atrio–ventricular valve (asterisks; M,O) and fins (arrowheads; M,O). Bars: C–E,I–P, 200 µm; A,B, 500 µm.
While we cannot rule out influence by Wnt/β-catenin-independent functions of BCL9 and Pygo, our results are consistent with their joint requirement for β-catenin-dependent signaling and emphasize that disconnecting β-catenin from the BCL9–Pygo module during both mouse and zebrafish development does not cause systemic perturbation of canonical Wnt signaling but has selective impact on isolated cell types, including heart formation.
Different cardiac lineages are sensitive to mutations in BCL9 and Pygo
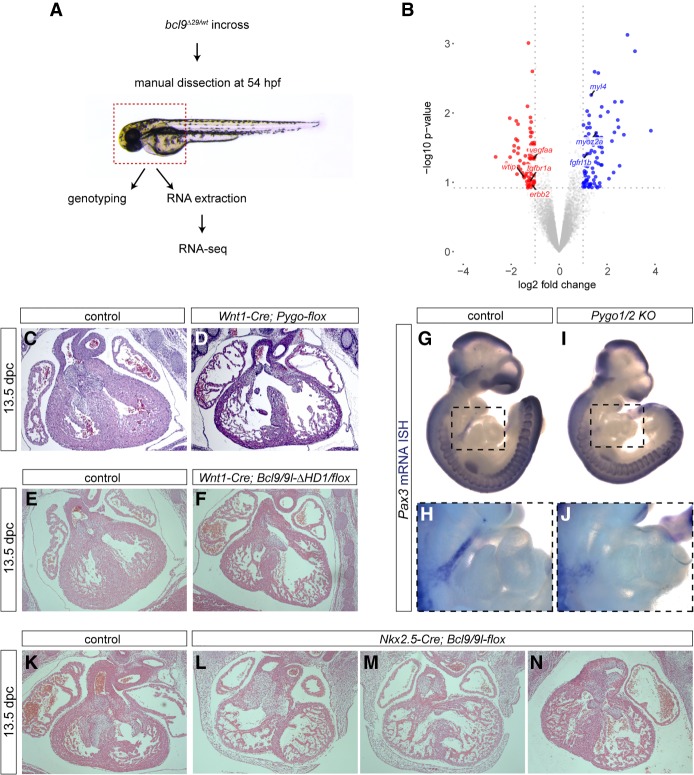
β-Catenin-dependent target gene control is required at different stages of vertebrate heart development, including the initiation of the cardiac program within the lateral plate mesoderm (LPM), chamber formation, and valve development (Hurlstone et al. 2003; Armstrong and Bischoff 2004; Gessert and Kühl 2010; Bosada et al. 2016). Transcriptome analysis of anterior embryo tissues (including the heart, pharyngeal arches, pectoral fins, and craniofacial structures) (Fig. 5A) from bcl9Δ29 mutant zebrafish at 54 hpf compared with wild-type siblings detected deregulation of 157 genes (83 genes down, fold change <0.5; 74 genes up, fold change >2.5) (Fig. 5B; Supplemental Fig. S9). The deregulated genes fall within different annotated gene expression categories, including the heart, LPM, and neural crest (Supplemental Tables S1, S2).
Figure 5.
The Bcl9–Pygo complex acts in neural crest and cardiac heart progenitor cells. (A) Schematic representation of the RNA sequencing (RNA-seq) approach used for bcl9Δ29 mutant zebrafish embryos. RNA extraction was performed on manually dissected anterior structures of 54-hpf embryos as indicated by the dashed box. (B) Comparing three independent biological replicates revealed 157 differentially expressed (74 up-regulated and 83 down-regulated) genes between wild-type and bcl9Δ29 mutants as represented in the volcano plot. Genes associated with cardiac development are highlighted. (C–F) Hematoxylin/eosin staining of heart sections of control (C,E) compared with Wnt1-Cre; Pygo1/2-flox (D) and Bcl9/9l-ΔHD1/flox (F) 13.5-dpc siblings. (G–J) Pax3 expression in 10.5-dpc embryos in the region of branchial arches: view of whole-mount mRNA ISH of control (G) compared with Pygo1/2 knockout (KO; I) siblings. H and J are magnified insets of the regions marked with dashed squares in G and I, respectively. (K–N) Hematoxylin/eosin staining of heart sections of control (K) compared with Nkx2.5-Cre; Bcl9/9l-flox (L–N) 13.5-dpc siblings.
In mammals and chickens, cardiac neural crest (CNC) cells contribute to OFT development and septation (Waldo et al. 2005; Bradshaw et al. 2009). We tested the requirement of the PYGO/BCL9 complex in CNC cells by combining the Pygo-flox and the BCL9-ΔHD1 strains with the neural crest-specific Wnt1-Cre driver. Pygo1flox/flox; Pygo2flox/flox;Wnt1-CreTg/* (referred to here as Wnt1-Cre;Pygo-flox) and Bcl9ΔHD1/flox;Bcl9l ΔHD1/flox;Wnt1-CreTg/* (referred to here as Wnt1-Cre;Bcl9/9l-ΔHD1/flox) displayed embryonic lethality at 13.5/14.5 dpc and featured heart malformations that morphologically recapitulated the constitutive loss of Pygo1/2 (Fig. 5C–F). The expression of the CNC marker Pax3 (Bradshaw et al. 2009) was significantly decreased in migrating CNC cells at 10.5 dpc in Pygo1/2 knockout embryos (Fig. 5G–J). These results indicate that BCL9 and PYGO loss in the CNC is sufficient to perturb the OFT, valves, and cardiac septation.
To test for the contribution of these proteins in LPM-derived heart progenitors, we combined the Bcl9/9l-flox conditional strain (Deka et al. 2010; Cantù et al. 2014) with Nkx2.5-Cre, in which Cre recombinase is active in the expression domain of the cardiac homeobox gene Nkx2.5 at early cardiac crescent stages (∼7.5 dpc) (Stanley et al. 2002). Nkx2.5-Cre-mediated Bcl9/9l recombination led to embryonic lethality accompanied by cardiac malformations, including thinner myocardium (Fig. 5K–N). The effect on the compact myocardium upon Bcl9/9l mutations is consistent with the role of canonical Wnt signaling in driving cardiomyocyte proliferation while preventing terminal maturation (Naito et al. 2006; Gessert and Kühl 2010).
Taken together, these results establish that the BCL9–PYGO complex connected to β-catenin is required in both migrating CNC cells and early mesodermal Nkx2.5-expressing cardiac progenitors, two key lineages whose interplay is crucial during mammalian heart development (Brade et al. 2013).
The tripartite β-catenin–BCL9–Pygo complex controls cardiac regulators
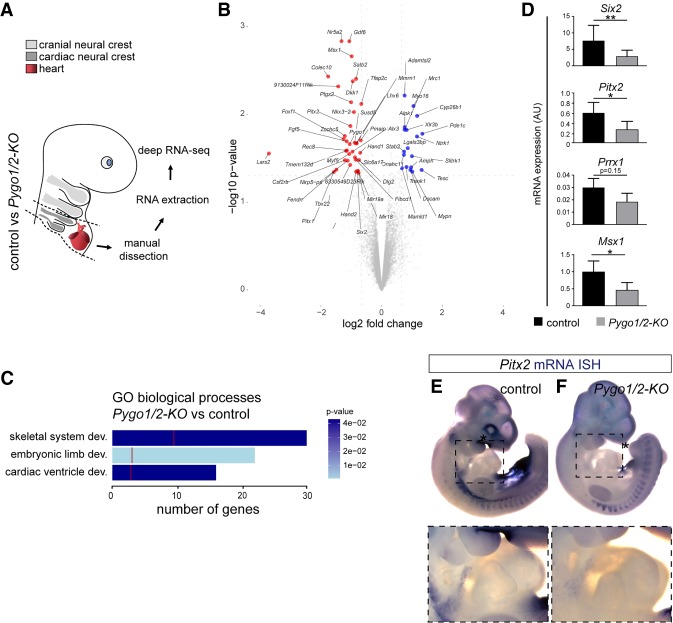
We next sought to gain insight into the transcriptional changes occurring in the developing cardiac lineages upon perturbation of the β-catenin–BCL9–Pygo module. RNA sequencing (RNA-seq) of the developing heart tube and adjacent branchial arches 3–6 (through which CNC cells migrate toward the loping heart) from constitutive Pygo1/2 mutant mice and control siblings at 10.5 dpc (n = 4) (Fig. 6A) revealed a discrete set of 65 deregulated genes, 44 of which were down-regulated and 21 of which were up-regulated (P-value < 0.05; fold change <0.66 and >1.5) (Fig. 6B). Gene ontology (GO) analysis associated a broader group of the most deregulated genes with cardiac ventricle development, embryonic limb morphogenesis, and skeletal system development, recapitulating the biological processes perturbed upon bcl9 mutation in zebrafish (Fig. 6C). Notably, the down-regulated genes comprised several encoding for mesodermal and neural crest transcription factors crucial for heart development, including Pitx2, Hand2, Msx1, and Prrx1 (Fig. 6B,C; Kioussi et al. 2002; Chen et al. 2007; Andersen et al. 2013; Ocaña et al. 2017). We confirmed the down-regulation of the most relevant genes by quantitative RT–PCR (qRT–PCR) and ISH (Fig. 6D–F). RNA-seq of Pygo1/2 mutant hearts at 12.5 dpc—when septation, valve formation, and myocardium proliferation and thickening have occurred (Gessert and Kühl 2010)—showed more profound gene expression changes consistent with progressive deterioration of cardiac development (533 genes down; 1064 genes up; P-value < 0.05; n = 3) (Supplemental Table S3).
Figure 6.
The Pygo–BCL9–β-catenin transcriptional complex controls the expression of a heart-specific genetic program. (A) Schematic representation of the RNA-seq approach used for Pygo1/2 knockout (KO) mutant mouse embryos. RNA extraction was performed on manually dissected branchial arches 3–6 and hearts of 10.5-dpc embryos, as indicated by the dashed outline. (B) Volcano plot depicting the set of up-regulated (23) and down-regulated (43) genes in Pygo1/2 knockout (KO) mouse embryos with a fold change of <0.66 and >1.5. P < 0.05. n = 4. (C) GO-based categorization of a broader group consisting of the 500 most deregulated genes (based on P-value) indicates the biological processes affected. The total bar length indicates the number of genes that categorize to the indicated GO term, the red line indicates the expected number of genes in a random deep sequencing analysis, and the color of the bar indicates the P-value for the gene enrichment in our analysis. (D) qRT-PCR validation of a subset of target genes. mRNA expression level was calculated based on Gapdh and is shown as arbitrary units (AU). (*) P < 0.05; (**) P < 0.01. (E,F) Whole-mount ISH in 10.5-dpc control (E) and Pygo1/2 knockout (KO; F) mouse embryos shows tissue-specific (branchial arches) down-regulation of the target gene Pitx2.
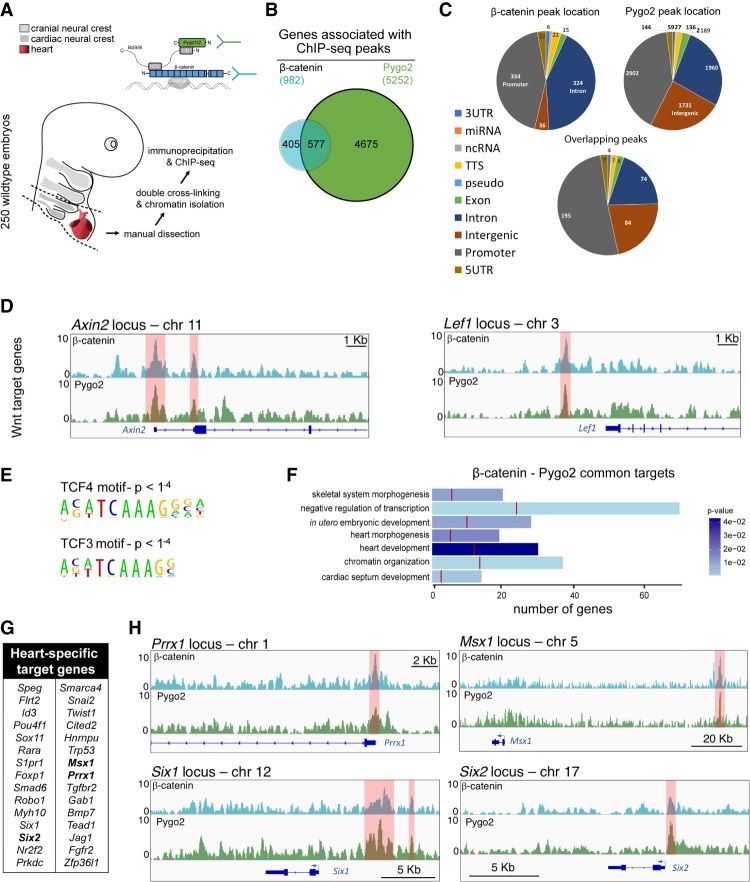
To chart the genomic locations that are bound by the β-catenin–BCL9–PYGO complex, we performed chromatin immunoprecipitation (ChIP) followed by sequencing (ChIP-seq) in duplicate for β-catenin and PYGO2 at 10.5 dpc from dissected pharyngeal arches 3–6 and the developing heart (Fig. 7A). ChIP-seq analysis assigned a total of 982 genes associated with loci occupied by β-catenin and 5252 genes associated with loci occupied by PYGO2 (Fig. 7B); the broader chromatin association of PYGO2 is in line with the known role of this factor as a widely deployed chromatin code reader (Bienz 2006; Fiedler et al. 2015) and its Wnt-independent functions (Cantù et al. 2013). Nonetheless, the genome-wide peak distribution was similar between β-catenin and PYGO2 (Fig. 7C), and their co-occupancy occurred at 59% of all β-catenin target genes (577 out of 982) (Fig. 7B,C), including at those loci found in the vicinity of canonical Wnt targets such as Axin2 and Lef1 (Fig. 7D). These peaks correspond to previously described WREs (Hovanes et al. 2001; Jho et al. 2002) and display the consensus binding sequence of the TCF/LEF transcription factors as anticipated for β-catenin-responsive elements (Fig. 7E). Therefore, while systemic canonical Wnt target genes retain most of their activity in Pygo and Bcl9 mutants, these data combined with the tissue-specific loss of Axin2 expression in Bcl9/9l mutants (Fig. 3G–L) indicate a tissue-specific contribution from the β-catenin–BCL9–PYGO complex in branchial arches and heart cells. GO analysis of target genes that were associated with β-catenin/PYGO2-co-occupied regions (based on proximity to known transcriptional start sites [TSSs]) revealed enrichment for genes involved in heart morphogenesis, valve formation, cardiac chamber septation, and skeletal development (Fig. 7F,G). The intersecting data of the RNA-seq and ChIP-seq experiments converged on a short list of heart-associated regulatory genes that are potentially controlled by the β-catenin–BCL9–PYGO complex, including Msx1, Prrx1, Six1, and Six2 (Fig. 7H).
Figure 7.
PYGO2 and β-catenin co-occupy regulatory regions of Wnt target and heart-specific genes. (A) Schematic representation of the ChIP-seq approach. Branchial arches 3–6 and hearts of 10.5-dpc wild-type embryos were dissected manually as indicated by the dashed outline. Immunoprecipitation was performed for β-catenin and PYGO2. (B) Venn diagram depicting the overlap between β-catenin and PYGO2 target genes. (C) Genomic distribution of β-catenin, PYGO2 and overlapping peak locations indicates prominent binding to regulatory regions found within promoters or introns. (D) β-Catenin and PYGO2 ChIP-seq peaks on the WREs of the prototypical target genes Axin2 (left panel) and Lef1 (right panel). Red columns indicate the position of previously described WREs. (E) The binding motif of TCF/LEF transcription factors is present within β-catenin/PYGO2 overlapping peaks. (F) GO analysis of common β-catenin and PYGO2 target genes reveals the direct regulation of processes associated with heart development. The total bar length indicates the number of genes that categorize to the indicated GO term, the red line indicates the expected number of genes in a random deep sequencing analysis, and the color of the bar indicates the P-value for the gene enrichment in our analysis. (G) A list of the 30 direct PYGO2/β-catenin target genes that have been implicated previously in heart development. (H) β-Catenin and PYGO2 ChIP peaks (red columns) within the regulatory regions of genes involved in cardiac development and found down-regulated upon Pygo2 loss (see also Fig. 6). ChIP-seq custom tracks were visualized in the Integrative Genomic Viewer genome browser.
Collectively, our data support the notion that a subset of heart-specific transcriptional regulators is controlled by BCL9 and Pygo as a consequence of their context-dependent interaction with β-catenin.
Discussion
Numerous proteins have been shown to function with nuclear β-catenin, but only a few are considered universal core components of the canonical Wnt pathway. Contrary to the mandatory requirement in Drosophila (Fiedler et al. 2015; van Tienen et al. 2017), genetic evidence in mammals has questioned the significance and involvement of BCL9-mediated tethering of the histone reader Pygo to β-catenin in Wnt target gene control (Song et al. 2007; Cantù et al. 2014, 2017). In parallel, in vivo evidence for BCL9 and Pygo function has suggested divergent functions for these proteins in different model organisms: While overexpression and morpholino-based experiments in zebrafish and Xenopus have assigned β-catenin-dependent functions for BCL9 and Pygo proteins already during gastrulation, loss-of-function studies during mouse development have uncovered mainly Wnt-independent phenotypes (Brembeck et al. 2004; Kennedy et al. 2010; Cantù et al. 2014). To date, no clear consensus of their function during vertebrate development has emerged. Here, by combining genetic and chemical loss-of-function studies in both zebrafish and mice, we revealed a conserved contribution of BCL9 and Pygo proteins as context-dependent regulators of Wnt/β-catenin signaling in vertebrates. The phenotypes resulting from abrogating their function imply that the Pygo–BCL9–β-catenin complex acts as a context-dependent transcriptional regulator during tissue-specific processes, in particular during heart development. Our in vivo data clarify, consolidate, and extend previous work on BCL9 and Pygo protein function. Additionally, our results implicate perturbed BCL9/9L function as potentially causative for human CHD, as it occurs in copy number variants of the chromosome region 1q21.1.
Previous work and our results confirm that various genetic loss-of-function permutations of BCL9 and PYGO proteins do not perturb gastrulation in mice (Figs. 2,3; Cantù et al. 2014). In zebrafish, we generated mutants with a premature stop codon between the HD1 and HD2 coding sequences of bcl9 and bcl9l; these mutations uncouple BCL9/9L–PYGO1/2 from engaging with β-catenin. We did not detect any gastrulation defects as reported previously for morpholino knockdown of Xenopus XBcl9 and zebrafish bcl9l (Supplemental Fig. S1; Brembeck et al. 2004; Kennedy et al. 2010). Double-mutant bcl9/bcl9l zebrafish and specific chemical interruption of the BCL9/9L–β-catenin interaction by LH-4-20 (Hoggard et al. 2015) phenocopy the bcl9Δ29 mutant phenotype (Fig. 4; Supplemental Fig. S6). We cannot rule out that potentially high levels of maternal BCL9 are sufficient for gastrulation stage functions; we further cannot rule out genetic compensation during gastrulation for bcl9 by bcl9l and vice versa (Rossi et al. 2015) despite the absence of such an effect at later stages in the double mutants (Fig. 1).
Both zebrafish and mouse embryos with perturbed BCL9 and Pygo maintain broad transcriptional activity of β-catenin (Figs. 3, 4). Nonetheless, following our observation of cardiac defects in interaction-perturbing Bcl9 and Pygo mutants, transcriptome analysis (Fig. 6) and chromatin occupancy of β-catenin and PYGO (a challenging experiment in vivo for β-catenin due to its indirect DNA binding) (Fig. 7) uncovered a collection of cardiac and pharyngeal genes sensitive to disrupting the β-catenin–BCL9–Pygo complex. Among these, we found common pan-β-catenin target genes, including Axin2 and Lef1. Importantly however, we predominantly uncovered genes associated with tissue-specific functions (Fig. 7). Our data support a model of canonical Wnt target gene expression by context-dependent nuclear β-catenin interactions and define BCL9 and Pygo proteins as selectively required β-catenin interactors in vertebrate heart development. Of note, cardiac defects in mice, such as atrio–ventricular valve malformations, lead to embryonic lethality between 13.5 and 16.5 dpc (Ranger et al. 1998; Combs and Yutzey 2009), suggesting that the cardiac malformations could indeed cause the lethality of Pygo1/2 mutant and Bcl9/9l-ΔHD1 and embryos. The limb and additional skeletal phenotypes observed in our mutants deserve detailed future analysis. Our findings further emphasize that BCL9 and Pygo cannot be strictly defined as core components of the canonical Wnt pathway; whether their mandatory contribution to canonical Wnt signaling in Drosophila represents an ancestral function or a specialization warrants further investigation.
Of note, in nonsyndromic sporadic cases of human CHD (with or without additional extracardiac phenotypes), copy number gains and losses have repeatedly been found in the genomic region 1q21.1 that includes the BCL9 locus (Christiansen et al. 2004; Mefford et al. 2008; Brunet et al. 2009; Greenway et al. 2009; Soemedi et al. 2012; Tomita-Mitchell et al. 2012; Dolcetti et al. 2013). Copy number variants (CNVs) that lead to either loss or gain of BCL9 copies could perturb the sensitive balance of the PYGO-interacting β-catenin coactivator function of BCL9 during human heart development. Furthermore, mutations in BCL9L, including changes in the HD1, have been genetically linked in a small pedigree to heterotaxia (HTX) with congenital cardiac malformations including ventricular and atrial septal defects (Saunders et al. 2012). Our genetic and functional data reveal a causative connection between varying Wnt/β-catenin signaling levels by mutations in Bcl9 and Pygo with developmental heart defects in both zebrafish and mice; therefore, our findings indicate that CNVs of, or mutations in, BCL9 and BCL9L might contribute to the etiology of human CHD through tissue-specific perturbation of canonical Wnt signaling.
Materials and methods
Zebrafish husbandry and transgenic strains
All zebrafish embryos were raised and maintained in E3 medium at 28.5°C without light cycle essentially as described previously (Westerfield 2007). All experiments were performed on embryos up to 5 dpf and older larvae kept only for raising mating pairs in agreement with procedures mandated by the veterinary office of the University of Zürich and the Canton of Zürich. Embryo staging was done according to morphological characteristics corresponding to hours post-fertilization or days post-fertilization as described previously (Kimmel et al. 1995). Previously established transgenic zebrafish lines used for this study included Tg(-6.35drl:EGFP) (Mosimann et al. 2015), Tg(-6.35drl:mCherry) (Sánchez-Iranzo et al. 2018), Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 (Moro et al. 2012), and TgBAC(hand2:EGFP) (Yin et al. 2010).
CRISPR–Cas9 mutagenesis in zebrafish
CRISPR–Cas9 mutagenesis was essentially performed as described in Burger et al. (2016). Oligo-based sgRNA templates (Bassett et al. 2013) were generated by PCR amplification using the invariant reverse primer 5′-AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTT-3′ and forward primers of the sequence 5′-GAAATATTTAGGTGACACTATA-(N20–22)-GTTTTAGAGCTAGAAATAGC-3′, with N representing the 20 nucleotides of the sgRNA target sequence plus up to 2 Gs at the 5′ end for successful T7 in vitro transcription. sgRNA (plus added 5′ Gs in brackets) sequences were (1) bcl9 (5′-[G]GCCTGACTGCGCTACTTTAG-3′), (2) bcl9l (5′-[G]GATTTAGGTGTGCCAATCGG-3′), (3) pygo1 (5′-GGACTTCCCAGTAGCAGCCC-3′), and (4) pygo2 (5′-[G]GCCGATGGTTGACCACCTGG-3′).
Crispants were raised to adulthood and crossed to wild type to make F1 germline mutants. All analyses and experiments were taken out on F2 mutant generations and beyond. Genotyping primers were designed to amplify target regions of mutated alleles that were subsequently analyzed via sequencing (for pygo1Δ1) or high-percentage gel electrophoresis (for bcl9Δ29), or allele-specific primers were designed to bind to mutated versus wild-type sequences specifically (for bcl9lΔ4 and pygo1Δ5). The primers used were (1) bcl9 (5′-GGTGGAAAGCCCCAACTCC-3′ [fwd] and 5′-CGTGTGCCAACTGCTGGTGG-3′ ]rev]), (2) bcl9l (5′-CACTTGCAGGTGCTGCATGG-3′ [fwd] and 5′-CTTTGAGATTTAGGTGTGCCGG-3′ [rev]), (3) pygo1 (5′-CACTTTTACTGACCCCCACAC-3′ [fwd] and 5′-GGACTTCCCAGTAGCAGGA-3′ [rev]), and (4) pygo2 (5′-GCCCAGAGAAAAAGAAGAGG-3′ [fwd] and 5′-GCTGTCCACTTCCAGGTCC-3′ [rev]). Genotyping results were analyzed and alleles visualized using CrispRVariantsLite (Lindsay et al. 2016). The final mutant designations are bcl9Δ29 (zh714), bcl9lΔ4 (zh715), pygo1Δ5 (zh716), and pygo2Δ1 (zh717).
Chemical treatments
Wild-type and Tg(7xTCF-Xla.Siam:nlsmCherry)ia5 embryos and embryos obtained from bcl9Δ29 heterozygous incrosses were treated with LH-2-40 to globally perturb Bcl9ΔHD2–β-catenin interaction at the respective developmental stages. LH-2-40, originally called compound 22 as referred to in Hoggard et al. (2015), was kindly provided by the laboratory of Dr. Haitao Ji (Moffitt Cancer Center, Tampa, FL). Single-use LH-2-40 stocks were kept at a concentration of 100 mM in DMSO at −80°C and thawed and diluted in E3 to a working concentration indicated in individual experiments directly before administration to the embryos.
Alcian blue staining
Wild-type, bcl9, and pygo1/2 mutant zebrafish embryos were fixed in 4% paraformaldehyde (PFA) overnight at 4°C and, after washing in 0.1% PBS-Tween (PBST), stained in Alcian blue staining solution (0.1 g of Alcian blue, 70 mL of ethanol, 30 mL of glacial acetic acid) overnight at room temperature. Embryos were washed in ethanol, transferred through an ethanol series to PBST, and subsequently bleached in hydrogen peroxide (3% H2O2 in 1% KOH in PBS) for 1 h or until pigments of specimens became transparent.
Zebrafish whole-mount ISH
First strand complementary DNA (cDNA) was generated from pooled zebrafish RNA isolated from different developmental stages using SuperScript III first strand synthesis kit (Invitrogen). DNA templates were generated using first strand cDNA as a PCR template and the following primers: vcana (5′-TTACGTATGCAGCCTTCTCG-3′ and 5′-GGTTCATGGGGTAACTGTGG-3′), myh6 (5′-ACGGATCAAGTAAGCGAAGG-3′ and 5′-GTTCCTCCAGTTCGTTTTGC-3′), gata4 (5′-GAGCTGCGTCTTACGAGTCC-3′ and CAGACTGGCTCTCCTTCTGC-3′), and spry4 (5′-ACTGATGAGGACGAGGAAGG-3′ and 5′-GACTCGGAATCCTTCAGTGG-3′). For in vitro transcription initiation, the T7 RNA polymerase promoter 5′-TAATACGACTCACTATAGGG-3′ was added to the 5′ ends of reverse primers. The DNA template for nkx2.5 was amplified using T3 and T7 primers. PCR reactions were performed under standard conditions using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific). RNA probes were generated via overnight incubation at 37°C using 20 U/µL T7 RNA polymerase (Roche) and digoxigenin (DIG)-labeled dNTPs (Roche). The resulting RNA was precipitated in lithium chloride and ethanol.
Embryos were fixed in 4% PFA overnight at 4°C, transferred into 100% methanol, and stored at −20°C. ISH of whole-mount zebrafish embryos was performed essentially according to standard protocols (Thisse and Thisse 2008).
Immunostaining on dissected zebrafish hearts
Hearts from 120-hpf zebrafish embryos were dissected in Tyrode's solution [136 mM NaCl, 5.4 mM KCl, 1 mM MgCl2 × 6H2O, 5 mM D(+) glucose, 10 mM HEPES, 0.3 mM Na2HPO4 × 2H2O, 1.8 mM CaCl2 × 2H2O at pH 7.4] with 20 mg/mL BSA and fixed with Shandon Glyo-Fixx (Thermo Fisher Scientific, 9990920) for 20 min at room temperature. Hearts were blocked in blocking buffer containing 5% normal goat serum, 20 mg/mL BSA, 1% DMSO, and 0.1% Tween-20 in PBS followed by overnight incubation at 4°C with primary antibodies diluted in blocking buffer. Primary antibodies used were mouse anti-MHC (1:50; Developmental Studies Hybridoma Bank, MF20 supernatant) and rabbit anti-PKC-ζ (1:100; Santa Cruz Biotechnology, sc-216). Hearts were washed three times in blocking buffer, incubated with Alexa-conjugated secondary antibodies for 4 h and DAPI in PBS for 20 min at room temperature, washed again three times with blocking buffer, and mounted in the ProLong Gold anti-fade reagent (Thermo Fisher Scientific, P36934).
Microscopy and data processing
Bright-field, basic fluorescence, and ISH imaging was performed using a Leica M205FA equipped with a DFC450 C camera. Detailed fluorescent embryo imaging of reporter transgenics was performed by single-plane illumination microscopy (SPIM) with a Zeiss Lightsheet Z.1 microscope with a plan-apochromat 20×/1.0 water objective. Prior to imaging, embryos were embedded in a rod of 1% low-melting agarose in E3 with 0.016% ethyl 3-aminobenzoate methanesulfonate salt (Tricaine, Sigma) in a 50-µL glass capillary. Heartbeat was viably stopped with 30 mM 2,3-butanedione monoxime (BDM; Sigma, B0753). Embryos and their hearts were imaged from opposing sides, and the views were combined into a single image. Imaging of dissected hearts was performed with a Leica SP8 confocal microscope using a plan-apochromat CS2 63×/1.3 glycerol objective.
Image processing was done with Leica LAS, Zeiss Zen Black, ImageJ/Fiji, and Adobe Photoshop and Illustrator CS6 according to image-preserving guidelines to ensure unbiased editing of the acquired image data. Quantitative data analysis was performed using Graphpad Prism 5.0. Data are presented as mean ± SEM (unpaired t-test with Welch correction). A lowercase “n” denotes the number of embryos, while an uppercase “N” signifies the number of replicates. To perform SPIM on embryonic mouse heart, tissue samples were extracted at 13.5 dpc, embedded in low-melting agarose, dehydrated, and cleared using benzyl alcohol/benzyl benzoate (1:2). Three-dimensional optical sectioning of the anatomy of the samples was performed with a Qls-Scope lightsheet microscopy system (Planelight S.L.) by measuring autofluorescence with a 5× objective where the signal was optimal (in this case, exciting at 532 nm and collecting emission at 590 nm ± 40 nm). Samples were imaged from two opposing views and combined to form a single volumetric image with an isotropic voxel size of 2.6 µm.
Mouse lines
Knock-in mutants in Bcl9 and Bcl9l were generated by standard techniques, as described previously (Cantù et al. 2014). Briefly, the targeting vector was electroporated into BA1 (C57BL/6 × 129/SvEv) hybrid embryonic stem cells. After selection with the antibiotic G418, surviving clones were expanded for PCR and Southern blotting analyses to confirm recombinant embryonic stem cell clones. Mouse embryonic stem cells harboring the knock-in allele were microinjected into C57BL/6 blastocysts. Resulting chimeras were bred to wild-type C57BL/6N mice to generate F1 heterozygous offspring. Neo cassette excision was obtained by crossing heterozygous knock-in animals with mice expressing Flp recombinase. All mouse experiments were performed in accordance with Swiss guidelines and approved by the Veterinarian Office of the Kanton of Zurich, Switzerland.
Histological analysis
Embryos between 9.5 and 14.5 dpc were fixed overnight in 4% PFA at 4°C, dehydrated, and embedded in paraffin. Sections were stained with hematoxylin and eosin for histological analysis. The same paraffin-embedded material was sectioned under RNase-free conditions for mRNA ISH.
Mouse whole-mount mRNA ISH
Whole-mount ISH was performed as described previously (Constam and Robertson 2000). The probes for Pax3 and Pitx2 were kindly provided by Dr. Joerg Huelsken and Dr. Daniel Constam, respectively. DIG-labeled probes (Roche) were detected by enzymatic color reaction using alkaline phosphatase-conjugated anti-DIG Fab fragments (1:1000; Roche) and BM purple alkaline phosphatase substrate (Roche). DIG-labeled RNA probes were detected with peroxidase-conjugated anti-DIG Fab fragments (1:500; Roche) followed by fluorescence detection using Tyramide signal amplification (PerkinElmer). Mouse antisense RNA probes were as described in the Supplemental Material. Quantitative mRNA ISH for Axin2 was performed using single-molecule mRNA ISH via RNAscope 2.5 (ACD) according to the manufacturer's instruction. Mean intensity measurements were performed using ImageJ.
Intracardiac injection of India ink
For the analysis of the cardiovascular system, India ink was injected into the left ventricles of mouse embryos at 14.5 dpc using a finely drawn glass pipette right after embryonic dissection, when the heart was still beating. The embryos were subsequently fixed in 4% PFA, and hearts were dissected for the anatomic examination.
Zebrafish RNA-seq
For zebrafish samples (polyA selection), zebrafish embryos from bcl9Δ29/wt crosses were dissected at 54 hpf. A cut was made between the anterior part of the embryo (containing the head, pharyngeal, and cardiac structures) and the posterior part starting from the beginning of the yolk extension. The anterior tissues were directly snap-frozen in liquid nitrogen in single tubes of PCR eight-tube strips. The posterior trunks and tails were transferred in 50 mM NaOH, and genomic DNA was extracted with alkaline lysis. Single embryos were genotypes using the target sequence primers listed above, and PCR products were separated through high-percentage gel electrophoresis, leading to a separation of the wild-type allele and mutant allele with the following outcome: (1) a low running band in bcl9Δ29 mutants, (2) a high running band in wild-type, and (3) two bands in bcl9Δ29/wt heterozygous mutants. All snap-frozen anterior parts of bcl9Δ29 mutants and of wild type were pooled in two separate tubes, and RNA isolation was performed with the RNeasy Plus minikit (Qiagen). The whole procedure was repeated for a total of three clutches, making three independent replicates.
The TruSeq mRNA stranded kit from Illumina was used for the library preparation with 250 ng of total RNA as input. The libraries were 50 bases sequenced on an Illumina HiSeq 2500 sequencer. The quality control of the resulting reads was done with FastQC and the reads were mapped to the University of California at Santa Cruz (UCSC) Danio rerio danRer10 genome with the TopHat version 2.0.11 software. For differential expression analysis, the gene features were counted with HTSeq version 0.6.1 (htseq-count) on the UCSC danRer10 gene annotation. The normalization and differential expression analyses were performed with R/Bioconductor package EdgeR version 3.14. The P-values of the differentially expressed genes were corrected for multiple testing error with a 5% false discovery rate (FDR) using Benjamini-Hochberg. The volcano plot was done selecting the genes that have >10 counts per million (CPM) in at least three samples, P-value < 0.12, and absolute value of log fold change >1. STRING analysis was performed on differentially expressed genes implicated in cardiac development in the literature (see Supplemental Tables S1, S2; permalink to analysis: http://bit.ly/2wmba7z).
Mouse RNA-seq
Developing heart tubes and adjacent branchial arches 3–6 were collected from 10.5-dpc mutant and control embryos (N= 4). The Illumina TruSeq stranded total RNA library preparation kit with RiboZero was used for the library preparation with 300 ng of total RNA as input. The libraries were 100 bases sequenced on an Illumina HiSeq 4000 sequencer. The quality control of the resulting reads was done with FastQC, and the reads were mapped to the UCSC Mus musculus mm10 genome with TopHat version 2.0.11 software. For differential expression analysis, the gene features were counted with HTSeq version 0.6.1 (htseq-count) on the UCSC mm10 gene annotation. The normalization and differential expression analyses were performed with R/Bioconductor package EdgeR version 3.14. The P-values of the differentially expressed genes are corrected for multiple testing error with a 5% FDR using Benjamini-Hochberg.
qRT–PCR
Quantitative real-time SYBR Green-based RT–PCR reactions were performed in triplicate and monitored with the ABI Prism 7900HT system (Applied Biosystem). Gapdh expression was used as internal reference. Statistical significance was determined based on unpaired t-test. Primers sequences are available on request.
ChIP
Approximately 200 branchial arches of embryonic day 10.5 mouse embryos were dissected and dissociated to a single-cell suspension. Cells were cross-linked in 20 mL of PBS for 40 min with the addition of 1.5 mM ethylene glycol-bis(succinimidyl succinate) (Thermo Scientific) for protein–protein cross-linking and 1% formaldehyde for the last 20 min of incubation to preserve DNA–protein interactions. The reaction was blocked with glycine, and the cells were subsequently lysed in 1 mL of HEPES buffer (0.3% SDS, 1% Triton-X 100, 0.15 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES). Chromatin was sheared using Covaris S2 (Covaris) for 8 min with the following setup: duty cycle: maximum, intensity: maximum, cycles/burst: maximum, and mode: power tracking. The sonicated chromatin was diluted to 0.15% SDS and incubated overnight at 4°C with anti-IgG (Santa Cruz Biotechnology), anti-PYGO2 (Novus Biological, NBP1-46171), and anti-β-catenin (Santa Cruz Biotechnology, sc-7199). The beads were washed at 4°C with wash buffer 1 (0.1% SDS, 0.1% deoxycholate, 1% Triton X-100, 0.15 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES), wash buffer 2 (0.1% SDS, 0.1% sodium deoxycho-late, 1% Triton X-100, 0.5 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES), and wash buffer 3 (0.25 M LiCl, 0.5% sodium deoxycholate, 0.5% NP-40, 1 mM EDTA, 0.5 mM EGTA, 20 mM HEPES) and, finally, twice with Tris EDTA buffer. The chromatin was eluted with 1% SDS and 0.1 M NaHCO3, decross-linked by incubation for 5 h at 65°C with 200 mM NaCl, extracted with phenol-chloroform, and ethanol-precipitated. The immunoprecipitated DNA was used as input material for DNA deep sequencing. The ChIP-seq experiment was performed in duplicate.
Data analysis—peak calling
All sequenced reads were mapped using the tool for fast and sensitive read alignment (Bowtie 2; http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) onto the UCSC mm10 reference mouse genome. The command “findPeaks” from the HOMER tool package (http://homer.salk.edu/homer) was used to identify enriched regions in the β-catenin immunoprecipitation samples using the “-style = factor” option (routinely used for transcription factors with the aim of identifying the precise location of DNA–protein contact). Input and IgG samples were used as enrichment normalization controls. Peak calling parameters were adjusted as follows: L = 4 (filtering based on local signal) and F = 4 (fold change in target experiment over input control). Annotation of peak positions (i.e., the association of individual peaks to nearby annotated genes) was obtained by the all-in-one program called “annotatePeaks.pl.” Finally, the HOMER command “makeUCSCfile” was used to produce bedGraph-formatted files that can be uploaded as custom tracks and visualized in the UCSC (http://genome.ucsc.edu) or Integrative Genomic Viewer (https://software.broadinstitute.org/software/igv) genome browsers.
Immunofluorescence
FFPE sections were blocked with 5% heat-inactivated goat serum, 5% BSA, and 0.1% Tween in PBS and incubated overnight at 4°C with the following antibodies: mouse anti p53 5E2 (NovusBio), rabbit anti Sox9 (Millipore), Troponin type2 (NovusBio), Acta2, and GFP (Aves). Slides were then incubated with a fluorescently labeled secondary antibody (Alexa 488 goat anti-mouse, Alexa 555 goat anti-rabbit, or Alexa 594 goat anti-chicken; 1:500). Nuclei were stained with DAPI (1:1000; Sigma).
Accession codes
ChIP-seq and RNA-seq reads were deposited in the Gene Expression Omnibus (GEO) repository under the accession codes GSE108240 for zebrafish mRNA-seq data, GSE110782 for mouse mRNA-seq data, and GSE110781 for mouse ChIP-seq data.
Supplementary Material
Acknowledgments
We thank Sibylle Burger, Seraina Bötschi, and Eliane Escher for technical support in zebrafish husbandry and mouse genotyping; the laboratory of Dr. Stephan Neuhauss for zebrafish support; Dr. Haitao Ji and his team for kindly providing us with LH-2-40/compound 22 for our studies; Dr. Sandra Varum Tavares for help with neural crest analyses; the Center for Microscopy and Image Analysis (ZMB) at University of Zürich (UZH) for imaging support; and the members of the Mosimann and Basler laboratories for constructive input. This work was supported by a Swiss National Science Foundation (SNSF) professorship (PP00P3_139093), SNSF R'Equip grant 150838 (Lightsheet Fluorescence Microscopy), a Marie Curie Career Integration grant from the European Commission (CIG PCIG14-GA-2013-631984), the Canton of Zürich, the UZH Foundation for Research in Science and the Humanities, and the Swiss Heart Foundation to C.M.; the SSNF to K.B.; and grants from the Forschungskredit of UZH to C.C. J.R. acknowledges support from the Ministry of Economy and Competitiveness (MINECO; FIS2016-77892-R).
Author contributions: C.C., A.F., and D.Z. designed and performed the experiments, interpreted the data, and wrote the manuscript. E.C., S.B., L.K., and A.B. assisted with and coordinated zebrafish mutant generation and characterization. E.M.C. performed bioinformatics analyses. K.D.P. performed SPIM-based live imaging and data analyses. T.V. assisted with mouse breeding and genotyping. A.B. and G.H. helped with the design of the work and critically revised the manuscript. J.R. performed and analyzed the mouse SPIM images. N.V. and M.A. performed the initial analyses on Pygo1/2 mutant mice. K.M.M.-A. and D.P. performed and analyzed dissected zebrafish heart experiments. K.B. and C.M. supervised and assisted the research teams, interpreted results, and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.315531.118.
References
- Andersen TA, Troelsen Kde LL, Larsen LA. 2013. Of mice and men: molecular genetics of congenital heart disease. Cell Mol Life Sci 71: 1327–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong EJ, Bischoff J. 2004. Heart valve development: endothelial cell signaling and differentiation. Circ Res 95: 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu J-L. 2013. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep 4: 220–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienz M. 2006. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci 31: 35–40. [DOI] [PubMed] [Google Scholar]
- Bosada FM, Devasthali V, Jones KA, Stankunas K. 2016. Wnt/β-catenin signaling enables developmental transitions during valvulogenesis. Development 143: 1041–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brade T, Pane LS, Moretti A, Chien KR, Laugwitz KL. 2013. Embryonic heart progenitors and cardiogenesis. Cold Spring Harb Perspect Med 3: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw L, Chaudhry B, Hildreth V, Webb S, Henderson DJ. 2009. Dual role for neural crest cells during outflow tract septation in the neural crest-deficient mutant Splotch2H. J Anat 214: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, Birchmeier W. 2004. Essential role of BCL9-2 in the switch between β-catenin's adhesive and transcriptional functions. Genes Dev 18: 2225–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Armengol L, Heine D, Rosell J, García-Aragonés M, Gabau E, Estivill X, Guitart M. 2009. BAC array CGH in patients with velocardiofacial syndrome-like features reveals genomic aberrations on chromosome region 1q21.1. BMC Med Genet 10: 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger A, Lindsay H, Felker A, Hess C, Anders C, Chiavacci E, Zaugg J, Weber LM, Catena R, Jinek M, et al. 2016. Maximizing mutagenesis with solubilized CRISPR–Cas9 ribonucleoprotein complexes. Development 143: 2025–2037. [DOI] [PubMed] [Google Scholar]
- Cantù C, Valenta T, Hausmann G, Vilain N, Aguet M, Basler K. 2013. The Pygo2–H3K4me2/3 interaction is dispensable for mouse development and Wnt signaling-dependent transcription. Development 140: 2377–2386. [DOI] [PubMed] [Google Scholar]
- Cantù C, Zimmerli D, Hausmann G, Valenta T, Moor A, Aguet M, Basler K. 2014. Pax6-dependent, but β-catenin-independent, function of Bcl9 proteins in mouse lens development. Genes Dev 28: 1879–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantù C, Pagella P, Shajiei TD, Zimmerli D, Valenta T, Hausmann G, Basler K, Mitsiadis TA. 2017. A cytoplasmic role of Wnt/β-catenin transcriptional cofactors Bcl9, Bcl9l, and Pygopus in tooth enamel formation. Sci Signal 10: 1–11. [DOI] [PubMed] [Google Scholar]
- Chen YH, Ishii M, Sun J, Sucov HM, Maxson RE. 2007. Msx1 and Msx2 regulate survival of secondary heart field precursors and post-migratory proliferation of cardiac neural crest in the outflow tract. Dev Biol 308: 421–437. [DOI] [PubMed] [Google Scholar]
- Christiansen J, Dyck JD, Elyas BG, Lilley M, Bamforth JS, Hicks M, Sprysak KA, Tomaszewski R, Haase SM, Vicen-Wyhony LM, et al. 2004. Chromosome 1q21.1 contiguous gene deletion is associated with congenital heart disease. Circ Res 94: 1429–1435. [DOI] [PubMed] [Google Scholar]
- Combs MD, Yutzey KE. 2009. Heart valve development: regulatory networks in development and disease. Circ Res 105: 408–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constam DB, Robertson EJ. 2000. Tissue-specific requirements for the proprotein convertase furin/SPC1 during embryonic turning and heart looping. Development 127: 245–254. [DOI] [PubMed] [Google Scholar]
- Deka J, Wiedemann N, Anderle P, Murphy-Seiler F, Bultinck J, Eyckerman S, Stehle J-C, André S, Vilain N, Zilian O, et al. 2010. Bcl9/Bcl9l are critical for Wnt-mediated regulation of stem cell traits in colon epithelium and adenocarcinomas. Cancer Res 70: 6619–6628. [DOI] [PubMed] [Google Scholar]
- Dolcetti A, Silversides CK, Marshall CR, Lionel AC, Stavropoulos DJ, Scherer SW, Bassett AS. 2013. 1q21.1 Microduplication expression in adults. Genet Med 15: 282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler M, Graeb M, Mieszczanek J, Rutherford TJ, Johnson CM, Bienz M. 2015. An ancient Pygo-dependent Wnt enhanceosome integrated by chip/LDB-SSDP. Elife 4: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessert S, Kühl M. 2010. The multiple phases and faces of Wnt signaling during cardiac differentiation and development. Circ Res 107: 186–199. [DOI] [PubMed] [Google Scholar]
- Greenway SC, Pereira AC, Lin JC, Depalma SR, Samuel J, Mesquita SM, Ergul E, Conta JR, Korn JM, Steven A, et al. 2009. De novo copy number variants identify new genes and loci in isolated, sporadic tetralogy of Fallot. Nat Genet 41: 931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmans R, Städeli R, Basler K. 2005. Pygopus and legless provide essential transcriptional coactivator functions to armadillo/β-catenin. Curr Biol 15: 1207–1211. [DOI] [PubMed] [Google Scholar]
- Hoggard LR, Zhang Y, Zhang M, Panic V, Wisniewski JA, Ji H. 2015. Rational design of selective small-molecule inhibitors for β-catenin/B-cell lymphoma 9 protein–protein interactions. J Am Chem Soc 137: 12249–12260. [DOI] [PubMed] [Google Scholar]
- Hovanes K, Li TW, Munguia JE, Truong T, Milovanovic T, Lawrence Marsh J, Holcombe RF, Waterman ML. 2001. β-Catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet 28: 53–57. [DOI] [PubMed] [Google Scholar]
- Hurlstone AFL, Haramis A-PG, Wienholds E, Begthel H, Korving J, van Eeden F, Cuppen E, Zivkovic D, Plasterk RHA, Clevers H. 2003. The Wnt/β-catenin pathway regulates cardiac valve formation. Nature 425: 633–637. [DOI] [PubMed] [Google Scholar]
- Jho E, Zhang T, Domon C, Joo C, Freund J, Costantini F. 2002. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 22: 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MW, Cha S-W, Tadjuidje E, Andrews PG, Heasman J, Kao KR. 2010. A co-dependent requirement of xBcl9 and Pygopus for embryonic body axis development in Xenopus. Dev Dyn 239: 271–283. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310. [DOI] [PubMed] [Google Scholar]
- Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, et al. 2002. Identification of a Wnt/Dvl/β-catenin → Pitx2 pathway mediating cell-type-specific proliferation during development. Cell 111: 673–685. [DOI] [PubMed] [Google Scholar]
- Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Züllig S, Basler K. 2002. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear β-catenin-TCF complex. Cell 109: 47–60. [DOI] [PubMed] [Google Scholar]
- Li B, Rheaume C, Teng A, Bilanchone V, Munguia JE, Hu M, Jessen S, Piccolo S, Waterman ML, Dai X, et al. 2007. Developmental phenotypes and reduced Wnt signaling in mice deficient for pygopus 2. Genesis 45: 318–325. [DOI] [PubMed] [Google Scholar]
- Lindsay H, Burger A, Biyong B, Felker A, Hess C, Zaugg J, Chiavacci E, Anders C, Jinek M, Mosimann C, et al. 2016. CrispRVariants charts the mutation spectrum of genome engineering experiments. Nat Biotechnol 34: 701–702. [DOI] [PubMed] [Google Scholar]
- Mefford HC, Sharp AJ, Baker C, Itsara A, Jiang Z, Buysse K, Huang S, Maloney VK, Crolla JA, Baralle D, et al. 2008. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med 359: 1685–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro E, Ozhan-Kizil G, Mongera A, Beis D, Wierzbicki C, Young RM, Bournele D, Domenichini A, Valdivia LE, Lum L, et al. 2012. In vivo Wnt signaling tracing through a transgenic biosensor fish reveals novel activity domains. Dev Biol 366: 327–340. [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. 2009. β-Catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev Mol Cell Biol 10: 276–286. [DOI] [PubMed] [Google Scholar]
- Mosimann C, Panáková D, Werdich AA, Musso G, Burger A, Lawson KL, Carr LA, Nevis KR, Sabeh MK, Zhou Y, et al. 2015. Chamber identity programs drive early functional partitioning of the heart. Nat Commun 6: 8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, Kikuchi A, Komuro I. 2006. Developmental stage-specific biphasic roles of Wnt/β-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci 103: 19812–19817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R, Clevers H. 2017. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169: 985–999. [DOI] [PubMed] [Google Scholar]
- Ocaña OH, Coskun H, Minguillón C, Murawala P, Tanaka EM, Galcerán J, Muñoz-Chapuli R, Nieto MÁ. 2017. A right-handed signalling pathway drives heart looping in vertebrates. Nature 549: 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranger AM, Grusby MJ, Hodge MR, Gravallese EM, de la Brousse FC, Hoey T, Mickanin C, Baldwin HS, Glimcher LH. 1998. The transcription factor NF-ATc is essential for cardiac valve formation. Nature 392: 186–190. [DOI] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, Stainier DYR. 2015. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524: 230–233. [DOI] [PubMed] [Google Scholar]
- Sánchez-Iranzo H, Galardi-Castilla M, Minguillón C, Sanz-Morejón A, González-Rosa JMJM, Felker A, Ernst A, Guzmán-Martínez G, Mosimann C, Mercader N. 2018. Tbx5a lineage tracing shows cardiomyocyte plasticity during zebrafish heart regeneration. Nat Commun 9: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, Andraws N, Patterson ML, Krivohlavek LA, Fellis J, et al. 2012. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med 4: 154ra135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soemedi R, Topf A, Wilson IJ, Darlay R, Rahman T, Glen E, Hall D, Huang N, Bentham J, Bhattacharya S, et al. 2012. Phenotype-specific effect of chromosome 1q21.1 rearrangements and GJA5 duplications in 2436 congenital heart disease patients and 6760 controls. Hum Mol Genet 21: 1513–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N, Schwab KR, Patterson LT, Yamaguchi T, Lin X, Potter SS, Lang RA. 2007. pygopus 2 has a crucial, Wnt pathway-independent function in lens induction. Development 134: 1873–1885. [DOI] [PubMed] [Google Scholar]
- Städeli R, Basler K. 2005. Dissecting nuclear Wingless signalling: recruitment of the transcriptional co-activator Pygopus by a chain of adaptor proteins. Mech Dev 122: 1171–1182. [DOI] [PubMed] [Google Scholar]
- Stanley EG, Biben C, Elefanty A, Barnett L, Koentgen F, Robb L, Harvey RP. 2002. Efficient cre-mediated deletion in cardiac progenitor cells conferred by a 3′UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int J Dev Biol 46: 431–439. [PubMed] [Google Scholar]
- Thisse C, Thisse B. 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc 3: 59–69. [DOI] [PubMed] [Google Scholar]
- Tomita-Mitchell A, Mahnke DK, Struble CA, Tuffnell ME, Stamm KD, Hidestrand M, Harris SE, Goetsch MA, Simpson PM, Bick DP, et al. 2012. Human gene copy number spectra analysis in congenital heart malformations. Physiol Genomics 44: 518–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley FM, Cliffe A, Bienz M. 2004a. Pygopus and Legless target Armadillo/β-catenin to the nucleus to enable its transcriptional co-activator function. Nat Cell Biol 6: 626–633. [DOI] [PubMed] [Google Scholar]
- Townsley FM, Thompson B, Bienz M. 2004b. Pygopus residues required for its binding to Legless are critical for transcription and development. J Biol Chem 279: 5177–5183. [DOI] [PubMed] [Google Scholar]
- van Tienen LM, Mieszczanek J, Fiedler M, Rutherford TJ, Bienz M, Labhart T, Desplan C, Hursh D, Jones T, Bejsovec A, et al. 2017. Constitutive scaffolding of multiple Wnt enhanceosome components by Legless/BCL9. Elife 6: 477–488. [Google Scholar]
- Waldo KL, Hutson MR, Stadt HA, Zdanowicz M, Zdanowicz J, Kirby ML. 2005. Cardiac neural crest is necessary for normal addition of the myocardium to the arterial pole from the secondary heart field. Dev Biol 281: 66–77. [DOI] [PubMed] [Google Scholar]
- Westerfield M. 2007. The zebrafish book: a guide for the laboratory use of zebrafish (Danio rerio), 5th ed. University of Oregon Press, Eugene. [Google Scholar]
- Wisniewski JA, Yin J, Teuscher KB, Zhang M, Ji H. 2016. Structure-based design of 1,4-dibenzoylpiperazines as β-catenin/B-cell lymphoma 9 protein–protein interaction inhibitors. ACS Med Chem Lett 7: 508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C, Kikuchi K, Hochgreb T, Poss KD, Stainier DYR. 2010. Hand2 regulates extracellular matrix remodeling essential for gut-looping morphogenesis in zebrafish. Dev Cell 18: 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XJ, Liu YD, Dai ZM, Zhang X, Yang XQ, Li Y, Qiu M, Fu J, Hsu W, Chen YP, et al. 2014. BMP-FGF signaling axis mediates Wnt-induced epidermal stratification in developing mammalian skin. PLoS Genet 10: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.