In this study, Su et al. investigate how post-translational modifications of Myc that affect stability and oncogenic activity regulate its function. They show that Ser62 phosphorylation and PIN1-mediated isomerization of MYC dynamically regulate the spatial distribution of MYC in the nucleus, promoting its association with the inner basket of the nuclear pore in response to proliferative signals, where it recruits the histone acetyltransferase GCN5 to bind and regulate local gene acetylation and expression, thus providing new insights into how post-translational modification of MYC controls its spatial activity.
Keywords: GCN5, MYC, nuclear pore complex, PIN1, Ser62 phosphorylation, transcriptional regulation
Abstract
The transcription factor MYC (also c-Myc) induces histone modification, chromatin remodeling, and the release of paused RNA polymerase to broadly regulate transcription. MYC is subject to a series of post-translational modifications that affect its stability and oncogenic activity, but how these control MYC's function on the genome is largely unknown. Recent work demonstrates an intimate connection between nuclear compartmentalization and gene regulation. Here, we report that Ser62 phosphorylation and PIN1-mediated isomerization of MYC dynamically regulate the spatial distribution of MYC in the nucleus, promoting its association with the inner basket of the nuclear pore in response to proliferative signals, where it recruits the histone acetyltransferase GCN5 to bind and regulate local gene acetylation and expression. We demonstrate that PIN1-mediated localization of MYC to the nuclear pore regulates MYC target genes responsive to mitogen stimulation that are involved in proliferation and migration pathways. These changes are also present at the chromatin level, with an increase in open regulatory elements in response to stimulation that is PIN1-dependent and associated with MYC chromatin binding. Taken together, our study indicates that post-translational modification of MYC controls its spatial activity to optimally regulate gene expression in response to extrinsic signals in normal and diseased states.
The proto-oncogenic transcription factor MYC has a broad range of gene targets that control many cellular behaviors, including metabolism, proliferation, and morphology, the malfunctions of which contribute to tumorigenesis and cancer progression (Dang 2012; Kress et al. 2015). Gain-of-function and loss-of-function in vivo studies demonstrate a critical role for MYC in the induction of cell growth and proliferation, stem, and migratory properties as well as microenvironment programming, and inactivation of MYC can induce tumor collapse and tumor microenvironment normalization (Kortlever et al. 2017; for review, see Soucek and Evan 2010; Gabay et al. 2014). Efforts to target MYC for cancer therapeutics are under way but suffer from an incomplete understanding of the regulation of MYC expression and mechanisms that control its specific activities.
MYC protein levels and activity are in part regulated by sequential phosphorylation events at Ser62 (S62) and Thr58 (T58) (Sears 2004; Hann 2006; Vervoorts et al. 2006). In response to growth stimulation, RAS-induced kinases and cyclin-dependent kinases (CDKs) phosphorylate MYC at S62, which is associated with MYC stabilization and activation (Sears et al. 2000; Hydbring et al. 2010). The subsequent T58 phosphorylation by GSK3β then promotes proteasome degradation of MYC through the pathway of the E3 ubiquitin ligase Fbw7 (Gregory and Hann 2000; Welcker et al. 2004; Yada et al. 2004). These phosphorylation events on the 58-TPPLSP-63 motif lead to MYC's interaction with PIN1 (Yeh et al. 2004). As the only identified phosphorylation-dependent prolyl isomerase, PIN1 catalyzes proline isomerization (trans to cis or cis to trans, depending on equilibrium and other restraining factors) of pS/pT-P motifs, influencing protein conformation to alter protein function (Liou et al. 2011). Most known PIN1-mediated isomerization events promote tumor growth and drug resistance (Zhou and Lu 2016). In the case of MYC, PIN1 catalyzed P63 both trans to cis, promoting the oncogenic activity of Ser62 phosphorylated (pS62) MYC and its transcriptional control of specific genes (Farrell et al. 2013), and cis to trans, increasing PP2A-mediated S62 dephosphorylation and MYC turnover in response to T58 phosphorylation (Yeh et al. 2004). This dual function of PIN1 may provide a molecular clock, controlling the timing of MYC activation/inactivation. In addition to this temporal control of MYC activity, recent reports suggest that the S62/T58/FBW7 MYC post-translational regulation pathway is associated with specific subnuclear localization, where pS62MYC is prevalent at the nuclear periphery, and FBW7-mediated ubiquitin-dependent degradation of MYC occurs in the nucleolus (Welcker et al. 2004; Myant et al. 2015; Sun et al. 2015). Further insight into the spatial and temporal regulation of MYC and whether these are mechanistically linked under physiological conditions and in diseased states is currently lacking.
The spatial organization of chromatin and gene expression is associated with specific nuclear architecture (Lanctôt et al. 2007; Zhou et al. 2011). In mammalian cells, the nuclear interior contains euchromatin and active transcription; the nuclear periphery, especially the lamina-associated domain (LAD), is enriched for condensed chromatin and repressive transcription (Guelen et al. 2008; Pombo and Dillon 2015). However, although also localized to the nuclear periphery, the regions near the nuclear pore complex (NPC) exclude heterochromatin and are permissive for transcription (Blobel 1985; Krull et al. 2010; Beck and Hurt 2017). Consistent with the idea that the NPC is involved in regulating active gene transcription, many of the NPC components (nucleoporins [Nups]) in yeast and Drosophila are found to be associated with active genes or regulatory elements (Casolari et al. 2004; Capelson 2010; Kalverda et al. 2010; Liang et al. 2013; Pascual-Garcia et al. 2017). Interestingly, a recent study has shown that pS62MYC is enriched at the nuclear periphery in proximity with Lamin A/C (Myant et al. 2015). However, which compartment of the nuclear periphery is involved in MYC's function and how this regulates transcription and cellular functions remain to be elucidated.
In this study, we investigated the link between the temporal activation of MYC through Ser62 phosphorylation and PIN1-mediated isomerization and the spatial nuclear distribution of MYC in cancer and normal cells under varied growth conditions. Using proximity ligation assay (PLA) and superresolution stochastic optical reconstruction microscopy (STORM) imaging, we found that pS62MYC associates with the basket of the NPC. PIN1-mediated proline isomerization of MYC promoted the recruitment of MYC and corecruitment of the histone acetyltransferase (HAT) GCN5 to the nuclear pore basket, leading to nearby histone acetylation and gene activation in response to growth stimuli. Using ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing), RNA-seq (RNA sequencing), ATAC-seq (assay for transposase-accessible chromatin [ATAC] using sequencing), and DNA FISH (fluorescence in situ hybridization), we demonstrate that this PIN1-mediated subnuclear localization of Ser62 phosphorylated MYC is associated with global chromatin accessibility changes and the expression of a group of genes involved in proliferation and migration pathways. Together, this work provides novel insights into the dynamic spatial control of MYC's gene regulatory activity responsive to environmental signals.
Results
MYC associates with the nuclear pore basket
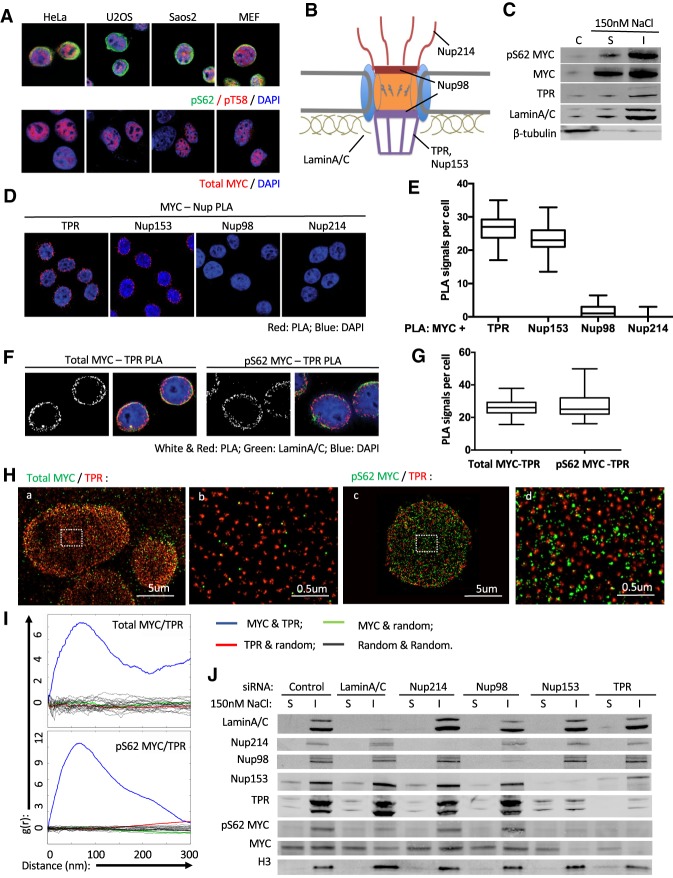
Previous studies suggest an enrichment of pS62MYC at the nuclear periphery (Myant et al. 2015). Consistent with this report, using antibodies validated to be specific against pS62MYC (Supplemental Fig. S1A,B; Zhang et al. 2012; Helander et al. 2015; Myant et al. 2015), we found a substantial number of cells showing nuclear rim-like distribution of pS62MYC in vitro and in vivo (Fig. 1A; Supplemental Fig. S1A,C–F). Notably, the pattern of pS62 is distinct from the phosphorylated Thr58 (pT58) MYC or total MYC signal, which showed more diffuse nucleoplasmic staining in all cells (Fig. 1A; Supplemental Fig. S1A,C). The enrichment of pS62MYC at the nuclear periphery is supported by the presence of pS62MYC in the nuclear-insoluble fraction that includes lamina and the nuclear pore basket component TPR (Fig. 1B,C). We speculated an involvement of the NPC in MYC localization at the nuclear periphery, suggested by an early electron microscopy study that visualized MYC localized at the nuclear pore (Royds et al. 1992). To examine the possibility of MYC association with the nuclear pore, we conducted PLA with confocal microscopy to view interaction between MYC and various Nups representing different components of the NPC (Fig. 1B,D). Using antibodies against TPR, Nup98, Nup153, and Nup214 showing specific nuclear peripheral staining (Supplemental Fig. S2A), we observed robust PLA signals of MYC association with TPR and Nup153 (pore basket) but not signals of MYC association with Nup98 (inner rings) or Nup214 (cytoplasmic filaments) (Fig. 1D,E). We also performed MYC coimmunoprecipitation experiments and found that both Nup153 and TPR were coprecipitated with MYC, while Nup98, Nup214, and Lamin A/C did not show appreciable pull-down (Supplemental Fig. S2B). These data indicate that MYC and pS62MYC (Fig. 1F,G; Supplemental Fig. S1E) associate with the basket of the NPC.
Figure 1.
pS62MYC associates with the nuclear pore basket. (A) Immunofluorescence (IF) staining through confocal imaging (Zeiss, LSM 880) of pS62MYC (red; top) together with pT58MYC (green; top) and total MYC (red; bottom) in HeLa, Saos2, and U2OS cells and mouse embryonic fibroblasts (MEFs). Phospho-antibody specificity is shown in Supplemental Figure S1A. Single-channel staining is shown in Supplemental Figure S1C. The pT58 and total MYC staining are consistent in all cells; the percentages of nuclear peripheral pS62 staining across cells in each line are quantified in Supplemental Figure S1D. (B) A schematic of the NPC with representative nuclear porins labeled. (C) Western blot of subcellular distribution of pS62MYC and total MYC in cytoplasmic (C), nuclear-soluble (S), and nuclear-insoluble (I) fractions of HeLa cells by 150 nM NaCl nuclear extraction. Lamin A/C and TPR represent nuclear markers; β-tubulin represents a cytoplasmic marker. (D) PLA of MYC association with nuclear porins (Nups) in HeLa cells. The PLA signals (red) are overlaid with DAPI (blue) from confocal imaging. (E) Quantification of PLA signals in D; the whiskers represent 5%–95% intervals, and the box represents median and 25%–75% intervals of PLA signals per cell from 50 cells per condition. Data are representative of three independent experiments. (F) PLA of total MYC and pS62MYC with TPR (red) in HeLa cells. (Green) Lamin A/C; (blue) DAPI. (G) Quantification similar to E of PLA signals in F. (H) STORM analysis of colocalization of total MYC and pS62MYC with TPR. Total MYC (green; panel a) and pS62MYC (green; panel c) with TPR (red) at a focal plane near the bottom of the nucleus. (Panels b, d) Zoom on a part of each left image. (I) Plots of pair correlation functions showing the density variation of a total MYC or pS62MYC signal on the Y-axis as a function [g(r)] of radial distance (in nanometers) on the X-axis from a TPR signal (blue lines). Shown also are the plots of MYC or pS62MYC against random signal (green), TPR against random signal (red), and random signal against random signal (gray). (J) Western blot of the indicated proteins showing subnuclear distribution of pS62MYC and total MYC in nuclear-soluble (S) and nuclear-insoluble (I) fractions of HeLa cells by 150 nM NaCl nuclear extraction with control or siRNA knockdown of the indicated nuclear peripheral components. Histone 3 (H3) marks the insoluble fraction.
To visualize the detail of MYC protein at the nuclear periphery, we took advantage of the superresolution STORM imaging technology to improve the resolution to ∼20 nm. We costained pS62 or total MYC with TPR and took images at the nuclear equator and the bottom of the nucleus. At the focal plane of the equator, both MYC and pS62MYC signals not only overlapped with TPR at the nuclear periphery (Supplemental Fig. S2Ca-d) but also distributed toward the nucleoplasmic side of the TPR signals (Supplemental Fig. S2C-b and d). At the bottom plane (Fig. 1H, panels a–d), individual nuclear pores are well resolved, and the colocalization of both MYC and pS62MYC with TPR could be more clearly assessed. In both cases, spatial pattern analysis with pair correlation function indicated significant colocalization compared with random distributions (Fig. 1I; Nickerson et al. 2014). A subpopulation of total MYC (Fig. 1I, top) appeared at the same distance (∼60 nm) away from TPR as pS62MYC (Fig. 1I, bottom), suggesting that this subpopulation is pS62MYC. Of note, the peak at ∼60 nm indicates that TPR and MYC (pS62) are in close proximity and not more than 60 nm apart (actual distance is affected by the large sizes of the detection antibodies and the finite localization precision of the fluorescent molecules), which is consistent with the results from PLA (40–50 nm). Another subpopulation of total MYC, but not pS62MYC, can be found further away (>200 nm) (Fig. 1I, top) from the center of TPR, suggesting that non-Ser62 phosphorylated MYC is located further away from the nuclear pore and does not associate closely with TPR.
To further evaluate the relevance of various nuclear architecture components in MYC's association with the nuclear periphery, we knocked down Nups and Lamin A/C to see whether this affected the presence of MYC in the nuclear-insoluble fraction (Fig. 1J; Supplemental Fig. S2E). Consistent with our PLA results and superresolution imaging (Fig. 1D,E,H,I), depletion of the pore basket subunits Nup153 and TPR significantly reduced the levels of pS62MYC and total MYC in the insoluble fraction, whereas knockdown of Nup98 and Nup214 had little effect. Notably, we did not observe an accumulation of pS62MYC or total MYC in the nuclear-soluble fraction upon its loss from the insoluble fraction, which is in agreement with previously published and our unpublished observations that MYC in the nuclear-soluble fraction is less stable and thus rapidly turned over (Tworkowski et al. 2002; Myant et al. 2015). In addition, knockdown of Lamin A/C also modestly reduced pS62MYC and total MYC levels in the insoluble fraction, suggesting a potential role for the nuclear lamina in storing MYC, consistent with a previous report (Myant et al. 2015), although we cannot rule out disruption of the NPC structure with the lamin knockdown. We also could not tease out whether Nup153 or TPR is more important for MYC's association with the basket, as depletion of TPR affected Nup153 levels, and depletion of Nup153 interfered with the level and distribution of TPR, supporting the tight structural and functional association of the two proteins (Hase and Cordes 2003; Krull et al. 2004; Rajanala and Nandicoori 2012). Given the critical roles of TPR in nuclear pore structure and nuclear peripheral chromatin organization (Krull et al. 2004, 2010), we focused on TPR for the following localization and functional characterizations.
Ser62 phosphorylation is required for MYC association with the nuclear pore
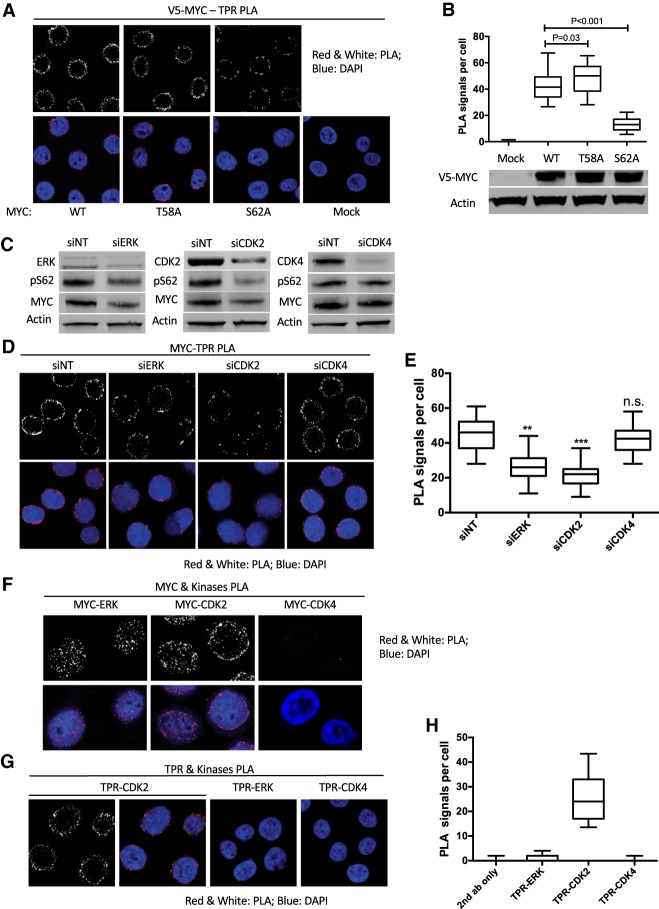
The enrichment of pS62MYC at the nuclear periphery and our discovery that pS62MYC associates with the basket of the NPC led us to test whether Ser62 phosphorylation is important for MYC's association with the NPC. We transfected HeLa cells with constructs expressing V5-tagged wild-type MYC or MYC phosphorylation mutants S62A and T58A and compared their associations with TPR via V5-TPR PLA. While S62A MYC lacks phosphorylation on Thr58 and Ser62 due to the hierarchical nature of GSK3β-mediated Thr58 phosphorylation, T58A MYC has increased Ser62 phosphorylation due to the inability of PP2A to dephosphorylate T58A MYC (Lutterbach and Hann 1994; Yeh et al. 2004; Arnold and Sears 2006; Hann 2006). The PLA signal from S62A MYC was significantly decreased compared with wild-type or T58A MYC (Fig. 2A,B). Additionally, we tested the association of S62A MYC with TPR in a coimmunoprecipitation experiment and observed reduced coprecipitation of S62A MYC compared with wild-type MYC with anti-TPR (Supplemental Fig. S2D). Taken together, these data suggest that Ser62 phosphorylation facilitates MYC interaction with TPR.
Figure 2.
Ser62 phosphorylation is required for MYC association with the nuclear pore. (A) PLA of V5-MYC–TPR association in transfected HeLa cells expressing equal amounts of V5 tagged wild-type, T58A, and S62A MYC. The mock transfection was used as negative control. (B) Quantification of A showing median PLA signals per cell from 50 cells. The whiskers represent 5%–95% intervals, and the box represents median and 25%–75% intervals. Data are representative of three independent experiments. Western blot showing the expressions of the MYC constructs. (C) Western blots of pS62MYC, total MYC, and the indicated kinases in HeLa cells transfected with nontargeting (NT) control or siRNAs specific to the indicated kinases. Actin was used as a loading control. (D) PLA of MYC–TPR association in HeLa cells transfected with siRNAs targeting the indicated kinases. (E) Quantification of D as in B showing PLA signals per cell. (F) PLA of MYC association with the indicated kinases in HeLa cells. (G) PLA of TPR association with the indicated kinases in HeLa cells. (H) Quantification of G as in B showing PLA signals per cell. P-values are shown for relevant significant comparisons. (**) P < 0.01; (***) P < 0.001.
To test upstream kinases that might affect MYC association with the NPC, we focused on ERK and CDK2, which have shown a strong specificity in phosphorylating MYC at Ser62 and increasing MYC stability (Lutterbach and Hann 1994; Pulverer et al. 1994; Sears et al. 2000; Hydbring et al. 2010). Knockdown of ERK or CDK2 through siRNA reduced Ser62 phosphorylation and total MYC as expected (Fig. 2C; Supplemental Fig. S2F), and this was coupled with a significant reduction in endogenous MYC–TPR PLA (Fig. 2D,E), suggesting that ERK and CDK2 are upstream kinases involved in Ser62 phosphorylation of MYC and MYC–TPR association. As a control, depletion of CDK4, which is a downstream target of MYC (Mateyak et al. 1999; Hermeking et al. 2000; Marval et al. 2004), did not change MYC Ser62 phosphorylation levels or the MYC–TPR PLA signal (Fig. 2C–E). Consistent with the kinase specificity, we detected robust PLA signals, indicating interactions of MYC with both ERK and CDK2 but not with CDK4 (Fig. 2F). Intriguingly, the MYC–ERK PLA signals were found ubiquitously throughout the nucleus, whereas the MYC–CDK2 PLA signals were enriched in the nuclear periphery (Fig. 2F). We speculated that while ERK may phosphorylate Ser62 in the nuclear interior, CDK2 might associate with TPR and phosphorylate MYC at the NPC. Indeed, we found a robust PLA signal of TPR–CDK2 but only a low TPR–ERK PLA signal and background levels with TPR–CDK4 PLA (Fig. 2G,H). This interaction of CDK2 with TPR is consistent with the previous identification of TPR as a CDK2 substrate through mass spectrometry (Chi et al. 2008). Together, these results suggest that ERK and CDK2 facilitate MYC interaction with the NPC by phosphorylating MYC at Ser62 and that, while ERK can phosphorylate MYC in the nuclear interior, CDK2 may phosphorylate MYC directly at the NPC.
PIN1-mediated isomerization promotes pS62MYC association with the nuclear pore
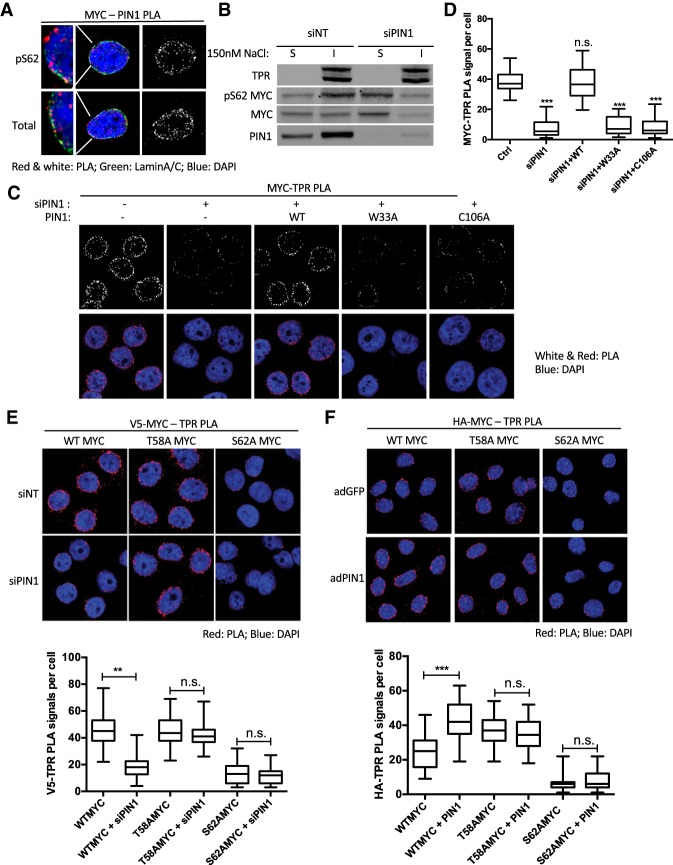
PIN1 catalyzes pSer62–Pro63 trans–cis isomerization, where the peptidyl proly bond is initially in trans conformation following ERK or CDK-mediated phosphorylation (Brown et al. 1999; Weiwad et al. 2000; Zhou 2000). This isomerization increases MYC DNA-binding, transcriptional, and oncogenic activity (Farrell et al. 2013; Helander et al. 2015). PIN1 subsequently catalyzes pSer62–Pro63 back to the trans conformation, depending on secondary phosphorylation at Thr58, and this facilitates PP2A-mediated dephosphorylation of pSer62 and MYC degradation (Yeh et al. 2004). This dynamic regulation is thought to tightly control MYC activity. To test whether PIN1 plays a role in pS62MYC interaction with the NPC, we first looked at the association of PIN1 with total MYC and pS62MYC by PLA. Interestingly, we found a substantial amount of PLA signal localized to the nuclear periphery in both MYC–PIN1 and pS62MYC–PIN1 PLA (Fig. 3A). Notably, there were also PLA speckles in the nuclear interior, suggesting that PIN1 regulation of MYC may occur in multiple nuclear compartments. We examined the effects of knocking down PIN1 expression via siRNA on the level of MYC in the nuclear-insoluble and nuclear-soluble fractions. The expression of both total and pS62MYC in the nuclear-insoluble fraction decreased upon PIN1 knockdown, while their levels increased in the soluble fraction (Fig. 3B; Supplemental Fig. S3A), suggesting that PIN1 is important for the localization of MYC to the insoluble compartment that includes chromatin, lamin, and the NPC, while it facilitates MYC turnover in the soluble fraction, consistent with our prior findings (Yeh et al. 2004). To analyze the role of PIN1 on MYC association with the NPC, we knocked down PIN1 and found a significant reduction of MYC–TPR PLA signals (Fig. 3C,D). Importantly, MYC interaction with TPR can be rescued by wild-type PIN1 but not the substrate-binding mutant (W33A) (Liu et al. 2008) or catalytic-deficient mutant (C109A) (Winkler et al. 2000) of PIN1 (Fig. 3C,D; Supplemental Fig. S3B), indicating that PIN1's catalytic activity is required to facilitate MYC–TPR interaction.
Figure 3.
PIN1-mediated isomerization promotes pS62MYC association with the nuclear pore. (A) PLA of PIN1 and total MYC or pS62MYC in HeLa cells. The far left panels show the detailed colocalization of PIN1–MYC PLA with the nuclear envelope marker Lamin A/C. (B) Western blot of subcellular fractionation of pS62MYC and total MYC in nuclear-soluble (S) and nuclear-insoluble (I) fractions of HeLa cells transfected with nontargeting (NT) control or PIN1 targeted siRNAs and extracted by 150 nM NaCl. (C) PLA of MYC–TPR association in HeLa cells transfected with nontargeting or PIN1 siRNAs and the indicated PIN1 expression constructs. The expression of relevant proteins is shown in Supplemental Figure S3B. (D) Quantification of C showing box plots of PLA signals. The whiskers represent 5%–95% intervals, and the box represents median and 25%–75% intervals of PLA signals per cell from 50 cells per condition. Data are representative of three independent experiments. (E) PLA of V5-MYC–TPR association in HeLa cells transfected with nontargeting or PIN1 siRNAs and V5-tagged wild-type, T58A, and S62A MYC expression constructs. Quantification box plots are as in D. The expression of relevant proteins and grayscale images are shown in Supplemental Figure S3D and F, respectively. (F) PLA of HA-MYC–TPR association in primary MEF cells with Cre-dependent ROSA knock-in-expressed HA-tagged wild-type, T58A, and S62A MYC. Cells were coinfected with adenovirus Cre for MYC expression and GFP (adGFP) or PIN1 (adPIN1). Quantification box plots are as in D. The expression of relevant proteins and grayscale images were shown in Supplemental Figure S3E and G, respectively. P-values are shown for relevant significant comparisons. (**) P < 0.01; (***) P < 0.001.
To examine the role of Ser62 and Thr58 phosphorylation in PIN1's regulation of the association of MYC with the NPC, we first transfected HeLa cells with constructs expressing V5-tagged wild-type, T58A, and S62A MYC and examined the interaction of PIN1 with these constructs through V5-PIN1 PLA. Similar to previous reports demonstrating that PIN1 does not recognize S62A MYC and has a reduced interaction with and effect on T58A MYC, where T58A MYC target gene binding as measured by quantitative ChIP (qChIP) is not as significantly enhanced by PIN1 as it is for wild-type MYC (Yeh et al. 2004; Farrell et al. 2013), both T58A and S62A MYC showed a significant reduction in interaction with PIN1 compared with wild-type MYC by PLA (Supplemental Fig. S3C). We then knocked down PIN1 to test whether the various V5-MYC–TPR interactions are affected. Consistent with the positive role of PIN1 in the endogenous MYC–TPR interaction (Fig. 3C), the PLA signal between V5-MYC wild type and TPR significantly decreased with PIN1 knockdown (Fig. 3E; Supplemental Fig. S3F). In contrast, T58A MYC–TPR PLA was resistant to PIN1 depletion, and S62A MYC–TPR PLA remained at baseline level (Fig. 3E; Supplemental Fig. S3F). To further test the role of PIN1 on different forms of MYC interaction with TPR, we took advantage of primary mouse embryonic fibroblasts (MEFs) generated from ROSA-LSL-Myc knock-in mice, which, once induced by Cre, express HA-tagged wild-type, T58A, or S62A MYC at physiological levels (Wang et al. 2011). When we coexpressed knock-in HA-tagged MYC by infection with adenovirus-driven Cre (adCre) and PIN1 with adPIN1, wild-type MYC–TPR association was up-regulated. In contrast, T58A MYC interaction with TPR was already strong and not further affected by PIN1, while S62A MYC again showed minimal interaction that was not affected by PIN1 (Fig. 3F; Supplemental Fig. S3G). Taken together, these data demonstrate that PIN1 promotes MYC association with the NPC in a pSer62-dependent manner, while the interaction of T58A MYC, which has more constitutive Ser62 phosphorylation, with the NPC is less dependent on PIN1.
MYC recruitment to the NPC promotes formation of a TPR–CDK2–PIN1–MYC–GCN5 complex
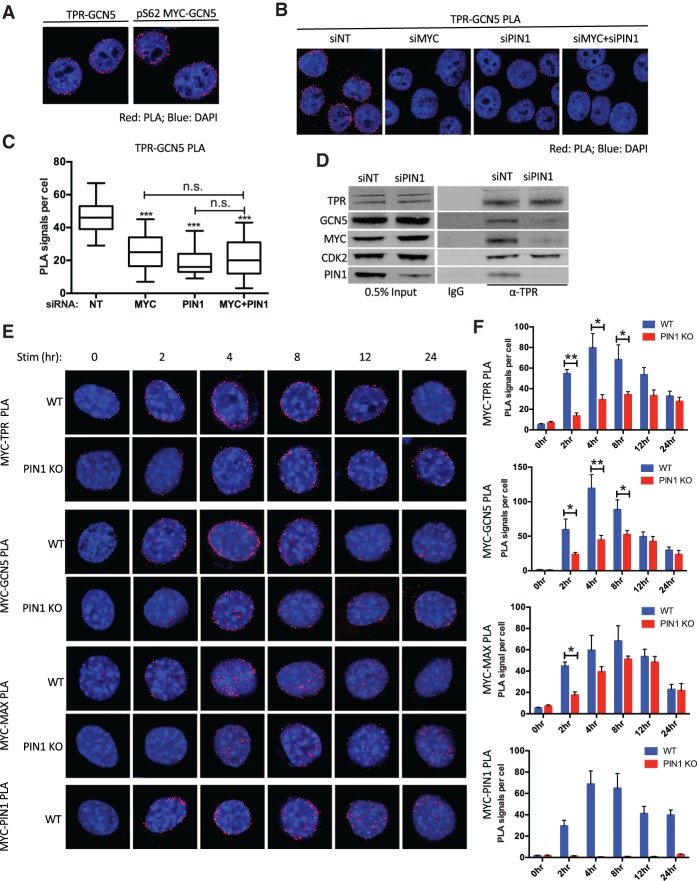
In yeast and higher eukaryotes, the nuclear pore basket helps to maintain a transcriptionally permissive microenvironment (Mendjan et al. 2006; Taddei et al. 2006; Krull et al. 2010), which is potentially mediated by the HAT SAGA (Spt–Ada–GCN5 acetyltransferase) complex (Cabal et al. 2006; Luthra et al. 2007). The catalytic subunit of SAGA, GCN5, forms a complex with MYC and cooperates with MYC for gene activation (Flinn et al. 2002; Kenneth et al. 2007; Martínez-Cerdeño et al. 2012). In addition, the interaction between MYC and GCN5 is enhanced by PIN1 function (Farrell et al. 2013). Therefore, we tested whether GCN5 is involved in the MYC–TPR interaction. We first examined TPR–GCN5 interaction through PLA in HeLa cells and found a robust PLA signal at the nuclear periphery (Fig. 4A, left), consistent with the association of GCN5 with Mlp1/2 (the yeast homolog of TPR) in yeast (Luthra et al. 2007). In accordance with the pS62MYC–TPR association (Fig. 1F) and GCN5-TPR association, a substantial proportion of pS62MYC–GCN5 PLA signal also resided at or close to the nuclear periphery (Fig. 4A, right; Supplemental Fig. S4A). We also detected robust tertiary PLA (Söderberg et al. 2006) of TPR–pS62MYC–GCN5 (Supplemental Fig. S4B), suggesting that pS62MYC and GCN5 exist together in a complex with TPR.
Figure 4.
PIN1 and MYC recruit GCN5 to the NPC, and PIN1 facilitates mitogen-induced MYC–MAX–GCN5 association with the NPC. (A) PLA of GCN5 association with TPR and pS62MYC in HeLa cells. (B) PLA of TPR–GCN5 association in HeLa cells transfected with control nontargeting (NT) and MYC and PIN1 targeted siRNAs as indicated. (C) Quantification of B showing PLA signals per cell from 50 cells per condition. The whiskers represent 5%–95% intervals, and the box represents median and 25%–75% intervals. Data are representative of three independent experiments. (D) Western blots of the indicated proteins coimmunoprecipitated with TPR using a TPR-specific antibody (a-TPR) in HeLa cells transfected with nontargeting and PIN1 targeted siRNAs as indicated. IgG control immunoprecipitation and 0.5% volume input are also shown. (E, from top to bottom) PLA of MYC association with TPR, GCN5, MAX, and PIN1 in wild-type and PIN1 knockout primary MEFs at the indicated time points after serum stimulation. Serum starvation and stimulation were conducted as in Supplemental Figure S4. (F) Quantification of E showing the average number of PLA signals per cell from 50 cells per condition. The error bars indicate standard errors of three independent experiments. P-values are shown for relevant significant comparisons. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Given the important role of PIN1 in the association of pS62MYC with the NPC and the known role of MYC in binding GCN5, we hypothesized that both MYC and PIN1 are required for efficient GCN5–TPR interaction. To test this hypothesis, we examined the PLA signal between TPR and GCN5 following knockdown of MYC and/or PIN1 (Fig. 4B,C; Supplemental Fig. S4C). Individual depletion of MYC or PIN1 reduced ∼40% of the TPR–GCN5 interaction; however, simultaneous depletion of both MYC and PIN1 did not further decrease the PLA signal (Fig. 4B,C), consistent with PIN1 facilitating MYC's recruitment to the NPC and MYC's corecruitment of GCN5.
Since we detected TPR interactions with CDK2, MYC, PIN1, and GCN5 via PLA, we asked whether we could also detect these interactions via coimmunoprecipitation experiments and what the effects of PIN1 knockdown would be on the associations. In agreement with the PLA results, we coimmunoprecipitated CDK2, MYC, GCN5, and PIN1 using a TPR-specific antibody. Depletion of PIN1 via siRNA substantially reduced the interaction of MYC and GCN5 with TPR but had little effect on CDK2 association with TPR (Fig. 4D). The TPR coimmunoprecipitation data suggest a hierarchy of the assembly of these proteins at the NPC: TPR-associated CDK2 phosphorylates MYC at Ser62, providing a recognition site for phosphorylation-directed PIN1-mediated MYC isomerization, which promotes and/or stabilizes MYC's interaction with TPR and facilitates the recruitment of GCN5.
PIN1 facilitates mitogen-induced MYC–MAX–GCN5 association with the NPC
PIN1 is viewed as a “molecular timer” because it adds a second level of control to signaling events involving MAPK- and CDK-mediated phosphorylation of S/T–P motifs in proteins responding to environmental change (Lu et al. 2007). A role for PIN1 in facilitating the response to growth signals was demonstrated by the phenotype of MEFs derived from PIN1 knockout mice, which proliferate normally under asynchronous conditions but show defects in recovering from serum deprivation upon serum restimulation (Fujimori et al. 1999). Serum stimulation following growth arrest also induces MYC expression through up-regulation of its mRNA and Ser62 phosphorylation-mediated protein stabilization (Kelly et al. 1983; Lutterbach and Hann 1994; Sears et al. 1999, 2000; Frank et al. 2001). This up-regulation of MYC is critical for mitogen-induced cell proliferation, and primary MYC knockout cells exhibit growth arrest (de Alboran et al. 2001). Therefore, we investigated whether mitogen stimulation involves PIN1-mediated recruitment of MYC to the NPC.
We serum-starved primary PIN1 wild-type and knockout MEFs generated from isogenic sibling embryos for 48 h to render the cells quiescent and then stimulated the cells by changing to serum-complete medium. We confirmed that the PIN1 knockout MEFs show a reduced proliferative response to serum starvation and restimulation (Supplemental Fig. S4D,E; Fujimori et al. 1999) and then characterized protein expression and PLA interaction signals at the indicated time points (Fig. 4E,F; Supplemental Fig. S4F). The expression of TPR, GCN5, and CDK2 was similar in wild-type and PIN1 knockout MEFs; all three gradually increased over the time course (Supplemental Fig. S4F). The levels of pS62MYC and total MYC in wild-type MEFs increased during the first 4–8 h and then declined to almost baseline levels, similar to previous reports (Kelly et al. 1983; Yeh et al. 2004). In contrast, pS62MYC and total MYC sustained high levels of expression even at 24 h in the PIN1 knockout MEFs, consistent with prior data and a defect in the MYC degradation pathway due to loss of PIN1 (Yeh et al. 2004).
Aligning with MYC expression in wild-type cells, the association between MYC and TPR peaked at 4 h after stimulation (Fig. 4E,F). MYC–GCN5 interaction similarly peaked at 4 h in the wild-type cells, and, importantly, this interaction was primarily at the nuclear periphery (Fig. 4E,F; Supplemental Fig. S4G, peri vs. center, 4 h). The interaction between MYC and PIN1 also peaked at the 4-h time point and was also localized to the periphery (Fig. 4E,F; Supplemental Fig. S4G). The early and robust inductions of MYC–TPR and MYC–GCN5 PLA signals at the nuclear periphery were significantly blunted in PIN1 knockout cells, supporting the important role of PIN1 in promoting MYC–GCN5–TPR association following mitogen stimulation in primary MEFs. The data in MEFs are consistent with what we observed in HeLa cells, although HeLa cells showed constant strong associations between MYC, GCN5, and TPR, probably due to the hyper-Ser62 phosphorylation status in cancerous settings (Junttila et al. 2007; Niemelä et al. 2012; Zhang et al. 2012).
MYC binds DNA with its partner protein, MAX (Littlewood et al. 1992; Nair and Burley 2003). We therefore analyzed the spatial distribution of MYC associated with MAX during the same serum stimulation time course. Notably, the induction of MYC–MAX PLA signal occurred at both the nuclear periphery and the interior during early time points in the wild-type cells (Fig. 4E,F; Supplemental Fig. S4G). In PIN1 wild-type cells, there was an overall decrease in MYC–MAX association in the first 8 h (Fig. 4E,F), with the peripheral MYC–MAX PLA signal at early time points showing the most decrease relative to wild-type cells (Supplemental Fig. S4G). By comparing the interaction patterns of MYC–MAX with MYC–TPR, MYC–PIN1, and MYC–GCN5, it appears that a portion of MYC–MAX is involved in the early response MYC–GCN5–PIN1–TPR interactions at the NPC but that MYC–MAX is also present in the nuclear interior following serum induction.
MYC–GCN5 localization to the NPC activates resident target genes
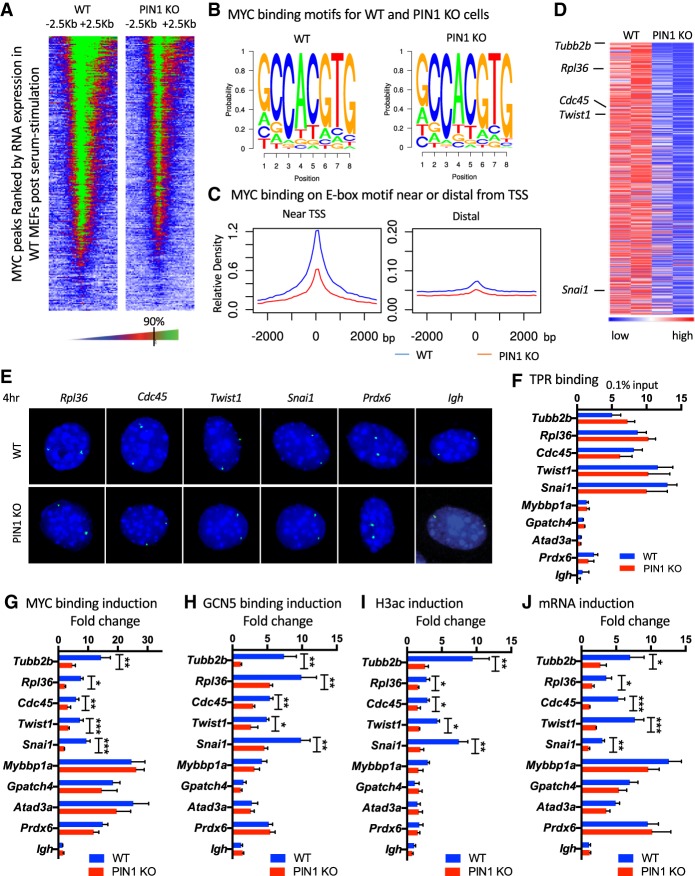
So far, we demonstrated that in response to serum stimulation, the association of MYC with the nuclear pore is inefficient in PIN1 knockout cells. This defect also seems likely to affect MYC DNA-binding activity, as suggested by the reduction of MYC–MAX at the nuclear periphery. To globally examine MYC DNA binding and identify genes potentially affected by this mechanism, we performed MYC ChIP-seq in wild-type and PIN1 knockout MEFs at 4 h after serum stimulation, when expression levels of MYC were similar, but differences in the MYC–TPR PLA were large (Fig. 4E,F; Supplemental Fig. S4F). Our MYC ChIP-seq analysis identified 4877 peaks for MYC in wild-type cells and 3987 peaks in PIN1 knockout cells that are common between biological replicates (irreproducible discovery rate [IDR] <0.05) (Supplemental Table 1). Global analysis of MYC binding centered at transcriptional start sites (TSSs) ± 2.5 kb correlates with the expression of genes, as expected (Fig. 5A; Lin et al. 2012a; Nie et al. 2012); however, we observed an overall reduction in binding in the PIN1 knockout cells. We performed motif analysis and found an enrichment of the canonical E-box sequence in the MYC-binding peaks for both wild-type and PIN1 knockout cells (Fig. 5B), indicating that sequence selection is not affected by PIN1 loss. To examine the effects of PIN1 loss on both promoter-proximal and promoter-distal MYC E-box binding, we analyzed global MYC-binding density at canonical E-box sites within 2 kb of a TSS and >10 kb from a TSS. We observed enriched MYC binding proximal to the TSS, and, with PIN1 loss, there was an overall decrease in MYC binding at both proximal and distal E-boxes (Fig. 5C). Together, these data indicate that PIN1 regulation of MYC DNA binding does not affect motif selection, but PIN1 loss does decrease overall MYC occupancy at both promoters and distal elements.
Figure 5.
PIN1 facilitates MYC DNA binding at E-box sites, and PIN1-mediated MYC–GCN5 localization to the NPC activates resident target genes. (A) Heat map of MYC binding at TSSs (midline of the heat map) of all expressed genes ranked by their expression in wild-type cells. The color scale is indicated at the bottom, where the 90 percentile is marked at the black bar. (B) Motifs enriched at the center of MYC-binding peaks in wild-type and PIN1 knockout MEFs. (C) Averaged MYC-binding density profiles in a 5-kb window centered on E-box motif (CACGTG) sets near (<2 kb) the TSS or distal to (>10 kb) the TSS in wild-type (blue) or PIN1 knockout (red) MEFs. MYC binding is scaled based on total sequencing depth and the number of E-box regions per set to make the scales comparable between near and distal plots. (D) Heat map of genes showing more MYC binding at the TSS in wild-type than PIN1 knockout MEFs (>1.5 fold) (Supplemental Table 2). Genes were ranked by average fold change of wild-type/knockout in two biological replicates. MYC-binding levels are scaled for each row (gene) independently, with red indicating high binding and blue representing low binding. University of California at Santa Cruz (UCSC) genome browser tracks for the indicated representative genes are shown in Supplemental Figure S5C. (E) Representative DNA FISH images of the indicated gene loci (green) in wild-type and PIN1 knockout primary MEFs at the 4-h time point in response to serum stimulation. The 0 time point is shown in Supplemental Figure S6A. (F) qChIP of TPR binding to the indicated PIN1-dependent (Tubb2b, Cdc45, Rpl36, Twist1, and Snai1) and PIN1-independent (Mybbp1a, Gpatch4, Atad3a, and Prdx6) MYC-bound target genes in wild-type (blue) and PIN1 knockout (red) MEFs at 4 h of serum stimulation. (G) qChIP of MYC binding to the indicated target genes in wild-type (blue) and PIN1 knockout (red) MEFs. Fold change between 0 and 4 h of serum stimulation is graphed. Primary data are shown in Supplemental Figure S7A. (H) qChIP of GCN5 binding to the indicated target genes in wild-type (blue) and PIN1 knockout (red) MEFs. Fold change between 0 and 4 h of serum stimulation is graphed. Primary data are shown in Supplemental Figure S7B. (I) qChIP of H3ac levels of the indicated target genes in wild-type (blue) and PIN1 knockout (red) MEFs. Fold change between 0 and 4 h of serum stimulation is graphed. Primary data are shown in Supplemental Figure S7C. (J) RT–PCR of the mRNA of the indicated genes in wild-type (blue) and PIN1 knockout (red) MEFs. Fold change at 4 h of serum stimulation is graphed. For F–J, P-values are shown for relevant significant comparisons. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. The error bars indicate standard errors from three independent experiments.
To associate MYC binding with neighboring genes, we focused on genes for which MYC was bound within 1 kb upstream of or downstream from the TSS. Consistent with an overall reduction in MYC binding 4 h after serum stimulation in PIN1 knockout MEFs, across replicates, we observed more genes associated with increased MYC binding in wild-type versus knockout cells (Supplemental Fig. S5A; Supplemental Table 2, wild type vs. knockout: 630 up and 560 down). Using gene set enrichment analysis (GSEA), we found several pathways enriched for MYC-bound genes in wild-type versus PIN1 knockout MEFs, including cell motion, polysome pathways, and protein acetylation (Supplemental Fig. S5B; Supplemental Table 3). This is in agreement with the increased cell proliferation and increased histone acetylation following serum stimulation in wild-type versus PIN1 knockout cells (Supplemental Fig. S4D–F) as well as with published works showing a role for PIN1 and MYC in cell migration (Smith et al. 2008; Cho et al. 2010; Matsuura et al. 2010; Zhao et al. 2013; Luo et al. 2014; Zhu et al. 2016).
Among the genes with enriched MYC binding in wild-type cells, we chose to investigate the spatial relationships by DNA FISH of two genes encoding transcription factors involved in cell motility (Twist1 and Snai1) and two cell proliferation genes (Cdc45 and Rpl36) (Fig. 5D; Supplemental Fig. S5C). We hypothesized that, in wild-type cells, these genes would localize to the nuclear pore at the nuclear periphery, where PIN1 actively facilitates MYC localization during the early response to stimulation, and that, in PIN1 knockout cells, these genes would either localize away from the periphery or near the periphery but have reduced binding by MYC and its cofactors in the early response window. Across the cell population, we observed that the FISH signals at 0 and 4 h of serum stimulation for all four of these genes were enriched at the nuclear periphery in both wild-type and PIN1 knockout MEFs (Fig. 5E; Supplemental Fig. S6A,B), which supports the model in which reduced MYC DNA binding in PIN1 knockout cells is due to defects in MYC recruitment to nuclear peripheral targets. Interestingly, the FISH signal of Prdx6, a gene bound similarly by MYC in wild-type and PIN1 knockout cells (Supplemental Fig. S5C; Supplemental Table 2), showed a dispersed pattern across the nuclear radius (Fig. 5E; Supplemental Fig. S6A,B). As a control for nuclear peripheral localized genes, the lamin-associated Igh gene (Kosak et al. 2002; Yang et al. 2005; Lin et al. 2012b; Harr et al. 2015) was present almost exclusively (>75% in both wild-type and PIN1 knockout cells) (Fig. 5E; Supplemental Fig. S6B) at the nuclear periphery. We further tested the association of TPR with MYC-bound genes affected by loss of PIN1 (PIN1-dependent MYC targets), including Tubb2b, Cdc45, Rpl36, Twist1, and Snai1 (Fig. 5D), and MYC-bound genes not affected by loss of PIN1 (PIN1-independent MYC targets), including Mybbp1a, Gpatch4, Atad3a, and Prdx6 (Supplemental Table 2). In support of our hypothesis and the DNA FISH that we conducted, the PIN1-dependent MYC target genes associated with TPR, as measured by TPR qChIP, while the PIN1-independent MYC targets did not (Fig. 5F). Importantly, Igh, although localized at the nuclear periphery, showed no interaction with TPR, consistent with its presence in LADs.
To test the hypothesis that loss of PIN1 impairs the functions of MYC and GCN5 at these TPR-associated genes, we performed a series of qChIP experiments using primers for the promoters of the above PIN1-dependent MYC targets and PIN1-independent MYC targets. In wild-type cells, MYC and GCN5 occupancy at all of the MYC targets were induced at 4 h (Fig. 5G,H; Supplemental Fig. S7A,B). However, on PIN1-dependent MYC targets, these inductions were blunted in PIN1 knockout cells. Consistent with these results, induction of histone acetylation at Tubb2b, Cdc45, Rpl36, Twist1, and Snai1 (detected by qChIP with a pan H3ac antibody) was reduced in the PIN1 knockout cells, while, on the genes with strong MYC binding regardless of PIN1 (including Mybbp1a, Gpatch4, Atad3a, and Prdx6), H3ac was high in the starved condition, and the induction was lower and unaffected by PIN1 (Fig. 5I; Supplemental Fig. S7C). Aligning with the induction of H3ac level, which marks transcriptionally active chromatin, the mRNA of Tubb2b, Cdc45, Rpl36, Twist1, and Snai1 was also induced to a greater degree in the wild-type versus PIN1 knockout cells, while the PIN1-independent MYC targets were induced regardless of PIN1 status (Fig. 5J). As a negative control, the lamin-associated Igh gene exhibited virtually no MYC or GCN5 binding and no mRNA induction and was depleted of H3ac during the stimulation process (Fig. 5G–J; Supplemental Fig. S7A–C), consistent with its transcriptionally repressed state in MEF cells (Kosak et al. 2002; Yang et al. 2005; Lin et al. 2012b; Harr et al. 2015). To further test whether the transcriptional defects of these MYC targets were due to the loss of PIN1-mediated regulation on MYC, we re-expressed T58A and S62A MYC in PIN1 knockout cells through adenovirus infection and found that ectopic expression of MYC T58A, which strongly associates with the nuclear pore independent of PIN1 (Fig. 3E,F), but not S62A, which showed little association with the nuclear pore, rescued the serum-induced MYC binding, H3ac, and mRNA expression in the PIN1-dependent MYC target genes (Supplemental Fig. S8B,D,E), but, in the PIN1-independent MYC target genes, T58A and S62A behaved similarly. This suggests that MYC with the ability to interact with the nuclear pore mediates the serum-stimulated regulation of PIN1-dependent target genes, supporting our hypothesis that PIN1 promotes MYC regulation of target genes at the nuclear pore in a pS62-dependent manner.
Together, these data support a model in which, in response to extracellular stimuli, PIN1 is critical for the efficient recruitment of MYC to the NPC, which, by corecruiting GCN5, increases histone acetylation and transcriptional activity of NPC-localized genes.
PIN1 facilitates MYC-driven transcriptional activation and chromatin opening in response to serum stimulation
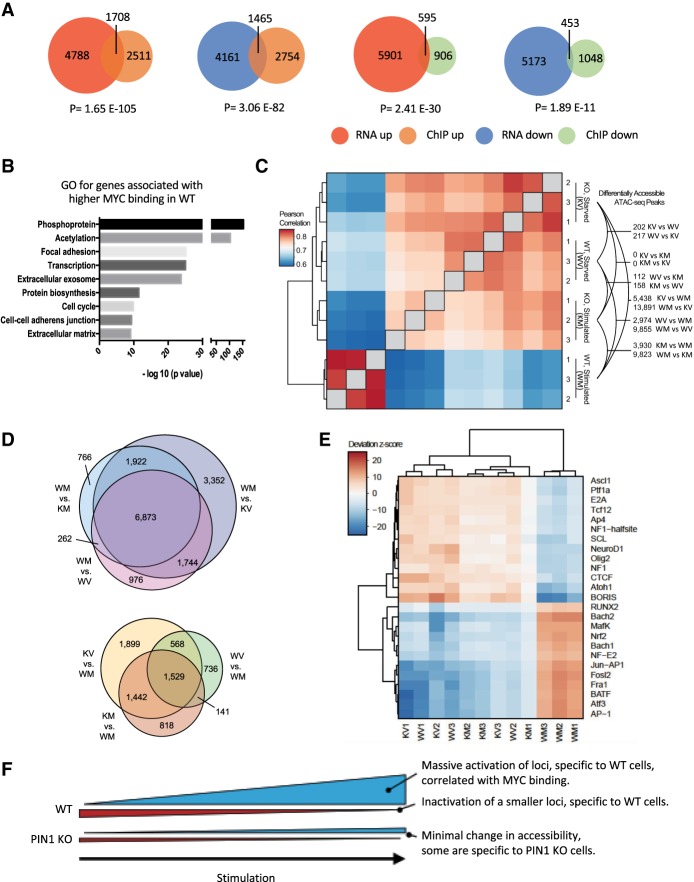
We next used RNA-seq (Supplemental Table 4) to globally assess the PIN1-regulated transcriptional program and its correlation with MYC binding. We statistically evaluated the intersection between the RNA-seq data and the MYC ChIP-seq data using a Fisher's exact test (Supplemental Tables 5, 6). We found the most significant intersection (n = 1708; P = 1.65 × 10−105) in the RNA up-regulated in wild-type cells and MYC binding up-regulated in wild-type cells, suggesting that PIN1-dependent MYC binding is associated primarily with transcriptional activation (Fig. 6A). We also found a large group of genes (n = 1465; P = 3.02 × 10−82) with MYC binding up-regulated in wild-type cells and RNA down-regulated in wild-type cells, suggesting that PIN1-dependent MYC binding may also associate with transcriptional repression, although not as significantly as activation. Using gene ontology (GO) analysis on the significantly changed genes (up-regulated and down-regulated in RNA) intersecting with MYC binding up in wild-type cells (PIN1-dependent MYC binding), we found phosphoprotein, acetylation, focal adhesion, and cell cycle among the most significant GO terms (Fig. 6B; Supplemental Table 7), which is consistent with the pathways enriched with genes showing higher MYC binding in wild-type cells (Supplemental Fig. S5B).
Figure 6.
PIN1 facilitates MYC-driven transcriptional activation and chromatin opening in response to serum stimulation. (A) Intersections of RNA-seq ([RNA up] RNA level up-regulated in wild-type compared with PIN1 knockout; [RNA down] RNA level down-regulated in wild-type compared with PIN1 knockout) and MYC ChIP-seq. (ChIP up) MYC binding up-regulated in wild-type compared with PIN1 knockout; (ChIP down) MYC binding down-regulated in wild-type compared with PIN1 knockout. P-value was calculated using Fisher's exact test. (B) GO analysis using DAVID for genes significantly up-regulated and down-regulated with higher MYC binding in wild-type cells. A more complete list of GO terms is shown in Supplement Table 7. (C) Hierarchical clustering of pairwise correlations of ATAC-seq libraries. Technical replicates cluster tightly, with the major branch splitting out wild-type stimulated cells from all others. The number of differentially accessible (DA) loci between groups is denoted at the right. (D) Intersections of differentially accessible loci for all wild-type differentially accessible peak sets (top) and peaks specifically active in starved or PIN1 knockout cells (bottom). (E) Motif accessibility deviation z-scores for those with a variability score of ≥5. Notably, wild-type cells after stimulation also cluster separately, as with the global signal comparison in C. (F) Model of chromatin accessibility changes in response to stimulation in wild-type and PIN1 knockout cells.
To globally assess the PIN1-dependent changes in regulatory element usage in response to stimulation, we used ATAC-seq (Corces et al. 2017), which produced a union peak set comprised of 197,785 loci. Broken down by condition, we observed a robust regulatory response to stimulation in wild-type cells that is almost completely lacking in PIN1 knockout cells (Fig. 6C). Upon serum stimulation, a large number of regulatory elements become activated (wild-type stimulated [WM] vs. wild-type starved [WV]; n = 9855 differentially accessible, q-value ≤0.001; fold change ≥2) (Materials and Methods) that were not present or had very little ATAC-seq signal under starved conditions in wild-type cells. These sites were highly enriched for MYC ChIP-seq peaks in stimulated cells (P = 2.11 × 10−194, cumulative hypergeometric test [CHT]) (Materials and Methods; Supplemental Fig. S9A,B). Additionally, the highest enriched motifs in the differentially accessible sites opening in stimulated wild-type cells are for the AP1–ATF family of transcriptional regulators (AP1, Atf3, BATF, Fosl2, Fra1, and Jun; P ≤ 1 × 10−100, CHT; false discovery rate [FDR] 5%) (Supplemental Table 8) involved in stimulus response (Lopez-Bergami et al. 2010), suggesting an interplay between MYC and this family of transcription factors. Furthermore, the majority of these loci (6873 out of 9855; 69.7%) was differentially accessible in common across all of the wild-type stimulated comparisons (Fig. 6D, top Venn diagram), representing a core PIN1-dependent activation module.
Under starved conditions, there was very little difference between the PIN1 knockout and wild-type cells, with only ∼200 differentially accessible loci in either sample (Fig. 6C, KV vs. WV and WV vs. KV); however, unlike wild-type, the PIN1 knockout cells lack the robust response to stimulation without any differentially accessible loci reaching statistical significance (Fig. 6C, KM vs. KV). This observation is also present in the unsupervised clustering of samples, where the first major branch splits out stimulated PIN1 wild-type cells from all other conditions, which are highly correlated with one another (Fig. 6C).
We next focused on the set of loci that was differentially accessible in starved wild-type cells versus their stimulated counterpart (n = 2974) (Fig. 6C, WV vs. WM), as these represent sites that become deactivated or closed during stimulation and were not significantly enriched for MYC ChIP-seq peaks (FDR > 0.05) (Supplemental Fig. S9B). One of the most significantly enriched motifs was for Atf4 (P = 4.74 × 10–108, CHT; FDR < 0.05), a API/ATF family member involved in cell stress response (Kilberg et al. 2009; Wortel et al. 2017). The majority ([568 + 1529] out of 2974; 70.5%) of these is also active in the starved PIN1 knockout cells (Fig. 6D, bottom Venn diagram, KV vs. WM), with a smaller number present in both starved and stimulated PIN1 knockout cells (1529 out of 2974; 51.4%) (Fig. 6D, KV vs. WM and KM vs. WM). This suggests that a number of loci are active in starved cells regardless of PIN1 status that deactivate upon stimulation; however, in the absence of PIN1, only roughly half of the loci deactivate.
One of the advantages of assessing chromatin accessibility is that it is agnostic as to which DNA-binding protein is responsible for the modulation of open chromatin. We can therefore use a database of DNA-binding motifs and determine whether there is increased accessibility at sites containing that motif compared with the rest of the samples (Schep et al. 2017). As expected, the majority of motifs showed little variability, since many are not expressed in these cells; however, a subset showed high variance (Supplemental Fig. S9C). We calculated deviation scores for each sample for each of these motifs, which revealed a strict separation of motifs as being active in either wild-type cells under stimulation or any of the other samples, resulting in the first branch of hierarchical clustering delineating these sets and conditions (Fig. 6E). As expected, a number of the motifs that were significantly enriched in the wild-type activation module differentially accessible peak set (Supplemental Table 8) also exhibited higher global accessibility in this analysis (e.g., AP1) (Fig. 6E). We also observed a number of motifs that have decreased global accessibility in the starved and/or PIN1 knockout conditions, including NF1, which is a negative regulator of Ras signaling (Ratner and Miller 2015), and CTCF, which functions to organize chromatin architectures (Ong and Corces 2014; Ghirlando and Felsenfeld 2016). It is worth noting that the MYC motif did not exhibit high variability by this analysis (variability = 2.75); however, this is largely due to the high number of MYC motifs in the genome, a minority of which MYC will be bound to in any given population, hence our prior targeted analysis of MYC ChIP-seq peaks.
Overall, the accessible chromatin landscape reveals a substantial PIN1-dependent response to stimulation that is correlated with MYC binding along with other factors that are typically associated with proliferation and stimulus response (Fig. 6E,F; Supplemental Fig. S9B). Some loci become closed or less accessible after stimulation, half of which are PIN1-dependent—far fewer than those that are activated. Interestingly, similar to the trend of PIN1-dependent landscape changes of chromatin under early stimulation conditions, we found a large number of genes that were induced in wild-type cells by stimulation for which the response was blunted in PIN1 knockout cells (Supplemental Fig. S9D; Supplemental Table 9).
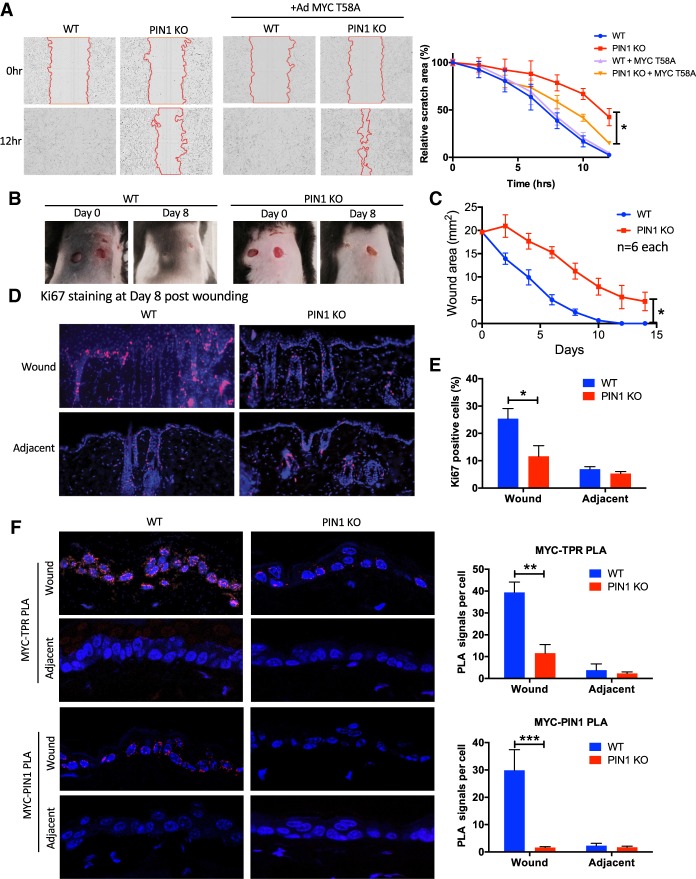
PIN1-facilitated wound healing involves MYC–NPC association
To further investigate the role of PIN1-regulated MYC–NPC-associated gene regulation in physiology, we performed in vitro and in vivo wound healing assays. Using an in vitro scratch assay on the wild-type and PIN1 knockout MEFs, we observed a delay in migration into the scratch area in PIN1 knockout MEFs (Fig. 7A). This defect was largely rescued by expressing the MYC T58A mutant (Fig. 7A), a form of MYC that can interact with the nuclear pore independently of PIN1 (Fig. 3E,F), suggesting that post-translational regulation of MYC NPC localization may at least partially mediate PIN1 function in migration in vitro. To further examine PIN1's function in vivo, we conducted skin punch wound healing assays in wild-type and PIN1 knockout mice. It took ∼10 d for wild-type mice to fully heal from the punch biopsy on the back, whereas PIN1 knockout mice were still not fully healed at 15 d (Fig. 7B,C). At day 8, the wild-type mice exhibited thickened epidermis at the wound area, whereas the epidermis at the wound area of the PIN1 knockout mice was mostly only a single-cell layer (Supplemental Fig. S10A). Immunofluorescent analysis of the day 8 skin showed a strong reduction of E-cadherin- and Integrin-Β1-expressing cells at both the wound and adjacent epidermis in PIN1 knockout mice (Supplemental Fig. S10B). Studies have shown that E-cadherin promotes collective cell migration in vivo (Cai et al. 2014) and that Integrin-Β1 mediates keratinocyte migration and epidermal stem cell maintenance (Kim et al. 1992; Jones and Watt 1993; Georges-Labouesse et al. 1996). Additionally, the PIN1 knockout mice showed fewer Ki67-positive cells at the wound area (Fig. 7D,E), suggesting that defects in both proliferation and migration in PIN1 knockout mice contribute to the impaired wound healing. To examine whether MYC interaction with the nuclear pore may be involved in this process, we performed PLA of MYC with TPR and PIN1. In wild-type mice, both MYC–TPR and MYC–PIN1 associations were induced in the nuclear periphery selectively in the wound area (Fig. 7F); in contrast, MYC association with TPR was significantly reduced in the wound area of PIN1 knockout mice (Fig. 7F). Together, these data suggest that PIN1-mediated MYC–TPR association facilitates proper wound healing.
Figure 7.
Pin1 facilitated wound healing involves increased proliferation and MYC-NPC association. (A) Representative phase contrast images of scratch assay of wild-type and PIN1 knockout primary MEFs with or without adenovirus-expressed T58A MYC. Shown are the boundaries of the scratches (red) at the indicated time points. Shown at the right is the quantification of the scratch area over time from live-cell imaging (IncuCyte) relative to the beginning (100%) with the indicated conditions. (B) Wound pictures of wild-type and PIN1 knockout mice after skin punch biopsy at the indicated time points, representing one of three mice for each genotype. (C) Quantification of the average wound area from three pairs of matched siblings, two wounds each. Error bars represent standard deviation of six replicates. (D) IF of Ki67 (proliferation marker) of the wound and adjacent (∼1.5 mm away from the wound) skin samples at day 8 after skin punch biopsy. (E) Quantification of the percentage of Ki67-positive cells for D from six wounds and adjacent 1-mm2 areas. The error bars indicate standard errors of six wound or adjacent areas from three mice. (F, from top to bottom) PLA of MYC association with TPR and PIN1 in wild-type and PIN1 knockout mouse wounds and adjacent skin FFPE samples at day 8 after skin punch biopsy. Shown at the right is the quantification of average PLA signals per cell from all six wounds and adjacent areas. The error bars indicate standard errors from three mice. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Discussion
Although the nuclear peripheral localization of MYC was observed many years ago, both the significance and mechanistic regulation remained unclear (Eisenman et al. 1985; Vriz et al. 1992). Early reports suggested that both the nuclear pore and the lamina were involved in MYC's association with the nuclear periphery (Winqvist et al. 1984; Royds et al. 1992). In a recent study, it was shown that the PP2A inhibitor CIP2a increases pS62MYC association with the Lamin A/C-associated structures (LAS) (Myant et al. 2015). The LAS compartment is likely to include the nuclear pore components, since the nuclear pore is embedded in the lamina meshwork, and Myant et al. (2015) suggested this possibility in their discussion. The NPC is composed of three scaffold rings and peripheral elements, including the cytoplasmic filaments and the pore basket (Beck and Hurt 2017). Here we demonstrate that pS62MYC associates selectively with the pore basket proteins TPR and NUP153. The PLA and STORM analyses indicate a close proximity (<60 nm) between the nuclear pore basket and the adjacent MYC; however, we could not tease out whether the association is through direct protein–protein interaction at this point, and future studies are needed to clarify this. The association between MYC and TPR is regulated by Ser62 phosphorylation, which can be mediated by ERK and CDK2. Despite a reported role of ERK at the nuclear pore (Vomastek et al. 2008), we did not detect it interacting with TPR. Instead, we identified CDK2 association with the nuclear pore basket and MYC, suggesting that CDK2 may directly phosphorylate MYC at the NPC (Chi et al. 2008), while ERK phosphorylation of Ser62 likely occurs in the nuclear interior, which can also then facilitate MYC's recruitment to the NPC.
Ser62 phosphorylation of MYC promotes its interaction with the prolyl isomerase PIN1, which facilitates MYC promoter binding, target gene regulation, and oncogenic activity as well as MYC turnover (Yeh et al. 2004; Farrell et al. 2013). We demonstrate that PIN1's catalytic activity facilitates the association between Ser62 phosphorylated MYC and TPR in both HeLa cells and primary MEFs. Consistent with yeast studies indicating GCN5 association with the nuclear pore basket (Cabal et al. 2006; Luthra et al. 2007), we also demonstrate that PIN1 recruitment of MYC facilitates the recruitment of GCN5 to TPR. However, CDK2 binding to TPR was irrespective of PIN1, providing a picture of assembly at the nuclear pore basket involving PIN1-regulated MYC–GCN5 association with CDK2–TPR. In primary MEFs where MYC expression is normally tightly regulated for transient activation, we observed that the induction of MYC coincided with its association with the nuclear pore. GCN5 similarly localized to the nuclear periphery with MYC at early time points following serum stimulation. In PIN1 knockout MEFs, these inductions were severely impaired, suggesting a role for PIN1-regulated MYC–GCN5-mediated control of genes localized near the nuclear pore basket. Interestingly, the MYC T58A mutant, which has constitutive Ser62 phosphorylation and increased oncogenic activity (Yeh et al. 2004; Hemann 2005; Wang et al. 2011) independent of PIN1 (Yeh et al. 2004; Farrell et al. 2013), showed enhanced interaction with TPR in MEFs irrespective of PIN1. Although we could not directly address how the Ser62 phosphorylation status translates into the structural information around Pro63 that is amenable to isomerization by PIN1, we think that fixed phosphorylation at S62, as in the case of T58A, might lock MYC in the cis conformation that is insensitive to PIN1 and conformationally favorable for association with the NPC. In contrast to the MEFs, in cancerous HeLa cells that express high pS62MYC and PIN1, MYC wild-type robustly interacted with TPR to a level similar to that of MYC T58A, suggesting that deregulated association of MYC with the NPC in cancer cells could contribute to MYC's oncogenic function.
The isomerase PIN1 is a unique molecular timer thought to be involved in cells’ response to acute environmental changes, which may explain why PIN1 knockout MEFs proliferate normally in culture but have defects in re-entering the cell cycle upon starvation and restimulation. These phenotypes are mimicked in Cyclin E1/E2 double-knockout MEFs (Geng et al. 2003), suggesting that PIN1 is involved in CDK2/Cyclin E regulation of G1/S cell cycle and that this is connected to our results that PIN1 and CDK2 promote MYC activity at the nuclear pore. However, the CDK2 knockout cells have minor S-phase re-entering defects, which is possibly due to the compensating effect of CDK1 and other kinases (Berthet et al. 2003; Ortega et al. 2003). It is thus interesting to examine whether CDK1 can compensate for CDK2 with respect to MYC S62 phosphorylation status as well as its subnuclear localization. Aside from the CDK2/Cyclin E pathway, PIN1 has been shown to control numerous cancer-driving pathways (for review, see Zhou and Lu 2016). We tried to tease out MYC-specific effects of PIN1 by intersecting MYC ChIP-seq and RNA-seq data in post-stimulation MEFs and found that MYC target genes regulated by PIN1 were involved in processes such as cell migration and proliferation (two pathways that are critical for wound healing), fitting our finding that PIN1 knockout MEFs and mice exhibit defects in in vitro and in vivo wound healing assays. It has been demonstrated that gene expression programs induced by serum reflect the wound healing role of fibroblasts and are shared by tumor cells and tumor-associated fibroblasts (Chang et al. 2004). In particular, MYC promotes TGF-β-induced expression of SNAIL, which facilitates epithelial-to-mesenchymal transition (Smith et al. 2008), and this was down-regulated in PIN1 knockout MEFs. Therefore, it would be interesting to investigate the coordinating role of MYC and PIN1 in tumor progression and cancer metastasis settings.
Importantly, individually interrogated PIN1-regulated MYC-bound genes in these pathways localized to the nuclear periphery and associated with TPR. The pattern of localization of these genes was distinct from Igh, a gene that is associated with the lamina and transcriptionally repressed in MEFs (Kosak et al. 2002; Yang et al. 2005; Lin et al. 2012b; Harr et al. 2015), which showed stronger association with the nuclear periphery but did not interact with TPR. This difference could be explained by the more dynamic interaction of genes with the NPC (Brickner and Walter 2004; Brickner et al. 2007; Tan-Wong et al. 2009; Green et al. 2012). In addition, the initiating levels of H3ac at the NPC-associated genes were significantly higher than Igh, suggesting that the increase of H3ac on these genes in response to serum and dependent on PIN1 was an expansion of already existing euchromatin rather than a switch from completely heterochromatin. Nonetheless, there are other MYC target genes that are induced by serum stimulation but independent of PIN1 function that tend to be more localized to the nuclear interior, with high basal levels of H3ac and only a modest increase of H3ac with serum stimulation. Thus, PIN1's regulation of MYC during early time points following serum stimulation of MEFs appears to occur mainly at the nuclear pore-associated euchromatin on a set of MYC target genes involved in processes such as proliferation and migration, which represent integral fibroblast responses to extracellular signaling.
In support of the flexible histone acetylation status at the nuclear periphery, it was shown that global histone acetylation following treatment with a histone deacetylase (HDAC) inhibitor induced genomic reorganization with recruitment of promoter region, euchromatin domains, and differentially expressed genes to the nuclear pore (Brown et al. 2008). Furthermore, in mouse oligodendrocyte progenitor cells, silencing of MYC was associated with a decrease in the histone acetylation levels on target genes and the induction of premature nuclear peripheral chromatin compaction (Magri et al. 2014). However, MYC's effect on chromatin modification, can extend beyond the nuclear periphery regions to regulate global chromatin modifications and genomic architecture (Knoepfler 2006; Kieffer-Kwon et al. 2017). In our study, we found a large number of chromatin landscape changes measured by ATAC-seq in response to serum stimulation. Many newly accessible chromatin regions overlapped with MYC binding in wild-type cells but were not present in PIN1 knockout cells, suggesting a role for PIN1-mediated MYC binding in chromatin architecture changes, which can be elucidated further in future experiments using high-throughput chromatin conformation capture technologies (Hi-C and Hi-ChIP) (Schmitt et al. 2016; Davies et al. 2017).
The functional consequence of gene localization to the NPC has not been thoroughly studied. In yeast, genes interact with the nuclear pore constitutively to regulate glycolysis and ribosomal biogenesis or conditionally in response to environmental stimuli, such as nutrients shifts, heat shock, and mating pheromone treatment (Brickner and Walter 2004; Casolari et al. 2005; Cabal 2006; Dieppois et al. 2006; Taddei 2006). Active transcription at the nuclear pore is proposed to be a mechanism for rapid mRNA exportation (Blobel 1985; Rajanala and Nandicoori 2012), and this function is supported by identification of mRNA export factors in the complex anchoring genes to the NPC (Rodríguez-Navarro et al. 2004; Kurshakova et al. 2007). In addition, studies in yeast and Drosophila cells indicate that the NPC serves as a scaffold that anchors and connects poised promoters and enhancers, which allows robust reactivation of recently transcribed genes, a process known as transcriptional memory (Tan-Wong et al. 2009; Light and Brickner 2013; Pascual-Garcia et al. 2017). In agreement with this model, our study suggests that the nuclear pore basket-associated MYC coordinates transcriptional and epigenetic regulators to facilitate gene induction in response to extracellular stimulation and cell cycle re-entry.
In summary, our study addresses post-translational regulation of MYC localization within the nucleus. We characterized the association between MYC and the nuclear pore basket, which is tightly regulated by Ser62 phosphorylation and PIN1-mediated isomerization of MYC. Our study suggests that MYC proteins in the absence of PIN1, although they may have elevated expression level (Yeh et al. 2004), lack the conformation controlled by PIN1 to efficiently localize to certain nuclear compartments to facilitate the timely regulation of resident target genes in response to extracellular stimuli. This process appears to be important in wound healing and deregulated in cancer where high pS62MYC–GCN5–PIN1–TPR association was observed, suggesting a potential point of therapeutic intervention, particularly given that, in an unchallenged state, PIN1 knockout mice are viable. Together, we provide mechanistic insight into MYC subnuclear localization and its effects on gene regulation, illuminating our understanding of spatial control of gene expression in cellular responses to environmental changes that may contribute to phenotype plasticity in normal and diseased states.
Materials and methods
Cell culture, transfection, and adenoviral infection
Generation of the MEFs and the following serum stimulation experiments were performed as described previously (Yeh et al. 2004). The Cre-inducible MYC-expressing MEFs were derived from mice described previously (Wang et al. 2011), and HeLa, Saos2, and U2OS cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% standard fetal bovine serum (FBS), 2.5 mM L-glutamine, and 1× penicillin–streptomycin. Plating of cells was done to achieve 60%–80% confluency 24 h after split for transfection. For serum stimulation, passage two primary wild-type or PIN1 knockout MEFs were grown to density arrest. Cells were split into ∼50% confluence in 0.2% FBS medium and starved for 2 d. Cells were then serum-stimulated with 20% FBS medium for the indicated length of time.
All transfections were performed using Lipofectamine 2000 (Life technology). Total transfected DNA was held constant (unless indicated otherwise) by the addition of empty control plasmid and included 50 ng of CMV-b-gal to normalize for transfection efficiencies between experimental conditions. The Pin1 adenovirus (AdPin1) was generated as described previously (Yeh et al. 2004). The Cre adenovirus (AdCre) was obtained from ViroQuest. For adenovirus infection, AdPin1 or AdCre (multiplicity of infection [MOI] = 10) was added to ∼1 × 106 cells in 500 µL of serum-free DMEM. Infected cells were incubated for an additional 18 h before analysis.
Immunofluorecense
Cells plated on chamber slides were grown to 70% confluence, fixed with 4% PFA for 15 min, and blocked with blocking solution for 30 min followed by incubating the primary antibodies overnight at 4°C with shaking. The next day, samples were incubated with secondary antibodies for 1 h at room temperature, stained for DAPI for 5–10 min, and processed to confocal imaging (Zeiss, LSM880).
For tissue samples, formalin-fixed slides were dewaxed in xylene and rehydrated in decreasing concentrations of alcohol followed by washing twice in tap water. The slides were then incubated in antigen retrieval buffer (Lab Vision citrate buffer, Thermo Scientific) for 30 min at 99°C, cooled down to room temperature, and rinsed in distilled water (dH2O). Next, the slides were blocked for 15 min in 1.5% H2O2 solution in PBS, rinsed in dH2O, and rinsed once in Tris-buffered saline with Tween 20 (TBST) buffer. Samples were blocked with 1% bovine serum albumin (BSA) for 30 min, followed by primary antibody incubation (1:100) overnight at 4°C. The next day, after washing, samples were incubated with secondary antibodies for 1 h at room temperature in a dark chamber, stained for DAPI for 20 min, and mounted with ProLong Gold anti-fade mountant (Thermo Fisher).
Cell fractionation assay
The cell fraction assays were done based on previous protocols (Myant et al. 2015). For fractionation of cellular proteins to cytoplasmic, soluble nuclear, and insoluble nuclear fractions, cells were resuspended in 500 µL of buffer A (10 mM HEPES at pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTG, 0.1% NP-40, 1.5 mM MgCl2) and incubated for 15 min at 4°C with rotation. After homogenizing with B pestle douncing 12 times followed by spinning down, the supernatant (cytoplasmic fraction) was collected, and the pellet was resuspended in 125 µL of buffer B (20 mM HEPES at pH 7.9, 150 mM NaCl, 0.25 mM EGTA 1.5 mM, MgCl2, 10% glycerol) followed by rotation for 20 min at 4°C. The supernatant was collected as nuclear-soluble fraction. The pellet was resuspended in 250 µL of RIPA buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 0.5% DOC, 0.1% SDS, 1% NP-40) and sonicated, and supernatant was collected as insoluble nuclear fraction.
Coimmunoprecipitation and Western blotting
Coimmunoprecipitation was performed based on previously published protocols (Markiewicz et al. 2002). Briefly, rabbit IgG protein A beads were coupled to TPR-specific antibody by incubation for 12 h at 4°C in the presence of 1% BSA. Asynchronously growing cells were extracted with hypotonic solution containing 10 mM HEPES (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTG, 0.1% NP-40, 1.5 mM MgCl2, and protease inhibitors. After a 10-min incubation at 4°C with homogenization, samples were centrifuged for 5 min in an Eppendorf microcentrifuge to isolate nuclei. Nuclei were extracted with buffer containing 150 nM NaCl, and samples were centrifuged at 13,000 rpm for 5 min. Soluble fractions after dialysis to PBS/0.1% Triton X-100 were processed for immunoprecipitation by using the specified antibody coupled to 100 µL of rabbit IgG protein A or G beads. After a 2-h incubation at 4°C, beads were washed three times with 5 vol of PBS/0.1%Triton X-100 and prepared for gel electrophoresis and immunoblotting.
Cell lysates were run on SDS-PAGE gels and transferred to Immobilon-FL membranes (Millipore). The membranes were blocked with Odyssey blocking buffer (LI-COR Biosciences). Primary antibodies were diluted in 1:1 Odyssey blocking buffer–PBS with 0.05% Tween 20. Primary antibodies were detected with secondary antibodies labeled with the near-infrared fluorescent dyes IRDye800 (Rockland) and Alexa fluor 680 (Molecular Probes). Secondary antibodies were diluted 1:10,000 in 1:1 Odyssey blocking buffer–PBS with 0.05% Tween 20. Blots were scanned with an Odyssey infrared imager (LI-COR Biosciences) to visualize proteins, and bands were quantified using the LI-COR Odyssey software.
Plasmids and siRNA
Construction of expression plasmids CMV-empty, CMV-Myc, pDEST40-His/V5-c-Myc, pD40-His/V5-c-MycT58A, and pD40-His/V5-c-MycS62A have been described previously (Sears et al. 1997; Yeh et al. 2004). pDEST40 constructs encoding Xenopus Pin1 (wild type, WW mutant, and C109A mutant) cDNAs were described previously (Farrell et al. 2013).
siRNA was purchased from the following resources and performed according to the manufacturers: Lamin A/C (Sigenome, Dharmacon), NUP153 (Thermo Fisher, 137886), NUP214 (Santa Cruz Biotechnology, sc-106320), NUP98 (Thermo Fisher, AM1670), CDK4 (Santa Cruz Biotechnology, sc-29261), ERK (Cell Signaling Technology, 6560), CDK2 (Santa Cruz Biotechnology, sc-156139), MYC (Qiagen, SI00300902), and PIN1 (Dharmacon, OnTarget).
Antibodies
The following antibodies were used: MYC (monoclonal anti-pS62 [Abcam, ab78318] and polyclonal anti-pS62 [Zhang et al. 2012]), anti-pT58 (Abm, Y011034), anti-total (Santa Cruz Biotechnology, N262 and C33), PIN1 (Santa Cruz Biotechnology, sc-15340; Novas Biologicals, 2f2), TPR (Santa Cruz Biotechnology, sc121094 and sc67116), GCN5 (Santa Cruz Biotechnology, sc20698), CDK2 (Santa Cruz Biotechnology, sc-163), CDK4 (Santa Cruz Biotechnology, sc-160), ERK (CSG, 4695S) H3 (Upstate Biotechnology, 31560), H3ac (Millipore, 06-599), NUP153 (Abcam, ab24700), NUP98 (Santa Cruz Biotechnology, sc-74578), NUP214 (Abcam, ab70497), Lamin A/C (Santa Cruz Biotechnology, sc-7292, sc-2068, and sc-6215), V5 (Invitrogen, 1718556), and HA (Abm, G036).
cDNA preparation
Transfected HEK293 cells were collected in 1× PBS with 1 mM EDTA, and 5% of the cells were reserved for βgal assay and Western analysis. RNA was isolated from cells exhibiting transfection efficiencies within 5% of each other using TRIzol reagent from Invitrogen. cDNA was made using the M-MLV reverse transcriptase according to the manufacturer's protocol (Invitrogen). Immunomix Red (2×) from Bioline was used for PCR analysis of cDNA (see the Supplemental Material for primer sequence and thermocycler setup).
Quantitative RT–PCR (qRT–PCR) analysis
RNA was isolated from 293tr-V5-Axin1 cells collected in 1mL of TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Isolated RNA was DNase-treated in 100 mM MgCl2, 10 mM DTT, RNasin (Promega), and RNase-free DNase (Roche) for 15 min at 37°C and purified using RNeasy (Qiagen). cDNA was made from DNase-treated RNA using M-MLV reverse transcriptase (Invitrogen) according to the manufacturer's protocol with oligo dT primers. qRT–PCR analysis was done using the specified primers (Supplemental Fig. S11) and TBP control as designed by Applied Biosystems on a 7300 qRT–PCR machine (Applied Biosystems) according to the manufacturer's preset qRT–PCR cycle conditions.
STORM
The multicolor imaging of the two-color sample was imaged on a custom-built multispectral superresolution microscope (MSSRM) (Huang et al. 2018). Signals from the sample were collected through a 60× TIRF objective (Nikon 60× TIRF) and split between a positional channel and a spectral channel to enable simultaneous imaging of multiple fluorophores without having to use emission filters. The MSSRM setup achieves ∼12-nm spectral resolution and hence is able to distinguish single Dyomics 634 and Alexa fluor 647 molecules reliably, with spatial resolution of 20 nm.
MSSRM imaging of fluorescently stained cells was performed in PBS buffer supplemented with 1% β-mercaptoethanol (βME), 5 µg/mL glucose oxidase (Sigma, G2133-50 kU), 0.4 µg/mL catalase (Sigma, C100-50 MG), and 10% (w/v) glucose (Fisher Chemicals, D16-500). The EM-CCD was operated in frame transfer mode at 15 msec per frame with a gain setting of 300. Raw images were acquired using an open source micromanager software suite (https://micro-manager.org). Image analyses for extracting single-molecule localization spectra were all performed with custom MatLab (Mathworks) scripts as described previously. Coordinates of single molecules were grouped based on their peak emission wavelength. Images in each channel were then rendered separately and recombined with Fiji software (http://imagej.net/Fiji) into a composite image.
STORM data analysis
The two-channel images showed significant colocalization of MYC with NPC, as shown by the yellow color in Figure 1H and Supplemental Figure S2C. Pair correlation distribution of these two channels clearly showed that MYC and NPC molecules predominated in a cluster within a diameter of 100-nm range. Pair correlation distribution function was processed by a home-built MatLab/C package (Ripley 1977).
PLA
The PLA was performed following the manufacturer's protocol (Duolink, Sigma). Briefly, cells plated on chamber slides were grown to 70% confluence, fixed with 4% PFA for 15 min, and blocked with blocking solution for 30 min followed by incubation of the primary antibodies overnight at 4°C with shaking. The next day, samples were incubated with PLA probes for 1 h at room temperature and 1 h at 37°C followed by ligation for 1 h at 37°C. After a brief wash, amplifications were carried out for 2 h at 37°C (secondary antibodies were added if needed for counterstaining of the third protein) before staining for DAPI for 5–10 min followed by confocal imaging (Zeiss, LSM880).
For tissue samples, formalin-fixed slides were dewaxed in xylene and rehydrated in decreasing concentrations of alcohol followed by washing twice in tap water. The slides were then incubated in antigen retrieval buffer (Lab Vision citrate buffer, Thermo Scientific) for 30 min at 99°C, cooled down to room temperature, and rinsed in distilled water (dH2O). Next, the slides were blocked for 15 min in 1.5% H2O2 solution in PBS, rinsed in dH2O, and rinsed once in Tris-buffered saline with Tween 20 (TBST) buffer. Finally, the PLA was performed following the PLA protocol for cell staining from blocking slides by blocking solution.
Tertiary PLA for Supplemental Figure S4B was performed based on Söderberg et al. (2006). Primary antibodies against TPR, pS62MYC, and GCN5 were conjugated with probe oligonucleotide A (SH-AAAAAAAAAAGACGCTAATAGTTAAGACGCTT [UUU]), probe oligonucleotide B (SH-AAAAAAAAAAATGGCCGACTCACGAATT AGA [UUU]), and probe oligonucleotide C (SH-AAAAAAAAAATATGACAGAACTAGACACTCTT), respectively. Ligations were performed by adding connector oligonucleotide A (P-CTATTAGCGTCCAGTGAATGCGAGTCCGTCTAAGAGAGTAGTACAGCAGCCGTCAAGAGTGTCTA), connector oligonucleotide B (P-GTTCTGTCATATTTATCTAATTCGT), and connector oligonucleotide C (P-GAGTCGGCCATAGTATCCTTTAAGCGTCTTAA). Amplification was performed using /5Alexa594/CAGTGAATGCGAGTCCGTCT and analyzed by Zeiss LSM 880 confocal microscopy.
Nuclear radial distribution analysis
Single-nucleus images were processed in ImageJ to separate the signal (PLA or FISH) channels from the DAPI channels. The DAPI channel was processed in ImageJ to generate a distance map; the signals were filtered and analyzed by ImageJ through the “analyze particles” algorithm and then overlaid with the DAPI channel to measure the distance from the center of nucleus. The ImageJ macros and parameters are available on request. Finally, histograms were generated using Graphpad Prism.
ChIP-seq analysis
ChIP assays were performed as described previously (Farrell et al. 2013). Cells were cross-linked with formaldehyde to a final concentration of 1% in medium and incubated for 10 min at room temperature. Cells were collected in 1× phosphate-buffered saline (PBS) and 1 mM EDTA and pelleted by gentle centrifugation. The cells were resuspended in 700 µL of ChIP lysis buffer (0.1% SDS, 0.5% Triton X-100, 20 mM Tris-HCl at pH 8.1, 150 mM NaCl). The cell lysates were sonicated six times (output = 3.5; 30% duty cycle; 10 pulses) and then cleared by centrifugation at 14,000 rpm for 15 min at 4°C. Cell lysates were precleared with 50 µL of a 50% slurry of protein A beads for 1 h with rotation at 4°C. The lysates were again cleared by centrifugation at 14,000 rpm for 5 min at 4°C. Immunoprecipitations were performed with 2 µg of each antibody overnight at 4°C. Two micrograms of normal rabbit IgG or normal mouse IgG (Santa Cruz Biotechnology) was used as a negative control. The immunoprecipitates were washed six times with ChIP lysis buffer and twice with 1× Tris-EDTA (TE). Samples were rotated for 15 min at 4°C for each wash step. The immunoprecipitates were eluted from the beads with elution buffer (0.1 M NaHCO3, 1% SDS) by rotation for 15 min at room temperature. The elution products were transferred to new tubes, 5 M NaCl was added to a final concentration of 0.2 M, and samples were incubated overnight at 65°C. DNA was purified with the QIAquick PCR purification kit (Qiagen) and used for qPCR analysis with the specified primers.
ChIP-seq and the analysis were done based on the previously published method (Xie et al. 2011). ChIP data were filtered to remove sequences duplicated more than twice (the first two sequences were used). A sliding window of 500 base pairs (bp) was then used to call peaks with a peak FDR threshold of 0.05. Only peaks that were at least fourfold over background were accepted (using YS_Pin1_null_b1_ INPUT as background input). Peaks within 5 kb of satellite repeats were also rejected. “Consistency analysis of peak calling on replicates” (https://github.com/spundhir/idr) was applied to each pair of replicate ChIP peaks to select consistently detected peaks with an IDR FDR threshold of 0.05 for genome-wide peak calling or 0.10 for peaks around the TSS. These IDR peaks from both wild-type replicates and PIN1 knockout replicates were merged to give a set of enriched and reproducible peaks to further analyze. The original ChIP-seq data were then filtered to allow only sequence alignments in these regions. Counts were then normalized to total sequence counts in these regions. For annotation to genes and binding comparisons, sums of these filtered and normalized sequence counts were generated for 2-kb windows centered on the TSSs for RefSeq genes. DESeq2 software was used for differential analysis of unnormalized count data.
RNA-seq analysis
cDNA libraries were sequenced on the Illumina HiSeq 2000 as 101-bp single-end reads. Base calling was performed using Illumina RTA (version 1.13.48), and conversion to FASTQ was performed using CASAVA (version 1.8.2, Illumina). Reads were then trimmed to 44 bases, discarding the first 4 bases and then keeping the next 44 bases. Trimmed reads were aligned to the mm9 reference genome using Bowtie software (version 1.0.0) allowing up to three mismatches and requiring best unique matches. Custom R scripts were used to count tags that aligned to the exons of University of California at Santa Cruz (UCSC) RefSeq gene models. DESeq2 software was used for differential gene expression analysis.
ATAC-seq assay and analysis
ATAC-seq was performed using the Omni-ATAC protocol (Corces et al. 2017) without the use of digitonin. All PCR amplification was performed on a Bio-Rad CFX system for real-time monitoring, and reactions were pulled once exponential amplification was reached at 10 cycles. Libraries were sequenced on an Illumina NextSeq500. Reads were aligned using bwa mem followed by duplicate removal using SAMtools. Peaks were called using MACS2 with default parameters. We then constructed a site × sample count matrix that served as the basis for all subsequent analyses using the union peak set. Hierarchical clustering was performed on the Pearson correlation of the term frequency-normalized count matrix after binarization to the median threshold. Differential accessibility analysis was performed on the raw count matrix using a negative binomial test and setting the threshold for inclusion as a q-value ≤0.001 and a log2 fold change ≥1. Peak sets were intersected using Intervene. Motif enrichment analysis was performed using ChromVAR (Schep et al. 2017). To calculate significance of enrichment for distinct peak sets, we performed a CHT with the total set of ATAC-seq peaks, the set of differentially accessible peaks, the set of ChIP-seq peaks or motif sites that intersect with ATAC-seq peaks, and then the subset of those intersected peaks that are present in the differentially accessible peak set. We then performed an FDR test to only consider significantly enriched peak intersections if they meet an FDR threshold of 5%.
Additional bioinformatics analysis
GSEA was performed with 1000 permutations and default values for other parameters. Ranked lists were run against the C5_all gene sets from the MSigDB (http://www.broadinstitute.org/gsea/msigdb). All GSEA results are available in Supplemental Table 3. GO analysis was performed using default settings via DAVID (Huang et al. 2009). All GO results are available in Supplemental Table 7. For MYC-binding motif analysis, Peaks were called using a sliding window-based peak caller. Sex chromosomes were ignored, and tags duplicated more that twice were ignored. Some of the highest peaks appeared to be associated with artifacts and were removed. Reference sequence was obtained in a range ±25 bases around the center of mass of these peaks. These sequence fragments were filtered to remove any that spanned repetitive regions as flagged in the reference genome. The remaining sequence fragments were sorted based on the height of their associated peaks, and the top 500 of these were used as input to the Weeder de novo motif discovery tool (version 1.4.2). The following parameters were used with Weeder: “MM small M S 15” (Pavesi et al. 2001). The R Bioconductor software package seqLogo was used to generate sequence logos from the Weeder output. For averaged sequence density profiles at E-box sites, sums of filtered and normalized sequence counts in a 5-kb window centered on canonical CACGTG E-boxes were scaled based on the number of E-boxes.
In vitro and in vivo wound healing assays
In vivo cutaneous wound healing was performed as described previously (Chmielowiec et al. 2007). Three pairs of gender-matched isogenic wild-type and PIN1 knockout siblings at 6 wk were used for the experiment. For in vitro wound healing assays, ∼5000 cells were plated in each well of 96-well plates. Cells were treated with 10 µg/mL mitomysin C for 2 h before scratching. The wound areas were generated by WoundMaker (96-well, ESSEN BioScience). The closure of the wound was analyzed by IncuCyte live-cell analysis (ESSEN BioScience).
Data and statistics
Each PLA experiment included at least three independent biological replicates. PLA was quantified to illustrate the cell-to-cell variance (from >50 cells) in a representative experiment from at least three biologic replicates, since the variation of averages across experiments was less than cell-to-cell variance in all PLA experiments. To test for global correlation between ChIP-seq and RNA-seq data, we used the fisher.test() function in R to test a 2-by-2 contingency table, checking separately for over/underenrichment of up-regulated or down-regulated genes in our RNA-seq data (Supplemental Table 4) in the genes with enrichment of binding within 10 kb of the TSS for our wild-type Myc ChIP-seq versus Pin1 knockout data (Supplemental Table 5). Graphpad Prism was used to calculate average fold changes and standard errors. Statistical significance was calculated using a two-tailed t-test (P < 0.05 [*], P < 0.01 [**], and P < 0.001[***]) as indicated in the graphs. If no P-value is shown, the comparison was not relevant or not significant.
Data accessibility
RNA-seq, ChIP-seq, and ATAC-seq data has been deposited into Gene Expression Omnibus under accession number GSE109459.
Supplementary Material
Acknowledgments
We thank Dr. Matt Thayer's and Dr. Susan Olson's laboratories for help with FISH experiments. We thank Dr. Ellen Langer, Dr. Meghan Joly, and Dr. Brittany Allen-Petersen for comments on the manuscript. ChIP-seq and RNA-seq were performed by the Oregon Health and Science University (OHSU) Massively Parallel Sequencing Shared Resources. The OHSU Shared Resources are supported by Knight National Cancer Institute Cancer Center support grant P30CA069533. This study is supported by R35GM124704 to A.A.; RO1 CA186241 to M.-S.D. and R.C.S.; RO1 CA100855, RO1 CA196228, and U54 CA209988 to R.C.S.; and the Prospect Creek Foundation and Brenden-Colson Foundation to R.C.S.
Author contributions: Y.S. and R.C.S. designed the majority of the experiments. Y.S. conducted the majority of the experiments. C.P. and S.I. helped to design and analyze RNA-seq and ChIP-seq. K.T. and A.A. helped to design and analyze ATAC-seq. T.H. and X.N. helped to design and conduct the superresolution imaging experiments. A.C. helped with imaging quantification analysis. X.W. helped with mouse experiments. C.J.D. helped with cellular experiments. J.L. and M.-S.D. contributed to the writing and correction of the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.314377.118.
Freely available online through the Genes & Development Open Access option.
References
- Arnold HK, Sears RC. 2006. Protein phosphatase 2A regulatory subunit B56α associates with c-Myc and negatively regulates c-Myc accumulation. Mol Cell Biol 26: 2832–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Hurt E. 2017. The nuclear pore complex: understanding its function through structural insight. Nat Rev Mol Cell Biol 18: 73–89. [DOI] [PubMed] [Google Scholar]
- Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. 2003. Cdk2 knockout mice are viable. Curr Biol 13: 1775–1785. [DOI] [PubMed] [Google Scholar]
- Blobel G. 1985. Gene gating: a hypothesis. Proc Natl Acad Sci 82: 8527–8529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, Walter P. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol 2: e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee P-C, Widom J, Brickner JH. 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol 5: e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NR, Noble ME, Endicott JA, Johnson LN. 1999. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol 1: 438–443. [DOI] [PubMed] [Google Scholar]
- Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. 2008. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev 22: 627–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal GG. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441: 770–773. [DOI] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin J-C, Hurt EC, et al. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441: 770–773. [DOI] [PubMed] [Google Scholar]
- Cai D, Chen S-C, Prasad M, He L, Wang X, Choesmel-Cadamuro V, Sawyer JK, Danuser G, Montell DJ. 2014. Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell 157: 1146–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M. 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140: 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427–439. [DOI] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. 2005. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev 19: 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi J-T, van de Rijn M, Botstein D, Brown PO. 2004. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol 2: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y, Welcker M, Hizli AA, Posakony JJ, Aebersold R, Clurman BE. 2008. Identification of CDK2 substrates in human cell lysates. Genome Biol 9: R149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielowiec J, Borowiak M, Morkel M, Stradal T, Munz B, Werner S, Wehland J, Birchmeier C, Birchmeier W. 2007. c-Met is essential for wound healing in the skin. J Cell Biol 177: 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho KB, Cho MK, Lee WY, Kang KW. 2010. Overexpression of c-myc induces epithelial mesenchymal transition in mammary epithelial cells. Cancer Lett 293: 230–239. [DOI] [PubMed] [Google Scholar]
- Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, Satpathy AT, Rubin AJ, Montine KS, Wu B, et al. 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14: 959–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. 2012. MYC on the path to cancer. Cell 149: 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JOJ, Oudelaar AM, Higgs DR, Hughes JR. 2017. How best to identify chromosomal interactions: a comparison of approaches. Nat Methods 14: 125–134. [DOI] [PubMed] [Google Scholar]
- de Alboran IM, O'Hagan RC, Gärtner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. 2001. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14: 45–55. [DOI] [PubMed] [Google Scholar]
- de Marval PLM, Macias E, Rounbehler R, Sicinski P, Kiyokawa H, Johnson DG, Conti CJ, Rodriguez-Puebla ML. 2004. Lack of cyclin-dependent kinase 4 inhibits c-myc tumorigenic activities in epithelial tissues. Mol Cell Biol 24: 7538–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G, Iglesias N, Stutz F. 2006. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol 26: 7858–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman RN, Tachibana CY, Abrams HD, Hann SR. 1985. V-myc- and c-myc-encoded proteins are associated with the nuclear matrix. Mol Cell Biol 5: 114–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell AS, Pelz C, Wang X, Daniel CJ, Wang Z, Su Y, Janghorban M, Zhang X, Morgan C, Impey S, et al. 2013. Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol Cell Biol 33: 2930–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinn EM, Wallberg AE, Hermann S, Grant PA, Workman JL, Wright APH. 2002. Recruitment of Gcn5-containing complexes during c-Myc-dependent gene activation. Structure and function aspects. J Biol Chem 277: 23399–23406. [DOI] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. 2001. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev 15: 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori F, Takahashi K, Uchida C, Uchida T. 1999. Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G(0) arrest. Biochem Biophys Res Commun 265: 658–663. [DOI] [PubMed] [Google Scholar]
- Gabay M, Li Y, Felsher DW. 2014. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med 4: a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. 2003. Cyclin E ablation in the mouse. Cell 114: 431–443. [DOI] [PubMed] [Google Scholar]
- Georges-Labouesse E, Messaddeq N, Yehia G, Cadalbert L, Dierich A, Le Meur M. 1996. Absence of integrin α6 leads to epidermolysis bullosa and neonatal death in mice. Nat Genet 13: 370–373. [DOI] [PubMed] [Google Scholar]
- Ghirlando R, Felsenfeld G. 2016. CTCF: making the right connections. Genes Dev 30: 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green EM, Jiang Y, Joyner R, Weis K. 2012. A negative feedback loop at the nuclear periphery regulates GAL gene expression. Mol Biol Cell 23: 1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory MA, Hann SR. 2000. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol 20: 2423–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. 2008. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453: 948–951. [DOI] [PubMed] [Google Scholar]
- Hann SR. 2006. Role of post-translational modifications in regulating c-Myc proteolysis, transcriptional activity and biological function. Semin Cancer Biol 16: 288–302. [DOI] [PubMed] [Google Scholar]
- Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL. 2015. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J Cell Biol 208: 33–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase ME, Cordes VC. 2003. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell 14: 1923–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander S, Montecchio M, Pilstål R, Su Y, Kuruvilla J, Elvén M, Ziauddin JME, Anandapadamanaban M, Cristobal S, Lundström P, et al. 2015. Pre-anchoring of Pin1 to unphosphorylated c-Myc in a fuzzy complex regulates c-Myc activity. Structure 23: 2267–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT. 2005. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 436: 807–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, O'Connell BC, Mateyak MK, Tam W, Kohlhuber F, et al. 2000. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci 97: 2229–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Huang T, Phelps C, Wang J, Lin LJ, Bittel A, Scott Z, Jacques S, Gibbs SL, Gray JW, Nan X. 2018. Simultaneous multicolor single-molecule tracking with single-laser excitation via spectral imaging. Biophys J 114: 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hydbring P, Bahram F, Su Y, Tronnersjö S, Högstrand K, von der Lehr N, Sharifi HR, Lilischkis R, Hein N, Wu S, et al. 2010. Phosphorylation by Cdk2 is required for Myc to repress Ras-induced senescence in cotransformation. Proc Natl Acad Sci 107: 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PH, Watt FM. 1993. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell 73: 713–724. [DOI] [PubMed] [Google Scholar]
- Junttila MR, Puustinen P, Niemelä M, Ahola R, Arnold H, Böttzauw T, Ala-aho R, Nielsen C, Ivaska J, Taya Y, et al. 2007. CIP2A inhibits PP2A in human malignancies. Cell 130: 51–62. [DOI] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. 2010. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140: 360–371. [DOI] [PubMed] [Google Scholar]
- Kelly K, Cochran BH, Stiles CD, Leder P. 1983. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell 35: 603–610. [DOI] [PubMed] [Google Scholar]
- Kenneth NS, Ramsbottom BA, Gomez-Roman N, Marshall L, Cole PA, White RJ. 2007. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc Natl Acad Sci 104: 14917–14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon K-R, Nimura K, Rao SSP, Xu J, Jung S, Pekowska A, Dose M, Stevens E, Mathe E, Dong P, et al. 2017. Myc regulates chromatin decompaction and nuclear architecture during B cell activation. Mol Cell 67: 566–578.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilberg MS, Shan J, Su N. 2009. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab 20: 436–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JP, Zhang K, Chen JD, Wynn KC, Kramer RH, Woodley DT. 1992. Mechanism of human keratinocyte migration on fibronectin: unique roles of RGD site and integrins. J Cell Physiol 151: 443–450. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS. 2006. Myc influences global chromatin structure. EMBO J 25: 2723–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortlever RM, Sodir NM, Wilson CH, Burkhart DL, Pellegrinet L, Swigart LB, Littlewood TD, Evan GI. 2017. Myc cooperates with Ras by programming inflammation and immune suppression. Cell 171: 1301–1315.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296: 158–162. [DOI] [PubMed] [Google Scholar]
- Kress TR, Sabò A, Amati B. 2015. MYC: connecting selective transcriptional control to global RNA production. Nat Rev Cancer 15: 593–607. [DOI] [PubMed] [Google Scholar]
- Krull S, Thyberg J, Björkroth B, Rackwitz H-R, Cordes VC. 2004. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol Biol Cell 15: 4261–4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S, Dörries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. 2010. Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. EMBO J 29: 1659–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner D, Schultz P, Tora L, Georgieva SG. 2007. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J 26: 4956–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctôt C, Cheutin T, Cremer M, Cavalli G, Cremer T. 2007. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet 8: 104–115. [DOI] [PubMed] [Google Scholar]
- Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. 2013. Dynamic association of NUP98 with the human genome. PLoS Genet 9: e1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Brickner JH. 2013. Nuclear pore proteins regulate chromatin structure and transcriptional memory by a conserved mechanism. Nucleus 4: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. 2012a. Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, Murre C. 2012b. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol 13: 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou Y-C, Zhou XZ, Lu KP. 2011. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci 36: 501–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlewood TD, Amati B, Land H, Evan GI. 1992. Max and c-Myc/Max DNA-binding activities in cell extracts. Oncogene 7: 1783–1792. [PubMed] [Google Scholar]
- Liu T, Huang Y, Likhotvorik RI, Keshvara L, Hoyt DG. 2008. Protein never in mitosis gene A interacting-1 (PIN1) regulates degradation of inducible nitric oxide synthase in endothelial cells. Am J Physiol Cell Physiol 295: C819–C827. [DOI] [PubMed] [Google Scholar]
- Lopez-Bergami P, Lau E, Ronai Z. 2010. Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat Rev Cancer 10: 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KP, Finn G, Lee TH, Nicholson LK. 2007. Prolyl cis–trans isomerization as a molecular timer. Nat Chem Biol 3: 619–629. [DOI] [PubMed] [Google Scholar]
- Luo M-L, Gong C, Chen C-H, Lee DY, Hu H, Huang P, Yao Y, Guo W, Reinhardt F, Wulf G, et al. 2014. Prolyl isomerase Pin1 acts downstream of miR-200 to promote cancer stem-like cell traits in breast cancer. Cancer Res 74: 3603–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, Chaurasia S, Valentini SR, Corbett AH. 2007. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem 282: 3042–3049. [DOI] [PubMed] [Google Scholar]
- Lutterbach B, Hann SR. 1994. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol Cell Biol 14: 5510–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri L, Gacias M, Wu M, Swiss VA, Janssen WG, Casaccia P. 2014. c-Myc-dependent transcriptional regulation of cell cycle and nucleosomal histones during oligodendrocyte differentiation. Neuroscience 276: 72–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz E, Dechat T, Foisner R, Quinlan RA, Hutchison CJ. 2002. Lamin A/C binding protein LAP2α is required for nuclear anchorage of retinoblastoma protein. Mol Biol Cell 13: 4401–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Cerdeño V, Lemen JM, Chan V, Wey A, Lin W, Dent SR, Knoepfler PS. 2012. N-Myc and GCN5 regulate significantly overlapping transcriptional programs in neural stem cells. PLoS ONE 7: e39456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Sedivy JM. 1999. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol 19: 4672–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura I, Chiang K-N, Lai C-Y, He D, Wang G, Ramkumar R, Uchida T, Ryo A, Lu K, Liu F. 2010. Pin1 promotes transforming growth factor-β-induced migration and invasion. J Biol Chem 285: 1754–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. 2006. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 21: 811–823. [DOI] [PubMed] [Google Scholar]
- Myant K, Qiao X, Halonen T, Come C, Laine A, Janghorban M, Partanen JI, Cassidy J, Ogg E-L, Cammareri P, et al. 2015. Serine 62-phosphorylated MYC associates with nuclear lamins and its regulation by CIP2A is essential for regenerative proliferation. Cell Rep 12: 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SK, Burley SK. 2003. X-ray structures of Myc-Max and Mad-Max recognizing DNA: molecular bases of regulation by proto-incogenic transcription factors. Cell 112: 193–205. [DOI] [PubMed] [Google Scholar]
- Nickerson A, Huang T, Lin LJ, Nan X. 2014. Photoactivated localization microscopy with bimolecular fluorescence complementation (BiFC-PALM) for nanoscale imaging of protein–protein interactions in cells. PLoS One 9: e100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Hu G, Wei G, Cui K, Yamane A, Resch W, Wang R, Green DR, Tessarollo L, Casellas R, et al. 2012. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 151: 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemelä M, Kauko O, Sihto H, Mpindi J-P, Nicorici D, Pernilä P, Kallioniemi O-P, Joensuu H, Hautaniemi S, Westermarck J. 2012. CIP2A signature reveals the MYC dependency of CIP2A-regulated phenotypes and its clinical association with breast cancer subtypes. Oncogene 31: 4266–4278. [DOI] [PubMed] [Google Scholar]
- Ong C-T, Corces VG. 2014. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega S, Prieto I, Odajima J, Martín A, Dubus P, Sotillo R, Barbero JL, Malumbres M, Barbacid M. 2003. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet 35: 25–31. [DOI] [PubMed] [Google Scholar]
- Pascual-Garcia P, Debo B, Aleman JR, Talamas JA, Lan Y, Nguyen NH, Won KJ, Capelson M. 2017. Metazoan nuclear pores provide a scaffold for poised genes and mediate induced enhancer-promoter contacts. Mol Cell 66: 63–76.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, Mauri G, Pesole G. 2001. An algorithm for finding signals of unknown length in DNA sequences. Bioinformatics 17: S207–S214. [DOI] [PubMed] [Google Scholar]
- Pombo A, Dillon N. 2015. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 16: 245–257. [DOI] [PubMed] [Google Scholar]
- Pulverer BJ, Fisher C, Vousden K, Littlewood T, Evan G, Woodgett JR. 1994. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene 9: 59–70. [PubMed] [Google Scholar]
- Rajanala K, Nandicoori VK. 2012. Localization of nucleoporin Tpr to the nuclear pore complex is essential for Tpr mediated regulation of the export of unspliced RNA. PLoS ONE 7: e29921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner N, Miller SJ. 2015. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer 15: 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripley BD. 1977. Modelling spatial patterns. J R Stat Soc Series B Stat Methodol 39: 172–212. [Google Scholar]
- Rodríguez-Navarro S, Fischer T, Luo M-J, Antúnez O, Brettschneider S, Lechner J, Pérez-Ortín JE, Reed R, Hurt E. 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86. [DOI] [PubMed] [Google Scholar]
- Royds JA, Sharrard RM, Wagner B, Polacarz SV. 1992. Cellular localisation of c-myc product in human colorectal epithelial neoplasia. J Pathol 166: 225–233. [DOI] [PubMed] [Google Scholar]
- Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. 2017. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 14: 975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AD, Hu M, Ren B. 2016. Genome-wide mapping and analysis of chromosome architecture. Nat Rev Mol Cell Biol 17: 743–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears RC. 2004. The life cycle of C-myc: from synthesis to degradation. Cell Cycle 3: 1133–1137. [PubMed] [Google Scholar]
- Sears R, Ohtani K, Nevins JR. 1997. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol 17: 5227–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears R, Leone G, DeGregori J, Nevins JR. 1999. Ras enhances Myc protein stability. Mol Cell 3: 169–179. [DOI] [PubMed] [Google Scholar]
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 14: 2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AP, Verrecchia A, Fagà G, Doni M, Perna D, Martinato F, Guccione E, Amati B. 2008. A positive role for Myc in TGFβ-induced Snail transcription and epithelial-to-mesenchymal transition. Oncogene 28: 422–430. [DOI] [PubMed] [Google Scholar]
- Söderberg O, Gullberg M, Jarvius M, Ridderstråle K, Leuchowius K-J, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson L-G, et al. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods 3: 995–1000. [DOI] [PubMed] [Google Scholar]
- Soucek L, Evan GI. 2010. The ups and downs of Myc biology. Curr Opin Genet Dev 20: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X-X, He X, Yin L, Komada M, Sears RC, and Dai M-S. 2015. The nucleolar ubiquitin-specific protease USP36 deubiquitinates and stabilizes c-Myc. Proc Natl Acad Sci 112: 3734–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441: 774–778. [DOI] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441: 774–778. [DOI] [PubMed] [Google Scholar]
- Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. 2009. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev 23: 2610–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tworkowski KA, Salghetti SE, Tansey WP. 2002. Stable and unstable pools of Myc protein exist in human cells. Oncogene 21: 8515–8520. [DOI] [PubMed] [Google Scholar]
- Vervoorts J, Lüscher-Firzlaff J, Lüscher B. 2006. The ins and outs of MYC regulation by posttranslational mechanisms. J Biol Chem 281: 34725–34729. [DOI] [PubMed] [Google Scholar]
- Vomastek T, Iwanicki MP, Burack WR, Tiwari D, Kumar D, Parsons JT, Weber MJ, Nandicoori VK. 2008. Extracellular signal-regulated kinase 2 (ERK2) phosphorylation sites and docking domain on the nuclear pore complex protein Tpr cooperatively regulate ERK2-Tpr interaction. Mol Cell Biol 28: 6954–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriz S, Lemaitre JM, Leibovici M, Thierry N, Méchali M. 1992. Comparative analysis of the intracellular localization of c-Myc, c-Fos, and replicative proteins during cell cycle progression. Mol Cell Biol 12: 3548–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cunningham M, Zhang X, Tokarz S, Laraway B, Troxell M, Sears RC. 2011. Phosphorylation regulates c-Myc's oncogenic activity in the mammary gland. Cancer Res 71: 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiwad M, Kullertz G, Schutkowski M, Fischer G. 2000. Evidence that the substrate backbone conformation is critical to phosphorylation by p42 MAP kinase. FEBS Lett 478: 39–42. [DOI] [PubMed] [Google Scholar]
- Welcker M, Orian A, Grim JA, Eisenman RN, Clurman BE. 2004. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol 14: 1852–1857. [DOI] [PubMed] [Google Scholar]
- Winkler KE, Swenson KI, Kornbluth S, Means AR. 2000. Requirement of the prolyl isomerase Pin1 for the replication checkpoint. Science 287: 1644–1647. [DOI] [PubMed] [Google Scholar]
- Winqvist R, Saksela K, Alitalo K. 1984. The myc proteins are not associated with chromatin in mitotic cells. EMBO J 3: 2947–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortel IMN, van der Meer LT, Kilberg MS, van Leeuwen FN. 2017. Surviving stress: modulation of ATF4-mediated stress responses in normal and malignant cells. Trends Endocrinol Metab 28: 794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S. 2011. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J 30: 1473–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI. 2004. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J 23: 2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Riblet R, Schildkraut CL. 2005. Sites that direct nuclear compartmentalization are near the 5′ end of the mouse immunoglobulin heavy-chain locus. Mol Cell Biol 25: 6021–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, Hahn WC, Stukenberg PT, Shenolikar S, Uchida T, et al. 2004. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol 6: 308–318. [DOI] [PubMed] [Google Scholar]
- Zhang X, Farrell AS, Daniel CJ, Arnold H, Scanlan C, Laraway BJ, Janghorban M, Lum L, Chen D, Troxell M, et al. 2012. Mechanistic insight into Myc stabilization in breast cancer involving aberrant Axin1 expression. Proc Natl Acad Sci 109: 2790–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Jian W, Gao W, Zheng Y-X, Wang Y-K, Zhou Z-Q, Zhang H, Wang C-J. 2013. RNAi silencing of c-Myc inhibits cell migration, invasion, and proliferation in HepG2 human hepatocellular carcinoma cell line: c-Myc silencing in hepatocellular carcinoma cell. Cancer Cell Int 13: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XZ. 2000. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol Cell 6: 873–883. [DOI] [PubMed] [Google Scholar]
- Zhou XZ, Lu KP. 2016. The isomerase PIN1 controls numerous cancer-driving pathways and is a unique drug target. Nat Rev Cancer 16: 463–478. [DOI] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7–18. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Zhang H, Lang F, Liu G, Gao D, Li B, Liu Y. 2016. Pin1 promotes prostate cancer cell proliferation and migration through activation of Wnt/β-catenin signaling. Clin Transl Oncol 18: 792–797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq, ChIP-seq, and ATAC-seq data has been deposited into Gene Expression Omnibus under accession number GSE109459.