Here, Vuolo et al. investigated the mechanisms controlling the relative size of leaves compared with their lateral appendages (stipules). Using genetics, live imaging, and modeling, they demonstrate that the LATE MERISTEM IDENTITY1 (LMI1) homeodomain protein regulates stipule proportions via an endoreduplication-dependent trade-off that limits tissue size despite increasing cell growth.
Keywords: plant homeobox, leaf development, organ proportions, live imaging
Abstract
How the interplay between cell- and tissue-level processes produces correctly proportioned organs is a key problem in biology. In plants, the relative size of leaves compared with their lateral appendages, called stipules, varies tremendously throughout development and evolution, yet relevant mechanisms remain unknown. Here we use genetics, live imaging, and modeling to show that in Arabidopsis leaves, the LATE MERISTEM IDENTITY1 (LMI1) homeodomain protein regulates stipule proportions via an endoreduplication-dependent trade-off that limits tissue size despite increasing cell growth. LM1 acts through directly activating the conserved mitosis blocker WEE1, which is sufficient to bypass the LMI1 requirement for leaf proportionality.
How spatiotemporal coordination of cell and tissue growth contributes to plant and animal form is a key question in biology. Plant leaves are a powerful system to study growth and form because they show complex and diverse geometries that can be studied genetically. Leaf growth typically involves a phase of cell proliferation early in development followed by cell expansion associated with endoreduplication (Melaragno et al. 1993). However, developmental inputs into endoreduplication and how these shape leaf form remain largely unknown (Walker et al. 2000; Cookson et al. 2005; Massonnet et al. 2011). A key feature of leaf shape is the production of repeated marginal protrusions. These outgrowths vary from slight serrations in simple leaves to distinct leaflets in dissected leaves (Hay and Tsiantis 2006; Bilsborough et al. 2011; Hasson et al. 2011; Bar and Ori 2014; Vlad et al. 2014; Rast-Somssich et al. 2015). Stipules are another type of outgrowth that contribute considerably to macroevolutionary diversity in leaf form (Sinnott and Bailey 1914), a possibility also introduced by Darwin (1865); however, the mechanisms that influence stipule growth and development are poorly understood. Stipules typically flank the leaf base and vary in morphology from vestigial structures, as in Arabidopsis thaliana, to large leafy photosynthetic units, as in the pea. In A. thaliana, stipules initially comprise a significant proportion of the leaf primordium but only a small fraction of the mature leaf length (Fig. 1A–C). This indicates strong allometric regulation of leaf versus stipule growth, the genetic basis of which is unknown.
Figure 1.
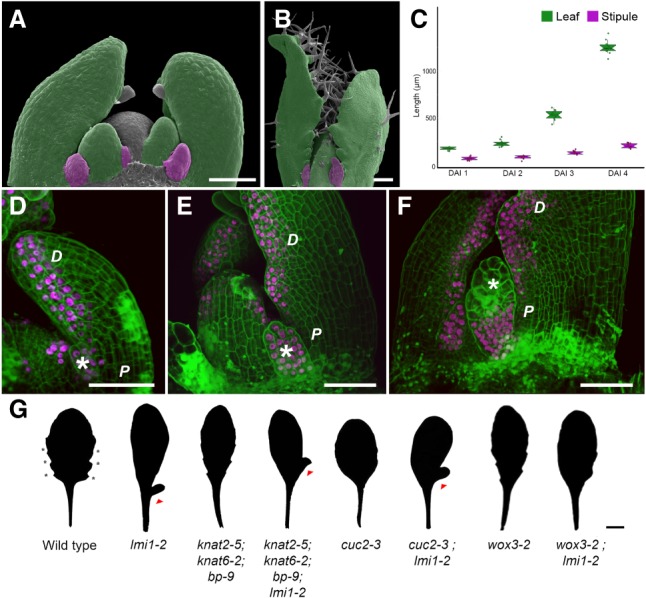
LATE MERISTEM IDENTITY1 (LMI1) is expressed in the distal leaf domain and stipule. (A–C) Wild-type leaves (green) and stipules (purple) in false-colored scanning electron micrographs at 2 d after leaf initiation (DAI) (A) and 8 DAI (B) and length (in micrometers) at successive DAI (C). n = 10 leaves. Mean ± SEM. (D–F) Confocal laser scanning microscopy (CLSM) of LMI1::LMI1:VENUS expression (magenta) in propidium iodide (PI)-stained (green) leaf primordia at 3 DAI (D), 5 DAI (E), and 7 DAI (F). n = 5 independent T2 lines. (Asterisks) Stipules; (D) distal leaf domains; (P) proximal leaf domains. Bars: A,B,D–F, 50 µm. (G) Silhouettes of adult leaves from wild-type and mutant plants. For penetrance scoring, see Supplemental Table 1. (Triangles) Ectopic lobes; (asterisks) serrations. Bar, 1 cm.
Results and Discussion
To identify molecular mechanisms required to yield correctly proportioned leaves, we investigated the HD-ZIP class I transcription factor LATE MERISTEM IDENTITY1 (LMI1) (Saddic et al. 2006), which regulates leaf growth in A. thaliana (Vlad et al. 2014). LMI1 expresses in the distal leaf margin (Fig. 1D–F), where serrations fail to form in lmi1 mutant leaves (Fig. 1G; Supplemental Fig. 1A–F; Saddic et al. 2006; Vlad et al. 2014; Vuolo et al. 2016). Conversely, LMI1 is not expressed in the proximal leaf margin (Fig. 1D–F), yet lmi1 leaves display ectopic lobes in this region of the leaf (Fig. 1G; Supplemental Fig. 1C,D,G–K; Saddic et al. 2006). The smooth distal margin of lmi1 leaves is consistent with LMI1 acting as a growth repressor (Vlad et al. 2014); for example, mutations in the local growth repressors REDUCED COMPLEXITY (RCO) and CUP-SHAPED COTYLEDON 2 (CUC2) lead to smoother leaves (Bilsborough et al. 2011; Vlad et al. 2014). However, the lobed proximal margin of lmi1 leaves is difficult to reconcile with LMI1 function. Surprisingly, lobe formation in lmi1 does not require CUC2 or KNOTTED-LIKE HOMEOBOX (KNOX) gene function, which are known regulators of lobe development (Fig. 1G; Supplemental Fig. 1K; Lincoln et al. 1994; Bilsborough et al. 2011; Hasson et al. 2011; Rast-Somssich et al. 2015). LMI1 expresses in developing stipules (Fig. 1D,E), eventually becoming restricted to the proximal part of mature stipules (Fig. 1F). Therefore, we hypothesized that LMI1 may act in stipules to limit their growth, leading to their excess growth into lobes in the lmi1 mutant. In this case, genetic ablation of stipules should remove ectopic lobes in lmi1 leaves. To test this idea, we used a stipule-less mutant of WUSCHEL-RELATED HOMEOBOX 3 (WOX3) (Shimizu et al. 2009) and found no lobed margins in wox3-2;lmi1-2 double mutants (Fig. 1G), strongly suggesting that the lobed margin in lmi1 leaves results from a transformation of the stipule into a leaf.
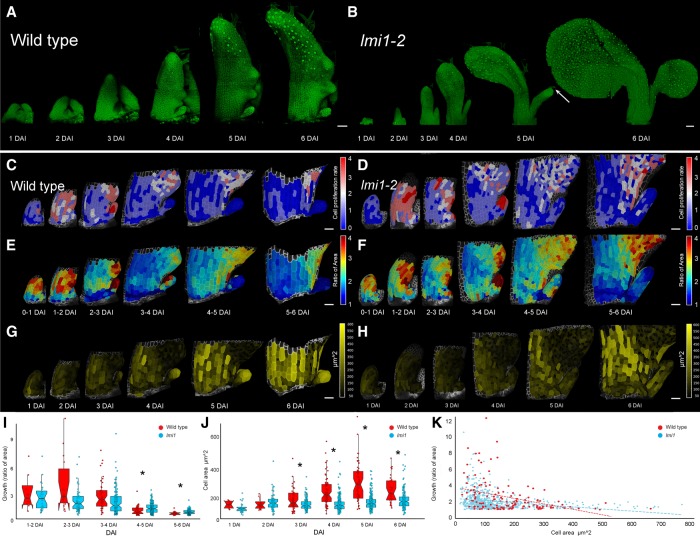
To verify whether basal lobes of lmi1 mutants are overgrown stipules and investigate the cellular basis of this transformation, we performed time-lapse imaging of growing leaves (Fig. 2A–H; Barbier de Reuille et al. 2015). Wild-type stipules show high rates of cell proliferation and growth starting 1 d after leaf initiation (DAI) (Fig. 2C,E,I). By 3 DAI, cell proliferation decreased dramatically, as did cell growth at 4 DAI (Fig. 2C,E,I). In contrast, cell proliferation and growth were maintained for longer in lmi1 stipules, with cells still dividing at 5–6 DAI (Fig. 2D), leading to smaller cells in lmi1 than wild-type stipules (Fig. 2G,H,J). In addition, clonal sectors derived from lineage tracing analysis were considerably more elongated in lmi1 stipules (Supplemental Fig. 2A–D). Thus, growth is higher in lmi1 than wild-type stipules from 3 DAI onward and proceeds for longer (Fig. 2E,F,I). Growth rate and cell area correlate negatively in these tissues, indicating that tissue growth reduces as cells enlarge (Fig. 2K). This correlation was weaker for lmi1 than wild-type stipules, suggesting that its underlying mechanism requires LMI1 (Fig. 2K).
Figure 2.
LMI1 represses tissue growth and cell proliferation in stipules. (A,B) Time-lapse CLSM of leaf 11 developing over 1–6 DAI in wild type (A) and lmi1-2 (B). Cells are outlined by PM-YFP expression. (Arrow) Trichome. (C–H) Cell proliferation rate (number of divisions; C,D), growth (ratio of areas; E,F), and cell area (G,H) quantified in wild-type (C,E,G) and lmi1-2 (D,F,H) leaf 11 time-lapse series. Scales are shown in heat maps. n = 5. (I,J) Quantification of cell growth (ratio of areas; I) and area (square micrometers; J) in wild-type (red) and lmi1-2 (blue) leaf 11. n = 3 time-lapse series; n > 50 cells. Mean ± SD. (*) P < 0.05, KS-test (I) and Student's t-test (J). (K) Cell area and growth values pooled for all DAI in wild-type (red; n = 139 cells) and lmi1-2 (blue; n = 690 cells) leaf 11. Dashed lines represent linear regressions. Bars: A–H, 50 µm.
Although LMI1 is expressed in at least two outer cell layers, stipules originate from epidermal founder cells (approximately one to five cells) (Supplemental Figs. 2A,C,E–G, 3A–C). In contrast, lmi1 stipules initiate from a larger number of founder cells (approximately seven to 12 cells epidermally) (Supplemental Fig. 3D–F) that can also include cells from internal layers (Supplemental Fig. 2H,I). This suggests that LMI1 represses stipule size at least in part by restricting stipule initial cells to the epidermis and limiting their number. We also observed stomata and trichome cells in lmi1 stipules, normally present only in the leaf and not in wild-type stipules (Fig. 2A,B). Therefore, in the absence of LMI1, more stipule initial cells are recruited to form a larger leaf-like outgrowth. In wild type, the stipule base is very narrow and stays attached to the boundary zone between the leaf base and adjacent tissue (Supplemental Fig. 2C). In contrast, the lmi1 stipule base grows together with adjacent petiole cells, progressively fusing the stipule with the leaf (Supplemental Figs. 1A–D,G–J, 2C,D). The extent of this fusion is variable, however, and the presence or absence of a lobe results from incomplete or complete fusion, respectively. The transformed stipules are more distally located (Fig. 1G) than their wild-type counterpart present at the leaf base, which is consistent with them initiating in a faster-growing context (Fig. 2E,F). lmi1 stipules also grow anisotropically, more akin to the leaf petiole than to wild-type stipules (Supplemental Fig. 2J–M). The length of lmi1 stipules reaches three times the length of wild-type stipules (Supplemental Fig. 4A,B), thus altering the allometric proportions of stipule to leaf in lmi1 leaves (Supplemental Fig. 4C,D). Overall, our findings suggest that LMI1 restricts stipule size by limiting founder cell recruitment and advancing cells from proliferative to expansive growth such that cell size increases but tissue growth is reduced.
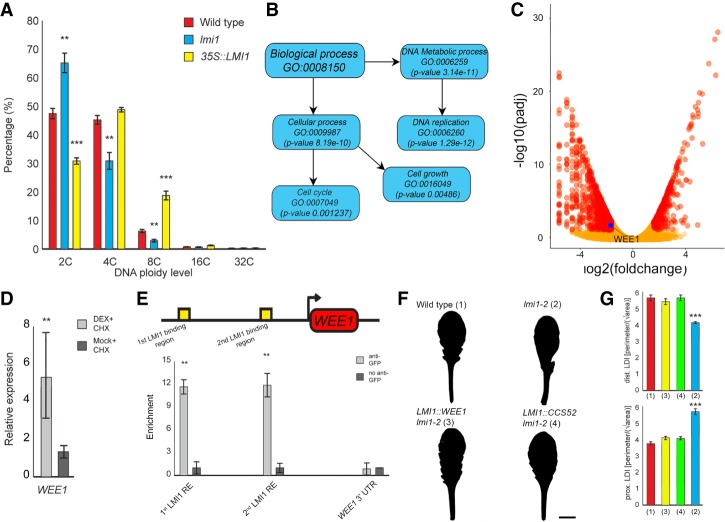
Endoreduplication counters cell proliferation, promotes cell enlargement, and can influence cell identity (Szymanski and Marks 1998; Bramsiepe et al. 2010; Maruyama et al. 2011; Roeder et al. 2012); therefore, we hypothesized that LMI1 might affect cell division, cell size, and organ identity in the stipule by promoting endoreduplication. Comparing DNA ploidy levels in wild-type, lmi1, and broadly expressing 35S::LMI1 plants (Fig. 3A), we found that the lmi1 mutant has 27% more 2C nuclei but 35% less 4C nuclei and 50% less 8C nuclei compared with wild type. In contrast, the 35S::LMI1 transgenic line shows almost 150% more 8C nuclei and 30% less 2C nuclei than wild-type samples. These observations indicate that LMI1 is necessary and sufficient to define the wild-type leaf endoreduplication profile (Fig. 3A). Consistent with these findings, leaf trichomes were excessively branched in 35S::LMI1 compared with wild type (Supplemental Fig. 5), a phenotype linked to increased endoreduplication (Walker et al. 2000). Additionally, we found that cell size and polytene regions (fused sister chromatids that form after endoreduplication) were reduced in the stipules and leaf margin of lmi1 (Supplemental Fig. 6), further indicating that LMI1 promotes endoreduplication in the leaf base. To explore cellular processes influenced by LMI1 at the whole-genome level, we used RNA sequencing (RNA-seq) to compare wild-type and lmi1 seedling transcriptomes. We found that differentially expressed genes were enriched for gene ontology (GO) terms related to the cell cycle, cell growth, and DNA replication (Fig. 3B), consistent with LMI1 promoting endoreduplication.
Figure 3.
LMI1 promotes endoreduplication by activating WEE1 expression. (A) Ploidy analysis by flow cytometry in wild-type (red), lmi1-2 (blue), and 35S::LMI1 (yellow) leaves. n = 5. Mean percentage ± SEM. (**) P < 0.01; (***) P < 0.001, ANOVA. (B) Subgroup of the GO categories enriched among differentially expressed genes (DEGs) between wild type and lmi1-2, derived from RNA-seq analysis. n = 3. (C) Volcano plot showing DEGs (red) and non-DEGs (orange) in lmi1-2 compared with wild-type seedlings, and the WEE1 gene with a putative LMI1-binding site (blue). (D) Quantitative RT–PCR (qRT–PCR) of WEE1 expression in lmi1-2;LMI1::LMI1:GR plants treated with dexamethasone (DEX) + cycloheximide (CHX) (gray) or mock + CHX (dark gray) 3 h after treatment. n = 3. Mean ± SEM. (**) P < 0.01, Student's t-test. (E, top) WEE1 gene model with upstream regions containing putative LMI1-binding sequences marked in yellow. The arrow indicates transcription start. (Bottom) ChIP-qRT–PCR (chromatin immunoprecipitation [ChIP] combined with qRT–PCR) after anti-GFP (gray) or control IgG (dark gray) pull-down in LMI1::LMI1:VENUS shows significant association of LMI1 with chromatin in regions containing the putative binding sites. n = 3. Mean ± SEM. (**) P < 0.0, Student's t-test1. WEE1 3′ untranslated region (UTR) was used as negative control. (F) Silhouettes of leaf 11 in wild-type, lmi1-2, lmi1-2;LMI1::WEE1, and lmi1-2;LMI1::CCS52 plants. n = 15 T2 lines. (G) Dissection indices of the distal domain (top graph) and proximal domain (bottom graph) in the lines in F, with relative numbers matching the ones in F. n = 10, each line. Bar, 1 cm.
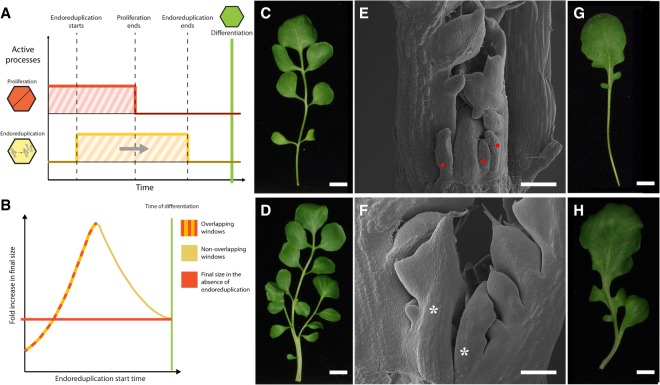
One of the key cell cycle genes showing reduced expression in lmi1 is WEE1 (Fig. 3C), which can inhibit mitosis, may promote endoreduplication (De Schutter et al. 2007; Gonzalez et al. 2007; Chevalier et al. 2011), and is expressed in both leaves and stipules (Supplemental Fig. 7). WEE1 is likely to be a direct target of LMI1, as it is transcriptionally up-regulated upon treatment with dexamethasone and the protein synthesis inhibitor cycloheximide in plants harboring a glucocorticoid-inducible LMI1 transgene (LMI1::LMI1-GR) (Fig. 3D). Consistent with this idea, we amplified WEE1 regulatory regions from chromatin immunoprecipitated using anti-GFP and found enrichment of LMI1:VENUS at two regions with predicted LMI1-binding sites (CAATwAT, where w is A or T) (Fig. 3E; Franco-Zorrilla et al. 2014). To determine whether WEE1 is critical for LMI1 function, we expressed LMI1::WEE1 in lmi1 mutants, which restored wild-type leaf shape, indicating that WEE1 expression suffices to bypass the requirement for LMI1 in leaf development (Fig. 3F,G). Although wee1 mutant leaves resemble wild type (De Schutter et al. 2007), this background was sufficient to ameliorate the growth repression caused by 35S::LMI1 (Vlad et al. 2014), underscoring the importance of WEE1 for LMI1 function (Supplemental Fig. 8). These findings suggest that LMI1-dependent endoreduplication regulates leaf form by locally restricting cell proliferation. To further test this idea, we expressed LMI1::CCS52, a known regulator of endoreduplication that is LMI1-independent (Fig. 3F; Cebolla et al. 1999), and found it sufficient to rescue both the lobed leaf and serration phenotype of lmi1 mutants (Fig. 3F,G). In addition, 35S::LMI1 plants developed smaller leaves (Supplemental Figs. 5A, 8D; Vlad et al. 2014). These results indicate that activating endoreduplication may limit final organ size. However, these findings are in contrast to previous reports showing that endoreduplication is associated with increased organ size in fruits and leaves (Melaragno et al. 1993; Gonzalez et al. 2007; Chevalier et al. 2011; Massonnet et al. 2011). We hypothesized that the interplay between cell proliferation and endoreduplication and the relative timing of their activation may be critical to determine final organ size. To formally examine this possibility, we constructed a minimal cell population model (Fig. 4A,B; Supplemental Fig. 9A,B; see the Supplemental Material for details). This model relates organ size to the timing of proliferation and endoreduplication within a finite window preceding differentiation. The model shows that endoreduplication leads to an increase in organ size except when activated very early. In this case, the decrease in cell number cannot be compensated for by the increase in cell size resulting from endoreduplication (Fig. 4B; Supplemental Fig. 9C,D). These results show that the timing of cells switching from proliferation to endoreduplication is critical and that early activation of endoreduplication may significantly reduce organ size.
Figure 4.
Modeling the regulation of organ size by endoreduplication and conservation of LMI1 function in Cardamine hirsuta. (A,B) Cell population model incorporating proliferation, endoreduplication, and differentiation. (A) The start time for endoreduplication (yellow line) is varied relative to the window of proliferation (orange line), and both processes are terminated by differentiation (green line). (B) Simulations showing fold increase of organ size as a function of endoreduplication start time relative to the size increase produced by proliferation alone (orange line; i.e., when Re = 0). The color of the curve indicates when windows of proliferation and endoreduplication overlap in time (yellow and orange) or occur sequentially (yellow only). (C–H) Leaves of wild-type (C,E), 35S::amiR-LMI1 (D,F), rco (G), and 35S::amiR-LMI1;rco (H) representative leaf 5 shown for n = 15 independent lines per genotype. Bars: C,D,G,H, 1 cm; E,F (scanning electron micrographs of developing leaves), 100 µm. (Red dots) Stipules; (asterisks) ectopic leaves.
By exploring the model's parameter space, we found that the start time of endoreduplication should have the largest effect on organ size in the context of highly proliferative tissues (i.e., when RS is large) (Supplemental Fig. 9D). In proliferative tissues, endoreduplication reduces cell division and, ultimately, cell number. For highly proliferative tissues, this loss exceeds what can be compensated for by the subsequent endoreduplication-driven increase in cell size. Therefore, the exclusion of LMI1 from the proximal leaf margin (Fig. 1E,F), a highly proliferative region (Kuchen et al. 2012; Vlad et al. 2014), should be crucial to maintain the correct size and shape of A. thaliana leaves. Specifically, the model predicts a reduction in final leaf size if endoreduplication is activated very early in this region of the leaf margin. To directly test this prediction, we fused LMI1 to three different promoters that express both earlier and more proximally than LMI1 in the leaf margin (RCO and CUC2) and throughout the leaf primordium (ASYMMETRIC LEAVES1 [AS1]) (Bilsborough et al. 2011; Hasson et al. 2011; Vlad et al. 2014). Leaf size was dramatically reduced in RCO::LMI1 (Vuolo et al. 2016), CUC2::LMI1, and AS1::LMI1 plants (Supplemental Fig. 10). In particular, AS1::LMI1 plants produced small bladeless leaves followed by arrested development (Supplemental Fig. 10C). These observations support the model predictions and highlight the importance of the precise regulation of LMI1 for attaining correct leaf form.
To understand whether LMI1 function is conserved between plants with different leaf shapes, we used an artificial microRNA (amir-LMI1) to silence expression of the orthologous LMI1 gene in Cardamine hirsuta, a relative of A. thaliana with dissected leaves (Hay and Tsiantis 2006). Fully dissected leaves replaced stipules in these transgenic plants (Fig. 4C–F; Supplemental Fig. 11), indicating that LMI1 function is conserved between crucifers with simple and dissected leaves. Furthermore, stipules of amiR-LMI1;rco plants were converted to simplified rco-like mutant leaves (Fig. 4G,H; Vlad et al. 2014). Therefore, LMI1 and RCO, which are tandemly duplicated genes, largely function divergently in the leaf, consistent with their distinct expression domains (Vlad et al. 2014). Our findings also raise the possibility that evolutionary tinkering with the LMI1 endoreduplication module, as described here, may underlie stipule transformations into leaf-like organs that are typical of many taxa (Darwin 1865; Sousa-Baena et al. 2018). For example, Tendrilless (Tl; pea LMI1) expression in pea leaves is absent from leafy stipules but present in bladeless tendrils, where it causes growth arrest, associated with increased endoreduplication (expression in tendrils) (Supplemental Fig. 12; also shown previously by Hofer et al. 2009). In conclusion, we demonstrated that spatially regulated expression of the LMI1 transcription factor influences organ proportions through an endoreduplication-mediated trade-off between cell and tissue growth. Our findings may help unify our understanding of the control of organ shape across multicellular eukaryotes. For example, in the developing vertebrate limb bud, the transcription factor GLI3R constrains digit size and number by limiting the pool of mesenchymal progenitors through directly repressing Cdk6, a regulator of the G1–S cell cycle transition (Lopez-Rios et al. 2012), highlighting the significance of local regulation of the cell cycle for controlling organ proportions. A future challenge will be to determine how broadly the growth trade-off that we identified here is used to control organ form and the degree to which it shaped organ diversity in different animals and plants (e.g., Sicard et al. 2014; Vlad et al. 2014; Andres et al. 2017).
Materials and methods
Plants were cultivated in growth chambers under long day (16-h light/8-h dark) or short day (8-h light/16-h dark) conditions. A. thaliana and C. hirsuta were transformed using Agrobacterium tumefaciens floral dip transformations as in Hay et al. (2014). Confocal microscopy was performed with a SP8 upright laser scanning confocal microscope with a long working distance water immersion objective (AP 20×/0.8 M27; Zeiss). Statistical analyses were performed with Excel and the R package. Promoter sequence analyses and transcription factor-binding site search were performed with MEME/MAST package. A detailed description of the Materials and Methods is in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank Bjorn Pieper for statistical consultation; Soeren Strauss for help with lineage tracing, using code shared by Aleksandra Sapala and Richard Smith, initially published in Barbier de Reuille et al. (2015); L. De Veylder and A. Schnitger for sharing materials and helpful discussions on cell cycle; and the Max Planck Institute for Plant Breeding Research-Genome Centre for RNA-seq. We also acknowledge U. Tartler and R. Berndtgen for technical assistance. This work was supported by Biotechnology and Biological Sciences Research Council grant BB/H011455/1 to M.T., Deutsche Forschungsgemeinschaft MorphoDynamics research unit FOR 2581 to M.T. and A.H., Marie Skłodowska-Curie fellowship IF-2015 703886 to A.R., EMBO Long-Term Fellowship ALTF 502-2015 to R.A.M., and a core grant from the Max Planck Society to M.T.
Author contributions: F.V. performed the majority of experiments. M.H. performed the chromatin immunoprecipitation and contributed to cloning and transgenic line generation. D.K. performed the time-lapse analysis. A.R. designed and wrote the cell population model and, together with D.K., quantified the time-lapse data and conducted lineage tracing. Y.W. constructed vectors and reporters for WEE1 expression and targeted LMI1 misexpression. D.V. conducted Cardamine experiments. P.H. performed the in situ experiment in Pisum sativum and contributed to imaging. M.D.G. performed the allometry analysis. X.G. analyzed the RNA-seq data. Z.Z. constructed the double mutants for cuc2-3;lmi1-2 and wox3-2;lmi1-2. A.P. provided training and assistance in flow cytometry experiments. R.A.M. conducted statistical analysis and helped with lineage tracing. A.H. contributed to data analysis. F.V., A.H., A.R., R.A.M., and M.T. wrote the manuscript with input from other authors. M.T. designed and directed the study.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.318212.118.
Freely available online through the Genes & Development Open Access option.
References
- Andres RJ, Coneva V, Frank MH, Tuttle JR, Samayoa LF, Han SW, Kaur B, Zhu L, Fang H, Bowman DT, et al. 2017. Modifications to a LATE MERISTEM IDENTITY1 gene are responsible for the major leaf shapes of Upland cotton (Gossypium hirsutum L.). Proc Natl Acad Sci 114: E57–E66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M, Ori N. 2014. Leaf development and morphogenesis. Development 141: 4219–4230. [DOI] [PubMed] [Google Scholar]
- Barbier de Reuille P, Routier-Kierzkowska AL, Kierzkowski D, Bassel GW, Schupbach T, Tauriello G, Bajpai N, Strauss S, Weber A, Kiss A, et al. 2015. MorphoGraphX: a platform for quantifying morphogenesis in 4D. Elife 4: 05864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilsborough GD, Runions A, Barkoulas M, Jenkins HW, Hasson A, Galinha C, Laufs P, Hay A, Prusinkiewicz P, Tsiantis M. 2011. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci 108: 3424–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramsiepe J, Wester K, Weinl C, Roodbarkelari F, Kasili R, Larkin JC, Hulskamp M, Schnittger A. 2010. Endoreplication controls cell fate maintenance. PLoS Genet 6: e1000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebolla A, Vinardell JM, Kiss E, Olah B, Roudier F, Kondorosi A, Kondorosi E. 1999. The mitotic inhibitor ccs52 is required for endoreduplication and ploidy-dependent cell enlargement in plants. EMBO J 18: 4476–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C, Nafati M, Mathieu-Rivet E, Bourdon M, Frangne N, Cheniclet C, Renaudin JP, Gévaudant F, Hernould M. 2011. Elucidating the functional role of endoreduplication in tomato fruit development. Ann Bot 107: 1159–1169. 10.1093/aob/mcq257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson SJ, Van Lijsebettens M, Granier C. 2005. Correlation between leaf growth variables suggest intrinsic and early controls of leaf size in Arabidopsis thaliana. Plant Cell Environ 28: 1355–1366. [Google Scholar]
- Darwin C. 1865. On the movements and habits of climbing plants. Bot J Linn Soc 9: 1–118. [Google Scholar]
- De Schutter K, Joubes J, Cools T, Verkest A, Corellou F, Babiychuk E, Van Der Schueren E, Beeckman T, Kushnir S, Inze D, et al. 2007. Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R. 2014. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci 111: 2367–2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Gévaudant F, Hernould M, Chevalier C, Mouras A. 2007. The cell cycle-associated protein kinase WEE1 regulates cell size in relation to endoreduplication in developing tomato fruit. Plant J 51: 642–655. 10.1111/j.1365-313X.2007.03167.x [DOI] [PubMed] [Google Scholar]
- Hasson A, Plessis A, Blein T, Adroher B, Grigg S, Tsiantis M, Boudaoud A, Damerval C, Laufs P. 2011. Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell 23: 54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Tsiantis M. 2006. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet 38: 942–947. [DOI] [PubMed] [Google Scholar]
- Hay AS, Pieper B, Cooke E, Mandakova T, Cartolano M, Tattersall AD, Ioio RD, McGowan SJ, Barkoulas M, Galinha C, et al. 2014. Cardamine hirsuta: a versatile genetic system for comparative studies. Plant J 78: 1–15. [DOI] [PubMed] [Google Scholar]
- Hofer J, Turner L, Moreau C, Ambrose M, Isaac P, Butcher S, Weller J, Dupin A, Dalmais M, Le Signor C, et al. 2009. Tendril-less regulates tendril formation in pea leaves. Plant Cell 21: 420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchen EE, Fox S, de Reuille PB, Kennaway R, Bensmihen S, Avondo J, Calder GM, Southam P, Robinson S, Bangham A, et al. 2012. Generation of leaf shape through early patterns of growth and tissue polarity. Science 335: 1092–1096. [DOI] [PubMed] [Google Scholar]
- Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. 1994. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6: 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rios J, Speziale D, Robay D, Scotti M, Osterwalder M, Nusspaumer G, Galli A, Hollander GA, Kmita M, Zeller R. 2012. GLI3 constrains digit number by controlling both progenitor proliferation and BMP-dependent exit to chondrogenesis. Dev Cell 22: 837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama R, Grevengoed E, Stempniewicz P, Andrew DJ. 2011. Genome-wide analysis reveals a major role in cell fate maintenance and an unexpected role in endoreduplication for the Drosophila FoxA gene Fork head. PLoS One 6: e20901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massonnet C, Tisne S, Radziejwoski A, Vile D, De Veylder L, Dauzat M, Granier C. 2011. New insights into the control of endoreduplication: endoreduplication could be driven by organ growth in Arabidopsis leaves. Plant Physiol 157: 2044–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW. 1993. Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5: 1661–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rast-Somssich MI, Broholm S, Jenkins H, Canales C, Vlad D, Kwantes M, Bilsborough G, Dello Ioio R, Ewing RM, Laufs P, et al. 2015. Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta. Genes Dev 29: 2391–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder AH, Cunha A, Ohno CK, Meyerowitz EM. 2012. Cell cycle regulates cell type in the Arabidopsis sepal. Development 139: 4416–4427. [DOI] [PubMed] [Google Scholar]
- Saddic LA, Huvermann B, Bezhani S, Su Y, Winter CM, Kwon CS, Collum RP, Wagner D. 2006. The LEAFY target LMI1 is a meristem identity regulator and acts together with LEAFY to regulate expression of CAULIFLOWER. Development 133: 1673–1682. [DOI] [PubMed] [Google Scholar]
- Shimizu R, Ji J, Kelsey E, Ohtsu K, Schnable PS, Scanlon MJ. 2009. Tissue specificity and evolution of meristematic WOX3 function. Plant Physiol 149: 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard A, Thamm A, Marona C, Lee YW, Wahl V, Stinchcombe JR, Wright SI, Kappel C, Lenhard M. 2014. Repeated evolutionary changes of leaf morphology caused by mutations to a homeobox gene. Curr Biol 24: 1880–1886. [DOI] [PubMed] [Google Scholar]
- Sinnott EW, Bailey IW. 1914. Investigations on the phylogeny of the angiosperms. 3. Nodal anatomy and the morphology of stipules. Am J Bot 1: 441–453. [Google Scholar]
- Sousa-Baena MS, Lohmann LG, Hernandes-Lopes J, Sinha NR. 2018. The molecular control of tendril development in angiosperms. New Phytol 218: 944–958. [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Marks MD. 1998. GLABROUS1 overexpression and TRIPTYCHON alter the cell cycle and trichome cell fate in Arabidopsis. Plant Cell 10: 2047–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlad D, Kierzkowski D, Rast MI, Vuolo F, Dello Ioio R, Galinha C, Gan X, Hajheidari M, Hay A, Smith RS, et al. 2014. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science 343: 780–783. [DOI] [PubMed] [Google Scholar]
- Vuolo F, Mentink RA, Hajheidari M, Bailey CD, Filatov DA, Tsiantis M. 2016. Coupled enhancer and coding sequence evolution of a homeobox gene shaped leaf diversity. Genes Dev 30: 2370–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JD, Oppenheimer DG, Concienne J, Larkin JC. 2000. SIAMESE, a gene controlling the endoreduplication cell cycle in Arabidopsis thaliana trichomes. Development 127: 3931–3940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.