Abstract
Significance: The Escherichia coli regulatory protein fumarate nitrate reduction (FNR) mediates a global transcriptional response upon O2 deprivation. Spanning nearly 40 years of research investigations, our understanding of how FNR senses and responds to O2 has considerably progressed despite a lack of structural information for most of that period. This knowledge has established the paradigm for how facultative anaerobic bacteria sense changes in O2 tension.
Recent Advances: Recently, the X-ray crystal structure of Aliivibrio fischeri FNR with its [4Fe-4S] cluster cofactor was solved and has provided valuable new insight into FNR structure and function. These findings have alluded to the conformational changes that may occur to alter FNR activity in response to O2.
Critical Issues: Here, we review the major features of this structure in context of previously acquired data. In doing so, we discuss additional mechanistic aspects of FNR function that warrant further investigation.
Future Directions: To complement the [4Fe-4S]-FNR structure, the structures of apo-FNR and FNR bound to DNA or RNA polymerase are needed. Together, these structures would elevate our understanding of how ligation of its [4Fe-4S] cluster allows FNR to regulate transcription according to the level of environmental O2.
Keywords: : FNR, transcription factor, O2 sensor, regulation, structure
O2 Sensing in Facultative Anaerobic Bacteria
It is well known that O2 availability exerts considerable influence on the physiology of most organisms. In the case of bacteria that thrive in both aerobic and anaerobic environments (facultative anaerobes), O2 tension dictates the use of distinct metabolic pathways usually directed to optimize energy conservation. Aerobic respiration is the more energetically favorable pathway, yielding maximal adenosine 5′-triphosphate (ATP) when O2 serves as terminal electron acceptor. Upon O2 deprivation, ATP is generated by alternative pathways, such as fermentation or anaerobic respiration, which utilizes alternative terminal electron acceptors (e.g., fumarate, nitrate, dimethyl sulfoxide, and trimethylamine N-oxide) (9, 16). The expression of stress response pathways is also affected by the presence of O2, which along with its toxic derivatives (e.g., hydrogen peroxide, superoxide, and hydroxyl radical) can inadvertently damage cellular components (28). Therefore, facultative anaerobes have developed strategies to sense abrupt changes in O2 concentration and rapidly respond by altering gene expression. Often, this regulation occurs through O2 sensing transcription factors that function to modify levels of messenger RNA transcripts.
Escherichia coli has served as a model organism for understanding O2 mediated transcriptional regulation. E. coli routinely cycles between environments of varying O2 tension, such as the anaerobic mammalian gut and aerobic niches outside the host, so it is not surprising that this enterobacterium has evolved an intricate transcriptional network to adapt to such changes. To directly sense O2, E. coli has exploited transcription factors containing protein cofactors that inherently react with O2, such as Fe-S clusters and Fe2+ [e.g., fumarate nitrate reduction (FNR), Fur] (4, 9, 16, 24). O2 may also be detected indirectly by monitoring changes in the quinone pool (e.g., ArcA) (9, 16). Perhaps the most extensively studied O2 sensing mechanism has been that of FNR, which mediates an adaptive response upon anaerobiosis. The findings of numerous studies to elucidate this mechanism have been the subject of many reviews (e.g., see references 13, 34, 72). Although these studies identified a [4Fe-4S]2+ cluster as central to FNR's ability to sense O2, the lack of a structure limited further mechanistic understanding.
Recently, Volbeda et al. solved the X-ray crystal structure of FNR from A. fischeri (which shares 84% of amino acid sequence identity with E. coli FNR) with the [4Fe-4S] cluster intact (Fig. 1A) (73). Not only does this structure support many of the earlier findings, it has also uncovered several exciting and novel facets of FNR function. Here, we highlight specific features of this structure and that of the previously solved FNR homolog FixK2 bound to DNA (8). In doing so, we dissect the major functional regions of FNR (Fe-S cluster binding, dimerization, DNA binding, and RNA polymerase (RNAP) interaction) and relate structural information with previously and newly acquired data. For ease of comparison, we will refer to amino acid residues using E. coli FNR numbering because of the wealth of FNR mutants that have been studied.
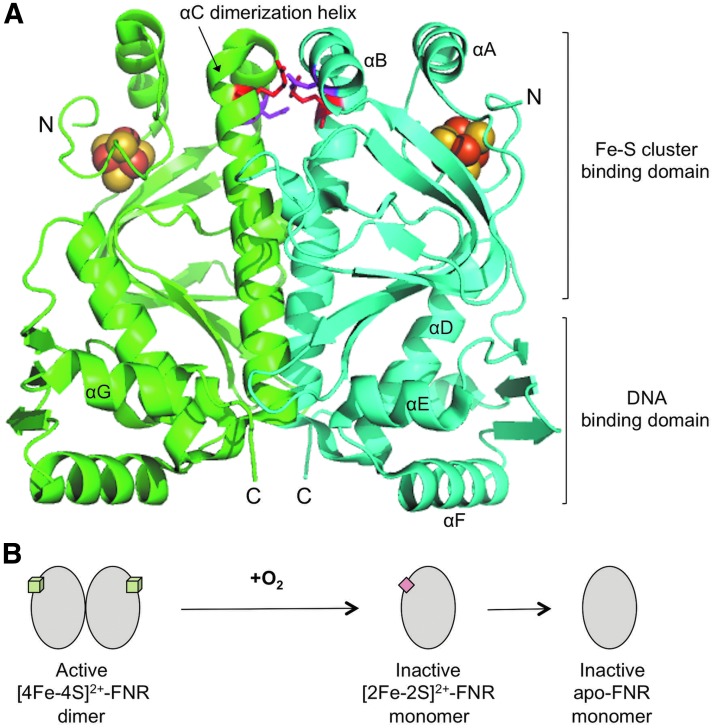
FIG. 1.
The role of the [4Fe-4S] cluster in FNR structure and function. (A) Crystal structure of [4Fe-4S]-FNR. The protein dimer is shown in cartoon representation, with individual protein subunits shown in cyan and green. Present in each subunit is a [4Fe-4S] cluster shown as yellow and orange spheres. Labeled are the N-terminal Fe-S cluster binding domain, the C-terminal DNA binding domain, and the seven α-helices present in FNR (designated with letters A–G), including the αC dimerization helix. Also shown are the side chains of residues R140 (red) and D130 (magenta), which form intersubunit salt bridges that contribute to dimerization. This figure was prepared with MacPyMOL using the structure available in the Research Collaboratory for Structural Bioinformatics PDB under the accession code 5E44 ref. (73). (B) Model for the regulation of FNR activity by O2. In the absence of O2, FNR contains a [Fe-4S]2+ cluster (green cube), which promotes subunit dimerization, site-specific DNA binding, and transcriptional regulation. When O2 is introduced, the [4Fe-4S]2+ cluster is rapidly converted to a [2Fe-2S]2+ cluster (red square), resulting in FNR inactivation through loss of dimerization. In aerobic cells, the [2Fe-2S]2+ cluster is further degraded, generating clusterless apo-FNR. FNR, fumarate nitrate reduction; PDB, protein data bank.
General Functional and Structural Properties of FNR
The first evidence to link a functional interaction between FNR and anaerobic respiration emerged from the discovery that fnr mutants were incapable of fumarate and nitrate reduction, hence the name FNR (38). This observation launched decades of research from several laboratories to define FNR's role in transcriptional regulation of respiratory pathways. More recently, genome-wide analyses have defined the scope of the direct FNR transcriptional network that encompasses ≥63 genes, many of which mediate adaptation to O2 limitation (12, 20, 22, 31, 56, 58, 63). For instance, FNR activates genes involved in anaerobic oxidation of carbon sources and reduction of alternative terminal electron acceptors and represses some genes specifically used in aerobic respiration. Although biochemical and genetic studies of E. coli FNR are reviewed here, it should be noted that FNR homologs are widely distributed in Proteobacteria and Bacilli and a core FNR regulon appears to be conserved across many facultative anaerobes (17, 37, 47).
The capacity of FNR to globally regulate gene expression depends on the presence of its [4Fe-4S]2+ cluster cofactor. The integrity of this cluster is intricately linked with FNR function since it promotes a protein conformation necessary for FNR dimerization, site-specific DNA binding, and transcriptional regulation (23, 32, 42). However, in the presence of O2, the [4Fe-4S]2+ cluster is rapidly converted to a [2Fe-2S]2+ cluster via sulfur-based oxidation, resulting in FNR inactivation through loss of dimerization (15, 30, 33, 60, 82). In aerobic cells, the [2Fe-2S]2+ cluster is further degraded, generating clusterless, apo-FNR (70) (Fig. 1B). Ultimately, due to sensitivity of the [4Fe-4S]2+ cluster toward O2, FNR-dependent transcriptional regulation occurs primarily during anaerobic growth despite similar FNR protein levels in aerobic and anaerobic cells (69).
Before the A. fischeri FNR structure, gaining mechanistic insight into how O2 mediated cluster conversion influenced the conformational alterations in FNR that must occur for regulated activity was somewhat challenging. It was known that FNR is structurally related to the cyclic adenosine monophosphate (cAMP) cAMP receptor protein (CRP) family of transcription factors whose members are broadly distributed in bacteria (37, 47, 65). These proteins characteristically contain an N-terminal effector binding domain that encompasses a β-roll motif, a long α-helix that mediates subunit dimerization, and a C-terminal DNA binding domain that comprises a helix-turn-helix (HTH) motif (Fig. 1) (79).
Structural similarities aside, individual CRP family members are distinguished by their respective effector molecules, which allow for adaptive responses to diverse environmental cues (e.g., cAMP, CO, O2, NO, 2-oxoglutarate, aromatic compounds) (37). The structures of CRP and other family members have been solved and have provided the initial framework for understanding how FNR operates as a transcription factor (6, 79). Despite the valuable information acquired from these structures, many questions regarding FNR activity remained. For instance, although most CRP family members are constitutive homodimers that bind DNA target sites upon association with their cognate effectors (26, 36, 79), FNR appears unique in that ligation of one [4Fe-4S]2+ cluster per subunit induces the protein dimerization that is required for DNA site recognition. Thus, the FNR structure has been highly anticipated to fully understand how FNR is mechanistically regulated by O2.
Ligation of the [4Fe-4S] Cluster Appears to Organize the FNR N-Terminal Region
A major question in the field has been how the N-terminal cluster binding domain of FNR differs from the analogous domain of other CRP family members. Although secondary structure of the effector binding domain is conserved within the CRP family, it is perhaps not surprising that the effector ligands and neighboring residues display significant variability, likely accounting for the broad range of effector molecules recognized by family members (37). Furthermore, FNR contains an N-terminal extension of 26 amino acid residues that are not present in CRP, and in vivo and in vitro experiments established that C20, C23, and C29 within this extension, along with C122 of the β-roll motif, serve as ligands for the [4Fe-4S]2+ cluster (48, 64, 67, 68). In addition, susceptibility of the N-terminal region to limited trypsin digestion suggested some conformational flexibility within this domain (55).
Although electron density is not visible for the first 18 residues, the X-ray crystal structure demonstrates that the FNR cluster binding region is near the protein surface and is indeed more disordered than the rest of the protein. In this region, the N-terminal residues of the protein (encompassing C20, C23, and C29) are unstructured and appear to partially wrap the [4Fe-4S] cluster and position it adjacent to the top of the β-roll motif (containing C122; Figs. 1 and 2). Although the β-roll and preceding αA helix are common to CRP family members, in FNR, these elements and the unstructured N-terminal residues form a topology that is similar to a protein fold shared by some Fe-S proteins (53). The structure also reveals that proximal to the [4Fe-4S] cluster is an unpredicted network of hydrophobic interactions involving residues of the αA, αB, and αC helices (I30, L34, L42, I45, I46, I124, I128, L129, I132, L139, I143; Fig. 2A). As discussed later, these interactions likely provide a signaling relay between O2 mediated cluster conversion and the loss of FNR dimerization.
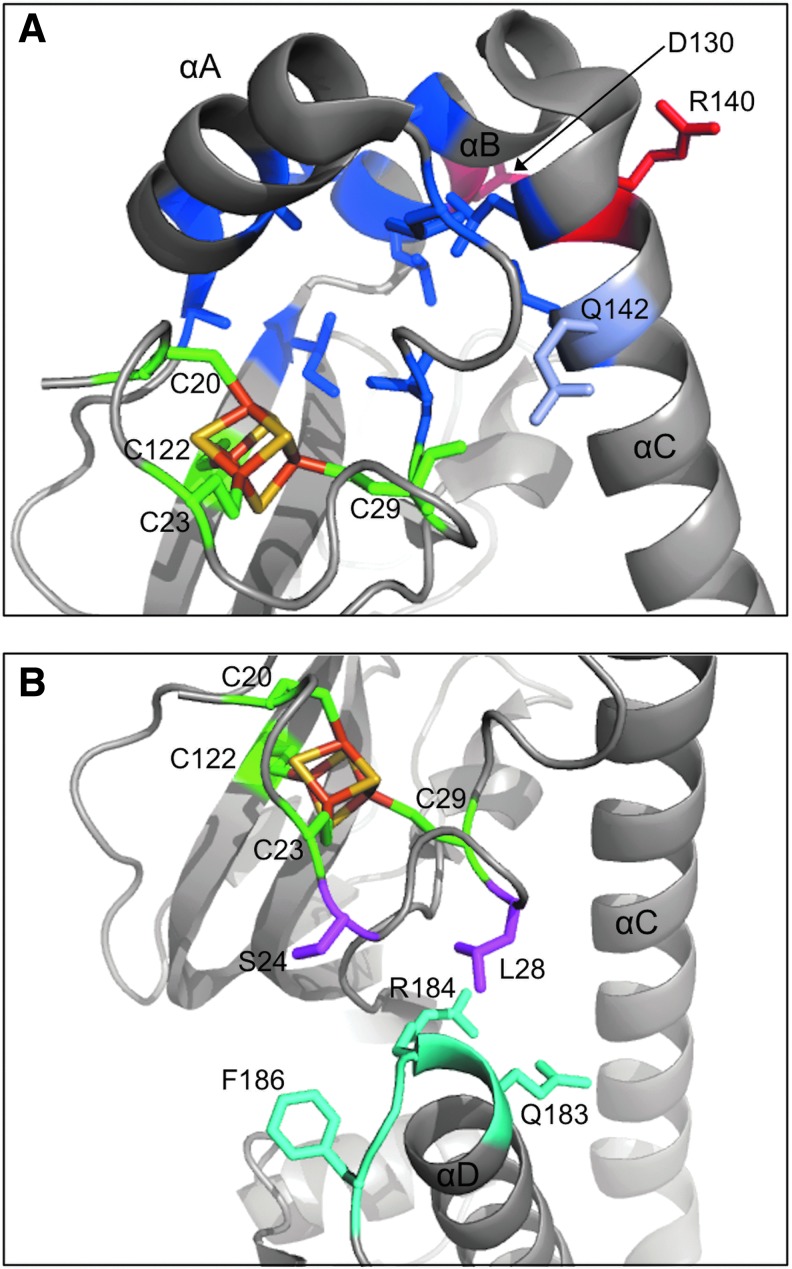
FIG. 2.
The [4Fe-4S] cluster binding region within FNR. The cluster is shown as orange and yellow sticks, and the protein is depicted in cartoon representation, with relevant α helices and amino acid side chains marked. Shown in green are the four cysteine cluster ligands (C20, C23, C29, and C122). (A) Proximal to the cluster is a patch of hydrophobic residues (shown in blue) that form a network of interactions. This hydrophobic patch likely provides communication between the cluster binding domain and the intersubunit R140–D130 salt bridges that contribute to dimerization (R140 and D130 are shown in red). (B) Facing away from the cluster are the side chains of residues S24 and L28 (magenta), which are proximal to residues 183–186 (cyan) of FNR AR1. AR, activating region.
In addition to [4Fe-4S]-FNR, Volbeda et al. solved the X-ray crystal structure of A. fischeri FNR with a partially degraded cluster. In this form, the electron density of the first 42 amino acids is not visible, suggesting that the cluster binding loop becomes disordered upon cluster degradation. Therefore, the [4Fe-4S]2+ cluster likely organizes this N-terminal region of FNR, but remains accessible to O2, a property presumed to be pivotal for FNR's physiological role.
The Structure Yields Additional Insight into Subunit Interactions That Are Necessary for FNR Dimerization
As discussed earlier, cluster binding is a major regulatory checkpoint for FNR since presence of the [4Fe-4S]2+ cluster dictates its oligomeric state. Only in the [4Fe-4S]2+ cluster-containing form is FNR capable of dimerization, which is required for affinity and specificity in DNA binding and hence transcriptional regulation (23, 32, 42). Conversely, keeping FNR apoprotein in a monomeric, inactive state under aerobic conditions is important for limiting gene expression to anaerobic conditions. Given the significance of this regulated oligomerization, biochemical and genetic studies were employed to characterize residues involved in subunit interaction. Specifically, the role of FNR residues 140–159 was evaluated by alanine mutagenesis as these correspond to the αC helix along which CRP dimerizes via a coiled coil motif (Fig. 1). It was found that mostly hydrophobic residues predicted to be on the same face of the αC helix (M144, M147, I151, I158) are important for dimerization (Fig. 3A). The isoleucine at position 151 appeared particularly compelling since substitution of this residue with alanine causes the most severe dimerization defects (54). In contrast, these studies also demonstrated that two negatively charged residues near the subunit interface deter dimerization of apo-FNR via subunit charge repulsion. Substitution of either D154 or to a lesser extent E150 with alanine results in dimeric, active FNR even in the absence of the cluster (55). Together, these findings suggested that cluster binding might induce a conformational rearrangement such that I151 would shield the negative charges of D154 and E150, thereby allowing energetically favorable subunit interactions between hydrophobic side chains. However, since the D154A substitution only partially rescues the I151A mutant phenotype, this indicated that I151 may have an additional role in FNR dimerization (54).
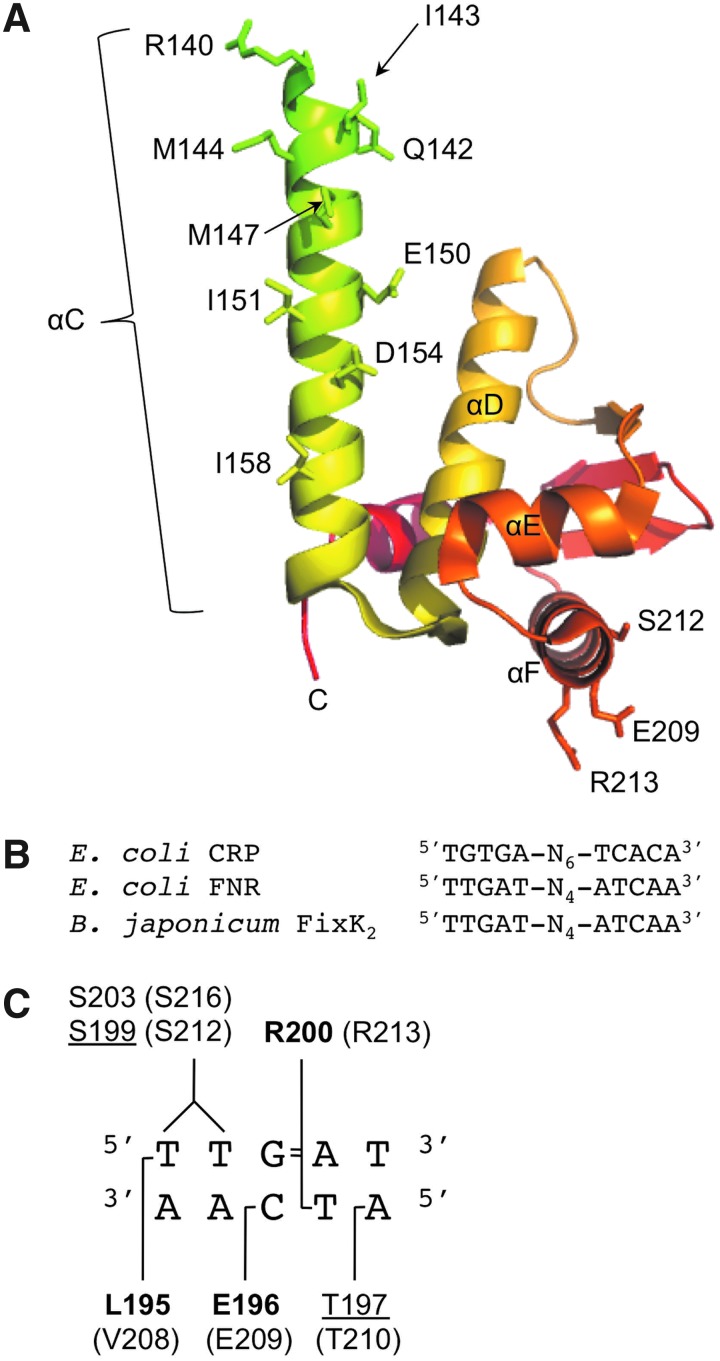
FIG. 3.
FNR residues important for dimerization and specific DNA recognition. (A) The FNR dimerization interface and DNA binding domain. Only shown are residues 140–246 of one FNR subunit, depicted in cartoon representation with relevant amino acid side chains shown as sticks. (B) The consensus DNA target sites recognized by CRP and FNR from E. coli, and FixK2 from B. japonicum. (C) The FixK2 residues involved in consensus half-site interactions. These residues form specific interactions with nucleotides (indicated in bold) or interact with the DNA phosphate backbone either directly or through water (underlined). Indicated in parentheses are residues at equivalent positions in E. coli FNR.
The structure has allowed refinement of this oligomerization model. As discussed by Volbeda et al., residues D154 and E150 from both subunits create a negatively charged pocket in [4Fe-4S]-FNR. Rather than shielding this negative charge as previously hypothesized, the I151 residues of each subunit establish van der Waals contacts. The proximity of these contacts to the inhibitory D154 residues may explain why I151 is especially critical for FNR oligomerization. Furthermore, the partial activity of the FNR-I151A-D154A double mutant suggests that even after removing negativity at the subunit interface, the van der Waals contacts between I151 residues make a significant contribution to maintaining dimer stability.
The structure has also clarified the role of R140 in FNR dimerization. A previous study showed that substitution of this positively charged residue with alanine, leucine, or glutamate results in substantial FNR activity defects (54). Consequently, it was hypothesized that R140, being at the start of the αC helix, may promote subunit interaction through formation of interhelical hydrogen bonds. Instead of making contacts at the coiled coil interface, however, the structure indicates that R140 of one subunit forms a salt bridge with D130 belonging to the αB helix of the opposite subunit (Figs. 1 and 2A) (73). Volbeda et al. propose that this salt bridge may be a critical determinant in the mechanism by which the [4Fe-4S]2+ cluster mediates FNR oligomerization. In light of this new structural information, it is surprising that when R140A is expressed at increased cellular levels, it exhibits activity similar to that of wild-type FNR (54). These findings suggest that at higher protein concentrations, the hydrophobic interactions and the I151 van der Waals contacts along the αC helix are sufficient to provide the energetic driving force needed for dimerization. However, at wild-type protein levels, the R140–D130 salt bridges, along with the aforementioned interactions, play a vital role in enabling the O2 sensitivity of the FNR monomer–dimer equilibrium.
The Structure Uncovers the Mechanism by Which O2 Mediated Cluster Conversion Leads to Loss of Dimerization
Before the structure, the cluster-induced conformational changes that propagate FNR oligomerization remained largely a mystery. Most CRP family members are constitutively dimeric. Furthermore, circular dichroism spectroscopy and limited proteolysis assays did not indicate any broad scale changes upon FNR cluster ligation (55). Indeed, these latter observations are corroborated by the structure, which suggests that conformational alterations may be more fine tuned. Volbeda et al. propose that the patch of hydrophobic interactions proximal to the [4Fe-4S]2+ cluster provides a communication relay between the cluster and the intramolecular R140–D130 salt bridges (Fig. 2A). Upon exposure of the [4Fe-4S]2+ cluster to O2, local rearrangements that take place to accommodate the resulting [2Fe-2S]2+ cluster may disseminate through this hydrophobic network to subsequently break the salt bridges. The resulting loss of binding energy would weaken stability at the top of the dimerization helix, which, in turn, may propagate unzipping of the coiled coil, and hence the loss of dimerization.
According to this model, disruption of the hydrophobic patch proximal to the [4Fe-4S]2+ cluster would likely alter FNR's ability to oligomerize. Consistent with this hypothesis is the phenotype of the previously characterized FNR-M143A mutant, which exhibits a significant dimerization defect (54). As shown in the structure, the residue at position 143 in A. fischeri FNR is an isoleucine that participates in the hydrophobic network (Figs. 2A and 3A) (73). In addition, the presence of a hydrophobic residue at this position is conserved among FNR homologs (54). Curiously, a different E. coli FNR mutant, FNR-Q142A, displayed slightly increased activity relative to wild-type FNR (54). Although the basis for this phenotype remains to be elucidated, it is tempting to speculate that Q142 may also play a role in linking [4Fe-4S]2+ cluster ligation and dimerization as the structure demonstrates that the Q142 side chain potentially interacts with the C29 cluster ligand main chain (Fig. 2A). Future work to solve the structures of [2Fe-2S]-FNR or apo-FNR will likely provide valuable information as to how this network of interactions is disrupted in the absence of the [4Fe-4S]2+ cluster, thereby inactivating FNR function.
The Structural Position of Residues S24 and L28 Emphasizes the Correlation Between Conformational Flexibility and Proper O2 Sensing
Tremendous insight into the functional form of FNR was delivered from genetic and biochemical characterization of FNR variants that retain activity in the presence of O2. In the case of substituting leucine at position 28 with histidine, it was found that the [4Fe-4S]2+ cluster was more resistant to O2 mediated conversion than wild-type FNR. Given its proximity to the cluster ligands, it was hypothesized that the imidazole ring of histidine may form a hydrogen bond with a cluster sulfide ion (3). A similar phenotype was observed upon substitution of S24 with F, R, H, W, or Y, suggesting that the presence of bulky amino acid side chains at this position may shield the [4Fe-4S]2+ cluster from O2 (29). However, based on the position of S24 and L28 in the FNR structure, which displays their side chains facing away from the [4Fe-4S] cluster, it is apparent that these residues affect cluster stability by a different mechanism (Fig. 2B).
Volbeda et al. propose that substitution of S24 or L28 with larger amino acids may hinder conformational flexibility of the cluster binding region such that increased rigidity would impede O2 mediated cluster destruction by restricting the rearrangement from binding a [4Fe-4S]2+ cluster to a [2Fe-2S]2+ cluster. In addition, S24 and L28 are in proximity to residues 183–186, which are part of an amino acid loop implicated in making contacts with α-C-terminal domain (CTD) of RNAP (Fig. 2B) (75, 76). The presence of smaller side chains at positions 24 and 28 is likely necessary to avoid steric hindrance with residues 183–186. Substitution of S24 or L28 may also establish new interactions with this amino acid stretch, potentially affecting plasticity of the structure. Ultimately, these findings suggest that for FNR, conformational flexibility of the cluster binding domain is important for its O2 sensing function, in addition to [4Fe-4S]2+ cluster accessibility.
Aside from the cluster ligands, the neighboring residues within the cluster-binding domain display variability even among FNR homologs. Considering the characteristics of the S24F and L28H mutants, this variability may account for the altered cluster sensitivities observed for FNR homologs, in which their clusters are either more susceptible (Azotobacter vinelandii CydR) or resistant (Neisseria meningitidis FNR) to destruction by O2 compared with E. coli FNR (18, 80). Indeed, it has been proposed that the cluster binding domain has evolved to tailor the appropriate O2 response according to the environmental niche (29).
Interestingly, although S24F and L28H mutants exhibit increased cluster stability, previous in vitro experiments have demonstrated that these variants display some defects in DNA binding and/or transcription activation (2, 57, 74). Together, these observations raise the question as to whether conformational flexibility is important for DNA and RNAP recognition, in addition to efficient O2 sensing. We address this question for the L28H mutant in further detail later in this review.
The FixK2-DNA Structure Has Broadened Our Understanding of FNR–DNA Interactions
Given that DNA binding is often a property among transcription factors that is critical for their function, several strategies have been used to map the contacts between the FNR HTH motif and its DNA recognition site (11, 41, 66). This work was initially guided by knowledge of CRP–DNA interactions, since primary and secondary structures of the HTH motif are relatively well conserved between CRP and FNR (65). In addition, these two proteins bind symmetrical DNA sites that share sequence similarity (Fig. 3B) (68). In fact, substitution of just two amino acids in the FNR–DNA binding domain with those at corresponding positions in CRP enables FNR to activate a CRP-dependent promoter (66). Both in vivo and in vitro assays revealed that FNR residues E209, S212, and R213 play a major role in DNA binding (41, 66). These residues belong to the αF helix of the HTH and were predicted to be surface exposed for making contacts with the DNA major groove. Indeed the FNR structure demonstrates that the side chains of E209, S212, and R213 are poised for potential DNA interactions (Fig. 3A) (73).
In recent years, the X-ray crystal structure of the protein–DNA complex containing the FNR homolog FixK2 from Bradyrhizobium japonicum was solved (8). Since the consensus DNA binding sequences for FNR and FixK2 are identical (Fig. 3B) (11), this cocrystal structure has provided deeper insight into putative FNR–DNA interactions. The FixK2–DNA complex reveals protein-induced DNA bending, a property presumed to be shared by FNR as suggested by DNA mobility shift assays and DNA sites hypersensitive to DNase I-mediated cleavage when FNR is present (51, 77, 83). The FixK2–DNA structure also demonstrates binding of each protein subunit to its cognate DNA half site by way of direct and indirect interactions. Specifically, the side chains of the E209 and R213 amino acid equivalents in FixK2 contact DNA bases through hydrogen bonds, whereas the side chain of the S212 equivalent makes interactions with the DNA phosphate backbone via water molecules (Fig. 3C). Additional FixK2–DNA contacts involve other residues of the αF helix, as well as nearby residues of the αE helix and the short loop between the dimerization αC helix and the αD helix. Based on this observation, it may be likely that the side chains of other FNR residues (e.g., R169, R197, T207, V208, T210, G216, G228) interact, either directly or through water, with DNA bases or the phosphate backbone. In the case of V208, the equivalent residue in FixK2 (L195) forms a hydrophobic interaction with the first T of the TTGAT half site (Fig. 3C). Interestingly, substitution of this valine in FNR with an arginine residue, which is present at the equivalent position in CRP, did not disrupt DNA binding. Rather, this FNR variant was able to activate both FNR- and CRP-dependent promoters (66). Together, these findings suggest that the hydrophobic interaction mediated by V208 may be critical in discriminating FNR and CRP recognition sites.
Although the FixK2–DNA complex has shed light on how FNR may specifically interact with DNA, differences in positioning of the HTH motif between these two proteins remain a possibility. Indeed, structures of other CRP family members suggest that upon effector binding, changes in αF helix orientation may occur to promote DNA recognition (79). FixK2 may be an exception since DNA binding activity is not controlled by the presence of an effector molecule, but rather the oxidation of a unique cysteine residue near its HTH motif (49). Furthermore, as already noted, FNR is unique in that cluster binding does not directly regulate DNA binding activity but instead induces the subunit dimerization required for DNA recognition. Thus, additional knowledge may be gained from an FNR–DNA cocrystal structure. This desired structure would not only confirm presumed FNR–DNA contacts, but it would also reveal alterations in protein conformation that may occur subsequent to DNA binding. These putative conformational changes may explain why FNR bound to DNA exhibited an increased rate of O2 mediated cluster degradation as compared with unbound FNR in vitro (14).
Structural Differences Between FNR and CRP Likely Contribute to Distinct Interactions with RNAP
For the CRP/FNR family of transcription factors, direct contacts with RNAP are necessary for transcription activation. Identifying these contacts has been the focus of several studies, which were guided by known CRP–RNAP interactions elucidated from experimental data and the CRP–α-CTD cocrystal structure (5, 10). Researchers established that like CRP, FNR contains three individual activating regions (ARs) that mediate contacts with RNAP (Fig. 4A). These include AR1, AR2, and AR3, predicted to recognize the α-CTD, the α-N-terminal domain, and σ70, respectively (43). Furthermore, the influence of each AR on transcription activation appears to depend on promoter architecture (Fig. 4B). At class I promoters in which the FNR recognition site is centered at −61.5 bp or further upstream of the +1 transcription start site (TSS), AR1–α-CTD interactions are required for transcription activation (44, 75, 76, 78). However, the mode of RNAP recognition differs for class II promoters, which constitute the most frequently occurring FNR-dependent promoters and contain the FNR target site ∼41.5 bp upstream of the TSS. At class II promoters, all three ARs have the potential to make contacts with RNAP, with AR1 and AR3 having predominant roles and AR2 making only a minor contribution (7, 39, 40, 45, 75, 77, 78). Consistent with this notion, the structure demonstrates that AR3 forms a surface exposed loop that is poised to make interactions with σ70 as previously predicted by alanine mutagenesis (40).
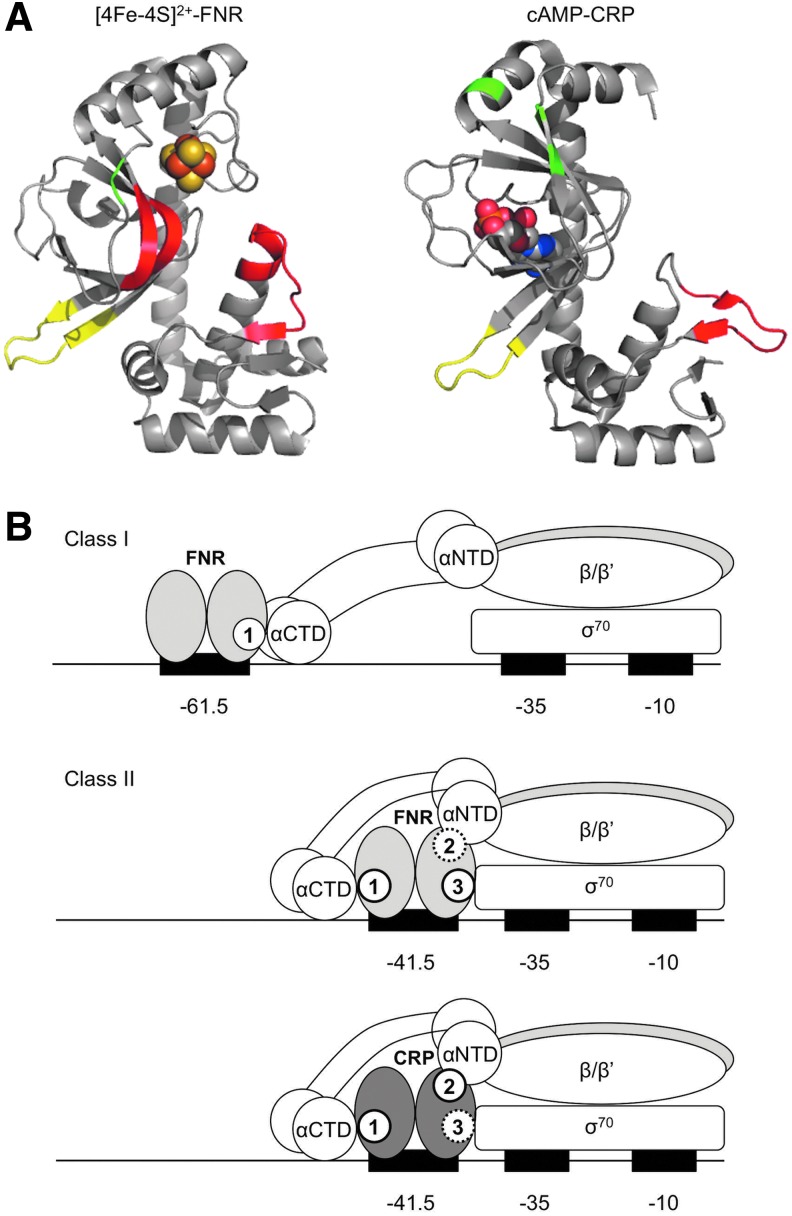
FIG. 4.
Developing a model for FNR transcription activation. (A) The ARs within FNR and CRP that are involved in interacting with RNAP. Shown are the monomeric crystal structures for [4Fe-4S]-FNR [PDB code 5E44, Ref. (73)] and cAMP-CRP [PDB code 1G6N, Ref. (59)] in cartoon representation with their respective cofactors shown as spheres and ARs highlighted: AR1 (red), AR2 (green), AR3 (yellow). (B) Promoters that are activated by FNR can be categorized into two main classes. At a class I promoter, the FNR binding site is centered ∼61.5 bases or further upstream of the transcriptional start site (+1), allowing FNR to make contacts with RNAP through AR1 in the downstream subunit. At a class II promoter, the FNR binding site is centered ∼41.5 bases upstream of the +1, and FNR is poised to make multiple contacts with RNAP through AR1 in the upstream subunit and AR2 and AR3 in the downstream subunit, although AR2 plays only a minor role. In contrast to that of FNR, in class II promoter activation mediated by CRP, AR2 plays a larger role and AR3 does not normally appear to be functional. cAMP-CRP, cyclic adenosine monophosphate-cAMP receptor protein.
Although some contacts with RNAP appear to be conserved between FNR and CRP, evidence suggests that significant functional differences exist for the ARs of these two transcription factors. For instance, in contrast to that of FNR, CRP AR2 appears to play a larger role and CRP AR3 does not normally appear to be functional in class II promoter activation unless a lysine at position 52 is mutated (10, 62). In addition, FNR–AR1 displays a significantly broader interacting surface than CRP (Fig. 4A). The ARs may also exhibit distinct transcription initiation mechanisms for FNR and CRP. CRP AR1 and AR2 enhance binding of RNAP to class II promoters and the rate of open complex formation, respectively (10). In contrast, FNR AR1 accelerates RNAP isomerization from a closed to an open complex at an FNR-dependent synthetic class II promoter (77). Although further work is needed to solve the FNR–RNAP cocrystal structure to elevate our understanding of the protein–protein interactions required for transcription activation, it is intriguing that the FNR cluster binding domain is in proximity to AR1, specifically the 183–186 amino acid stretch (Fig. 2B). It is possible that this proximity would permit communication between the cluster binding domain and AR1 upon cluster ligation. This is in contrast to CRP whose structure shows that the cAMP binding pocket is distally located from AR1 (Fig. 4A). This variation may at least partially explain the mechanistic differences between FNR and CRP transcription activation.
The Structure Reveals an Unexpected Link Between the Cluster Binding Domain and AR1
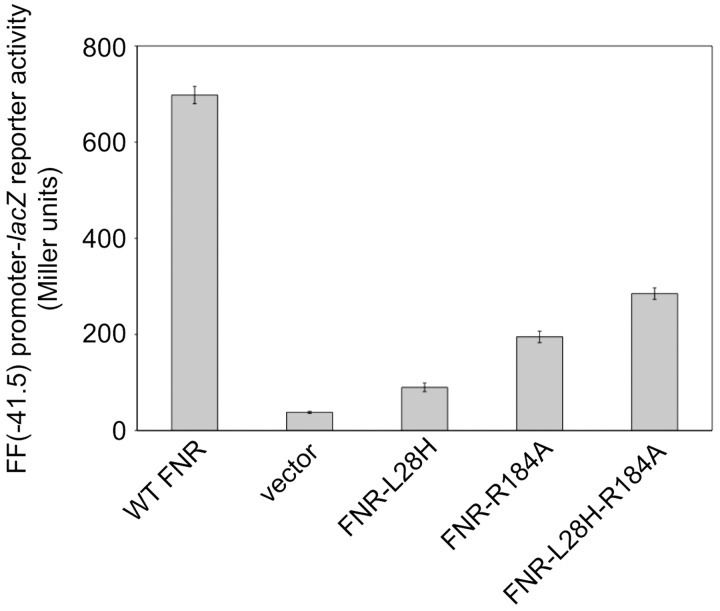
In a previous study, FNR residue R184 was proposed to be an AR1 determinant based on the observation that alanine substitution at this position resulted in decreased class II promoter activation but no change in DNA binding (75). According to the structure, the R184 side chain does not appear to be in a position to make direct contacts with RNAP (Fig. 2B). Notably, this residue is part of the 183–186 amino acid stretch that is in proximity to the cluster binding domain. As discussed earlier, this domain encompasses L28, which upon substitution with histidine results in increased [4Fe-4S]2+ cluster stability. Volbeda et al. hypothesize that a reduction in conformational flexibility of the cluster binding domain may account for this phenotype. Conformational inflexibility of the FNR-L28H mutant may also explain its general defect in activating transcription compared with wild-type [4Fe-4S]2+-FNR (3, 35, 57, 74). Data in Figure 5 show that anaerobic expression of a synthetic class II promoter is decreased nearly eightfold when FNR-L28H is present. Although this may be explained, in part, by a decrease in FNR-L28H DNA binding affinity, in vitro experiments from our laboratory indicate that this mutant is indeed less capable of activating transcription (2, 74). Interestingly, we found that introducing the R184A substitution results in an approximate threefold increase in FNR-L28H activity (Fig. 5). To explain this finding, we hypothesize that a histidine at position 28 may either interact or cause steric hindrance with R184, thereby affecting FNR-L28H transcription activation. By replacing R184 with a smaller amino acid, some conformational flexibility in the cluster binding region of FNR-L28H may be restored, thereby partially complementing the defect in activity. Together, these findings not only stress the significance of conformational flexibility in FNR's ability to carry out its physiological role, they also reveal an unexpected link between the cluster binding region and AR1 such that presence of the [4Fe-4S]2+ cluster may promote AR1 and RNAP interactions. This link may also explain why anaerobic transcription activation by the constitutive dimer mutant FNR-D154A is more than eightfold higher than that of double mutant variants (FNR-D154A-C23S and FNR-D154A-C122S) that lack the [4Fe-4S]2+ cluster (data not shown). Future investigation is needed to dissect the mechanism by which cluster binding may influence interactions between AR1 and RNAP.
FIG. 5.
The transcription activation defect displayed by the FNR-L28H mutant is partially restored by the R184A substitution. β-Galactosidase activity (given in Miller units) from the FNR-dependent synthetic class II FF(−41.5) promoter-lacZ reporter was measured in strains expressing wild-type FNR, FNR-L28H, FNR-R184A, or FNR-L28H-R184A from plasmid pET11a. These strains lack the chromosomal copy of fnr. Cultures were grown under anaerobic growth conditions in M9 minimal medium containing 0.2% glucose and 50 μg/mL ampicillin. Error bars represent the standard deviation of β-galactosidase activity measured from three independent strain isolates.
Future Directions
Despite the wealth of information provided by decades of research, many aspects of FNR function are still unresolved. This is largely true for the N-terminal cluster binding domain, in which electron density was not visible for the first 18 residues. In fact, residues within this amino acid stretch have been shown to play a critical role in regulating FNR activity. For instance, a mutant FNR lacking residues 2–15 surprisingly exhibited increased cluster stability toward O2 compared with the wild-type protein (81). It is possible that these residues are needed to maintain the cluster ligands in an orientation for appropriate O2 sensing. This region also encompasses one of the two target sequences in FNR that are recognized by the ClpXP protease (residues 5–11 and 249–250) (21). Although monomeric FNR is degraded by this protease, dimeric FNR escapes proteolysis (50). Thus, it is not clear how the ClpXP target sequences would be protected in the dimerized form. Addressing these questions will likely be challenging given the conformational flexibility of the FNR N-terminal region.
Future cocrystal structures will be helpful in further elucidating FNR contacts with RNAP, ClpXP, or with other proteins implicated in associating with FNR. For example, FNR must interact with carrier proteins of Fe-S biogenesis pathways that facilitate delivery of its [4Fe-4S]2+ cluster (52). FNR has also been implicated in binding the NarL transcription factor for activation of nirB (71). Lastly, there is evidence that at some promoters, repression of transcription depends on specific interactions between two tandem-bound FNR dimers (1, 25, 46). The latter point simultaneously stresses how an FNR–DNA cocrystal is likewise needed, not only for further defining FNR–DNA contacts but also because the mechanism(s) of FNR-mediated repression is not well understood.
Finally, we would greatly benefit from structures of FNR homologs from other bacteria. These structures would provide further insight as to how variation in the cluster binding domain might tune FNR's O2 sensing function to a particular bacterial niche. Furthermore, they would address the extent to which the cluster-induced dimerization mechanism proposed by Volbeda et al. is conserved. Indeed, variation of FNR regulation is readily apparent in Gram-positive Bacillus subtilis and Bacillus cereus, in which FNR utilizes different cluster ligands than that of E. coli FNR and is dimeric even in the apoprotein form (19, 27, 61). In summary, there is still much to discover regarding the versatility of the FNR transcription factor.
Abbreviations Used
- AR
activating region
- ATP
adenosine 5′-triphosphate
- cAMP
cyclic adenosine monophosphate
- CO
carbon monoxide
- CRP
cAMP receptor protein
- CTD
C-terminal domain
- DNA
deoxyribonucleic acid
- FNR
fumarate nitrate reduction
- HTH
helix-turn-helix
- NO
nitric oxide
- PDB
Protein Data Bank
- RNA
ribonucleic acid
- RNAP
RNA polymerase
- TSS
transcription start site
Acknowledgments
We are grateful to Dr. Derek Weber, whose work is presented in Figure 5, and to Dr. James Keck for his valuable insight into protein structure. This work was funded by NIH grant Nos. GM045844 and GM115894 to P.J.K.
References
- 1.Barnard AM, Green J, and Busby SJ. Transcription regulation by tandem-bound FNR at Escherichia coli promoters. J Bacteriol 185: 5993–6004, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates DM, Lazazzera BA, and Kiley PJ. Characterization of FNR* mutant proteins indicates two distinct mechanisms for altering oxygen regulation of the Escherichia coli transcription factor FNR. J Bacteriol 177: 3972–3978, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates DM, Popescu CV, Khoroshilova N, Vogt K, Beinert H, Münck E, and Kiley PJ. Substitution of leucine 28 with histidine in the Escherichia coli transcription factor FNR results in increased stability of the [4Fe-4S]2+ cluster to oxygen. J Biol Chem 275: 6234–6240, 2000 [DOI] [PubMed] [Google Scholar]
- 4.Beauchene NA, Myers KS, Chung D, Park DM, Weisnicht AM, Keleş S, and Kiley PJ. Impact of anaerobiosis on expression of the iron-responsive Fur and RyhB regulons. MBio 6: e01947–15, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, et al. Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science 297: 1562–1566, 2002 [DOI] [PubMed] [Google Scholar]
- 6.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, et al. The protein data bank. Nucleic Acids Res 28: 235–242, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blake T, Barnard A, Busby SJ, and Green J. Transcription activation by FNR: evidence for a functional activating region 2. J Bacteriol 184: 5855–5861, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet M, Kurz M, Mesa S, Briand C, Hennecke H, and Grutter MG. The structure of Bradyrhizobium japonicum transcription factor FixK2 unveils sites of DNA binding and oxidation. J Biol Chem 288: 14238–14246, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bueno E, Mesa S, Bedmar EJ, Richardson DJ, and Delgado MJ. Bacterial adaptation of respiration from oxic to microoxic and anoxic conditions: redox control. Antioxid Redox Signal 16: 819–852, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busby S. and Ebright RH. Transcription activation by catabolite activator protein (CAP). J Mol Biol 293: 199–213, 1999 [DOI] [PubMed] [Google Scholar]
- 11.Cherfils J, Gibrat JF, Levin J, Batut J, and Kahn D. Model-building of Fnr and FixK DNA-binding domains suggests a basis for specific DNA recognition. J Mol Recognit 2: 114–121, 1989 [DOI] [PubMed] [Google Scholar]
- 12.Constantinidou C, Hobman JL, Griffiths L, Patel MD, Penn CW, Cole JA, and Overton TW. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J Biol Chem 281: 4802–4815, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Crack JC, Green J, Thomson AJ, and Le Brun NE. Iron-sulfur clusters as biological sensors: the chemistry of reactions with molecular oxygen and nitric oxide. Accounts Chem Res 47: 3196–3205, 2014 [DOI] [PubMed] [Google Scholar]
- 14.Crack JC, Stapleton MR, Green J, Thomson AJ, and Le Brun NE. Influence of association state and DNA binding on the O2-reactivity of [4Fe-4S] fumarate and nitrate reduction (FNR) regulator. Biochem J 463: 83–92, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crack JC, Thomson AJ, and Le Brun NE. Mass spectrometric identification of intermediates in the O2-driven [4Fe-4S] to [2Fe-2S] cluster conversion in FNR. Proc Natl Acad Sci U S A 114: E3215–E3223, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiley PJ. and Donohue T. Global Responses of Bacteria to O2 Deprivation. In: Bacterial Stress Responses, edited by Storz G. and Hengge R. Washington, DC: ASM Press, 2011, pp. 175–189 [Google Scholar]
- 17.Dufour YS, Kiley PJ, and Donohue TJ. Reconstruction of the core and extended regulons of global transcription factors. PLoS Genet 6: e1001027, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards J, Cole LJ, Green JB, Thomson MJ, Wood AJ, Whittingham JL, and Moir JWB. Binding to DNA protects Neisseria meningitidis fumarate and nitrate reductase regulator (FNR) from oxygen. J Biol Chem 285: 1105–1112, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esbelin J, Jouanneau Y, and Duport C. Bacillus cereus Fnr binds a [4Fe-4S] cluster and forms a ternary complex with ResD and PlcR. BMC Microbiol 12: 125, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Federowicz S, Kim D, Ebrahim A, Lerman J, Nagarajan H, Cho BK, Zengler K, and Paisson B. Determining the control circuitry of redox metabolism at the genome-scale. PLoS Genet 10: e1004264, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flynn JM, Neher SB, Kim YI, Sauer RT, and Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell 11: 671–683, 2003 [DOI] [PubMed] [Google Scholar]
- 22.Grainger DC, Aiba H, Hurd D, Browning DF, and Busby SJ. Transcription factor distribution in Escherichia coli: studies with FNR protein. Nucleic Acids Res 35: 269–278, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green J, Bennett B, Jordan P, Ralph ET, Thomson AJ, and Guest JR. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem J 316: 887–892, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green J, Crack JC, Thomson AJ, and LeBrun NE. Bacterial sensors of oxygen. Curr Opin Microbiol 12: 145–151, 2009 [DOI] [PubMed] [Google Scholar]
- 25.Green J. and Marshall FA. Identification of a surface of FNR overlapping activating region 1 that is required for repression of gene expression. J Biol Chem 274: 10244–10248, 1999 [DOI] [PubMed] [Google Scholar]
- 26.Green J, Stapleton MR, Smith LJ, Artymiuk PJ, Kahramanoglou C, Hunt DM, and Buxton RS. Cyclic-AMP and bacterial cyclic-AMP receptor proteins revisited: adaptation for different ecological niches. Curr Opin Microbiol 18: 1–7, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruner I, Fradrich C, Bottger LH, Trautwein AX, Jahn D, and Hartig E. Aspartate 141 is the fourth ligand of the oxygen-sensing [4Fe-4S]2+ cluster of Bacillus subtilis transcriptional regulator Fnr. J Biol Chem 286: 2017–2021, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11: 443–454, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jervis AJ, Crack JC, White G, Artymiuk PJ, Cheesman MR, Thomson AJ, Le Brun NE, and Green J. The O2 sensitivity of the transcription factor FNR is controlled by Ser24 modulating the kinetics of [4Fe-4S] to [2Fe-2S] conversion. Proc Natl Acad Sci U S A 106: 4659–4664, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jordan PA, Thomson AJ, Ralph ET, Guest JR, and Green J. FNR is a direct oxygen sensor having a biphasic response curve. FEBS Lett 416: 349–352, 1997 [DOI] [PubMed] [Google Scholar]
- 31.Kang Y, Weber KD, Qiu Y, Kiley PJ, and Blattner FR. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol 187: 1135–1160, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khoroshilova N, Beinert H, and Kiley PJ. Association of a polynuclear iron-sulfur center with a mutant FNR protein enhances DNA binding. Proc Natl Acad Sci U S A 92: 2499–2503, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khoroshilova N, Popescu C, Münck E, Beinert H, and Kiley PJ. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc Natl Acad Sci U S A 94: 6087–6092, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiley PJ. and Beinert H. Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol Rev 22: 341–352, 1999 [DOI] [PubMed] [Google Scholar]
- 35.Kiley PJ. and Reznikoff WS. Fnr mutants that activate gene expression in the presence of oxygen. J Bacteriol 173: 16–22, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolb A, Busby S, Buc H, Garges S, and Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem 62: 749–795, 1993 [DOI] [PubMed] [Google Scholar]
- 37.Körner H, Sofia HJ, and Zumft WG. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol Rev 27: 559–592, 2003 [DOI] [PubMed] [Google Scholar]
- 38.Lambden PR. and Guest JR. Mutants of Escherichia coli K12 unable to use fumarate as an anaerobic electron acceptor. J Gen Microbiol 97: 145–160, 1976 [DOI] [PubMed] [Google Scholar]
- 39.Lamberg KE. and Kiley PJ. FNR-dependent activation of the class II dmsA and narG promoters of Escherichia coli requires FNR-activating regions 1 and 3. Mol Microbiol 38: 817–827, 2000 [DOI] [PubMed] [Google Scholar]
- 40.Lamberg KE, Luther C, Weber KD, and Kiley PJ. Characterization of activating region 3 from Escherichia coli FNR. J Mol Biol 315: 275–283, 2002 [DOI] [PubMed] [Google Scholar]
- 41.Lazazzera BA, Bates DM, and Kiley PJ. The activity of the Escherichia coli transcription factor FNR is regulated by a change in oligomeric state. Genes Dev 7: 1993–2005, 1993 [DOI] [PubMed] [Google Scholar]
- 42.Lazazzera BA, Beinert H, Khoroshilova N, Kennedy MC, and Kiley PJ. DNA binding and dimerization of the Fe-S-containing FNR protein from Escherichia coli are regulated by oxygen. J Biol Chem 271: 2762–2768, 1996 [DOI] [PubMed] [Google Scholar]
- 43.Lee DJ, Minchin SD, and Busby SJ. Activating transcription in bacteria. Annu Rev Microbiol 66: 125–152, 2012 [DOI] [PubMed] [Google Scholar]
- 44.Lee DJ, Wing HJ, Savery NJ, and Busby SJ. Analysis of interactions between Activating Region 1 of Escherichia coli FNR protein and the C-terminal domain of the RNA polymerase alpha subunit: use of alanine scanning and suppression genetics. Mol Microbiol 37: 1032–1040, 2000 [DOI] [PubMed] [Google Scholar]
- 45.Li B, Wing H, Lee D, Wu HC, and Busby S. Transcription activation by Escherichia coli FNR protein: similarities to, and differences from, the CRP paradigm. Nucleic Acids Res 26: 2075–2081, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marshall FA, Messenger SL, Wyborn NR, Guest JR, Wing H, Busby SJW, and Green J. A novel promoter architecture for microaerobic activation by the anaerobic transcription factor FNR. Mol Microbiol 39: 747–753, 2001 [DOI] [PubMed] [Google Scholar]
- 47.Matsui M, Tomita M, and Kanai A. Comprehensive computational analysis of bacterial CRP/FNR superfamily and its target motifs reveals stepwise evolution of transcriptional networks. Genome Biol Evol 5: 267–282, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melville SB. and Gunsalus RP. Mutations in fnr that alter anaerobic regulation of electron transport-associated genes in Escherichia coli. J Biol Chem 265: 18733–18736, 1990 [PubMed] [Google Scholar]
- 49.Mesa S, Reutimann L, Fischer HM, and Hennecke H. Posttranslational control of transcription factor FixK2, a key regulator for the Bradyrhizobium japonicum-soybean symbiosis. Proc Natl Acad Sci U S A 106: 21860–21865, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mettert EL. and Kiley PJ. ClpXP-dependent proteolysis of FNR upon loss of its O2-sensing [4Fe-4S] cluster. J Mol Biol 354: 220–232, 2005 [DOI] [PubMed] [Google Scholar]
- 51.Mettert EL. and Kiley PJ. Contributions of [4Fe-4S]-FNR and integration host factor to fnr transcriptional regulation. J Bacteriol 189: 3036–3043, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mettert EL, Outten FW, Wanta B, and Kiley PJ. The impact of O2 on the Fe-S cluster biogenesis requirements of Escherichia coli FNR. J Mol Biol 384: 798–811, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meyer J. Iron-sulfur protein folds, iron-sulfur chemistry, and evolution. J Biol Inorg Chem 13: 157–170, 2008 [DOI] [PubMed] [Google Scholar]
- 54.Moore LJ. and Kiley PJ. Characterization of the dimerization domain in the FNR transcription factor. J Biol Chem 276: 45744–45750, 2001 [DOI] [PubMed] [Google Scholar]
- 55.Moore LJ, Mettert EL, and Kiley PJ. Regulation of FNR dimerization by subunit charge repulsion. J Biol Chem 281: 33268–33275, 2006 [DOI] [PubMed] [Google Scholar]
- 56.Myers KS, Yan H, Ong IM, Chung D, Liang K, Tran F, Keleş S, Landick R, and Kiley PJ. Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet 9: e1003565, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Overton T, Reid EG, Foxall R, Smith H, Busby SJ, and Cole JA. Transcription activation at Escherichia coli FNR-dependent promoters by the gonococcal FNR protein: effects of a novel S18F substitution and comparisons with the corresponding substitution in E. coli FNR. J Bacteriol 185: 4734–4747, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Overton TW, Griffiths L, Patel MD, Hobman JL, Penn CW, Cole JA, and Constantinidou C. Microarray analysis of gene regulation by oxygen, nitrate, nitrite, FNR, NarL and NarP during anaerobic growth of Escherichia coli: new insights into microbial physiology. Biochem Soc T 34: 104–107, 2006 [DOI] [PubMed] [Google Scholar]
- 59.Passner JM, Schultz SC, and Steitz TA. Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 Å resolution. J Mol Biol 304: 847–859, 2000 [DOI] [PubMed] [Google Scholar]
- 60.Popescu CV, Bates DM, Beinert H, Münck E, and Kiley PJ. Mössbauer spectroscopy as a tool for the study of activation/inactivation of the transcription regulator FNR in whole cells of Escherichia coli. Proc Natl Acad Sci U S A 95: 13431–13435, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reents H, Gruner I, Harmening U, Bottger LH, Layer G, Heathcote P, Trautwein AX, Jahn D, and Härtig E. Bacillus subtilis Fnr senses oxygen via a [4Fe-4S] cluster coordinated by three cysteine residues without change in the oligomeric state. Mol Microbiol 60: 1432–1445, 2006 [DOI] [PubMed] [Google Scholar]
- 62.Rhodius VA. and Busby SJ. Transcription activation by the Escherichia coli cyclic AMP receptor protein: determinants within activating region 3. J Mol Biol 299: 295–310, 2000 [DOI] [PubMed] [Google Scholar]
- 63.Salmon K, Hung SP, Mekjian K, Baldi P, Hatfield GW, and Gunsalus RP. Global gene expression profiling in Escherichia coli K12. The effects of oxygen availability and FNR. J Biol Chem 278: 29837–29855, 2003 [DOI] [PubMed] [Google Scholar]
- 64.Sharrocks AD, Green J, and Guest JR. In vivo and in vitro mutants of FNR the anaerobic transcriptional regulator of E. coli. FEBS Lett 270: 119–122, 1990 [DOI] [PubMed] [Google Scholar]
- 65.Shaw DJ, Rice DW, and Guest JR. Homology between CAP and Fnr, a regulator of anaerobic respiration in Escherichia coli. J Mol Biol 166: 241–247, 1983 [DOI] [PubMed] [Google Scholar]
- 66.Spiro S, Gaston KL, Bell AI, Roberts RE, Busby SJ, and Guest JR. Interconversion of the DNA-binding specificities of two related transcription regulators, CRP and FNR. Mol Microbiol 4: 1831–1838, 1990 [DOI] [PubMed] [Google Scholar]
- 67.Spiro S. and Guest JR. Inactivation of the FNR protein of Escherichia coli by targeted mutagenesis in the N-terminal region. Mol Microbiol 2: 701–707, 1988 [DOI] [PubMed] [Google Scholar]
- 68.Spiro S. and Guest JR. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol Rev 6: 399–428, 1990 [DOI] [PubMed] [Google Scholar]
- 69.Sutton VR, Mettert EL, Beinert H, and Kiley PJ. Kinetic analysis of the oxidative conversion of the [4Fe-4S]2+ cluster of FNR to a [2Fe-2S]2+ cluster. J Bacteriol 186: 8018–8025, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sutton VR, Stubna A, Patschkowski T, Münck E, Beinert H, and Kiley PJ. Superoxide destroys the [2Fe-2S]2+ cluster of FNR from Escherichia coli. Biochemistry 43: 791–798, 2004 [DOI] [PubMed] [Google Scholar]
- 71.Tyson KL, Bell AI, Cole JA, and Busby SJ. Definition of nitrite and nitrate response elements at the anaerobically inducible Escherichia coli nirB promoter: interactions between FNR and NarL. Mol Microbiol 7: 151–157, 1993 [DOI] [PubMed] [Google Scholar]
- 72.Unden G, Achebach S, Holighaus G, Tran HG, Wackwitz B, and Zeuner Y. Control of FNR function of Escherichia coli by O2 and reducing conditions. J Mol Microbiol Biotechnol 4: 263–268, 2002 [PubMed] [Google Scholar]
- 73.Volbeda A, Darnault C, Renoux O, Nicolet Y, and Fontecilla-Camps JC. The crystal structure of the global anaerobic transcriptional regulator FNR explains its extremely fine-tuned monomer-dimer equilibrium. Sci Adv 1: e1501086, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Weber KD. The Role of FNR in Transcriptional Regulation in Escherichia coli. PhD Dissertation. University of Wisconsin-Madison, Madison, WI, 2004 [Google Scholar]
- 75.Weber KD, Vincent OD, and Kiley PJ. Additional determinants within Escherichia coli FNR activating region 1 and RNA polymerase alpha subunit required for transcription activation. J Bacteriol 187: 1724–1731, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams SM, Savery NJ, Busby SJ, and Wing HJ. Transcription activation at class I FNR-dependent promoters: identification of the activating surface of FNR and the corresponding contact site in the C-terminal domain of the RNA polymerase alpha subunit. Nucleic Acids Res 25: 4028–4034, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wing HJ, Green J, Guest JR, and Busby SJ. Role of activating region 1 of Escherichia coli FNR protein in transcription activation at class II promoters. J Biol Chem 275: 29061–29065, 2000 [DOI] [PubMed] [Google Scholar]
- 78.Wing HJ, Williams SM, and Busby SJ. Spacing requirements for transcription activation by Escherichia coli FNR protein. J Bacteriol 177: 6704–6710, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Won HS, Lee YS, Lee SH, and Lee BJ. Structural overview on the allosteric activation of cyclic AMP receptor protein. Biochim Biophys Acta 1794: 1299–1308, 2009 [DOI] [PubMed] [Google Scholar]
- 80.Wu G, Cruz-Ramos H, Hill S, Green J, Sawers G, and Poole RK. Regulation of cytochrome bd expression in the obligate aerobe Azotobacter vinelandii by CydR (Fnr). Sensitivity to oxygen, reactive oxygen species, and nitric oxide. J Biol Chem 275: 4679–4686, 2000 [DOI] [PubMed] [Google Scholar]
- 81.Yan A. and Kiley PJ. Dissecting the role of the N-terminal region of the Escherichia coli global transcription factor FNR. J Bacteriol 190: 8230–8233, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang B, Crack JC, Subramanian S, Green J, Thomson AJ, et al. Reversible cycling between cysteine persulfide-ligated [2Fe-2S] and cysteine-ligated [4Fe-4S] clusters in the FNR regulatory protein. Proc Natl Acad Sci U S A 109: 15734–15739, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ziegelhoffer EC. and Kiley PJ. In vitro analysis of a constitutively active mutant form of the Escherichia coli global transcription factor FNR. J Mol Biol 245: 351–361, 1995 [DOI] [PubMed] [Google Scholar]