Abstract
IFN-related pathways have not been studied in morphea, and biomarkers are needed. We sought to characterize morphea serum cytokine imbalance and IFN-related gene expression in blood and skin to address this gap by performing a case-control study of 87 participants with morphea and 26 healthy control subjects. We used multiplexed immunoassays to determine serum cytokine concentrations, performed transcriptional profiling of whole blood and lesional morphea skin, and used double-staining immunohistochemistry to determine the cutaneous cellular source of CXCL9. We found that CXCL9 was present at increased concentrations in morphea serum (P < 0.0001), as were other T helper type 1 cytokines. CXCL9 serum concentration correlated with the modified Localized Scleroderma Skin Severity Index (r = 0.44, P = 0.0001), a validated measure of disease activity. CXCL9 gene expression was also increased in inflammatory lesional morphea skin (fold change = 30.6, P = 0.006), and preliminary transcriptional profiling showed little evidence for IFN signature in whole blood. Double-staining immunohistochemistry showed CXCL9 co-localized with CD68+ dermal macrophages. In summary, inflammatory morphea is characterized by T helper type 1 cytokine imbalance in serum, particularly CXCL9, which is associated with disease activity. CXCL9 expression in lesional macrophages implicates the skin as the source of circulating cytokines. CXCL9 is a promising biomarker of disease activity in morphea.
INTRODUCTION
Morphea, also known as localized scleroderma, produces sclerosis of skin and underlying soft tissues and is thought to exist in the spectrum of scleroderma-like disorders (Fett and Werth, 2011; Kreuter, 2012). The best described of these is systemic sclerosis (SSc), which is characterized by immune dysregulation, vascular injury, and sclerosis affecting multiple organs (Ferri et al., 2002; Matucci-Cerinic et al., 2013). Dysregulation of IFN-inducible gene expression in peripheral blood in SSc has been shown in multiple cohorts, and related chemokines correlating with disease severity have been identified (Assassi et al., 2010; Gourh et al., 2009). Similar changes have been reported in the skin of SSc (Assassi et al., 2015; Hinchcliff et al., 2013). These findings have led authors to suggest that IFN-related chemokines may be a biomarker and therapeutic target in SSc. Increasing evidence suggests that although histologically similar, morphea and systemic sclerosis are different in terms of clinical presentation and underlying pathogenesis. This raises the question of whether similar dysregulation of the IFN pathway exists in morphea and, if present, whether there is any association with clinical features of morphea.
Biomarkers are needed in morphea. Although some patients present with clearly active and severe disease, many present with lesions in evolution, where the degree of activity is uncertain and the potential for extension of lesions is unknown. Because the goal of therapy is to shut down active disease to abrogate permanent sequelae such as limb length discrepancy, assessing disease activity is crucial to management. To date, the only tools available to the practitioner are clinical examination or musculoskeletal imaging, which are limited to the evaluation of visible erythema, new or extending lesions, or anatomic location and extent of the lesions.
Although numerous potential biomarkers have been reported in the sera of patients with morphea (IL-2, IL-4, IL-6, IL-8, IL-13, IP-10 [CXCL10], and tumor necrosis factor-α [TNF-α]), few have been systematically studied in a large cohort (Hasegawa et al., 2003; Ihn et al., 1994, 1995; Magee et al., 2013; Torok et al., 2015). Studies examining the association of these cytokines with validated measures of disease activity or severity are also sparse. These data indicate that cytokines may be promising biomarkers in morphea, but further study is needed.
To characterize IFN-regulated pathways and potential cytokine markers of disease activity and severity, we undertook a three-part study of participants in the Morphea in Adults and Children (MAC) cohort including (i) a case-control study to identify cytokines and chemokines that are dysregulated in morphea using multiplexed immunoassays and transcriptional profiling of serum and whole blood, respectively; (ii) longitudinal examination of serum cytokine concentrations during active and inactive disease; and (iii) determination of the cellular source of observed chemokines associated with morphea using polymerase chain reaction and immunohistochemistry.
RESULTS
Demographic and clinical information
This study included a total of 87 participants with morphea from the prospective MAC cohort and 26 healthy subjects selected for similarity in age, sex, and race. Reasons for exclusion were presence of other autoimmune diseases, malignancy, or acute/chronic infection, because these disorders have been linked to IFN-induced inflammation, or treatment with systemic immunosuppressants or photo-therapy in the 3 months preceding the baseline study visit. A total of 72 of the 87 study participants with morphea were included in the Luminex (Riverside, CA) immunoassay. Of these, 36 had follow-up serum available for longitudinal analysis (57 total additional samples). Participants were predominantly adult (89%), white (79%), and female (69%). The mean age at initial study visit was 50 ± 20 years. Most participants had linear or generalized subtypes, 49% of participants had deep involvement, and 28% had functional limitation secondary to morphea. Of participants with longitudinal serum available, 51% received methotrexate with or without corticosteroids, 31% received UVA1 photo-therapy, and 17% had only topical treatments or observation. Complete demographic and clinical information of all study participants is listed in Supplementary Table S1 online.
Cytokines are increased in morphea serum
To quantify serum concentrations of cytokines and growth factors, we used a Luminex assay, which included G-CSF, vascular endothelial growth factor, tumor necrosis factor (TNF)-α, RANTES, MIP-1α, MIP-1β, CXCL9, CXCL10, MCP-1, IL-8, IL-7, IL-6, IL-5, IL-4, IL-2 receptor (IL-2R), IL-2, IL-1β, IL-1 receptor antagonist, IL-17, IL-15, IL-13, IL-10, IFN-γ, IFN-α, hepatocyte growth factor, granulocyte macrophage colony-stimulating factor, epidermal growth factor, IL-12 (p40⁄p70), eotaxin, and fibroblast growth factor.
Seven out of eight T helper (TH) type 1-related cytokines available on the Luminex panel were increased in serum of participants with morphea compared with healthy control subjects (five out of eight after Bonferroni correction) including IL-2, IL-2R, IL-12, MIP-1α, TNF-α, CXCL9, and CXCL10 (P-values < 0.05) (Figure 1a, Table 1). Two of three (none with Bonferroni correction) TH2 cytokines, IL-4 (P = 0.04) and IL-13 (P = 0.008), were elevated in morphea The TH17-related cytokine IL-17 was not elevated in morphea. Cytokines known to be produced by macrophages, including G-CSF, IL-6, IL-8, and MCP-1 were also elevated in morphea sera (Table 1). Cytokines that were undetectable in over half of samples included IL-1B, IL-2, IL-4, IL-5, IL-6, IL-7, IL-10, IL-13, IL-15, IL-17, TNF-α, G-CSF, GM-CSF, vascular endothelial growth factor, and FGF-basic. Additional individual cytokine concentrations are available in Supplementary Figure S1 online.
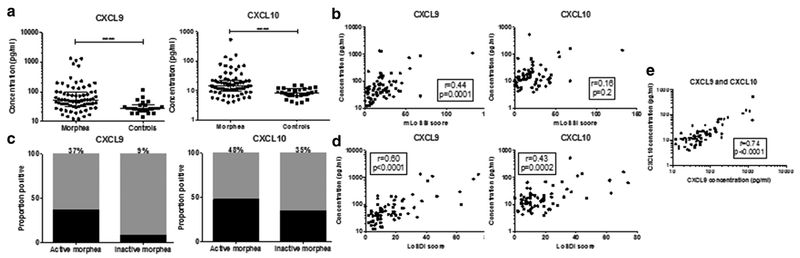
Figure 1. CXCL9 and CXCL10 are elevated in morphea serum.
(a) CXCL9 and CXCL10 are present at increased concentrations compared with control samples. (b, c) CXCL9 is increased in active morphea, decreases in inactive disease, and correlates with disease activity, but CXCL10 does not. (d) Both correlate with measures of disease damage. (e) CXCL9 concentrations correlate with CXCL10 concentrations. LoSDI, Localized Scleroderma Skin Damage Index; mLoSSI, modified Localized Scleroderma Skin Severity Index.
Table 1.
Median cytokine concentrations in participants with morphea and healthy control subjects
| Analyte (pg/ml) | Morphea Patients (n = 72) | Healthy Control Subjects (n = 26) | P-Value | ||
|---|---|---|---|---|---|
| Median | IQR | Median | IQR | ||
| TH1 cytokines | |||||
| IL-2 | 3.0 | 2.2–3.9 | 2.2 | 1.7–2.7 | 0.003 |
| IL-2R | 235.6 | 193.1–272.9 | 171.3 | 163.8–208.5 | <0.00011 |
| IL-12 | 249.0 | 217.1–302.1 | 208.1 | 190.1–236.5 | 0.00051 |
| MIP-1α | 43.61 | 37.8–57.1 | 37.5 | 34.8–42.1 | 0.00011 |
| TNF-α | 2.3 | 1.6–3.0 | 1.9 | 1.5–2.7 | 0.03 |
| CXCL9 | 52.7 | 32.1–99.9 | 27.5 | 23.4–35.4 | <0.00011 |
| CXCL10 | 14.3 | 9.7–22.4 | 8.5 | 6.9–11.6 | <0.00011 |
| IFN-γ | 22.4 | 1 8.4–24.8 | 21.06 | 18.6–23.9 | 0.77 |
| TH2 cytokines | |||||
| IL-4 | 10.3 | 8.8–12.0 | 8.9 | 8.7–10.3 | 0.04 |
| IL-13 | 23.0 | 13.1–33.8 | 16.3 | 5.3–22.8 | 0.008 |
| Other analytes of interest | |||||
| IL1-RA | 244.5 | 141.5–431.9 | 160.4 | 94.2–282.5 | 0.02 |
| IFN-α | 30.9 | 27.9–34.3 | 30.1 | 27.9–30.7 | 0.03 |
| G-CSF | 124.6 | 110.7–141.6 | 106.1 | 90.9–111.1 | 0.003 |
| IL-6 | 4.3 | 2.6–10.3 | 2.8 | 1.9–4.2 | 0.02 |
| IL-8 | 43.2 | 30.9–93.9 | 22.3 | 15.1–35.8 | 0.0004 |
| MCP-1 | 476.4 | 358.2–645.7 | 366.2 | 313.7–428.5 | 0.01 |
| IL-1β | 5.53 | 0.2–11.8 | 0.2 | 0.2–3.3 | 0.003 |
| RANTES | 5,717 | 4,715–7,493 | 8275 | 6086–9761 | 0.001 |
Abbreviations: IQR, interquartile range; TH1, T helper type 1; TH2, T helper type 2.
Indicates cytokines significant with Bonferroni correction.
To further assess which cytokines were increased in morphea, we determined a normal reference range, defined as mean of controls plus two standard deviations, for each cytokine and chemokine and performed frequency counts of numbers of participants with values above the reference range (termed positive). CXCL10 was positive in 44% of participants with morphea, CXCL9 was positive in 35%, IL-2R was positive in 35%, and IL-12 was positive in 29%. Other analytes positive in fewer participants included MIP-1α (26%), hepatocyte growth factor (24%), MIP-1β (22%), MCP-1 (21%), IL-4 (19%), and IL-13 (19%).
Receiver operating characteristic curves were generated for each cytokine that was present with greater frequency in morphea than control subjects. Only CXCL9 produced a curve with preset values of sensitivity greater than 70% and specificity greater than 70%. CXCL9 showed area under the curve of 0.76 (95% confidence interval = 0.66–0.86). With a positive cutoff defined at 36 pg/ml, CXCL9 concentrations have a sensitivity of 72.2% and a specificity of 80.8%.
Correlations with clinical outcome measures
We performed Spearman rank correlations to correlate cytokine levels with validated measures of disease activity and damage. Disease activity was ascertained based on the modified Localized Scleroderma Skin Severity Index (mLoSSI) (Arkachaisri et al., 2009), and disease damage was measured with the Localized Scleroderma Skin Damage Index (LoSDI) (Arkachaisri et al., 2010). CXCL9 concentrations correlated with mLoSSI (r = 0.44, P = 0.0001) and LoSDI scores (r = 0.60, < P 0.0001) (Figure 1b). Furthermore, 37% of participants with active morphea had CXCL9 concentrations greater than two standard deviations above the mean of the control subjects, compared with only 9% of patients with inactive morphea (Figure 1c). There was no correlation between CXCL10 and disease activity, but CXCL10 did correlate with LoSDI score (r = 0.43, P = 0.0002) (Figure 1d). Notably, 48% of participants with active disease had elevated CXCL10 concentrations, but 35% of participants with inactive disease also had elevated CXCL10 concentrations (Figure 1c). In addition, CXCL9 and CXCL10 concentrations correlated with each other (r = 0.74, P < 0.0001) (Figure 1e). IL-4 also showed correlation with mLoSSI (r = 0.47, P < 0.0001) and LoSDI (r = 0.38, P = 0.001). Cytokines with weaker correlations with LoSDI included IL-12 (r = 0.35, P = 0.003), IFN-α (r = 0.34, P = 0.004), and IL-2R (r = 0.30, P = 0.01). No correlations with disease activity or damage ¼ were observed for the other cytokines. Frequencies of positive values for each analyte in participants with active (n = 67) and inactive disease (n = 34) were calculated and are listed in Table 2.
Table 2.
Frequency of cytokine positivity in active and inactive morphea
| Analyte | Active morphea (n = 67), n (%) |
Inactive morphea (n = 34), n (%) |
||
|---|---|---|---|---|
| Increased frequency in active disease | ||||
| CXCL9 | 25 | (37) | 3 | (9) |
| IL-12 | 22 | (33) | 5 | (15) |
| MIP-lα | 18 | (27) | 4 | (12) |
| HGF | 17 | (25) | 1 | (3) |
| IL-4 | 14 | (21) | 2 | (6) |
| TNF-α | 10 | (15) | 0 | (0) |
| Similar frequency in active and inactive disease | ||||
| CXCL10 | 32 | (48) | 12 | (35) |
| IL-2R | 26 | (39) | 8 | (24) |
| IL-13 | 14 | (21) | 5 | (15) |
| MIP-1β | 14 | (21) | 5 | (15) |
| MCP-1 | 14 | (21) | 9 | (26) |
Abbreviation: TNF, tumor necrosis factor.
Subgroup analyses
In assessing participants who presented with functional impairment secondary to morphea compared with all others with morphea, CXCL10 was increased in those with functional impairment (median = 17.3 pg/ml vs. 13.4 pg/ml, P = 0.04). When comparing the generalized and linear subtypes, CXCL9 (65.2 pg/ml vs. 31.5 pg/ml, P = 0.002), IL-4 (11.1 pg/ml vs. 8.8 pg/ml, P = 0.03), TNF-α (2.3 pg/ml vs. 1.6 pg/ml, P = 0.005), and IL-8 (43.9 pg/ml vs. 32.5 pg/ml, P = 0.02) were all increased in generalized morphea. There were no differences in CXCL10 between those with the linear and generalized subtype. No other cytokine concentrations differed between the linear and generalized subtypes or in those with functional impairment. There were too few participants with plaque morphea to allow for comparisons.
Longitudinal data
To determine if cytokine concentrations improved with resolution of disease activity in a given patient, we included longitudinally collected serum samples from patients when disease was inactive, and, when applicable, when disease was inactive after treatment was discontinued, with a washout period of longer than 3 months to avoid potential confounding treatment effect. We performed the Wilcoxon signed-rank test comparing baseline visits (active disease) with early follow-up visits (inactive disease on treatment) and comparing baseline visits (active disease) with later follow-up visits (inactive disease, off treatment >3 months). We found that CXCL9, IL-2R, and IL-12 concentrations were all significantly higher in participants with active disease than with inactive disease during and after treatment discontinuation (n = 21 in each group). The median CXCL9 concentration at baseline was 44.7 pg/ml and decreased to 24.9 pg/ml (P = 0.0002) at follow-up with inactive disease while on treatment (methotrexate or phototherapy). After treatment was discontinued, we found that CXCL9 concentrations remained similar to levels on treatment (on treatment = 24.9 pg/ml vs. off treatment = 26.8 pg/ml) (see Supplementary Figure S2a online). We were unable to assess for changes in chemokine concentrations with disease recurrence because of a paucity of participants with disease flare. However, we were able to describe four patients who had recurrence of disease activity after a period of inactivity. Two had increases in CXCL9 levels with flare, one had persistently elevated CXCL9 whether disease was active or inactive, and one had persistently normal CXCL9 levels throughout follow-up. The median CXCL9 level for those (n = 4) at the time of flare was 53.8 pg/ml. Representative examples of CXCL9 changes and changes in disease activity scores over time are available in Supplementary Figure S2b–d.
To further investigate changes between the initial visit and follow-up visit, we calculated the percentage change in cytokine or chemokine concentration from initial visit to first follow-up visit (n = 36) [(Concentrationbaseline – Concentrationfollow-up)/Concentrationbaseline] and compared it with raw change in mLoSSI score (mLoSSIbaseline – mLoSSIfollow-up) using Spearman rho (r). We found correlations with CXCL9 (r = 0.37, P = 0.02), CXCL10 (r = 0.45, P = 0.006), IL-2R (r = 0.40, P = 0.02), and IL-4 (r = 0.38, P = 0.02).
Gene expression profiling
Whole blood.
To determine if cytokines detected in the sera were the result of dysregulated IFN gene expression in peripheral blood, we performed whole blood transcriptional profiling of 19 participants with active morphea and 11 matched healthy donors. Ten of those 19 were also included in the Luminex assay. There were no differentially expressed inflammatory genes in the blood of participants with morphea compared with healthy control subjects when using a fold change greater than 1.5 and raw P-value less than 0.05. The dataset is publicly available at Gene Expression Omnibus.
Lesional skin.
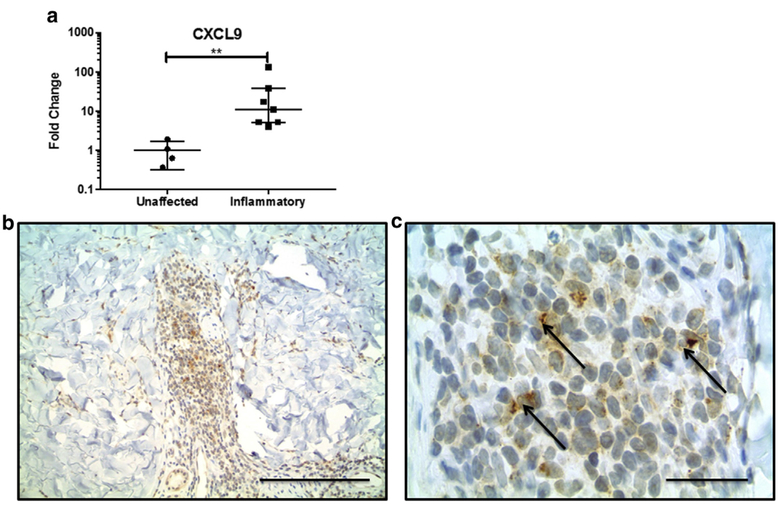
Because there was no transcriptional IFN signature in blood, we hypothesized that morphea lesional skin was the source of circulating CXCL9 and CXCL10, so we performed quantitative PCR of inflammatory morphea lesional skin: CXCL9 (fold change = 30.6, P = 0.006) and CXCL10 (fold change = 2.9, P = 0.004) were both up-regulated in lesional skin (Figure 2a).
Figure 2. CXCL9 is increased in inflammatory morphea skin.
(a) CXCL9 mRNA expression is increased in lesional skin compared with site-matched unaffected skin. (b, c) CXCL9 is present in dermal interstitial infiltrates and stains dendritic-appearing cells. Arrows indicate typical perinuclear cap staining pattern of CXCL9. Scale bar in b = 100 mm; scale bar in c = 25 mm.
Immunohistochemistry
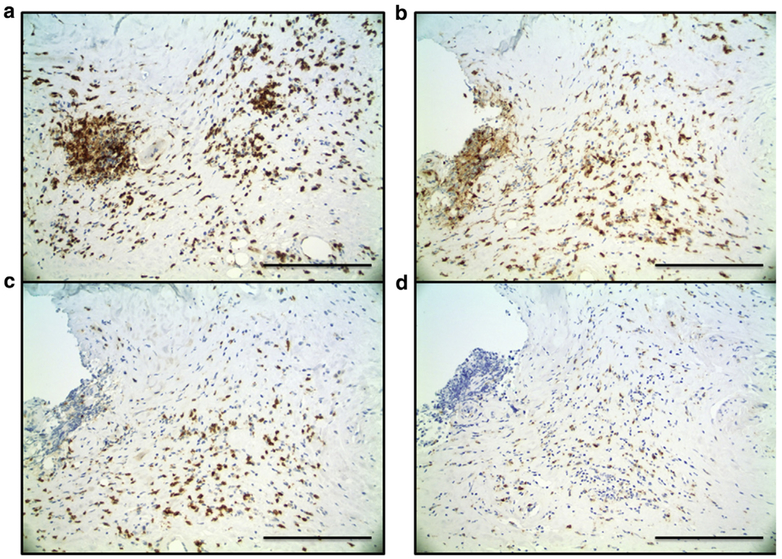
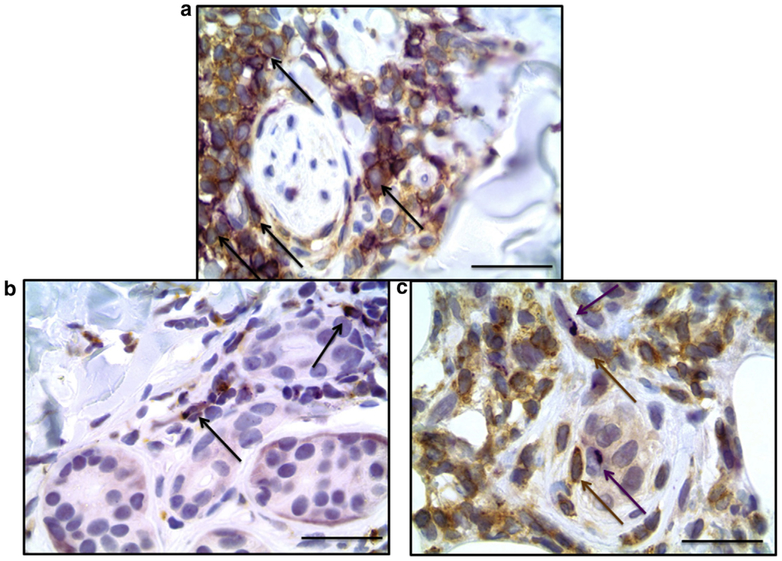
To investigate our hypothesis that CXCL9 is produced in the skin, we performed a pilot immunohistochemical study with single- and double-labeling experiments of punch biopsy skin specimens from three participants with active, inflammatory morphea to characterize the inflammatory infiltrate and investigate cell type-specific expression of CXCR3 and its ligands, specifically CXCL9 (Figure 2b). CD3+ and CD4+ T lymphocytes made up most cells in the inflammatory infiltrate in morphea (Figure 3a and b). CXCR3 stains produced a similar pattern and distribution of cells, and double-immunostaining assays confirmed co-localization of CXCR3 and CD4 within the same lymphocytes (Figure 4a). CXCR3 co-localization with markers of other lymphocyte populations, including CD8+ and CD20+ lymphocytes, was not definitively observed. CXCL9 was expressed by dendritic-appearing cells within perivascular, periadnexal, and inter-stitial infiltrates (Figure 2b and c) in a perinuclear pattern (a pattern previously described for CXCL9) (Ohtani et al., 2009).
Figure 3. Morphea skin, immunohistochemistry.
(a) CD3- and (b) CD4-positive cells make up most of the inflammatory infiltrate. (c) CD8+ lymphocytes and (d) CD68+ macrophages were present less frequently. Scale bar = 50 μm.
Figure 4. Morphea skin, double-staining immunohistochemistry.
(a) The inflammatory cell infiltrate was enriched with CXCR3+ cells that stained similar cells as CD4 (brown) and CXCR3 (purple) double immunostain. (b) CXCL9 co-localized with CD68. CXCL9 (purple) and CD68 (brown) double immunostain; arrows indicate double-staining. (c) CXCL9+ cells were in close proximity to CXCR3+ lymphocytes. CXCL9 (purple, arrow) and CXCR3 (brown, arrow). Scale bar = 25 μm.
Normal skin control samples showed absence of dermal staining for CXCL9 (see Supplementary Figure S3a and b online), whereas psoriasis and mycosis fungoides showed the typical perinuclear staining pattern for CXCL9 (see Supplementary Figure S3c–f). Because of the positive staining of dendritic-appearing cells, we focused our double-staining on cell types of that morphology. CXCL9 co-localized with CD68 in a subset of CD68+ macrophages in all three cases examined (Figure 4b). CXCL9 did not co-localize with CD34 (hematopoietic progenitor cells), CD123 (plasmacytoid dendritic cells), or smooth muscle actin (myoepithelial cells), suggesting that at least a subset of dermal macrophages produce CXCL9. We also found CXCR3+ expressing immune cells in close proximity to cells expressing CXCL9 (Figure 4c).
DISCUSSION
This study examined immune dysregulation in the skin and blood of patients with morphea. Our overarching objective was to determine whether there is dysregulation of the IFN pathway in active morphea and whether these changes could be used as a biomarker for active disease. Our results confirm the presence of dysregulation of IFN-related genes in the skin, particularly CXCL9 and CXCL10, which are the downstream chemokines of IFN-γ. Although gamma IFN serum concentration was not elevated to the same degree, this is strong evidence for TH1 dominance in morphea.
This is congruent with Torok et al., who reported that CXCL10 is elevated in pediatric patients with morphea (Magee et al., 2013; Torok et al., 2015). However, although they examined CXCL10, they did not examine CXCL9. We found that CXCL9 was more strongly correlated with disease activity and that CXCL10 may be a better indicator of overall severity as evidenced by correlations with LoSDI score and similar proportions of patients positive for CXCL10 whether they had active or inactive disease. In patients with hepatic fibrosis secondary to hepatitis C virus, CXCL10 has been associated with advanced fibrosis, whereas CXCL9 was elevated in those with advanced inflammation (Zeremski et al., 2009), which is analogous to our observations in skin. Further, those with functional impairment typically have densely fibrotic lesions and may be analogous to those with advanced hepatic fibrosis in that both have elevated CXCL10.
Although increased circulating CXCL9 and CXCL10 have been shown in SSc (Hasegawa et al., 2011; Liu et al., 2013; Rabquer et al., 2011), our results differ from those in SSc. First, increased expression of IFN-related genes in peripheral blood is well documented in SSc (Assassi et al., 2010; Bos et al., 2009; Duan et al., 2008; York et al., 2007). Further, in SSc, the IFN gene expression signature score of whole blood has been shown to correlate with plasma concentrations of IFN-inducible chemokines, including CXCL10 (Liu et al., 2013). In this context, the lack of any disease-specific transcriptional signatures in whole blood of morphea patients was unexpected. These results suggest that although morphea and SSc share histologic features, they may differ on a molecular level in that morphea could be a skin-directed process, whereas SSc clearly reflects systemic immune dysregulation.
We observed increased circulating levels of CXCL9 and CXCL10 IFN-related gene expression in the skin but little preliminary evidence for IFN-related gene expression in blood, implicating cutaneous production and elaboration of these chemokines. To further characterize the cells involved in this local skin production of chemokines, we performed double-staining immunohistochemistry, which showed CXCL9 co-localized with macrophages, implying that at least a subset of macrophages are the source of circulating CXCL9. We also found CXCR3+/CD4+ immune cells in close proximity to CXCL9+ macrophages, indicating that macrophage expression of CXCR3 ligands, including CXCL9, may enhance recruitment of TH1 CXCR3+-activated T lymphocytes into morphea lesional skin, as has been reported in other organs, including lung, liver, and synovium (Belperio et al., 2003; Shields et al., 1999; Tsubaki et al., 2005).
We based our study design on guidelines for validation of biomarker studies. Therefore, we included a control group that is similar in age, sex, and race; included participants with active and inactive disease representing a wide range of activity scores; and used a validated clinical outcome measure (Tektonidou and Ward, 2011). By these criteria, CXCL9 is a promising biomarker for disease activity in morphea. Specifically, CXCL9 showed high frequency in active morphea (37%) and similar frequency in inactive morphea as in healthy control subjects (9% morphea, 4% control subjects). We also found that CXCL9 correlated with clinical disease activity scores (r = 0.44, P = 0.0001) and had the best performance on the receiver operating characteristic curve. Taken together, CXCL9 may be a marker of disease activity, whereas CXCL10 is a marker of severity as related to sclerosis.
Limitations of this study include sample size, which limited detection of differences in morphea subgroups such as sub-types and pediatric versus adult-onset morphea. Additional studies are needed to more fully explore the role of other autoimmune conditions in addition to morphea on the behavior of CXCL9 and CXCL10, because these patients were excluded from this study. In addition, our analysis of the transcriptional profile of whole blood in morphea is preliminary, and although our results were largely negative, additional analysis is underway by our group to further confirm these observations. In the interim, the lack of transcriptional signatures in the whole blood of morphea should be considered a preliminary finding that requires confirmatory analysis.
Morphea patient serum shows evidence of immune dysregulation, particularly TH1 γ-IFN imbalance, which likely stems from cutaneous production of these cytokines by cutaneous macrophages. Preliminary results imply an absence of a transcriptional IFN signature in peripheral blood, which leads us to hypothesize that morphea may result from skin-directed immune dysregulation rather than a systemic one as in SSc. The chemokines CXCL9 and CXCL10 warrant confirmatory studies as biomarkers for disease activity and severity, respectively.
MATERIALS AND METHODS
Participants
The characteristics of the MAC cohort were described previously (Johnson and Jacobe, 2012). All participants or legal guardians provided written informed consent for inclusion in the MAC cohort, which was approved by the University of Texas Southwestern Medical Center Institutional Review Board. Participants were included if they were enrolled in the MAC cohort and they had serum, whole blood in PAXgene tubes (PreAnalytiX, Hombrechtikon, Switzerland), skin sample in RNALater (Qiagen, Frederick, MD), or a paraffin-embedded skin sample with documented corresponding clinical data available for analysis.
Clinical variables
Participant information included demographics, clinical information, and a detailed physical examination using predetermined case report forms of the MAC cohort. One dermatologist (HJ) examined all participants and classified them into one of five clinical subtypes as described by Laxer and Zulian (2006). Presence of deep involvement was defined as inflammatory infiltrate or sclerosis in the subcutis or other soft tissues on histologic examination or magnetic resonance imaging, or clinically apparent subcutaneous atrophy. Functional limitation included limitation in range of motion, limb length discrepancy, and contracture. All morphea lesions were scored using the Localized Scleroderma Cutaneous Assessment Tool, a validated outcome measure that includes components of disease activity and damage (Arkachaisri et al., 2009), the mLoSSI and LoSDI scores, respectively. Active disease was defined as a mLoSSI score greater than 3.
Serum cytokine determination
At study visits, blood was drawn into serum tubes, centrifuged at 2,500 r.p.m. for 10 minutes, and stored in aliquots at e80 °C. The Human Cytokine Magnetic 30-Plex Panel (Thermo Fisher Scientific, Walsham, MA) was used for serum cytokine determination using a Luminex MAGPIX system. The assay was performed at the University of Texas Southwestern Medical Center Microarray and Genomics Core Facility according to the manufacturer’s instructions. Briefly, 50 μl of serum was added to each well, and then incubated with antibodies against proteins of interest that are coupled to magnetic beads, biotinylated detection antibodies, and streptavidinphycoerythrin conjugates. Concentrations were determined by detecting the amount of R-phycoerythrin fluorescence and reported as continuous variables.
Immunohistochemistry
We performed immunohistochemical staining with single- and double-labeling experiments of lesional skin samples from three participants with inflammatory morphea. Formalin-fixed paraffin-embedded tissues (multitumor sandwich blocks) (Miller, 1993) were sectioned at 4 mm and mounted on adhesive slides. Briefly, primary antibody incubation with anti-CD4, anti-CD8, anti-CD20, anti-CD68, anti-CD34, anti-CD123, smooth muscle actin, and anti-CXCR3/CD183 and CXCL9 was performed. Then, a double-staining procedure with CXCL9 as the purple chromogen and all others as the brown chromogen was performed. Double-staining positivity was determined by two observers (GH and YR) as defined by the cytoplasm of a single cell staining positive for both purple and brown chromogens using various combinations of antibodies. A detailed protocol is available in the Supplementary Materials and Methods online.
Statistical analysis
Continuous variables are reported as medians and interquartile ranges. Categorical variables are reported as numbers and percentages. Cytokine values below the lower limit of detection were set to half of the lower limit of detection for analysis. We used the Mann-Whitney test to compare participants with morphea and healthy control subjects to determine which cytokine and chemokine concentrations were increased. Raw P-values less than 0.05 were considered significant. We indicated values that remained significant after Bonferroni correction (P = 0.0017) to account for multiple comparisons within the Luminex assay. We generated receiver operating characteristic curves for each analyte present at increased concentrations in morphea. We designated the mLoSSI as the primary outcome variable and the LoSDI as a secondary outcome and assessed for correlations between cytokines and measures of disease activity (mLoSSI) and damage (LoSDI) using Spearman rho (r) with significance defined as r > 0.3 and P-value < 0.05. All analyses were performed using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA).
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was supported in part by National Institutes of Health grant no. K23AR056303–5 and by the National Center for Advancing Translational Sciences of the National Institutes of Health under award Number TL1TR001104, and with support from UT-STAR, NIH/NCRR/NCATS grant number UL1TR000451. The content is solely the responsibility of the authors and does not necessarily represent the official views of UT-STAR, The University of Texas Southwestern Medical Center at Dallas and its affiliated academic and health care centers, the National Center for Research Resources, or the National Institutes of Health.
Abbreviations:
- IL-2R
IL-2 receptor
- LoSDI
Localized Scleroderma Skin Damage Index
- MAC
Morphea in Adults and Children
- mLoSSI
modified Localized Scleroderma Skin Severity Index
- SSc
systemic sclerosis
- TH
T helper
- TNF-α
tumor necrosis factor-α
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at www.jidonline.com, and at http://dx.doi.org/10.1016/j.jid.2017.04.008.
REFERENCES
- Arkachaisri T, Vilaiyuk S, Li S, O’Neil KM, Pope E, Higgins GC, et al. The localized scleroderma skin severity index and physician global assessment of disease activity: a work in progress toward development of localized scleroderma outcome measures. J Rheumatol 2009;36:2819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkachaisri T, Vilaiyuk S, Torok KS, Medsger TA Jr. Development and initial validation of the localized scleroderma skin damage index and physician global assessment of disease damage: a proof-of-concept study. Rheumatology 2010;49:373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assassi S, Mayes MD, Arnett FC, Gourh P, Agarwal SK, McNearney TA, et al. Systemic sclerosis and lupus: points in an interferon-mediated continuum. Arthritis Rheum 2010;62:589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assassi S, Swindell WR, Wu M, Tan FD, Khanna D, Furst DE, et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis. Arthritis Rheumatol 2015;67:3016–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belperio JA, Keane MP, Burdick MD, Lynch JP, Zisman DA, Xue YY, et al. Role of CXCL9/CXCR3 chemokine biology during pathogenesis of acute lung allograft rejection. J Immunol 2003;171:4844–52. [DOI] [PubMed] [Google Scholar]
- Bos CL, van Baarsen LG, Timmer TC, Overbeek MJ, Basoski NM, Rustenburg F, et al. Molecular subtypes of systemic sclerosis in association with anti-centromere antibodies and digital ulcers. Genes Immun 2009;10: 210–8. [DOI] [PubMed] [Google Scholar]
- Duan H, Fleming J, Pritchard DK, Amon LM, Xue J, Arnett HA, et al. Combined analysis of monocyte and lymphocyte messenger RNA expression with serum protein profiles in patients with scleroderma. Arthritis Rheum 2008;58:1465–74. [DOI] [PubMed] [Google Scholar]
- Ferri C, Valentini G, Cozzi F, Sebastiani M, Michelassi C, La Montagna G, et al. Systemic sclerosis: demographic, clinical, and serologic features and survival in 1,012 Italian patients. Medicine 2002;81:139–53. [DOI] [PubMed] [Google Scholar]
- Fett N, Werth VP. Update on morphea: part I. Epidemiology, clinical presentation, and pathogenesis. J Am Acad Dermatol 2011;64:217–28. [DOI] [PubMed] [Google Scholar]
- Gourh P, Agarwal SK, Divecha D, Assassi S, Paz G, Arora-Singh RK, et al. Polymorphisms in TBX21 and STAT4 increase the risk of systemic sclerosis: evidence of possible gene-gene interaction and alterations in Th1/Th2 cytokines. Arthritis Rheum 2009;60:3794–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Fujimoto M, Matsushita T, Hamaguchi Y, Takehara K, Sato S. Serum chemokine and cytokine levels as indicators of disease activity in patients with systemic sclerosis. Clin Rheumatol 2011;30:231–7. [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Sato S, Nagaoka T, Fujimoto M, Takehara K. Serum levels of tumor necrosis factor and interleukin-13 are elevated in patients with localized scleroderma. Dermatology 2003;207:141–7. [DOI] [PubMed] [Google Scholar]
- Hinchcliff M, Huang CC, Wood TA, Matthew Mahoney J, Martyanov V, Bhattacharyya S, et al. Molecular signatures in skin associated with clinical improvement during mycophenolate treatment in systemic sclerosis. J Invest Dermatol 2013;13:1979–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihn H, Sato S, Fujimoto M, Kikuchi K, Takehara K. Demonstration of inter-leukin 8 in serum samples of patients with localized scleroderma. Arch Dermatol 1994;130:1327–8. [PubMed] [Google Scholar]
- Ihn H, Sato S, Fujimoto M, Kikuchi K, Takehara K. Demonstration of interleukin-2, interleukin-4 and interleukin-6 in sera from patients with localized scleroderma. Arch Dermatol Res 1995;287:193–7. [DOI] [PubMed] [Google Scholar]
- Johnson W, Jacobe H. Morphea in adults and children cohort II: patients with morphea experience delay in diagnosis and large variation in treatment. J Am Acad Dermatol 2012;67:881–9. [DOI] [PubMed] [Google Scholar]
- Kreuter A Localized scleroderma. Dermatol Ther 2012;25:135–47. [DOI] [PubMed] [Google Scholar]
- Laxer RM, Zulian F. Localized scleroderma. Curr Opin Rheumatol 2006;18:606–13. [DOI] [PubMed] [Google Scholar]
- Liu X, Mayes MD, Tan FK, Wu M, Reveille JD, Harper BE, et al. Correlation of interferon-inducible chemokine plasma levels with disease severity in systemic sclerosis. Arthritis Rheum 2013;65:226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee KE, Kelsey CE, Kurzinski KL, Ho J, Mlakar LR, Feghali-Bostwick CA, et al. Interferon-gamma inducible protein-10 as a potential biomarker in localized scleroderma. Arthritis Res Ther 2013;15:R188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matucci-Cerinic M, Kahaleh B, Wigley FM. Review: evidence that systemic sclerosis is a vascular disease. Arthritis Rheum 2013;65:1953–62. [DOI] [PubMed] [Google Scholar]
- Miller R. Multitumor “sandwich” blocks in immunohistochemistry. Simplified method of preparation and practical uses. Appl Immunohistochem 1993;1: 156–9. [Google Scholar]
- Ohtani H, Jin Z, Takegawa S, Nakayama T, Yoshie O. Abundant expression of CXCL9 (MIG) by stromal cells that include dendritic cells and accumulation of CXCR3+ T cells in lymphocyte-rich gastric carcinoma. J Pathol 2009;217:21–31. [DOI] [PubMed] [Google Scholar]
- Rabquer BJ, Tsou PS, Hou Y, Thirunavukkarasu E, Haines GK 3rd, Impens AJ, et al. Dysregulated expression of MIG/CXCL9, IP-10/CXCL10 and CXCL16 and their receptors in systemic sclerosis. Arthritis Res Ther 2011;13:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields PL, Morland CM, Salmon M, Qin S, Hubscher SG, Adams DH. Chemokine and chemokine receptor interactions provide a mechanism for selective T cell recruitment to specific liver compartments within hepatitis C-infected liver. J Immunol 1999;163:6236–43. [PubMed] [Google Scholar]
- Tektonidou MG, Ward MM. Validation of new biomarkers in systemic autoimmune diseases. Nat Rev Rheumatol 2011;7:708–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok KS, Kurzinski K, Kelsey C, Yabes J, Magee K, Vallejo AN, et al. Peripheral blood cytokine and chemokine profiles in juvenile localized scleroderma: T-helper cell-associated cytokine profiles. Sem Arthritis Rheum 2015;45:284–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubaki T, Takegawa S, Hanamoto H, Arita N, Kamogawa J, Yamamoto H, et al. Accumulation of plasma cells expressing CXCR3 in the synovial sublining regions of early rheumatoid arthritis in association with production of Mig/CXCL9 by synovial fibroblasts. Clin Exp Immunol 2005;141:363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York MR, Nagai T, Mangini AJ, Lemaire R, van Seventer JM, Lafyatis R. A macrophage marker, Siglec-1, is increased on circulating monocytes in patients with systemic sclerosis and induced by type I interferons and toll-like receptor agonists. Arthritis Rheum 2007;56:1010–20. [DOI] [PubMed] [Google Scholar]
- Zeremski M, Dimova R, Brown Q, Jacobson IM, Markatou M, Talal AH. Peripheral CXCR3-associated chemokines as biomarkers of fibrosis in chronic hepatitis C virus infection. J Infect Dis 2009;200:1774–80. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.