Abstract
Cell-to-cell variations contribute to drug resistance with consequent therapy failure in cancer. Experimental techniques have been developed to monitor tumor heterogeneity, but estimates of cell-to-cell variation typically fail to account for the expected spatiotemporal variations during the cell growth process. To fully capture the extent of such dynamic variations, we developed a mechanistic mathematical model supported by in vitro experiments with an ovarian cancer cell line. We introduce the notion of dynamic baseline cell-to-cell variation, showing how the emerging spatiotemporal heterogeneity of one cell population can be attributed to differences in local cell density and cell-cycle. Manipulation of the geometric arrangement and spatial density of cancer cells revealed that given a fixed global cell density, significant differences in growth, proliferation, and paclitaxel-induced apoptosis rates were observed based solely on cell movement and local conditions. We conclude that any statistical estimate of changes in the level of heterogeneity should be integrated with the dynamics and spatial effects of the baseline system. This approach incorporates experimental and theoretical methods to systematically analyze biological phenomena, and merits consideration as an underlying reference model for cell biology studies that investigate dynamic processes affecting cancer cell behavior.
Keywords: Drug resistance, cell-to-cell baseline variation, spatial agent-based model, stochastic differential equation, local cell density
Quick Guide to Equations and Assumptions
We developed a spatial mathematical model of cancer cell growth in a two-dimensional spatial environment. The model is formulated as an agent-based model (ABM) that incorporates spatial aggregation/repulsion forces, as well as transitions through three cellular states: proliferation (P), apoptosis (A), and quiescence (Q). Cell movement is governed by a stochastic differential equation (SDE) with no constraints to a specific lattice, and transition rates between cellular compartments depend on a cell’s microenvironment via a local cell density (LCD) measurement. A more thorough description of the model is provided in the Supplementary Materials. An important feature of this work is that a cell interacts only with other cells in its local neighborhood, defined here as a sphere of radius εa centered at the particle’s position (Xk(t) for the kth particle at time t). We model cells as hard spheres with centers Xk(t) and fixed radius εr. Thus the LCD can be defined as
| (1) |
where is the number of distinct cells in the local neighborhood. The kth cell’s motility is then governed by a SDE combining both attractive and repulsive forces:
| (2) |
Here, Zk (t) describes the population LCD at time t, and Fa (Fr) models the attractive (repulsive) forces experienced by a cell due to the presence of neighboring cells. Both force terms are modeled deterministically as a gradient of symmetric functions, with distinct supports. The attractive force is chosen to account for the observed phenomenon of cellular aggregation and generally has a large support relative to Fr, while the repulsive force models volume exclusion. Lastly, random effects are included in the standard Wiener term σdWk(t), where σ denotes the mean free path. Randomness is included to account for the experimentally observed random walk-like motion by cells in low-density conditions. Note also that the movement is not constrained to a lattice, and that this framework is adapted from Morale et al. (1–3).
Cells also experience transitions between three compartments: P, A, and Q. These transition rates are determined by an individual LCD ρloc, defined as in (1). Cells in the Q compartment are neither dividing nor dying, but instead are in a resting phase. Often this state is referred to as G0, and is the compartment where all dividing cells originate. From Q, cells can either transition into compartments P or A. Cells in P are undergoing proliferation, i.e. progressing through the cell-cycle. Thus, each cell in P has a state variable a(t), the cell-cycle stage, which measures a cell’s progress through P. a(t) increases between discrete time steps t and t+Δt via the rate ω(ρloc, c):
| (3) |
Here, c is the spatially constant drug concentration, which acts to increase ω (and thus increase the cell cycle duration (CCD)). Note that the rate of cell-cycle progression ω is also dependent upon the LCD ρloc. We assume that due to LCD constraints, proliferating cells do not move through the cell-cycle at a constant speed, but instead decelerate as ρloc increases. More precisely, we assume a base length of μ (=15) h after which if a(t) > μ, division is successful and both mother and daughter cell reenter compartment Q. If the amount of time in culture spent in P exceeds a specified limit, the cell transitions to apoptosis, A. Cells undergoing apoptosis are destined to complete cell death; that is, once cells enter compartment A there are no transitions back to P or Q. Once completed, the cell is removed from the simulation. As in our previous report (4), the amount of time spent in A is dictated by an a prior gamma distribution, independent of any state variables. Transitions can be either explicit or implicit. Explicit transition rates are interpreted as probabilities per unit time, i.e. continuous Markov chain transition rates, while implicit rates depend on state variables specific to individual cells.
Introduction
Within an individual tumor, there are typically genotypic and phenotypic variations. This heterogeneity, due to both genetic and non-genetic alterations, can be either temporary or irreversible (5–10). Tumor heterogeneity has been identified as one of the causes of cancer therapy failure, contributing to drug resistance (11, 12). Great efforts have been made to identify and categorize the different sub-populations of cells within a tumor/patient, and to determine their importance in relation to treatment, with the hope of finding ways to efficiently target them. It is accepted that such an approach primarily aims to find genetically stable clones, and assumes that each clone consists mainly of a homogenous population of cells, with insignificant variations concerning the subject of study. Thus, the common goal is to focus on (and target) the identified genetic alterations. However, this approach does not take into account the importance of temporal changes that are not necessarily the result of genetic alterations (13). Determining, additionally, the extent of each clone’s plasticity would result in a more pragmatic treatment protocol.
It has been long recognized that a single clone of cells may have significant phenotypic variations, even concerning drug sensitivity (14, 15). Perhaps the most easily observed evidence of intrinsic non-genetic heterogeneity regarding drug response occurs in virtually every survival curve for cancer cells exposed to drugs, as killing curves have 2 key features: 1) a continuous curve, i.e., a gradual slope, 2) distinct residual cells that survive even after administration of high doses of the drug (Fig. 1A, Table S1). Different types of cell-to-cell variations have been experimentally observed for a single population in many complex cellular processes, such as duration of apoptosis (8, 16), cell size and age (17), and duration of cell-cycle (18). These variations occur in many organisms, generated by a variety of mechanisms that are based on stochastic and/or deterministic (primarily external) signals in a given cell population. In cancer studies, predictions of the disease dynamics are highly dependent on the way those expected baseline variations are evaluated, prior to any additional new alterations, both experimentally and theoretically. So far, the baseline variations have been reported in a limited way, as short-term observations. However normalization with the expected dynamic baseline cell-to-cell variation of the spatiotemporal growth process has not been included, and thus the current statistical approach that determines the reference model may produces false conclusions.
Figure 1.
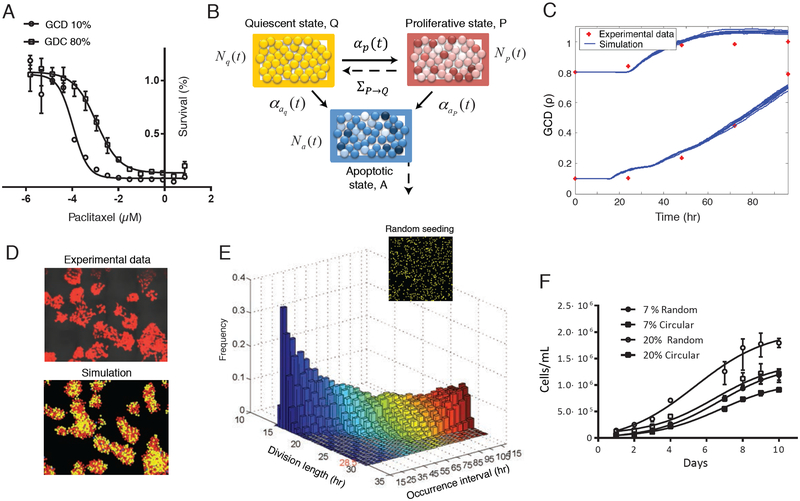
Cell-to-cell baseline growth variation. (A) GCD impacts drug sensitivity. Typical experimental survival curves demonstrating the short-term impact of 10% vs. 80% GCD on drug sensitivity. (B) Mechanistic mathematical model diagram with three compartments. Cells move stochastically through the three phases in ABM, with explicit rates represented by solid lines, and implicit rates given by dashed lines. (C) Experimental growth curves over 4 days in two initial conditions. To calibrate the math model, growth over 4 days was measured (red dots, with initial 10% vs. 80% GCD of cells that were seeded in random). Ten simulation results are plotted in blue. (D) Tracking spatiotemporal growth. To calibrate the math model, we tracked the spatiotemporal growth over 24hr using confocal microscopy (top). Including these data sets, we set the parameter space of the model (bottom; P in red, Q in yellow, and A in blue). (E) Simulate distribution of CCD as a function of LCD. Histogram showing CCDs based on the time interval of division. (F) Differences in growth depend on LCD. Variation in global growth based on variation in LCD. Growth curves with initial GCDs of approximately 7% and 20% demonstrate the dramatic variations in growth over 10 days between two seeding methods (Figures 1E, vs. 2A, top).
Cells are never isolated from their surroundings. Even in an experimental setting cells always grow as part of a population. As such, they are constantly exposed to random fluctuations in external signals. One such fluctuating signal comes from local cell density (LCD). LCD and the ability of cells to move can dramatically and dynamically change cell-to-cell variation. We hypothesized that the intrinsic spatiotemporal process of cell growth should result in intrinsic considerable cell-to-cell variations. These constitute intrinsic dynamic baseline variations that are characteristics of all cell populations. We argue that the initial global cell density (GCD) is not sufficient to determine the system’s baseline variations. To quantify and mechanistically understand the impact of spatial LCD and the ability of cells to move, we developed a spatial mathematical model of cancer dynamics that can be used to theoretically investigate if a specific set of local cellular principles can explain the observed distributions in cell-cycle duration (CCD), growth and drug response. We show that this mechanism of spatial LCD can explain in vitro experimental cell growth data. We experimentally tested two representative geometric seeding configurations from a range of low to high LCD. Both seeding configurations shared the same GCD. We seeded OVCAR-8 cells in a simple 2D culture system, and examined the spatiotemporal cell-to-cell differences in growth rate, proliferation, and paclitaxel-induced apoptosis percentages. Experimentally and theoretically, we demonstrate the extent of those cell-to-cell dynamic baseline variations, based solely on differences in geometrical seeding configurations, yet sharing the same global conditions. Understanding and quantifying these dynamic baseline variations can help to determine the significance of any alteration resulting from cell-to-cell variability, especially in cancer.
Materials and Methods
Growth Curve
Cells (OVCAR-8 parental human ovarian carcinoma cells) were seeded with culture medium (RPMI-1640 medium + 10%FBS + 100U/ml Penicillin Streptomycin + 2 mM Glutamine) in 100 mm dishes in three different GCDs (about 7%, 20%, 30%), with each density in two different geometrical configurations (Figures 1E, 2A). The OVCAR-8 cell line was obtained from the National Cancer Institute, NIH. It is part of the NCI-60 panel of cancer cell lines, which were authenticated by the NCI using short tandem repeat markers to confirm cell identity. The cells were passaged for approximately two weeks in our laboratory after resuscitation, and then used for experiments. While for the random seeding was mixed very well, the seeding pattern was 5 dots, spaced uniformly on the plate, containing 2/3×105, 2×105, and 10/3×105 cells per dot, respectively, to reach the desired global cell density. Two plates per condition were seeded. The plates were incubated at 37°C in humidified 5% CO2. Every 24hr, each plate was counted using a cellometer counter (Nexcelon) four times (Figure 2B, Table S2).
Figure 2.
Spatial cell-to-cell baseline variation in proliferation. (A) Simulated distribution of CCD (bottom) in cells grown in a circular seeding pattern (top). This histogram is parallel to the one shown in Fig 1E (top) but with the circular seeding configuration. (B) Experimental spatial proliferation. Using confocal microscopy imaging for the circular seeding pattern, after 48hr, we observed more proliferative cells in the surrounding area, as determined by ki67 staining. The circular configuration was formed using a cloning cylinder (top right). (C-F) Dynamic heterogeneity in proliferation. Using FACS to measure the Ki67 florescence intensity over 4 days for two geometric configurations, dynamic cell-to-cell variation can be observed in proliferation percentage and in transition between proliferation- to non-proliferation sates.
Time Series Imaging using Confocal Microscopy
A Zeiss 710 confocal microscope was used to image over time the entire well on an 8-well slide (ibidi), and form a 24-hr time series movie of cell movements and growth (several snapshots are provided in Figure S1). The tool employed on the Zeiss software was ‘Tiling’ which allowed the microscope to take a series of spatial-sequence snapshots of the well and combine them together to create a single whole matrix image with a wide view of hundreds of cells (OVCAR-8-DsRed2 cell line) (19). The plates were imaged using a Zeiss 710 confocal microscope with 20x objective, 10x z-stacks, and a 5×5 tile, to form two Time Series Imaging movies.
Proliferation Detection
Detecting Ki67 using FACS.
The percentage of Ki67-mediated cell proliferation as a function of local and global cell density was measured by a staining method using the Ki67 protocol using an Anti-human Ki67 FITC, Flow Cytometry Staining Buffer, and Foxp3 Fixation / Permeabilization solution (eBioscience) according to the manufacturer’s instructions. OVCAR-8 were seeded in 150 mm dishes in drug-free medium and incubated at 37° for 24hr to allow cells to attach. Three different global cell densities (7%, 20%, 33%) were seeded, each density in two different geometrical configurations. While the random formation was mixed very well, the circular formation was 15 dots containing 2/3 × 105, 2 × 105, and 10/3 × 105 cells per dot, respectively. The percentages of proliferation were then evaluated by FACS (BD LSRFortessa) every day for 4 days. Data from 10,000 gated events per sample were collected. Data were collected three times. For experiments with drug, after 24 hr with drug-free media, random and circular plates from each density were dosed with IC50 paclitaxel, 0.001 ± 6.20E-6 μM/L, and with IC50 × 4 paclitaxel. After 3 days of incubation with the drug, data were collected. See Table S3 for the results.
Detecting Ki67 using Confocal Microscopy.
Cells were seeded at the desired density in an 8-well plate (ibidi). To form a circular structure for imaging, cloning cylinders (purchased from Sigma-Aldrich) were placed using forceps in the center of each well before seeding. 4 × 104 cells per well were seeded and after 24 hr the cylinders were removed and the circular arrangement of the cells could be observed. After 48 hr, we followed the manufacturer’s instructions to add the primary antibody (see details in Supplementary Materials). 200 μl of PBS was added to each well prior to imaging. The plates were imaged using a Zeiss 710 confocal microscope with 20x objective, 10x z-stacks, and a 3 × 10 tile, to form Figure 3B.
Figure 3.
Cell-to-cell baseline variation in drug sensitivity. (A) Cells after paclitaxel treatment demonstrate changes in their cell-cycle phases. Ki67-positive cells represent proliferative cells, i.e., in their cell-cycle. To track those changes we monitored the median in Ki67 florescence intensity using all cells, and measured the percentage of the positive cells (after 4 days, 3 days with paclitaxel). These results are the average of three experiments (see Table S3). The differences between the two seeding methods were calculated in two ways: linear differences (i.e., median or percentage values of random seeded cells - values of circular seeded cells), and as a ratio given as a percentage (i.e., (ValueRand /Value −1)*100=x%)). (B) Simulated survival curves. Relative cell survival is computed after 3 days of treatment as a function of dosage (x-axis). Note that the dense geometries (S2 and S3) experience an overall greater survival rate, and thus are intrinsically drug-resistant. (C) Experimental apoptosis percentage of both geometrical configurations, using Annexin V assay. std= standard deviation.
Survival Assay (MTT)
Cell survival was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) viability assays. OVCAR-8 were randomly seeded at two different densities, 104 and 105 cells per well, in 24 well plates. After 24 hr, all cells were dosed with 20 mM Paclitaxel in 16 different dilutions (Figure 1A, Table S1). 300 μl of each dilution was added to each of the 24 wells. Once all the plates were dosed they were incubated for another 72hr. The MTT assay was performed according to the manufacturer’s instructions (Molecular Probes, Eugene, OR). Absorbance values were determined at 570 nm on a Spectra Max 250 spectrophotometer (Molecular Devices, Sunnyvale, CA). All MTT assays were performed three times in triplicate.
Apoptosis
Cells were seeded as in the Ki67 FACS experiments. The percentage of apoptosis-mediated cell death was measured by a double staining method using a Dead Cell Apoptosis Kit with Annexin V Alexa Fluor® 488 & PI (Invitrogen) according to the manufacturer’s instructions. The percentages of apoptosis and necrosis were then evaluated by FACS (BD LSRFortessa). Data from 10000 gated events per sample were collected. Cells in early stages of apoptosis were positively stained with Annexin V, whereas cells in late apoptosis and necrosis were positively stained with both Annexin V and PI. The number of cells positively stained with Annexin V was used to estimate the death rate. Data were collected three times; see Figure 3C and Table S4 for the results.
Results
Dynamic variation in cell-cycle duration
In this study, we focused on cell-to-cell dynamic baseline variations in relation to three key features of cancer cells: cell-cycle, growth, and drug sensitivity. First, we explored the variation between cells in their CCD, since this affects growth and drug sensitivity. We sought to identify a set of cellular principles that would explain our experimental patterns of OVCAR-8 growth, reproduce the common normal-like distribution of CCD with its parameters, and analyze the implications of spatial variations in proliferation rates. To study these issues, we took a mathematical modeling approach to study the entire growth and treatment processes as a function of both time and space. We hypothesized that the local spatial component significantly affects the characteristics of this duration distribution.
Our spatial mathematical model is formulated as an ABM that incorporates spatial aggregation/repulsion forces, as well as transitions through three cellular states (see Supplementary Materials, Fig. 1B). Cell movement is described via a stochastic differential equation, and transition rates are functions of LCD, ρloc. Using our in-silico model, we investigated the effect of imposing a deterministic cell-cycle structure based on the spatial location of the cell. In particular, we were interested in the distribution of the CCD as a function of time. The initial simulated GCD was ρ(0) = 10%, and the population of cells was allowed to grow until a time t such that a plate was fully covered, i.e., ρ(t) = 100%. The entire population of cells was initially assumed to be quiescent synchronized and randomly distributed in space. By calibrating the model with growth curves over 4 days (initial 10% and 80% GCD, Fig. 1C), and experimental time series imaging data (random, 10%) for a period of 24hr, we imaged a wide spatial range of the cultured plate with hundreds of cells (Fig. 1D, Fig.S1.A, Fig.S2.A). In this way, we were able to determine the parameter space of the model (see Supplementary Materials, Figures S1, S2).
Our in silico results clearly showed that the basic assumptions of an intrinsic CCD as a deterministic process with growth limitation based merely on LCD, can produce the normal-like distribution of CCD in randomly seeded cells. Moreover, as illustrated in Fig. 1E (where the CCD is modeled using equations (S.9)-(S.10)), a full 3D histogram for the lengths of successful divisions for 10 h windows in a single simulation demonstrated normal-like distributions that are functions of both time and space. For example, the first histogram labeled 15 on the “occurrence interval” axis denotes the distribution for all divisions occurring in simulation time t such that t ∈ [15, 25] hr. We observed a traveling wave of normal-like distributions, starting with short cell-cycles at low GCDs, which gradually increased as the population grew. Averaging over all time intervals yields a distribution with mean ≈ 23hr, similar to our previously observed experimental value (4) (24.41hr). Figure 1E illustrates the interesting temporal dynamics of the variance distribution (simulation), with a relative maximum variationoccurring at approximately t = 80 hr, i.e., at this point the plate reaches a 50%−70% value of GCD.
Local conditions create different global behavior
Since LCD affects the distributional dynamics, we experimentally tracked the growth of cells over time with different initial geometric configurations, while the initial total seeding number of cells held constant. Throughout this paper, we describe our work concerning two main geometrical configurations that represent the extreme cases in a range of commonly used seeding methods in any experimental cell culture laboratory (illustrated in Fig. 1E), which are random seeding (uniformly distributed across the plate), or circular seeding structure (can be achieved by direct application with a pipette, without mixing the plate, illustrated in Fig. 2A). Common growth curves with shared initial GCDs of about 7%, and 20% demonstrate the dramatic variations in growth over 10 days (Fig. 1F). The cells with an initial 20% GCD in random vs. patterned circular seeding grew about 70% faster (i.e., (# cellsRand /# cells −1)*100=70%) even during the first 4 days (Table S2). These results show that not only the known global, but also LCD conditions can produce distinctive cell-to-cell baseline variations in growth.
Given that cell spacing had major effects on growth, we used our mathematical model to ask how the previous distribution of CCD would vary when the system grew in a specific geometrical configuration of circular (Fig. 2A, top), instead of random seeding (initially with 10% GCD, as before). The simulation predicts a wide and stable CCD distribution, with the variation values of most time points approximately the same. Interestingly, a bi-normal distribution with two distinct average values was obtained (Fig. 2A, bottom). This reflects the two sub-populations in the P state, the slow growing cells closer to the center and the faster, more proliferative cells growing at the periphery, when the area occupied by each sub-population varies based on the GCD. In the case of low GCD, most cells are in a proliferative state, even ones closer to the center. However, they have different CCDs. On the other hand, once the GCD increases, most of the cells in the center move to a quiescent state, and the CCD for the rest of the P population changes based on the circular bands that the cells occupy. Thus, the two mean values of the bi-normal distribution approach one another, and may be considered as a single distribution at later stages of the process. This process can be seen in Movie S1, in which we simulated the spatiotemporal heterogeneity in CCD for a circular structure. Contact inhibition is well known to regulate the progression of the cell-cycle. The contribution of this work is mainly to mechanistically and systematically predict the entire process of cancer cell dynamics as a population, based on local interactions, beginning with low cell density, growing with or without geometrical constraints. For cancer cells, cell-to-cell interactions slow cellular growth, but in many cases do not necessarily completely stop cell divisions, leading to heterogeneity. The unknowns are the extent and implications of such dynamic heterogeneity.
Using confocal microscopy imaging, for the circular pattern, after 48 h, we observed more proliferative cells in the surrounding area, as determined by ki67 staining (Fig. 2B). Note that it took 48 h to reach this spatial distribution of proliferation. Prior to 48 h, most cells showed random spatial proliferation (data not shown), as was predicted by the simulated movie. We further measured the cell-to-cell differences in the cell population distribution of ki67 protein expression using flow cytometry, for two seeding patterns for 4 days at 3 different GCDs: 7%, 20% and 30%. Three key FACS measurements were closely studied, the percentage of Ki67 positive cells, the Ki67 median intensity of all cells (positive and negative), and the shape of the Ki67 intensity distribution. These three measurements include information about the entire population, proliferative, non-proliferative state, and the transition between them over time. On average, the Ki67 distributions (example plotted in Fig. 2C) in populations with circular seeding at low GCDs were wider than when cells were seeded randomly, as was predicted by the simulation. In higher GCDs, we can observe higher fluctuations in proliferation over time at the random case (Fig. 2D). We found that the ki67 percentages over 4 days for low (less than 30%) GCDs reach a maximum in random seeding, while a minimum was observed in circular seeding (Fig. 2E–F and Table S3); altogether, the highest variation in proliferation percentages between the two geometrical configurations, can reach up to 36% over time (e.g., (#Ki67PositiveCellsRand 72h/# Ki67PositiveCellscircular_72h-1)*100=36%). In other words, the baseline variations of both seeding methods are dynamic over time and the highest variations occur 72hr after seeding. This is a growth feature related to the cells’ adjusting process after initial seeding, which is also a function of local spatial constraints.
For cells that initially were seeded at low GCDs of less than 30%, there was a pattern that included the LCD and GCD. During the first 24hr, cells are mainly in the process of attaching themselves to the plate. During the second 24hr, i.e., 24–48h, cells mostly proliferate. During the third 24hr, i.e., 48–72hr, cells increase their movement to contact other close neighboring cells. During the fourth 24h, i.e., 72–96hr, the plate was much more dense, changing the initial geometry. As a result, cells that were locally dense act differently from the random case over time. These variations can be seen in the changes of median fluorescence intensity as a function of seeding methods with varying GCDs (Fig. 2E), and variations as a function of time (Table S3), or variations in Ki67 positive percentage as a function of seeding methods (Fig. 2F).
Taken together, this evidence implies that there are two main mechanisms that affect the population dynamic and thus its expected variations: the transition process between the P and Q states, and the CCD (for cells in P state). We showed that the fluctuation in the transition process between states is as profound as the variations in CCD, especially for the geometrical cases such as the circular rearrangement at low GCDs. However, in the case of randomly seeded cells, changes in the CCD are the main factors causing variations in spatial proliferation. These experimental results validate our understanding from the simulated predictions of a traveling wave vs. stable distribution of CCD illustrated in Figs. 1E and 2A, which show the different mechanisms of the expected variations.
Cell-to-cell baseline variation in drug-sensitive cells
Lastly, we examined whether or not our finding concerning the role of LCD and cell movement as an intrinsic inducer of spatial proliferation variation can also explain the cell-to-cell baseline variation in drug sensitivity. We found that both killing curve features, the continuous curve and the distinct residual surviving cells after high drug doses, can be largely explained based on the LCD mechanism, at least for chemotherapy drugs such as paclitaxel. Paclitaxel is a mitotic inhibitor and hence acts on cells in the proliferative compartment P. More precisely, its primary chemotherapeutic effect is on the cytoskeleton, targeting tubulin (20). During cell division, chromosomal microtubules are necessarily destabilized and broken down as the mother cell divides. Paclitaxel acts as a stabilizer for these microtubules, thus preventing metaphase spindle configuration. At high paclitaxel doses, the cell is unable to complete mitosis and remains at the mitotic checkpoint of the cell-cycle for a prolonged period of time. Eventually, prolonged activation of the mitotic checkpoint triggers apoptosis, due to unsuccessful cell division. Using FACS we quantified the fluorescence intensity of Ki67 staining for cells that were treated with an IC50 dose of paclitaxel for 72h. We observed that in cases of low GCD (less than 30%) there were differences in the median Ki67 values of the entire population (all cells included) between the two seeding methods: random vs. circular (Fig. 3A, Table S3). Randomly seeded cells expressed a much higher level of Ki67, consistent with increased proliferation (or at least staying in ‘cell-cycle’) at low GCDs. When high doses (IC50×4) were applied, much higher differences in the median occurred (Fig. 3A, first row). The range of median ratios for IC50 was 17%−47% (7% and 20% GCDs, respectively), while for IC50×4 the range was 73%−88%. In addition, when we examined the percentage of only Ki67 positive cells, we found the same trend, approximately a 10% range in linear differences (Fig. 3A, second row), and about 13% in their ratio. Again, with randomly seeded cells, there are more positive cells than occur with the circular seeding method.
Furthermore, we hypothesize that different geometrical configurations sharing the same GCD can produce variations in actual apoptosis rates, and this intrinsic resistance can be traced back to cell-cycle dynamics. In general, increasing GCD is a well-documented cause of decreasing drug sensitivity. An example can be seen in Fig. 1A (Table S1), where we performed MTT assays on initially randomly seeded cells with 10% vs. 80% GCD of cells, treated with paclitaxel. Killing curves showed the percentage of surviving cells after 72 h of treatment as a function of the drug dosage, normalized by non-treated cells growing in parallel. The observed increase in IC50 at high GCDs is slightly more than one order of magnitude. To examine our theory about significant variations due to fluctuations in LCD, we utilized our ABM to mechanistically study the killing curves for three different spatial seeding configurations: ρ1(0) = 10% (of GCD), and ρ2(0) = 80% in random structure, and ρ3(0) = 10% in circular structure. The mathematical model qualitatively showed that the randomly seeded plate can reach a higher relative survival percentage for most drug concentrations, and experience a more gradual decrease with increasing drug concentrations, i.e., a more gradual slope (Fig. 3B). The simulations also predicted that the compact configuration (i.e., circular structure) would exhibit the same trend as high GCD, meaning that the LCD can produce significant higher survival with a more gradually sloped curve and higher number of surviving cells after exposure to high drug concentrations than randomly seeded cells that share the same GCD of 10%.
The model could not predict differences for low drug concentrations, and was not sensitive enough to provide information about the observed IC50 variations. However, the model did provide direction based on the underlying dynamics to examine the general survival variation in a more detailed way, namely to focus on the relationship between processes related to CCD, the transition between the P and Q states, and the apoptosis rates as a function of the LCD. To experimentally quantify the cell-to-cell baseline variations in paclitaxel drug sensitivity, we further directly measured the apoptosis percentages under different local conditions: random and circular, using Annexin V staining, with the same GCD (20%). For the IC50 and IC50×4 settings, the variation between the random vs. circular structures reached 12–14% linear differences in actual apoptosis percentage, after normalizing each geometrical seeding method with their non-treated apoptosis rates (Fig. 3C, Table S4). Therefore, our results suggest that features related to the changes in growth (e.g., CCD, and transition between the P and Q states), and apoptosis rates are functions of the geometric cell configuration, i.e., LCD, in addition to GCD.
Discussion
Recently, Sandler et al. (21) addressed a central question concerning the origin of cell-to-cell variation in CCD, by studying the differences between generations of cells derived from a parental cell. The main debate is whether observed variation originates from stochastic or deterministic processes. While stochasticity, for instance, results from random bursts of gene expression or noisy partition of cellular components at division, deterministic characteristics may result from the robust design of the dynamic cellular system that can produce a stable oscillator. They concluded, based on an experimental setting with very low cell densities, that the cell-cycle is a deterministic and not a stochastic process. This leads to the conclusion that the internal noise, without high external noise, does not seem to add enough stochasticity to cause cell-to-cell variation in CCD.
Most experimental settings involve higher GCDs (30%−50%), yet have low LCDs (by randomly seeding the cells), especially in high throughput screening experiments. In our current study, we suggest a method to integrate the conclusions from single cell studies (such as those from Sandler et al.) with experimental conditions that include higher cell densities into a single unified model. This unified model can serve as a general reference model for cell biology studies that focus on dynamic features of single cells, population of cells, and the relationship between them. Theoretically and experimentally, we show that the LCD, in addition to the GCD, can lead to significant differences in spatial growth, proliferation, and paclitaxel-induced apoptosis rates. Another important aspect of this work is that it addresses a phenomenon that all cell biologists take for granted: killing curves are not discrete in nature, but rather are gradual, reflecting intrinsic heterogeneity in a cell population that can be attributed to differences in LCD and cell-cycle. Furthermore, our model allows us to systematically explain why, and specifically how, one population can lose synchronization much faster than another.
This type of mechanistic model can be used to determine if the observed system behavior or characteristics are the result of specific principles. For example, a future extension to our model could test the following questions: to what extent would additional internal noise (neglected here due to the Sandler et al. findings) change the population dynamics? Or what is the threshold level of collective internal noise that would begin to affect the entire system dynamics, in relation to spatial growth, proliferation, or drug response? Moreover, beyond a cell’s response to a drug, our model has implications concerning other cellular processes, such as adult stem cell regeneration. Lei et al. have recently suggested the origin and consequence of heterogeneity in healthy tissues (22). They showed that heterogeneous proliferation, when not all cells have the same probability of proliferation, increases robustness in the process of tissue regeneration and replacement of defective cells, and hence provides improved long-term performance.
In summary, a comprehensive reference model is essential in cell biology studies in order to determine significance. We stress the value of including in the reference model not only statistical tests, but also dynamic features of the process as an integral method.
Supplementary Material
Major Findings.
Cells are never isolated from their surroundings. Even in an experimental setting cells always grow as part of a population. As such, they are constantly exposed to random fluctuations in external signals. One such fluctuating signal comes from local cell density. Local cell density and the ability of cells to move can dramatically and dynamically change cell-to-cell variation. Tracking the intrinsic spatiotemporal process of cell growth reveals heterogeneity in crucial features, such as cell-cycle duration, growth, proliferation and apoptosis rates. These constitute intrinsic dynamic baseline variations that are characteristics of all cell populations. These baseline variations are needed to explain and predict cell heterogeneity and to determine the significance of any alteration resulting from cell-to-cell variability, especially in cancer.
Acknowledgements
We would like to thank George Leiman, Carol Cardarelli, and Susan Garfield, Langston Lim and Poonam Mannan, Karen Wolcott and Subhadra Banerjee, Mirit Aladjem, and Haiqing Fu for their assistance and for insightful discussions. This work was supported by the Intramural Research Program of the NIH and was also supported in part by a seed grant from the UMD-NCI Partnership for Cancer Technology. The work of DL was supported by the John Simon Guggenheim Memorial Foundation.
Support: This research was funded, in part, by the Intramural Research Program of the National Institutes of Health.
Footnotes
Conflicts of interest: None declared
References
- 1.Burger M, Capasso V, Morale D. On an aggregation model with long and short range interactions. Nonlinear Analysis-Real World Applications. 2007;8:939–58. [Google Scholar]
- 2.Morale D. Modeling and simulating animal grouping - Individual-based models. Future Generation Computer Systems. 2001;17:883–91. [Google Scholar]
- 3.Morale D, Capasso V, Oelschlager K. An interacting particle system modelling aggregation behavior: from individuals to populations. Journal of Mathematical Biology. 2005;50:49–66. [DOI] [PubMed] [Google Scholar]
- 4.Greene JM, Levy D, Fung KL, Souza PS, Gottesman MM, Lavi O. Modeling intrinsic heterogeneity and growth of cancer cells. J Theor Biol. 2015;367:262–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805:105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park SY, Gonen M, Kim HJ, Michor F, Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120:636–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanz-Moreno V, Gadea G, Ahn J, Paterson H, Marra P, Pinner S, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–23. [DOI] [PubMed] [Google Scholar]
- 8.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–44. [DOI] [PubMed] [Google Scholar]
- 10.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saunders NA, Simpson F, Thompson EW, Hill MM, Endo-Munoz L, Leggatt G, et al. Role of intratumoural heterogeneity in cancer drug resistance: molecular and clinical perspectives. EMBO Mol Med. 2012;4:675–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol. 2013;8:277–302. [DOI] [PubMed] [Google Scholar]
- 13.Pisco AO, Brock A, Zhou J, Moor A, Mojtahedi M, Jackson D, et al. Non-Darwinian dynamics in therapy-induced cancer drug resistance. Nat Commun. 2013;4:2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–22. [DOI] [PubMed] [Google Scholar]
- 15.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–6. [DOI] [PubMed] [Google Scholar]
- 16.Messam CA, Pittman RN. Asynchrony and commitment to die during apoptosis. Exp Cell Res. 1998;238:389–98. [DOI] [PubMed] [Google Scholar]
- 17.Tzur A, Kafri R, LeBleu VS, Lahav G, Kirschner MW. Cell growth and size homeostasis in proliferating animal cells. Science. 2009;325:167–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P, Robert L, Pelletier J, Dang WL, Taddei F, Wright A, et al. Robust growth of Escherichia coli. Curr Biol. 2010;20:1099–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brimacombe KR, Hall MD, Auld DS, Inglese J, Austin CP, Gottesman MM, et al. A dual-fluorescence high-throughput cell line system for probing multidrug resistance. Assay Drug Dev Technol. 2009;7:233–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowinsky EK, Donehower RC. Paclitaxel (taxol). N Engl J Med. 1995;332:1004–14. [DOI] [PubMed] [Google Scholar]
- 21.Sandler O, Mizrahi SP, Weiss N, Agam O, Simon I, Balaban NQ. Lineage correlations of single cell division time as a probe of cell-cycle dynamics. Nature. 2015;519:468–71. [DOI] [PubMed] [Google Scholar]
- 22.Lei J, Levin SA, Nie Q. Mathematical model of adult stem cell regeneration with cross-talk between genetic and epigenetic regulation. Proc Natl Acad Sci U S A. 2014;111:E880–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.