Abstract
Legionella pneumophila, the causative agent of Legionnaires' disease, is a waterborne bacterium mainly found in man-made water systems in close association with free-living amoebae and multispecies biofilms. Pseudomonas strains, originating from various environments including freshwater systems or isolated from hospitalized patients, were tested for their antagonistic activity towards L. pneumophila. A high amount of tested strains was thus found to be active. This antibacterial activity was correlated to the presence of tensioactive agents in culture supernatants. As Pseudomonas strains were known to produce biosurfactants, these compounds were specifically extracted and purified from active strains and further characterized using reverse-phase HPLC and mass spectrometry methods. Finally, all biosurfactants tested (lipopeptides and rhamnolipids) were found active and this activity was shown to be higher towards Legionella strains compared to various other bacteria. Therefore, described biosurfactants are potent anti-Legionella agents that could be used in the water treatment industry although tests are needed to evaluate how effective they would be under field conditions.
1. Introduction
Bacteria of the Legionella genus are Gram-negative natural inhabitants of freshwater environments. Among the 65 Legionella species referenced to date, L. pneumophila is the leading cause of severe pneumonia called Legionnaires' disease (LD). Furthermore the serogroup 1 is responsible for 82.9% of the cases in Europe [1] and for over 80% of the cases worldwide [2, 3]. L. pneumophila can colonize man-made water settings from natural water sources, being now considered as an opportunistic plumbing pathogen. Through the literature, we can notice that LD outbreaks have been linked to a variety of water sources like cooling towers, drinking water supply systems, spa pools, and even street cleaning trucks [4–6]. Multiplication of Legionella in those artificial water systems is highly facilitated by temperatures around 35°C and factors such as water stagnation, poor maintenance, no or reduced water disinfection and the presence of free-living protozoa feeding on biofilms [7, 8]. Biofilms have been identified as an ecological key niche in which L. pneumophila survives and stays in wait for its natural hosts, amoebae, and ciliates [9]. Indeed, protozoan predators are the natural hosts of L. pneumophila, and humans are accidental hosts as judged by the evidence that only a single and recent case of probable human-to-human transmission has been reported to date [10]. Following the uptake of L. pneumophila by phagocytic cells, this bacterium avoids lysosome mediated degradation and forms a unique replication-permissive compartment within its host cell, called the Legionella containing vacuole (LCV) [11]. After replication, they are able to evade LCV, escape from the spent host cell, and disseminate in the environment looking for new suitable hosts (for recent review see [12]).
In order to survive in water sources, L. pneumophila is facing other biological challengers aside from protozoa as represented by others microbial inhabitants. In 2008, a study screened 80 aquatic bacterial strains including 5 referenced strains and showed that 66.2% displayed antagonistic activity against L. pneumophila [13]. Interestingly, among the Pseudomonas genus (representing 75% of all tested strains), 72% were active. However, to date, the antagonistic molecules of interest remain uncharacterized. On the other hand, several authors have shown that many Pseudomonas species can produce biosurfactants which are surface-active compounds. Those compounds known to display various functional properties like in structural biofilm formation/cells dispersion also exhibit particularly lytic and growth-inhibitory activities against a broad range of microorganisms, including viruses, mycoplasmas, bacteria, fungi, and oomycetes [14–16]. Among the wide range of structurally different biosurfactants that have been identified to date, Pseudomonas species can produce glycolipids and lipopeptides [17, 18]. Lipopeptides are constituted by a lipid tail linked to a short cyclic or linear peptide moiety. Based on differences in the length and composition of the lipid moiety as well as in the type, number and configuration of the amino acids in the peptide chain were initially classified into four groups: Amphisin, syringomycin, tolaasin, and viscosin [19]. However, several lipopeptides produced by Pseudomonas spp. which were characterized later displayed structural features which differ from these archetypes like putisolvins, syringofactins, and orfamides that extend the initial classification [20–22]. The first lipopeptide described in the literature and which remains the best known biosurfactant to date is surfactin produced by many Bacillus strains [23]. Members of the surfactin family are constituted of a heptapeptide moiety linked to a β-hydroxylated fatty acid to form a cyclic lactone ring and display antiviral and antibacterial activities [24]. Recently, a surfactin mixture produced by the strain B. subtilis AM1 was found active against various Legionella strains, including nonpneumophila, pneumophila serogroup 1, and pneumophila nonserogroup 1 strains [25]. To the best of our knowledge, this is the first time that such an activity was demonstrated and this study remains the only one published so far. This activity was striking as several reviews have highlighted a limited activity of many lipopeptides against Gram-negative bacteria [14, 15, 17]. However, lipopeptides produced by Pseudomonas and Bacillus species have not been tested extensively for activity against other saprophytic bacteria, but mostly for activity against human pathogenic bacteria like Bacillus sp. or Mycobacterium sp. [14]. The surfactin mixture from B. subtilis AM1 was also able to break down existing biofilms of L. pneumophila [25] suggesting that it could represent a potent tool for the biological control of the pathogen in the water treatment industry.
Rhamnolipids are glycolipid secondary metabolites typically constituted of dimer of 3-hydroxy fatty acids linked through a beta glycosidic bond to a mono- or di-rhamnose moiety [26]. Up to now, more than one hundred rhamnolipids homologues have been discovered mainly in Pseudomonas species even if several bacteria belonging to other genera like Burkholderia or Acinetobacter were also reported to produce rhamnolipids [27]. These amphiphilic biodegradable molecules have been reported to be useful as biological control agents due to their intrinsic wide-ranging antimicrobial potency [28]. For example, the mixture (six homologues) extracted from the P. aeruginosa LBI strains displayed a high activity against many bacterial strains like Enterobacter aerogenes and Proteus mirabilis with MIC between 4 and 8 μg/ml [29]. Also, various bacterial species were sensitive to the P. aeruginosa 47T2 mixture (up to 11 homologues), Enterobacter aerogenes being by far the most sensitive one (MIC of 4 μg/ml) [30]. To date, except for surfactin which was already described for its anti-Legionella activity [25], neither lipopeptide nor rhamnolipid mixture was reported to be active against bacteria of the Legionella genus.
The aim of our study was to discover and characterize natural anti-Legionella compounds produced by Pseudomonas strains. Therefore, a bacterial collection with both clinical and environmental strains was built and screened to find Pseudomonas sp. with the capacity to inhibit the growth of L. pneumophila. Biomolecules responsible for this antagonistic activity were then purified by RP-HPLC and their chemical structures were elucidated by LC-MS-MS. Antimicrobial activities were determined against selected Gram-positive and Gram-negative indicator strains including many Legionella species.
2. Materials and Methods
2.1. Bacterial Strains
Pseudomonas strains used in this study are listed in Table 1 while other bacterial strains are listed in Tables 5 and 6. Pseudomonas strains were routinely cultured at 28°C for nonaeruginosa strains or 37°C for aeruginosa strains either on LB agar plates or in LB broth under shaking (180 rpm). LB was composed of 5 g/l Yeast extract, 10g/l Tryptone and 5 g/l NaCl. Legionella strains were cultured at 37°C either on buffered charcoal yeast extract (BCYE) agar plates or in buffered yeast extract (BYE) liquid medium under shaking (150 rpm). BYE was composed of 5 g/l N-(2-Acetamido)-2-aminoethanesulfonic acid, 10 g/l Yeast extract and pH 6.9. BCYE was made from BYE by adding 2 g/l Activated charcoal and 15 g/l Agar. L. pneumophila Lens CIP 108286 [31] was used as the main target for anti-Legionella activity assays. Other bacteria were grown either on Brain Heart Infusion (BHI; Fisher Scientific, Illkirch, France) agar plates or broth, at 30°C or 37°C, depending on the tested strain.
Table 1.
Anti-Legionella pneumophila activity of Pseudomonas strains.
| Pseudomonas strains | Anti-Legionella activity a | Reference/Source b | |
|---|---|---|---|
| Colony | Concentrated supernatant (15X) |
||
| P. aeruginosa 8H | +++ | +++ | EBI collection |
| P. aeruginosa CHA | +++ | +++ | [32] |
| P. aeruginosa UCBPP-PA14 | +++ | +++ | [33] |
| P. cepacia 4512 | - | - | EBI collection |
| P. sp DSS73 | +++ | ++ | [34] |
| P. fluorescens MFAD21c | +++ | + | [35] |
| P. fluorescens MFAH4a | +++ | + | [35] |
| P. fluorescens MFAO2 | +++ | ++ | [35] |
| P. fluorescens MFE01 | +++ | + | [36] |
| P. fluorescens PfA7b | +++ | + | LMSM collection |
| P. fulva 1324 | - | - | EBI collection |
| P. fulva B6 | - | - | EBI collection |
| P. fulva B8 | - | - | EBI collection |
| P. libanensis 4000 | - | - | EBI collection |
| P. otitidis 4014 | +++ | + | EBI collection |
| P. putida 1243 | - | - | EBI collection |
| P. putida 1312 | - | - | EBI collection |
| P. putida MFAF88 | ++ | + | [35] |
| P. putida MFAK14 | - | - | [35] |
| P. syringae MFAA66a | + | - | [35] |
| P. syringae MFAE88 | + | - | [35] |
The anti-Legionella activity of the concentrated supernatant for each strain grown in LB medium is the mean of three independent experiments.
a-: no activity.
+: zone of inhibition with a diameter < 4 mm.
++: zone of inhibition with a diameter between 4 and 8 mm.
+++: zone of inhibition with a diameter > 8 mm.
bEBI: Laboratoire Ecologie & Biologie des Interactions, UMR CNRS 7267, Université de Poitiers; LMSM: Laboratoire de Microbiologie Signaux et Microenvironnement, EA4312, Université de Rouen.
Table 5.
Antibacterial spectrum of lipopeptide mixtures produced by Pseudomonas strains.
| MIC of lipopeptide mixtures (mg/ml) | |||||||
|---|---|---|---|---|---|---|---|
| DSS73 | MFAD21c | MFAF88 | MFAH4a | MFAO2 | PfA7b | ||
| Gram positive | B. subtilis LMG 28342 | - | - | - | - | - | - |
| E. faecalis V583 | - | - | - | - | - | - | |
| L. ivanovii Li4pVS2 | - | - | - | - | - | - | |
| L. monocytogenes EGDe ATCC BAA-679 | - | - | - | - | - | - | |
|
| |||||||
| Gram negative | A. hydrophila LMG 2844 | - | - | - | - | - | - |
| E. coli LMG 2092 | - | - | - | - | - | - | |
| F. breve LMG 4011 | - | - | - | - | - | - | |
| K. pneumoniae 050283 | - | - | - | - | - | - | |
| P. aeruginosa LMG 1242 | - | - | - | - | - | - | |
| P. aeruginosa UCBPP-PA14 | - | - | - | - | - | - | |
| S. enterica J18 | - | - | - | - | - | - | |
| L. bozemanii ATCC 33217 | 0.09 | 0.14 | 0.61 | 0.19 | 0.38 | 0.20 | |
| L. dumoffii ATCC 33279 | 0.09 | 0.14 | 0.30 | 0.19 | 0.38 | 0.20 | |
| L. feeleii ATCC 35072 | 0.09 | 0.14 | 0.15 | 0.19 | 0.04 | 0.20 | |
| L. longbeachae ATCC 33484 | 0.18 | 0.14 | 0.30 | 0.19 | 0.19 | 0.20 | |
| L. micdadei ATCC 33218 | 0.18 | 0.27 | 0.30 | 0.19 | 0.19 | 0.20 | |
| L. pneumophila ATCC 33155 (Sg 3) | 0.18 | 0.27 | 0.61 | 0.38 | 0.38 | 0.40 | |
| L. pneumophila ATCC 33215 (Sg 6) | 0.36 | 0.27 | 0.61 | 0.38 | 0.38 | 0.40 | |
| L. pneumophila ATCC 33216 (Sg5) | 0.18 | 0.27 | 0.30 | 0.38 | 0.19 | 0.40 | |
| L. pneumophila ATCC Baa74 130b | 0.36 | 0.27 | 0.61 | 0.76 | 0.38 | 0.40 | |
| L. pneumophila Corby (Sg1) | 0.18 | 0.27 | 0.61 | 0.38 | 0.38 | 0.40 | |
| L. pneumophila Lens CIP 108286 (Sg 1) | 0.36 | 0.54 | 0.61 | 0.76 | 0.38 | 0.40 | |
-: no growth of inhibition was detected even with undiluted lipopeptide mixture (0.72 mg/ml for the DSS73 mixture, 1.08 mg/ml for the MFAD21c mixture, 0.76 mg/ml for the MFAH4a mixture, 0.76 mg/ml for the MFAO2 mixture, 1.6 mg/ml for the PfA7b mixture, and 1.22 mg/ml for the MFAF88 mixture).
Bacteria (106 CFU/ml) were incubated in BYE or BHI medium with two-fold dilutions of lipopeptide mixtures. Results correspond to the MIC after incubation for 24h or 96 h at 37°C depending on the tested strain and are the mean of three independent experiments.
Bacterial strains were obtained from various culture collections: ATCC: American Type Culture Collection, CIP: Collection Institut Pasteur, France, and LMG: BCCM/LMG Bacteria Collection, Ghent University, Belgium. The Corby strain was kindly provided by the Centre National de Référence des Légionelles (Lyon, France). Other strains were from the laboratory culture collection. Sg: serogroup.
Table 6.
Antibacterial spectrum of rhamnolipid mixtures produced by Pseudomonas strains.
| MIC of rhamnolipid mixtures ( µ g/ml) | |||||
|---|---|---|---|---|---|
| 8H | CHA | PA14 | 4014 | Commercial mixture | |
| A. hydrophila LMG 2844 | - | - | - | - | - |
| B. subtilis AM1 | 41.2 | 80 | 26.3 | 100 | - |
| E. faecalis V583 | 41.2 | - | - | - | - |
| E. coli DH5Y | 20.6 | - | - | - | - |
| F. breve LMG 4011 | 82.5 | - | - | - | - |
| K. pneumoniae 050283 | 165 | - | - | - | - |
| L. bozemanii ATCC 33217 | 0.2 | 6.2 | 1.6 | 5.2 | 7.8 |
| L. dumoffii ATCC 33279 | 0.4 | 12.5 | 0.4 | 5.2 | 15.6 |
| L. feeleii ATCC 35072 | 0.2 | 6.2 | 0.4 | 3.9 | 11.7 |
| L. longbeachae ATCC 33484 | 0.4 | 12.5 | 3.3 | 7.9 | 13.1 |
| L. micdadei ATCC 33218 | 0.03 | 3.1 | 3.3 | 3.9 | 15.6 |
| L. pneumophila ATCC 33155 (Sg 3) | 0.2 | 12.5 | 3.3 | 15.8 | 8.8 |
| L. pneumophila ATCC 33215 (Sg 6) | 0.2 | 6.2 | 0.4 | 7.9 | 3.9 |
| L. pneumophila ATCC 33216 (Sg5) | 0.05 | 25 | 1.6 | 7.9 | 19.5 |
| L. pneumophila ATCC Baa74 130b | 0.05 | 12.5 | 0.4 | 7.9 | 15.6 |
| L. pneumophila Corby (Sg1) | 0.2 | 12.5 | 3.3 | 5.2 | 11.7 |
| L. pneumophila Lens CIP 108286 (Sg1) | 0.4 | 12.5 | 1.6 | 10.5 | 11.7 |
| L. ivanovii Li4pVS2 | 41.2 | - | - | - | - |
| L. monocytogenes EGDe ATCC BAA-679 | 41.2 | - | - | - | - |
| P. aeruginosa LMG 1242 | - | - | - | - | - |
| P. aeruginosa PA14 | 82.5 | - | - | - | - |
| S. enterica J18 | 82.5 | - | - | - | - |
| S. aureus ATCC 29213 | - | 805 | 52.5 | 50 | - |
-: no growth of inhibition was detected even with undiluted rhamnolipid mixture (660 µg/ml for the 8H mixture, 1610 µg/ml for the CHA extract, 105 µg/ml for the PA14 extract, 100 µg/ml for the 4014 extract, and 250 µg/ml for the commercial mixture).
Bacteria (106 CFU/ml) were incubated in BYE or BHI medium with two-fold dilutions of rhamnolipid mixtures. Results correspond to the MIC after incubation for 24h or 96 h at 37°C depending on the tested strain and are the mean of three independent experiments.
Bacterial strains were obtained from various culture collections: ATCC: American Type Culture Collection, CIP: Collection Institut Pasteur, France, and LMG: BCCM/LMG Bacteria Collection, Ghent University, Belgium. The Corby strain was kindly provided by the Centre National de Référence des Légionelles (Lyon, France). Other strains were from the laboratory culture collection. Sg: serogroup.
2.2. Culture Conditions and Reagents
Rhamnolipids production was achieved by cultivating Pseudomonas strains at 30°C for 96 h under shaking (180 rpm) in a mineral salt medium (MSM) with mannitol (20 g.l−1) as the only carbon source [41]. For lipopeptides production, Pseudomonas strains were grown at 17°C for 96 h under shaking (180 rpm) in MSM medium supplemented with glucose (20 g.l−1).
A commercial solution of rhamnolipids was purchased from Sigma-Aldrich (Reference R95DD; Sigma-Aldrich, St. Louis, MO, USA). Rhamnolipids were dissolved in a water/acetonitrile (ACN) (65:35, v/v) mixture with 4 mM ammonium acetate at a final concentration of 5 mg.ml−1. Stock solutions were stored at 4°C and freshly diluted in sterile distilled water prior to each experiment. All other reagents were purchased from Sigma-Aldrich (Saint-Louis, MO, USA) unless stated otherwise.
2.3. In Vitro Antibacterial Assays
2.3.1. Spot on Lawn Assay
The target strain, L. pneumophila Lens, was spread (100 μl at OD600 = 0.1) onto a BCYE agar plate. Then, 10 μl of overnight cultures of each Pseudomonas strain was spotted onto the surface of the agar plate before incubation for 96 h at 28°C or 37°C. An inhibition area, around the producing strain, revealed the antibacterial activity.
2.3.2. Well Diffusion Assay
A L. pneumophila Lens suspension (100 μl at OD600 = 0.1) was spread onto a BCYE agar plate. Wells were punched into the agar and filled with 100 μl of 15 times concentrated supernatants of active Pseudomonas strains. Plates were then incubated 96 h at 37°C. Antibacterial activity was revealed by a zone of inhibition around the well.
2.3.3. Determination of Minimum Inhibitory Concentrations
Minimum inhibitory concentrations (MICs) of biosurfactants towards various bacterial strains were measured according to the dilution method detailed elsewhere [42]. MIC was defined as the lowest concentration of biosurfactant required to totally inhibit the growth of a selected strain after a chosen incubation period (24 h or 96 h), depending on the tested strain.
2.4. Detection of Biosurfactants
2.4.1. Drop Collapse Test
A drop of 50 μl of each active Pseudomonas strain supernatant, containing 2.5 μl of a 20 mg. ml−1 methylene blue solution, was placed on a piece of parafilm. Drops containing biosurfactants collapse, whereas nonsurfactant-containing drops remain stable. In this study, concentrated LB broth (15X) was used as a negative control while surfactin produced by Bacillus subtilis LMG 28342 [25] was used as a positive control.
2.4.2. Amino Acids and Sugars Detection
The peptide moiety of lipopeptides was detected using a 0.25% ninhydrin solution (prepared in acetone and acetic acid). A sample volume of 10 μl was spotted onto a silica plate (Saint-Louis, MO, USA). The plate was then sprayed with ninhydrin and heated at 105°C until development of a purple color. Surfactin produced by Bacillus subtilis LMG 28342 [25] was used as a positive control. The sugar moiety of rhamnolipids was detected using a 0.15% anthrone solution. A sample volume of 10 μl was spotted onto a silica plate and the plate was then sprayed with anthrone. The presence of sugars was revealed by the development of a blue color. Glucose and a commercial solution of rhamnolipids were both used as positive controls.
2.5. Extraction of Biosurfactants
2.5.1. Lipopeptides
Bacteria were removed from MSM culture medium by centrifugation (10,000 x g, 30 min, 4°C) and the resulting supernatant was sterilized by filtration through a 0.22 μm syringe filter (Sarstedt AG & Co. KG, Germany). Then, the cell-free supernatant was extracted three times with ethyl acetate (1:1, v/v). The collected organic fractions were evaporated under vacuum, and the residue was dissolved in 5 ml of H2O/ACN (3:2, v/v). The resulting solution was termed “Lipopeptides raw extract”.
2.5.2. Rhamnolipid
Bacteria were removed from MSM culture medium by centrifugation (10,000 x g, 30 min, 4°C) and the resulting supernatant was sterilized by filtration through a 0.22 μm syringe filter (Sarstedt AG & Co. KG, Germany). The supernatant pH was then adjusted to 3 (using 1M HCl). The resulting acidified cell-free supernatant was extracted three times with ethyl acetate (1:1, v/v) and the collected organic fractions were evaporated under vacuum. The crude extract was then dissolved in 5 ml of H2O/ACN (3:2, v/v) and the solution was named “Rhamnolipids raw extract”.
2.6. Purification of Biosurfactants
2.6.1. Lipopeptides
The “Lipopeptides raw extracts” were diluted in a H2O/ACN (1:1, v/v) mixture with 0.2% formic acid and separated by reverse-phase HPLC. Separation was conducted on a Chromolith® SpeedROD RP-18e reverse-phase HPLC column (4.6 x 50 mm) (Merck Millipore, Billerica, MA, USA) with a Dionex P680 HPLC pump, fitted with a Dionex UltiMate 3000 detector. Elution was monitored at 205 nm, 214 nm and 280 nm. Separation was carried out using a H2O/ACN/formic acid 0.2% (v/v) solvent system. After an initial 2 min wash with 60% ACN, elution was achieved in 23 min at a flow rate of 0.8 ml min−1 with an 18 min linear gradient from 60 to 100% ACN, followed by a 5 min wash with 100% ACN. All the collected fractions were lyophilized and stored at -20°C for further studies.
2.6.2. Rhamnolipids
The “Rhamnolipids raw extracts” were diluted in a H2O/ACN (1:1, v/v) mixture with 4 mM ammonium acetate and separated by reverse-phase HPLC similarly as described above for lipopeptides. Separation was carried out using a H2O/ACN/4mM ammonium acetate solvent system. After an initial 4 min wash with 35% ACN, elution was achieved in 31 min at a flow rate of 0.4 ml min−1 with a 5 min linear gradient from 35% to 50% ACN, 50% ACN for 6 min, followed by a linear 20 min gradient from 50% to 90% ACN. Fractions were collected every minute, lyophilized and stored at -20°C for further studies.
2.7. Mass Spectrometry Analyses of Biosurfactants
The molecular masses of biosurfactants were determined by electrospray ionization mass spectrometry (ESI-MS) with a Xevo Q-TOF (Waters, Milford, MA, USA) mass spectrometer. Samples were suspended in 50% ACN/0.2% formic acid (v/v). LC-MS mass spectra were performed, in positive mode for lipopeptides and negative mode for rhamnolipids, with a cone voltage ramping from 20 to 40 V. The spray voltage was set to 3.0 kV, the source temperature to 120°C and the desolvation temperature to 450°C. The LC separation was conducted using the same column and gradient as for HPLC analyses indicated previously except for the flow rate which was reduced to 0.5 ml min−1. LC-MS/MS mass spectra were performed in the MSE mode (Waters, Milford, MA, USA). Briefly, in the MSE mode, mass spectrometric scans alternate all along the experiment between low (10 V) and high (ramping from 30 to 60 V) fragmentation energy delivering for the same LC separation two chromatograms corresponding to MS and MS/MS analyses, respectively.
3. Results
3.1. Screening of a Pseudomonas Collection
Twenty-one Pseudomonas sp. strains of environmental or clinical origin and representing eight different species were screened for antagonistic activity against L. pneumophila Lens using a spot on lawn assay. As presented in Table 1, an inhibition zone was observed around all colonies of P. aeruginosa (3/3), P. fluorescens (5/5), and, with a smaller diameter, P. syringae (2/2). P. otitidis 4014 and P. sp. DSS73 also displayed a large zone of inhibition. Surprisingly, only one of the 4 P. putida strains tested appeared to be active against L. pneumophila Lens. Therefore, these strains inhibited L. pneumophila Lens growth via the production of, at least, one diffusible active compound. On the contrary, no zone of inhibition was observed around colonies of all tested P. fulva strains (3/3) as well as P. cepacia 4512 and P. libanensis 4000. These strains did not secrete any anti-Legionella compound or in too low concentration to be detected.
In order to demonstrate that antimicrobial compounds were effectively secreted by all the active strains, their culture supernatants were also tested against L. pneumophila Lens by well diffusion assay on BCYE plates (Table 1). Crude supernatants did not show any antibacterial activity but after being concentrated 15 times, an anti-Legionella activity, related to a zone of inhibition around the well containing samples, was observed. Anti-Legionella compounds were indeed secreted by active bacteria cultivated in broth medium, but at quite low amounts.
3.2. Chemical Nature of Active Compounds
Different members of the genus Pseudomonas are known to produce biosurfactants with antimicrobial activities [15, 28]. To check if the anti-Legionella activity could be attributed to biosurfactants, their presence in the culture supernatant was determined by the drop collapse test. This qualitative test is indicative of the surface-active and wetting activities [43] and it represents an indirect measurement of surface activity of a biosurfactant. Interestingly, the drops of culture supernatant for all inactive strains remained stable, whereas the culture supernatant of all active strains induced a drop collapse (Table 2). These results indicate that the anti-Legionella compounds secreted by Pseudomonas strains were correlated to the presence of biosurfactants. Therefore, putative biosurfactants were extracted from culture supernatants using ethyl acetate as previously described [41, 44]. The subsequent extracts were found active against L. pneumophila when tested using well diffusion assay (Table 1). On the contrary, similar extracts obtained from inactive strains did not display any activity. Moreover, all aqueous extracts that correspond to the remaining cell-free supernatants after extraction with ethyl acetate were found inactive too.
Table 2.
Determination of the chemical nature of active compounds produced by Pseudomonas strains.
| Pseudomonas strains | Drop collapse assay | Ethyl acetate extract activity | Coloration by anthrone | Coloration by ninhydrin |
|---|---|---|---|---|
| P. aeruginosa 8H | + | + | + | - |
| P. aeruginosa CHA | + | + | + | - |
| P. aeruginosa UCBPP-PA14 | + | + | + | - |
| P. cepacia 4512 | - | - | - | - |
| P. sp DSS73 | + | + | - | + |
| P. fluorescens MFAD21c | + | + | - | + |
| P. fluorescens MFAH4a | + | + | - | + |
| P. fluorescens MFAO2 | + | + | - | + |
| P. fluorescens MFE01 | + | + | - | + |
| P. fluorescens PfA7b | + | + | - | + |
| P. fulva 1324 | - | - | - | - |
| P. fulva B6 | - | - | - | - |
| P. fulva B8 | - | - | - | - |
| P. libanensis 4000 | - | - | - | - |
| P. otitidis 4014 | + | + | + | - |
| P. putida 1243 | - | - | - | - |
| P. putida 1312 | - | - | - | - |
| P. putida MFAF88 | + | + | - | + |
| P. putida MFAK14 | - | - | - | - |
| P. syringae MFAA66a | + | + | - | + |
| P. syringae MFAE88 | + | + | - | + |
+: three independent assays give a positive result.
-: three independent assays give a negative result.
The extracts obtained from the three P. aeruginosa strains and P. otitidis 4014 were found positively colored by anthrone, but not ninhydrin, revealing the presence of glucidic compounds whereas all the other active extracts were only colored by ninhydrin (Table 2). These results indicated that tested P. aeruginosa strains could produce rhamnolipids as well as P. otitidis 4014. Other active strains were found to produce biosurfactants containing peptidic moieties, probably lipopeptides. Finally, inactive strains did not produce any glucidic or peptidic biosurfactants as indicated by the lack of coloration (Table 2).
3.3. Purification and Identification of Lipopeptides
P. fluorescens (MAFD21c, DSS73, MFAO2, PfA7b, and MFAH4a) and P. putida MFAF88 were grown in MSM medium [44] and lipopeptides were extracted with ethyl acetate. For each strain, RP-HPLC chromatograms of ethyl acetate extracts displayed at least two peaks (data not shown) which could correspond to lipopeptides, according to Janek and coworkers [44]. In parallel, extracts were analyzed by LC-MS and LC-MS/MS in order to characterize active molecules. Results of these analyses are summarized in Table 3. All of the active fractions were found to contain a molecule displaying a molecular mass already described in the literature (Table 3) except for the P. sp. DSS73 fraction with a retention time of 11.9 min. The latter one molecular mass, which was named Amphisin-like, is reduced by 14 Da as compared to Amphisin and could correspond to the replacement of a leucine residue by a valine in the peptidic part of the molecule. The other lipopeptides produced by this strain correspond to Amphisin and Tensin which were also found in the P. fluorescens MFAO2 extract (Table 3). P. putida MFAF88 lipopeptides were identified as Putisolvin I and II. Massetolide E and Viscosin were identified in the P. fluorescens PfA7b extract. Finally, P. fluorescens MHA4a and MFAD21c were both found to produce PPZPM-1a and PPZPM-2a. Finally, full 1:1 identification is likely for the various lipopeptides, but must await further confirmation through chemical or genomic sequence analysis (Supplementary Table S1).
Table 3.
Pseudomolecular ions and main product ions obtained in positive mode from ESI-MSn (n=1, 2) analyses of purified active HPLC fractions from ethyl acetate extract of Pseudomonas sp. cell-free supernatants.
| Producing strain | Name | [M+H] + | Main product ions in MS-MS (m/z) | R t [min] | Relative area (%) | Anti-L. pneumophila activity | Reference |
|---|---|---|---|---|---|---|---|
| P. putida MFAF88 | Putisolvin I | 1380.7 | 454; 567; 863; 927; 1040; 1169 | 14.7 | 76.6 | + | [20] |
| Putisolvin II | 1394.7 | 1054; 1183 | 15.7 | 23.4 | - | ||
|
| |||||||
| P. fluorescens PfA7b | Massetolide E | 1112.6 | 496; 595; 829; 700 | 13.9 | 54.8 | + | [37] |
| Viscosin/Massetolide F | 1126.6 | 496; 595; 714; 843 | 14.8 | 45.2 | + | ||
|
| |||||||
| P. fluorescens MFAD21c | PPZPM-1a | 1253.8 | 284; 496; 609; 970; 1235 | 11.2 | 61.7 | + | [38] |
| PPZPM-2a | 1239.8 | 284; 496; 595; 956; 1221 | 12.2 | 38.3 | + | ||
|
| |||||||
| P. fluorescens MFAH4a | PPZPM-1a | 1253.8 | 284; 496; 609; 970; 1235 | 11.2 | 53.1 | + | [38] |
| PPZPM-2a | 1239.8 | 284; 496; 595; 956; 1221 | 12.1 | 46.9 | + | ||
|
| |||||||
| P. sp DSS73 | Amphisin | 1395.6 | 482; 595; 997; 1112 | 12.5 | 78.4 | + | [39, 40] |
| Amphisin-like | 1384.7 | 482; 595; 997; 1098 | 11.9 | 18.2 | - | ||
| Tensin | 1409.5 | 482; 595; 1011; 1026 | 13.2 | 3.4 | - | ||
|
| |||||||
| P. fluorescens MFAO2 | Amphisin | 1395.6 | 482; 595; 997; 1112 | 12.7 | 6.1 | - | [39, 40] |
| Tensin | 1409.8 | 482; 595; 1011; 1026 | 13.2 | 93.9 | + | ||
To estimate the proportion between lipopeptides contained in each extract, the relative quantity for each molecule was measured (Table 3). For some strains, in our conditions, lipopeptides were produced in similar amounts (P. fluorescens PfA7b and P. fluorescens MFAH4a) whereas for most of strains (P. fluorescens MFAO2, MFAD21c, P. sp. DSS73, and P. putida MFAF88) proportions of lipopeptides produced were really dissimilar. Thus, proportions of Amphisin, Amphisin-like and Tensin produced by the strain DSS73 were 78.4%, 18.2% and 3.4%, respectively, whereas for P. fluorescens MFAO2, the biosurfactant mixture was composed of 6.1% of Amphisin and 93.9% of Tensin.
3.4. Purification and Identification of Rhamnolipids
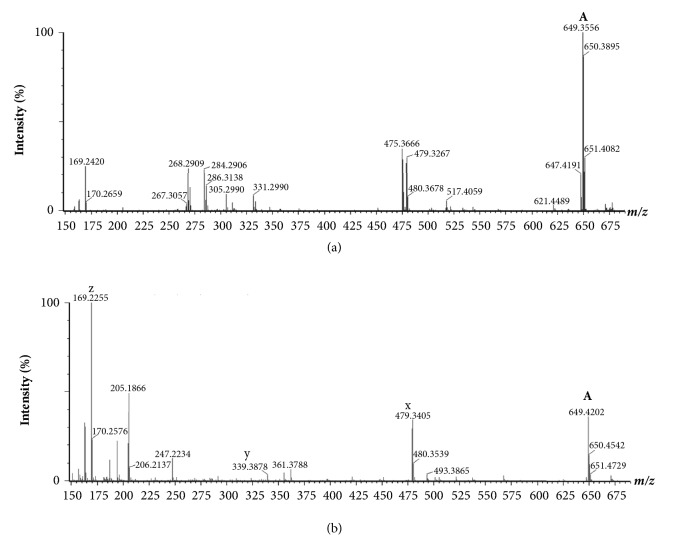
To characterize the chemical structure of rhamnolipids produced by P. aeruginosa strains (8H, CHA and UCBPP-PA14) and P. otitidis 4014, bacteria were cultivated in MSM broth containing mannitol as the only carbon source [41]. Rhamnolipids extracted with ethyl acetate from culture supernatants were then separated using RP-HPLC. Because rhamnolipids have no UV-absorptive properties, fractions were blind collected every minute during the elution and then tested against L. pneumophila. In parallel, extracts were analyzed by LC-MS and LC-MS/MS in order to characterize active molecules. All fractions, which were found active against Legionella, contained rhamnolipids with at least two congeners. Rhamnolipids consist of one or two units of rhamnose linked to one or two hydroxylated fatty acid with C8 to C12 chains, which could be saturated or not. The molecular masses of pseudomolecular ions and characteristic fragments observed, respectively, in MS and MS/MS spectra are listed in Table 4. Several types of rhamnolipids congeners of molecular masses in the range m/z 473-703, depending on their number of rhamnose residues and the length of their fatty acid chains, were observed. Structural characterization was achieved using MS/MS spectra by detection of characteristic fragments [29] and an example is detailed on Figures 1 and 2. The m/z of the pseudomolecular ion [M-H]− of component A is 649 Da (Figure 1(a)). Thus, the parent ion at m/z 649 was fragmented by MS/MS and showed daughter ions at m/z 479 (x), 309, 339 (y), and 169 (z) (Figure 1(b)). Fragment at m/z 309 is characteristic of di-rhamnolipids. Indeed, it corresponds to the di-rhamnosyl residue (Figure 2). The fragment at m/z 339 corresponds to the lipid moiety composed of two hydroxylated fatty acids containing ten carbons (C10). The fragment at m/z 479 results from the rupture of the ester bond between the two fatty acids. This fragment is characteristic of di-rhamnolipids carrying a C10 hydroxylated fatty acid directly linked to the carbohydrate part of the molecule. Taken together, the component A is a di-rhamnolipid carrying two C10 hydroxylated fatty acids (Rha-Rha-C10 -C10) (Figure 2).
Table 4.
Chemical composition of rhamnolipid mixtures produced by Pseudomonas strains.
| Rhamnolipid structure | [M-H]− (m/z) |
Ion fragments (m/z) |
Pseudomonas strain | |||
| PA14 | 8H | CHA | 4014 | |||
|
| ||||||
| Mono-rhamno-di-lipid | ||||||
|
| ||||||
| Rha-C8-C8 | 447 | Nd | + | - | - | - |
|
| ||||||
| Rha-C10-C8:1 | 473 | 327 | - | - | - | + |
|
| ||||||
| Rha-C8:1-C10 | 473 | 333 | - | - | - | + |
|
| ||||||
| Rha-C10-C8 | 475 | 333 | + | - | + | + |
|
| ||||||
| Rha-C8-C10 | 475 | 305 | + | - | + | + |
|
| ||||||
| Rha-C10-C10 | 503 | 339, 333, 169 | + | - | + | + |
|
| ||||||
| Rha-C10-C12:1 | 529 | 333, 195 | + | - | + | - |
|
| ||||||
| Rha-C12:1-C10 | 529 | 359, 169 | + | - | + | - |
|
| ||||||
| Rha-C10-C12 | 531 | 333 | + | - | - | - |
|
| ||||||
| Rha-C12-C10 | 531 | 361 | + | - | - | - |
|
| ||||||
| Di-rhamno-mono-lipid | ||||||
|
| ||||||
| Rha-Rha-C8 | 451 | Nd | + | - | + | - |
|
| ||||||
| Rha-Rha-C10 | 479 | Nd | + | - | + | - |
|
| ||||||
| Rha-Rha-C12:2 | 503 | Nd | + | - | + | - |
|
| ||||||
| Di-rhamno-di-lipid | ||||||
|
| ||||||
| Rha-Rha-C8:1-C8:1 | 589 | 449, 309, 279, 140 | - | - | - | + |
|
| ||||||
| Rha-Rha-C8-C8 | 593 | 451, 142 | + | + | + | - |
|
| ||||||
| Rha-Rha-C10-C8 | 621 | 479, 142 | + | + | + | + |
|
| ||||||
| Rha-Rha-C8-C10 | 621 | 451, 169 | + | + | + | + |
|
| ||||||
| Rha-Rha-C10-C10:1 | 647 | 479, 337, 309, 167 | - | + | - | - |
|
| ||||||
| Rha-Rha-C8-C12:1 | 647 | 451 | + | - | - | - |
|
| ||||||
| Rha-Rha-C12:1-C8 | 647 | 505 | + | - | - | - |
|
| ||||||
| Rha-Rha-C10-C10 | 649 | 479, 339, 309, 169 | + | + | + | + |
|
| ||||||
| Rha-Rha-C11-C10 | 665 | 494, 309, 169 | + | + | - | - |
|
| ||||||
| Rha-Rha-C10-C11 | 665 | 479, 309, 184 | + | + | - | - |
|
| ||||||
| Rha-Rha-C10-C12:1 | 675 | 479, 365, 309, 195 | + | + | + | + |
|
| ||||||
| Rha-Rha-C12:1-C10 | 675 | 505, 365, 309, 169 | + | + | + | + |
|
| ||||||
| Rha-Rha-C10-C12 | 677 | 479, 367, 309, 197 | + | + | + | + |
|
| ||||||
| Rha-Rha-C12-C10 | 677 | 507, 367, 309, 169 | + | + | + | + |
|
| ||||||
| Rha-Rha-C12-C12:1 | 703 | 507, 311 | + | - | - | - |
|
| ||||||
| Rha-Rha-C12:1-C12 | 703 | 505, 311 | + | - | - | - |
+: presence of the rhamnolipid in the extract.
-: absence of the rhamnolipid in the extract.
Nd: not detected.
Figure 1.
MS (a) and MS/MS (b) spectra of the RP-HPLC fraction eluted at 19 min from P. aeruginosa CHA extract.
Figure 2.
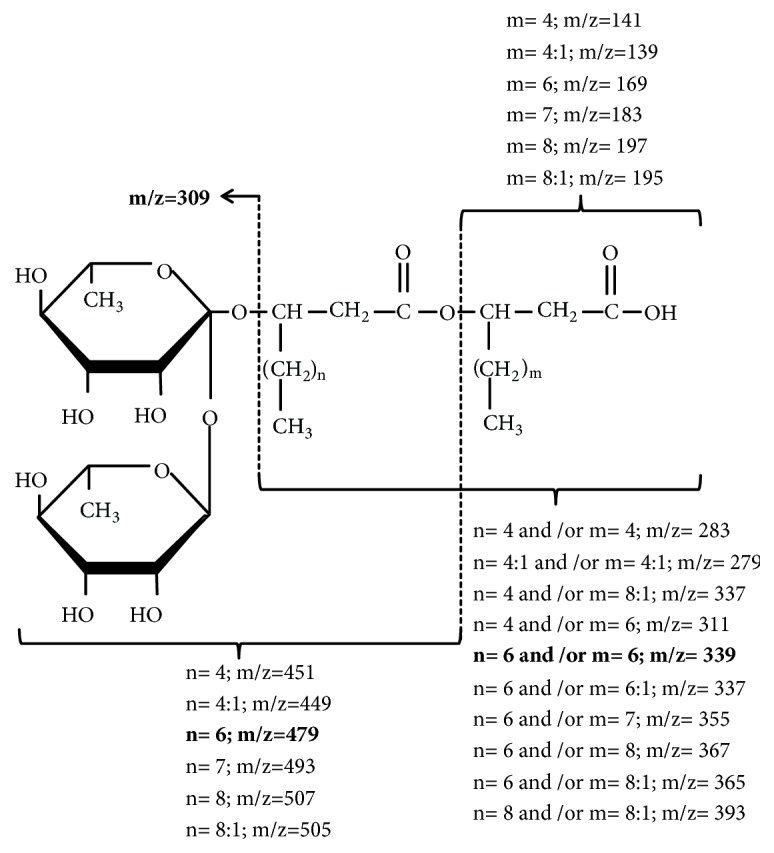
Fragmentation of di-rhamnolipids observed in MS/MS analysis. The m/z of the fragments as a function of the structures of fatty acids is indicated.
Up to 29 rhamnolipids homologues containing one or two rhamnose residues linked to one or two hydroxylated fatty acids were identified in Pseudomonas culture supernatants (Table 4). P. aeruginosa PA14 produced the highest number of homologues (25) and the lowest was found for P. aeruginosa 8H (11). Only 7 rhamnolipid homologues (Rha-C10 C8; Rha-C10 C10; Rha-Rha-C10 C8; Rha-Rha-C10 C10; Rha-Rha-C10 C12:1; Rha-Rha-C10 C12; Rha-Rha-C10) were produced by the 3 strains. Interestingly, P. aeruginosa 8H secreted only di-rhamnosyl carrying two fatty acids species.
Finally, the orcinol reaction revealed that extracts contained rhamnose amounts of 79.95 ± 16.61 mg/ml for P. aeruginosa 8H, 66.84 ± 3.43 mg/ml for P. aeruginosa CHA, 10.99 ± 4.18 mg/ml for P. aeruginosa UCBPP-PA14 and 4.07 ± 1.56 mg/ml for P. otitidis 4014. It has to be noted that the rhamnolipid content of P. aeruginosa 8H extract is overestimated because it contains only di-rhamnosyl rhamnolipids contrarily to the other strains extracts.
3.5. Anti-Legionella Activity of Biosurfactants
3.5.1. Lipopeptides
Firstly, HPLC fractions corresponding to identified lipopeptides were collected, concentrated and tested against L. pneumophila. All fractions were found active except those corresponding to the molecules with the lowest proportions, Putisolvin II (23.4%), Amphisin-like (18.2%), Tensin in DSS73 extract (3.4%) and Amphisin in MFAO2 extract (6.1%) (Table 3). Because Amphisin and Tensin were found active when obtained in larger amounts from other extracts, we supposed that Tensin from the strain DSS73, Putisolvin II and Amphisin-like appeared inactive because of their low concentration. It has to be noted that it is impossible to quantify the amounts of lipopeptides using classical colorimetric methods. To confirm this proposal, fractions corresponding to each of these two molecules were obtained from more than twenty HPLC runs and pooled before being concentrated. In both cases, fractions were found active against L. pneumophila. In conclusion, all the lipopeptides produced by Pseudomonas species were found to be active against L. pneumophila.
Secondly, the antagonistic potency of the purified lipopeptide mixtures was determined against various bacterial strains previously used in antibacterial assays [25, 45, 46]. Because it was difficult to quantify the lipopeptide content of extracts, activities were expressed as a function of the first twofold dilution of the extract which totally inhibited the growth of the target bacteria (Table 5). All of the mixtures were more or less active against Legionella species. However, mixtures were not active against the other Gram-negative or Gram-positive bacteria tested so far. Thus, these results highly suggest a specific activity of lipopeptide mixtures against bacteria of the Legionella genus. The P. fluorescens MFAH4a extract seemed to be the less active one, with activities observed only for undiluted to four times diluted solutions. On the contrary, P. fluorescens PfA7b extract appeared to be the most antibacterial extract (Table 5).
3.5.2. Rhamnolipids
MICs of the rhamnolipid mixtures produced by Pseudomonas strains cultured in MSM broth were determined against the same collection of bacterial strains used for lipopeptides. Results are given in Table 6. Extracts were found highly active against all the Legionella sp. tested with low MIC values between 0.027 and 25 μg/ml. The mixture produced by P. aeruginosa 8H was the most effective against Legionella sp, with the lowest MIC values. On the opposite, the extract originating from P. aeruginosa CHA culture supernatant and commercial rhamnolipid mixture displayed 10 to 100-fold higher MICs.
Interestingly, rhamnolipid mixtures were found less or not active against other tested bacteria. B. subtilis appeared sensitive to the rhamnolipid extracts but not to commercial mixture. The P. aeruginosa 8H extract displayed activity against most of the other bacteria tested but not S. aureus. In contrast, S. aureus was found sensitive to P. aeruginosa CHA (52.5 μg/ml) extracts. Finally, the P. aeruginosa 8H extract appears about 2 to 10 times more active than the one from P. aeruginosa PA14 and displays an anti-Legionella activity from 5 to 50 times higher than both P. aeruginosa CHA and the commercial mixture. Taken together, these results indicate that Legionella sp. are particularly sensitive to the rhamnolipid mixtures produced by Pseudomonas sp.
In order to evaluate the activity of various homologues of rhamnolipids, HPLC fractions obtained from the four producing strains were tested against L. pneumophila and then their content was analyzed by LC-MS in order to identify all the rhamnolipid species (Data not shown). All fractions were found more or less active against Legionella according to their amount of rhamnolipids, estimated from their peaks area in LC-MS chromatograms (data not shown). However, all these fractions were found to contain at least two rhamnolipid homologues. Consequently, even if many of these molecules are undoubtedly active against Legionella, it is not possible to affirm which one is effectively active against Legionella.
4. Discussion
Artificial water settings provide suitable conditions for growth and multiplication of waterborne pathogens including L. pneumophila. In those nutrient-poor environments, L. pneumophila is able to interact positively with other microorganisms to obtain the nutrients it requires to survive [9, 47]. Although much work has been conducted on the stimulation of Legionella growth by other microorganisms, little work has been done on the negative interactions that occur between Legionella bacteria and other microorganisms in man-made water systems. Compilation of latest findings shows that many bacterial genera isolated from drinking water pipes were able to inhibit the growth of Legionella species [13, 48–50]. While active isolated strains were taxonomically diverse, bacteria belonging to the Pseudomonas genus were always found or tested. Another constant of those studies is the lack of molecular identification of active compounds that have been thought to be bacteriocins or bacteriocin-like substances [13, 48].
To further investigate the chemical nature of those anti-Legionella compounds, a Pseudomonas sp. collection, comprising both environmental and clinical strains, was defined and screened. Among the 21 tested strains, 14 were active against L. pneumophila (66.7%). This result is in good agreement with the data published by Guerrieri and coworkers as they found 72% of active Pseudomonas strains in their collection [13]. The anti-Legionella compounds secreted by Pseudomonas strains were then correlated to the presence of biosurfactants, as Pseudomonas members are well known to produce many biosurfactants with antimicrobial activities [15, 28].
Lipopeptides constitute a specific class of microbial secondary metabolites produced by a wide range of microorganisms. Moreover, those produced by Bacillus and Pseudomonas species are the most studied by far [17]. Here, all active fractions purified from five P. fluorescens and one P. putida ethyl acetate extracts contained various already known lipopeptides. According to the literature, only the molecule with a molecular mass of 1383 Da is original. Thus, this lipopeptide was named Amphisin-like as its molecular mass is 14 Da lower than the molecular mass of Amphisin [39]. We also found lipopeptides belonging to the orfamide group named PPZPMs, a group of CLPs thought to be the missing link between the viscosin and Amphisin groups due to the number of amino acids forming the cyclic moiety [22, 38]. Lipopeptides are mainly characterized by highly structural diversity and are considered as multifunctional microbial tools. Indeed, they exhibit a very wide range of biological activities including lytic and growth-inhibitory activities against a broad range of microorganisms [14]. In particular, many authors have reported antibacterial activities (for review see [17]). Usually, Gram-negative bacteria are poorly inhibited by lipopeptides whereas Gram-positive bacteria appear more susceptible [15, 17, 19]. To date, only one lipopeptide was reported to be active against Legionella species [25]. It corresponds to surfactin, a well characterized lipopeptide produced by Bacillus species. However, all the lipopeptides tested were found active against Legionella species. It is the first time, to our knowledge, that Pseudomonas biosurfactants were shown to be active against Legionella. Interestingly, other Gram-negative and Gram-positive bacteria tested were insensitive to those compounds used at a similar concentration. In many studies, when available, MIC values of lipopeptides against bacteria ranged from less than 10 μg/ml (massetolide A and viscosin against M. tuberculosis) to around 1 mg/L (milkisin against S. enterica) [37, 51]. Thus, MIC determined in this study were in a similar concentration range. Nevertheless, Legionella species appeared more sensitive to surfactin than to Pseudomonas lipopeptides as MIC values were lower (1-4 μg/ml) [25]. Interestingly, Loiseau and coworkers showed that nonlegionella bacterial strains tested were resistant to surfactin, even at the highest tested concentration (265 μg/mL). Taken together, those data highlight a very specific sensitivity of Legionella bacteria to lipopeptides.
Interestingly, rhamnolipids extracts were active against all the Legionella tested, whatever their species. The percentage of di-rhamno-di-lipid in each mixture was higher than those of monorhamno-di-lipid and di-rhamno-monolipid. This result is in good agreement with previous observation of Arino and coworkers [52]. Interestingly, the P. aeruginosa 8H extract, which is the more active, contained the lower number of rhamnolipid congeners as compared to other extracts. It could be related to the higher concentration of each rhamnolipid molecule in the extract. Indeed, in all cases, the rhamnolipid content was evaluated as a function of its rhamnose content. Thus, the mean concentration of each congener is higher for P. aeruginosa 8H which contained 11 different rhamnolipids than those of P. aeruginosa CHA (16 congeners), PA14 (25 congeners) and P. otitidis 4014 (13 congeners). However, this cannot explain the higher activity of P. aeruginosa PA14 as compared to P. aeruginosa CHA. The other main difference of P. aeruginosa 8H extract as compared to the others corresponds to its content restricted to di-rhamnosyl species which could then be related to its higher anti-Legionella activity. Moreover, this P. aeruginosa 8H extract was found to be the most active towards no-Legionella strains except S. aureus and B. subtilis for which the P. aeruginosa PA14 and P. otitidis 4014 extracts were found the most active. On the other hand, the insensitivity of S. aureus to the P. aeruginosa 8H extract could be related to its particular content restricted to di-rhamnosyl species. Indeed, the comparison of the rhamnolipids congeners produced by P. aeruginosa PA14 (this study), CHA (this study), AT10 [53], LBI [29] and RL 47T2 [30] did not reveal the presence of a specific anti-Staphylococcus aureus compound. However, all these extracts contained monorhamnosyl species which could be postulated to exert this specific activity. Strikingly, Legionella species were shown to be highly sensitive to rhamnolipid mixtures as MIC values were quite low (0.03-19.5 μg/ml) while throughout the literature, many sensitive bacterial species displayed higher MIC values [27, 28]. Does Legionella bacteria possess some specificity that could explain this high sensitivity? As these compounds are membrane active, maybe a part of the answer is hidden in the composition of the cell envelope. Indeed, Legionella are also highly sensitive to detergents (SDS, Tween 80, Triton X-100…) or detergent-like molecules such as antimicrobial peptides [25, 45, 54, 55]. Thus, the phospholipid composition of Legionella cell envelope as well as the membrane thickness, the fluidity, the presence of phospholipids clusters and even composition of the lipopolysaccharide could be key parameters involved in Legionella sensitivity towards membrane-active compounds [56].
5. Conclusions
In this study, we showed, for the first time, that biological challengers present in the microenvironment of Legionella such as Pseudomonas bacteria exhibit antagonistic activity because of the production of various biosurfactants species. These compounds are known to be multifunctional biomolecules with many depicted potential biotechnological applications including their use as antimicrobials [57]. Thus, the wide sensitivity of Legionella species to rhamnolipids and lipopeptides make biosurfactants promising tool for their biological control in water treatment industry although experimental data are needed to evaluate how effective biosurfactants would be in real conditions.
Acknowledgments
The authors gratefully acknowledge S. Rebuffat for technical assistance in the identification of P. fluorescens MFAD21c and MFAF4a lipopeptides. This study was funded by 4 grants. C. Loiseau was supported by a grant from the French Minister of Research. This work was partly supported by an internal funding of the CNRS UMR 7267 Laboratory and partly granted by the following 2015–2020 programs: the State-Region Planning Contracts (CPER) and the European Regional Development Fund (FEDER).
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that there are no conflicts of interest regarding the publication of this paper.
Authors' Contributions
Clémence Loiseau and Emilie Portier contributed equally to this work.
Supplementary Materials
Table S1: lipopeptide structures detected in active HPLC fractions from ethyl acetate extract of Pseudomonas sp. cell-free supernatants according to their pseudomolecular ion masses as described in the literature.
References
- 1.Beauté J. Legionnaires’ disease in Europe, 2011 to 2015. Eurosurveillance. 2017;22(27) doi: 10.2807/1560-7917.ES.2017.22.27.30566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu V. L., Plouffe J. F., Pastoris M. C., et al. Distribution of legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: An international collaborative survey. The Journal of Infectious Diseases. 2002;186(1):127–128. doi: 10.1086/341087. [DOI] [PubMed] [Google Scholar]
- 3.Newton H. J., Ang D. K. Y., Van Driel I. R., Hartland E. L. Molecular pathogenesis of infections caused by Legionella pneumophila. Clinical Microbiology Reviews. 2010;23(2):274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falkinham J. O., Hilborn E. D., Arduino M. J., Pruden A., Edwards M. A. Epidemiology and ecology of opportunistic premise plumbing pathogens: Legionella pneumophila, Mycobacterium avium, and Pseudomonas aeruginosa. Environmental Health Perspectives. 2015;123(8):749–758. doi: 10.1289/ehp.1408692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lesnik R., Brettar I., Höfle M. G. Legionella species diversity and dynamics from surface reservoir to tap water: From cold adaptation to thermophily. The ISME Journal. 2016;10(5):1064–1080. doi: 10.1038/ismej.2015.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valero N., De Simón M., Gallés P., et al. Street cleaning trucks as potential sources of Legionella pneumophila. Emerging Infectious Diseases. 2017;23(11):1880–1882. doi: 10.3201/eid2311.161390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashbolt N. J. Environmental (saprozoic) pathogens of engineered water systems: Understanding their ecology for risk assessment and management. Pathogens. 2015;4(2):390–405. doi: 10.3390/pathogens4020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrison L. E., Kunz J. M., Cooley L. A., et al. Vital Signs: Deficiencies in Environmental Control Identified in Outbreaks of Legionnaires' Disease-North America, 2000-2014. American Journal of Transplantation. 2016;16(10):3049–3058. doi: 10.1111/ajt.14024. [DOI] [PubMed] [Google Scholar]
- 9.Abdel-Nour M., Duncan C., Low D. E., Guyard C. Biofilms: The stronghold of Legionella pneumophila. International Journal of Molecular Sciences. 2013;14(11):21660–21675. doi: 10.3390/ijms141121660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Correia A. M., Ferreira J. S., Borges V. Probable person-to-person transmission of Legionnaires' disease. The New England Journal of Medicine. 2016;374(5):497–498. doi: 10.1056/nejmc1505356. [DOI] [PubMed] [Google Scholar]
- 11.Bärlocher K., Welin A., Hilbi H. Formation of the Legionella replicative compartment at the crossroads of retrograde trafficking. Frontiers in Cellular and Infection Microbiology. 2017;7:p. 482. doi: 10.3389/fcimb.2017.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliva G., Sahr T., Buchrieser C. The Life Cycle of L. pneumophila: Cellular Differentiation Is Linked to Virulence and Metabolism. Frontiers in Cellular and Infection Microbiology. 2018;8:1–12. doi: 10.3389/fcimb.2018.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guerrieri E., Bondi M., Sabia C., De Niederhäusern S., Borella P., Messi P. Effect of bacterial interference on biofilm development by Legionella pneumophila. Current Microbiology. 2008;57(6):532–536. doi: 10.1007/s00284-008-9237-2. [DOI] [PubMed] [Google Scholar]
- 14.Raaijmakers J. M., de Bruijn I., Nybroe O., Ongena M. Natural functions of lipopeptides from Bacillus and Pseudomonas: more than surfactants and antibiotics. FEMS Microbiology Reviews. 2010;34(6):1037–1062. doi: 10.1111/j.1574-6976.2010.00221.x. [DOI] [PubMed] [Google Scholar]
- 15.Inès M., Dhouha G. Lipopeptide surfactants: Production, recovery and pore forming capacity. Peptides. 2015;71:100–112. doi: 10.1016/j.peptides.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Pamp S. J., Tolker-Nielsen T. Multiple roles of biosurfactants in structural biofilm development by Pseudomonas aeruginosa. Journal of Bacteriology. 2007;189(6):2531–2539. doi: 10.1128/JB.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mnif I., Ghribi D. Lipopeptides biosurfactants: Mean classes and new insights for industrial, biomedical, and environmental applications. Biopolymers. 2015;104(3):129–147. doi: 10.1002/bip.22630. [DOI] [PubMed] [Google Scholar]
- 18.Irorere V. U., Tripathi L., Marchant R., McClean S., Banat I. M. Microbial rhamnolipid production: a critical re-evaluation of published data and suggested future publication criteria. Applied Microbiology and Biotechnology. 2017;101(10):3941–3951. doi: 10.1007/s00253-017-8262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raaijmakers J. M., De Bruijn I., De Kock M. J. D. Cyclic lipopeptide production by plant-associated Pseudomonas spp.: Diversity, activity, biosynthesis, and regulation. Molecular Plant-Microbe Interactions. 2006;19(7):699–710. doi: 10.1094/MPMI-19-0699. [DOI] [PubMed] [Google Scholar]
- 20.Kuiper I., Lagendijk E. L., Pickford R., et al. Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Molecular Microbiology. 2004;51(1):97–113. doi: 10.1046/j.1365-2958.2003.03751.x. [DOI] [PubMed] [Google Scholar]
- 21.Berti A. D., Greve N. J., Christensen Q. H., Thomas M. G. Identification of a biosynthetic gene cluster and the six associated lipopeptides involved in swarming motility of Pseudomonas syringae pv. tomato DC3000. Journal of Bacteriology. 2007;189(17):6312–6323. doi: 10.1128/JB.00725-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross H., Stockwell V. O., Henkels M. D., Nowak-Thompson B., Loper J. E., Gerwick W. H. The Genomisotopic Approach: A Systematic Method to Isolate Products of Orphan Biosynthetic Gene Clusters. Chemistry & Biology. 2007;14(1):53–63. doi: 10.1016/j.chembiol.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Arima K., Kakinuma A., Tamura G. Surfactin, a crystalline peptidelipid surfactant produced by Bacillus subtilis: Isolation, characterization and its inhibition of fibrin clot formation. Biochemical and Biophysical Research Communications. 1968;31(3):488–494. doi: 10.1016/0006-291X(68)90503-2. [DOI] [PubMed] [Google Scholar]
- 24.Ongena M., Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends in Microbiology. 2008;16(3):115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Loiseau C., Schlusselhuber M., Bigot R., Bertaux J., Berjeaud J.-M., Verdon J. Surfactin from Bacillus subtilis displays an unexpected anti-Legionella activity. Applied Microbiology and Biotechnology. 2015;99(12):5083–5093. doi: 10.1007/s00253-014-6317-z. [DOI] [PubMed] [Google Scholar]
- 26.Soberón-Chávez G., Lépine F., Déziel E. Production of rhamnolipids by Pseudomonas aeruginosa. Applied Microbiology and Biotechnology. 2005;68(6):718–725. doi: 10.1007/s00253-005-0150-3. [DOI] [PubMed] [Google Scholar]
- 27.Chen J., Wu Q., Hua Y., Chen J., Zhang H., Wang H. Potential applications of biosurfactant rhamnolipids in agriculture and biomedicine. Applied Microbiology and Biotechnology. 2017;101(23-24):8309–8319. doi: 10.1007/s00253-017-8554-4. [DOI] [PubMed] [Google Scholar]
- 28.Abdel-Mawgoud A. M., Lépine F., Déziel E. Rhamnolipids: Diversity of structures, microbial origins and roles. Applied Microbiology and Biotechnology. 2010;86(5):1323–1336. doi: 10.1007/s00253-010-2498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benincasa M., Abalos A., Oliveira I., Manresa A. Chemical structure, surface properties and biological activities of the biosurfactant produced by Pseudomonas aeruginosa LBI from soapstock. Antonie van Leeuwenhoek-Journal of Microbiology. 2004;85(1):1–8. doi: 10.1023/B:ANTO.0000020148.45523.41. [DOI] [PubMed] [Google Scholar]
- 30.Haba E., Pinazo A., Jauregui O., Espuny M. J., Infante M. R., Manresa A. Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnology and Bioengineering. 2003;81(3):316–322. doi: 10.1002/bit.10474. [DOI] [PubMed] [Google Scholar]
- 31.Cazalet C., Jarraud S., Ghavi-Helm Y., et al. Multigenome analysis identifies a worldwide distributed epidemic Legionella pneumophila clone that emerged within a highly diverse species. Genome Research. 2008;18(3):431–441. doi: 10.1101/gr.7229808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toussaint B., Delic-Attree I., Vignais P. M. Pseudomonas aeruginosa contains an IHF-like protein that binds to the algD promoter. Biochemical and Biophysical Research Communications. 1993;196(1):416–421. doi: 10.1006/bbrc.1993.2265. [DOI] [PubMed] [Google Scholar]
- 33.Rahme L., Stevens E., Wolfort S., Shao J., Tompkins R., Ausubel F. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268:1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 34.Groboillot A., Portet-Koltalo F., le Derf F., Feuilloley M. J. G., Orange N., Poc C. D. Novel application of cyclolipopeptide amphisin: Feasibility study as additive to remediate polycyclic aromatic hydrocarbon (PAH) contaminated sediments. International Journal of Molecular Sciences. 2011;12(3):1787–1806. doi: 10.3390/ijms12031787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duclairoir Poc C., Verdon J., Groboillot A., et al. Original Research Article Airborne fluorescent pseudomonads: What potential for virulence? International Journal of Current Microbiology and Applied Sciences (IJCMAS) 2014;3(8):708–722. [Google Scholar]
- 36.Decoin V., Barbey C., Bergeau D., et al. A type VI secretion system is involved in Pseudomonas fluorescens bacterial competition. PLoS ONE. 2014;9(2) doi: 10.1371/journal.pone.0089411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerard J., Lloyd R., Barsby T., Haden P., Kelly M. T., Andersen R. J. Massetolides A-H, antimycobacterial cyclic depsipeptides produced by two pseudomonads isolated from marine habitats. Journal of Natural Products. 1997;60(3):223–229. doi: 10.1021/np9606456. [DOI] [PubMed] [Google Scholar]
- 38.Weisshoff H., Hentschel S., Zaspel I., Jarling R., Krause E., Pham T. L. H. PPZPMs - A novel group of cyclic lipodepsipeptides produced by the Phytophthora alni associated strain Pseudomonas sp. JX090307 - The missing link between the viscosin and amphisin group. Natural Product Communications (NPC) 2014;9(7):989–996. [PubMed] [Google Scholar]
- 39.Sørensen D., Nielsen T. H., Christophersen C., Sørensen J., Gajhede M. Cyclic lipoundecapeptide amphisin from Pseudomonas sp. Strain DSS73. Acta Crystallographica Section C: Crystal Structure Communications. 2001;57(9):1123–1124. doi: 10.1107/S0108270101010782. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen T. H., Thrane C., Christophersen C., Anthoni U., Sørensen J. Structure, production characteristics and fun gel antagonism of tensin—a new antifungal cyclic lipopeptide from Pseudomonas fluorescens strain 96.578. Journal of Applied Microbiology. 2000;89(6):992–1001. doi: 10.1046/j.1365-2672.2000.01201.x. [DOI] [PubMed] [Google Scholar]
- 41.Déziel E., Lépine F., Dennie D., Boismenu D., Mamer O. A., Villemur R. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1999;1440(2-3):244–252. doi: 10.1016/S1388-1981(99)00129-8. [DOI] [PubMed] [Google Scholar]
- 42.Verdon J., Berjeaud J.-M., Lacombe C., Héchard Y. Characterization of anti-Legionella activity of warnericin RK and delta-lysin I from Staphylococcus warneri. Peptides. 2008;29(6):978–984. doi: 10.1016/j.peptides.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 43.Youssef N. H., Duncan K. E., Nagle D. P., Savage K. N., Knapp R. M., McInerney M. J. Comparison of methods to detect biosurfactant production by diverse microorganisms. Journal of Microbiological Methods. 2004;56(3):339–347. doi: 10.1016/j.mimet.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 44.Janek T., Łukaszewicz M., Rezanka T., Krasowska A. Isolation and characterization of two new lipopeptide biosurfactants produced by Pseudomonas fluorescens BD5 isolated from water from the Arctic Archipelago of Svalbard. Bioresource Technology. 2010;101(15):6118–6123. doi: 10.1016/j.biortech.2010.02.109. [DOI] [PubMed] [Google Scholar]
- 45.Schlusselhuber M., Humblot V., Casale S., et al. Potent antimicrobial peptides against Legionella pneumophila and its environmental host, Acanthamoeba castellanii. Applied Microbiology and Biotechnology. 2015;99(11):4879–4891. doi: 10.1007/s00253-015-6381-z. [DOI] [PubMed] [Google Scholar]
- 46.Verdon J., Coutos-Thevenot P., Rodier M.-H., et al. Armadillidin H, a glycine-rich peptide from the terrestrial crustacean Armadillidium vulgare, displays an unexpected wide antimicrobial spectrum with membranolytic activity. Frontiers in Microbiology. 2016;7 doi: 10.3389/fmicb.2016.01484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor M., Ross K., Bentham R. Legionella, protozoa, and biofilms: Interactions within complex microbial systems. Microbial Ecology. 2009;58(3):538–547. doi: 10.1007/s00248-009-9514-z. [DOI] [PubMed] [Google Scholar]
- 48.Cotuk A., Dogruoz N., Zeybek Z., Kimiran-Erdem A., Ilhan-Sungur E. The effects of Pseudomonas and Aeromonas strains on Legionella pneumophila growth. Annals of Microbiology. 2005;55(3):219–224. [Google Scholar]
- 49.Toze S., Sly L. I., MacRae I. C., Fuerst J. A. Inhibition of growth of Legionella species by heterotrophic plate count bacteria isolated from chlorinated drinking water. Current Microbiology. 1990;21(2):139–143. doi: 10.1007/BF02091832. [DOI] [Google Scholar]
- 50.Solimini A. G., Cottarelli A., Marinelli L., De Giusti M. Factors influencing persistence of Legionella pneumophila serogroup 1 in laboratory cocultures. BMC Microbiology. 2014;14(1):1–8. doi: 10.1186/s12866-014-0249-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schlusselhuber M., Godard J., Sebban M., et al. Characterization of Milkisin, a Novel Lipopeptide With Antimicrobial Properties Produced By Pseudomonas sp. UCMA 17988 Isolated From Bovine Raw Milk. Frontiers in Microbiology. 2018;9 doi: 10.3389/fmicb.2018.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arino S., Marchal R., Vandecasteele J.-P. Identification and production of a rhamnolipidic biosurfactant by a Pseudomonas species. Applied Microbiology and Biotechnology. 1996;45(1-2):162–168. doi: 10.1007/s002530050665. [DOI] [Google Scholar]
- 53.Abalos A., Pinazo A., Infante M. R., Casals M., García F., Manresa A. Physicochemical and antimicrobial properties of new rhamnolipids produced by Pseudomonas aeruginosa AT10 from soybean oil refinery wastes. Langmuir. 2001;17(5):1367–1371. doi: 10.1021/la0011735. [DOI] [Google Scholar]
- 54.Verdon J., Falge M., Maier E., et al. Detergent-like activity and a-helical structure of warnericin RK, an anti-Legionella peptide. Biophysical Journal. 2009;97(7):1933–1940. doi: 10.1016/j.bpj.2009.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marchand A., Verdon J., Lacombe C., Crapart S., Héchard Y., Berjeaud J. M. Anti-Legionella activity of staphylococcal hemolytic peptides. Peptides. 2011;32(5):845–851. doi: 10.1016/j.peptides.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 56.Berjeaud J.-M., Chevalier S., Schlusselhuber M., et al. Legionella pneumophila: The paradox of a highly sensitive opportunistic waterborne pathogen able to persist in the environment. Frontiers in Microbiology. 2016;7 doi: 10.3389/fmicb.2016.00486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santos D. K. F., Rufino R. D., Luna J. M., Santos V. A., Sarubbo L. A. Biosurfactants: Multifunctional biomolecules of the 21st century. International Journal of Molecular Sciences. 2016;17(3):1–31. doi: 10.3390/ijms17030401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: lipopeptide structures detected in active HPLC fractions from ethyl acetate extract of Pseudomonas sp. cell-free supernatants according to their pseudomolecular ion masses as described in the literature.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.