Abstract
Genomic variants associated with inherited cardiac conditions yet detected incidentally (‘secondary findings’) are likely to arise with increasing frequency as genome sequencing transitions into clinical practice. Since genotyping has until recently been directed by clinical diagnosis, assessment and management of individuals found to harbour such a variant as a secondary finding is unclear. Here we illustrate some diagnostic and psychosocial complexities of inherited cardiac condition secondary findings, exemplified by disclosure of a pathogenic variant in KCNQ1, associated with long QT syndrome, to a healthy male enrolled in diagnostic genome sequencing as an unaffected relative. This early case represents a shift from ‘phenotype-to-genotype’ to ‘genotype-to-phenotype’; we describe clinical evaluation, family history and a qualitative research interview with the secondary finding recipient, discuss the role of specialist services in variant interpretation, genetic counselling and clinical assessment, and some challenges of realising improved health outcomes following disclosure of a secondary finding.
Journal Subject Terms: Ethics and Policy, Arrhythmias
Keywords: genome-wide analysis, long QT syndrome, psychology and behavior, public policy, ethics, secondary findings
Secondary Findings and Inherited Cardiac Conditions
Genome sequencing (GS) is an approach for investigation of suspected monogenic conditions, and for population studies exploring the contribution of genomic variation to health and disease.1 Screening for, and feedback of ‘secondary findings’ (SF)—variants believed to be associated with serious Mendelian health conditions unrelated to the indication for sequencing—are a subject of ongoing debate. Unresolved issues include participant/patient autonomy, clinical utility, and justice.2 Studies of a range of patients, participants and the public find widespread support for disclosure of SF that relate to potentially actionable disease,3 although in practice a smaller majority choose return of SF than when asked hypothetically.4,5 Alongside clinical genome sequencing programmes, such as the UK 100,000 Genomes Project (www.genomicsengland.co.uk), an increasing number of initiatives are beginning to consider the handling of SF in ‘healthy’ sequenced populations.
A range of outcomes of SF disclosure are expected to inform policy around SF, including disease-variant association and behavioural and psychosocial impacts; in advance of this evidence, diverse guidelines and policies have emerged.6 For example, the American College for Medical Genetics and Genomics (ACMG) recommend screening for variants implying risk of potentially life-threatening disease for which intervention is available, in all individuals undergoing clinical genome sequencing.7 The ACMG recommendation includes a benchmark list of genes, the majority of which are associated with either inherited cardiac disease, which can present as fatal arrhythmia at any age, or cancer predisposition. Inherited cardiac conditions (ICC), including hypertrophic cardiomyopathy, dilated cardiomyopathy, arrhythmia right ventricular cardiomyopathy and long QT syndrome (LQTS) are relatively common (in aggregate around 1 in 250). These conditions are usually autosomal dominant and genetically heterogeneous, with variable penetrance and expressivity within and between families. Genetic testing and family screening, clinical evaluation based on cardiac imaging and ECG, and risk stratification for sudden cardiac death, are well established in clinical practice.8 Individuals at risk of sudden cardiac death can be managed by lifestyle advice, medical therapy and/or implantation of a cardiac defibrillator.
Informed variant interpretation underpins clinical genetic testing and is considered key to SF screening and feedback policy;2 current practice is aided by large population datasets such as the exome aggregation consortium cohort (ExAC) and the genome aggregation database (gnomAD),9 and by efforts to establish consistency in classification.10,11 However, until now, genetic testing in inherited disease has been directed by phenotype,10 and existing genotype-phenotype data come almost exclusively from individuals and families with manifest disease. In this setting, the prior probability that a potentially pathogenic variant in a relevant gene is actually pathogenic is high and the variant in question is, by definition, penetrant at least in the proband. In contrast, phenotype correlation with genotype in unselected populations is largely unknown and there are indications that penetrance and interpretability of variants may be lower; population prevalence of variants previously considered pathogenic and penetrant is much higher then would be compatible with known disease prevalence.12
Clinical Case
Genomic Analysis
BJ enrolled in GS via a purpose-designed protocol ‘Molecular Genetic Studies of Individuals and Families at Risk of Inherited Disease’ (MGAC, REC reference 13/WM/0466),[13] with the aim of identifying a cause for his child’s rare disease. SF policy in MGAC uses an opt-in approach based on the original ACMG gene list,7 offered to adult participants. Genomic analysis was targeted to genes associated with the primary condition, and ACMG gene list.
A deletion of 5 base pairs in exon 3 of KCNQ1 was detected in BJ’s sample. This variant, (NM_000218.2 c.573_577del p.R192Cfs*91, genomic location Chr11:hg19:g.2591953_2591957), creates a frame shift with new reading frame ending in a stop codon 91 amino acids downstream. KCNQ1 encodes the alpha subunit of the slowly activating voltage-gated potassium ion channel and contributes up to 49% of putative pathogenic variants in genetically confirmed LQTS cases.14 Approximately 20% of KCNQ1 variants are predicted to lead to haploinsufficiency, which is a known mechanism of disease associated with a lower risk of cardiac events in patients with LQTS.15 Using current guidelines, supplemented by in-house data from over 1500 LQTS gene tests, our accredited NHS laboratory interpreted the variant as highly likely pathogenic. The variant was present in four individuals with LQTS in the Oxford cohort (with limited segregation data) and has been reported in families and individuals with LQTS16,17,18 and as a homozygous variant in individuals with Jervell and Lange-Nielsen syndrome.17 The variant is present in gnomAD (release 2.0.1) at frequency 0.0016%.
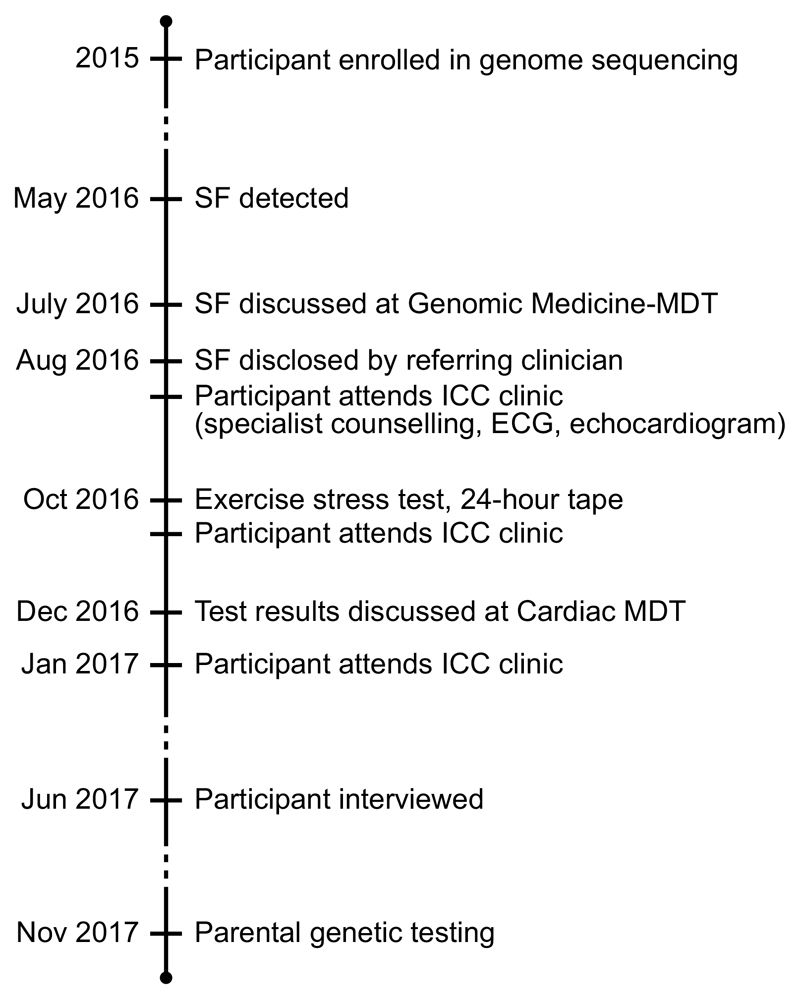
The University Hospital Genomic Medicine multidisciplinary team agreed that the variant should be reported. BJ was informed by his child’s clinical geneticist by pre-arranged web discussion, and referred to a specialist ICC clinic for genetic counselling and clinical assessment (Figure 1).
Figure 1.
Timeline of events. SF: secondary finding; MDT: multidisciplinary team; ECG: electrocardiogram; ICC: inherited cardiac condition
Clinical Assessment
BJ is an apparently healthy male aged 39, with no significant medical history. His occupation is moderately physically demanding in a regulated environment; recreationally, he is a competitive track cyclist who trains intensively one to four hours, six days per week.
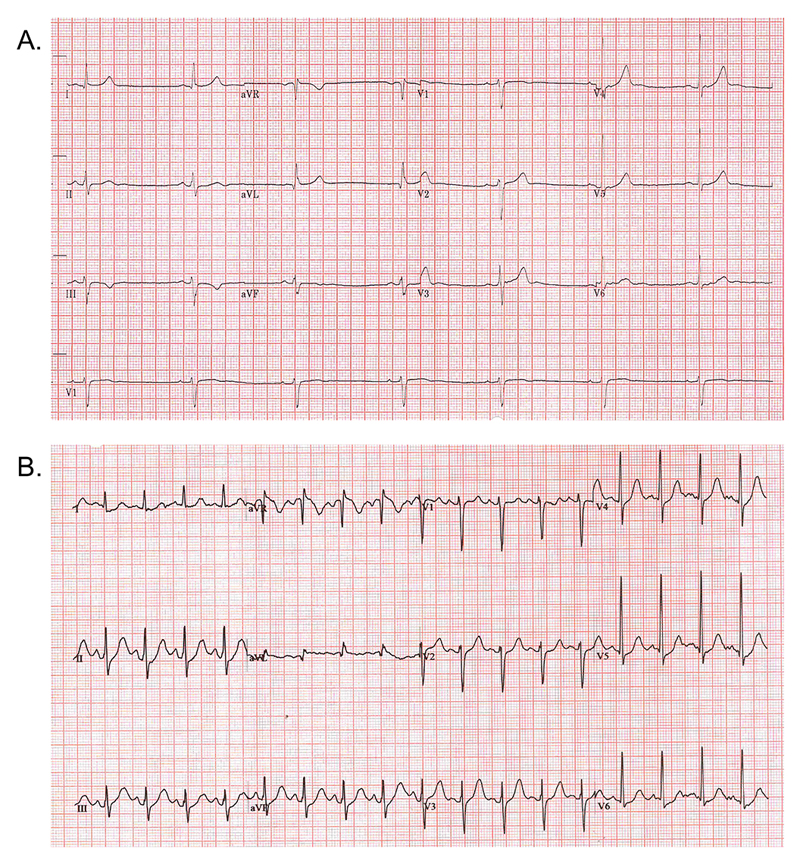
ECG showed sinus (athletic) bradycardia, rate 37 beats per minute with normal axis and normal conduction. Broadly normal pattern of repolarisation, but with a prominent U wave. Absolute QT value 498, QTc using Bazett formula 390 (Figure 2A). A 24-hour Holter monitor showed QT intervals corrected to within the normal range. During an exercise stress test, BJ achieved two minutes of stage VII of Bruce protocol with no arrhythmias. Peak heart rate response low (164bpm), and heart rate slowing delayed in recovery. QT interval did not show expected shortening during exercise, and was prolonged in recovery: 4 minutes into recovery (a time point proposed to have sensitivity and specificity for detecting manifestations of LQT1 genotype) QTc was just over 400ms at heart rate 105bpm, QTc using Bazett formula 529 (Figure 2B). Cardiac multidisciplinary team discussion concluded that these changes are consistent with a LQT1 phenotype, albeit with normal QT interval at rest.
Figure 2.
Participant electrocardiograms: A. Resting 12 lead; B. 4 minutes into recovery after exercise stress test.
In light of these results, BJ was advised to: moderate his training, limiting to around three quarters of peak work rate; not compete; avoid potentially QT prolonging medication, and begin a non-cardioselective beta blocker.
Family History
BJ is one of three healthy offspring, all of whom have children, of living parents. BJ was unaware of any syncope, sudden or unexplained death in the family. Cascade genetic counselling and testing was offered to BJ’s parents; his mother reported the death of her parent—who took prescription barbiturates—during sleep aged mid-40s. BJ’s mother tested positive for the variant. She is asymptomatic and has a normal resting ECG. Further clinical investigations and cascade testing in other relatives are ongoing.
Psychological, Behavioural and Financial Impacts
Following disclosure, a semi-structured interview was undertaken exploring understanding, perception and behaviours (consent under MGAC protocol). The interview guide was based on psychosocial literature on genetic risk,19 and clinical experience. The interview was audio-recorded and transcribed verbatim. The transcript was analysed thematically.20
BJ’s initial reaction to the disclosure was disbelief. The suggestion of a cardiac condition was incompatible with his perception of self, and conflicted with his level of fitness and lack of symptoms. BJ described an episode of acute distress between disclosure and specialist clinic appointment. Although his recall of electing to receive SF was partial, he had clear recall of the disclosure conversation and subsequent discussions in the specialist clinic. While acknowledging that sudden cardiac death can occur in young individuals, he was sceptical of his own potential risk. He considered that the SF had impacted him primarily through the implication that training and competitive cycling presented additional, yet unquantifiable, risk. He feared a sudden unheralded collapse while cycling, comparing that with the perceived controllability of avoiding a collision.
BJ did not regret his decision to receive SF and appreciated that the disclosure had occurred after he had already enjoyed many years’ competing. Throughout the interview, BJ displayed reiterative, personally inconclusive evaluation of his risk; he described contacting sports scientists and visiting online forums. This was apparently driven by a sense of responsibility to his family. His provisional acceptance of his risk of LQTS manifested in selective adherence to clinical recommendations: he had moderated cycling but, despite intentions at the time of interview, had not started taking protective medication.
Subsequent to cardiac evaluation, BJ was unable to obtain critical illness cover; attempts to purchase life cover for an increased mortgage were ultimately successful.
Discussion
This case represents an early example of evaluating a genomic secondary finding. The variant and gene in question were anticipated to be relatively straightforward within the spectrum of inherited heart condition SF, as the variant was well characterised and LQT1 is relatively easy to diagnose and treat. However, the case generated significant diagnostic and psychosocial challenges. Initial clinical assessment by resting ECG was reassuring; a phenotype consistent with the variant was discovered only through subsequent specialist assessment. Similarly, initial family history elicitation was reassuring; the grandparent’s death—suspicious for LQTS—only came to light on cascade screening. Interventions that might ordinarily be well tolerated, avoidance of extreme exertion and taking beta blockers, proved problematic.
Genomic variant interpretation is typically informed by phenotype and family history in the context of a clinical diagnosis;10 evidence to inform interpretation in unselected populations is awaited. Further, at present, there are no guidelines on workup that should follow identification of a potentially pathogenic SF. Several large-scale projects are beginning to generate SF with consent or considering approaches to disclosure where no specific consent exists, with the result that return of SF will become frequent. The clinical utility of genetic testing depends on positive clinical, psychosocial and behavioural outcomes.21 If clinical utility underlies the rationale for search and disclosure of SF, a relationship with clinical risk of ICC must be established. Examination of electronic medical records found no excess of disease expression (by ECG) in individuals harbouring a putatively pathogenic cardiac arrhythmia gene variant12 but further data are required, specifically specialist cardiac evaluation and family history collection.
For realisation of clinical utility, ascertainment of risk must be followed by consistent risk reduction actions: adherence to screening and clinical recommendations, and informing relatives. The Health Belief Model (HBM)22 conceptualises factors involved in taking action to mitigate disease risk: personal susceptibility, severity of disease consequence and wider life impacts, benefit of taking action, and perceived or experienced barriers to that action. It appears that BJ perceives the seriousness of his diagnosis to be high but remained, at interview, unsure about his susceptibility given his prolonged endurance training and lack of symptoms. For BJ, barriers include reluctance to take beta blockers and moderation of participation in sport from which he derives multiple benefits. Reassuringly, emerging data on psychological impacts of SF disclosure,23 including LQTS-associated variants24 suggest that recipients do not experience distress and anxiety; however it will be important to understand factors affecting adherence in SF recipients. Rosenstock22 suggests that conflicting motives of avoidance may result when the factors outlined in the HBM are finely balanced. The individual may then vacillate between options, and/or experience fear and anxiety. Thus, the absence of an unambiguous disease phenotype might complicate assimilation of SF, and may result in requirement for psychosocial support.
Conclusions
A genotype-driven approach to identification of patients with ICC may detect at-risk individuals and allow clinical and/or lifestyle management that is potentially life-saving. However, this approach presents new challenges for clinical management and genetic counselling, as well as for patients. In order to inform guidelines and practice, systematic collection and curation of data from the return of SF is required. Gene and variant-specific evidence from unselected populations is needed to inform estimates of penetrance and understand the phenotypic spectrum of variants discovered as SF. The immediate impact of disclosure on the recipient highlights the need to provide timely clinical review and specialist genetic counselling; wider exploration of the impact of disclosure on recipients should be undertaken. This case highlights some challenges of realising improved health outcomes following disclosure of secondary findings, and provides insights into reasons for which clinical recommendations may not always be followed after disclosure of genomic secondary findings.
Acknowledgments
We thank BJ and his family for their participation, Dr Andrea Nemeth, Dr John Taylor and other members of the Oxford University Hospitals NHS Foundation Trust/University of Oxford Genomic Medicine MDT and the OUHFT cardiac MDT.
Sources of Funding: MPM was funded by the Rhodes Trust and the Radcliffe Department of Medicine. EO and JCT are funded by NIHR Oxford Biomedical Research Centre. HW acknowledges support from an NIHR Senior Investigator Award. The study is funded in part by the Wellcome Trust/Department of Health as part of the Health Innovation Challenge Fund (R6-388 and WT 100127). The views expressed in this manuscript are those of the authors and not necessarily of the Wellcome Trust or Department of Health.
Footnotes
Disclosures: None.
References
- 1.Gonzaga-Jauregui C, et al. Human genome sequencing in health and disease. Annu Rev Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ormondroyd E, et al. “Not pathogenic until proven otherwise”: perspectives of UK clinical genomics professionals toward secondary findings in context of a Genomic Medicine Multidisciplinary Team and the 100,000 Genomes Project. Genet Med. 2017 doi: 10.1038/gim.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackley MP, et al. Stakeholder views on secondary findings in whole-genome and whole-exome sequencing: a systematic review of quantitative and qualitative studies. Genet Med. 2017;19:283–93. doi: 10.1038/gim.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wynn J, et al. Impact of Receiving Secondary Results from Genomic Research: A 12-Month Longitudinal Study. J Genet Couns. 2017 doi: 10.1007/s10897-017-0172-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackley MP, et al. Views of rare disease participants in a UK whole-genome sequencing study towards secondary findings: a qualitative study. Eur J Hum Genet. 2018 Feb 13; doi: 10.1038/s41431-018-0106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knoppers BM, et al. Return of genetic testing results in the era of whole-genome sequencing. Nat Rev Genet. 2015;16:553–9. doi: 10.1038/nrg3960. [DOI] [PubMed] [Google Scholar]
- 7.Green RC, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–74. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ackerman MJ, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Europace. 2011;13:1077–109. doi: 10.1093/europace/eur245. [DOI] [PubMed] [Google Scholar]
- 9.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly MA, et al. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen's Inherited Cardiomyopathy Expert Panel. Genet Med. 2018 doi: 10.1038/gim.2017.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Driest SL, et al. Association of Arrhythmia-Related Genetic Variants With Phenotypes Documented in Electronic Medical Records. JAMA. 2016;315:47–57. doi: 10.1001/jama.2015.17701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ormondroyd E, et al. Insights from early experience of a Rare Disease Genomic Medicine Multidisciplinary Team: a qualitative study. Eur J Hum Gen. 2017 doi: 10.1038/ejhg.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Napolitano C, et al. Genetic Testing in the Long QT Syndrome. Development and Validation of an Efficient Approach to Genotyping in Clinical Practice. JAMA. 2005;294:2975–2980. doi: 10.1001/jama.294.23.2975. [DOI] [PubMed] [Google Scholar]
- 15.Ruwald MH, et al. Stop-codon and C-terminal nonsense mutations are associated with a lower risk of cardiac events in patients with long QT syndrome type 1. Heart Rhythm. 2016 Jan;13:122–31. doi: 10.1016/j.hrthm.2015.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapplinger JD, et al. Spectrum and prevalence of mutations from the first 2,500 consecutive unrelated patients referred for the FAMILION long QT syndrome genetic test. Heart Rhythm. 2009;6:1297–303. doi: 10.1016/j.hrthm.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tranebjaerg L, et al. Jervell and Lange-Nielsen syndrome: a Norwegian perspective. Am J Med Genet. 1999;89:137–46. [PubMed] [Google Scholar]
- 18.Choi G, et al. Spectrum and frequency of cardiac channel defects in swimming-triggered arrhythmia syndromes. Circulation. 2004;110:2119–24. doi: 10.1161/01.CIR.0000144471.98080.CA. [DOI] [PubMed] [Google Scholar]
- 19.Rolland JS, Williams JK. Toward a biopsychosocial model for 21st-century genetics. Fam Process. 2005;44:3–24. doi: 10.1111/j.1545-5300.2005.00039.x. [DOI] [PubMed] [Google Scholar]
- 20.Braun V, Clarke V. Using thematic analysis in psychology. Qual Res Psych. 2006;3:77–101. doi: 10.1191/1478088706qp063oa. [DOI] [Google Scholar]
- 21.Grosse SD, Khoury MJ. What is the clinical utility of genetic testing? Genet Med. 2006;8:448–50. doi: 10.1097/01.gim.0000227935.26763.c6. [DOI] [PubMed] [Google Scholar]
- 22.Rosenstock IM. Historical Origins of the Health Belief Model. Health Educ Monogr. 1974;2 doi: 10.1177/109019817400200403. [DOI] [PubMed] [Google Scholar]
- 23.Lewis KL, et al. Participant use and communication of findings from exome sequencing: a mixed-methods study. Genet Med. 2016;18:577–83. doi: 10.1038/gim.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haukkala A, et al. The return of unexpected research results in a biobank study and referral to health care for heritable long QT syndrome. Public Health Genom. 2013;16:241–50. doi: 10.1159/000354105. [DOI] [PubMed] [Google Scholar]