Abstract
Pleiotrophin (PTN) is a potent glycosaminoglycan-binding cytokine important in neural development, angiogenesis and tissue regeneration. Much of its activity is attributed to its interactions with the chondroitin sulfate (CS) proteoglycan, receptor type protein tyrosine phosphatase ζ (PTPRZ). However, there is little high resolution structural information on interactions between PTN and CS, nor is it clear why the C-terminal tail of PTN is necessary for signaling through PTPRZ even though it does not contribute to binding heparin. We determined the first structure of PTN and analyzed its interactions with CS. Our structure shows PTN possesses large basic surfaces on both of its structured domains and residues in the hinge segment connecting the domains have significant contacts with the C-terminal domain. Our analysis of PTN-CS interactions showed the C-terminal tail of PTN is essential for maintaining stable interactions with CSA, the type of CS commonly found on PTPRZ. These results offer the first possible explanation of why truncated PTN missing the C-terminal tail is unable to signal through PTPRZ. NMR analysis of PTN’s interactions with CS revealed that the C-terminal domain and hinge of PTN make up the major CS binding site in PTN, and that removal of the C-terminal tail weakened the site’s affinity for CSA, but not for other high sulfation density CS.
Keywords: Glycosaminolgycan, glycosaminoglycan-binding protein, cytokine, NMR
Introduction
Pleiotrophin (PTN) is a potent mitogenic cytokine that acts through several proteoglycan receptors. First isolated in neural tissues, PTN plays an active role in the development of the central nervous system, but is also produced during tissue repair and regeneration.[1–4] In addition, expression of PTN is elevated in a number of cancer cell lines, and the expression level often correlates with metastatic abilities of these cancer cells. [5–13] However, PTN expression can also be beneficial: studies have shown that PTN helps to maintain hematopoietic stem cells, triggers tissue repair during ischemia and provides protective effects on neural tissues during addiction. [14–20]
The two most well-established PTN receptors are the heparan sulfate proteoglycan N-syndecan and the chondroitin sulfate proteoglycan receptor type protein tyrosine phosphatase ζ (PTPRZ). [21, 22] N-syndecan is believed to be crucial to PTN’s activity during neural development while PTPRZ has been associated with PTN’s ability to facilitate cell proliferation and growth under both normal and pathological situations.[4, 5, 22, 23] For these receptors, PTN’s affinity for the glycosaminoglycan (GAG) portion of the receptors is considered vital to its activity [21, 24].
GAGs are linear sulfated polysaccharides consisted of repeating disaccharide units. One of the residues in the disaccharide unit is always an amino sugar in the form of either N-acetylglucosamine (GlcNAc) or N-acetylgalactosamine (GalNAc), while the other sugar is usually an uronic acid sugar such as iduronic acid (IdoA) or glucuronic acid (GlcA). In heparan sulfate (HS) or heparin, the amino sugar is always a glucosamine and majority of uronates are IdoA. Chondroitin sulfate (CS), on the other thand, contains only GalNAc and GlcA. Despite their simple polysaccharide structure, GAGs are extremely complex because of their large size and the semi-random modifications of the sugars through sulfation. In heparin and HS, IdoA can be sulfated at the 2-O position, and glucosamine in HS and heparin can be sulfated at the 2-N, 3-O, and 6-O positions. In CS, GalNAc can be sulfated at both the 4-O and 6-O positions while 2-O-sulfated GlcA can also be found. Because GAGs rely on these sulfate groups to attract proteins such as PTN in order to modulate signaling, the sulfation pattern and density of GAGs are important determinants of their biological activity.
Although the structure of PTN’s homologue, midkine, is well characterized [25, 26], high resolution structure of PTN does not yet exist. Kilpelainen et al. were the first to conduct structural investigations on PTN at pH 4.7. Their results showed that, similar to midkine, PTN possessed two thrombospodin type-1 repeat (TSR) domains as well as unstructured N- and C-termini.[27] The study determined secondary structures of PTN as well their tertiary arrangement, but it stopped short of producing a high resolution structure of PTN. Subsequent studies by the same group also investigated the interactions of PTN with heparin, which is an analog of HS from N-syndecan.[28] Through titrations with heparin oligosaccharides and ELISA analysis, they demonstrated both TSR domains are needed to maintain high heparin affinity and that the C-terminal TSR domain (CTD) has higher affinity for intact heparin than the N-terminal TSR domain’s (NTD). However, the C-terminal tail of PTN, despite having a large number of basic amino acids, does not contribute significantly to heparin binding. [28]
In contrast to heparin, very little information is available regarding PTN’s interactions with CS, the type of GAG found on PTPRZ. Given their connection to cell proliferation and growth [4, 29, 30], the role of PTN-CS interactions in facilitating PTPRZ signaling is crucial to understanding the mechanism of PTN-dependent metastasis and stem cell maintenance. Several studies have measured PTN’s affinities for various forms of CS. In particular, these studies showed that PTN prefers oversulfated CS structures enriched in 4,6-O-disulfated GalNAc (CSE) or 2-O-sulfate-GlcUA-6-O-sulfate-GalNAc (CSD) rather than CS containing only 4-O-sulfated GalNAc (CSA), and affinity of the interaction is also dependent on the size of CS ligand.[31–34] However, no experimental information on how PTN domains contribute to binding CS is available. Furthermore, studies have also shown that truncated PTN missing the C-terminal tail (residues 115 to 136) can be found in vivo in significant quantities, but is incapable of signaling through PTPRZ [35–37]. In addition, peptides derived from the C-terminal tail of PTN can inhibit PTPRZ-dependent cell migration.[38] This implies the C-terminal tail, which does not contribute to heparin binding, plays an important role in interactions with PTPRZ. However, the nature of its role continues to be unclear.
To answer some of these questions, we have determined the solution structure of PTN at pH 6.0 and characterized its interactions with CSA and CSE fragments. Our structure is consistent with the secondary and tertiary structural information deduced by Kilpelainen et al.[27]. However, our data also indicate that part of the hinge between the TSR domains interacts with CTD, placing the hinge in position to participate in GAG binding. Our characterization of PTN-CS interactions using ELISA showed, unlike PTN’s interactions with heparin, truncated PTN missing the C-terminal tail had significantly lower affinities for CSA than wild type PTN. These results demonstrate, for the first time, that the lower CS affinity of truncated PTN may contribute to these mutants’ inability to signal via PTPRZ. CS titrations of PTN, monitored using solution NMR, revealed that CTD of PTN have much higher affinities for CSE dp6 (degree of polymerization 6, or hexasaccharides) than CSA dp8. However, removal of the C-terminal tail weakened CTD’s affinity for CSA dp8, but not CSE dp6, providing additional molecular insights into contributions of PTN domains to CS binding.
Results
Structure of PTN.
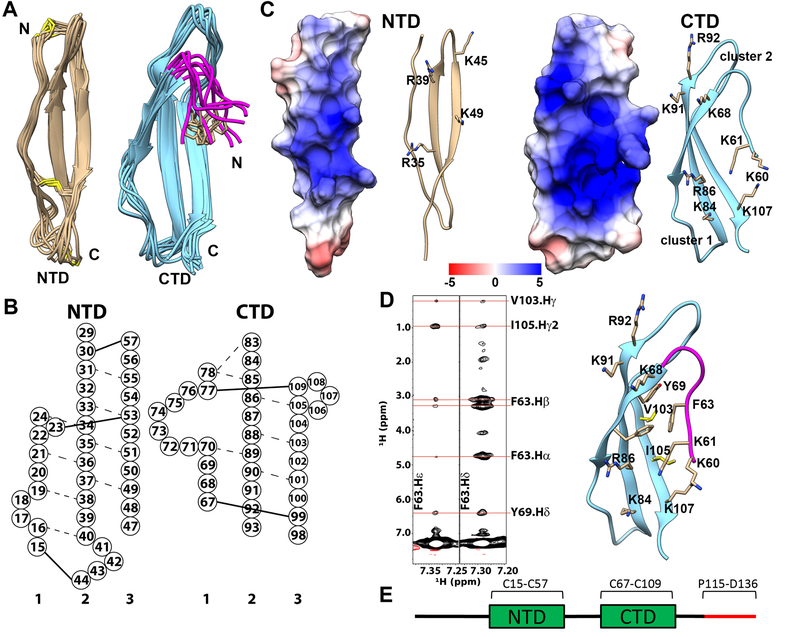
To date, structures of midkine from both human and zebra fish have been determined.[25, 26] The high resolution structure of PTN, however, has not been reported. Here we report the solution structure of human PTN at pH 6.0. In agreement with the previous study of PTN [27], we found PTN is comprised of two TSR domains flanked by unstructured termini. Because no inter-domain NOE was detected between CTD and NTD, the two domains appear to be largely independent of each other and do not have a well-defined inter-domain orientation. Ten cystines are found in the protein and all have Cβ chemical shifts above 40 ppm. This indicates all cystines exist in the oxidized form [39], and is consistent with the fact that five disulfide bonds exist in the protein. Figure 1A shows the superimposed NTDs and CTDs of ten lowest energy structures. Table 1 shows NMR structural statistics for the structure. One thing to note is that less than 150 long range NOEs were observed in PTN, lower than other proteins of similar size. This is because PTN lacks significant hydrophobic packing, which is often the main source of long range NOE contacts in proteins.
Figure 1.
Structure of PTN. A) Superimpositions of NTD and CTD/hinge from the ensemble of 10 lowest energy structures. Disulfide bonds are represented in yellow. Hinge segment is shown in magenta. Side chain of F63 is shown in beige. B) Schematic depiction of the β-sheet structures of PTN. Residues are represented by circles labeled with residue numbers. Dashed lines represent observed NOE between amide protons. Solid lines represent disulfide bonds. C) Electrostatic potential mapping of NTD and CTD/hinge. The surface depiction of these domains are shown in the same orientation as the ribbon diagrams to their right. The unit of potential is in kT/e. D) Left: strips from aromatic 13C-edited NOESY showing NOE cross peaks between aromatic protons of residue F63 in the hinge and Y69, V103, I105 in CTD. Right: structural details of interactions between hinge and CTD. E) Schematic of the PTN sequence. The C-terminal tail is shown in red.
Table 1.
Structural statistics for ensemble of PTN structures.
| No. of NOE-based distance constraints | |
| Total | 749 |
| Intraresidue (i = j) | 263 |
| Sequential (|i – j| = 1) | 295 |
| Medium-range (1 < |i – j| < 5) | 44 |
| Long-range | 147 |
| NOE constraints per restrained residue | 5.9 |
| No. of dihedral angle constraints | 125 |
| No of H-N RDC used | |
| NTD | 28 |
| CTD | 31 |
| Total no. of structures computed | 100 |
| No. of structures used | 10 |
| Residual constraint violationsa | |
| No. of distance violations per structure | |
| 0.1–0.5 Å | 29.6 |
| >0.5 Å | 0.3 |
| No. of dihedral angle violations per structure | |
| 1–10° | 4.8 |
| >10° | 0.1 |
| No. of RDC violations per structure, > 0.5 Hz | 0 |
| Experimental vs. Calculated RDC Correlation R | 0.99 |
| RMSD | |
| backbone atoms | |
| NTDb | 0.6 Å |
| CTDc | 0.7 Å |
| heavy atoms | |
| NTDb | 1.2 Å |
| CTDc | 1.6 Å |
| Ramachandran plot summary from Procheckd (%) | |
| Most favored regions | 91.9 |
| Additionally allowed regions | 8.1 |
| Generously allowed regions | 0.0 |
| Disallowed regions | 0.0 |
a Calculated for all constraints for the given residues, using sum over r−6.
b Residues with sum of phi and psi order parameters [61] > 1.8. Ordered residue are residues 17 to 23, 33 to 40, 46 to 53.
c Residues with sum of phi and psi order parameters [61] > 1.8. Ordered residue are residues 66 to 72, 82 to 92, 102 to 109.
d Residues selected on basis of dihedral angle order parameter [61], with sum of phi and psi order parameters 1.8. Selected residue ranges are the same as the ordered residue ranges.
In both TSR domains, second and third strands of the domain contain more β strand structure than the first strand. The unstructured nature of strand one in both domains can be partially attributed to the disulfide bond distribution of the protein (Figure 1B). In particular, we found that in order for C23 and C53 to be joined by a disulfide bond, strand one of NTD must be twisted since a continuous β strand in strand one will place the side chains of the two cystines on opposing sides of the β sheet. In CTD, cross strand amide proton contacts between residues 78 and 85 as well as between residues 70 and 89 have been observed in 15N-edited NOESY. This is the result of a disulfide bond between residues C77 and C109, which creates a large loop in the second half of strand one in CTD and disrupting the β strand structures in one half of the first strand. The topology of disulfide bonds in PTN also differs from that of midkine. Specifically, the first disulfide bond in midkine connects strands one and two of NTD [25, 26]. However, the corresponding disulfide bond between residues C15 and C44 in PTN connects strands one and three.
Because GAG’s interactions with protein is mostly mediated by electrostatic interactions, basic amino acid clusters are often potential GAG-binding sites. Besides strings of lysine residues found on N- and C-termini, PTN contains two basic amino acid clusters in CTD and one cluster in NTD. One CTD cluster is made up of residues K68, K91 and R92, which corresponds to cluster 2 in midkine [25] (Figure 1C). The other cluster, located on the opposite side of the β sheet as cluster 2, is equivalent to cluster 1 in midkine and includes residues K84, R86 and K107. Although the two clusters are located on opposite sides of the β-sheet, the curvature in the sheet places them in close proximity to one another. Electrostatic potential mapping shows they form a single basic surface. The basic residues R35, R39 and K49 can also be found on one side of the β-sheet in NTD, but the distribution of these residues in the NTD is sparser than residues in the two CTD clusters. The NTD basic amino acid cluster is equivalent to cluster 3 identified in zebra fish midkine [26] (Figure 1C).
Although there are no NOE contacts between the TSR domains, definitive NOE cross peaks between CTD and residues in the hinge segment connecting the two TSR domains (residues 58 to 66) can be observed. Specifically, aromatic protons of F63 can be seen contacting side chains of Y69, V103 and I105 (Figure 1D). Together with L90, these residues form a small hydrophobic cluster. These interactions have not been reported in studies of midkine structures, but may have important consequences on GAG-binding because the bend formed in the hinge places two basic amino acids, K60 and K61, close to cluster 1 of CTD. Figure 1A shows the positions of the hinge segments in the ensemble of the ten lowest energy structures when CTD is superimposed. Because F63 is the only residue with significant contacts with CTD and the tolerance in these distance constraints are relatively large, the hinge shows larger positional variation than the TSR domains.
CS Interactions with wild type and C-terminal truncated PTN.
Although high resolution characterizations of PTN’s interactions with heparin exist [28], the same level of information is not available for PTN’s interactions with CS, the type of GAG found on PTPRZ. Nevertheless, biophysical characterization of the effect of CS size and sulfation density on PTN’s CS affinity has been carried out [31, 33]. In this study, we characterize the interactions of PTN with native and size-defined CS using NMR and ELISA. We are particularly interested in determining the CS-binding sites in PTN and in understanding how the C-terminal tail may control PTN’s affinity for CS.
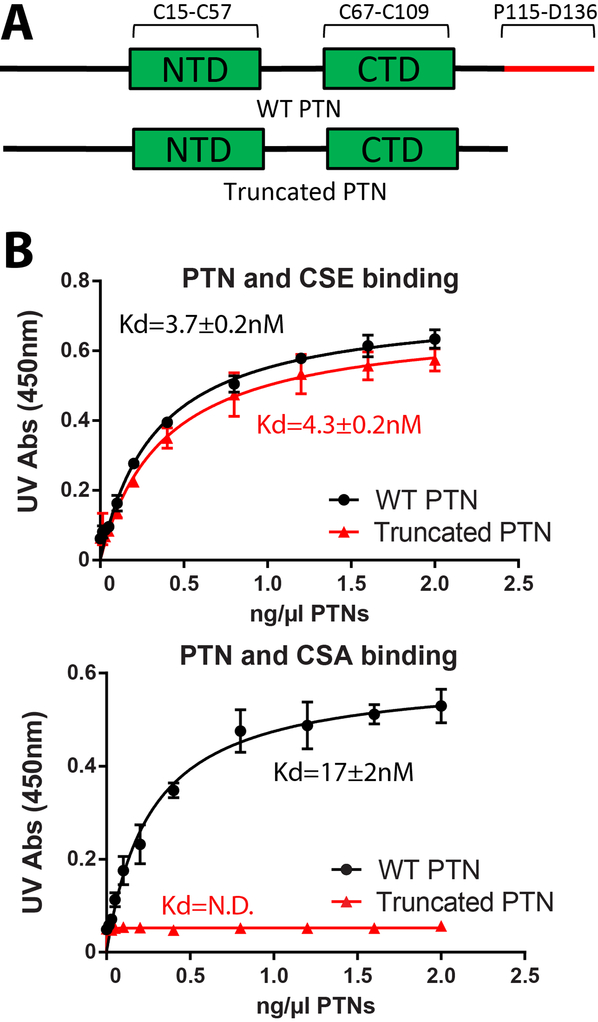
We first characterized PTN’s affinity for different CS variants using ELISA. In particular, we measured the affinities of both wild type PTN and C-terminal truncated PTN (residues 1 to 114) for immobilized native CSA and CSE. We are interested in knowing whether weakened interactions between truncated PTN and CS is the reason behind its inability to activate PTPRZ. Results for these assays are shown in Figure 2. Specifically, the assays showed the C-terminal tail of PTN was not necessary for binding CSE. Both native and C-terminal truncated PTN bound to CSE with Kd in the four to five nM range. However, truncated PTN failed to bind to CSA entirely while wild type PTN binds CSA with high affinity (Kd ~ 17 nM). These data indicate that the C-terminal tail of PTN is crucial for maintaining strong interactions with CSA, the type of CS commonly found on PTPRZ.
Figure 2.
CSA and CSE ELISA of wild type and truncated PTN. A) CSE ELISA of wild type and C-terminal truncated PTN. Both proteins bound CSE with high affinity. B) CSA ELISA of wild type and C-terminal truncated PTN. Only wild type PTN bound CSA with measurable affinity. Error bars represent standard deviations among three independent experiments (n=3).
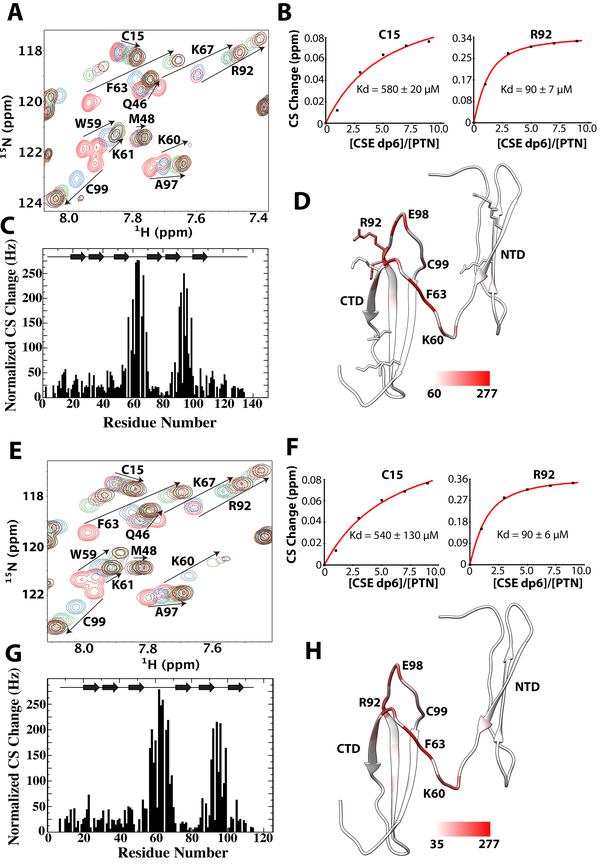
To determine the CS binding sites of PTN, we conducted NMR studies of PTN’s interactions with size defined CSE and CSA. We were unable to use native polysaccharides in the NMR studies because the signal broadening and precipitation induced by native CS polymers’ interactions with PTN makes extraction of useful information impossible. In addition, previous studies have shown that the size of CS fragments used here is sufficient for binding [33]. We first titrated native PTN with CSE dp6. The overlay of PTN’s 15N-edited HSQCs at different CSE dp6 concentrations and the magnitude of chemical shift changes for each residue are shown in Figure 3A. The HSQCs show CSE dp6 was able to induce chemical shift change in residues throughout the protein. However, residues K60, Q62, F63, G64, C67 in the hinge region and K91, R92, N96, E98, C99 in CTD showed the largest chemical shift changes upon binding CSE (Figure 3C). On the other hand, residues in NTD and the C-terminal tail showed little or no change in chemical shifts. In addition, most residues demonstrating large chemical shift changes are also close to the basic amino acid clusters in CTD. This implies CTD maybe a major CS binding site in PTN. Binding curves of different residues showed CTD and hinge residues bound CSE dp6 with a Kd of ~90 μM. However, Kd values extracted from residues in NTD were significantly higher (Kd ~ 550 μM) (Figure 3B). This discrepancy indicates GAG affinities of different domains in PTN are different. Residues in the C-terminal tail showed little or no change in chemical shifts in the presence of CSE dp6. We also conducted similar CSE dp6 titration using C-terminal truncated PTN. The results of the titration are shown in Figure 3 (panels E to H). The titration showed that removal of the C-terminal tail had no effect on CSE dp6 affinity of CTD and NTD. In addition, CSE dp6 perturbed the same set of residues in truncated and wild type PTN. These results imply that the C-terminal tail of PTN is inconsequential to binding CSE, in agreement with the result of ELISA.
Figure 3.
Titration of wild type and C-terminal truncated PTN with CSE dp6 A) 15N-edited HSQC overlay of wild type PTN at different concentrations of CSE dp6. Residues with large chemical shift changes are labeled and their movements are illustrated with arrows. B) Normalized CSE dp6-induced chemical shift changes of amide proton and nitrogen for each residue in wild type PTN. Schematic illustration of PTN’s secondary structure is shown on the top of the plot. C) Binding curves of residues C15 (NTD) and R92 (CTD) used to calculate the CSE dp6 binding Kd of each domain. D) Magnitudes of chemical shift perturbation mapped onto the ribbon diagram of PTN. Coloring scale is shown on the bottom. E) 15N-edited HSQC overlay of C-terminal truncated PTN (residues 1 to 114) at different concentrations of CSE dp6. F) Normalized CSE dp6-induced chemical shift changes of amide proton and nitrogen for each residue in truncated PTN. G) Binding curves of residues C15 (NTD) and R92 (CTD) used to calculate the CSE dp6 binding Kd of each domain. H) Magnitudes of chemical shift perturbation mapped onto the ribbon diagram of PTN. Coloring scale is shown on the bottom.
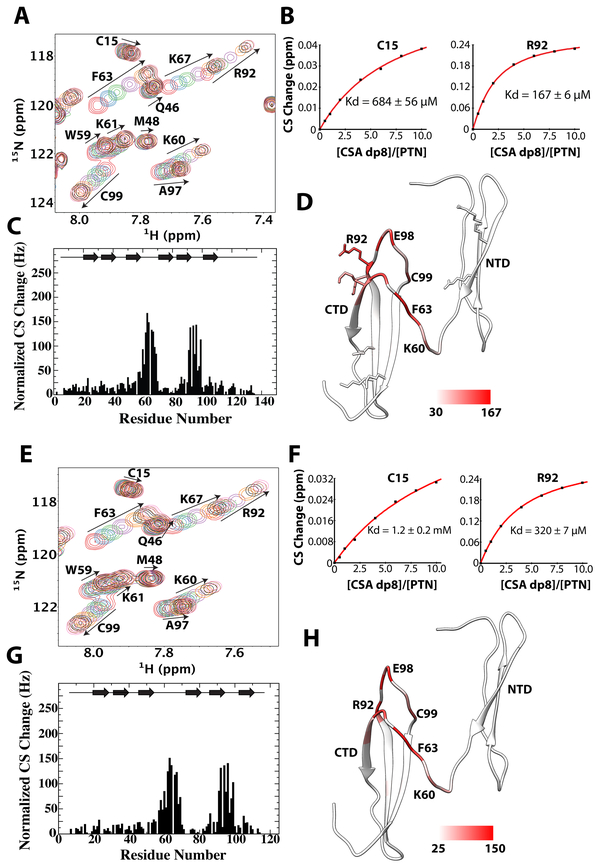
To elucidate contributions of PTN domains to CSA binding, we titrated both wild type and C-terminal truncated PTN with CSA dp8. Because several studies have already shown that PTN’s affinity for CSA is considerably lower than its affinity for CSE [31, 32], we used a larger CSA fragment for these titrations to ensure Kd values are within measureable range. Figure 4A shows the overlay of 15N-HSQCs taken during the titration. Chemical shift perturbation analysis showed the residues perturbed by CSE and CSA are similar, indicating the residues responsible for binding CSE are also binding CSA. Although CSA dp8 was not able to induce the large chemical shift changes observed in CSE titrations, accurate Kd values were still extracted for several residues. Consistent with previous studies, our titrations showed that PTN’s affinity for CSA is significantly lower than that of CSE. In particular, Kd values extracted from CTD residues has increased from 90 μM for CSE to 170 μM for CSA. Surprisingly, NTD’s Kd for CSA dp8 is only slightly larger than its Kd for CSE dp6 (Figure 4B). Identical CSA dp8 titrations were also performed on C-terminal truncated PTN (Figures 4E to 4H). In the absence of the C-terminal tail, CTD residues bound CSA dp8 with a Kd of only ~ 320 μM. Similarly, the CSA dp8 binding Kd of NTD also doubled to 1.2 mM. This indicates removal of the C-terminal tail has a significant effect on PTN’s affinity for CSA dp8 and is consistent with the ELISA results. It should be noted that truncated PTN’s inability to bind native CSA in ELISA does not contradict the results of NMR titrations. The discrepancy can be explained by the successive washes required in ELISA, which prevents weak interactions between truncated PTN and CSA from being detected by ELISA.
Figure 4.
Titration of wild type and C-terminal truncated PTN with CSA dp8. A) 15N-edited HSQC overlay of wild type PTN at different concentrations of CSA dp8. Residues with large chemical shift changes are labeled and their movements are illustrated with arrows. B) Normalized CSA dp8-induced chemical shift changes for each residue in wild type PTN. Schematic illustration of PTN’s secondary structure is shown on the top of the plot. C) Binding curves of residues C15 (NTD) and R92 (CTD) used to calculate the CSA dp8 binding Kd of each domain. D) Magnitudes of chemical shift perturbation mapped onto the ribbon diagram of PTN. Coloring scale is shown on the bottom. E) 15N-edited HSQC overlay of C-terminal truncated PTN (residues 1 to 114) at different concentrations of CSA dp8. F) Normalized CSA dp8-induced chemical shift changes of amide proton and nitrogen for each residue in C-terminal truncated PTN. G) Binding curves of residues C15 (NTD) and R92 (CTD) used to calculate the CSA dp8 binding Kd of each domain. H) Magnitudes of chemical shift perturbation mapped onto the ribbon diagram of PTN. Coloring scale is shown on the bottom.
Determining CS-binding site of PTN using paramagnetic CSE and CSA ligands
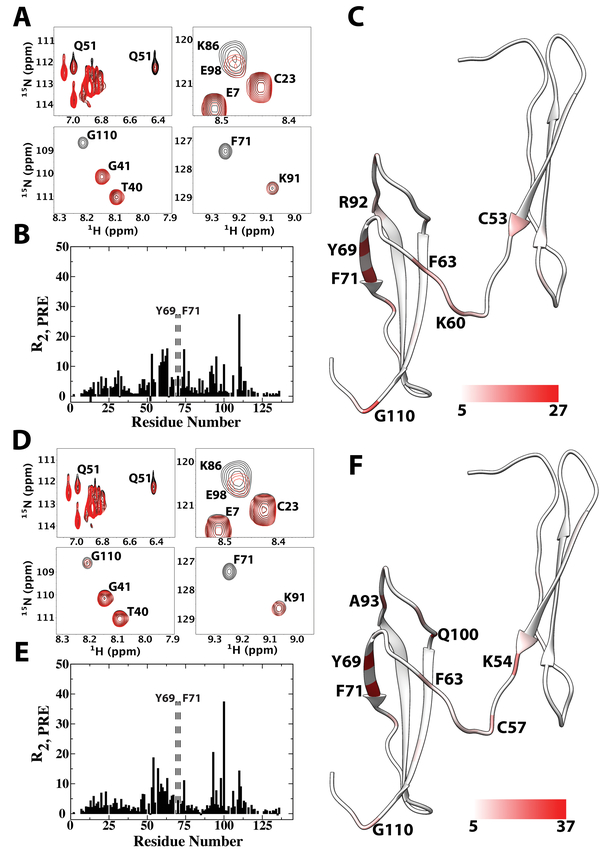
Although magnitudes of ligand-induced chemical shift changes are usually a reliable predictor of ligand binding sites, certain type of interactions, especially dynamic interactions with heterogeneous ligand binding conformations, may produce relatively small chemical shift changes despite the high affinity of the interaction. To confirm the binding sites identified using chemical shift perturbation analysis are involved in CS binding, we also studied the interactions of PTN with CSE dp6 and CSA dp8 ligands functionalized with the paramagnetic compound TEMPO (2,2,6,6-Tetramethylpiperidin-1-yl)oxyl, or (2,2,6,6-tetramethylpiperidin-1-yl)oxidanyl) at the reducing end. TEMPO is a stable nitroxide radical. The unpaired electron enhances relaxation of the NMR signal in a distance-dependent manner. This paramagnetic relaxation enhancement (PRE) can be quantified for each atom by either comparing the intensities of the atom’s NMR signal in the presence of oxidized and reduced radicals, or by directly measuring the difference in the atom’s transverse relaxation time (R2,PRE) in the presence of reduced and oxidized radicals. The magnitude of PRE experienced by an atom is directly correlated with the average distance between the paramagnetic center and the atom. Figure 5A shows selective areas from the HSQC overlay of full length PTN in the presence of either oxidized or reduced TEMPO-labeled CSE dp6 ligand. The ratio of ligand to protein is only 0.25:1. Under such conditions, only selective signals showed severe perturbations due to the fast kinetics of the interactions and the small amount of ligand used. R2,PRE analysis shows residue G110 experienced the greatest PRE, and signals for residues K54, Y69 and F71 are only visible after the radical has been reduced, indicating these residues are also close to the reducing end of the ligand. Y69, F71 and G110 are found in CTD whereas K54 is in NTD. Although many perturbed CTD residues are close to cluster 2, G110 are close to cluster 1. This means no single GAG-binding conformation can simultaneously account for all perturbations. The sulfation pattern heterogeneity in the CSE dp6 sample, which is known to result in heterogeneous binding orientations in protein-GAG interactions [40], is a possible explanation for this observation. In the NTD, although only side chains of NTD residues (residues Q51, W18 and W20) showed significant PRE, backbone HN atoms of NTD residues, with exception of K54, are only lightly perturbed, consistent with NTD’s lower GAG affinity. Residues in the C-terminal tail also showed no significant perturbation, indicating the reducing end of the ligand is not near it. We also repeated the experiments using TEMPO-labeled CSA dp8. Because of PTN’s weaker affinity for CSA, we increased the molar ratio of ligand-to-protein to 0.5:1 in this experiment. As a result, the magnitude of PRE due to CSA dp8 is larger than that of the CSE dp6 experiment. The PRE perturbation pattern produced by CSA dp8 is similar to that of CSE dp6 (Figures 5D to 5F), indicating the two ligands interacted with PTN in a similar manner. One notable difference between the CSA dp 8 and CSE dp6 data is that G110, located near cluster 1, experienced proportionally smaller PRE when CSA dp8 is the ligand. On the other hand, Q100, located near cluster 2, experienced proportionally larger PRE with the CSA dp8 ligand than with CSE dp6. This indicates CSA dp8 prefers a binding orientation that places its reducing end at cluster 2. Overall, the results are in good agreement with the chemical shift perturbation analysis and confirm that CTD and the hinge constitute the strong CS binding site of PTN. A control experiment carried out using 4-hydroxy-TEMPO showed there is no non-specific interactions between the TEMPO moiety and PTN under the condition of the PRE experiments.
Figure 5.
PRE perturbation on wild type PTN by TEMPO-labeled CSE-dp6 and CSA dp8. A) Sections from 15N-edited HSQC overlays of PTN in the presence of oxidized (red) and reduced (black) TEMPO-labeled CSE dp6. B) Quantitative R2,PRE values experienced by backbone amide protons in the presence of TEMPO-labeled CSE dp6. Residues whose amide proton signals are only visible after reduction of TEMPO are indicated by dashed rectangular boxes and labeled. C) TEMPO-labeled CSE dp6 R2,PRE values mapped onto the ribbon diagram of PTN. Coloring scale is shown on the bottom. D) Sections from 15N-edited HSQC overlays of PTN in the presence of oxidized (red) and reduced (black) TEMPO-labeled CSA dp8. E) Quantitative R2,PRE values experienced by backbone amide protons in the presence of TEMPO-labeled CSA dp8. Residues whose amide proton signals are only visible after reduction of TEMPO are indicated by dashed rectangular boxes and labeled. F) TEMPO-labeled CSA dp8 R2,PRE values mapped onto the ribbon diagram of PTN. Coloring scheme is shown on the bottom.
HADDOCK Models of the CTD-CSE Dp6 Complex
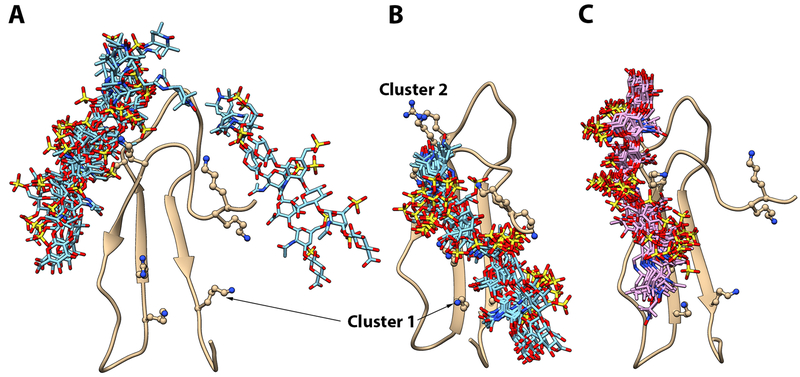
Using the chemical shift and the PRE perturbation data, we constructed models of PTN’s CTD complexed to TEMPO-labeled CSE dp6 using the docking program HADDOCK. We chose to focus on the CTD and hinge rather than intact PTN because this segment of PTN has the strongest affinity for CS, therefore is most likely to have stable interactions with CSE dp6 ligands. The chemical shift perturbation data were used to generate highly ambiguous distance constraints between the ligand and CTD while the PRE data were used to define unambiguous distance constraints between the paramagnetic center in TEMPO and HN atoms of perturbed residues. Although exact distances cannot be obtained because radical contents of the ligands are not known, it is certain that the distance between the paramagnetic center and the most perturbed HN atoms are most likely less than 15 Å. This fact was used to define the upper distance limit between HN atoms of residues Y69, F71 and G110 and the paramagnetic center in TEMPO. Our chemical shift perturbation data point to cluster 2 of CTD as a major GAG binding site and PRE perturbations show the reducing end of the protein can be close to both cluster 1 and cluster 2. We initially attempted to construct the model using these unfiltered data. However, instead of producing two distinct populations, HADDOCK chose to place the ligand between cluster 1 and cluster 2, which does not agree with the PRE data. We then attempted to construct the two populations separately by using PRE distance contraints to cluster 1 and cluster 2 in separate runs. Figure 6A shows the ensemble of 10 lowest energy structures obtained when only PRE distance constraints to cluster 2 were used along with the chemical shift perturbation data. The ligand in these models are found at two binding sites. Majority of the ligands appears to interact with cluster 2 residues in CTD. However, two models place the ligand close to the hinge. Neither conformation alone is completely consistent with the chemical shift perturbation data, but the ensemble average is in agreement with the chemical shift perturbation data. On the other hand, cluster 1, which has a strong electronegative surface, did not appear to have any interaction with the ligand. We believe this was because the docking was biased by the lack of chemical shift perturbation to cluster 1 residues. However, chemical shift perturbation does not always reflect interaction with ligands. In particular, the rigid structures surrounding cluster 1 may not allow large chemical shift changes in the presence of CSE. We then repeated the docking after adding cluster 1 residues as active residues. The result was a highly converged ensemble with the non-reducing end of the ligand interacting with cluster 1 and reducing end of the ligand near cluster 2 so that the TEMPO tag is close to the HN atoms of residues Y69 and F71 (Figure 6B). Figure 6C shows the ensemble of structures obtained when only PRE distance constraints to cluster 1 (residue G110) were used with the chemical shift perturbation data. In this ensemble, the non-reducing end of the CSE ligand interacts mostly with cluster 2. The reducing end of the ligand extends into cluster 1 and the TEMPO tag is placed close to G110, consistent with the PRE data. It should be noted that many sulfates in the CSE ligand are pointing away from the protein. However, ionic interaction between sulfates and basic amino acids is known to be crucial to GAG-protein interactions, therefore this maybe an artifact of incomplete ligand conformation sampling or incorrect handling of the electrostatics by HADDOCK.
Figure 6.
Models of CTD-CSE dp6 complex. A) Ensemble of 10 lowest energy models of CTD-CSE dp6 complex constructed using unfiltered experimental data. B) Ensemble of 10 lowest energy models of CTD-CSE dp6 complex after adding cluster 1 residues as active residues and using PRE distance constraints to cluster 2 only. C) Ensemble of 10 lowest energy models of CTD-CSE dp6 complex after using PRE distance constraints to cluster 1 only. In each ensemble, protein is superimposed and ribbon diagram of only one copy is shown. TEMPO-labeled CSE dp6 ligand is shown in the bond representation and basic amino acids in clusters 1 and 2 are shown in the ball-and-stick representation.
Discussion
In this study, we have determined the structure of PTN and characterized its interactions with CS. The structure of PTN is homologous to that of known midkine structures. However, the distribution of basic amino acids on PTN’s NTD is different from that of midkine. Specifically, Iwasaki et al. did not identify any basic amino acid clusters in NTD of human midkine [25] while Lim et al. showed two basic amino acid clusters exist in the NTD of zebra fish midkine [26]. Cluster 3 in zebra fish midkine’s NTD is similar to the cluster in NTD of PTN, but the cluster in PTN also includes an additional basic amino acid, R39. This difference in the number of basic amino acid clusters in NTD may explain why NTD of zebra fish midkine showed the largest changes when titrated with heparin [26], while CS-induced chemical shift changes from NTD of PTN are far smaller than those in the hinge region and the CTD (Figures 2 & 3). PTN’s CTD and NTD also have significantly different affinities for CS, which were not noted in heparin binding studies involving zebra fish midkine [26], but is consistent with results of previous investigations of PTN-heparin interactions [28, 41]. In particular, CTD’s apparent Kds of interaction for both CSA and CSE are four times smaller than that of NTD. Our results also show that the C-terminal half of the hinge can interact with residues in CTD through hydrophobic interactions. These interactions places the two lysines in the hinge (residues K60 and K61) close to cluster 1 of CTD. The fact that residues in the hinge were highly perturbed by CSE both in terms of chemical shift changes as well as paramagnetic effects indicate that the hinge is close to the bound GAG and may play a crucial role in PTN’s interaction with CS. Similar observations were also made in the characterization of midkine-heparin interactions by Lim et al.[26] Our postulate is that during GAG binding, the hinge most likely folds along the CTD, allowing residues K60 and K61 in the hinge to become part of cluster 1 in CTD.
The C-terminal tail of PTN is necessary for PTPRZ signaling [35, 36], and peptides derived from the C-terminal tail can inhibit PTN activity [38]. However, previous studies of midkine and PTN showed the C-terminal tail of neither protein is necessary for binding heparin [26, 28]. But no study has examined the role of the C-terminal tail on binding CS, the type of GAG found on PTPRZ. Work presented here shows that, although the C-terminal tail plays a minimal role in binding the highly sulfated CSE, it is vital in maintaining stable interactions with CSA. Since more than 90 % of disaccharides in PTPRZ’s GAG chains are CSA disaccharides [42], this finding demonstrates PTN’s C-terminal tail is a crucial mediator of PTN’s interaction with PTPRZ. In light of these results, previous observations that C-terminal truncated PTN cannot initiate PTPRZ signaling can now be attributed, at least in part, to truncated PTN’s inability to interact strongly with CS chains on PTPRZ. Our NMR titrations also provided some molecular insight into the mechanism of the phenomenon. In particular, the titrations showed removal of the C-terminal tail reduced CTD’s and NTD’s affinity for CSA dp8 measurably, implying that the C-terminal tail of PTN acts synergistically with the TSR domains to facilitate interactions of CSA. However, because residues in the C-terminal tail exhibited neither large CS-induced chemical shift changes nor any significant PRE effects when treated with TEMPO-labeled CS, the mechanism of the synergisitic interactions between the domains is unclear. One possibility is that the C-terminal tail’s affinity for short CS ligands used in the titrations is too weak to be observed, but its interactions with long, native CS polysaccharides maybe significantly higher because of avidity effects. PTN is known to have many disparate binding partners, and it has long been suspected that the domains of PTN may confer specificity for different partners. Our results show that domains of PTN are also responsible for conferring specificity for GAGs. This regulation maybe crucial in determining the receptor specificity of PTN.
Results from this study confirmed that PTN’s affinities for different types of CS vary greatly. This is may have significant consequences on the activity of PTN. In particular, CSC, a variant of CS containing mostly 6-O-sulfated GalNAc are expressed at higher levels in the CNS during early stage neural development [43]. Although not studied here, CSC has been shown to have higher affinity for PTN than CSA [24]. Therefore, presence of CSC during early development will significantly enhance interactions between PTN and PTPRZ. This is consistent with the fact that PTN expression is highest during early stage neural development. Furthermore, CSE has been shown to be enriched in various tumors [44–46] and activity of GalNAc 4-O-sulfate 6-O-sulfotransferase, the enzyme responsible for making CSE, is also elevated in metastatic cancer cells [47]. These CSE containing proteoglycans can serve as high-affinity receptors for PTN, and may explain the improved proliferation and migration of these cancer cells during metastasis.
Interestingly, the C-terminal tail is also known to interact with other PTN receptors. In particular, the C-terminal tail is known to associate with integrins [38]. In fact, integrins have shown to be a crucial part of PTN-induced cell migration and transformation mechanism [23]. Therefore, it is conceivable that removing the C-terminal tail may also inhibit PTN-integrin interactions. There are also reports that PTN can interact with unglycosylated PTPRZ [24, 42]. Because of the acidic nature of the PTPRZ core protein, it is conceivable that PTN may be attracted to the core protein. This implies that the C-terminal tail of PTN may also play a role in modulating stable interactions with the PTPRZ core protein.
Experimental Procedures
Expression and Purification of PTN and PTN Short.
The open reading frame for human wild type PTN and C-terminus truncated PTN (residues 1 to 114) were obtained from DNASU (dnasu.asu.edu) and cloned into the pET-15b vector. For expression, the plasmids were transformed into Origami B (DE3) (Novagen) and grown in M9 medium at 37o C to an OD600 of 0.8, at which time the bacteria were induced with 0.25mM IPTG and incubated overnight at room temperature. The cultures were harvested and lysed using 1 mg/mL lysozyme and sonication. Protein was extracted from supernatant through Heparin-affinity chromatography with a 5 mL HiTrap heparin column (GE Life Sciences) and a NaCl gradient of 0 M to 1.5 M at pH 7.5. Production of isotopically labeled wild type and C-terminal truncated (residues 1 to 114) PTN was attained by supplementing M9 medium with 15NH4Cl and/or 13C glucose.
Biotinylation of PTN.
PTN was biotinylated via EDC-mediated coupling chemistry. Specifically, biotinylation was carried out in 100 μL reactions containing 26μM protein, 5.2 mM Amine-PEG-biotin (Thermo-Fisher) and 2.6 mM EDC in 100 mM MES (pH 5.5). The reactions were incubated for 30 min at room temperature and buffer exchanged into PBS buffer to remove excess labels. After buffer exchange, amount of biotinylated protein was quantified using a biotin quantitation assay kit (Thermo-Fisher) with biotinylated horseradish peroxidase as a standard. Biotinylation efficiency was approximately 0.5 – 1%.
Semi-Synthesis of Chondroitin Sulfate E.
Chondroitin Sulfate E (CSE) was synthesized through chemical sulfation of Chondroitin Sulfate A (CSA) according the procedure in Cai et al.[48] Briefly, 3.5 g of CSA was dissolved in 90 mL of formamide with 5 g of trimethylamine sulfur trioxide. The reaction was stirred vigorously and kept at 60o C under N2 gas for 24 hours. After completion of the reaction, the product was dialyzed for two days in water before lyophilization.
Preparation of Size-Defined and Paramagnetic CSA & CSE Ligands.
Size defined CSA and CSE fragments were obtained through partial enzymatic digestion of native polysaccharides. Specifically, 0.5 g of CSA from porcine trachea was digested with 50 mg of type V hyaluronidase from sheep testes in 20 mL of 50 mM NaH2PO4, pH 6.0, 150 mM NaCl buffer at 37o C for two days. 0.1 g of CSE was digested using 0.8 U of chondroitinase ABC (Sigma Aldrich) in 12 mL of a digestion buffer (50 mM Tris pH 8.0, 60 mM NaAc, 50 μg/mL kanamycin, 0.01% NaN3) at 37o C for 24 hours. Kanamycin was added to prevent bacterial growth during digestion. All fragments were separated with a 2.5 cm × 175 cm size exclusion chromatography column (Bio-Rad Biogel P10) with a flow rate of 0.2 mL/min. Fractions containing identical sized fragments were combined and subsequently desalted, and lyophilized. NMR analysis shows the main component (~ 80%) in CSA dp8 contains three 4-O-sulfated GalNAc and one 6-O-sulfated GalNAc. No 2-O-sulfated GlcA was found in CSA. The CSE dp6 contains mostly two components. One component (40 %) contains entirely 4,6-O-disulfated GalNAc. The other component (40%) has one 6-O-sulfated GalNAc and two 4,6-O-sulfated GalNAc. There was no 2-O-sulfated GlcA in CSE. CSE and CSA dp6 were functionalized with 4-amino-TEMPO through reductive amination using procedures similar to those outlined in Morgan et al. [49] Briefly, 2 mg of CSE or CSA dp6 were reductively aminated in 400 μL of H2O containing 0.3 M of 4-amino-TEMPO and 20 mM NaCNBH3. To prevent reduction of the radical, the reaction was carried out at neutral pH and incubated at 65 ° C overnight.
Collection and Analysis of NMR Data.
NMR data for PTN was collected on Bruker 600 MHz Avance III HD spectrometers equipped with a Prodigy probe. PTN samples used for data collection contained 0.4–1 mM PTN in 10 mM MES and 150 mM NaCl (pH 6.0). 13C/15N-labeled PTN was used for collection of HNCO, HNCACO, HNCACB, and HNCOCACB spectra for backbone assignment. Side chain proton assignments were obtained using a combination of HCCONH, CCONH, HCCHTOCSY, 13C-methyl detected HCCHTOCSY [50] and 13C- or 15N-edited NOESY experiments. Aromatic proton and carbon assignments were obtained using HBCBCGCDHD, HBCBCGCDCEHE and aromatic 13C-edited NOESY experiments. Aligned sample in a 7% positive charged polyacrylamide gel was also used to obtain HN residual dipolar couplings (RDCs). All data processing was performed with NMRPipe [51] and analyzed using NMRView. [52] The numbering scheme for residues was based on mature PTN, starting with G1 after the signal peptide. Measurements of 1H R2,PRE from TEMPO-labeled CSE dp6 were carried out on a 200 μM wildtype PTN samples containing either 50 μM TEMPO-tagged CSE dp6 or 100 μM TEMPO-tagged CSA dp8. The paramagnetic contribution to the transverse relaxation was measured as the difference between 1H T2 before and after reduction of the TEMPO radical with 10 molar equivalents of ascorbate.
Structure Calculation.
The secondary structure prediction program TALOS [53] was used to predict the backbone torsion angle restraints of PTN using chemical shift data. 13C-, and 15N-edited NOESY spectra were assigned manually to determine as many unambiguous long- and short-range contacts as possible. Partially assigned NOESY peak lists and RDC data were used in the structure calculation program CYANA. [54] The resulting structure and constraints were used to further refine the structure using XPLOR-NIH. [55] The ten best structures out of 100 calculated in the calculations were used. Table 1 lists the final constraints used in the refinement of the structures. NTD and CTD RDCs were refined using separate and independent tensors. There was no RDC violation over 0.5 Hz. Root mean square deviation between experimental and calculated RDCs were 0.29 ± 0.07 Hz for NTD and 0.16 ± 0.05 Hz for CTD. Electrostatic surface potential of the TSR domains was calculated using the program APBS [56]. The calculation assumed the ionic strength of the solution was 0.15 M. The dielectric constant of solvent and solute was 78.5 and 10, respectively. The potential is specified in the unit of kT/e, where k is the Boltzmann constant, T is the temperature and e is the charge of the electron.
Titrations of Wild Type and Truncated PTN with CSA & CSE.
Titrations of wild type and C-terminal truncated PTN with CSE dp6 and CSA dp8 were performed on the Bruker 600 MHz Avance III spectrometer. Titration of wild type PTN with CSE dp6 was performed by adding aliquots of 40 mM CSE dp6 stock solution to a 125 μM PTN sample to produce final CSE dp6 concentrations of 0.125, 0.375, 0.625, 0.875, and 1.125 mM. CSA dp8 titration of wild type PTN was performed by adding aliquots of 8.5 mM CSA dp8 to a 190 μM sample to final concentrations of 0.09, 0.26, 0.51, 0.77, 1.11, 1.45, and 1.87 mM CSA dp8. Titration of C-terminal truncated PTN with CSE dp6 and CSA dp8 were carried out with 100 μM PTN in 10 mM MES, 150 mM NaCl (pH 6.0). Aliquots of CSE dp6 from a 40 mM stock solution were added to reach concentrations of 0.1, 0.3, 0.5, 0.7, and 0.9 mM. CSA dp8 from a 20 mM stock solution was added to reach concentrations of 0.05, 0.1, 0.2, 0.4, 0.6, 0.8, and 1 mM. At each increment in the titrations, a 15N-edited HSQC spectrum was acquired. Observed chemical shift changes in the 1H and 15N dimensions were measured and normalized to a single value using the equation: δ = [ΔδH2 + (0.17 × ΔδN)2]1/2, in which ΔδH and ΔδN represent chemical shift changes in the 1H and 15N dimensions in ppm units. Proton chemical shifts were referenced to internal DSS, and 13C and 15N resonances were referenced indirectly to DSS, using the absolute frequency ratios. Kd values were extracted by fitting plots of normalized chemical shift changes vs. ligand concentration using the 1:1 binding model fitting feature in xcrvfit (http://www.bionmr.ualberta.ca/bds/software/xcrvfit/). It should be noted that the model used in the fitting does take into consideration the depletion of ligands during the titration.
Immobilized CSA and CSE ELISA of PTN.
CSA and CSE were attached to microwell plates through covalent crosslinking. To accomplish this, amine binding plates (G-Biosciences) were first treated with 100 μL of 100 μg/mL poly-L-lysine (Sigma). CSE or CSA were then covalently linked to poly-L-lysines by treating each well with 100 μL of 250 μg/mL CSE or CSA, 1mM EDC, 10μM NHS and 100mM MES (pH5.5). The reactions were incubated overnight at room temperature. To carry out the ELISA, 5% BSA in PBS was used to block the plate for 1.5 hours at room temperature first. The plate was washed with TBST 3 times before 100 μL of PTN samples containing protein concentrations ranging from 2.5μg/ml to 0.015μg/ml were added to the wells. After 1 hour incubation, the wells were washed with TBST and incubated with Streptavidin-HRP in PBS for 1 hour in room temperature. To quantify the amount of PTN in each well, 100μl 1-Step Ultra TMB ELISA Substrate (Thermo-Fisher) was added to each well. The reaction was stopped by adding 100 μl 0.1 M HCl and 450nm absorbance of each well was measured. The Kds for the titrations were calculated using the Langmuir Isotherm with the assumption that the protein-to-ligand ration in the complex is one-to-one. Specifically, the plot of A450 vs. CS concentration is fitted with the equation A450=([CS]*Amax)/(Kd+[CS]), where Amax is the maximum absorbance possible. Each ELISA was repeated at least three times for reproducibility, and triplicate of each data point were included each time.
Modeling of the CTD-CSE dp6 Complex Using HADDOCK.
Models of the CTD-CSE dp6 complex was constructed with HADDOCK 2.2 [57] starting with the structure of CTD and hinge. Force field parameters of the ligand were derived from monosaccharide parameters distributed with CNS 1.3 [58] and parameters for sulfate groups derived from the work by Huige et al. [59] Parameters for the TEMPO tag was calculated using the PRODRG server [60]. Residues showing strong chemical shift perturbations in CSE dp6 titrations were designated as active residues and solvent exposed residues nearby were designated passive residues. Unambiguous upper distance constraints of 20 Å were specified between the paramagnetic center in TEMPO and HN atoms showing strong PRE perturbation (residues Y69, F71 and G110). The ligand and residues in the hinge region were allowed to be fully flexible in the docking. The docking followed the default HADDOCK protocol with 1000 structures calculated in the initial round. 200 structures with the lowest energy were then used in a second round of refinement. Finally, all second round structures were refined in explicit solvents.
Acknowledgement
We want to thank Dr. Brian Cherry for maintenance of the spectrometers, Dr. Ashli Morgan for assistance in ELISA, and Shaghayegh Mazoury for preparing size-defined CS ligands. This work was funded by the NIGMS grant R21GM118339.
Abbreviations:
- CS
chondroitin sulfate
- CSA
chondroitin sulfate A
- CSE
chondroitin sulfate E
- CTD
C-terminal TSR domain
- EDC
1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide
- GAG
glycosaminoglycan
- GalNAc
N-acetylgalactosamine
- GlcA
glucuronic acid
- GlcNAc
N-acetylglucosamine
- HSQC
heteronuclear single quantum coherence spectroscopy
- HS
heparan sulfate
- IdoA
iduronic acid
- NTD
N-terminal TSR domain
- PTN
pleiotrophin.
- PTPRZ
receptor type protein tyrosine phosphatase ζ
- TSR
Thrombospondin type-1 repeat.
- TBST
Tris-buffered saline with Tween 20.
Footnotes
Database: Coordinates of the ensemble of ten PTN structures are deposited in RCSB with the accession number 2n6f. Chemical shifts assignments and structural constraints are deposited in BMRB with the accession number 25762.
Reference
- 1.Yeh HJ, He YY, Xu J, Hsu CY & Deuel TF (1998) Upregulation of pleiotrophin gene expression in developing microvasculature, macrophages, and astrocytes after acute ischemic brain injury, J Neurosci. 18, 3699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li YS, Milner PG, Chauhan AK, Watson MA, Hoffman RM, Kodner CM, Milbrandt J & Deuel TF (1990) Cloning and expression of a developmentally regulated protein that induces mitogenic and neurite outgrowth activity, Science. 250, 1690–4. [DOI] [PubMed] [Google Scholar]
- 3.Silos-Santiago I, Yeh HJ, Gurrieri MA, Guillerman RP, Li YS, Wolf J, Snider W & Deuel TF (1996) Localization of pleiotrophin and its mRNA in subpopulations of neurons and their corresponding axonal tracts suggests important roles in neural-glial interactions during development and in maturity, J Neurobiol. 31, 283–96. [DOI] [PubMed] [Google Scholar]
- 4.Himburg HA, Harris JR, Ito T, Daher P, Russell JL, Quarmyne M, Doan PL, Helms K, Nakamura M, Fixsen E, Herradon G, Reya T, Chao NJ, Harroch S & Chute JP (2012) Pleiotrophin regulates the retention and self-renewal of hematopoietic stem cells in the bone marrow vascular niche, Cell Rep. 2, 964–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatziapostolou M, Delbe J, Katsoris P, Polytarchou C, Courty J & Papadimitriou E (2005) Heparin affin regulatory peptide is a key player in prostate cancer cell growth and angiogenicity, Prostate. 65, 151–8. [DOI] [PubMed] [Google Scholar]
- 6.Jager R, Noll K, Havemann K, Pfluger KH, Knabbe C, Rauvala H & Zugmaier G (1997) Differential expression and biological activity of the heparin-binding growth-associated molecule (HB-GAM) in lung cancer cell lines, Int J Cancer. 73, 537–43. [DOI] [PubMed] [Google Scholar]
- 7.Mentlein R & Held-Feindt J (2002) Pleiotrophin, an angiogenic and mitogenic growth factor, is expressed in human gliomas, J Neurochem. 83, 747–53. [DOI] [PubMed] [Google Scholar]
- 8.Vacherot F, Caruelle D, Chopin D, Gil-Diez S, Barritault D, Caruelle JP & Courty J (1999) Involvement of heparin affin regulatory peptide in human prostate cancer, Prostate. 38, 126–36. [DOI] [PubMed] [Google Scholar]
- 9.Wellstein A, Fang WJ, Khatri A, Lu Y, Swain SS, Dickson RB, Sasse J, Riegel AT & Lippman ME (1992) A heparin-binding growth factor secreted from breast cancer cells homologous to a developmentally regulated cytokine, J Biol Chem. 267, 2582–7. [PubMed] [Google Scholar]
- 10.Zhang L, Mabuchi T, Satoh E, Maeda S, Nukui H & Naganuma H (2004) Overexpression of heparin-binding growth-associated molecule in malignant glioma cells, Neurol Med Chir (Tokyo). 44, 637–43; discussion 644–5. [DOI] [PubMed] [Google Scholar]
- 11.Zhang N, Zhong R, Wang ZY & Deuel TF (1997) Human breast cancer growth inhibited in vivo by a dominant negative pleiotrophin mutant, J Biol Chem. 272, 16733–6. [DOI] [PubMed] [Google Scholar]
- 12.Weber D, Klomp HJ, Czubayko F, Wellstein A & Juhl H (2000) Pleiotrophin can be rate-limiting for pancreatic cancer cell growth, Cancer Res. 60, 5284–8. [PubMed] [Google Scholar]
- 13.Kong Y, Bai PS, Nan KJ, Sun H, Chen NZ & Qi XG (2012) Pleiotrophin is a potential colorectal cancer prognostic factor that promotes VEGF expression and induces angiogenesis in colorectal cancer, Int J Colorectal Dis. 27, 287–98. [DOI] [PubMed] [Google Scholar]
- 14.Gramage E, Herradon G, Martin YB, Vicente-Rodriguez M, Rojo L, Gnekow H, Barbero A & Perez-Garcia C (2013) Differential phosphoproteome of the striatum from pleiotrophin knockout and midkine knockout mice treated with amphetamine: correlations with amphetamine-induced neurotoxicity, Toxicology. 306, 147–56. [DOI] [PubMed] [Google Scholar]
- 15.Herradon G & Perez-Garcia C (2014) Targeting midkine and pleiotrophin signalling pathways in addiction and neurodegenerative disorders: recent progress and perspectives, Br J Pharmacol. 171, 837–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vicente-Rodriguez M, Gramage E, Herradon G & Perez-Garcia C (2013) Phosphoproteomic analysis of the striatum from pleiotrophin knockout and midkine knockout mice treated with cocaine reveals regulation of oxidative stress-related proteins potentially underlying cocaine-induced neurotoxicity and neurodegeneration, Toxicology. 314, 166–73. [DOI] [PubMed] [Google Scholar]
- 17.Gramage E & Herradon G (2011) Connecting Parkinson’s disease and drug addiction: common players reveal unexpected disease connections and novel therapeutic approaches, Curr Pharm Des. 17, 449–61. [DOI] [PubMed] [Google Scholar]
- 18.Herradon G & Ezquerra L (2009) Blocking receptor protein tyrosine phosphatase beta/zeta: a potential therapeutic strategy for Parkinson’s disease, Curr Med Chem. 16, 3322–9. [DOI] [PubMed] [Google Scholar]
- 19.Marchionini DM, Lehrmann E, Chu Y, He B, Sortwell CE, Becker KG, Freed WJ, Kordower JH & Collier TJ (2007) Role of heparin binding growth factors in nigrostriatal dopamine system development and Parkinson’s disease, Brain Res. 1147, 77–88. [DOI] [PubMed] [Google Scholar]
- 20.Taravini IR, Chertoff M, Cafferata EG, Courty J, Murer MG, Pitossi FJ & Gershanik OS (2011) Pleiotrophin over-expression provides trophic support to dopaminergic neurons in parkinsonian rats, Mol Neurodegener. 6, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raulo E, Chernousov MA, Carey DJ, Nolo R & Rauvala H (1994) Isolation of a neuronal cell surface receptor of heparin binding growth-associated molecule (HB-GAM). Identification as N-syndecan (syndecan-3), J Biol Chem. 269, 12999–3004. [PubMed] [Google Scholar]
- 22.Meng K, Rodriguez-Pena A, Dimitrov T, Chen W, Yamin M, Noda M & Deuel TF (2000) Pleiotrophin signals increased tyrosine phosphorylation of beta beta-catenin through inactivation of the intrinsic catalytic activity of the receptor-type protein tyrosine phosphatase beta/zeta, Proc Natl Acad Sci U S A. 97, 2603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikelis C, Sfaelou E, Koutsioumpa M, Kieffer N & Papadimitriou E (2009) Integrin alpha(v)beta(3) is a pleiotrophin receptor required for pleiotrophin-induced endothelial cell migration through receptor protein tyrosine phosphatase beta/zeta, FASEB J. 23, 1459–69. [DOI] [PubMed] [Google Scholar]
- 24.Maeda N, Nishiwaki T, Shintani T, Hamanaka H & Noda M (1996) 6B4 proteoglycan/phosphacan, an extracellular variant of receptor-like protein-tyrosine phosphatase zeta/RPTPbeta, binds pleiotrophin/heparin-binding growth-associated molecule (HB-GAM), J Biol Chem. 271, 21446–52. [DOI] [PubMed] [Google Scholar]
- 25.Iwasaki W, Nagata K, Hatanaka H, Inui T, Kimura T, Muramatsu T, Yoshida K, Tasumi M & Inagaki F (1997) Solution structure of midkine, a new heparin-binding growth factor, EMBO J. 16, 6936–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim J, Yao S, Graf M, Winkler C & Yang D (2013) Structure-function analysis of full-length midkine reveals novel residues important for heparin binding and zebrafish embryogenesis, Biochem J. 451, 407–15. [DOI] [PubMed] [Google Scholar]
- 27.Kilpelainen I, Kaksonen M, Kinnunen T, Avikainen H, Fath M, Linhardt RJ, Raulo E & Rauvala H (2000) Heparin-binding growth-associated molecule contains two heparin-binding beta -sheet domains that are homologous to the thrombospondin type I repeat, J Biol Chem. 275, 13564–70. [DOI] [PubMed] [Google Scholar]
- 28.Raulo E, Tumova S, Pavlov I, Pekkanen M, Hienola A, Klankki E, Kalkkinen N, Taira T, Kilpelainen I & Rauvala H (2005) The two thrombospondin type I repeat domains of the heparin-binding growth-associated molecule bind to heparin/heparan sulfate and regulate neurite extension and plasticity in hippocampal neurons, J Biol Chem. 280, 41576–83. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Pinera P, Berenson JR & Deuel TF (2008) Pleiotrophin, a multifunctional angiogenic factor: mechanisms and pathways in normal and pathological angiogenesis, Curr Opin Hematol. 15, 210–4. [DOI] [PubMed] [Google Scholar]
- 30.Perez-Pinera P, Chang Y & Deuel TF (2007) Pleiotrophin, a multifunctional tumor promoter through induction of tumor angiogenesis, remodeling of the tumor microenvironment, and activation of stromal fibroblasts, Cell Cycle. 6, 2877–83. [DOI] [PubMed] [Google Scholar]
- 31.Maeda N, Fukazawa N & Hata T (2006) The binding of chondroitin sulfate to pleiotrophin/heparin-binding growth-associated molecule is regulated by chain length and oversulfated structures, J Biol Chem. 281, 4894–902. [DOI] [PubMed] [Google Scholar]
- 32.Mizumoto S, Fongmoon D & Sugahara K (2013) Interaction of chondroitin sulfate and dermatan sulfate from various biological sources with heparin-binding growth factors and cytokines, Glycoconjugate journal. 30, 619–32. [DOI] [PubMed] [Google Scholar]
- 33.Li F, Nandini CD, Hattori T, Bao X, Murayama D, Nakamura T, Fukushima N & Sugahara K (2010) Structure of pleiotrophin- and hepatocyte growth factor-binding sulfated hexasaccharide determined by biochemical and computational approaches, J Biol Chem. 285, 27673–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizumoto S, Murakoshi S, Kalayanamitra K, Deepa SS, Fukui S, Kongtawelert P, Yamada S & Sugahara K (2013) Highly sulfated hexasaccharide sequences isolated from chondroitin sulfate of shark fin cartilage: insights into the sugar sequences with bioactivities, Glycobiology. 23, 155–68. [DOI] [PubMed] [Google Scholar]
- 35.Lu KV, Jong KA, Kim GY, Singh J, Dia EQ, Yoshimoto K, Wang MY, Cloughesy TF, Nelson SF & Mischel PS (2005) Differential induction of glioblastoma migration and growth by two forms of pleiotrophin, J Biol Chem. 280, 26953–64. [DOI] [PubMed] [Google Scholar]
- 36.Mathivet T, Mazot P & Vigny M (2007) In contrast to agonist monoclonal antibodies, both C-terminal truncated form and full length form of Pleiotrophin failed to activate vertebrate ALK (anaplastic lymphoma kinase)?, Cell Signal. 19, 2434–43. [DOI] [PubMed] [Google Scholar]
- 37.Bernard-Pierrot I, Delbe J, Caruelle D, Barritault D, Courty J & Milhiet PE (2001) The lysine-rich C-terminal tail of heparin affin regulatory peptide is required for mitogenic and tumor formation activities, J Biol Chem. 276, 12228–34. [DOI] [PubMed] [Google Scholar]
- 38.Mikelis C, Lamprou M, Koutsioumpa M, Koutsioubas AG, Spyranti Z, Zompra AA, Spiliopoulos N, Vradis AA, Katsoris P, Spyroulias GA, Cordopatis P, Courty J & Papadimitriou E (2011) A peptide corresponding to the C-terminal region of pleiotrophin inhibits angiogenesis in vivo and in vitro, J Cell Biochem. 112, 1532–43. [DOI] [PubMed] [Google Scholar]
- 39.Sharma D & Rajarathnam K (2000) 13C NMR chemical shifts can predict disulfide bond formation, J Biomol NMR. 18, 165–71. [DOI] [PubMed] [Google Scholar]
- 40.Deshauer C, Morgan AM, Ryan EO, Handel TM, Prestegard JH & Wang X (2015) Interactions of the Chemokine CCL5/RANTES with Medium-Sized Chondroitin Sulfate Ligands, Structure. 23, 1066–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ori A, Free P, Courty J, Wilkinson MC & Fernig DG (2009) Identification of heparin-binding sites in proteins by selective labeling, Mol Cell Proteomics. 8, 2256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maeda N, He J, Yajima Y, Mikami T, Sugahara K & Yabe T (2003) Heterogeneity of the chondroitin sulfate portion of phosphacan/6B4 proteoglycan regulates its binding affinity for pleiotrophin/heparin binding growth-associated molecule, J Biol Chem. 278, 35805–11. [DOI] [PubMed] [Google Scholar]
- 43.Rauch U, Gao P, Janetzko A, Flaccus A, Hilgenberg L, Tekotte H, Margolis RK & Margolis RU (1991) Isolation and characterization of developmentally regulated chondroitin sulfate and chondroitin/keratan sulfate proteoglycans of brain identified with monoclonal antibodies, J Biol Chem. 266, 14785–801. [PubMed] [Google Scholar]
- 44.Vallen MJ, Schmidt S, Oosterhof A, Bulten J, Massuger LF & van Kuppevelt TH (2014) Primary ovarian carcinomas and abdominal metastasis contain 4,6-disulfated chondroitin sulfate rich regions, which provide adhesive properties to tumour cells, PLoS One. 9, e111806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.ten Dam GB, van de Westerlo EM, Purushothaman A, Stan RV, Bulten J, Sweep FC, Massuger LF, Sugahara K & van Kuppevelt TH (2007) Antibody GD3G7 selected against embryonic glycosaminoglycans defines chondroitin sulfate-E domains highly up-regulated in ovarian cancer and involved in vascular endothelial growth factor binding, The American journal of pathology. 171, 1324–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugahara KN, Hirata T, Tanaka T, Ogino S, Takeda M, Terasawa H, Shimada I, Tamura J, ten Dam GB, van Kuppevelt TH & Miyasaka M (2008) Chondroitin sulfate E fragments enhance CD44 cleavage and CD44-dependent motility in tumor cells, Cancer Res. 68, 7191–9. [DOI] [PubMed] [Google Scholar]
- 47.Ito Y, Watanabe M, Nishizawa T, Omachi T, Kobayashi T, Kasama S, Habuchi O & Nakayama J (2007) The utility of formalin-fixed and paraffin-embedded tissue blocks for quantitative analysis of N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase mRNA expressed by colorectal cancer cells, Acta histochemica et cytochemica. 40, 53–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai C, Solakyildirim K, Yang B, Beaudet JM, Weyer A, Linhardt RJ & Zhang F (2012) Semi-synthesis of chondroitin sulfate-E from chondroitin sulfate-A, Carbohydr Polym. 87, 822–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morgan AM & Wang X (2015) Structural Mechanisms Underlying Sequence-Dependent Variations in GAG Affinities of Decorin Binding Protein A, a Borrelia burgdorferi Adhesin, Biochem J. [DOI] [PubMed] [Google Scholar]
- 50.Permi P, Tossavainen H & Hellman M (2004) Efficient assignment of methyl resonances: enhanced sensitivity by gradient selection in a DE-MQ-(H)CC(m)Ht (m)-TOCSY experiment, J Biomol NMR. 30, 275–82. [DOI] [PubMed] [Google Scholar]
- 51.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J & Bax A (1995) Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes, Journal of Biomolecular Nmr. 6, 277–293. [DOI] [PubMed] [Google Scholar]
- 52.Johnson BA (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules, Methods Mol Biol. 278, 313–52. [DOI] [PubMed] [Google Scholar]
- 53.Shen Y, Delaglio F, Cornilescu G & Bax A (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts, J Biomol Nmr. 44, 213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guntert P (2004) Automated NMR structure calculation with CYANA, Methods Mol Biol. 278, 353–78. [DOI] [PubMed] [Google Scholar]
- 55.Schwieters CD, Kuszewski JJ, Tjandra N & Clore GM (2003) The Xplor-NIH NMR molecular structure determination package, J Magn Reson. 160, 65–73. [DOI] [PubMed] [Google Scholar]
- 56.Baker NA, Sept D, Joseph S, Holst MJ & McCammon JA (2001) Electrostatics of nanosystems: application to microtubules and the ribosome, Proc Natl Acad Sci U S A. 98, 10037–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dominguez C, Boelens R & Bonvin AM (2003) HADDOCK: a protein-protein docking approach based on biochemical or biophysical information, J Am Chem Soc. 125, 1731–7. [DOI] [PubMed] [Google Scholar]
- 58.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T & Warren GL (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination, Acta crystallographica Section D, Biological crystallography. 54, 905–21. [DOI] [PubMed] [Google Scholar]
- 59.Huige CJM & Altona C (1995) Force-Field Parameters for Sulfates and Sulfamates Based on Ab-Initio Calculations - Extensions of Amber and Charmm Fields, J Comput Chem. 16, 56–79. [Google Scholar]
- 60.Schuttelkopf AW & van Aalten DM (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes, Acta crystallographica Section D, Biological crystallography. 60, 1355–63. [DOI] [PubMed] [Google Scholar]
- 61.Hyberts SG, Goldberg MS, Havel TF & Wagner G (1992) The solution structure of eglin c based on measurements of many NOEs and coupling constants and its comparison with X-ray structures, Protein Sci. 1, 736–51. [DOI] [PMC free article] [PubMed] [Google Scholar]