Abstract
Phototrophic microbial mats dominated terrestrial ecosystems for billions of years, largely causing, through cyanobacterial oxygenic photosynthesis, but also undergoing, the great oxidation event (GOE) at ca. 2.5 Ga. Taking a space-for-time approach based on the universality of core metabolic pathways expressed at ecosystem level, we studied gene content and co-occurrence networks in high-diversity metagenomes from spatially close microbial mats along a steep redox gradient. The observed functional shifts suggest that anoxygenic photosynthesis was present but not predominant under early Precambrian conditions, being accompanied by other autotrophic processes. Our data also suggest that, in contrast to general assumptions, anoxygenic photosynthesis largely expanded in parallel to the subsequent evolution of oxygenic photosynthesis and aerobic respiration. Finally, our observations might represent space-for-time evidence that the Wood-Ljungdahl carbon fixation pathway dominated phototrophic mats in early ecosystems, whereas the Calvin cycle likely evolved from pre-existing variants before becoming the dominant contemporary form of carbon fixation.
Phototrophic microbial mats were the forests of the past. Fossil stromatolites, their fossil remnants, constitute the oldest reliable traces of life on Earth1,2. These microbial ecosystems dominated shallow aquatic and terrestrial habitats before large multicellular organisms expanded ca. 550 Ma ago3–5. As such, early Archaean mats, likely built by anoxygenic photosynthetic bacteria3, witnessed the atmospheric oxygen rise that occurred at 2.4-2.5 Ga (Great Oxidation Event, GOE). It is widely assumed that the GOE was promoted by the evolution of oxygenic photosynthesis in the cyanobacterial lineage3 (including the phylogenetic ancestors of extant cyanobacteria), although oxygen derived from atmospheric water photolysis or released from the Earth’s mantle might have also contributed6. Today, in addition to several coastal settings, microbial mats are restricted to a few, mostly extreme (e.g. hot or salty) environments7, where they are not outcompeted, and are commonly considered as analogs of major Precambrian ecosystems3,8,9. The idea that phototrophic mats prior to the GOE were built by anoxygenic photosynthetic bacteria and later by cyanobacteria is actually simplistic, since these stratified microbial communities are phylogenetically and metabolically diverse. Microbial diversity studies of both calcifying10–13 and non-calcifying7,14–18 phototrophic mats reveal a wide variety of members from the three domains of life, although most often bacterial and sometimes also archaeal lineages dominate. Metagenomic analyses show a variety of potential associated metabolic functions, including oxygenic and anoxygenic photosynthesis, sulfate reduction and sulfur oxidation19–23, which are consistent with the steep redox gradients that these communities both endure and contribute to maintain24.
Yet, how good are modern microbial mats as analogs of past ecosystems? From a phylogenetic perspective, there is a severe limit to actualism (the idea that the present is the key to the past) because biological evolution is at work. Species and lineages are not static but change through time and, with them, their phenotypic properties, including most metabolic abilities. This is further complicated by the prevalence of horizontal gene transfer, especially among prokaryotes and involving metabolism-related genes25. Even long-distance (e.g. bacteria-to-archaea) transfers affecting genes involved in key metabolic processes, such as aerobic respiration, have repeatedly occurred26. Thus, despite some general trends27 and the notable exception of oxygenic photosynthesis (exclusive of cyanobacteria and their plastid derivatives) and possibly methanogenesis (exclusive of some archaea, and perhaps ancestral to the domain), inferring ancient functions from contemporary microbial diversity data might be risky. Nonetheless, from the perspective of core metabolism the situation is radically different. Despite the incredible diversity of life28, only a handful of core metabolic processes is known. Cell bioenergetics is universally sustained by the generation of electrochemical potential across biological membranes (though using a wide variety of electron donors and acceptors) and/or fermentative substrate-level phosphorylation29,30. Likewise, only seven pathways of C fixation are known31,32. Even if new C fixation pathways (or variants thereof) might still be discovered in novel candidate divisions, their number will likely remain low. At the same time, classical ecologists observe functional redundancy in ecosystems33, at least at some level34. Such observations were long inaccessible for microbial communities but recent metagenomic analyses suggest that functional stability exists across various microbial communities despite high taxonomic variability35,36. This reinforces the idea that metabolic phenotypes are reliably seen at the level of ecosystems37. Consequently, using a pathway-centric approach opens the possibility to retrace early core metabolisms from modern microbial mats subjected to environmental conditions similar to those prevailing in the past.
Since we cannot directly observe historical changes in past microbial communities, applying a space-for-time approach might be a reasonable alternative. Space-for-time substitution modeling is widely used in ecology to infer past or future trajectories of ecosystems from contemporary spatial patterns38 and can be applied to genomic variation moving beyond species-level variation39. Because modern phototrophic mats are stratified following redox gradients where oxygen is rapidly depleted and nitrate, sulfide and methane increase with depth9, they might in principle be thought to represent relevant model systems to carry out space-for-time substitution studies of core metabolism. However, the physical separation of different functional mat layers is challenging and, most importantly, the presence of oxygen in the atmosphere and upper mat layers can significantly affect the nature and availability of redox pairs and organics in deeper layers. Therefore, ideally, the best-suited model systems would be microbial mats that are as a whole exposed to an anoxic-oxic gradient mimicking the evolution of the Earth’s atmosphere. With this aim, we use here metagenomic approaches to study the metabolic potential of several microbial mats uniquely located spatially close (few cm away) in a small shallow pond along a strong vertical redox gradient, from oxic down to oxygen-deprived waters.
Results and discussion
We studied microbial mats from a shallow pond (LLA9) located in the Salar de Llamara (Atacama Desert, Chile) along vertical redox, salinity (3.1-8.7%) and temperature (28-53°C) gradients17 (Supplementary Fig. 1). We collected mat samples (with replicates) at four different depths: LLA9-A and LLA9-B (oxic zone), LLA9-C (transition zone) and LLA9-D (anoxic zone)17. Interestingly, mat D was hot (53-54°C). Although solar irradiation and the absence of water mixing below the chemocline might contribute, the temperature increase seemed largely due to heat production by the actively growing thick mat.
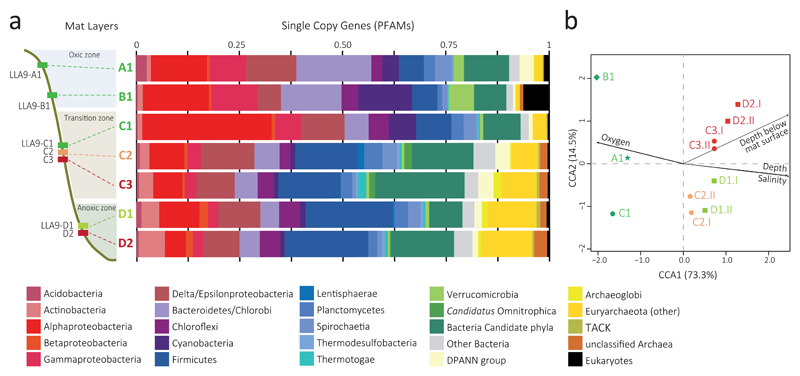
We generated metagenomes for mat samples, including replicates for the most diverse (LLA9-C2/C3, LLA9-D1/D2; Supplementary Table 1)17. Estimates of average coverage as a function of sequencing effort suggested that they were rather complete (Supplementary Fig. 2). Average GC content appeared bimodal for LLA9-A1/B1 but tended to unimodal and increased in deeper mats (Supplementary Fig. 3a-b). Average GC values were higher than 50%, consistent with the idea that more complex environments, with more competition, correlate with higher GC content40. Proteobacteria, Acidobacteria and PVC-supergroup genomes had higher GC content (Supplementary Fig. 3c). We identified clusters of orthologous genes (COGs), PFAMs and KEGG orthologs (KOs)(Supplementary Tables 2-4). To characterize mat microbial diversity, we used conserved marker genes usually present in single copy in sequenced genomes (universal single copy genes). Based on their phylogenetic affiliation (COGs and PFAMs; Supplementary Tables 5-6), we inferred a wide microbial diversity (Fig. 1a and Supplementary Fig. 4), as reflected by high Shannon (3.68-4.61) and Simpson (0.97-0.98) indices (Supplementary Table 1), confirming previous metabarcoding analyses17. As expected, prokaryotes largely dominated; eukaryotes, mainly photosynthetic organisms (Bacillariophyta, Chlorophyta), were essentially found in the uppermost mats. Although bacteria dominated, archaea were abundant in the deepest layers representing up to ca. 20% of annotated sequences (Fig. 1a). Euryarchaeota were particularly profuse, followed by DPANN members and occasionally Thorarchaeota (Asgard), while Crenarchaeota and Thaumarchaeota had minor proportions (Supplementary Fig. 4b). Bacteria were highly diverse. Proteobacteria (especially Alpha-, Gamma- and Deltaproteobacteria), was the most abundant phylum together with Firmicutes in the deepest layers. Lineages of photosynthetic bacteria were abundant in mats LLA9-A1, B1 and C1. Although cyanobacteria were present, members of Chloroflexi, some Chlorobi and likely photosynthetic Alpha/Gammaproteobacteria were collectively more numerous (Fig. 1). These lineages, including cyanobacteria (in minor proportions), were also present in the photosynthetic D1 layer and in deeper, in principle not photosynthetically active (between 2 and 10 cm depth), LLA9-C2/C3 and D2 layers. In the anoxic D1, cyanobacteria affiliated to Oscillatoriales, many of which can use H2S as electron donor for photosynthesis to cope with fluctuating redox gradients41, but Chloroflexi were relatively more abundant (most Bacteroidetes/Chlorobi did not affiliate to known photosynthetic lineages). The presence of typical photosynthetic lineages in deeper layers (carefully collected to avoid cross-contamination and in replicates) may indicate that their decay during burial is low under the prevailing anoxic and salty conditions. Unlike in most classically studied ecosystems, candidate bacterial phyla were remarkably abundant (12-27% bacteria; Fig. 1) and diverse, Patescibacteria being dominant (Supplementary Fig. 4c). Many of these lineages presumably include parasites or episymbionts28,42,43. Patescibacteria are most likely strict fermenters, lacking the tricarboxylic acids (TCA) cycle and electron transport chain components44. Based on known functions for described phylogenetic groups27, cyanobacteria, eukaryotic algae and many Chloroflexi, Chlorobi and Alpha/Gammaproteobacteria lineages are likely responsible for most primary production in these phototrophic mats, whereas most other bacterial and archaeal lineages are likely heterotrophic and intervene at different steps in the degradation cascade of organic matter.
Fig. 1.
Microbial diversity in microbial mat metagenomes along a redox gradient in a pond from the Salar de Llamara (Atacama Desert, Chile). (a) Overall community composition of metagenomic assemblies inferred from the phylogenetic assignment of PFAMs corresponding to 237 single copy genes. Layers LLA9-C2, C3, D1 and D2 correspond to merged metagenomes (see Supplementary Fig. 4a for individual replicates). Relatively abundant Candidatus Omnitrophica and Archaeoglobi are highlighted independently from their respective bacterial and archaeal clades. Detailed classification of archaea and bacterial candidate divisions is shown in Supplementary Fig. 4b-c. (b) Canonical correspondence analysis (CCA) plot based on normalized COG frequencies in individual metagenome datasets showing the similarity between replicate metagenomes and how the different metagenomes correlate with local environmental conditions (see also Supplementary Fig. 5).
To compare the functional potential of metagenomes and see whether local environmental conditions correlate with functional shifts, we applied multivariate statistical analyses based on COGs and KOs. Canonical correspondence analysis (CCA) of normalized COG (4,717 COGs; Fig. 1b and Supplementary Fig. 5a) and KO (12,082 KOs; Fig. S5c) frequencies in individual metagenomes recurrently grouped replicate datasets closely. Mats A1-B1-C1, correlating with exposure to oxic surface conditions, aligned on axis 1, which explained most of the variance (73.3%), although they separated on axis 2 (Fig. 1b). Mat layers LLA9-C2-D1 and C3-D2 respectively grouped in two CCA quadrants (Fig. 1b, Supplementary Fig. 5c). Clustering analysis of COGs and KOs yielded very similar results (Supplementary Fig. 5b and d), which were also similar to those of 16S rDNA-based operational taxonomic unit frequencies17.
To further compare functional properties, we carried out co-occurrence network analyses on the three categories of mats grouped by CCA analyses. For simplicity, we named these networks Upper Mat Layers (UML; A1/B1/C1), ‘Middle’ Mat Layers (MML; C2/D1) and Bottom Mat Layers (BML; C3/D2). Although D1 is the upper photosynthetic layer of the anoxic zone mat, it clustered with the transition-zone mat middle layer based on COGs and KOs (Fig. 1b; Supplementary Fig. 5), suggesting more shared metabolic traits (which networks might reveal). COG-based co-occurrence networks were extremely complex (‘hairballs’; Supplementary Fig. 6). Nonetheless, the UML network was more compact, MML was composed of two highly anti-correlated modules and BML exhibited an intermediate topology. Interestingly, many orthologous genes without known function were abundant and highly inter-connected in the three networks (Supplementary Fig. 6), suggesting important core functions. COGs involved in anoxygenic photosynthesis, sulfur oxidation (SOX system), fermentation and several C fixation pathways (Wood-Ljungdahl, 3-hydroxyproprionate, 3-hydroxypropionate/4-hydroxybutyrate cycles) were also relatively abundant and connected in the three networks. Because global networks were complex, we next focused on a selection of diagnostic genes involved in primary energy, N and C fixation processes (Supplementary Table 7; Fig. 2).
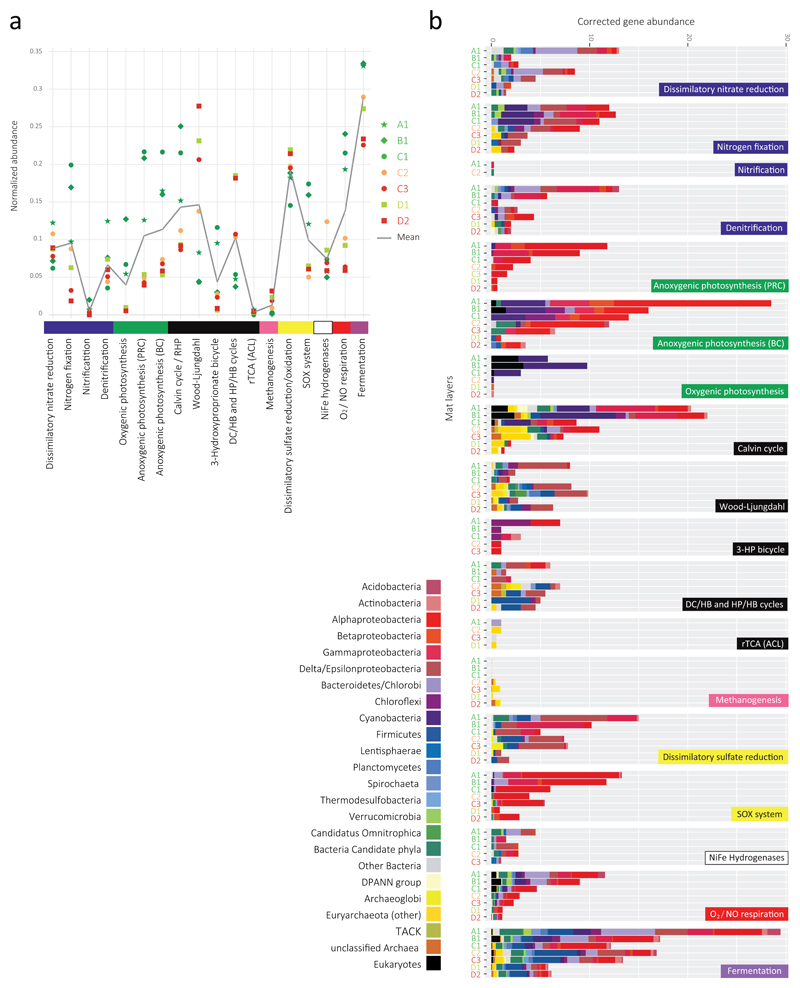
Fig. 2.
Major energy and carbon metabolisms in Llamara microbial mats based on diagnostic genes. (a) Normalized abundance of diagnostic KEGG orthologs (KOs) involved N and S cycling (mostly dissimilatory), photosynthesis, C fixation pathways, Ni-Fe hydrogenases, oxygen respiration (cytochrome oxidases) and fermentation, as corrected by the presence of single-copy genes by the MUSiCC software70. (b) Stacked bar histogram showing the abundance of metabolic genes corrected by the total number of diagnostic KOs selected per pathway. BC, bacteriochlorophylls; most of these genes corresponded to green non-sulfur bacteria (Chloroflexi). PRC, photoreaction center; most of these genes affiliated to purple bacteria (Proteobacteria). RHP, reductive hexulose-phosphate pathway; DC/HB, dicarboxylate/hydrobutyrate; HP/HB, 3-hydroxyproprionate/4-hydrobutyrate; rTCA, reverse tricarboxylic acid cycle; ACL, ATP-dependent citrate lyase.
We compared the normalized abundance of metabolic genes across the different metagenomes. Fermentation was the most abundant energy-generating process followed by sulfate reduction, aerobic respiration, sulfide/sulfur oxidation, dissimilatory nitrate reduction and H-dependent redox reactions (Fig. 2a). Different bacterial and archaeal phyla contributed to those functions (Fig. 2b). Genes for anoxygenic photosynthesis were more abundant than those for oxygenic photosynthesis in all mats. Cyanobacteria and photosynthetic eukaryotes contributed oxygenic photosynthesis-related genes, whereas Chloroflexi (better detected using bacteriochlorophyll synthesis genes) and diverse Proteobacteria accounted for anoxygenic photosynthesis-related genes (Fig. 2b). Regarding C fixation pathways, present in diverse (including candidate) phyla, the Wood-Ljungdahl (reductive acetyl-CoA) pathway and the Calvin cycle dominated, followed by the dicarboxylate/hydroxybutyrate (DC/HB) and 3-hydroxyproprionate/4-hydrobutyrate (HP/HB) cycles, and the 3-hydroxypropionate bicycle. Calvin and 3-hydroxypropionate cycles were most abundant in upper mats, whereas Wood-Ljungdahl and DC/HB-HP/HB cycles dominated in deeper mats/mat layers. Wood-Ljungdahl is considered the most ancestral pathway of C fixation45, sometimes together with the reverse TCA (rTCA) cycle31,46. ATP-dependent citrate lyase (ACL), deemed diagnostic for rTCA, is virtually absent from our mats. However, we cannot rule out the possibility that the core TCA cycle, highly represented in our mats (Supplementary Fig. 7), is operating in reverse using the classical citrate lyase (reverse oxidative TCA, roTCA), as recently shown for thermophilic sulfur-reducing bacteria47,48. Thermophilic sulfate reduction indeed occurs in mat D, mainly by Deltaproteobacteria, Firmicutes and Archaeoglobi (Fig. 2b). Deltaproteobacteria, many of which appear to fix C using the Wood-Ljungdahl pathway, seem major sulfate-reducers in upper mats, although other phyla are also involved, including candidate phyla. Sulfur oxidation is largely contributed by anoxygenic photosynthetic Alphaproteobacteria (and little-abundant Chlorobi in A1/B1), using H2S as electron donor. Regarding the N cycle, dissimilatory nitrate reduction and denitrification (nitrite reduction to N2) are, like N fixation, important processes, especially in upper mats. By contrast, nitrification barely occurs (and only in mats from the oxic zone; Fig. 2a). Dominant N fixers shifted from Cyanobacteria and phototrophic Alpha/Gammaproteobacteria to non-phototrophic Deltaproteobacteria, Firmicutes and Methanomicrobia below the chemocline. We only found a few methanogenesis marker genes (Fig. 2b) in the deeper mats, belonging to Thermoplasmatales (likely Methanomassilicoccales) and Methanomicrobia. This suggests that, as in other microbial mats and sediments9, methanogenesis mostly occurs deeper.
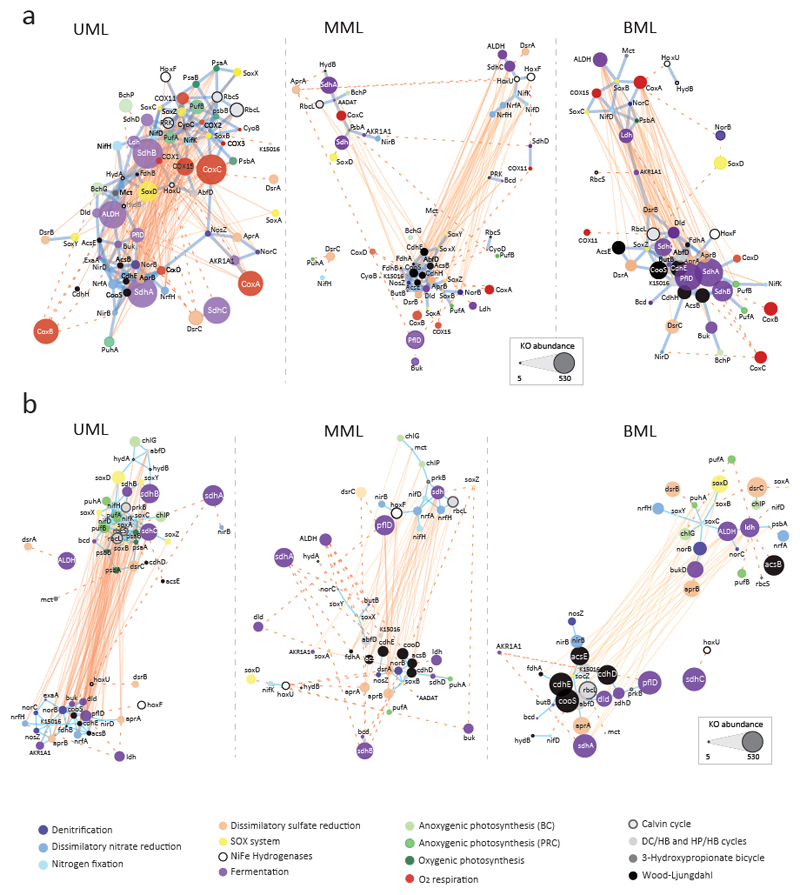
Co-occurrence networks reconstructed with core metabolic functions (KOs) reflected similar, albeit simplified, topologies compared with global COG networks (Fig. 3; Supplementary Fig. 8). The oxic UML mats (A1/B1/C1) appear more connected. By contrast, MML (C2/D1) comprises two anti-correlated modules plus a third module exhibiting less correlations (all negative) with them. BML (C3/D2) comprises two anti-correlated modules positively connected through one fermentation-related protein (Fig. 3a). Although the biological interpretation of gene networks must be cautious, positively correlated modules might imply preference for similar local conditions or potential synergistic interactions between specific metabolisms (syntrophy, metabolic cascading). Because oxygen is a key determinant in redox gradients, we also reconstructed networks excluding genes involved in aerobic respiration (essentially cytochrome oxidase genes; Supplementary Table 7) to see whether and how this affected the observed patterns of potential metabolic interactions. Interestingly, these genes appear responsible for the high connectivity in UML. Indeed, when excluded, the UML network splits in two clearly anti-correlated modules; one includes most genes involved in oxygenic and anoxygenic photosynthesis, N fixation, the Calvin cycle and sulfur oxidation whilst the other connects nitrate reduction, denitrification and Wood-Ljungdahl-related genes (Fig. 3b). By contrast, MML and BML network topologies are not significantly impacted, showing the same two highly anticorrelated modules (plus the small loose anticorrelated module in MML). One of these modules is enriched in Wood-Ljungdahl, fermentation and, sometimes, denitrification genes (Fig. 3). In apparent paradox, RuBisCO genes, typically involved in the Calvin cycle, appear in the BML module connecting Wood-Ljungdahl with many fermentative enzymes. This module displays a clear anaerobic core, being always strongly anti-correlated with oxygen-related enzymes in mats (Fig. 3). Many of these genes affiliate to candidate phyla and, especially in deep mats, archaea (Fig. 2). Many archaea are known to contain a RuBisCO form involved in nucleoside synthesis49,50. Moreover, some RuBisCO-containing archaea also possess phosphoribulokinase and perform the newly described reductive hexulose-phosphate pathway (RHP) for C fixation, which differs only in a few steps from the Calvin cycle32. Indeed, it has been proposed that the photosynthetic Calvin–Benson cycle may have originated from a primitive carbon metabolic pathway utilizing RuBisCO, such as the archaeal RHP pathway, by replacement of some steps without release of carbon32. RuBisCO might have even first worked as an oxygenase before evolving its carboxylase activity51. The presence of oxygen-type cytochrome oxidases in deep, anoxic mats/mat layers, although less abundant than in surface, seems also puzzling. They might result from the progressive burial of aerobic organisms before decay (LLA9-C2, C3, D2), from the presence of microaerophilic microbes using trace oxygen levels in LLA9-D1 and/or, hypothetically, microbes using as final electron acceptor nitric oxide (NO) generated in deep mats during denitrification (Fig. 2). Cytochrome oxidases belong to the same superfamily as NO reductases and it has been proposed that, in the early Archaean, NO, instead of O2, was the terminal electron acceptor for the cytochrome oxidase/NO reductase family before later evolution by subfunctionalization52.
Fig. 3.
Co-occurrence networks of diagnostic KOs involved in major energy and carbon fixation metabolic pathways in Llamara microbial mats from pond LLA9. Networks were built including (a) and excluding (b) cytochrome oxidase genes (O2/NO respiration). Upper Mat Layers (UML) networks derive from metagenomes LLA9-A1, B1 and C1. ‘Middle’ Mat Layers (MML) networks derive from metagenomes LLA9-C2.I, C2.II, D1.I and D1.II. Bottom Mat Layers (BML) networks derive from metagenomes C3.I, C3.II, D2.I and D2.II. Each node represents a KO; node size is proportional to abundance. Node color alludes to the type of metabolic pathway (color code indicated). Edges depict KO co-occurrence in mat layers; reddish edges mean a negative correlation (R < -0.7) and blue edges depict positive correlations (R > 0.7). The distance between nodes is proportional to R values; edges shown in dotted lines have been shortened for visualization purposes (real size networks are shown in Supplementary Fig. 8). Network properties are detailed in Supplementary Table 8.
Because i) iron is a key component of photoreaction centers and electron transporters during photosynthesis, ii) some anoxygenic photosynthesizers can use Fe2+ as electron donor53 and iii) Fe2+ was very abundant before the GOE (which also marked the transition from a ferrous to a sulfidic ocean)3, we also studied genes involved in iron uptake and reduction. Hierarchical networks showed a prominent position of iron uptake regulation in UML. Iron uptake positively correlated with both oxygenic and anoxygenic photosynthesis but negatively with sulfur oxidation. N fixation and nitrite and sulfite/sulfate reduction appeared also important in UML but were only indirectly connected to iron uptake (Supplementary Fig. 9a). Iron uptake regulation was also prominent in MML, but unrelated to photosynthesis (Deltaproteobacteria; Supplementary Fig. 9b). Finally, iron uptake was absent from co-occurrence networks in BML, consistent with the decrease in photosynthesis-related processes in these layers. Sulfate reduction and H-related redox reactions were the more connected and abundant processes in BML (Supplementary Fig. 9c). Given the prevalence of sulfur reduction/oxidation processes, our mats appear to reflect more the conditions of early sulfidic environments3. Iron reduction was present in all mats at moderate levels, and exhibited many connections with other metabolic activities in BML.
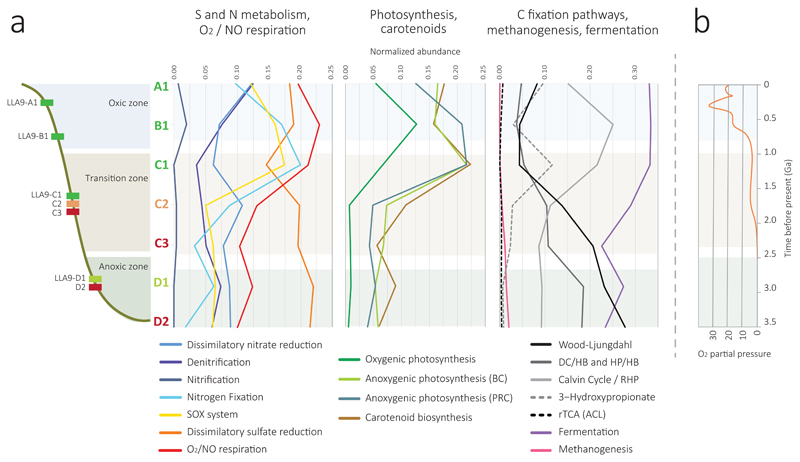
The normalized abundance of diagnostic genes in mat metagenomes along the vertical redox gradient shows marked shifts (Fig. 4a). In mats/layers exposed to oxic conditions (LLA9-A1, B1, C1), aerobic respiration, sulfur oxidation and N fixation genes were more abundant than in deeper, anoxic mats. Likewise, oxygenic photosynthesis genes increased in oxic mat layers, but were almost negligible in deeper mats. Surprisingly, anoxygenic photosyntheses-related genes were more diverse (Fig. 2b) and relatively much more abundant in oxic than in anoxic layers (Fig. 4a), along with a slight fermentation rise and a strong increase in Calvin cycle-related enzymes. The latter likely reflects the succession from the RHP pathway associated to some anaerobic metabolisms to the Calvin-Benson cycle typical of oxygenic photosynthesizers. In addition, C fixation pathways show a remarkable inversion in the deepest, anoxic part of the redox gradient as compared with oxygen-exposed mats, with dicarboxylate/hydrobutyrate and 3-hydroxiproprionate/4-hydrobutyrate and, most importantly, Wood-Ljungdahl pathways becoming more abundant. These metabolic shifts were noticeable within mat C located at the transition zone, which could support the idea that functional changes within a single mat displaying an inner redox gradient might reflect historical metabolic transitions. However, the comparison of mat LLA9-C with mat LLA9-D suggests that that inference is not as straightforward. Indeed, despite LLA9-D is the thickest mat, apparently highly active, and exhibiting conspicuous dark green areas in the upper sampled layer (D1), the normalized (Fig. 4) and net (Fig. 2 and Supplementary Fig. 10) abundance of anoxygenic photosynthesis-related genes was very limited in comparison with LLA9-C and upper mats. Why? The fact that the two replicate metagenomes show very similar results suggests that this observation is not due to local subsampling heterogeneity. Because this was the more glutinous mat and high polysaccharide content usually lowers DNA yield during purification, DNA-extraction bias could be partially invoked. However, this does not really explain the selectivity against photosynthetic organisms since heterotrophic and fermentative organisms are intimately embedded (consuming) in the exopolymeric matrix. It might also be that the anoxygenic photosynthesis genes are highly divergent in LLA9-D or, more speculatively, that new anoxygenic photosynthesis variants are at play. The most likely explanation is that mat LLA9-D is extremely phylogenetically (Fig. 1) and metabolically diverse, such that the relative proportion of anoxygenic photosynthesis genes is low in a huge (meta)genomic diversity. This suggests that, despite high solar irradiation, anoxygenic photosynthesis in this mat is a relatively minor process and that other autotrophic metabolisms operate in parallel and sustain primary production. A low efficiency of anoxygenic photosynthesis in this layer might partly correlate with the mat thermogenicity. Local heat production might come from fermentative processes54 (but these seem even higher in upper, non –or less– thermogenic mats), or simply be due to a partial uncoupling with the electron transport chain, photon-derived energy being dissipated in the form of heat. Applying a space-for-time logic, this could suggest that anoxygenic photosynthesis was less active in early Archaean phototrophic mats growing in anoxic atmospheric conditions, as has been previously suggested55, raising in addition the intriguing possibility that early microbial mats were thermogenic. Anoxygenic photosynthesis would have further evolved and got optimized in parallel to oxygenic photosynthesis. Part of this evolution might have dealt with the development of encoded abilities to cope with reactive oxygen species (ROS). One such mechanisms is provided by alternative oxidases, which are non-energy conserving terminal oxidases best studied in mitochondria and chloroplasts but present in many bacteria, notably cyanobacteria and diverse Proteobacteria, playing key roles in ROS management, thermogenesis and homeostasis56. Interestingly, alternative oxidase (AOX) genes are only detected in upper Llamara mats, associated to cyanobacteria and chloroplasts, but also to several Proteobacteria, including anoxygenic photosynthesizers (Supplementary Table 7 and Fig. 11).
Fig. 4.
Distribution of key energy and carbon fixation metabolisms in microbial mats across a redox gradient and, likely, time. (a) Spatial distributions of diagnostic genes (KOs) for selected core energy and carbon fixation pathways in microbial mats across a redox gradient in the Salar de Llamara. Carotenoid biosynthesis genes were plotted with photosynthesis-related genes. Plots represent mean normalized abundances of diagnostic KOs for each kind of metabolism corrected by single-copy gene abundance by MUSiCC70. The Calvin-Benson enzymes detected in the anoxic zone likely correspond to the new reductive hexulose-phosphate (RHP) pathway32. (b) Space-for-time metabolic transitions (indicated by background colors) represented on a plot of atmospheric oxygen concentrations along the Earth history. O2/NO respiration correspond to cytochrome oxidase genes. BC, bacteriochlorophylls; PRC, photoreaction center. DC/HB, dicarboxylate/hydrobutyrate; HP/HB, 3-hydroxyproprionate/4-hydrobutyrate; rTCA, reverse tricarboxylic acid cycle; ACL, ATP-dependent citrate lyase.
Taking into account the universal conservation of core metabolic functions and assuming that a space-for-time approach can be applied, the metabolic shifts observed in metagenomes of spatially close phototrophic mats across this redox gradient may well represent core metabolic transitions that occurred across the GOE. Our data support two major hypotheses. First, anoxygenic photosynthesis was relatively modestly abundant (perhaps with limited efficiency) in early phototrophic mats under global anoxic conditions; counterintuitively, anoxygenic photosynthesis diversified, appearing in various phylogenetic lineages, and became more prolific in parallel to oxygenic photosynthesis. This might have been partly due to the evolution of adaptive mechanisms to cope with reactive oxygen species. Second, the Wood-Ljungdahl pathway was the early dominant carbon fixation pathway, accompanied to a lesser extent by the dicarboxylate/hydrobutyrate and 3-hydroxiproprionate/4-hydrobutyrate pathways; its primacy was then supplanted by the Calvin cycle as photosyntheses evolved, increasing their ecological success. The Calvin cycle likely evolved from predating variants potentially resembling the archaeal RHP pathway. Further comparative analyses of core metabolic pathways in phototrophic mats from similar contextual environments should help to validate and refine these ideas.
Methods
Sample collection, DNA purification and sequencing
Mat samples were collected in March 2012 in a small pond (LLA9) of the Salar de Llamara, in the North of the Atacama Desert (21°16'7.37"S, 69°37'4.01"W), as previously described17. Mats were collected in pond LLA9 along a steep vertical gradient spanning ca. 30 cm depth, with a chemocline at 25 cm, which was accompanied by salinity (3.1-8.7%) and temperature (28-53°C) gradients (Supplementary Fig. 1). This mat was highly irradiated, the Salar de Llamara being located at 750 m of altitude in one of the driest deserts of the planet17. We collected mat fragments (with replicates) of ca. 10 x 15 cm of surface, and up to 10 cm depth at four different depths: LLA9-A and LLA9-B at increasing depth in the oxic zone, LLA9-C in the transition zone and LLA9-D in the anoxic zone. Mats A and B were thinner (1-3 cm), mat A having poor consistency, and were therefore not subsampled (equally referred to as A1, B1). Mats C and D were much thicker (7-10 cm) and were subsampled in three (C1-C3) and two (D1-D2) broad sub-layers, C1 and D1 comprising all the observable photosynthetic layers which, in the case of D1, displayed green pinnacles of 2-3 cm high at the surface (Supplementary Fig. 1). Mat D was hot (53-54°C within the mat). Temperature decreased below mat LLA9-D to 30°C (Supplementary Fig. 1). Physicochemical parameters (conductivity, oxygen and temperature) were measured using a multi-parameter probe Hanna HI9828. Mat samples were fixed in ethanol (>70%) and stored at -20°C until DNA extraction. DNA was extracted using the Power Biofilm™ DNA Isolation Kit (MoBio, Carlsbad, CA, USA) according to manufacturer’s instructions. Duplicate mat subsamples were collected from distant ends of each mat sample. For each duplicate, the collected material was thoroughly mixed prior to DNA purification; several independent purification reactions per duplicate were performed in parallel and then pooled to minimize potential biases due to sample and/or process heterogeneity. Total DNA yield ranged from 0.6 (LLA9-C3) to 3.1 µg (LLA9-D1). DNA libraries for Illumina paired-end sequencing were prepared for each sample without any amplification step. DNA from LLA9-D1, LLA9-D2 and LLA9-B1 metagenomic libraries were sequenced using Illumina HiSeq2000 v3 (2x100 bp paired-end reads) by Beckman Coulter Genomics (Danvers, MA, USA). DNA from LLA9-A1, LLA9-C1, LLA9-C-2, LLA9-C-3 and LLA9-D-1 metagenomic libraries were sequenced using Illumina HiSeq2500 (2x125 bp paired-end reads) by Eurofins Genomics (Ebersberg, Germany). DNA from LLA9-C2, LLA9-C3, LLA9-D1 and LLA9-D2 duplicate samples were also sequenced in an independent run using Illumina HiSeq2500 (2x125 bp paired-end reads) by Eurofins Genomics (Ebersberg, Germany). Replicate mat samples are noted as, e.g., D1.I and D1.II. The total number of paired-end reads per metagenome ranged from ca. 43 to 120 million, i.e., 4.3–12.0 Gbp per library and orientation (forward and reverse). Various statistics of the 11 generated metagenomes, as well as merged replicates are given in Supplementary Table 1. Estimates of average coverage as a function of sequencing effort suggested that Llamara metagenomes were rather complete (70-92%; Supplementary Fig. 2a); merged replicate metagenomes exhibited a slight coverage increase as compared to individual metagenomes (Supplementary Fig. 2b, Supplementary Table 1).
Prediction and affiliation of rRNA genes
The metagenomic reads were mined for 16S rRNA genes with the EMIRGE software57. Statistics regarding the total number of reads and paired-end sequences per sample, the number of predicted 16S rRNA genes and the average sequence lengths retained are presented in Supplementary Table 1.
Assembly and annotation
The level of coverage of the community achieved by each metagenomic dataset was estimated and projected using Nonpareil version 2.4 with default parameters58 and after preprocessing the reads with Trimmomatic and a minimum Phred quality score of 3059. For each metagenomic dataset, the reads were assembled into contigs using stringent criteria to facilitate gene prediction. Forward and reverse reads were assembled using MEGAHIT (version 1.3.060 with default parameters but a minimum length of 200 bp for the assembled contigs and a starting kmer size of 23 up to 93 with an increasing step of 10. Gene prediction was performed on the newly assembled contigs using Prokka61. For functional annotation purposes, reads from replicate metagenomes were merged for assembly with the same above-mentioned parameters (LLA9-C2.m, LLA9-C3.m, LLA9-D1.m and LLA9-D2.m in Supplementary Table 1). For taxonomic affiliation, we compared the amino acid sequence of our metagenome predicted genes to a home-made non-redundant protein database (RefSeq nr release 74; March, 2017 + a customized database of manually added Candidate Phyla) through the DIAMOND software (version 0.7.9.5862). For subsequent analyses, we retained only the best hit to represent each annotated gene, with a minimum amino acid identity of 50% over at least 80% of the query length. For each defined best hit of an annotated gene, their taxid was retrieved through NCBI e-fetch via an ad hoc Perl script. Various statistics regarding contig assembly and annotation are provided in Supplementary Table 1. Predicted clusters of orthologous genes (COGs), PFAMs and KEGG orthologs (Supplementary Methods; Supplementary Tables 2-4) were used to characterize and compare the different metagenomes. COGs were assigned by profile hidden Markov model (profile HMM) searches using the hmmsearch program of the HMMER3 package63. For every COG, a multiple sequence alignment of bona fide representative sequences were generated using the Muscle program64 and, then, the corresponding Hidden Markov Model was built using the hmmerbuild program, also provided in the HMMER3 package63. The cut-off E-value in the hmmsearch process varies largely for every COG. For each COG, we defined a high confidence cut-off E-value value as the highest E-value (smallest bit score) observed for the members of that COG. None of the COG cut-off E- values was greater than 1e-10. Additionally, all PFAMs (Pfam-A) were predicted with hmmersearch tool from HMMER (version 3.1b165 and KEGG orthologs (KOs) were assigned via GhostKOALA web server66. Abundance matrices for six ribosomal protein PFAMs were used to calculate diversity (Shannon and Simpson), evenness (Pielou) and richness (Chao1) indices using the Vegan package in R. The distribution of COGs, PFAMs and KOs identified in the different Llamara metagenomic assemblies can be found in Supplementary Tables 2-4. COGs, PFAMs and KOs were given a taxonomic assignation via their best hit’s taxid. A subset of 40 COGs corresponding to single copy gene families universally distributed in prokaryotic genomes67 was initially used to characterize the phylogenetic structure of the communities (Supplementary Table 5), which was very similar to community structure derived from 16S/18S rRNA gene metabarcoding analyses17. In addition, we also mined for this purpose 237 single copy genes (PFAMs) previously used to characterize the diversity of archaea42 and bacteria68 in the Llamara metagenomic assemblies (Supplementary Table 6). These yielded a comparable community structure to that observed by 16S/18S rRNA gene metabarcoding17 and single-copy genes (COGs; Fig. 1 and Supplementary Fig.4).
Mining of diagnostic metabolic genes
Orthologous protein-coding genes exclusively involved in one particular energy or carbon metabolic pathway in the KEGG database (KOs69 were considered as diagnostic for that pathway (Supplementary Table 7). For example, for the Calvin cycle, only the two RuBisCO subunits and phosphoribulokinase (PRK) were considered as diagnostic. We looked for diagnostic KOs involved in all known pathways for C fixation, oxygenic and anoxygenic photosynthesis (biosynthesis of bacteriochlorophylls and/or genes for the photosystem reaction centers) (Supplementary Table 7) and fermentation35. In the case of green non-sulfur anoxygenic photosynthesis, we used as diagnostic only the genes involved in the last two steps of the bacteriochlorophyll a/b biosynthesis to follow KEGG annotation, although this bacteriochlorophyll is present in small concentrations in other anoxygenic photosynthetic organisms (e.g. green sulfur phototrophs). In addition, we looked for diagnostic genes for energy metabolism involved in N and S cycling (i.e. dissimilatory nitrate reduction, nitrification, denitrification, dissimilatory sulfate reduction, SOX system) and nitrogen fixation. Genes were assigned to major taxa as described previously and gene abundance was graphed in stack bars for comparison (Fig. 2). To be able to estimate the relative abundance of diagnostic metabolic genes within metagenomic assemblies regardless of the taxa involved, and to compare it across metagenomic datasets, we corrected KO abundances by that of single-copy genes with the program MUSiCC (Metagenomic Universal Single-Copy Correction)70. Total and average diagnostic gene abundances are used in Figs. 2 and 4.
Statistical analyses
Statistical analyses were conducted with the R software71 (http://cran.r-project.org). The comparison of the taxonomic distribution inferred from protein-coding marker genes with results obtained either by mining of 16S rRNA genes in metagenomic reads or by amplicon sequencing17 was done using Bray-Curtis dissimilarity distances. They were calculated on frequencies of high-rank bacterial and archaeal taxa using the 'Vegan' R package (version 2.0-1072) with no prior transformation of the data. Raw counts of high-rank taxa corresponding to replicate samples were pooled before the computation of Bray-Curtis distances shown in Fig. 1 (see de-replicated frequencies in Supplementary Fig. 4a). The influence of the environmental conditions on the functional capacities of the different Llamara metagenomes was estimated by Canonical Correspondence Analysis (CCA). They were conducted using a Euclidean matrix containing a set of environmental factors (depth below water level, depth below mat surface, temperature, oxygen concentration and salinity) and a matrix of Bray-Curtis distances based on the normalized abundance of individual COGs and KOs (as corrected by MUSiCC). CCAs were carried out with the Vegan package in R, wherein sample ordinations were constrained and co-plotted by environmental parameters with significance using an Analysis of Variance (ANOVA) with 999 permutations (P < 0.001 for both KOs and COGs). For KOs, CCA global inertia was 68.13%; 62.7% for axis CCA1 and 19.6% for axis CCA2. For COGs, CCA global inertia was 56.3%, axis CCA1, 73.3% and axis CCA2, 14.5%. Clustering of COGs and KOs was also performed with ad hoc scripts in R and visualized in heatmaps (Supplementary Fig. 5). Regardless of whether CCA or clustering for heatmaps are performed, and regardless the use of KOs or COGs, mat layers always showed the same broad clusters. These were noted as Upper Mat Layers (UML), including LLA9-A1, LLA9-B1 and LLA9-C1 (in CCA, only along axis CCA1); ‘Middle’ Mat Layers (MML), including LLA9-C2.I, LLA9-C2.II, LLA9-D1.I and LLA9-D1.II; and Bottom Mat Layers (BML), including LLA9-C3.I, LLA9-C3.II, LLA9-D2.I and LLA9-D2.II.
Metabolic network reconstruction
Co-occurrence networks involving energy and carbon fixation pathways were performed on metagenomes of microbial mats that grouped according to their COG and KO metabolic similarity based on CCA and clustering analyses (Figs. 2 and Supplementary Fig. 5), namely, upper mat layers (UML, 3 metagenomes), correlating with oxygen, ‘middle’ mat layers (MML, four metagenomes) and bottom mat layers (BML, four metagenomes), correlating with depth, temperature and salinity. Initially, we reconstructed networks based on COGs (Supplementary Fig. 6). Given their complexity, we also reconstructed networks based on two different sets of diagnostic genes (105 KOs; 50 PFAMs) (Supplementary Table 7). The abundance of these diagnostic genes was first arranged in matrices for UML, MML and BML groups. Low frequency genes (less than 5% for PFAMs and less than 1% for KOs) were removed from each matrix. The new matrices were used to reconstruct the correlation and p-values matrices with SparCC73. Ten iterations were used to estimate the median correlation of each pair and the statistical significance of the correlations was calculated by bootstrapping with 500 iterations. Correlations were then sorted according to their statistical significance; we retained only those with p < 0.001 and R > 0.7 or R < -0.7. Networks were built using ad hoc scripts in R and visualized with the aid of the igraph package (http://igraph.org/) and Cytoscape74. Taxa affiliations at phylum level were assigned to each node (this information is collectively summarized in Fig. 2b and, for dominant groups, is indicated at the level of PFAMs in Supplementary Fig. 9). Network properties are given in Supplementary Table 8. Networks were visualized with Cytoscape either in perfused forced directed layout based on their correlation values (e.g. Fig. 4) or as hierarchical networks (e.g. Supplementary Fig. 9), where upper nodes have higher degree and betweenness.
Supplementary Material
Acknowledgments
We thank Ricardo Rodríguez de la Vega for discussions about bioinformatics tools, Jonathan Friedman for advice on SparCC, Alejandro Abdala for helping with MySQL database reconstruction and Enrique Merino for help in COG assignation. We also thank Ismael Aracena (Departamento de Medio Ambiente, Sociedad Química y Minera, Chile), José M. López-García (Instituto Geológico y Minero de España) and Juan M. García-Ruiz’s team (Universidad de Granada) for sampling access, georeferencing, and company during the field trip. This research was funded by the European Research Council Grant no. 322669 to P.L.G. under the European Union’s Seventh Framework Program.
Footnotes
Data availability
The sequence datasets of the Llamara metagenomes have been deposited in GenBank with the BioProject accession code PRJNA438773 (corresponding to BioSample accessions SAMN08688543 to SAMN08688553). All codes generated for this study are available at Gitlab https://gitlab.com/DeemTeam/LLA9-Metagenomes. To facilitate color identification in figures, numbers used to build all histograms are provided in the supplementary file Tables_for_Figures.xlsx
Author contributions
A.G.P. and A.S. performed the bioinformatic analyses, analyzed the data and wrote a preliminary draft of the manuscript; Y.Z. supervised initial metagenome assembly and annotation; P.D. helped with software installation and use; D.M. and P.L.G. collected the samples and conceived the study; P.L.G. prepared the samples for sequencing, supervised the work, provided hypotheses and wrote the final manuscript.
Competing interests
Authors declare no competing interests.
References
- 1.Allwood AC, Walter MR, Kamber BS, Marshall CP, Burch IW. Stromatolite reef from the Early Archaean era of Australia. Nature. 2006;441:714–718. doi: 10.1038/nature04764. [DOI] [PubMed] [Google Scholar]
- 2.Nutman AP, Bennett VC, Friend CR, Van Kranendonk MJ, Chivas AR. Rapid emergence of life shown by discovery of 3,700-million-year-old microbial structures. Nature. 2016;537:535–538. doi: 10.1038/nature19355. [DOI] [PubMed] [Google Scholar]
- 3.Knoll AH, Bergmann KD, Strauss JV. Life: the first two billion years. Philos Trans R Soc Lond B Biol Sci. 2016;371 doi: 10.1098/rstb.2015.0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamilton TL, Bryant DA, Macalady JL. The role of biology in planetary evolution: cyanobacterial primary production in low-oxygen Proterozoic oceans. Environ Microbiol. 2016;18:325–340. doi: 10.1111/1462-2920.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lenton TM, Daines SJ. Matworld - the biogeochemical effects of early life on land. The New phytologist. 2017;215:531–537. doi: 10.1111/nph.14338. [DOI] [PubMed] [Google Scholar]
- 6.Andrault D, et al. Large oxygen excess in the primitive mantle could be the source of the Great Oxygenation Event. Geochem Pers Lett. 2018;6:5–10. [Google Scholar]
- 7.Bolhuis H, Cretoiu MS, Stal LJ. Molecular ecology of microbial mats. FEMS Microbiol Ecol. 2014;90:335–350. doi: 10.1111/1574-6941.12408. [DOI] [PubMed] [Google Scholar]
- 8.Des Marais DJ. Microbial mats and the early evolution of life. Trends Ecol Evol. 1990;5:140–144. doi: 10.1016/0169-5347(90)90219-4. [DOI] [PubMed] [Google Scholar]
- 9.Paerl HW, Pinckney JL, Steppe TF. Cyanobacterial-bacterial mat consortia: examining the functional unit of microbial survival and growth in extreme environments. Environ Microbiol. 2000;2:11–26. doi: 10.1046/j.1462-2920.2000.00071.x. [DOI] [PubMed] [Google Scholar]
- 10.Souza V, et al. An endangered oasis of aquatic microbial biodiversity in the Chihuahuan desert. Proc Natl Acad Sci U S A. 2006;103:6565–6570. doi: 10.1073/pnas.0601434103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goh F, et al. Determining the specific microbial populations and their spatial distribution within the stromatolite ecosystem of Shark Bay. ISME J. 2009;3:383–396. doi: 10.1038/ismej.2008.114. [DOI] [PubMed] [Google Scholar]
- 12.Myshrall KL, et al. Biogeochemical cycling and microbial diversity in the thrombolitic microbialites of Highborne Cay, Bahamas. Geobiology. 2010;8:337–354. doi: 10.1111/j.1472-4669.2010.00245.x. [DOI] [PubMed] [Google Scholar]
- 13.Saghaï A, et al. Metagenome-based diversity analyses suggest a significant contribution of non-cyanobacterial lineages to carbonate precipitation in modern microbialites. Front Microbiol. 2015;6:797. doi: 10.3389/fmicb.2015.00797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris JK, et al. Phylogenetic stratigraphy in the Guerrero Negro hypersaline microbial mat. ISME J. 2013;7:50–60. doi: 10.1038/ismej.2012.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez AB, et al. Microbial diversity in sediment ecosystems (evaporites domes, microbial mats, and crusts) of hypersaline Laguna Tebenquiche, Salar de Atacama, Chile. Front Microbiol. 2016;7:1284. doi: 10.3389/fmicb.2016.01284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiel V, et al. The dark side of the Mushroom Spring microbial mat: life in the shadow of chlorophototrophs. I. Microbial diversity based on 16S rRNA gene amplicons and metagenomic sequencing. Front Microbiol. 2016;7:919. doi: 10.3389/fmicb.2016.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saghaï A, et al. Unveiling microbial interactions in stratified mat communities from a warm saline shallow pond. Environ Microbiol. 2017;19:2405–2421. doi: 10.1111/1462-2920.13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berlanga M, Palau M, Guerrero R. Functional stability and community dynamics during spring and autumn seasons over 3 years in Camargue microbial mats. Front Microbiol. 2017;8:2619. doi: 10.3389/fmicb.2017.02619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruvindy R, White RA, 3rd, Neilan BA, Burns BP. Unravelling core microbial metabolisms in the hypersaline microbial mats of Shark Bay using high-throughput metagenomics. ISME J. 2016;10:183–196. doi: 10.1038/ismej.2015.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casaburi G, Duscher AA, Reid RP, Foster JS. Characterization of the stromatolite microbiome from Little Darby Island, The Bahamas using predictive and whole shotgun metagenomic analysis. Environ Microbiol. 2016;18:1452–1469. doi: 10.1111/1462-2920.13094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saghaï A, et al. Comparative metagenomics unveils functions and genome features of microbialite-associated communities along a depth gradient. Environ Microbiol. 2016;18:4990–5004. doi: 10.1111/1462-2920.13456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thiel V, Hugler M, Ward DM, Bryant DA. The dark side of the Mushroom Spring microbial mat: life in the shadow of chlorophototrophs. II. Metabolic functions of abundant community members predicted from metagenomic analyses. Front Microbiol. 2017;8:943. doi: 10.3389/fmicb.2017.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mobberley JM, et al. Organismal and spatial partitioning of energy and macronutrient transformations within a hypersaline mat. FEMS Microbiol Ecol. 2017;93:e00133–00116. doi: 10.1093/femsec/fix028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupraz C, Visscher PT. Microbial lithification in marine stromatolites and hypersaline mats. Trends Microbiol. 2005;13:429–438. doi: 10.1016/j.tim.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 25.Gogarten JP, Townsend JP. Horizontal gene transfer, genome innovation and evolution. Nat Rev Microbiol. 2005;3:679–687. doi: 10.1038/nrmicro1204. [DOI] [PubMed] [Google Scholar]
- 26.López-García P, Zivanovic Y, Deschamps P, Moreira D. Bacterial gene import and mesophilic adaptation in archaea. Nat Rev Microbiol. 2015;13:447–456. doi: 10.1038/nrmicro3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martiny JB, Jones SE, Lennon JT, Martiny AC. Microbiomes in light of traits: A phylogenetic perspective. Science. 2015;350 doi: 10.1126/science.aac9323. aac9323. [DOI] [PubMed] [Google Scholar]
- 28.Hug LA, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 29.Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- 30.Schoepp-Cothenet B, et al. On the universal core of bioenergetics. Biochim Biophys Acta. 2013;1827:79–93. doi: 10.1016/j.bbabio.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 31.Fuchs G. Alternative pathways of carbon dioxide fixation: insights into the early evolution of life? Annu Rev Microbiol. 2011;65:631–658. doi: 10.1146/annurev-micro-090110-102801. [DOI] [PubMed] [Google Scholar]
- 32.Kono T, et al. A RuBisCO-mediated carbon metabolic pathway in methanogenic archaea. Nature Communications. 2017;8:14007. doi: 10.1038/ncomms14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubbell SP. Neutral theory in community ecology and the hypothesis of functional equivalence. Funct Ecol. 2005;19:166–172. [Google Scholar]
- 34.Loreau M. Does functional redundancy exist? Oikos. 2004;104:606–611. [Google Scholar]
- 35.Louca S, et al. High taxonomic variability despite stable functional structure across microbial communities. Nat Ecol Evol. 2016;1:15. doi: 10.1038/s41559-016-0015. [DOI] [PubMed] [Google Scholar]
- 36.Louca S. Probing the metabolism of microorganisms. Science. 2017;358:1264–1265. doi: 10.1126/science.aar2000. [DOI] [PubMed] [Google Scholar]
- 37.Braakman R, Smith E. The compositional and evolutionary logic of metabolism. Physical biology. 2013;10 doi: 10.1088/1478-3975/10/1/011001. 011001. [DOI] [PubMed] [Google Scholar]
- 38.Blois JL, Williams JW, Fitzpatrick MC, Jackson ST, Ferrier S. Space can substitute for time in predicting climate-change effects on biodiversity. Proc Natl Acad Sci U S A. 2013;110:9374–9379. doi: 10.1073/pnas.1220228110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fitzpatrick MC, Keller SR. Ecological genomics meets community-level modelling of biodiversity: mapping the genomic landscape of current and future environmental adaptation. Ecol Lett. 2015;18:1–16. doi: 10.1111/ele.12376. [DOI] [PubMed] [Google Scholar]
- 40.Rocha EP, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- 41.Grim SL, Dick GJ. Photosynthetic versatility in the genome of Geitlerinema sp. PCC 9228 (formerly Oscillatoria limnetica 'Solar Lake'), a model anoxygenic photosynthetic cyanobacterium. Front Microbiol. 2016;7:1546. doi: 10.3389/fmicb.2016.01546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 43.Nelson WC, Stegen JC. The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front Microbiol. 2015;6:713. doi: 10.3389/fmicb.2015.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wrighton KC, et al. Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science. 2012;337:1661–1665. doi: 10.1126/science.1224041. [DOI] [PubMed] [Google Scholar]
- 45.Ragsdale SW, Pierce E. Acetogenesis and the Wood-Ljungdahl Pathway of CO2 Fixation. Biochim Biophys Acta. 2008;1784:1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braakman R, Smith E. The emergence and early evolution of biological carbon-fixation. PLoS Comput Biol. 2012;8:e1002455. doi: 10.1371/journal.pcbi.1002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nunoura T, et al. A primordial and reversible TCA cycle in a facultatively chemolithoautotrophic thermophile. Science. 2018;359:559–563. doi: 10.1126/science.aao3407. [DOI] [PubMed] [Google Scholar]
- 48.Mall A, et al. Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science. 2018;359:563–567. doi: 10.1126/science.aao2410. [DOI] [PubMed] [Google Scholar]
- 49.Sato T, Atomi H, Imanaka T. Archaeal type III RuBisCOs function in a pathway for AMP metabolism. Science. 2007;315:1003–1006. doi: 10.1126/science.1135999. [DOI] [PubMed] [Google Scholar]
- 50.Wrighton KC, et al. RubisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. ISME J. 2016;10:2702–2714. doi: 10.1038/ismej.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ślesak I, Ślesak H, Kruk JC. RubisCO early oxygenase activity: a kinetic and evolutionary perspective. BioEssays. 2017;39 doi: 10.1002/bies.201700071. 1700071-n/a. [DOI] [PubMed] [Google Scholar]
- 52.Ducluzeau AL, et al. Was nitric oxide the first deep electron sink? Trends Biochem Sci. 2009;34:9–15. doi: 10.1016/j.tibs.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 53.Bryant DA, Frigaard NU. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 2006;14:488–496. doi: 10.1016/j.tim.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Norman AG, Richards LA, Carlyle RE. Microbial Thermogenesis in the decomposition of plant materials: Part I. An adiabatic fermentation apparatus. J Bacteriol. 1941;41:689–697. doi: 10.1128/jb.41.6.689-697.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Canfield DE, Rosing MT, Bjerrum C. Early anaerobic metabolisms. Philos Trans R Soc Lond B Biol Sci. 2006;361:1819–1834. doi: 10.1098/rstb.2006.1906. discussion 1835-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.May B, Young L, Moore AL. Structural insights into the alternative oxidases: are all oxidases made equal? Biochem Soc Trans. 2017;45:731–740. doi: 10.1042/BST20160178. [DOI] [PubMed] [Google Scholar]
- 57.Miller CS, Baker BJ, Thomas BC, Singer SW, Banfield JF. EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biol. 2011;12:R44. doi: 10.1186/gb-2011-12-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodriguez RL, Konstantinidis KT. Nonpareil: a redundancy-based approach to assess the level of coverage in metagenomic datasets. Bioinformatics. 2014;30:629–635. doi: 10.1093/bioinformatics/btt584. [DOI] [PubMed] [Google Scholar]
- 59.Cox MP, Peterson DA, Biggs PJ. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 61.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 62.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 63.Mistry J, Finn RD, Eddy SR, Bateman A, Punta M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013;41:e121. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- 66.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 67.Creevey CJ, Doerks T, Fitzpatrick DA, Raes J, Bork P. Universally distributed single-copy genes indicate a constant rate of horizontal transfer. PLoS One. 2011;6:e22099. doi: 10.1371/journal.pone.0022099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Campbell JH, et al. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci U S A. 2013;110:5540–5545. doi: 10.1073/pnas.1303090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manor O, Borenstein E. MUSiCC: a marker genes based framework for metagenomic normalization and accurate profiling of gene abundances in the microbiome. Genome Biol. 2015;16:53. doi: 10.1186/s13059-015-0610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2017. v. http://www.r-project.org. [Google Scholar]
- 72.Vegan. Community Ecology Package. R package version 1.17-9. 2011 http://CRAN.R-project.org/package=vegan.
- 73.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.