Abstract
Diatoms are one of the most abundant and diverse groups of phytoplankton and play a major role in marine ecosystems and Earth’s biogeochemical cycles. Here we combine DNA metabarcoding data from the Tara Oceans Expedition with palaeoenvironmental data and phylogenetic models of diversification to analyse the diversity dynamics of marine diatoms. We reveal a primary effect of pCO2 variation on early diatom diversification, followed by a major burst of diversification in the late Eocene, after which diversification is chiefly affected by sea level, an influx of silica availability, and competition with other planktonic groups. Our results demonstrate a remarkable heterogeneity of diversification dynamics across diatoms and suggest that a changing climate will favor some clades at the expense of others.
Fossil evidence suggests that diatoms originated in the late Jurassic, but remained rather sparse until the final rifting of Pangaea during the Cretaceous (1, 2). At this time, there was an influx of nutrients to the marine world, owing to increased continental erosion, which favored the diversification of large-celled marine phytoplankton, such as diatoms (3). The subsequent drawdown in CO2 (4) and opening of the Southern Ocean gateways, including Drake Passage (5) and the Tasman Gateway (6), resulted in a dynamic presence of continental ice sheets (7), marking the late Eocene (LE) greenhouse-icehouse transition, which fossil evidence shows positively affected planktonic species diversity (8). The particular success of diatoms throughout the Cenozoic has been attributed to an expanded bioavailability of silica from increased silicate rock weathering (9) and terrestrial grassland expansion (10, 11), to an influx of nutrient-rich seawater into the South Atlantic brought by the Antarctic Circumpolar Current (12, 13), and generally to conditions in a cool, low-CO2 planet particularly favorable to diatoms that allowed them to outcompete other eukaryotic phytoplankton (14). There is, however, disagreement about the precise pattern of diversification since the Jurassic, as well as its environmental influences (15, 2, 11). Here we take a molecular approach to studying diatom diversification through time, which allows us to account for heterogeneity in diversification dynamics across phylogenetic clades, as well as cryptic species, of which there are many in diatoms (16). While fossil-based analyses have served as the primary means of reconstructing diversification dynamics in marine microorganisms (17, 18), our approach adds a phylogenetic dimension to the study of the interplay between species evolution and the biotic and abiotic drivers of diversification.
Results and Discussion
We used a unique diatom phylogeny built by combining an extensive DNA metabarcoding dataset of eukaryotic plankton generated from the Tara Oceans expedition (19, 20) (Supplementary Data 1), a robust backbone phylogeny of diatoms constructed with sequences from the Protist Ribosomal Reference database (21) (Fig. 1, Supplementary Data 2), and fossil divergence time estimates from previous work (Supplementary Table 1). We produced four maximum clade credibility (MCC) phylogenies that corresponded to different alignment algorithms and tree construction procedures. Each of them was constructed from a set of 26 phylogenies reflecting uncertainty in the placement of fossil calibrations (Supplemental Fig. 1, Supplementary Data 3). The phylogeny includes 19,197 Operational Taxonomic Units (OTUs) at 97% sequence identity (Fig. 1, Supplementary Data 4), which includes more than 100 genera and represents all major diatom classes (Supplementary Data 5). It has few unresolved polytomies (< 1% of all branching events) and robust support at most nodes (Supplemental Fig. 2). Using Bayesian fits to sample abundance distributions (22), we estimated that the 19,197 OTUs represent ∼ 10% of total extant diversity (Supplemental Fig. 3).
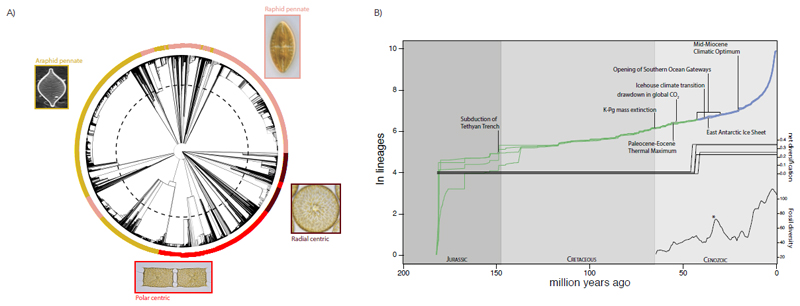
Fig. 1. Phylogenetic diversification in diatoms.
(A) The backbone phylogeny constructed from 3163 sequences from the Protist Ribosomal Database (21) (grey) embedded in the ultrametric diatom phylogeny (black) of 19,197 diatom OTUs derived from 220,018 globally-derived samples of V9-18S ribotypes (using the MAFFT+FastTree build). The colors around the tips correspond to the morphotypes of the diatom OTUs as inferred from the backbone sequences. A representative image of each morphotype is shown. The dashed line at 40 Ma indicates the shift in diversification our analyses identify. (B) Lineage-through-time plots (88) and net diversification rates estimated for the four versions of the phylogeny. A shift in rate is detected at 40±4 Ma, in the Late Eocene. The green segment of the lineage-through-time plots indicates the pre-LE period (which grossly corresponds to a ÒgreenhouseÓ Earth) and the blue segment the post-LE period (which grossly corresponds to an ÒicehouseÓ Earth) and the Earth). Range-through estimates of fossil diversity during the Cenozoic are shown (10, 11) with an asterisk denoting an estimated fossil diversity peak at the Eocene-Oligocene boundary (26). Dates for key climatic and geological events are noted: subduction of Tethyan trench (89), K-Pg mass extinction (90), drawdown of CO2 (4), Paleocene-Eocene thermal maximum (91), icehouse climate transition (8, 51), East Antarctic icesheet formation (12), opening of Southern Ocean gateways (13, 6, 92), and mid-Miocene climatic optimum (87).
We used diversification-rates-through-time analyses (23) applied to the global diatom phylogeny to identify significant events in the evolutionary history of diatoms. Previous work has shown conflicting estimates of the effects of the K-Pg mass extinction, with estimates of survival ranging from 37 – 84% of all diatom species (24, 25). Likewise, diversification dynamics at the Eocene-Oligocene boundary (33.9 million years ago, Ma) are debated: diatom diversity either dropped sharply after a diversity peak at the boundary (26) or increased steadily until the present (11). We found no major effect on diversification rates of either the K-Pg mass extinction or the Eocene-Oligocene transition, but a single significant shift in the LE at 40 ± 4 Ma (depending on the MCC tree considered), owing to an increase in net diversification and decrease in relative extinction (Fig. 1, Supplementary Table 2). This shift is broadly consistent with diversity curves reconstructed from fossil diatoms (9, 11) (Fig. 1B).
We sectioned the global phylogeny into a single tree (hereafter referred to as the pre-LE tree) dating from the crown (estimated at 186 Ma) to the evidential shift at 40 Ma and into multiple subtrees from the shift to the present (post-LE trees). This allowed us to analyze pre-Cenozoic diatom diversification dynamics and, over the last 40 million years, to consider the individual dynamics of a large set of clades (128 phylogenies with more than 30 tips). We applied time-dependent (27) and environment-dependent (28, 29) diversification models to these pre- and post-LE trees. We tested specific classical hypotheses about the role of silica weathering, grassland expansion (here reflected by declines in land plant diversity), pCO2 and δ13C trends, and sea level and temperature changes, as well as interactions with other plankton groups through consumption (e.g. by ostracods) or competition (e.g. with radiolarians, coccolithophores, foraminifera, green algae, red algae) in shaping diatom biodiversity (30) (Supplemental Fig. 4).
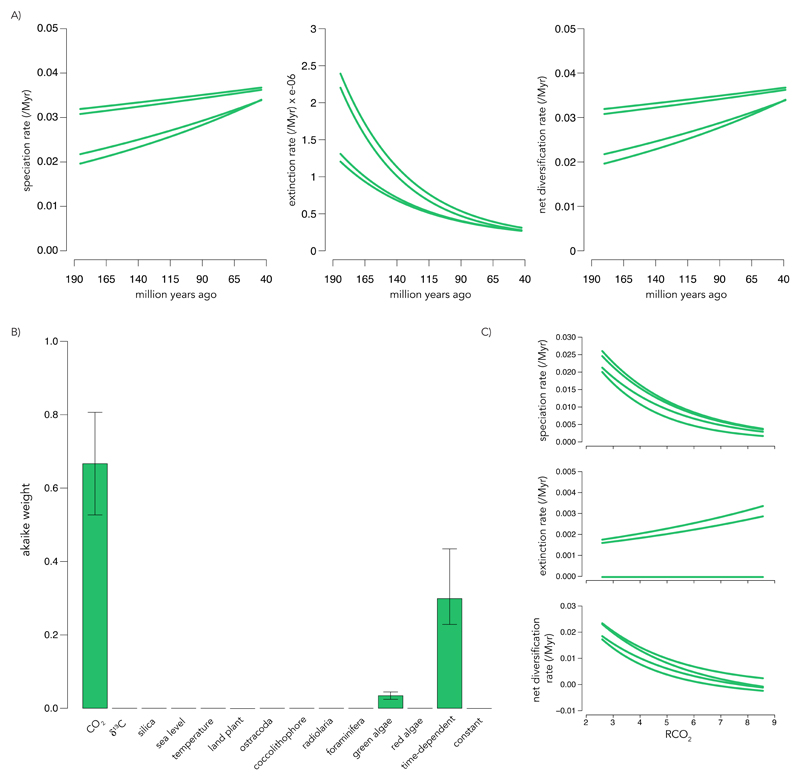
Prior to the Cretaceous, diatom fossils are rare, possibly due to having only lightly silicified frustules (2). Therefore, little is known about their diversification dynamics during the pre-Eocene greenhouse climate. We identify an increase in net diversification rate at this time as a result of exponentially increasing speciation rates and exponentially decreasing extinction rates (Fig. 2A). During this period, pCO2 is the main environmental factor affecting diversification (Fig. 2B). We find a negative effect of pCO2 on speciation rates, and a positive (or no) effect on extinction rates (depending on the build of the phylogeny), which results in an overall negative effect on net diversification rates (Fig. 2C). We find time-dependency is the second best-supported model and little support for any influence of predators or competitors on diatom diversification (Fig. 2B). The result that pCO2 is the primary dependency during this period is robust to uncertainties in our estimates of extant diversity (Supplemental Fig. 5), although support for an effect of green algae increases (above time-dependency) when we consider the upper or lower bounds of estimated extant diversity (Supplemental Fig. 5B,E).
Fig. 2. Diatom diversification dynamics in the Jurassic and Cretaceous.
(A) Time-dependent speciation, extinction, and net diversification rate for the pre-LE diatom OTU phylogeny. Each slope is inferred from one of the four versions of the phylogeny. (B) Median Akaike weights for environment-dependent models fit to the pre-LE phylogenies. Error bars indicate the minimum and maximum support across all versions of the phylogeny. (C) Speciation, extinction, and net diversification rates as a function of RCO2 (ratio of pCO2 relative to the present) for all versions of the phylogeny. Results obtained using median diversity estimates; results with lower and upper bounds of diversity estimates are given in Supplemental Fig. 5.
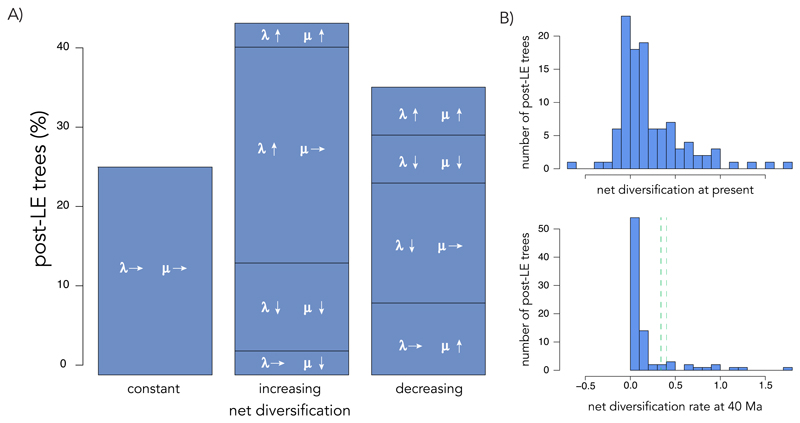
Diversification dynamics from the LE to the present reveal considerable heterogeneity across diatom clades (Fig. 3A, Supplemental Fig. 6A,B). 42% of the clades show an increase in net diversification towards the present, while 34% show a decrease and 24% have constant rates. These dynamics contrast sharply with those observed in other eukaryotes, where the dominating pattern is either declining (31, 32) or constant-rate diversification (33), and suggest that the Cenozoic provided a favorable environment for the diversification of diatoms. Estimates of net diversification rates at 40 Ma show a sharp increase in diatom diversification in the LE in some clades and a drop in others; and estimates at present show that 69% of the clades are expanding while the rest are on a trajectory of diversity decline (i.e., negative net diversification rate at present, Fig. 3B).
Fig. 3. Time-dependent diatom diversification dynamics from the late Eocene.
(A) Median percentage of post-LE trees supported by models of constant, increasing, and decreasing net diversification. Arrows represent the time-variation in speciation (λ) and extinction (µ) rates: up-arrow, increasing; down-arrow, decreasing; right-arrow, constant. (B) Histogram of net diversification rate at present (top) and at 40 Ma (bottom) for all post-LE trees. The green dashed lines show the range of net diversification rates across the four versions of the pre-LE tree at 40 Ma. Results obtained using median diversity estimates; results with lower and upper bound of diversity estimates are given in Supplemental Fig. 6.
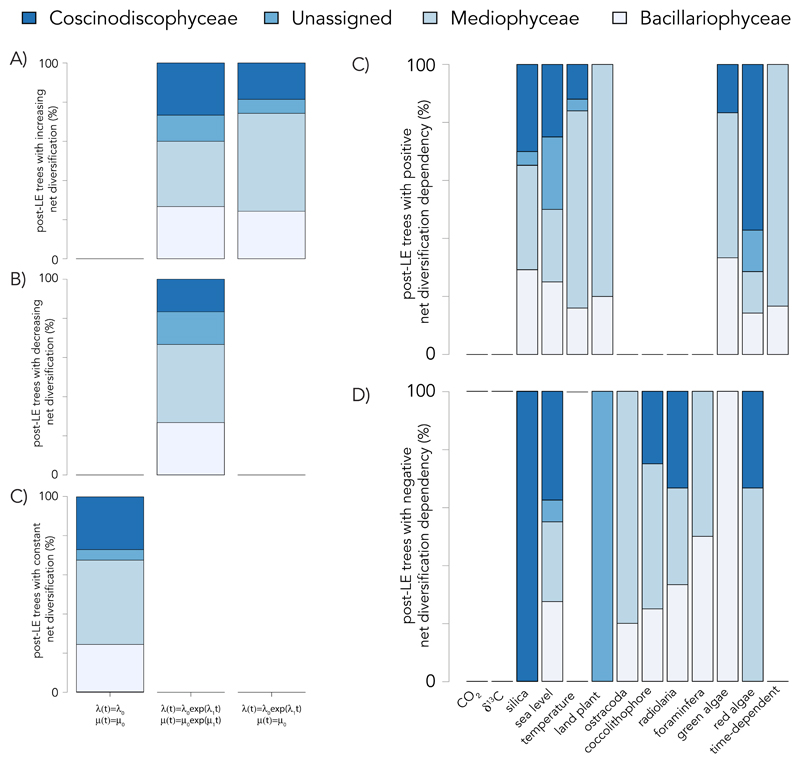
The main drivers of diversification from the LE to the present are very diverse across diatom clades (Fig. 4). There is no single biotic or abiotic driver: diversification patterns in different post-LE trees are dependent on different drivers, and the nature of those dependencies is not uniformly positive or negative across clades (Fig. 4A-C, Supplemental Fig. 6C,D). This suggests that, not only have contemporaneous clades been influenced by various biotic and abiotic factors, but that some have adapted distinct evolutionary strategies in response to the same factor.
Fig. 4. Environment-dependent diatom diversification dynamics from the late Eocene.
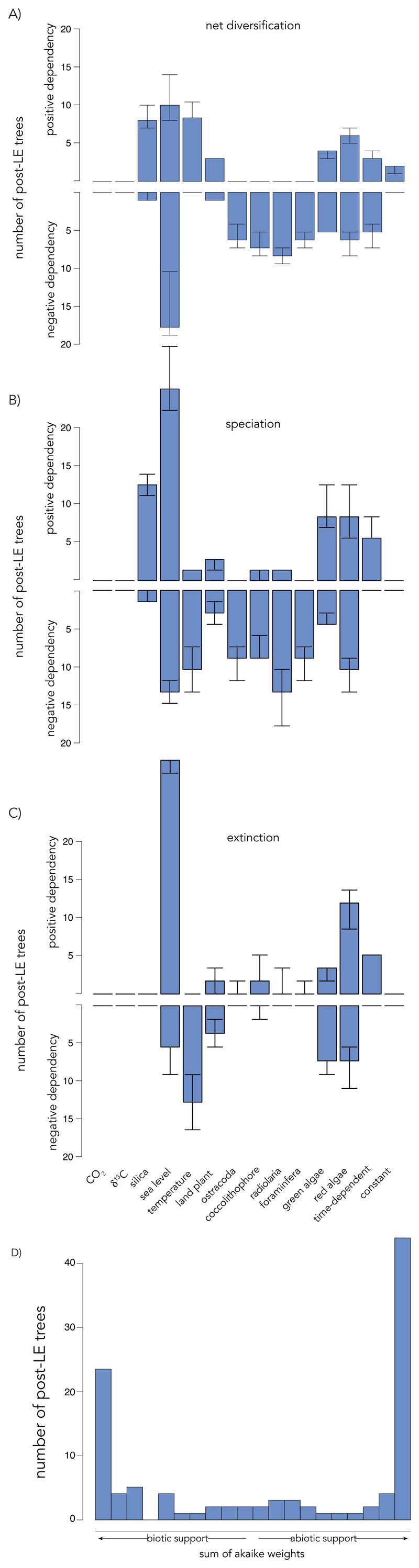
Median number of post-LE trees best fit by each environment-dependent, time-dependent, and constant-rate model showing either a positive or negative dependency of (A) net diversification, (B) speciation, or (C) extinction. Error bars indicate the minimum and maximum number of trees for all versions of the phylogeny. (D) Histogram across all post-LE trees of the cumulative support, as measured by the sum of Akaike weights, for abiotic models. Trees with sums falling on the right side of the histogram show high relative support for abiotic models, whereas trees with sums falling on the left side show high relative support for biotic models.
Diatoms are well known for their obligate requirement for silicic acid and so it has been hypothesized that silica bioavailability has had a major influence on their diversification (9). We tested this in three ways. Silica weathering, the most direct measure of silica bioavailability to diatoms over time, is best supported in 9% of post-LE clades (Fig. 4A). Land plant diversity, an inverse proxy for the expansion of terrestrial grasses which has led to the dissolution of silica-based phytoliths in coastal sediments (2), is best supported in 4% of post-LE clades with both positive and negative dependencies on speciation and extinction. Radiolarian diversity, which fossil analyses have found to have either an antagonistic effect (34) or no effect at all (9) on diatom diversity, is best supported in 8% of post-LE clades, with a negative effect on diversification consistent with expectations of competition for silicic acid availability. Together, these three factors contributing to ocean silica biovailability are best supported in 21% of post-LE diatom clades. These results are consistent with fossil analyses (26) and suggest that silica bioavailability, which is vital to diatom survival and influences diversification in some clades, has not been the sole or even the principal driver of diatom diversification over the last 40 million years.
Sea level change appears as the most important single driver of diatom diversification over the last 40 million years: it is best supported in 27% of all post-LE clades (Fig. 4A-C). The nature of the dependency is not consistent, however, with as many clades negatively rather than positively affected by high seawater levels. This may explain why diatom, compared to dinoflagellate and coccolithophore, fossil diversity has not been found to parallel peaks in sea level (2). Because sea level affects many aspects of the marine biome, it is likely that different diatom clades with distinct ecologies have responded to different aspects of sea level change, resulting in different dependencies. We also find substantial support for diversification dependencies on temperature changes (Fig. 4A-C), which are often used as a general indicator of climate change (35) and more specifically as a proxy for ocean productivity and stratification (36). The positive temperature-dependencies are driven by negative dependencies on speciation, which is consistent with expectations of diatom success in colder climates, but negative dependencies on extinction, as well. Variables associated with the carbon cycle (δ13C and pCO2), which have been suggested to be coupled to diatom diversity during the Cenozoic (11), were not supported in any post-LE clades (although, this rose to a few clades when upper and lower estimates of extant diversity were considered, Supplementary Fig. 6).
While pCO2-dependence seemingly played no primary role in diatom diversification over the last 40 million years (Fig. 4A-C), it played a key role in early diatom diversification (Fig. 2B,C). It is difficult, however, to disentangle the so-called drive-response nature of this negative relationship. The negative relationship between pCO2 and net diversification is consistent with previous conclusions on Cenozoic fossil data, which attributed the drawdown of atmospheric CO2 to the considerable role diatoms play in inorganic carbon fixation (diatoms as the drive) (26). More generally, the early diversification of eukaryotic phytoplankton likely contributed to the depletion of pCO2 beginning in the late Jurassic (1). However, increased net diversification of diatoms as a function of decreasing pCO2 may instead reflect a direct or indirect effect of pCO2 (pCO2 as the drive): higher speciation (and/or lower extinction) can occur as pCO2 levels decrease towards the phytoplankton productivity-diversity optimum (37); they can also occur under an increasingly cool ocean, with amplified latitudinal thermal gradients that result in a turbulent environment for which diatoms are well adapted (14), and with the presence of icy coasts that are also favorable to diatoms (38). The absence of correlation between pCO2 and clade diversification after the LE suggests that pCO2 is the drive (otherwise the success of diatoms after the LE would continue to precipitate pCO2 down), but that other drivers, such as sea level change and interspecific competition, became more prominent as pCO2 levels dropped off.
Interactions with other planktonic groups have been hypothesized to regulate diatom diversity dynamics (14). We show a negative effect of coccolithophores, radiolarians, and foraminifera on diatom diversification over the last 40 million years (Fig. 4A-C), as may be expected from competitive effects. Red and green algae show both positive and negative effects on diversification depending on the diatom clade, which belies the complex co-evolutionary history of these algal groups (39). We also show a negative effect of ostracods, which may reflect benthic-pelagic coupling (40), specifically linking sinking diatoms to benthic ostracods for consumption. Therefore, interactions with other planktonic groups have played significant roles in recent diatom diversification.
The evolutionary contexts in which these factors influence diversification are framed in the Court Jester Hypothesis (41), which attributes rates of diversification to global changes in climatic or geologic events, and the Red Queen Hypothesis (42), which suggests that diversification rates are primarily affected by interspecific interactions. Biotic factors find little support in the pre-LE tree (although, when the upper and lower estimates of extant diversity are used in the analyses, green algal diversity finds more support) (Fig. 4A-C, Supplementary Fig. 5C,D). After the LE, however, there is no clear partiality for diatom clades to be dependent on abiotic or biotic factors, with 50% and 44% of clades best supported by each, respectively (Fig. 4A-C), which is inconsistent with the supposition that abiotic drivers operate at large (i.e., million-year) temporal scales and biotic at small (i.e., thousand-year) ones (41, 43). Nor do we find any difference in the magnitude of the dependencies on abiotic versus biotic factors (T = 0.50, P = 0.617). Furthermore, clades tend to show either a strong cumulative support for abiotic models (47% clades with a cumulative support for abiotic > 0.8) or for biotic models (30% clades with a cumulative support for abiotic < 0.2) compared to a shared support between the two (Fig. 4D). Taken together, these results suggest that, while the Court Jester and Red Queen hypotheses are not mutually exclusive for understanding the evolution of life, we may expect one or the other to predominate at certain periods or in certain clades.
Finally, we find no significant differences in the patterns of speciation, extinction, or net diversification among the major diatom classes (Fig. 5A), Coscinodiscophyceae (polar centrics), Mediophyceae (multipolar centrics), and Bacillariophyceae (pennates), based on comparisons of time-integrated rates on the 0–40 Ma period (F < 5, P > 0.05); nor do we find any significant difference in the distribution of environment-dependencies across classes (D ≤ 0.5, P > 0.05) (Fig. 5B). While this classification scheme (44, 45, 46) is contended (47, 48) and does not distinguish between araphid and raphid pennates, the pervasive patterns of diversification and dependencies across all morphotypes suggest that new ecological opportunities that appeared during the LE, rather than any morphological invention, was of primary importance in allowing diatoms to diversify into new niches and adapt to new environmental pressures.
Fig. 5. Diversification dynamics among diatom classes.
The percentage of post-LE trees with (A) increasing, (B) decreasing, or (C) constant-rate diversification best fit by different time-dependent processes or with (D) positive or (E) negative environment-dependences that could be assigned to Coscinodiscophyceae, Mediophyceae, Bacillariophyceae, or could not be definitively assigned. Median percentages are shown from analyses across all versions of the phylogeny.
Conclusion
We recognize that phylogenetic-based diversification analyses have their limitations, including the difficulty of estimating extinction (49, 29). They are also fundamentally dependent on the robustness and completeness of the phylogenetic data, which remains a major challenge in groups as diverse as diatoms. Our study relies on a single marker; it also relies exclusively on diatom samples from the ocean euphotic zone and is therefore biased against diatom diversity at different ocean depths and in freshwater. In this respect, the comparison with fossil data that is not exclusively planktonic (e.g. ostracods and radiolarians) is not ideal. Additionally, phylogenetic approaches for testing the effect of paleoenvironments on diversification depend on the datation of both the environmental variables and the phylogenies, which both have uncertainties. The phylogenetic time calibration, in particular, relies on fossil date estimates. Despite these uncertainties, ambitious, global-scale metabarcoding surveys, such as those provided by the Tara Oceans project, begin to allow us to apply to the microbial world tools that have been key to our understanding of the evolution of macroorganisms. The consistency of our results across all phylogenetic builds and their general accordance with fossil-based work underpin the utility of using large metabarcoding datasets to infer broad-scale macroevolutionary patterns. Future work will ideally go further in the integration of molecular and fossil data (50).
Our phylogenetic analysis of diatom diversification suggests that events that happened in the LE – much more so than the K-Pg mass extinction, the Eocene-Oligocene transition, the expansion of grasslands, or gross morphological change – have huge implications for the evolutionary diversification of diatoms. During the LE the main drivers of diversification changed, from a dominating effect of pCO2 throughout the Cretaceous to more heterogeneous dependencies in the last 40 million years, including a marked effect of seawater levels, silica bioavailability, and competition with other planktonic groups. Which particular events drove this shift in diatom diversification in the LE is not clear. This period marked the greenhouse-icehouse transition with a complex association of tectonic and climatic effects that our study cannot disentangle, including the opening of Drake Passage and the Tasman Gateway, the onset of Antartic Circumpolar Current, the expansion of the cryosphere, the cooling of the Southern Ocean and more generally of the Earth, and the influx of nutrient-rich Pacific seawater into the South Atlantic (6, 7, 4). The low, less variable pCO2 levels correspondent with the icehouse Earth initiated in the LE transformed the oceans into a cool environment (51) advantageous to diatoms (14). That the shift in diatom diversification occurred ∼ 6 million years prior to the Eocene-Oligocene boundary suggests that the rapid drawdown of pCO2, waning sea level, and grassland expansion of the LE, which together introduced dynamic ice caps near the poles (7, 52), lengthened coastlines (53), and inundated the ocean with silicic acid (54, 55), were sufficient to provide diatoms with new niches to spur speciation and dampen extinction. The opening of Drake Passage, in particular, which brought an influx of nutrient-rich Pacific seawater into the South Atlantic, may have allowed diatoms to diversify into new niches and adapt to new ecological and environmental pressures and instituted this age of high abundance and cosmopolitanism for diatoms. This comports with ecological and fossil data showing a proclivity for diatoms in polar and coastal regions (20) and the evolutionary success of diatoms in a silica-rich environment (9). Insofar as macroevolutionary conclusions can inform short-term predictions for climate change, we expect that ocean acidification, global increases in sea level and temperature, and anticipated mass extinctions of marine life will have a variegated effect on diatom biodiversity and will favor some clades at the expense of others.
Materials and Methods
Constructing the diatom backbone phylogeny
We downloaded all small subunit (SSU) ribosomal RNA sequences taxonomically assigned to diatoms (Bacillariophyta) from the Protist Ribosomal Reference database (PR2, accessed June 2017, (21)). We obtained 3,163 sequences and used them to construct a backbone diatom phylogeny (using Bolidomonas pacifica as the outgroup). We aligned these sequences using two alignment schemes: (i) the L-INS-i algorithm in MAFFT v.7 (56); and (ii) CLUSTALW v.2 (57). In each case, we imposed a stringent gap penalty (= 60) and subsequently trimmed the alignment using trimAl (58). This resulted in a 1408 and 1411 nucleotide-long alignment. Next, we used jModelTest (59, 60) to identify the substitution model, among a set of five, with which to construct the tree from each alignment. Based on corrected Akaike Information Criterion (AICc) scores (61), the best-fit model for both alignments was GTR. As different tree construction methods have unique strengths and weaknesses (62), we used two tree construction methods on each alignment: (i) RaXML v.8 (63) using the BFGS method to optimise GTR rate parameters with the maximum likelihood+bootstrap approach; and (ii) FastTree 2 (64) with the GTR model. We therefore generated a total of four backbone phylogenies: MAFFT+RaXML, MAFFT+FastTree, CLUSTAL+RaXML, CLUSTAL+FastTree (Supplementary Data 3). We recapitulate the sequence of divergence of major diatom morphological clades (radial centrics, polar centrics, and araphid and raphid pennates) on the backbone (Figure 1A) (45).
Retrieving diatom OTUs from Tara samples
We used the global metabarcoding data (EBI accession number PRJEB16766) generated from 1046 biological samples collected from 146 sampling locations across the global ocean euphotic zone during the Tara Oceans expeditions (65, 66, 67). These samples represent a major extension of the samples from (19, 20). Out of these stations, 17% were located within 20 km of the coast, where diatoms dominate phytoplankton communities. We retrieved the sequences using 85% sequence identity to the V9 reference sequences database; this threshold was chosen based on percentage of conserved positions in diatom V9 sequences (20). We obtained 2,220,000 V9-18S ribosomal DNA diatom sequences. Of these, 220,018 represented unique diatom sequences, each of which was given a taxonomic assignment at at least the class level by performing a global similarity with V9 reference sequences. The 220,018 unique diatom sequences were clustered into biologically meaningful operational taxonomic units (OTUs) at a 97% sequence similarity threshold using the default parameters of the Uclust software v1.2.22q (68). This resulted in 19,197 OTUs. We conducted downstream analyses on these 19,197 OTUs, using the longest sequence within each OTU as the representative sequence and the taxonomic assignment as that of the most abundant V9 sequence (Supplementary Data 5).
Constructing the diatom OTU phylogeny
We constructed a phylogeny for the diatom OTUs by combining molecular data from the backbone phylogeny (Supplementary Data 2) and the diatom OTUs (Supplementary Data 5). Again, we used multiple approaches at the step of the construction to account for the possible shortcomings of different approaches, particularly when dealing with many short sequences. These included two alignment schemes: (i) we aligned the diatom OTUs to the MAFFT backbone alignment with MAFFT using the –addfragments option and a stringent gap penalty (= 60); and (ii) we aligned the diatom OTUs using CLUSTALW, setting the CLUSTALW backbone alignment to -PROFILE1 and the diatom OTUs to -PROFILE2, using a stringent gap penalty (= 60). We constructed non-ultrametric phylogenies for each alignment using a GTR model in FastTree 2 (64). For each alignment, we constructed two phylogenies: (i) for the MAFFT alignment, we constructed a phylogeny where the topology was constrained using either the MAFFT+RaXML or MAFFT+FastTree backbone; and (ii) for the CLUSTALW alignment, we constructed a phylogeny where the topology was constrained using either the CLUSTALW+RaXML or CLUSTALW+FastTree backbone. In each case, we constrained the topology using the perl script TreetoConstraints referenced in (64).
Time-calibrating the diatom OTU phylogeny
We dated the PR2+OTU phylogeny with PATHd8 v1.9.8 (69) using 13 calibration points (Supplementary Data 3), including 10 estimates of lineage divergence dates from fossil data, a secondary estimate for the crown age of Chaetoceros (70), and two secondary estimates for diatom crown age. The fossil estimates were placed at the crown and stem of each corresponding taxonomic group to give the maximum range of plausible dates of appearance. The crown of each taxonomic group was defined as the most recent common ancestor node of all sequences assigned to that group. The secondary calibrations for diatom crown age were taken from two phylogenetic studies (1, 71). We generated 26 scenarios based on maximum and minimum ages for fossil calibrations and the two alternative diatom crown ages. We then removed all PR2 lineages from the phylogeny and resolved polytomies (< 1% of all branching events) by randomly assigning an order of descent using the R function multi2di (72) and using an arbitrarily small branch-length of 10e-3. In total, we constructed 104 phylogenies (26 per backbone) and compiled a maximum clade credibility (MCC) phylogeny using TreeAnnotator (73) for each backbone (Supplementary Data 3). We computed the Jensen-Shannon index distances between the spectral density profiles of the 104 reconstructed phylogenies and clustered them using hierarchical and k-medoids clustering (74). The distances among phylogenies reconstructed from the same backbone were considerably lower than the distances between phylogenies reconstructed from different backbones (Supplemental Figure 1). Therefore, analyses were run on the four MCC phylogenies only.
Estimating extant diversity
Fitting diversification models to phylogenetic data requires accounting for the number of missing data (75). In order to estimate the total extant diversity (i.e., total number of extant OTUs) of diatoms, we followed the Bayesian approach of (22). This approach is based on extrapolating sampled taxa abundance distributions. We computed a single sampled taxa abundance distribution by pooling all sequences from each OTU across all individual samples. The approach of (22) is parametric and requires assuming a specific shape for the global scale taxa abundance distribution. We used the log-normal and the Sichel distributions, both of which are routinely used to describe microbial taxa abundance distributions (76, 77, 78, 22). Following (22), we ran three MCMC chains, each of which included 250,000 steps and a burn-in period of 100,000; this has been shown to be sufficient for convergence (22). We used non-informative prior distributions, the parameters of which were found by executing trial MCMC runs until the acceptance ratios reached 0.5 in fewer than 4000 iterations. The diversity estimate was computed as the median value of the last 150,000 steps in the three chains; we also outputted 95% confidence intervals. The total number of OTUs was estimated to be This is higher than that previously reported (20). The difference in diversity estimates may be explained by our use of both a different OTU clustering algorithm and a more complete dataset: our dataset included over twice as many samples, sampling stations, and unique diatom sequences.
The diversification analyses reported in the main text correspond to diversity estimates using the median value for the probability distribution model with the lowest deviance information criterion (79). We also used a range of diversity estimates corresponding to the 95% confidence intervals found with the 2 distributions. The sampling fraction in our analyses was computed as the ratio of all sampled OTUs and the global scale diversity estimate.
Identifying natural shifts in diversification in the diatom phylogeny
We searched the phylogeny for natural shifts in diversification using the bd.shifts.optim function in the R package TreePar (23). We searched the entire timespan of the phylogeny at 2-million-year intervals for the likelihood of shifts in diversification and up to eight mass extinction events, setting the sampling parameter to 0.11 to account for undersampling in the tree (as estimated above). We used a 2-million-year interval because it provided the most resolution while keeping the computation time reasonable.
Sectioning the phylogeny at 40 Ma
In order to analyze the diversification of diatoms before and after the diversification shift at 40 Ma, we sliced the diatom phylogeny into two sections using the treeSlice function from the R package phytools (80) and our own code: from the crown of the phylogeny until the late Eocene (LE); and from the LE until the present. We call the sets of resulting trees the pre-LE tree and post-LE trees, respectively. We obtained 285 post-LE trees. We calculated total extant diversity in each post-LE tree as above. The median sampling fraction across all post-LE trees is similar to that of the full phylogeny (Supplemental Figure 3).
Taxonomic assignment for the post-LE trees
We gave a taxonomic assignment to each OTU as outlined above. The most resolved level for which taxonomy assignment was available for all OTUs was at the class level (Coscinodiscophyceae, Mediophyceae, and Bacillariophyceae). We therefore classified each post-LE tree at the class level. Each post-LE was assigned a class if at least 50% of its tips corresponded to one of the class-level taxonomies; otherwise, the post-LE tree was classified as ”unassigned”. The taxonomy scheme of the PR2 database is different from that of the V9 reference database, as the latter follows the CMB taxonomic classification (44); for this reason, the morphotype designation (radial centric, polar centric, araphid pennate, raphid pennate) was only available for the PR2 sequences. Before removing the PR2 sequences from the time-calibrated phylogeny, we assigned a morphotype to each OTU tip based on the morphotype of its closest PR2 sequence. This morphotyped phylogeny is shown in Fig. 1.
Fitting time-dependent models
We fit time-dependent models of diversification to each post-LE tree with more than 30 tips (this resulted in 128 post-LE trees), using the RPANDA function fit_bd (81) conditioned on stem age. We computed the sampling fraction as the ratio of sampled OTUs in a tree and its total estimated extant diversity. We set speciation to be a constant or exponential function of time; and extinction to be zero, constant, or an exponential function of time (27). We selected the best-fit model as that with the lowest corrected (on number of tips) AIC score.
In order to fit time-dependent models to the pre-LE tree, we modified the fit_bd function in RPANDA to properly compute the likelihood of a tree sliced in the past. To confirm it worked properly, we simulated 1000 birth-death trees with time-dependent speciation (λ (t) = 0.075 *e0.05t) and constant extinction (µ(t) = 0.05) for 50 million years and then sliced them at 15 million years in the past (average initial species richness = 3110). We inferred the speciation and extinction rates of the sliced trees using the new function and tested model selection against a constant-rate birth-death model. We repeated these analyses on the same trees jackknifed at 10%, 40%, and 70% of total tips to confirm that our codes were also accurate in the presence of undersampling (Supplemental Fig. 7). For analysis of the pre-LE tree, we computed the sampling fraction at present as the ratio of all sampled OTUs and the total extant diversity of the diatom phylogeny. We fit time-dependent models as above using the new function and conditioned the fit on crown age. We selected the best-fit model as that with the lowest AICc.
The direction of the time-dependency, as it pertains to net diversification rather than just speciation or extinction, was determined based on whether the net diversification slope (obtained from a linear regression of the estimated net diversification rate through time) for the best-fit parameters trended positive or negative towards the present.
To test for an effect of taxonomy on the different time-dependent diversification patterns, we used a one-way ANOVA to compare the speciation, extinction, and net diversification rates among clades of each taxonomic class. We measured speciation, extinction, and net diversification rates as the time-integrated rates on the 0-40 Myrs period (e.g., for time-integrated speciation) using the best supported model.
Fitting environment-dependent models
We fit environment-dependent models of diversification to all post-LE trees with more than 30 tips using the RPANDA function fit_env (28, 81, 29). We also fit environment-dependent models to the pre-LE tree using a modified version of the fit_env function adjusted in order to accommodate time-sliced trees (see above). We fit speciation and extinction rates as exponential and linear functions of the palaeoenvironmental curves, accounting for missing taxa by applying the relevant sampling fraction as above. For all trees, we included three abiotic variables – pCO2 based on direct proxy reconstructions (82, 83), δ13C (84), temperature (pH-adjusted and computed as deviations from present-day temperature) (82, 83), and sea level (based on backstripping) (53) – and six diversity curves extracted from fossil occurrence data – land plants, red algae (Rhodophyceae), green algae (Chlorophyte and Charophyta), coccolithophore, ostracoda, foraminifera, and radiolaria. Fossil data were compiled from the Neptune Database (85, 10) and diversity curves were estimated at the genus level using shareholder quorum subsampling (86) at two-million-year bins. While the foraminifera data from the Neptune database are planktonic (85, 10), the radiolarian and foraminifera data include both planktonic and benthic taxa. This sampling is not ideal, but in the absence of a purely planktonic fossil record, it is useful for reflecting broad trends in global diversity. For post-LE trees, we additionally included a curve for silica weathering ratio (9), which only includes data as far back as 67 million years ago, and used better resolved curves for δ13C (11), state-dependent pCO2 (87), and temperature (36). Curves were normalized to avoid biases (29) and truncated to the appropriate time-periods. See Supplemental Fig. 4 for plotted curves. For the pre-LE tree, we computed Akaike weights for the model fits; for the post-LE trees, best-fit models were selected by AICc scores, as above.
The cumulative support of abiotic versus biotic models was estimated using Akaike weights. For each post-LE tree, we calculated the Akaike weights for the 6 abiotic (pCO2, δ13C, temperature, silica weathering ratio, sea level, and land plant diversity, which is an inverse proxy for silica transport into the ocean) and the 6 biotic (fossil diversity curves for red algae, green algae, coccolithophore, ostracoda, radiolaria, and foraminifera) variables. The support of each model type (i.e., abiotic or biotic) was calculated for each post-LE tree as the sum of Akaike weights for the models of that type. We used a t-test to estimate significant differences between speciation-rate dependencies in biotic verus abiotic models using both the actual and absolute values of the inferred dependency parameters.
To test for an effect of taxonomic class on the environment-dependent diversification patterns, we used a Kolmogorov-Smirnov test to estimate whether the distribution of environmental dependencies across taxonomic classes was significantly different between any of the classes. We did this separately for positive and negative dependencies.
Supplementary Material
Acknowledgments
We would like to thank Julien Clavel, Guilhem Sommeria Klein, Odile Maliet, Marc Manceau, and Olivier Missa for helpful comments on the manuscript. EL would like to thank Evan Charles for helpful discussion. Funding was provided through a European Research Council Consolidator grant (ERC-CoG-PANDA) attributed to HM. CB acknowledges funding from the ERC Advanced Award Diatomite (294823), the LouisD Foundation, and the French Government ”Investissements d’Avenir” programmes MEMO LIFE (ANR-10-LABX-54), PSL* Research University (ANR-1253 11-IDEX-0001-02), and OCEANOMICS (ANR-11-BTBR-0008). This article is contribution number 80 of the Tara Oceans project.
Footnotes
Competing interests The authors report no competing interests for this work. The views expressed are those of the authors and should not be construed to represent the positions of the US Army or the Department of Defense.
Data Availability All data are included as Supplementary Data or EBI accession numbers are provided.
Contributions E.L., H.M., and C.B. conceived the study. E.L. analysed the data. L.B., S.M., and C.B. contributed data. E.L. and H.M. wrote the manuscript. C.B. contributed substantially to revisions.
References
- [1].Katz ME, Finkel ZV, Grzebyk D, Knoll AH, Falkowski PG. Evolutionary Trajectories and Biogeochemical Impacts of Marine Eukaryotic Phytoplankton. Annual Review of Ecology, Evolution, and Systematics. 2004 Dec;35:523–556. [Google Scholar]
- [2].Kooistra WH, Gersonde R, Medlin LK, Mann DG. {CHAPTER} 11 - the origin and evolution of the diatoms: Their adaptation to a planktonic existence. In: Falkowski PG, Knoll AH, editors. Evolution of Primary Producers in the Sea. Burlington: Academic Press; 2007. pp. 207–249. [Google Scholar]
- [3].Katz ME, Miller KG, Wright JD, Wade BS, Browning JV, Cramer BS, Rosenthal Y. Stepwise transition from the eocene greenhouse to the oligocene icehouse. Nature Geosci. 2008 May;1:329–334. [Google Scholar]
- [4].Baatsen M, von der Heydt A, Kliphuis M, Viebahn J, Dijkstra H. Multiple states in the late eocene ocean circulation. Global and Planetary Change. 2018;163:18–28. [Google Scholar]
- [5].Livermore R, Hillenbrand C-D, Meredith M, Eagles G. Drake Passage and Cenozoic climate: An open and shut case?: DRAKE PASSAGE AND CENOZOIC CLIMATE. Geochemistry, Geophysics, Geosystems. 2007 Jan;8:n/a–n/a. [Google Scholar]
- [6].Bijl PK, Bendle JAP, Bohaty SM, Pross J, Schouten S, Tauxe L, Stickley CE, McKay RM, Röhl U, Olney M, Sluijs A, et al. Eocene cooling linked to early flow across the tasmanian gateway. Proceedings of the National Academy of Sciences. 2013;110(24):9645–9650. doi: 10.1073/pnas.1220872110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Carter A, Riley TR, Hillenbrand C-D, Rittner M. Widespread antarctic glaciation during the late eocene. Earth and Planetary Science Letters. 2017;458:49–57. [Google Scholar]
- [8].Miller KG, Wright J, Fairbanks RG. Unlocking the ice house: Oligocene-miocene oxygen isotopes, eustasy, and margin erosion. Journal of Geophysical Research: Solid Earth. 1991;96(B4):6829–6848. [Google Scholar]
- [9].Cermeño P, Falkowski PG, Romero OE, Schaller MF, Vallina SM. Continental erosion and the Cenozoic rise of marine diatoms. Proceedings of the National Academy of Sciences of the United States of America. 2015 Apr;112:4239–4244. doi: 10.1073/pnas.1412883112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Spencer-Cervato C. The cenozoic deep sea microfossil record: explorations of the dsdp/odp sample set using the neptune database. Palaeontologia electronica. 1999;2(2):270. [Google Scholar]
- [11].Lazarus D, Barron J, Renaudie J, Diver P, Türke A. Cenozoic Planktonic Marine Diatom Diversity and Correlation to Climate Change. PLoS ONE. 2014 Jan;9:e84857. doi: 10.1371/journal.pone.0084857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Barker PF. Scotia sea regional tectonic evolution: implications for mantle flow and palaeocirculation. Earth-Science Reviews. 2001;55(1–2):1–39. [Google Scholar]
- [13].Scher HD, Martin EE. Timing and Climatic Consequences of the Opening of Drake Passage. Science. 2006 Apr;312:428–430. doi: 10.1126/science.1120044. [DOI] [PubMed] [Google Scholar]
- [14].Falkowski PG, Katz ME, Knoll AH, Quigg A, Raven JA, Schofield O, Taylor FJR. The evolution of modern eukaryotic phytoplankton. Science (New York, N.Y.) 2004 Jul;305:354–360. doi: 10.1126/science.1095964. [DOI] [PubMed] [Google Scholar]
- [15].Finkel ZV, Katz ME, Wright JD, Schofield OME, Falkowski PG. Climatically driven macroevolutionary patterns in the size of marine diatoms over the Cenozoic. Proceedings of the National Academy of Sciences of the United States of America. 2005 Jun;102:8927–8932. doi: 10.1073/pnas.0409907102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kooistra WH, Sarno D, Balzano S, Gu H, Andersen RA, Zingone A. Global Diversity and Biogeography of Skeletonema Species (Bacillariophyta) Protist. 2008 Apr;159:177–193. doi: 10.1016/j.protis.2007.09.004. [DOI] [PubMed] [Google Scholar]
- [17].Ezard THG, Aze T, Pearson PN, Purvis A. Interplay between changing climate and species’ ecology drives macroevolutionary dynamics. Science (New York, N.Y.) 2011 Apr;332:349–351. doi: 10.1126/science.1203060. [DOI] [PubMed] [Google Scholar]
- [18].Yasuhara M, Tittensor DP, Hillebrand H, Worm B. Combining marine macroecology and palaeoecology in understanding biodiversity: microfossils as a model. Biological Reviews. 2017;92(1):199–215. doi: 10.1111/brv.12223. [DOI] [PubMed] [Google Scholar]
- [19].de Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R, Lara E, Berney C, Le Bescot N, Probert I, Carmichael M, et al. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015 May;348 doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- [20].Malviya S, Scalco E, Audic S, Vincent F, Veluchamy A, Poulain J, Wincker P, Iudicone D, de Vargas C, Bittner L, Zingone A, et al. Insights into global diatom distribution and diversity in the world’s ocean. Proceedings of the National Academy of Sciences of the United States of America. 2016 Mar;113:E1516–1525. doi: 10.1073/pnas.1509523113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Guillou L, Bachar D, Audic S, Bass D, Berney C, Bittner L, Boutte C, Burgaud G, de Vargas C, Decelle J, Del Campo J, et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Research. 2013 Jan;41:D597–604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Quince C, Curtis TP, Sloan WT. The rational exploration of microbial diversity. The ISME journal. 2008 Oct;2:997–1006. doi: 10.1038/ismej.2008.69. [DOI] [PubMed] [Google Scholar]
- [23].Stadler T. Mammalian phylogeny reveals recent diversification rate shifts. Proceedings of the National Academy of Sciences of the United States of America. 2011 Apr;108:6187–6192. doi: 10.1073/pnas.1016876108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Harwood DM. Geological Society of America Memoirs. Vol. 169. Geological Society of America; 1988. Upper Cretaceous and lower Paleocene diatom and silicoflagellate biostratigraphy of Seymour Island, eastern Antarctic Peninsula; pp. 55–130. [DOI] [Google Scholar]
- [25].Jordan R, Stickley CE. Diatoms as indicators of paleoceanographic events. Cambridge University Press; 2010. Jan, [Google Scholar]
- [26].Rabosky DL, Sorhannus U. Diversity dynamics of marine planktonic diatoms across the Cenozoic. Nature. 2009 Jan;457:183–186. doi: 10.1038/nature07435. [DOI] [PubMed] [Google Scholar]
- [27].Morlon H, Parsons TL, Plotkin JB. Reconciling molecular phylogenies with the fossil record. Proceedings of the National Academy of Sciences of the United States of America. 2011 Sep;108:16327–16332. doi: 10.1073/pnas.1102543108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Condamine FL, Rolland J, Morlon H. Macroevolutionary perspectives to environmental change. Ecology Letters. 2013 May;:72–85. doi: 10.1111/ele.12062. [DOI] [PubMed] [Google Scholar]
- [29].Lewitus E, Morlon H. Detecting environment-dependent diversifcation from phylogenies: a simulation study and some empirical illustrations. Systematic Biology. 2017 doi: 10.1093/sysbio/syx095. [DOI] [PubMed] [Google Scholar]
- [30].Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth’s biogeochemical cycles. Science (New York, N.Y.) 2008 May;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- [31].Phillimore AB, Price TD. Density-dependent cladogenesis in birds. PLoS biology. 2008 Mar;6:e71. doi: 10.1371/journal.pbio.0060071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].McPeek MA. The ecological dynamics of clade diversification and community assembly. The American Naturalist. 2008 Dec;172:E270–284. doi: 10.1086/593137. [DOI] [PubMed] [Google Scholar]
- [33].Hedges SB, Marin J, Suleski M, Paymer M, Kumar S. Tree of life reveals clock-like speciation and diversification. Molecular biology and evolution. 2015 Apr;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lazarus DB, Kotrc B, Wulf G, Schmidt DN. Radiolarians decreased silicification as an evolutionary response to reduced Cenozoic ocean silica availability. Proceedings of the National Academy of Sciences of the United States of America. 2009 Jun;106:9333–9338. doi: 10.1073/pnas.0812979106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Elderfield H, Ferretti P, Greaves M, Crowhurst S, McCave I, Hodell D, Piotrowski A. Evolution of ocean temperature and ice volume through the mid-pleistocene climate transition. Science. 2012;337(6095):704–709. doi: 10.1126/science.1221294. [DOI] [PubMed] [Google Scholar]
- [36].Zachos JC, Dickens GR, Zeebe RE. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature. 2008 Jan;451:279–283. doi: 10.1038/nature06588. [DOI] [PubMed] [Google Scholar]
- [37].Vallina SM, Follows MJ, Dutkiewicz S, Montoya JM, Cermeno P, Loreau M. Global relationship between phytoplankton diversity and productivity in the ocean. Nature Communications. 2014 Jul;5:4299 EP. doi: 10.1038/ncomms5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Armbrust EV. The life of diatoms in the world’s oceans. Nature. 2009;459(7244):185. doi: 10.1038/nature08057. [DOI] [PubMed] [Google Scholar]
- [39].Dorrell RG, Smith AG. Do red and green make brown?: Perspectives on plastid acquisitions within chromalveolates. Eukaryotic Cell. 2011;10(7):856–868. doi: 10.1128/EC.00326-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gooday AJ. Biological responses to seasonally varying fluxes of organic matter to the ocean floor: a review. Journal of Oceanography. 2002;58(2):305–332. [Google Scholar]
- [41].Barnosky AD. Distinguishing the effects of the red queen and court jester on miocene mammal evolution in the northern rocky mountains. Journal of Vertebrate Paleontology. 2001 Mar;21:172–185. [Google Scholar]
- [42].Van Valen L. A new evolutionary law. Evolutionary Theory. 1973;1:1–30. [Google Scholar]
- [43].Benton MJ. The red queen and the court jester: Species diversity and the role of biotic and abiotic factors through time. Science. 2009;323(5915):728–732. doi: 10.1126/science.1157719. [DOI] [PubMed] [Google Scholar]
- [44].Medlin LK, Kaczmarska I. Evolution of the diatoms: V. morphological and cytological support for the major clades and a taxonomic revision. Phycologia. 2004;43(3):245–270. [Google Scholar]
- [45].Sims PA, Mann DG, Medlin LK. Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia. 2006 Jul;45:361–402. [Google Scholar]
- [46].M LK, S Shinya, M DG, K WHCF. Molecular evidence confirms sister relationship of ardissonea, climacosphenia, and toxarium within the bipolar centric diatoms (bacillariophyta, mediophyceae), and cladistic analyses confirm that extremely elongated shape has arisen twice in the diatoms1. Journal of Phycology. 2008;44(5):1340–1348. doi: 10.1111/j.1529-8817.2008.00560.x. [DOI] [PubMed] [Google Scholar]
- [47].Williams DM, Kociolek JP. Pursuit of a natural classification of diatoms: History, monophyly and the rejection of paraphyletic taxa. European Journal of Phycology. 2007;42(3):313–319. [Google Scholar]
- [48].Theriot EC, Cannone JJ, Gutell RR, Alverson AJ. The limits of nuclear encoded ssu rdna for resolving the diatom phylogeny. European journal of phycology. 2009 Aug;44:277–290. doi: 10.1080/09670260902749159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rabosky DL. Challenges in the estimation of extinction from molecular phylogenies: A response to Beaulieu and O’Meara: BRIEF COMMUNICATION. Evolution. 2016 Jan;70:218–228. doi: 10.1111/evo.12820. [DOI] [PubMed] [Google Scholar]
- [50].Hunt G, Slater G. Integrating paleontological and phylogenetic approaches to macroevolution. Annual Review of Ecology, Evolution, and Systematics. 2016;47(1):189–213. [Google Scholar]
- [51].DeConto RM, Pollard D. Rapid cenozoic glaciation of antarctica induced by declining atmospheric co2. Nature. 2003 Jan;421:245 EP. doi: 10.1038/nature01290. [DOI] [PubMed] [Google Scholar]
- [52].Tripati A, Darby D. Evidence for ephemeral middle eocene to early oligocene greenland glacial ice and pan-arctic sea ice. Nature Communications. 2018;9(1):1038. doi: 10.1038/s41467-018-03180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Miller KG. The Phanerozoic Record of Global Sea-Level Change. Science. 2005 Nov;310:1293–1298. doi: 10.1126/science.1116412. [DOI] [PubMed] [Google Scholar]
- [54].Richter FM, Rowley DB, DePaolo DJ. Sr isotope evolution of seawater: the role of tectonics. Earth and Planetary Science Letters. 1992;109(1–2):11–23. [Google Scholar]
- [55].Misra S, Froelich PN. Lithium isotope history of cenozoic seawater: changes in silicate weathering and reverse weathering. Science. 2012;335(6070):818–823. doi: 10.1126/science.1214697. [DOI] [PubMed] [Google Scholar]
- [56].Katoh K, Standley DM. Mafft multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, McWilliam H, Valentin F, Wallace I, Wilm A, Lopez R, Thompson J, et al. Clustal w and clustal x version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- [58].Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009 Aug;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003 Oct;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- [60].Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012 Jul;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Akaike H. A new look at the statistical model identification. IEEE Transactions on Automatic Control. 1974 Dec;19:716–723. [Google Scholar]
- [62].Liu K, Linder CR, Warnow T. RAxML and FastTree: comparing two methods for large-scale maximum likelihood phylogeny estimation. PloS One. 2011;6(11):e27731. doi: 10.1371/journal.pone.0027731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 2014 May;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PloS One. 2010 Mar;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Pesant S, Not F, Picheral M, Kandels-Lewis S, Le Bescot N, Gorsky G, Iudicone D, Karsenti E, Speich S, Troublé R, Dimier C, et al. Open science resources for the discovery and analysis of tara oceans data. Scientific Data. 2015 May;2:150023 EP. doi: 10.1038/sdata.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].De Vargas C, Audic S, C Tara Oceans Consortium, P Tara Oceans Expedition Total V9 rDNA information organized at the OTU level for the Tara Oceans Expedition (2009-2012) 2017 [Google Scholar]
- [67].Bork P, Bowler C, de Vargas C, Gorsky G, Karsenti E, Wincker P. Tara oceans studies plankton at planetary scale. Science. 2015;348(6237):873–873. doi: 10.1126/science.aac5605. [DOI] [PubMed] [Google Scholar]
- [68].Edgar RC. Search and clustering orders of magnitude faster than blast. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- [69].Britton T, Anderson CL, Jacquet D, Lundqvist S, Bremer K. Estimating divergence times in large phylogenetic trees. Systematic Biology. 2007 Oct;56:741–752. doi: 10.1080/10635150701613783. [DOI] [PubMed] [Google Scholar]
- [70].Kooistra WH, Medlin LK. Evolution of the diatoms (Bacillariophyta). IV. A reconstruction of their age from small subunit rRNA coding regions and the fossil record. Molecular Phylogenetics and Evolution. 1996 Dec;6:391–407. doi: 10.1006/mpev.1996.0088. [DOI] [PubMed] [Google Scholar]
- [71].Sorhannus U. A nuclear-encoded small-subunit ribosomal RNA timescale for diatom evolution. Marine Micropaleontology. 2007 Oct;65:1–12. [Google Scholar]
- [72].Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics (Oxford, England) 2004 Jan;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- [73].Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 2012 Aug;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lewitus E, Morlon H. Characterizing and Comparing Phylogenies from their Laplacian Spectrum. Systematic Biology. 2016 May;65:495–507. doi: 10.1093/sysbio/syv116. [DOI] [PubMed] [Google Scholar]
- [75].Morlon H, Schwilk DW, Bryant JA, Marquet PA, Rebelo AG, Tauss C, Bohannan BJM, Green JL. Spatial patterns of phylogenetic diversity. Ecology Letters. 2011 Feb;14:141–149. doi: 10.1111/j.1461-0248.2010.01563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Sichel HS. On a Distribution Representing Sentence-Length in Written Prose. Journal of the Royal Statistical Society. Series A (General) 1974;137(1):25–34. [Google Scholar]
- [77].Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proceedings of the National Academy of Sciences of the United States of America. 2002 Aug;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hong S-H, Bunge J, Jeon S-O, Epstein SS. Predicting microbial species richness. Proceedings of the National Academy of Sciences of the United States of America. 2006 Jan;103:117–122. doi: 10.1073/pnas.0507245102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Spiegelhalter DJ, Best NG, Carlin BP, van der Linde A. Bayesian measures of model complexity and fit. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2002 Oct;64:583–639. [Google Scholar]
- [80].Revell LJ, Graham Reynolds R. A new Bayesian method for fitting evolutionary models to comparative data with intraspecific variation. Evolution; International Journal of Organic Evolution. 2012 Sep;66:2697–2707. doi: 10.1111/j.1558-5646.2012.01645.x. [DOI] [PubMed] [Google Scholar]
- [81].Morlon H, Lewitus E, Condamine FL, Manceau M, Clavel J, Drury J. RPANDA: an R package for macroevolutionary analyses on phylogenetic trees. Methods in Ecology and Evolution. 2016 May;7:589–597. [Google Scholar]
- [82].Royer DL, Berner RA, Montañez IP, Tabor NJ, Beerling DJ. Co˜ 2 as a primary driver of phanerozoic climate. GSA today. 2004;14(3):4–10. [Google Scholar]
- [83].Mayhew PJ, Jenkins GB, Benton TG. A long-term association between global temperature and biodiversity, origination and extinction in the fossil record. Proceedings Biological Sciences / The Royal Society. 2008 Jan;275:47–53. doi: 10.1098/rspb.2007.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Hannisdal B, Peters SE. Phanerozoic Earth system evolution and marine biodiversity. Science (New York, N.Y.) 2011 Nov;334:1121–1124. doi: 10.1126/science.1210695. [DOI] [PubMed] [Google Scholar]
- [85].Lazarus D. Neptune: A marine micropaleontology database. Mathematical Geology. 1994 Oct;26:817–832. [Google Scholar]
- [86].Alroy J. Geographical, environmental and intrinsic biotic controls on Phanerozoic marine diversification: Controls on phanerozoic marine diversification. Palaeontology. 2010 Nov;53:1211–1235. [Google Scholar]
- [87].Hansen J, Sato M, Russell G, Kharecha P. Climate sensitivity, sea level and atmospheric carbon dioxide. Philosophical Transactions. Series A, Mathematical, Physical, and Engineering Sciences. 2013 Oct;371:20120294. doi: 10.1098/rsta.2012.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Nee S, Mooers AO, Harvey PH. Tempo and mode of evolution revealed from molecular phylogenies. Proceedings of the National Academy of Sciences. 1992;89(17):8322–8326. doi: 10.1073/pnas.89.17.8322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].der Voo RV, Spakman W, Bijwaard H. Tethyan subducted slabs under india. Earth and Planetary Science Letters. 1999;171(1):7–20. [Google Scholar]
- [90].Schulte P, Alegret L, Arenillas I, Arz JA, Barton PJ, Bown PR, Bralower TJ, Christeson GL, Claeys P, Cockell CS, Collins GS, et al. The chicxulub asteroid impact and mass extinction at the cretaceous-paleogene boundary. Science. 2010;327(5970):1214–1218. doi: 10.1126/science.1177265. [DOI] [PubMed] [Google Scholar]
- [91].Kennett JP, Stott LD. Abrupt deep-sea warming, palaeoceanographic changes and benthic extinctions at the end of the palaeocene. Nature. 1991;353(6341):225. [Google Scholar]
- [92].Yasuhara M, Hunt G, Okahashi H. Quaternary deep-sea ostracods from the north-western pacific ocean: global biogeography and drake-passage, tethyan, central american and arctic pathways. Journal of Systematic Palaeontology. 2017;0(0):1–20. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.