Abstract
Collective cell chemotaxis, the directed migration of cell groups along gradients of soluble chemical cues, underlies various developmental and pathological processes. Here we use neural crest cells, a migratory embryonic stem cell population whose behavior has been likened to malignant invasion, to study collective chemotaxis in vivo. Studying Xenopus and zebrafish, we show that the neural crest exhibits a tensile actomyosin ring at the edge of the migratory cell group that contracts in a supracellular fashion. This contractility is polarized during collective cell chemotaxis: it is inhibited at the front but persists at the rear of the cell cluster. The differential contractility drives directed collective cell migration ex vivo and in vivo through intercalation of rear cells. Thus, in neural crest cells, collective chemotaxis works by rear wheel drive.
Directed migration orchestrates events in development, homeostasis and disease (1–4). Most long-range directed migration in vivo occurs by chemotaxis (2, 4–9), in which cells follow gradients of soluble chemical cues. This has been best understood in individually migrating cells, whereby several mechanisms have been proposed (10–13), but less studied during collective migration.
In collective migration, leader cells have dynamic actin-based protrusions (Fig. 1A, darker red) (1, 6), form contacts with follower cells and with the extracellular matrix, and are responsive to chemotactic signals (3, 14, 15). Here, we ask whether cells at the group’s rear (Fig. 1A, dotted square) may contribute to collective cell chemotaxis. To investigate the mechanism of collective chemotaxis ex vivo and in vivo, we studied Xenopus and zebrafish cranial neural crest, an embryonic cell population that undergoes collective cell migration (6, 16) in a manner similar to cancer cells (17), unlike neural crest of other species or in the trunk, where less is known about the collectiveness (18). Although contact inhibition of locomotion and cluster confinement (19, 20) are needed for cephalic neural crest directional movement in Xenopus and zebrafish, they are not sufficient, as collective chemotaxis toward SDF1 is essential for long-range directed movement (6).
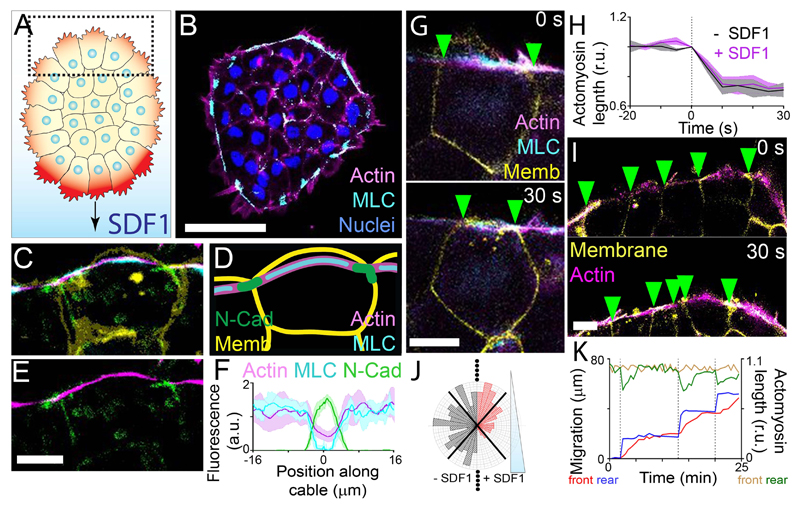
Fig. 1. Xenopus neural crest clusters exhibit a contractile actomyosin ring.
(A) Neural crest with protrusions (red) at the edge undergoes chemotaxis to SDF1. SDF1 stabilizes the protrusions at the front (darker red) (7). Dotted square: rear cells. (B) Immunofluorescence of a neural crest explant in the absence of SDF1. MLC: myosin light chain. Scale bar, 50 μm. (C to E) Immunofluorescence of a cell at the edge of a neural crest explant (C and E) and diagram (D). Memb: membrane. Scale bar, 10 μm. (F) Protein fluorescence levels (means ± SEM) along the actin cable. Position 0 μm represents the cell contact. n = 8 cells. (G) Spontaneous contraction of the actomyosin cable. Green arrowheads: cell-cell contacts. Scale bar, 10 μm. (H) Actomyosin length (means ± SEM) measured over time. Contractions start at 0 s. n = 20 cells. (I) Multicellular contraction of the actomyosin cable. Scale bar, 10 μm. (J) Distribution of actomyosin contractility at different angles without (-SDF1) or with (+SDF1) an SDF1 gradient. n = 150 contractions. (K) Relative actomyosin length at the front (brown line) and rear (green line) of a cluster, and the position of the front (red line) and rear (blue line) of the cluster.
Imaging of fluorescently-tagged actin and myosin in neural crest explants revealed the presence of a multicellular actomyosin ring localized at the periphery of the cell group, in both the absence and presence of an SDF1 gradient (Fig. 1B; fig. S1, A and B). Enrichment of N-Cadherin near the actomyosin cable at the cell junction (Fig. 1, C to F; fig. S1, C to E) suggests this cable is supracellular. Pre-migratory neural crest and neural crest overexpressing E-Cadherin, but not N-Cadherin, have internalized myosin localization, rather than myosin at the cluster periphery (fig. S1, F to J), suggesting that the switch of cadherin expression during EMT may be required for the formation of the actomyosin cable.
To determine whether the actomyosin cable is contractile, we performed laser photoablation of the structure, resulting in recoil of both the actomyosin cable and cell-cell junctions (fig. S2, A and B), followed by the cable’s reformation (fig. S2, C and D). To assess contractility, we measured actomyosin length and we found frequent shortening (Fig. 1, G and H), independent of SDF1. These contractions were multicellular as adjacent cells contracted synchronously (Fig. 1I; fig S2E). A second ablation in a nearby cell after an initial ablation resulted in reduced actomyosin recoil (fig. S2, F and G), indicating that tension of the cable is transmitted between cells. Unlike epithelial cells, where the presence of an actomyosin cable seems to inhibit protrusion formation (21), this does not happen in mesenchymal neural crest cells (fig. S2, H and I).
Whilst exposure to SDF1 gradients did not affect the magnitude of actomyosin contractions (Fig. 1H), contractions occurred less frequently in front cells during collective chemotaxis without affecting cells at the rear (Fig. 1J; fig S3A). A similar inhibition of front contractions was observed with the chemoattractant, PDGF-A (22) (fig. S3B). Mechanistically, this contractility gradient is likely setup by SDF1 activation of Rac1 in front cells, which inhibits RhoA and myosin phosphorylation (fig. S4). Uniform SDF1, unlike the SDF1 gradient, did not inhibit contractility (fig. S5A), suggesting that the cluster responds to the chemotactic gradient instead to absolute SDF1 levels. This was further supported by the observation that rear contractility (fig. S5B) and cluster speed (fig. S5C) were unchanged when clusters were closer to the chemoattractant source, where higher SDF1 levels should be present.
To explore the connection between the asymmetric actomyosin contraction and collective chemotaxis, we simultaneously measured the position of front and rear cells of explants during migration, as well as the length of the actomyosin cable at the front and rear. Pulsatile contraction of the cable at the rear (Fig. 1K, green lines; fig. S6A) coincided with the forward movement of the rear (Fig. 1K, blue lines; fig. S6A). Both events immediately preceded the movement of the front of the cluster (Fig. 1K, red lines; fig. S6A). A similar local contraction precedes a short forward movement in the absence of SDF1 (fig. S6, B and C), but with no long-range directed movement. Together, these results suggest that supracellular actomyosin contractility at the rear may drive collective cell chemotaxis.
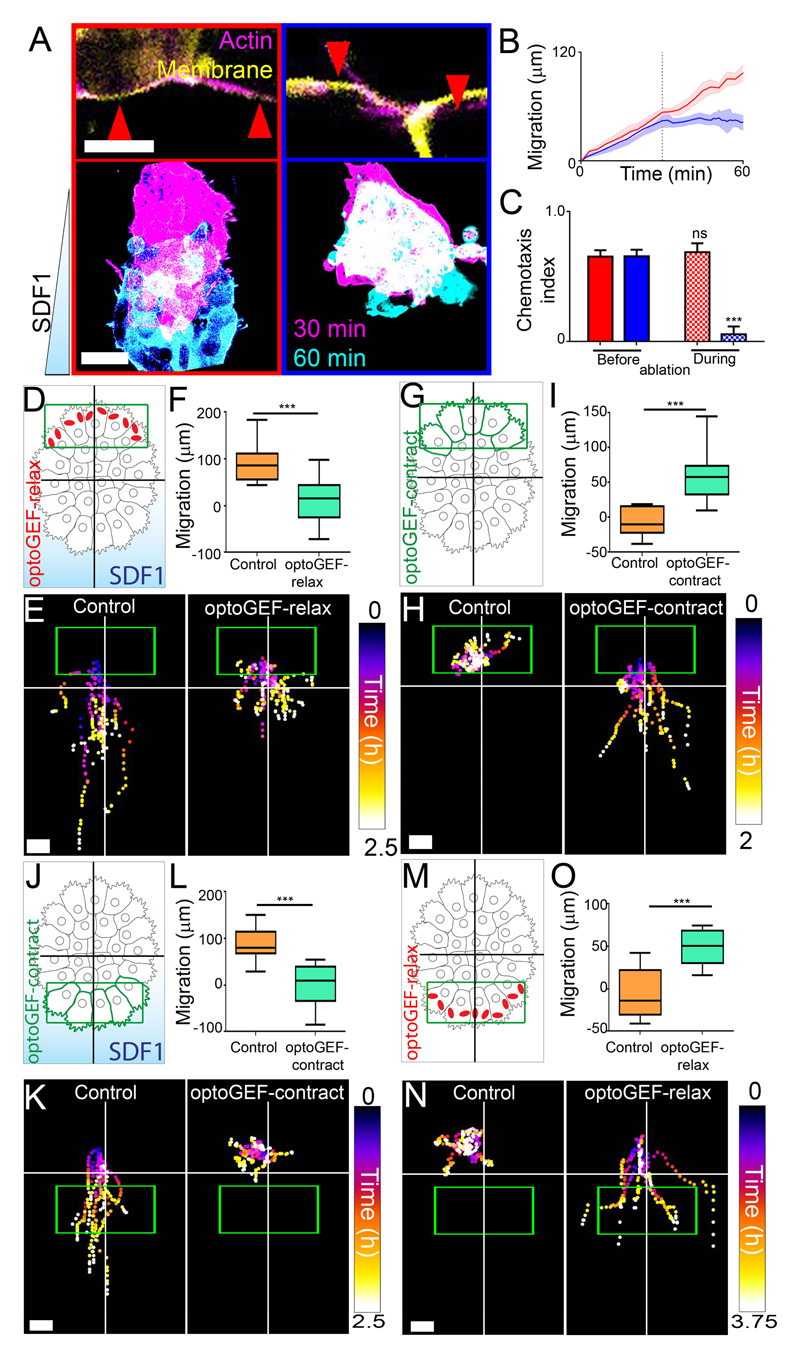
We tested the role of rear contractility of the actomyosin ring on collective chemotaxis by performing laser ablation. Chemotaxis was impaired by ablation of the actomyosin ring in rear cells, but not by equivalent ablations in front cells, or other control ablations (Fig. 2, A to C; fig. S7), suggesting the necessity of a rear supracellular actomyosin cable for chemotaxis. To test the requirement of rear contractility, we used an optogenetic system (23, 24) to either increase (optoGEF-contract) or decrease (optoGEF-relax) contractility and myosin phosphorylation in the actomyosin cable upon illumination with low doses of blue light (fig. S8). No effect was observed on cell protrusions (fig. S9), focal adhesions (fig. S10), cell dispersion (fig. S11) or phosphorylation of myosin located basally outside the cable (fig. S8K) upon illumination in the conditions of our assay. We first tested whether high contractility at the rear is necessary for collective chemotaxis, by photoactivating optoGEF-relax at the rear of migrating clusters exposed to SDF1 (Fig. 2D). Inhibition of contractility in rear cells (Fig. 2D) impaired chemotaxis (Fig. 2, E and F). By contrast, inhibition of contractility in front cells failed to affect collective chemotaxis (fig. S12). To determine whether rear contractility is sufficient to drive collective cell migration, we activated contractility in rear cells in the absence of SDF1 (Fig. 2G). Whilst control neural crest did not exhibit directional migration, activated neural crest moved forward, away from the region of photoactivation (Fig. 2, H and I).
Fig. 2. Rear contractility is necessary and sufficient for collective chemotaxis of Xenopus neural crest.
(A) Above, examples of two neighboring cells with ablations (red arrowheads). Scale bar, 10 μm. Below, images of explants exposed to SDF1 gradients during ablations between the indicated times. For A to C, red: front actomyosin cable ablation; blue: rear actomyosin cable ablation. Scale bar, 50 μm. (B) Position of the front of explants during chemotaxis (means ± SEM); dashed line indicates when ablations begin. n = 6-8 clusters. (C) Chemotaxis index (means ± SEM) of clusters. n = 6-8 clusters. ***P ≤ 0.001 (two-tailed Student’s t-test); ns, not significant. (D to O) Experimental setup for treated explants (D, G, J and M), representative cluster tracks (E, H, K and N) and the distance migrated (means ± SEM) over times as indicated in Methods (F, I, L and O). n = 10-23 clusters (F), n = 10-11 clusters (I), n = 14-18 clusters (L), n = 11-12 clusters (O). ***P ≤ 0.001 (two-tailed Student’s t-test). Scale bar, 40 μm (E and K); 20 μm (H and N). Green box: initial illumination area; cross: initial cluster position. Top of all pictures is the rear.
To test whether SDF1-dependent inhibition of contractility in front cells is required for collective chemotaxis, we activated contractility in front cells of migrating clusters exposed to SDF1 (Fig. 2J); this repressed chemotaxis (Fig. 2, K and L), suggesting that low front contractility is essential for collective chemotaxis. Finally, we asked whether front inhibition of contractility by SDF1 was sufficient to generate directed migration. We inhibited front contractility in the absence of SDF1 (Fig. 2M), which resulted in directional migration (Fig. 2, N and O). These optogenetic treatments affected contractility (fig. S13) (23) and not cell motility (fig. S14). Together, these results suggest that collective migration requires greater contractility at the rear than at the front of the cell cluster.
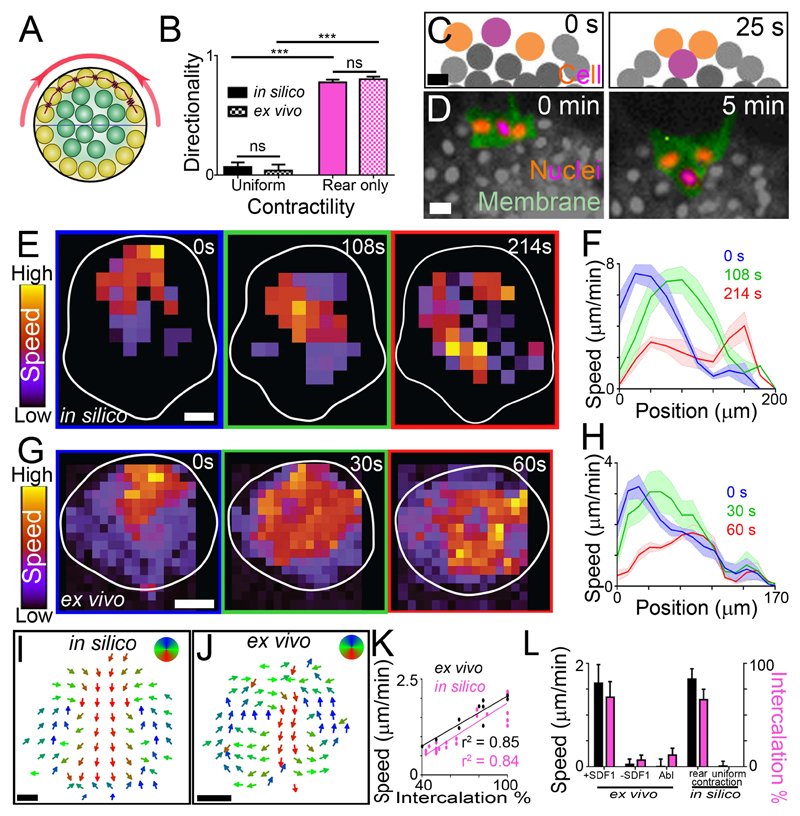
To understand how rear cell contractility might drive directed collective cell migration, we implemented a cell-centered computational model of a cell group with contractile edge cells (Methods; Fig. 3A; fig. S15, A to C). Cells interact through a soft core repulsion and mid-range attraction; to model contractions, cells at the edge (either around the cluster or at the rear) periodically attract one another with additional force (Fig. 3A, red springs). Similar to the ex vivo data, only simulations with rear but not with uniform contractility were able to migrate forward (Fig. 3B; fig. S15D, and Movie S1). Other migration parameters were comparable between in silico and ex vivo clusters (fig. S15, E and F). Unexpectedly, analysis of cell movements in silico revealed that rear cells in contractile regions intercalated forward, into the cell group (Fig. 3C). As predicted by the model, we found an equivalent intercalation at the rear of neural crest clusters (Fig. 3D; fig S15G). Furthermore, our simulations predicted that the effect of this local cell rearrangement is spread through the whole cell group such that when the cluster’s rear contracts, the rear cells intercalate, triggering a wave of cell movement that propagates from the rear towards the front of the cluster (Fig. 3, E and F). A similar wave was observed ex vivo (Fig. 3, G and H), as predicted by the model. This suggests that rear cell intercalation might push cells forward progressively over time, following rear contractions. Averaging cell movement over time and subtracting cluster movement reveals an intra-cluster flow of cells in silico, whereby rear cell intercalation causes a drift forwards through the middle of the group and cells at the front and sides move backwards, replacing rear cells (Fig. 3I). This was then confirmed to occur ex vivo too (Fig. 3J). Consistent with this mechanism driving cluster movement, we found a positive correlation between the speed of ex vivo and in silico clusters during collective migration and the amount of rear cell intercalation (Fig. 3, K and L). Non-migratory ex vivo and in silico clusters had low intercalation; and migratory clusters had comparable cluster speeds (Fig. 3L). We observed that contractions were normally accompanied of relaxation events (fig. S16A, green and red bars); however we showed that ex vivo and in silico clusters were able to migrate directionally independent of the level of rear relaxation (fig. S16, A and B, and movie S2). Altogether, these results suggest that rear contractility drives collective cell migration by inducing cell intercalation which pushes the group forward.
Fig. 3. Modelling contractility-driven collective migration.
(A) Illustration of the computational model cluster. Yellow: edge cells; green: internal cells; red: contraction; horizontal line: distinction between front and rear, with rear outer cells contracting (red spring). (B) Directionality (means ± SEM) of clusters. n = 10 clusters. ***P ≤ 0.001 (two-tailed Student’s t-test); ns, not significant. (C and D) Intercalation of a rear cell (purple) between two adjacent cells (orange) in silico (C) and ex vivo (D) during directional migration. Scale bar, 20 μm. (E to H) Wave of contraction. Speed heat map during migration in silico (E) and ex vivo (G). Speed profile (means ± SEM) from clusters in silico (F) and ex vivo (H) at different times during directional migration. Position 0 μm represents the rear of the cluster; position 200 μm and 170 μm (F and H, respectively) represents the front of the cluster. n = 5 clusters. Scale bar, 40 μm. (I and J) Direction of intra-cluster cell movements shown from time-averaged cell tracks in silico (I) and PIV ex vivo (J) after subtracting cluster movement. n = 5 clusters. Scale bar, 40 μm. (K) Cluster speed and rear cell intercalation during migration. (L) Cluster speed (means ± SEM) and rear cell intercalation (means ± SEM) of clusters. Abl: laser ablation of the actomyosin ring in rear cells. n = 6-21 clusters. ***P ≤ 0.001 (two-tailed Student’s t-test); ns, not significant. Top of all pictures is the rear.
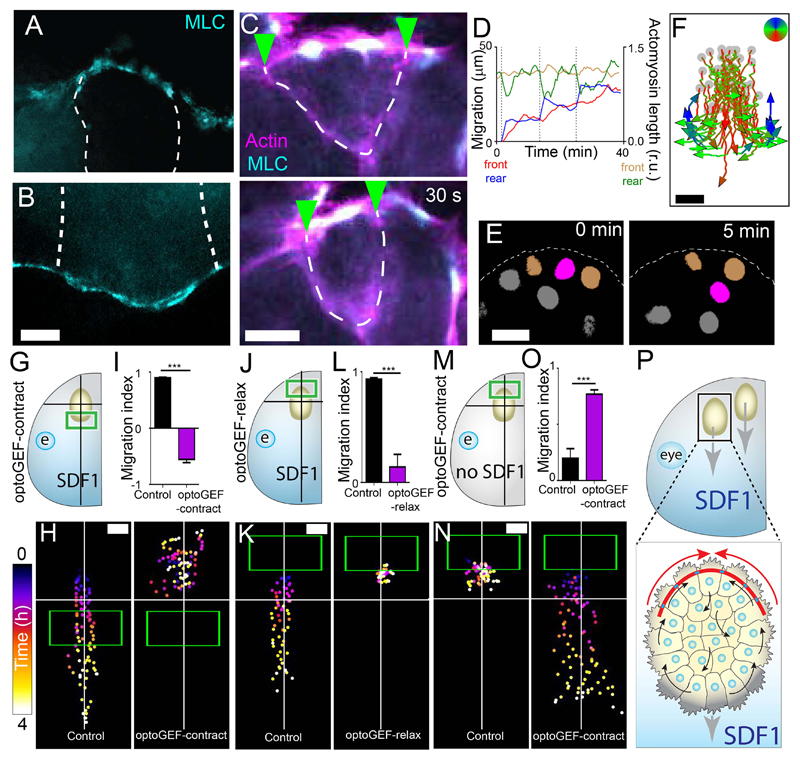
Next, we analyzed whether this model of collective cell chemotaxis explains in vivo migration of neural crest cells. Similar to ex vivo, an actomyosin cable is present at the edge of the neural crest in both Xenopus (Fig. 4, A and B; fig. S17, A and B) and zebrafish (fig. S18, A and B). Live imaging of the actomyosin cable shows that it is a contractile structure in vivo in both Xenopus (Fig. 4C; fig. S17C) and zebrafish (fig. S18, C and D) and contracts more often at the rear of the neural crest stream than at the front (fig. S17D). Like ex vivo, rear contractility precedes forward movement of the cluster in vivo (Fig. 4D). Less phospho-myosin was present at the front than at the rear at the beginning of migration (fig. S17, E to H; fig. S19). To identify whether individual neural crest cells flowed through clusters, as predicted from in silico and ex vivo results, we tracked live cells during migration. In both Xenopus and zebrafish, cells that were initially at the rear of the group indeed intercalated forward during migration (Fig. 4E; fig. S20, A and B). Similar to the ex vivo and in silico data, subtracting cluster movement to in vivo cell tracks reveals an intra-cluster flow (Fig. 4F; fig. S20C). This suggests that rear contractility might be driving neural crest migration in vivo.
Fig. 4. Actomyosin drives collective chemotaxis in vivo in Xenopus.
(A and B) Immunofluorescence of the rear (A) and front (B) of the Xenopus neural crest stream. MLC: myosin light chain; dashed lines: cell-cell contacts between neural crest cells. Scale bar, 10 μm. (C) Contraction of the actomyosin cable of Xenopus neural crest in vivo; green arrowheads: cell-cell contacts; dashed line: cell edges. Scale bar, 10 μm. (D) Actomyosin length at the front (brown line) and rear (green line) of a Xenopus cluster in vivo, and the position of the front (red line) and rear (blue line) of the cluster. (E) Intercalation of a rear cell (purple) between two adjacent cells (orange) in in vivo. Scale bar, 20 μm. (F) Tracks of rear neural crest cells in vivo after subtracting the cluster movement. Scale bar, 30 μm. Grey dots: initial cell positions. (G to N) Experimental design of treated Xenopus embryos (G, J and M), representative tracks of neural crest clusters (H, K and N) and migration index (means ± SEM) (I, L and O). Green box: initial illumination area; cross: starting position of the explant. n = 10 clusters. ***P ≤ 0.001 (two-tailed Student’s t-test). Scale bar, 50 μm. (P) The model: collective cell chemotaxis is driven by actomyosin contractility at the rear (red arrows). Top of all pictures is the rear.
To test whether rear contractility is required for neural crest migration in vivo, we grafted neural crest expressing optoGEF-contract or optoGEF-relax into wild-type Xenopus embryos. Activation of contractility at the front of the stream (Fig. 4, G to I, and movie S3) or inhibition at the rear (Fig. 4, J to L, and movie S4) impaired neural crest migration, indicating that greater contractility at the rear than the front was necessary for migration in the embryo. Neural crest grafted into host embryos lacking SDF1 failed to migrate but activation of contractility at the rear of such grafts rescued migration (Fig. 4, M to O, and movie S5), demonstrating that high actomyosin contractility at the rear can drive directed collective migration in vivo. We conclude that rear contractility, as produced by a supracellular actomyosin cable, can drive collective cell chemotaxis in vivo (Fig. 4P).
Theory of active gels show how anisotropies in viscoelastic materials can generate rotating flows similar to the cellular flows described here (25, 26). In addition, physicists have proposed that cells can move by using tangential retrograde movement of their surfaces (27) and that this movement is more energetically efficient that other modes of swimming (28). However, only recently such surface retrograde propulsion has been described for the migration of single cells (29). Our work identifies an equivalent surface retrograde propulsion for collective cell migration, suggesting that the whole cluster behaves as a “supracell”.
It is likely that for in vivo collective chemotaxis, rear actomyosin contractility works together with protrusions at the front to drive migration. Interestingly, peripheral actomyosin has been similarly observed in the collective migration of other cell types including cancer cells (30, 31), suggesting other cell types may migrate under similar principles.
Supplementary Material
One Sentence Summary.
A rear engine drives collective chemotaxis in mesenchymal cells.
Acknowledgments
We thank G. Charras, L. Cramer, C. Stern, B. Stramer and D. Wilkinson for critical reading of the manuscript and members of the Mayor laboratory for discussions. We thank M. Tada, M. Meyer and E. Sahai for providing us with vectors and antibodies, and E. Scarpa for preliminary data.
Funding: This study was supported by grants from the Medical Research Council (M010465 and J000655 to R.M.), Biotechnology and Biological Sciences Research Council (M008517 to R.M.) and Wellcome Trust (102489/Z/13/Z Wellcome Trust PhD fellowship to A. Shellard) and by a Marie Curie Fellowship (329968 to A. Szabó).
Footnotes
Author contributions: Conceptualization, R.M.; Methodology, A.Sh., A.Sz., X.T., R.M.; Software: A.Sz.; Resources: R.M.; Writing – original draft: A.Sh, R.M.; Writing – review and editing: A.Sh., A.Sz., X.T., R.M.; Supervision – R.M.; Project administration: R.M.; Funding acquisition: R.M.
Competing interests: Authors declare no competing interests.
Data and materials availability: All data is available in the main text or the supplementary materials.
References
- 1.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nature Reviews Molecular Cell Biology. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 2.Roca-Cusachs P, Sunyer R, Trepat X. Mechanical guidance of cell migration: lessons from chemotaxis. Current Opinion in Cell Biology. 2013;25:543–549. doi: 10.1016/j.ceb.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Mayor R, Etienne-Manneville S. The front and rear of collective cell migration. Nature Reviews Molecular Cell Biology. 2016;17:97–109. doi: 10.1038/nrm.2015.14. [DOI] [PubMed] [Google Scholar]
- 4.Haeger A, Wolf K, Zegers MM, Friedl P. Collective cell migration: guidance principles and hierarchies. Trends in Cell Biology. 2015;25:556–566. doi: 10.1016/j.tcb.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Majumdar R, Sixt M, Parent CA. New paradigms in the establishment and maintenance of gradients during directed cell migration. Current Opinion in Cell Biology. 2014;30:33–40. doi: 10.1016/j.ceb.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Theveneau E, et al. Collective Chemotaxis Requires Contact-Dependent Cell Polarity. Developmental Cell. 2010;19:39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilmour D, Rembold M, Leptin M. From morphogen to morphogenesis and back. Nature. 2017;541:311–320. doi: 10.1038/nature21348. [DOI] [PubMed] [Google Scholar]
- 8.Haas P, Gilmour D. Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Developmental Cell. 2006;10:673–680. doi: 10.1016/j.devcel.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 9.Dona E, et al. Directional tissue migration through a self-generated chemokine gradient. Nature. 2013;503:285. doi: 10.1038/nature12635. [DOI] [PubMed] [Google Scholar]
- 10.Welch MD. Cell Migration, Freshly Squeezed. Cell. 2015;160:581–582. doi: 10.1016/j.cell.2015.01.053. [DOI] [PubMed] [Google Scholar]
- 11.Paluch EK, Raz E. The role and regulation of blebs in cell migration. Current Opinion in Cell Biology. 2013;25:582–590. doi: 10.1016/j.ceb.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petrie RJ, Yamada KM. Multiple mechanisms of 3D migration: the origins of plasticity. Current Opinion in Cell Biology. 2016;42:7–12. doi: 10.1016/j.ceb.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Insall RH. Understanding eukaryotic chemotaxis: a pseudopod-centred view. Nature Reviews Molecular Cell Biology. 2010;11:453–458. doi: 10.1038/nrm2905. [DOI] [PubMed] [Google Scholar]
- 14.Cai D, et al. Mechanical Feedback through E-Cadherin Promotes Direction Sensing during Collective Cell Migration. Cell. 2014;157:1146–1159. doi: 10.1016/j.cell.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang XB, He L, Wu YI, Hahn KM, Montell DJ. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nature Cell Biology. 2010;12:591–U154. doi: 10.1038/ncb2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaucka M, et al. Analysis of neural crest-derived clones reveals novel aspects of facial development. Science Advances. 2016;2 doi: 10.1126/sciadv.1600060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerosuo L, Bronner-Fraser M. What is bad in cancer is good in the embryo: Importance of EMT in neural crest development. Seminars in Cell & Developmental Biology. 2012;23:320–332. doi: 10.1016/j.semcdb.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theveneau E, Mayor R. Collective Cell Migration of the Cephalic Neural Crest: The Art of Integrating Information. Genesis. 2011;49:164–176. doi: 10.1002/dvg.20700. [DOI] [PubMed] [Google Scholar]
- 19.Carmona-Fontaine C, et al. Contact inhibition of locomotion in vivo controls neural crest directional migration. Nature. 2008;456:957–961. doi: 10.1038/nature07441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szabo A, et al. In vivo confinement promotes collective migration of neural crest cells. Journal of Cell Biology. 2016;213:543–555. doi: 10.1083/jcb.201602083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reffay M, et al. Interplay of RhoA and mechanical forces in collective cell migration driven by leader cells. Nature Cell Biology. 2014;16:217. doi: 10.1038/ncb2917. [DOI] [PubMed] [Google Scholar]
- 22.Bahm I, et al. PDGF controls contact inhibition of locomotion by regulating N-cadherin during neural crest migration. Development. 2017;144:2456–2468. doi: 10.1242/dev.147926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valon L, Marin-Llaurado A, Wyatt T, Charras G, Trepat X. Optogenetic control of cellular forces and mechanotransduction. Nature Communications. 2017;8 doi: 10.1038/ncomms14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uroz M, et al. Regulation of cell cycle progression by cell–cell and cell–matrix forces. Nature Cell Biology. 2018;20:646–654. doi: 10.1038/s41556-018-0107-2. [DOI] [PubMed] [Google Scholar]
- 25.Kruse K, Joanny JF, Julicher F, Prost J, Sekimoto K. Asters, vortices, and rotating spirals in active gels of polar filaments. Physical Review Letters. 2004;92 doi: 10.1103/PhysRevLett.92.078101. [DOI] [PubMed] [Google Scholar]
- 26.Voituriez R, Joanny JF, Prost J. Generic phase diagram of active polar films. Physical Review Letters. 2006;96 doi: 10.1103/PhysRevLett.96.028102. [DOI] [PubMed] [Google Scholar]
- 27.Purcell EM. LIFE AT LOW REYNOLDS-NUMBER. American Journal of Physics. 1977;45:3–11. [Google Scholar]
- 28.Leshansky AM, Kenneth O, Gat O, Avron JE. A frictionless microswimmer. New Journal of Physics. 2007;9 [Google Scholar]
- 29.O'Neill PR, et al. Membrane Flow Drives an Adhesion-Independent Amoeboid Cell Migration Mode. Developmental Cell. 2018;46:9. doi: 10.1016/j.devcel.2018.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hidalgo-Carcedo C, et al. Collective cell migration requires suppression of actomyosin at cell-cell contacts mediated by DDR1 and the cell polarity regulators Par3 and Par6. Nature Cell Biology. 2011;13:49–U123. doi: 10.1038/ncb2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zajac O, et al. Tumour spheres with inverted polarity drive the formation of peritoneal metastases in patients with hypermethylated colorectal carcinomas. Nature Cell Biology. 2018;20:296. doi: 10.1038/s41556-017-0027-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.