Abstract
As part of large on-going vaccine impact studies in Fiji and Mongolia, we identified 25/2750 (0.9%) of nasopharyngeal swabs by microarray that were positive for Streptococcus pneumoniae contained pneumococci with a divergent 33F capsular polysaccharide locus (designated ‘33F-1’). We investigated the 33F-1 capsular polysaccharide locus to better understand the genetic variation and its potential impact on serotyping results. Whole genome sequencing was conducted on ten 33F-1 pneumococcal isolates. Initially, sequence reads were used for molecular serotyping by PneumoCaT. Phenotypic typing of 33F-1 isolates was then performed using the Quellung reaction and latex agglutination. Genome assemblies were used in phylogenetic analyses of each gene in the capsular locus to investigate genetic divergence. All ten pneumococcal isolates with the 33F-1 cps locus typed as 33F by Quellung and latex agglutination. Unlike the reference 33F capsule locus sequence, DNA microarray and PneumoCaT analyses found that 33F-1 pneumococci lack the wcjE gene, and instead contain wcyO with a frameshift mutation. Phylogenetic analyses found the wzg, wzh, wzd, wze, wchA, wciG and glf genes in the 33F-1 cps locus had higher DNA sequence similarity to homologues from other serotypes than to the 33F reference sequence. We have discovered a novel genetic variant of serotype 33F, which lacks wcjE and contains a wcyO pseudogene. This finding adds to the understanding of molecular epidemiology of pneumococcal serotype diversity, which is poorly understood in low and middle-income countries.
Introduction
Streptococcus pneumoniae (the pneumococcus) is a Gram-positive pathogenic bacterium and a leading cause of community-acquired pneumonia [1]. Pneumococci are classified by serotype, defined by an antigenically-distinct polysaccharide capsule. Capsule biosynthesis is encoded by the capsular polysaccharide (cps) locus within the pneumococcal genome. High levels of genetic diversity within this locus has resulted in over 90 pneumococcal serotypes described to date.
The pneumococcal capsule is the target for currently licensed vaccines, which only include a subset of serotypes. Although pneumococcal conjugate vaccines (PCVs) have been successful in reducing carriage and disease caused by the targeted serotypes, a rise in carriage and disease caused by serotypes not included in these vaccines is commonly observed (serotype replacement) [2,3]. To precisely monitor vaccine impact and disease surveillance, accurate tools for pneumococcal serotyping are required.
Molecular approaches to serotyping pneumococci rely on existing knowledge of cps loci. Data on pneumococcal cps loci from low- and middle-income countries (LMICs) are relatively limited, which can impact serotyping results. For example, we recently described a novel genetic variant of pneumococcal serotype 11A in Fiji. Genetically, the cps locus of these isolates is most closely related to the 11F cps locus, with only a few minor nucleotide changes resulting in the production of 11A capsule [4].
Among the replacing serotypes post-PCV introduction, serotype 33F has become a concern world-wide. Serotype 33F is commonly reported among the predominant serotypes not included in PCVs causing invasive disease following vaccine introduction [5–7]. The increased invasive disease caused by serotype 33F has warranted its inclusion in two new vaccine formulations, which are in development by Merck [8]. In this study, we describe a novel 33F cps locus identified in Fiji and Mongolia by investigating the genetic basis of the variation in this locus and the potential impact this may have on serotyping results.
Materials and methods
Nasopharyngeal swab collection and screening for pneumococci
As part of ongoing programs in the Asia-Pacific region measuring pneumococcal vaccine impact, nasopharyngeal swabs from healthy participants in Fiji, and children diagnosed with pneumonia in Mongolia were collected in accordance with WHO recommendations [9]. Ethical approval for the study in Fiji was granted from the Fiji National Research ethics review committee and The University of Melbourne Human research ethics committee. Ethical approval for the study in Mongolia was granted from the ethics committee associated with The Ministry of Health in Mongolia and the Royal Children’s Hospital in Melbourne. Written consent for study participants was provided by parents/guardians. Following collection, the swabs were placed in 1 ml skim milk, tryptone, glucose, and glycerol media [10] and stored at -80°C. Samples were screened for the presence of pneumococci by conducting quantitative PCR (qPCR) on DNA extracted from 100 μl aliquots of the swabs using the pneumococcal lytA gene as a target as previously described [11].
Molecular serotyping by microarray
Molecular serotyping of pneumococci was performed by DNA microarray. An aliquot of the nasopharyngeal swab was inoculated onto Horse Blood Agar supplemented with gentamicin (5 μg/ml), to select for pneumococci, and incubated overnight at 37°C with 5% CO2. For plates with α-hemolytic growth, the bacterial growth was collected using 1 ml PBS, pelleted by centrifugation and stored at -30°C. DNA was extracted from thawed bacterial pellets using the QIAcube HT with the QIAamp 96 DNA QIAcube HT Kit (Qiagen) with the inclusion of a pre-treatment lysis step whereby 180 μl lysis buffer (20 mM TrisHCl, 2 mM EDTA, 1% Triton X-100, 2 mg/ml RNase A, 20 mg/ml lysozyme) was added to the bacterial pellet and incubated at 37ºC for 60 min. The remaining extraction procedure was as per the manufacturer’s instructions. This DNA was then used for microarray as described previously [12]. In brief, 200 ng of DNA was labelled with Cy3 or Cy5 using the Genomic DNA ULS Labeling Kit (Agilent Technologies) and incubated at 85°C for 30 min. The labelled pneumococcal DNA was incubated with Senti-SPv1.5 microarray slides (BUGS Bioscience) overnight at 65°C rotating at 20 rpm. Microarray slides were washed, scanned, and analyzed using the Agilent microarray scanner and feature extraction software. Serotype calls were analyzed by Senti-NET software (BUGS Bioscience) using Bayesian-based algorithms.
Bacterial isolates
The S. pneumoniae isolates used in this study were purified from ten nasopharyngeal swabs containing 33F-1 from Fiji and Mongolia on selective media as described above. Isolates were confirmed as S. pneumoniae with microarray and whole genome sequencing.
Whole genome sequencing and molecular typing
For whole genome sequencing, DNA was extracted from pure cultures using the Wizard SV genomic DNA purification system (Promega) with some modifications. Briefly, pneumococcal cultures were pre-treated with a lysis solution containing 5 mM EDTA, 3 mg/ml lysozyme and 37.5 μg/ml mutanolysin in TE buffer and incubated at 37°C for 2 h. Proteinase K was added to a final concentration of 1 mg/ml and samples were incubated at 55°C for 1 h. Following incubation, 200 μl of nuclear lysis buffer and 5 μl of RNase (final concentration of 40 μg/ml) were added and samples were incubated at 80°C for 10 min. The remaining extraction procedure was performed as per the manufacturer’s instructions. Eluted DNA was sequenced in 2 x 300 bp paired end reads on the MiSeq platform. Using the Geneious 11.0.4 software package [13], sequence reads were trimmed with BBDuk and de novo assembled using SPAdes. The capsule loci were annotated within Geneious using a database consisting of capsule loci from the 90 serotypes described by Bentley et al. [14]. Sequence reads were also used for molecular typing with PneumoCaT [15].
Sequence analysis
Pairwise alignments were using either MUSCLE or Clustal Omega. Phylogenetic analyses were performed for each 33F-1 cps gene using MEGA 7 [16]. For each gene, the phylogenetic analysis included a representative 33F-1 sequence as well as homologues from all other serotypes containing that gene as described by Bentley et al. [14], where Genbank accession numbers are provided. DNA sequences were aligned using MUSCLE and the alignments were used to generate maximum likelihood trees based on the Tamura-Nei model. Phylogenetic relationships were statistically analyzed by bootstrapping (1000 replicates). The 33F-1 cps loci have been deposited in Genbank (accession no. MH256127, MH256128, MH256129, MH256130, MH256131, MH256132, MH256133, MH256134, MH256135, MH256136).
Quellung and latex agglutination serotyping
Quellung serotyping was performed as described previously [17]. A saline suspension of pneumococci was prepared from an overnight culture. Using an inoculation loop, 1 μl was placed on a microscope slide and mixed with 1 μl of antisera from the Statens Serum Institut (SSI) (http://www.ssi.dk/ssidiagnostica). The sample was then viewed under the microscope (x400 magnification). A positive reaction was defined as an enlargement or ‘swelling’ of cells, with serotype call based on the reaction profile with each typing sera. For latex agglutination, latex reagents were prepared with SSI typing sera [18] and testing performed as previously described [19]. The bacterial suspension and latex reagent (10 μl of each) were mixed on a glass slide. The slide was then incubated on an orbital shaker for 2 min at ~140 rpm. A positive reaction was defined by the presence of visible agglutination. The SSI factor sera used for serotyping of 33F-1 and 33F strains were 33b, 33e, 33f, 6a and 20b.
Results
In our studies evaluating pneumococcal vaccine impact in Fiji and Mongolia, we have used DNA microarray as a molecular approach to serotype pneumococci contained within nasopharyngeal swabs. DNA microarray uses 15,000 oligonucleotides that are spotted onto glass slides and recognize each capsule gene from the 90+ serotypes. Labelled pneumococcal DNA is allowed to hybridize to the oligonucleotides so that pneumococcal serotype can be inferred. From 2750 swabs that contained pneumococci 25 (0.9%) contained pneumococci that typed as ‘33F-like’ (hereby referred to as ‘33F-1’). Ten of these samples were selected and the 33F-1 pneumococci were isolated for further analysis (Table 1).
Table 1. Pneumococcal 33F-1 isolates used in this study.
| MLST | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isolate | Source | Country of isolation | aroE | ddl | gdh | gki | recP | spi | xpt | Sequence type |
| PMP1348 | Nasopharynx of healthy child (2–7 years old) | Fiji | 2 | 18 | 5 | 23 | 18 | 42 | 3 | 13802a |
| PMP1349 | Nasopharynx of healthy child (5–8 weeks old) | Fiji | 2 | 18 | 5 | 23 | 18 | 42 | 3 | 13802a |
| PMP1351 | Nasopharynx of healthy child (12–23 months old) | Fiji | 2 | 18 | 5 | 23 | 18 | 42 | 3 | 13802a |
| PMP1352 | Nasopharynx of healthy child (12–23 months old) | Fiji | 2 | 18 | 5 | 23 | 18 | 42 | 3 | 13802a |
| PMP1353 | Nasopharynx of healthy child (5–8 weeks old) | Fiji | 2 | 18 | 5 | 23 | 18 | 42 | 3 | 13802a |
| PMP1379 | Nasopharynx of healthy child (12–23 months old) | Fiji | 2 | 18 | 5 | 23 | 18 | 42 | 3 | 13802a |
| PMP1380 | Nasopharynx of healthy child (12–23 months old) | Fiji | 2 | 18 | 5 | 23 | 18 | 42 | 3 | 13802a |
| PMP1383 | Nasopharynx of healthy adult | Fiji | 2 | 18 | 5 | 23 | 18 | 42 | 3 | 13802a |
| PMP1386 | Nasopharynx of child with pneumonia | Mongolia | 2 | 18 | 5 | 29 | 16 | 42 | 3 | 673 |
| PMP1387 | Nasopharynx of child with pneumonia | Mongolia | 2 | 18 | 5 | 29 | 16 | 42 | 3 | 673 |
aNovel sequence type identified in this study.
Compared to the expected results for serotype 33F, microarray reported the wciG, glf and wcjE genes in the nasopharyngeal swabs containing these isolates as ‘absent/divergent’. In addition, the wcyO gene was also detected, which has not been reported in the serotype 33F cps locus previously. To investigate the impact of the divergent 33F-1 cps locus on other molecular approaches to serotyping, we sequenced the genomes of all ten isolates and ran the sequence reads through the PneumoCaT pipeline [15]. PneumoCaT uses wcjE to differentiate 33A from 33F, as this gene contains a frameshift mutation in 33F, resulting in a lack of WcjE-mediated O-acetylation of the 33F capsular polysaccharide [20]. Consistent with microarray, PneumoCaT typed all isolates as 33F and was unable to detect wcjE. Phenotypic serotyping methods (Quellung and latex agglutination) also typed these isolates as 33F (S1 Table and S1 Fig).
Following investigation of the 33F-1 cps locus, it was evident that not only did all ten isolates lack wcjE, the locus contained wcyO at this position. The wcyO gene encodes an acetyltransferase and mediates the same modification as wcjE (6-O-acetylation of galactose) [21]. The wcyO open reading frame from all 33F-1 isolates contained a frameshift mutation. The wcyO gene in 33F-1 pneumococci from Fiji had a single T insertion whereas this gene in isolates from Mongolia contained a single A deletion (Fig 1). These frameshift mutations were also confirmed by Sanger sequencing and were not present in traditional wcyO-containing isolates (serotypes 34 and 39) from Fiji (S2 Fig).
Fig 1. Comparison of the wcyO open reading frames of 33F-1 sequences to a representative serotype 34 sequence.
Only a selected portion of the DNA sequence is shown. Numbers refer to the position number in the serotype 34 sequence with an in-frame wcyO gene.
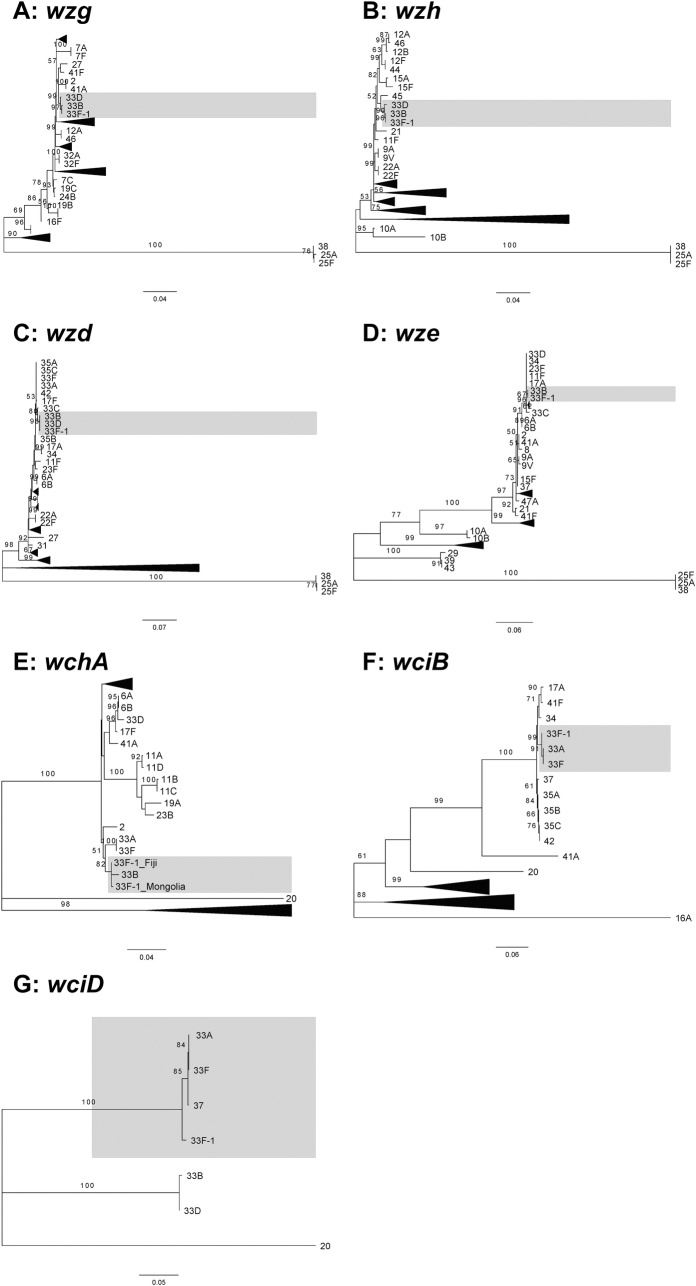
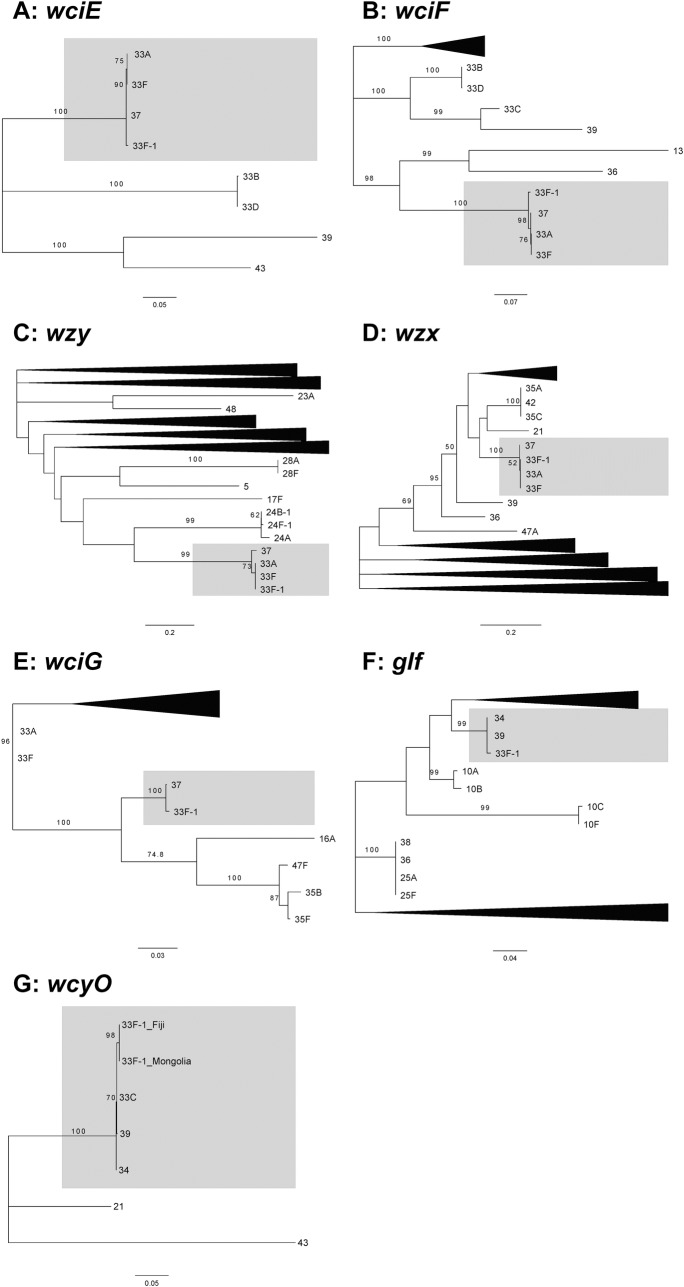
In addition to the differences in wcjE and wcyO, microarray detected some divergence in other genes in the 33F-1 cps locus compared to the reference 33F sequence. To gain a better understanding of the relationships of the 33F-1 cps genes to homologues from other serotypes we performed phylogenetic analyses for each gene. In support of the pairwise alignments (S2 Table), the 33F-1 wciB, wciD, wciE, wciF, wzy and wzx genes clustered with 37/33A/33F sequences (Fig 2F and 2G and Fig 3A–3D). In contrast, 33F-1 wzg, wzh, wzd, wze and wchA clustered with serotype 33B sequences (Fig 2A–2E), wciG with serotype 37 (Fig 3E), glf with serotypes 34 and 39 (Fig 3F) and wcyO with 33C, 34 and 39 (Fig 3G). All branches had strong statistical support (>85% bootstrap score from 1000 replicates for all genes, except wze with a 67% bootstrap score for the 33F-1/33B branch).
Fig 2. Maximum likelihood phylogenetic trees of 33F-1 cps genes (wzg-wciD) with homologues from all other serotypes.
As all genes except wchA were identical in all 33F-1 isolates only one sequence is included as a representative. Un-collapsed trees are provided in S5 Fig. Tree for wciC is not shown as this gene is only present in serotypes 33F, 33A and 37, which all have over 98% DNA sequence identity to the 33F-1 sequence. DNA sequences were aligned using MUSCLE and trees were constructed using the Tamura-Nei model in MEGA 7. Only bootstrap values above 50% are shown.
Fig 3. Maximum likelihood phylogenetic trees of 33F-1 cps genes (wciE-wcyO) with homologues from all other serotypes.
As all genes except wcyO were identical in all 33F-1 isolates only one sequence is included as a representative. Un-collapsed trees are provided in S5 Fig. DNA sequences were aligned using MUSCLE and trees were constructed using the Tamura-Nei model in MEGA 7. Only bootstrap values above 50% are shown.
Discussion
Pneumococcus is a highly successful pathogen, in part due to the high level of capsule diversity, resulting in over 90 serotypes each with unique antigenic properties. Even small differences in the cps locus can have biologically relevant consequences. Serotypes 33F and 33A have the same cps locus, except that 33F has a wcjE gene containing a frameshift mutation rendering it non-functional [22]. Using DNA microarray, we identified a high degree of genetic divergence in the capsule DNA sequence of some serotypes in Fiji and Mongolia. We characterized a serotype 33F variant (33F-1) that has the same genes as the canonical 33F and 33A cps loci, except it possesses wcyO instead of wcjE. Interestingly, in the 33F-1 variants wcyO is predicted to encode a truncated protein due to a frameshift mutation. These frameshift mutations suggest a loss of 6-O-acetylation in 33F-1 capsular polysaccharide as the truncated protein would unlikely be functional. Interestingly, the same variant has been simultaneously identified in the Global Pneumococcal Sequencing Project in other countries (van Tonder et al, unpublished), demonstrating 33F-1 pneumococci are not restricted to Fiji and Mongolia.
Although Quellung and latex agglutination serotyping supports the notion that the 33F-1 cps locus encodes a 33F capsule, it is important to note that sera used in these methods are polyclonal and there is potential for closely related serotypes to cross-react. In addition, the sera may not recognize all relevant epitopes in the capsule. Although there is no genetic difference that would result in an obvious antigenic change and no serological differences were detected in our study, we cannot eliminate the possibility that differences between 33F and 33F-1 capsules exist. Pairwise alignment of the amino acid sequences of 33F and 33F-1 capsular biosynthesis proteins showed high levels of similarity (over 96% identity for all proteins) (S3 Fig). The exceptions were the divergent WciG (85.2% identity) and Glf (94.6% identity). Despite reduced amino acid similarity of the 33F-1 WciG and Glf proteins to 33F homologues, we hypothesise that they would likely mediate the same modifications. This is supported in other serotypes where these enzymes have similar levels of variation but still perform the same function [14,20].
The frameshift mutations in isolates from Fiji and Mongolia have both occurred within homopolymeric regions (Fig 1 and S2 Fig). Such regions are prone to slipped-strand mispairing, whereby errors made during DNA replication can result in the insertion or deletion of a nucleotide [23]. We postulate that the frameshift mutations in the 33F-1 wcyO genes are the result of slipped-strand mispairing events.
This is the first report identifying the wcyO acetyltransferase gene in the 33F cps locus, and it is also the first report of a naturally occurring frameshifted allele of wcyO. The fact that the mutation type and location differ between isolates from Fiji and Mongolia demonstrates this mutation event has occurred on at least two independent occasions. Whether the mutation of wcyO is due to selective pressure to inactivate a disadvantageous gene or due to a lack of selective advantage to maintain it remains to be investigated. Previously, mutations have been identified in other pneumococcal capsule acetyltransferase genes including wciG [24] and wcjE [22,25,26]. Serotype 11E, which lacks WcjE-mediated acetylation can evade opsonophagocytosis more efficiently compared to 11A (which possess WcjE-mediated acetylation) [25]. Pneumococci expressing 33F capsules, which lack WcjE-mediated acetylation, exhibit enhanced survival during drying compared to serotype 33A (with intact WcjE-mediated acetylation) [27]. Laboratory constructed wciG mutants in serogroup 33 isolates were more susceptible to opsonophagocytosis, and displayed increased adherence and biofilm formation [27]. It is plausible that mutation of wcyO in the 33F-1 pneumococci may serve a similar purpose, however this requires further investigation.
Within the 33F-1 cps locus we identified 7/15 genes that exhibit higher DNA sequence similarity to homologues from other serotypes rather than 33F. Both glf and wcyO are similar to sequences from serotypes 34 and 39 (and 33C for wcyO) (Fig 3F and 3G) and wzg, wzh, wzd, wze and wchA similar to sequences from 33B (Fig 2A–2E). Recombination of the pneumococcal capsule genes resulting in mosaic cps loci such as that of 33F-1 have been reported previously [28,29]. Alignment of the 33F and 33F-1 cps loci support this by showing higher sequence divergence across the wzg through to the 5’ half of wchA, as well as in the second half of wciG and the 5’and 3’ ends of glf, suggesting these may be the recombination sites (S4 Fig). Although it is difficult to infer the direction of horizontal transfer of these genes, the mosaic nature of the 33F-1 cps locus would suggest an ancestral 33A/F cps locus was the recipient of these genes. The 33F-1 isolates in our study were either MLST ST673 or ST13802 (Table 1). Interrogation of the PubMLST database (as of 16th September 2018) shows that ST673 is primarily associated with serotypes 33A/F and not in any wcyO-associated serotypes. Likewise, although ST13802 is a novel sequence type described in this study, the most similar MLST profiles (five or more allele matches to ST13802) were either 33A/F serotypes or serotypes that do not possess wcyO. These data support the notion that the ancestral strain(s) from which the 33F-1 cps locus arose likely possessed a 33A/F cps locus. However, it is important to note that the reverse scenario (a wcyO-associated serotype acquiring 33A/F cps genes) is possible and further genetic analyses are needed.
Interestingly, a serogroup 33 related cps locus has been identified in Streptococcus oralis subsp. tigurinus strain Az_3a [30]. This cps locus possessed the same genes as the 33F-1 locus with variable DNA identity (<77% with the 33F-1 wzg, wzh, wzd, wze, wchA and wciB genes, >96% for wciC, wciD, wciE, wciF and wzy genes, and 85–90% for wzx, wciG and glf genes, S2 Table). The higher DNA identity of 33F-1 cps genes with homologues from other pneumococcal serotypes suggests the Az_3a cps locus may have evolved independently of the 33F-1 locus. In contrast to 33F-1, the wcyO gene in Az_3a is in frame and most similar to the pneumococcal serotype 21 homologue (DNA identity 86.8% with serotype 21 wcyO compared to 74.5% with 33F-1 wcyO). The existence of a divergent 33F-1 cps locus with a functional wcyO raises interesting questions around why this gene has been inactivated in 33F-1 pneumococci but remains intact in a non-pneumococcal streptococcal species.
This study describes a novel genetic basis for pneumococcal serotype 33F. Serotype 33F is a replacing serotype in invasive disease following vaccine introduction [5–7]. The public health importance of 33F is reflected in that it has been included in two upcoming vaccine formulations (PCV15 and PCV24) [8]. In addition, there is increasing popularity in molecular serotyping approaches and it is therefore important to identify genetic variants, which have the potential to impact serotyping results. This is particularly important for the implementation of such methods in LMICs, where there is limited understanding of the pneumococcal cps loci. The data gained from this study will be used to update genetic typing tools for more accurate typing of serotype 33F in LMICs.
Supporting information
(DOCX)
Pairwise alignments of each 33F-1 gene with homologues from other serotypes were performed using MUSCLE.
(XLSX)
Latex reagents were prepared using SSI antisera (33b, 33e, 33f, 6a and 20b) as described in the materials and methods. A positive reaction was defined by the presence of clumping and a reduction in background turbidity.
(DOCX)
These sequences were aligned to a representative sequence from serotype 34 strain 676/74 (Genbank accession no. CR931703), serotype 39 strain 203/40 (Genbank accession no. CR931711) and representative wcyO sequences from serotype 34 and 39 isolates from Fiji using Clustal Omega. Red sequence indicates the 3’end of glf, blue indicates wcyO, and the highlighted regions denote the frameshift mutation site in the 33F-1 sequences. Identical nucleotides are denoted by an asterisk.
(DOCX)
Identical, conserved and semi-conserved residues are denoted by the symbols ‘*’, ‘:’ and ‘.’, respectively.
(DOCX)
Identical nucleotides are noted by ‘|’ and differences are highlighted by ‘:’.
(DOCX)
DNA sequences were aligned using MUSCLE and trees were constructed using the Tamura-Nei model in MEGA 7. Only bootstrap values above 50% are shown.
(DOCX)
Acknowledgments
We thank the participants, their families and villages; Fiji Ministry of Health and Medical Services and the Ministry of Health in Mongolia. We also thank all study staff involved in recruitment, swab collection and laboratory analyses, including Suuri Bujinlham, Tupou Ratu, Silivia Mantanitobua, Evelyn Tuivaga and Mere Guanivalu.
Data Availability
All data are publically available with the exception of the microarray data. The DNA microarray data have not been made available for ethical reasons because they contain personal information on the study participants. Sequences have been deposited in Genbank (accession no. MH256127, MH256128, MH256129, MH256130, MH256131, MH256132, MH256133, MH256134, MH256135, MH256136).
Funding Statement
This study was supported by the Bill & Melinda Gates Foundation (OPP1126272 and OPP1084341); Gavi, the Vaccine Alliance; and the Department of Foreign Affairs and Trade of the Australian Government and Fiji Health Sector Support Program (FHSSP). CS holds a NHMRC Career Development Fellowship and a veski Inspiring Women Fellowship. SM received a Robert Austrian Research Award in Pneumococcal Vaccinology funded by Pfizer. This work was also supported by the Victorian Government's Operational Infrastructure Support Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 2009;374:893–902. 10.1016/S0140-6736(09)61204-6 [DOI] [PubMed] [Google Scholar]
- 2.Mulholland K, Satzke C. Serotype replacement after pneumococcal vaccination. Lancet (London, England) 2012;379:1387; author reply 1388–9. 10.1016/S0140-6736(12)60588-1 [DOI] [PubMed] [Google Scholar]
- 3.Weinberger DM, Malley R, Lipsitch M. Serotype replacement in disease after pneumococcal vaccination. Lancet 2011;378:1962–73. 10.1016/S0140-6736(10)62225-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manna S, Ortika BD, Dunne EM, Holt KE, Kama M, Russell FM, et al. A novel genetic variant of Streptococcus pneumoniae serotype 11A discovered in Fiji. Clin Microbiol Infect 2018;24:428.e1–428.e7. 10.1016/j.cmi.2017.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balsells E, Guillot L, Nair H, Kyaw MH. Serotype distribution of Streptococcus pneumoniae causing invasive disease in children in the post-PCV era: A systematic review and meta-analysis. PLoS One 2017;12:e0177113 10.1371/journal.pone.0177113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, Bennett NM, et al. Sustained Reductions in Invasive Pneumococcal Disease in the Era of Conjugate Vaccine. J Infect Dis 2010;201:32–41. 10.1086/648593 [DOI] [PubMed] [Google Scholar]
- 7.Hicks LA, Harrison LH, Flannery B, Hadler JL, Schaffner W, Craig AS, et al. Incidence of Pneumococcal Disease Due to Non–Pneumococcal Conjugate Vaccine (PCV7) Serotypes in the United States during the Era of Widespread PCV7 Vaccination, 1998–2004. J Infect Dis 2007;196:1346–54. 10.1086/521626 [DOI] [PubMed] [Google Scholar]
- 8.McFetridge R, Meulen AS, Folkerth SD, Hoekstra JA, Dallas M, Hoover PA, et al. Safety, tolerability, and immunogenicity of 15-valent pneumococcal conjugate vaccine in healthy adults. Vaccine 2015;33:2793–9. 10.1016/j.vaccine.2015.04.025 [DOI] [PubMed] [Google Scholar]
- 9.Satzke C, Turner P, Virolainen-Julkunen A, Adrian P V., Antonio M, Hare KM, et al. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: Updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine 2013;32:165–79. 10.1016/j.vaccine.2013.08.062 [DOI] [PubMed] [Google Scholar]
- 10.O’Brien KL, Bronsdon MA, Dagan R, Yagupsky P, Janco J, Elliott J, et al. Evaluation of a medium (STGG) for transport and optimal recovery of Streptococcus pneumoniae from nasopharyngeal secretions collected during field studies. J Clin Microbiol 2001;39:1021–4. 10.1128/JCM.39.3.1021-1024.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvalho M da GS, Tondella ML, McCaustland K, Weidlich L, McGee L, Mayer LW, et al. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 2007;45:2460–6. 10.1128/JCM.02498-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Satzke C, Dunne EM, Porter BD, Klugman KP, Mulholland EK, Vidal JE, et al. The PneuCarriage Project: A Multi-Centre Comparative Study to Identify the Best Serotyping Methods for Examining Pneumococcal Carriage in Vaccine Evaluation Studies. PLoS Med 2015;12 10.1371/journal.pmed.1001903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012;28:1647–9. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2006;2:0262–9. 10.1371/journal.pgen.0020031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapatai G, Sheppard CL, Al-Shahib A, Litt DJ, Underwood AP, Harrison TG, et al. Whole genome sequencing of Streptococcus pneumoniae: development, evaluation and verification of targets for serogroup and serotype prediction using an automated pipeline. PeerJ 2016;4:e2477 10.7717/peerj.2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 2016;33:1870–4. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habib M, Porter BD, Satzke C. Capsular Serotyping of Streptococcus pneumoniae Using the Quellung Reaction. J Vis Exp 2014. 10.3791/51208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ortika BD, Habib M, Dunne EM, Porter BD, Satzke C. Production of latex agglutination reagents for pneumococcal serotyping. BMC Res Notes 2013;6:49 10.1186/1756-0500-6-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porter BD, Ortika BD, Satzke C. Capsular Serotyping of Streptococcus pneumoniae by Latex Agglutination. J Vis Exp 2014:51747 10.3791/51747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geno KA, Gilbert GL, Song JY, Skovsted IC, Klugman KP, Jones C, et al. Pneumococcal capsules and their types: Past, present, and future. Clin Microbiol Rev 2015;28:871–99. 10.1128/CMR.00024-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bush CA, Yang J, Yu B, Cisar JO. Chemical Structures of Streptococcus pneumoniae Capsular Polysaccharide Type 39 (CPS39), CPS47F, and CPS34 Characterized by Nuclear Magnetic Resonance Spectroscopy and Their Relation to CPS10A n.d. 10.1128/JB.01731-14 [DOI] [PMC free article] [PubMed]
- 22.Mavroidi A, Aanensen DM, Godoy D, Skovsted IC, Kaltoft MS, Reeves PR, et al. Genetic relatedness of the Streptococcus pneumoniae capsular biosynthetic loci. J Bacteriol 2007;189:7841–55. 10.1128/JB.00836-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol 1987;4:203–21. 10.1093/oxfordjournals.molbev.a040442 [DOI] [PubMed] [Google Scholar]
- 24.Geno KA, Bush CA, Wang M, Jin C, Nahm MH, Yang J. WciG O-Acetyltransferase Functionality Differentiates Pneumococcal Serotypes 35C and 42. J Clin Microbiol 2017;55:2775–84. 10.1128/JCM.00822-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brady AM, Calix JJ, Yu J, Geno KA, Cutter GR, Nahm MH. Low invasiveness of pneumococcal serotype 11A is linked to ficolin-2 recognition of O-acetylated capsule epitopes and lectin complement pathway activation. J Infect Dis 2014;210:1155–65. 10.1093/infdis/jiu195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calix JJ, Brady AM, Du VY, Saad JS, Nahm MH. Spectrum of pneumococcal serotype 11A variants results from incomplete loss of capsule O-acetylation. J Clin Microbiol 2014;52:758–65. 10.1128/JCM.02695-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spencer BL, Saad JS, Shenoy AT, Orihuela CJ, Nahm MH. Position of O-acetylation within the capsular repeat unit impacts the biological properties of pneumococcal serotypes 33A and 33F. Infect Immun 2017;85:e00132–17. 10.1128/IAI.00132-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salter SJ, Hinds J, Gould KA, Lambertsen L, Hanage W, Antonio M, et al. Variation at the capsule locus, cps, of mistyped and non-typable Streptococcus pneumoniae isolates. Microbiol (United Kingdom) 2012;158:1560–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Tonder AJ, Bray JE, Quirk SJ, Haraldsson G, Jolley KA, Maiden MCJ, et al. Putatively novel serotypes and the potential for reduced vaccine effectiveness: capsular locus diversity revealed among 5405 pneumococcal genomes. Microb Genomics 2016;2:000090 10.1099/mgen.0.000090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skov Sørensen UB, Yao K, Yang Y, Tettelin H, Kilian M. Capsular Polysaccharide Expression in Commensal Streptococcus Species: Genetic and Antigenic Similarities to Streptococcus pneumoniae. MBio 2016;7:e01844–16. 10.1128/mBio.01844-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Pairwise alignments of each 33F-1 gene with homologues from other serotypes were performed using MUSCLE.
(XLSX)
Latex reagents were prepared using SSI antisera (33b, 33e, 33f, 6a and 20b) as described in the materials and methods. A positive reaction was defined by the presence of clumping and a reduction in background turbidity.
(DOCX)
These sequences were aligned to a representative sequence from serotype 34 strain 676/74 (Genbank accession no. CR931703), serotype 39 strain 203/40 (Genbank accession no. CR931711) and representative wcyO sequences from serotype 34 and 39 isolates from Fiji using Clustal Omega. Red sequence indicates the 3’end of glf, blue indicates wcyO, and the highlighted regions denote the frameshift mutation site in the 33F-1 sequences. Identical nucleotides are denoted by an asterisk.
(DOCX)
Identical, conserved and semi-conserved residues are denoted by the symbols ‘*’, ‘:’ and ‘.’, respectively.
(DOCX)
Identical nucleotides are noted by ‘|’ and differences are highlighted by ‘:’.
(DOCX)
DNA sequences were aligned using MUSCLE and trees were constructed using the Tamura-Nei model in MEGA 7. Only bootstrap values above 50% are shown.
(DOCX)
Data Availability Statement
All data are publically available with the exception of the microarray data. The DNA microarray data have not been made available for ethical reasons because they contain personal information on the study participants. Sequences have been deposited in Genbank (accession no. MH256127, MH256128, MH256129, MH256130, MH256131, MH256132, MH256133, MH256134, MH256135, MH256136).