Abstract
Dendritic cell (DC)-based cytotoxic T lymphocyte (CTL) epitope vaccines are effective to induce CTL responses but require complex ex vivo DC preparation and epitope-loading. To take advantage of DC-based epitope vaccines without involving the ex vivo procedures, we aimed to develop carriers to directly load CTL epitopes onto DCs in vivo. Here, we first engineered a carrier consisting of a hydrophilic polypeptide, immune-tolerant elastin-like polypeptide (iTEP) and a substrate peptide of matrix metalloproteinases-9 (sMMP). The iTEP was able to solubilize CTL epitopes. CTL epitopes were connected to the carrier, iTEP-sMMP, through sMMP so that the epitopes can be cleaved from the carrier by MMP-9. iTEP-sMMP was found to release its epitope payloads in the DC culture media, which contained MMP-9 released from DCs. iTEP-sMMP allowed for the direct loading of CTL epitopes onto the surface MHC class I complexes of DCs. Importantly, iTEP-sMMP resulted in greater epitope presentation by DCs both in vitro and in vivo than a control carrier that cannot directly load epitopes. iTEP-sMMP also induced 2-fold stronger immune responses than the control carrier. To further enhance the direct epitope-loading strategy, we furnished iTEP-sMMP with an albumin-binding domain (ABD) and found the new carrier, ABD-iTEP-sMMP, had greater lymph node (LN) accumulation than iTEP-sMMP. ABD-iTEP-sMMP also resulted in greater immune responses than iTEP-sMMP by 1.5-fold. Importantly, ABD-iTEP-sMMP-delivered CTL epitope vaccine induced stronger immune responses than free CTL epitope vaccine. Taken together, these carriers utilized two physiological features of DCs to realize direct epitope-loading in vivo: the accumulation of DCs in LNs and MMP-9 released from DCs. These carriers are a potential substitute for DC-based CTL epitope vaccines.
Keywords: CTL vaccine, direct epitope-loading, albumin-binding domain, dendritic cell, matrix metalloproteinases-9
1. Introduction
Cytotoxic T lymphocyte (CTL) vaccines are promising and relatively safe modules to prevent and treat a wide variety of diseases and have been used clinically [1–4]. However, the efficacy of CTL vaccines for diseases like cancer is not satisfactory. To date, only two prophylactic [5, 6] and one therapeutic [7] cancer vaccines have been approved for clinical use. Among CTL vaccines under development, dendritic cell (DC)-based CTL vaccines are outstanding for their potency [8–10]. In fact, the only approved therapeutic cancer vaccine, Provenge (Sipuleucel-T), is a DC vaccine [7].
Clinically tested DC vaccines are generated by inducing monocyte-derived DCs from patients’ peripheral blood mononuclear cells (PBMCs) and loading the DCs with tumor cell lysates, protein antigens, or CTL epitopes ex vivo [11–13]. The loaded DCs are then infused back to the same patients to stimulate CTL responses [14]. When cell lysates and proteins are used as loading antigens, DCs need to process these antigens and extract CTL epitopes from the antigens before presenting the epitopes with the MHC class I complexes on the DC surface. When CTL epitopes are used, these epitopes can be directly loaded onto the MHC class I complexes on the DC surface, which bypasses antigen processing steps [14, 15]. This direct epitope-loading, often termed as epitope pulsing, happens because there are empty MHC class I complexes on the DC surface due to the dissociation of originally bound epitopes. Consequently, epitopes surrounding DCs can bind to the empty MHC class I complexes. The direct epitope-loading is an efficient approach to generate DC vaccines as it bypasses multiple inefficient steps of the intracellular antigen cross-presentation [16, 17]. Indeed, these epitope-loaded DC vaccines were found effective to induce CTL responses in both preclinical and clinical studies [18–20]. However, the epitope-loaded DC vaccines are hindered by several intrinsic limitations that are associated with DC vaccines in general: the requirement of autologous DCs, the complicated and costly ex vivo processing, and insufficient DC migration to lymph nodes (LNs) [14, 21–24]. These limitations seriously compromised the applicability of epitope-loaded DC vaccines.
It is possible to take advantage of the epitope-loaded DC vaccines while avoiding their limitations if one can directly load CTL epitopes onto DCs in vivo. The simplest idea is to administer free CTL epitopes as vaccines. Theoretically, these epitopes can be loaded onto MHC class I complexes on the DC surface in vivo. However, free epitopes have stability issues due to their small sizes and can be loaded onto cells other than DCs as long as these cells have MHC class I complexes [25]. To address the issues of size and undesired loading, we have developed a carrier that increases the size of CTL epitopes and specifically releases the epitopes in the proximity of DCs utilizing matrix metalloproteinases-9 (MMP-9) secreted by DCs [26]. We have proved that this carrier enabled direct epitope-loading onto the MHC class I complexes on the DC surface in vitro. The carrier and the epitope-loading strategy embedded in the carrier remain to be validated in vivo.
To achieve efficient CTL epitope-loading in vivo, it is required that not only CTL epitopes are released around DCs but also the epitopes are delivered to the proximity of DCs in the first place. An increased epitope accumulation in lymph nodes (LNs) would promote the access of the epitopes to DCs because DCs are abundant in LNs [27, 28]. Albumin-binding molecules preferentially accumulate in LNs [29–31] and, thus, have been used to deliver epitopes to LNs [32, 33]. Among reported albumin-binding molecules, an albumin-binding domain (ABD) that has 46 residues has been proven safe and effective in preclinical and clinical studies [34–36]. Particularly, the ABD can be easily incorporated into peptide-based vaccines and vaccine carriers through genetic engineering [37]. Therefore, it is intriguing to know whether the ABD would enhance the aforementioned carriers that have the direct epitope-loading capacity.
Here, we specially designed and generated a polypeptide-based carrier for a low-immunogenic melanoma CTL epitope that is strongly hydrophobic, pTRP2 (sequence: SVYDFFVWL). The carrier was designed to be very hydrophilic to solubilize pTRP2. The carrier was found to release CTL epitopes in an MMP-mediated fashion in DC culture medium, and load CTL epitopes directly onto the cell surface MHC class I complexes. In vivo, the carrier promoted epitope-loading by 3-fold and enhanced the pTRP2-specific immune responses by 2-fold. Further, the incorporation of ABD into this carrier promoted the LN accumulation of the carrier by 4-fold and further strengthened the pTRP2-specific immune responses by 1.5-fold. Last, pTRP2 delivered by the ABD-incorporated new carrier induced greater pTRP2-specific CTL responses than free pTRP2, a clinically used melanoma vaccine. Taken together, it is feasible to directly load CTL epitopes onto the MHC class I complexes on DCs in vivo, and the loading is boosted by carriers that are able to spatially control epitope accumulation and release.
2. Results
2.1. iTEP-sMMP was able to load its epitope payload directly onto MHC class I complexes on cell surface
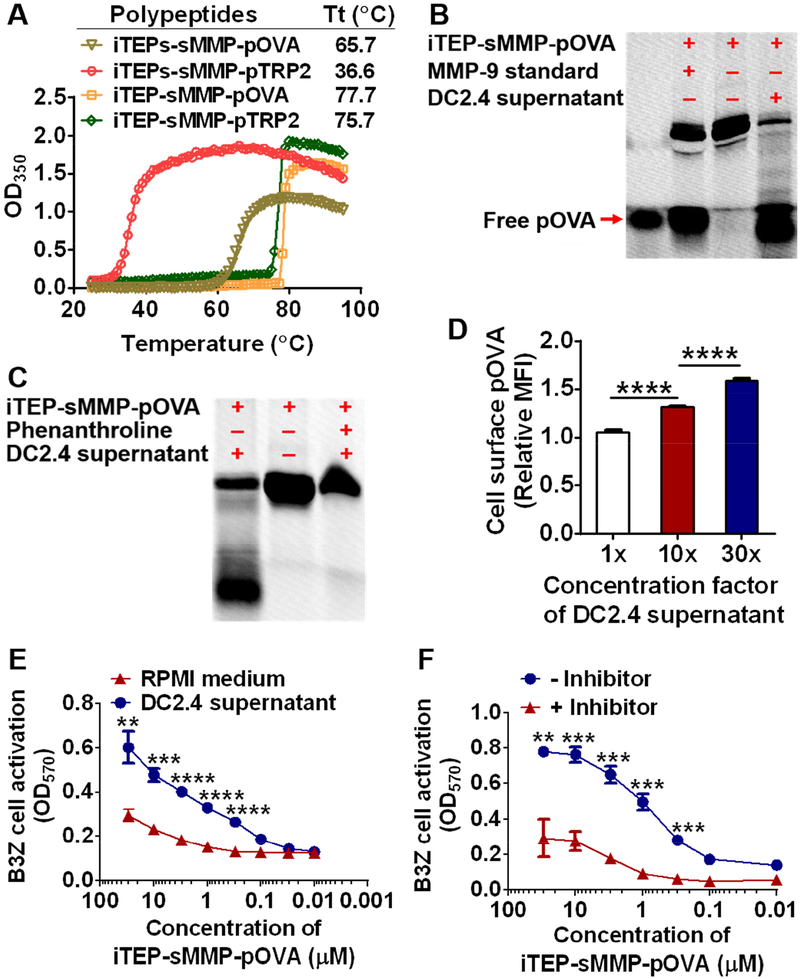
pTRP2, the model vaccine of this study, is a cancer vaccine used in clinical trials [12, 38, 39] (NCT00022438, NCT01456104) yet has low immunogenicity, thus, it is significant to use the strategy of direct epitope-loading to improve the immunogenicity of pTRP2. Our previously reported the carrier for direct epitope-loading, (GVGVPG)35-(GVLPGVG)16-PLGLAG [26], termed as iTEPs-sMMP, is not suitable for the delivery of pTRP2 because it cannot solubilize pTRP2 at a physiological temperature (Table 1, Fig. 1A). In view of this issue, we designed a new iTEP-based carrier that has a longer hydrophilic segment than the previous carrier. This new carrier, (GVGVPG)70-(GVLPGVG)16-PLGLAG, has a hydrophilic iTEP (GVGVPG)70 [40], a hydrophobic iTEP (GVLPGVG)16 [40], and a MMP-9 substrate peptide, PLGLAG. The carrier is termed as iTEP-sMMP. The fusion polypeptide of pTRP2 and iTEP-sMMP, namely iTEP-sMMP-pTRP2, is soluble at a physiological temperature (Table 1, Fig. 1A). iTEP-sMMP-pTRP2 has a transition temperature (Tt) between its soluble phase and aggregate phase of 75.7 ± 0.6 °C at 40 μM concentration in PBS.
Table 1. Sequences of the polypeptides used in this study.
| Polypeptides | Sequences (from N- to C-terminus)a | Molecular mass (kDa) | Tt (°C)b |
|---|---|---|---|
| iTEP-sMMP-pOVA | (GVGVPG)70-(GVLPGVG)16-PLGLAG-SIINFEKL | 41.6 | 77.7 ± 0.7 |
| iTEP-sMMP-pTRP2 | (GVGVPG)70-(GVLPGVG)16-PLGLAG-SVYDFFVWL | 41.8 | 75.7 ± 1.1 |
| iTEP-pOVA | (GVGVPG)70-(GVLPGVG)16-GGGGGG-SIINFEKL | 41.2 | 79.8 ± 0.3 |
| iTEP-pTRP2 | (GVGVPG)70-(GVLPGVG)16-GGGGGG-SVYDFFVWL | 41.6 | 76.7 ± 0.3 |
| ABD-iTEP-sMMP-pOVA | LAEAKVLANRELDKYGVSDFYKRLINKA | 46.9 | 53.2 ± 0.2 |
| KTVEGVEALKLHILAALP-(GVGVPG)70-(GVLPGVG)16-PLGLAG-SIINFEKL | |||
| ABD-iTEP-sMMP-pTRP2 | LAEAKVLANRELDKYGVSDFYKRLINKA | 47.2 | 47.6 ± 0.3 |
| KTVEGVEALKLHILAALP-(GVGVPG)70-(GVLPGVG)16-PLGLAG-SVYDFFVWL | |||
| iTEPs-sMMP-pOVAc | (GVGVPG)35-(GVLPGVG)16-PLGLAG-SIINFEKL | 26.1 | 65.7 ± 0.4 |
| iTE Ps-sM M P-pTRP2c | (GVGVPG)35-(GVLPGVG)16-PLGLAG-SVYDFFVWL | 26.3 | 36.6 ± 0.7 |
The subscripts were the repeating numbers of the peptide in parentheses.
The Tt of each polypeptide was shown as the mean ± standard deviation (SD) of three independent measurements.
pOVA and pTRP2 were fused with a previously reported carrier, iTEPs-sMMP that was capable of direct epitope-loading [26].
Figure 1.
iTEP-sMMP-pOVA enabled direct loading of pOVA onto MHC class I complexes on the cell surface. (A) The representative turbidity plots over temperature for four polypeptides, iTEP-sMMP-pOVA, iTEP-sMMP-pTRP2, iTEPs-sMMP-pOVA and iTEPs-sMMP-pTRP2. The turbidity was shown as the optical density at 350 nm (OD350). (B) A photo of a SDS-PAGE gel showed that a small peptide can be cleaved from iTEP-sMMP-pOVA in the presence of DC2.4 culture supernatant or MMP-9. Fluorescein-labeled pOVA was loaded into the leftmost lane. iTEP-sMMP-pOVA alone or its incubation mixtures with MMP-9 or the cell culture supernatant were loaded into the other lanes. The lower bands in these lanes represented cleavage products which had a similar size as pOVA. The upper bands in these lanes represent intact iTEP-sMMP-pOVA. (C) A photo of an SDS-PAGE gel showed that the cleavage of iTEP-sMMP-pOVA by DC2.4 culture supernatant was inhibited by phenanthroline. iTEP-sMMP-pOVA and its treatment conditions were indicated on the top of the gel photo. (D) Relative median fluorescence intensity (MFI) of B16-F10 cells after they were first pulsed by the incubation mixture of iTEP-sMMP-pOVA and DC2.4 culture supernatant and then stained with an antibody specific to pOVA/MHC class I complexes on cell surface. The relative MFI was generated by normalizing the observed MFI of each sample against the MFI of B16-F10 cells that were pulsed with DC2.4 cell culture supernatant and stained. (E) A plot of optical density at 570 nm (OD570) of B3Z cells showed that the directly loaded pOVA on B16-F10 surface can activate B3Z cells. Different concentrations of iTEP-sMMP-pOVA were first incubated with DC2.4 supernatant (1×) or RPMI medium overnight. The incubation mixtures were then used to pulse B16-F10 cells before the B16-F10 cells were mixed with B3Z cells. The extent of B3Z cell activation was reflected as OD570. (F) An OD570 plot of B3Z cell showed that the activation of B3Z cells was inhibited when phenanthroline was added into the incubation mixture of iTEP-sMMP-pOVA and DC2.4 supernatant. The other procedures were same as described in (E). Data were shown as mean ± SD. Data in (D) were analyzed by one way ANOVA with Bonferroni post-test. Data in (E) and (F) were analyzed by Student’s t test. **P < 0.01, ***P < 0.001, ****P < 0.0001.
Since the tools are not available to in vitro characterize the epitope-loading and T cell activation of pTRP2 delivered by iTEP-sMMP, a surrogate epitope, pOVA (sequence: SIINFEKL), was used for the in vitro characterizations of iTEP-sMMP, although the in vivo effect of iTEP-sMMP-pTRP2 was eventually examined. The strategy of direct epitope-loading relies on the presence of CTL epitopes in the proximity of DCs. We intended to utilize MMP-9 secreted from DCs (Figure S1) to cleave epitopes from iTEP-sMMP around DCs. We found that incubation between iTEP-sMMP-pOVA and the supernatant of DC2.4 cell culture or MMP-9 resulted in a small peptide product similar to free pOVA according to the sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis (Fig. 1B), suggesting that pOVA might be released from iTEP-sMMP-pOVA in the presence of the DC supernatant or MMP-9. In contrast, an incubation of iTEP-sMMP-pOVA alone did not result in that small peptide. Further, MMP-9 in the supernatant are likely responsible for cleavage because 1) the substrate of MMP-9, PLGLAG, connects the epitope and the carrier by design, and 2) phenanthroline, an inhibitor of MMP [41], drastically hindered the cleavage evidenced by a clear reduction of the band that represented the cleavage product (Fig. 1C).
Next, we examined whether iTEP-sMMP-pOVA led to direct loading of pOVA onto MHC class I complexes on the cell surface. In this examination, we utilized B16-F10 cells that have MHC class I complexes on their surface but do not have the cross-presentation capacity as they are not professional antigen-presenting cells. Thus, if exogenous epitopes are presented by B16-F10 cells with the MHC class I complexes, these epitopes must be directly loaded onto the complexes. We found that, after B16-F10 cells were pulsed with the incubation mixture of iTEP-sMMP-pOVA and the DC culture supernatant, the cells presented pOVA with MHC class I complexes (Fig. 1D). Further, when more concentrated supernatants were used for the incubation, more pOVA/MHC class I complexes were detected on B16-F10 cell surface. These results supported that 1) iTEP-sMMP-pOVA can result in direct loading of pOVA onto the cell surface MHC class I complexes, and 2) the DC culture supernatant, very likely the MMPs in the supernatant, enabled the epitope release and the consequent loading. We also found that the directly loaded pOVA was biologically functional because it activated its restricted T cell clone, B3Z cells [42] (Fig. 1E). In this experiment, two kinds of incubation mixture, 1) iTEP-sMMP-pOVA with the DC2.4 culture supernatant, and 2) iTEP-sMMP-pOVA with the RPMI medium, were first used to pulse B16-F10 cells, respectively; then, the treated B16-F10 cells were used to activate B3Z cells. We found that the first mixture resulted in significantly greater activation of B3Z cells than the second mixture when the concentrations of iTEP-sMMP-pOVA used in the experiments were greater than 0.3 μM (Fig. 1E). We also found the activation was concentration-dependent on iTEP-sMMP-pOVA. Further, the activation was significantly diminished when phenanthroline was added into the incubation mixture of iTEP-sMMP-pOVA and the DC culture supernatant (Fig. 1F). This conclusion was valid when the concentrations of iTEP-sMMP-pOVA used in the experiment were greater than 0.3 μM.
Taken together, CTL epitopes can be cleaved from iTEP-sMMP-pOVA in the supernatant of DC culture; iTEP-sMMP can result in direct loading of epitopes onto the MHC class I complexes on cell surface; the loaded epitopes were functional.
2.2. iTEP-sMMP led to greater epitope presentation than a carrier that was dependent on cross-presentation
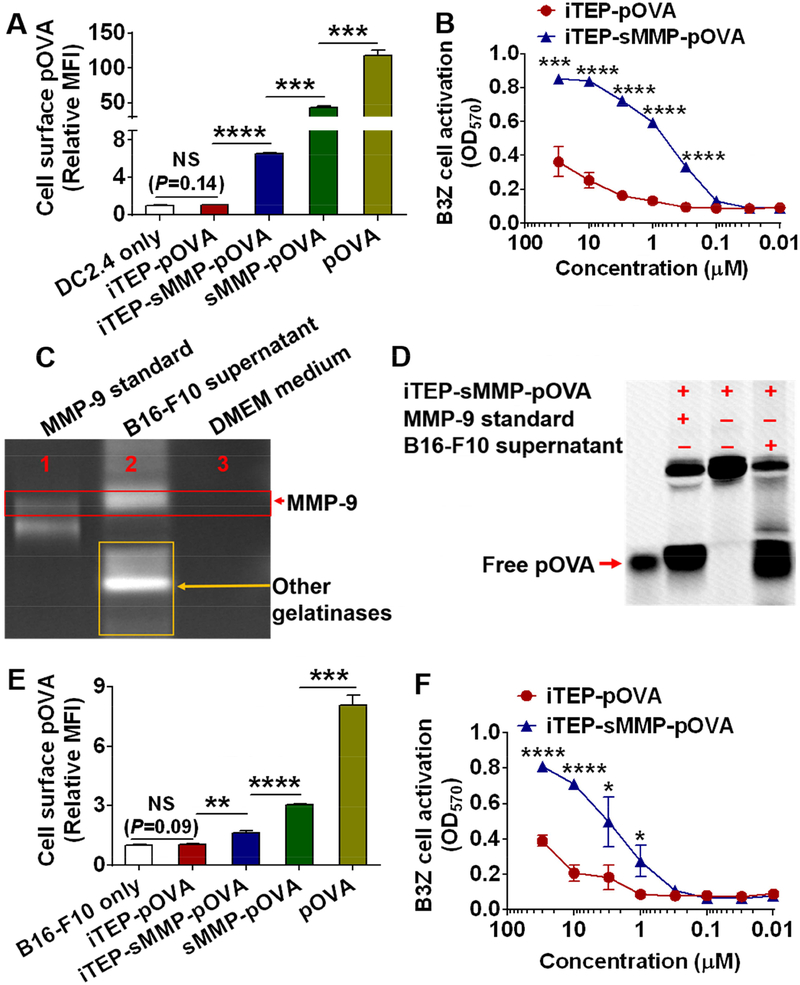
CTL epitope vaccine carriers may be categorized into two types based on the epitope presentation process the carriers utilize. The first type, like iTEP-sMMP, facilitates the direct loading as well as the cross-presentation of its epitope payloads onto the MHC class I complexes on DCs. The second type is solely dependent on the cross-presentation pathway of DCs to present its epitope payloads. The second type is unable to release epitopes outside of DCs and to directly load the epitopes onto MHC class I complexes. It is important to know whether iTEP-sMMP results in greater epitope presentation than the second type of carriers, which will determine the type of carriers we should use to deliver epitope vaccines. We first answered this question through in vitro studies. We compared iTEP-sMMP-pOVA with iTEP-pOVA (Table 1), a vaccine that was dependent on the cross-presentation. iTEP-pOVA did not contain the sMMP and was not expected to release pOVA outside of DCs. An incubation of iTEP-sMMP-pOVA with DC2.4 cells resulted in 6-fold more pOVA presentation by DCs than an incubation of iTEP-pOVA with DC2.4 cells (Fig. 2A and Fig. S2). Indeed, the presentation result of iTEP-pOVA was not statistically different to that of the DC only incubation (P = 0.14). At the same time, the peptides pOVA and sMMP-pOVA resulted in much higher presentation than iTEP-sMMP-pOVA and iTEP-pOVA. Consistently, the incubation of iTEP-sMMP-pOVA with DC2.4 cells also led to greater B3Z cell activation than the incubation of iTEP-pOVA with DC2.4 cells when the DCs collected from these incubations were used for the B3Z cell activation assay (Fig. 2B). Together, these results showed that iTEP-sMMP-pOVA was more effective than iTEP-pOVA to render DCs to present pOVA.
Figure 2.
iTEP-sMMP-pOVA resulted in greater pOVA presentation on the cell surface than iTEP-pOVA in vitro. (A) Relative MFI of DC2.4 cells. The cells were first incubated with pOVA, sMMP-pOVA, iTEP-sMMP-pOVA, iTEP-pOVA, or cell culture medium. The cells were then stained with an antibody specific to pOVA/MHC class I complexes on the cell surface. The relative MFI was generated by normalizing observed MFI of each sample against the MFI of DC2.4 cells that were incubated with cell culture medium. (B) An OD570 plot of B3Z cell indicated that iTEP-sMMP-pOVA generated more pOVA epitopes on DC2.4 surface than iTEP-pOVA. (C) A gel photo of the gelatin zymography assay showed that the culture supernatant of B16-F10 cells contained MMP-9. The top bright band indicated where MMP-9 migrated and cleaved gelatin. The additional bright bands in Lane 2 implied that the culture supernatant of B16-F10 cells contained other gelatinases. (D) A photo of a SDS-PAGE gel showed that a small peptide can be cleaved from iTEP-sMMP-pOVA in the presence of B16-F10 cell culture supernatant. When B16-F10 cells were used in the assay, iTEP-sMMP-pOVA still generated more pOVA epitopes onto the cell surface than iTEP-pOVA based on the flow cytometry assay (E) and B3Z cell activation assay (F). Experiments were conducted in triplicate. Data were shown as mean ± SD and analyzed by Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, NS = not significant.
Since iTEP-sMMP can both directly load epitopes onto cell surface MHC class I complexes and load the epitopes to the complexes through the cross-presentation, it was not clear whether the superior epitope presentation result of iTEP-sMMP-pOVA over iTEP-pOVA was due to the direct loading mechanism or the cross-presentation mechanism. To investigate this question, we utilized B16-F10 cells as antigen presenting cells for our antigen presentation and B3Z cell activation studies because B16-F10 cells do not have the cross-presentation pathway and hence can only support direct epitope-loading. On the other hand, B16-F10 cells secrete MMP-9 (Fig. 2C) and the supernatant of B16-F10 cell culture can cleave iTEP-sMMP-pOVA (Fig. 2D). The incubation of iTEP-sMMP-pOVA with B16-F10 cells resulted in 1.5-fold more pOVA presentation by B16-F10 cells than the incubation of iTEP-pOVA with the cells (Fig. 2E and Fig. S2); the incubation of iTEP-sMMP-pOVA with B16-F10 cells also induced 2-fold higher B3Z cell activation than the incubation of iTEP-pOVA with B16-F10 cells (Fig. 2F) when the concentrations of the vaccines were greater than 1 μM. Meanwhile, the epitope presentation result of iTEP-pOVA was not statistically different than that of the B16-F10 only incubation (P = 0.09; Fig. 2E). These results suggested that the direct epitope-loading mechanism contributed to the greater epitope presentation of iTEP-sMMP-pOVA.
2.3. iTEP-sMMP enhanced epitope presentation in vivo
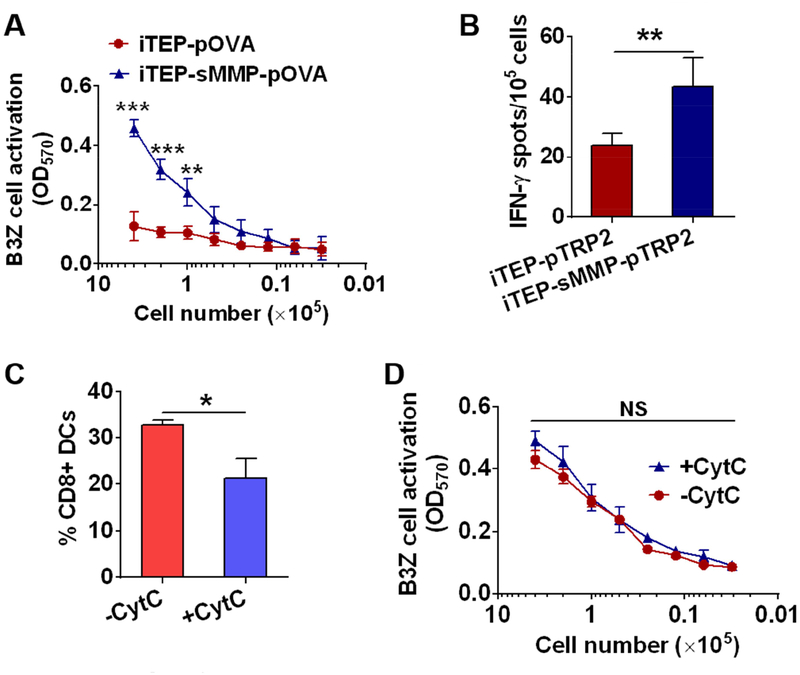
After we established that iTEP-sMMP resulted in greater epitope presentation than iTEP in vitro, we compared these two carriers for epitope presentation in vivo. We expected that iTEP-sMMP would facilitate the direct loading of its epitope payloads onto MHC class I complexes on the DC surface in vivo, because 1) primary DCs collected from mice secreted MMP-9 (Fig. S3A), and 2) the culture supernatant of these DCs cleaved iTEP-sMMP-pOVA and generated a peptide fragment that was same as the peptide fragment generated by MMP-9 (Fig. S3B). To execute the comparison between iTEP-sMMP and iTEP in vivo, we subcutaneously injected mice with iTEP-sMMP-pOVA or iTEP-pOVA and collected primary DCs from spleens and draining LNs (inguinal and axillary LNs) 24 h after the injections. Then, we used these DCs for B3Z cell activation assay and used the B3Z activation data to infer the extent that these DCs presented pOVA. When 1×105 to 4×105 DCs were used for the B3Z assay, DCs from iTEP-sMMP-pOVA treated mice resulted in significantly stronger B3Z cell activation than DCs from iTEP-pOVA treated mice (Fig. 3A). When less than 1×105 DCs were used, there was no difference in B3Z activation between the two sources of DCs. These results suggested that iTEP-sMMP-pOVA resulted in greater pOVA presentation by DCs than iTEP-pOVA in vivo.
Figure 3.
iTEP-sMMP-pOVA resulted in greater pOVA presentation by DCs than iTEP-pOVA in vivo. (A) An OD570 plot of B3Z cells showed that DCs from iTEP-sMMP-pOVA treated mice induced higher B3Z cell activation than DCs from iTEP-pOVA treated mice (n=3). 24 h after the injection of the vaccines, DCs were collected from the spleen and draining LNs of mice and cultured with B3Z cells to examine the pOVA presentation on the DC surface. (B) iTEP-sMMP-pTRP2 induced stronger pTRP2-specific immune responses than iTEP-pTRP2 in vivo (n=5). Mice were immunized with the vaccines, and the splenocytes were collected to examine pTRP2-specific immune responses. (C) Injection of CytC reduced CD8+ DCs in mice (n=3). Mice were subcutaneously injected with 5 mg CytC on each side of the tail base every 12 h for total of 4 injections. 24 h after the last injection, DCs from the spleen and draining LNs were collected to determine the fraction of CD8+ DCs by flow cytometry. (D) An OD570 plot of B3Z cell indicated that the pOVA presentation of iTEP-sMMP-pOVA in mice (n=3) was not affected by the injection of CytC. Data were shown as mean ± SD and analyzed by Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, NS = not significant.
Next, we examined whether iTEP-sMMP-delivered CTL epitopes induced stronger antigen-specific immune responses than iTEP-delivered epitopes. For this experiment, we returned to use pTRP2 as the model epitope and generated iTEP-sMMP-pTRP2 and iTEP-pTRP2 (Table 1). At 10 days after the second immunization of mice with iTEP-sMMP-pTRP2 or iTEP-pTRP2, we collected splenocytes from these mice. Then, we quantified the pTRP2-restricted immune responses using an IFN-γ-based enzyme-linked immunospot (ELISPOT) assay [43]. We used the ELISPOT data to gauge pTRP2-specific immune responses. We found that iTEP-sMMP-pTRP2 induced about 2-fold stronger pTRP2-specific immune responses than iTEP-pTRP2 (mean spot number: 43.6 ± 9.3 spots versus 23.8 ± 4.1 spots per 105 splenocytes, P < 0.01, Fig. 3B).
Since iTEP-sMMP and its epitope payloads can be internalized by DCs as a whole, thus, the epitopes delivered by iTEP-sMMP can be directly loaded onto the MHC class I complexes on DC surface or loaded onto the complexes through the antigen cross-presentation pathway. iTEP-sMMP-pOVA was found more effective than iTEP-pOVA to generate pOVA/MHC class I complexes on DC surface in vivo (Fig. 3A). However, it was not clear whether the edge of iTEP-sMMP-pOVA was due to its direct epitope-loading capacity or its other differences to iTEP-pOVA. It is possible that iTEP-sMMP-pOVA has more efficient cross-presentation than iTEP-pOVA. To elucidate the contribution of the direct epitope-loading in vivo, we performed the in vivo epitope-loading study in the presence of cytochrome c (CytC), an inhibitor of CD8+ DCs [44]. CytC can selectively deplete CD8+ DCs and compromise the cross-presentation pathway because CD8+ DCs are main effector DCs for the antigen cross-presentation [45]. We found that while CytC reduced the CD8+ DC fraction among all DCs from 32.8 ± 1.0% to 21.3 ± 4.3% (Fig. 3C and Fig. S3C), the presentation of pOVA from iTEP-sMMP-pOVA was not affected by the CytC treatment (Fig. 3D). These data implied that the pOVA presentation resulted from iTEP-sMMP-pOVA was mainly attributed by the direct epitope-loading capacity of iTEP-sMMP-pOVA instead of antigen cross-presentation, although we cannot completely rule out the contribution of cross-presentation.
2.4. Binding with albumin boosted the epitope presentation effect of iTEP-sMMP.
It is beneficial to the direct epitope-loading function of iTEP-sMMP if more iTEP-sMMP accumulate in the proximity of DCs in vivo. DCs are abundant in LNs, thus, an increased accumulation of iTEP-sMMP in LNs may boost its opportunity to be present around DCs in LNs. Subsequently, the increased accumulation in LNs may lead to the increased presentation of epitopes by DCs that are delivered by iTEP-sMMP. To further enhance the direct epitope-loading capacity of iTEP-sMMP, we generated a new carrier (Table 1) that contained iTEP-sMMP and ABD, a protein domain that binds to mouse serum albumin (MSA) and tends to accumulate in LNs [37].
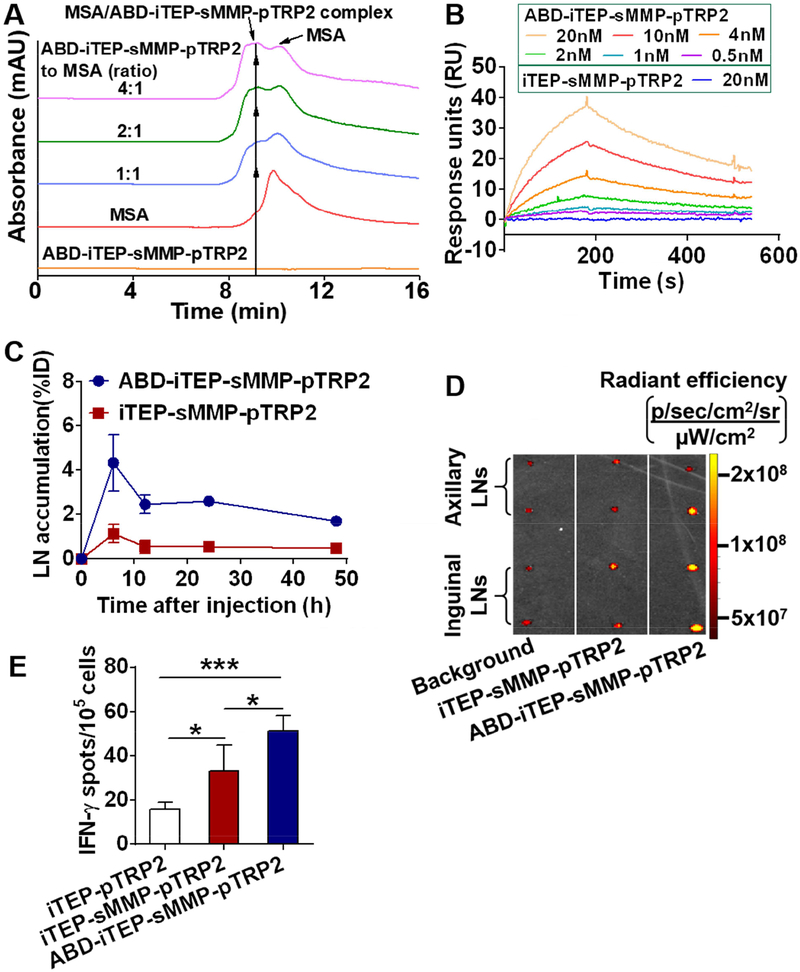
First, we confirmed that these ABD-containing vaccines bound to MSA (Fig. 4A–B and Fig. S4). Specifically, the incubation of ABD-iTEP-sMMP-pTRP2 and MSA resulted in a new peak showing at 9.2 min on size exclusion chromatography (SEC). In contrast, MSA showed as a peak at 9.9 min on the chromatogram; and ABD-iTEP-sMMP-pTRP2, by itself, did not show any prominent peak due to its low extinction coefficient (9970 M−1 cm−1 of ABD-iTEP-sMMP-pTRP2 versus 46030 M−1 cm−1 of MSA at 280 nm). In addition, the new peak resulting from the incubation mixtures increased with the increment of the ratio of ABD-iTEP-sMMP-pTRP2 to MSA; while the peak of MSA decreased with the increment of the ratio (Fig. 4A), implying the new peak belonged to the complex between ABD-iTEP-sMMP-pTRP2 and MSA. According to the surface plasmon resonance (SPR) analysis (Fig. 4B), the binding affinity (KD) between ABD-iTEP-sMMP-pTRP2 and MSA was 3.80 ± 0.06 nM, which was similar to a reported KD between free ABD and MSA, 1.24 ± 0.01 nM [34]. Meanwhile, iTEP-sMMP-pTRP2 showed no binding with MSA (Fig. 4B).
Figure 4.
ABD-iTEP-sMMP increased accumulation in LNs and improved immune responses toward epitopes it delivered. (A) An overlay of SEC chromatographs of ABD-iTEP-sMMP-pTRP2, MSA, and their mixtures showed the binding between ABD-iTEP-sMMP-pTRP2 and MSA. The mixtures were incubated overnight before analysis. ABD-iTEP-sMMP-pTRP2 had no apparent absorbance at 280 nm. (B) SPR sensorgrams of ABD-iTEP-sMMP-pTRP2 and iTEP-sMMP-pTRP2 after they were flowed over a chip surface immobilized with MSA. (C) LN accumulation of ABD-iTEP-sMMP-pTRP2 and iTEP-sMMP-pTRP2. The data were expressed as percentage of injected dose, %ID, (n=3). (D) ex vivo imaging of draining LNs from mice that were injected with PBS (background), Alex Fluor 488-labeled iTEP-sMMP-pTRP2 and Alex Fluor 488-labeled ABD-iTEP-sMMP-pTRP2. The unit of radiant efficiency is [p/sec/cm2/sr]/[μW/cm2]. (E) ABD-iTEP-sMMP-pTRP2 induced higher immune responses than iTEP-sMMP-pTRP2 and iTEP-pTRP2. Mice (n=4) were immunized with the vaccines as indicated. The splenocytes were collected to check the pTRP2-specific immune responses using the IFN-γ ELISPOT assay. Data were shown as mean ± SD and analyzed by one-way ANOVA with Bonferroni post-test. *P < 0.05, ***P < 0.001.
We next compared the LN accumulation of ABD-iTEP-sMMP-pTRP2 and iTEP-sMMP-pTRP2. Within 48 h after subcutaneous injection of the two vaccines, the area under the curve (AUC) of ABD-iTEP-sMMP-pTRP2 in draining LNs was 4-fold more than the AUC of iTEP-sMMP-pTRP2 (115.61 %ID.h versus 26.98 %ID.h) (Fig. 4C). Additionally, we confirmed the LN accumulation difference between ABD-iTEP-sMMP-pTRP2 and iTEP-sMMP-pTRP2 through ex vivo imaging of LNs. Here, mice were injected with either Alexa Flour 488-labelled ABD-iTEP-sMMP-pTRP2 or Alexa Flour 488-labelled iTEP-sMMP-pTRP2. Six hours after the injection, the mean fluorescence signal of LNs from the ABD-iTEP-sMMP-pTRP2 treated mice was about 4.5 times stronger than the mean fluorescence signal of LNs from the iTEP-sMMP-pTRP2 treated mice (8.05 ± 5.39 ×107 versus 1.72 ± 0.71×107 after subtraction of the background signal 4.34 ± 0.52×107, Fig. 4D). Together, these data demonstrated that ABD-iTEP-sMMP-pTRP2 had greater LN accumulation as compared to iTEP-sMMP-pTRP2.
Lastly, we investigated whether ABD-iTEP-sMMP-pTRP2 would induce stronger pTRP2-restricted immune responses than iTEP-sMMP-pTRP2. We used the IFN-γ-based ELISPOT assay to gauge the immune responses and found that ABD-iTEP-sMMP-pTRP2-induced immune responses were 1.5-fold greater than those of iTEP-sMMP-pTRP2, (mean spot number: 51.3 ± 6.8 spots versus 33.3 ± 11.6 spots per 105 splenocytes, P < 0.05, Fig. 4E). At the same time, the immune responses induced by iTEP-sMMP-pTRP2 were 2-fold greater than those of iTEP-pTRP2 (mean spot number: 33.3 ± 11.6 spots versus 16.0 ± 2.8 spots per 105 splenocytes, P < 0.05, Fig. 4E). Taken together, the carrier with both sMMP and ABD, ABD-iTEP-sMMP-pTRP2, most effectively boosted the immune responses of epitope payloads.
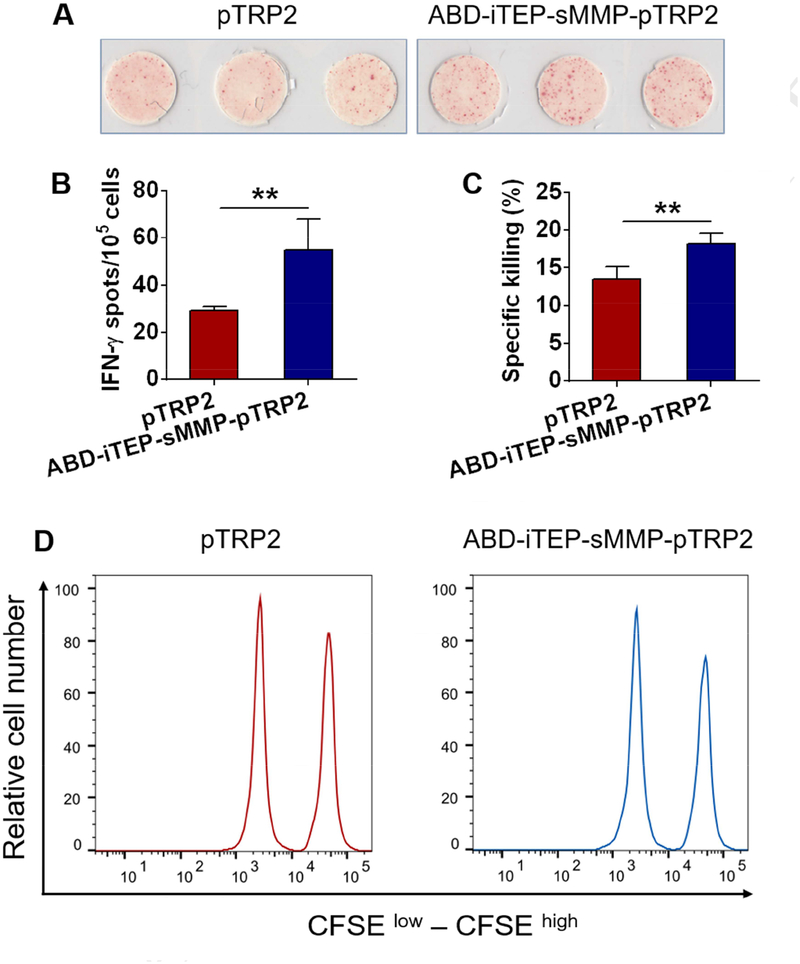
Free CTL epitopes can be directly loaded onto the MHC class I complexes on DC surface and they are commonly tested in CTL vaccine trails and combination immunotherapy trials [46, 47]. Thus, it is of clinical interest to examine whether ABD-iTEP-sMMP-pTRP2, the most effective vaccines that we have developed to enable direct epitope-loading, is more potent than a clinically used, free CTL epitope, pTRP2 peptide in inducing CTL responses. To this end, we immunized mice with ABD-iTEP-sMMP-pTRP2 or pTRP2 peptide and compared their pTRP2-specific immune responses using IFN-γ-based ELISPOT assay. We found that ABD-iTEP-sMMP-pTRP2 induced about 2-fold greater immune responses than pTRP2 (mean spot number: 54.7 ± 13.3 spots versus 29.3 ± 1.5 spots per 105 splenocytes, P < 0.01, Fig. 5A and 5B). We also evaluated the CTL responses using an in vivo CTL-mediated killing assay. Examination of CTL responses by this assay would answer how well the ABD-iTEP-sMMP-pTRP2 enahnced immune responses help to eliminate cells that present pTRP-2 epitope including melanoma cells. Specifically, mice were first immunized with monophosphoryl lipid A (MPLA), a clinically used vaccine adjuvant [48], together with ABD-iTEP-sMMP-pTRP2 or pTRP2. Ten days after immunization, mice were injected with pTRP2-pulsed target cells and the specific killing of target cells were analyzed 16 hours later. We found that mice immunized with ABD-iTEP-sMMP-pTRP2 eliminated more target cells than mice immunized with pTRP2 peptide (Fig. 5C and 5D), indicating that ABD-iTEP-sMMP-pTRP2 indcued stronger CTL responses than pTRP2 peptide. These results suggested that ABD-iTEP-sMMP-pTRP2 had the potential to improve CTL epitope vaccines tested in clinical trials.
Figure 5.
The direct epitope-loading vaccine ABD-iTEP-sMMP-pTRP2 was more efficient than a clinically used epitope vaccine pTRP2. Representative image (A) and statistical plot (B) of ELISPOT assay showed that ABD-iTEP-sMMP-pTRP2 induced greater immune responses than pTRP2 peptide. Mice (n=3) were immunized with ABD-iTEP-sMMP-pTRP2 or pTRP2. The pTRP2-specific immune responses were characterized by the IFN-γ ELISPOT assay. (C) ABD-iTEP-sMMP-pTRP2 resulted in a greater pTRP2-specific cell killing effect than pTRP2. Mice (n=5) were immunized with ABD-iTEP-sMMP-pTRP2 or pTRP2. The pTRP2-specific CTL responses were then examined through the in vivo CTL-mediated specific killing assay. (D) Representative flow cytometry plots showed the in vivo pTRP2-specific cell killing effects. Data were shown as mean ± SD and analyzed by Student’s t test. **P < 0.01.
3. Discussion and Conclusion
DC vaccines are a very effective module to induce CTL responses and have achieved clinical successes, such as Provenge. However, DC vaccines require complicated ex vivo manufacturing. The typical manufacturing procedures include steps like inducing DCs from patients’ PBMCs, ex vivo antigen loading to the DCs, and infusion of the DCs back to patients [11, 14, 15]. Further, these tedious procedures have to be repeated each time for each treatment. For example, a course of Provenge requires three infusions of DCs and hence three labor-intensive preparations of the vaccines. Each preparation takes 2–3 days [49, 50]. These hurdles of producing DC vaccines put a dramatic price tag on the vaccines. One course of Provenge treatment costs about $93,000 [22, 23]. Another barrier of DC vaccines is the strict requirement of an autologous DC source: DCs used to treat a patient have to be collected from the same patient. This requirement makes the scale-up of DC vaccines a significant challenge. In contrast to DC vaccines, the direct epitope-loading vaccines we developed can be easily produced. CTL epitopes were fused to the carriers through genetic engineering. The entire vaccines were produced and reproduced using Escherichia coli repeatedly and homogeneously at high yields (~200 mg/L). The vaccines were easily purified through cycling the reversible phase transition of the vaccines, thanks to the phase transition property of the iTEP in the vaccines [40]. The scalable and straightforward production procedures of the vaccines make them a potentially inexpensive and timesaving alternative for DC vaccines.
iTEP-sMMP-pOVA led to more pOVA presentation by DCs than iTEP-pOVA, both in vitro and in vivo. As an extrapolation of this result, vaccines with the direct epitope-loading capability, such as iTEP-sMMP-pOVA, are arguably more potent than the vaccines that solely rely on the antigen cross-presentation, such as iTEP-pOVA. The edge of direct epitope-loading vaccines is probably due to their decreased dependence on the cross-presentation by DCs. The cross-presentation is not an efficient pathway due to several limiting steps including antigen internalization by DCs, antigen transportation from endosomal compartments to cytosol inside DCs, as well as antigen degradation and epitope release by DC proteasomes [16, 17, 51]. To date, the majority of CTL vaccine carriers were developed for delivery of the vaccines into DCs and the subsequent cross-presentation [32, 52–54]. Based on our results, these carriers force the vaccines into the inefficient cross-presentation pathway and hence limit the overall effectiveness of the vaccines by design. Our results, on the other hand, call for a fresh thought and strategy for CTL vaccine delivery, that is the delivery of vaccines to the proximity of DCs instead of inside DCs.
iTEP-sMMP-pOVA was designed to directly load pOVA onto the MHC class I molecules on DC surface. However, the possibility exists that iTEP-sMMP-pOVA is internalized by DCs and the pOVA from iTEP-sMMP-pOVA is processed and cross-presented by DCs. Therefore, it is important to explore whether the superior epitope presentation results of iTEP-sMMP-pOVA over iTEP-pOVA are due to direct epitope-loading, or more efficient cross-presentation for iTEP-sMMP-pOVA over iTEP-pOVA, or both. To this end, we took advantage of B16-F10 cells that cannot cross-present exogenous vaccines, and proved that the direct epitope-loading was one pathway utilized by iTEP-sMMP-pOVA. The incubation of iTEP-sMMP-pOVA and B16-F10 cells led to pOVA presentation by the cells, while the incubation of iTEP-pOVA and B16-F10 cells led to no presentation as compared to untreated cells (Fig. 2E). More importantly, when CD8+ DCs, the cells capable of antigen cross-presentation were reduced by one third, we did not observe significant reduction of epitope presentation by DCs from iTEP-sMMP-pOVA-treated mice. These results suggested that direct epitope-loading was at least one working mechanism of iTEP-sMMP-pOVA to boost epitope presentation, and possibly the primary working mechanism of iTEP-sMMP-pOVA to boost epitope presentation in vivo. It is acknowledgeable that these results were not sufficient to rule out the contribution from cross-presentation of iTEP-sMMP-pOVA. However, it is possible to delineate the contributions of direct epitope-loading and cross-presentation more precisely by using murine models that stringently lack the cross-presentation capacity, such as TAP1-deficient mice [55] and, more recently, Rab43-deficient mice [56].
The direct epitope-loading vaccine, iTEP-sMMP-pTRP2, induced stronger pTRP2-specific immune responses than iTEP-pTRP2. The advantage of iTEP-sMMP-pTRP2 was further expanded by equipping the vaccine with albumin-binding capacity. Here, the vaccine was fused with ABD so that the vaccine can bind with albumin. Previously, it was shown that ABD enabled preferential accumulation of its fusion proteins in LNs over other organs [37]. Consistently, the ABD, in this study, also enabled ABD-iTEP-sMMP-pTRP2 to bind with albumin and increased its LN accumulation by 4-fold. The LN accumulation is significant because LNs are rich for DCs and greater LN accumulation of vaccines increases the exposure of the vaccines to DCs. In this study, the ABD, instead of other albumin-binding molecules, was used with iTEP-sMMP-pTRP2 because 1) it possesses a high binding affinity with albumin (3.80 ± 0.06 nM) and 2) it, as a protein domain, can be easily fused to iTEP-sMMP-pTRP2 with high fidelity through genetic engineering. Taken together, the ABD is a practical and effective tool to boost direct epitope-loading of CTL epitopes. Finally, it is demonstrated that the direct epitope-loading vaccine ABD-iTEP-sMMP-pTRP2 induced stronger immune responses inclduing the CTL-mediated killing effect than pTRP2, which suggested the potential clinical application of the direct epitope-loading vaccine in the future.
In summary, we developed a vaccine delivery system that directly loaded epitopes to the DC surface in vivo and enhanced CTL responses of a poorly-immunogenic CTL epitope. These direct-epitope loading vaccines may become an attractive substitution for DC vaccines.
4. Materials and Methods
4.1. Animals and cell lines
Four to 8-week old, female C57BL/6 mice were purchased from Charles River Laboratories. The mice were kept under the approved protocol by the University of Utah Institutional Animal Care and Use Committee (IACUC). DC2.4 cell, which was kindly provided by Dr. Kenneth Rock (University of Massachusetts), is a dendritic cell line with H-2Kb of MHC class I molecules. DC2.4 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 1% nonessential amino acids, 10 mM HEPES, 50 μM 2-mercaptoethanol, 100 U mL−1 penicillin and 100 μg mL−1 streptomycin. B3Z cell, which was kindly provided by Dr. Nilabh Shastri (University of California, Berkeley), is a T cell hybridoma of which the receptor specifically recognizes H-2Kb/SIINFEKL complexes. B3Z cells were cultured with RPMI 1640 medium supplemented with 10% FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 50 μM 2-mercaptoethanol, 100 U mL−1 penicillin and 100 μg mL−1 streptomycin. B16-F10 cells (American Type Culture Collection) were cultured with DMEM medium supplemented with 10% FBS, 100 U mL−1 penicillin and 100 μg mL−1 streptomycin. All cells were cultured in 37 °C with 5% CO2. In the detection of cell surface pOVA by flow cytometry and B3Z cell activation assays, cells were cultured with the medium mentioned above but without FBS in order to avoid the impact of MMPs present in FBS.
4.2. Production and purification of recombinant polypeptides
The iTEP-based polypeptides were expressed in Escherichia coli. and purified through inverse transition cycling using the previously reported method [40, 57]. The endotoxin in the polypeptides was subsequently removed using the published method [58]. The level of endotoxin in the polypeptides was controlled under 0.25 EU mg−1 for immune-related assays.
4.3. Characterization of the transition temperature (Tt)
Because of the reversible phase transition property of iTEP-based polypeptides, each polypeptide has a Tt. To characterize the Tt, each polypeptide was prepared to 40 μM in PBS and the solution was added to a cuvette. When the polypeptide solution was heated from 25 to 95 °C, the optical density at 350 nm (OD350) of the solution was continuously monitored using a UV-visible spectrophotometer equipped with a temperature controller (Varian Instruments). The curve between OD350 and temperature was fitted using Sigmoidal dose-response nonlinear regression method in GraphPad (version 6.01). The maximum first derivative of the curve was identified as Tt.
4.4. Preparation of supernatant of cell culture
Bone marrow derived dendritic cells (BMDCs) were prepared using the previously described method [59]. BMDCs, DC2.4 cells and B16-F10 cells were cultured in 100 mm cell culture dishes with corresponding medium. When cells reached 80% confluence, the culture medium was removed and the cells were washed gently with PBS for three times. Then 10 mL RPMI medium (without any supplement, for BMDCs and DC2.4 cells) and DMEM medium (without any supplement, for B16-F10 cells) were added to the dishes. Cells were cultured in the medium for 24 h before the supernatant was collected. The supernatant was concentrated based on the needs of experiments by using Amicon Ultra-4 centrifugal filter devices (10,000 molecular weight cutoff).
4.5. Gelatin zymography
The presence of MMP-9, also called gelatinase B, in the cell culture supernatant was assessed by gelatin zymography as previously reported method [60]. 10 μL of concentrated (30×) cell culture supernatant was loaded into one well of the gelatin-embedded sodium dodecyl sulfate (SDS) polyacrylamide gel. 25 ng recombinant mouse MMP-9 standard (BioLegend) was loaded as the positive control. The gel was run at 120 V for 1.5 h. The gel was then washed with renaturing buffer (2.5% Triton X-100 in water) for 30 minutes and incubated in developing buffer (50 mM Tris-HCl, pH 7.6, 10 mM CaCl2, 150 mM NaCl, 0.02% Brij 35) for 16 h at 37 °C. Next, the gel was stained with Coomassie Blue solution and destained with water. The clear bands on the gel represented the location of the gelatinases.
4.6. Polypeptides cleavage by MMP-9
iTEP-sMMP-pOVA was labelled with NHS-fluorescein (Thermo Fisher Scientific) using the manufacturer’s protocol. 10 μg labelled polypeptide was incubated with 10 μL concentrated (30×) cell culture supernatant or 100 ng recombinant mouse MMP-9 standard at 37 °C for overnight. For inhibition assay, the cell culture supernatant was treated with 30 mM 1,10-Phenanthroline (Sigma-Aldrich) for 2 h before the labelled polypeptide was added for overnight incubation. After incubation, the mixture was loaded to run SDS polyacrylamide gel electrophoresis (SDS-PAGE). The fluorescent image of the gel was captured using FluorChem FC2 imaging system (Alpha Innotech). The image was further analyzed by ImageJ [61].
4.7. Detection of cell surface pOVA by flow cytometry assay
1×105 DC2.4 or B16-F10 cells were pulsed with 5 μM cleaved-vaccines for 2 h or incubated with 5 μM intact vaccines for overnight in FBS free medium. The vaccine concentration is 5 uM. The cells were collected and stained with PE anti-mouse H-2Kb/SIINFEKL antibody (clone: 25-D1.16, BioLegend) and 4′,6-diamidino-2-phenylindole (DAPI). Cells were then analyzed by flow cytometry using BD FACS Canto (BD Biosciences) to quantify the pOVA epitopes bound to MHC class I on cell surface. 50,000 events were collected for analysis using FlowJo software. The results were shown as median fluorescence intensity (MFI) relative to the MFI of control cells.
4.8. Detection of cell surface pOVA by B3Z cell activation assay
DC2.4 and B16-F10 cells (75,000 cells/well) were pulsed with the cleaved-vaccines for 2 h or incubated with the intact vaccines for overnight in FBS free medium. The medium was then removed and 75,000 B3Z cells/well were added to co-culture for overnight. Next, the medium was removed and lysis buffer (100 mM 2-mercaptoethanol, 9 mM MgCl2, 0.2% Triton X-100 and 0.15 mM chlorophenol red-β-D-galactopyranoside) was added to incubate at 37 °C for 4 h. Then the absorbance at 570 nm of each well was measured using the Infinite M1000 pro microplate reader (Tecan) with 635 nm as a referential wavelength. The extent of B3Z cell activation was indicated by the optical density at 570 nm (OD570). For the inhibition assay, DC2.4 culture supernatant (1×) was treated with 30 mM 1,10-Phenanthroline for 2 h before iTEP-sMMP-pOVA was added and incubated for overnight at 37 °C. B16-F10 cells were then pulsed with the mixture for 2 h. Next, B3Z cells were added to co-culture for overnight to check the cell activation as described above.
4.9. Detection of in vivo pOVA presentation by B3Z cell activation assay
B3Z assay was used to detect the pOVA epitopes presentation in vivo. Mice were injected with 1.5 nmol vaccines at each side of the tail base. 24 h later, spleen and draining LNs were collected and prepared to single cell suspension. After lysis of red blood cells, DCs were separated from the cell suspension using the OptiPrep (Axis-Shield) density barrier centrifugation as described before [62]. The enriched DCs were plated with 2-time series dilution as indicated in the figures. 1×105/well B3Z cells were added to coculture with DCs for overnight. The activation of B3Z cells was examined as mentioned above to determine the pOVA presentation on DC surface.
4.10. Depletion of cross-presenting DCs in vivo by cytochrome c (CytC)
Mice were subcutaneously injected with 5 mg CytC (Sigma-Aldrich) on each side of the tail base every 12 h for 4 injections in total. 24 h after the last injection, cell suspensions from the spleen and draining LNs were prepared to enrich DCs as described above. The enriched DCs were then stained with PerCP anti-mouse CD11c (clone: N418, BioLegend) and FITC anti-mouse CD8α (clone: 53–6.7, BioLegend) to determine the fraction of CD8+ DCs by flow cytometry. To detect the pOVA epitopes presentation in mice that were treated with CytC, iTEP-sMMP-pOVA was injected subcutaneously at one hour after the last injection of CytC. 24 h later, DCs from the mouse spleen and draining LNs were collected to do B3Z activation assay as described above.
4.11. Size exclusion chromatography (SEC)
30 μM mouse serum albumin (MSA) was incubated with 30, 60 and 120 μM ABD-iTEP-sMMP-pOVA or ABD-iTEP-sMMP-pTRP2 at 4 °C for overnight. 100 μ L of the mixture was then taken to elute through the size exclusion column (diameter: 9.4 mm, length: 250 mm, particle diameter: 6.0 μm) equipped in an Agilent 1260 infinity liquid chromatography system (Agilent Technologies). The samples were eluted at a flow rate of 1 mL min−1 using 1× PBS. Absorbance spectra of each sample was monitored at 280 nm.
4.12. Surface plasmon resonance (SPR) assay
The SPR assay was conducted by using the MASS-1 SPR instrument (Sierra Sensors). MSA was immobilized on a SPR affinity sensor (High Capacity Amine, Sierra Sensors). Various concentrations of ABD-iTEP-sMMP-pTRP2 and iTEP-sMMP-pTRP2 were injected at a constant flow rate of 25 μL min−1 for 3 min. The running buffer (10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.05% Tween-20) was then injected and flowed for 10 min. At the end of each cycle, the surface of the sensor was regenerated with two injections of 25 μL of 10 mM HCl. The one-to-one Langmuir binding model with mass transport limitations was used to calculate the overall binding affinity (KD) between ABD-iTEP-sMMP-pTRP2 and MSA with the MASS-1 analysis software (Sierra Sensors).
4.13. Lymph node accumulation study
ABD-iTEP-sMMP-pTRP2 and iTEP-sMMP-pTRP2 were labelled with Alexa Fluor 488 5-SDP ester (Invitrogen) following the manufacturer’s protocol. 1.5 nmol labelled ABD-iTEP-sMMP-pTRP2 or iTEP-sMMP-pTRP2 was subcutaneously injected at each side of the tail base of the mice. At 6, 12, 24 and 48 h after injection, inguinal and axillary LNs were collected. Next, the LNs were homogenized and sonicated in PBS. The samples were then centrifuged and the supernatant were collected to measure the fluorescent intensity (excitation 494 nm, emission 518 nm). Based on the standard curves, the fluorescent intensity was converted to the amount of ABD-iTEP-sMMP-pTRP2 and iTEP-sMMP-pTRP2. The accumulation of ABD-iTEP-sMMP-pTRP2 and iTEP-sMMP-pTRP2 in LNs was expressed as percentage of injected dose, %ID.
4.14. Lymph node imaging
At 6 h after injection of ABD-iTEP-sMMP-pTRP2 and iTEP-sMMP-pTRP2, inguinal and axillary LNs were collected from the mice and imaged (excitation 465 nm, emission 520 nm, exposure 20 seconds) using the IVIS Spectrum (Caliper Life Sciences).
4.15. IFN-γ enzyme-linked immunospot (ELISPOT) assay
C57BL/6 mice were immunized with 1.5 nmol vaccines at each side of the tail base on day 0 and boosted on day 7. At day 17, mice were sacrificed and splenocytes were collected to perform IFN-γ ELISPOT assay. Splenocytes were cultured in complete RPMI medium (RPMI medium supplemented with 10% FBS, 2 mM L-glutamine, 50 μM 2-mercaptoethanol, 100 U Ml−1 penicillin and 100 μg mL−1 streptomycin) containing 10 μg mL−1 pTRP2 peptide (sequence: SVYDFFVWL) for 48 h. 2×105 cells were then seeded to the filter plate (Millipore) coated with anti-mouse IFN-γ antibody (clone: R4–6A2, Biolegend) and cultured for 24 h. The cells were then removed and the bound IFN-γ was detected by adding biotinylated anti-mouse IFN-γ antibody (clone: XMG1.2, Biolegend). Then horseradish peroxidase avidin (Avidin-HRP) and 3-amino-9-ethylcarbazole (AEC) were added to visualize the spots. The spots on the plate membrane were scanned and counted by ImageJ [61].
4.16. In vivo CTL-mediated specific killing assay
The assay was designed according to published methods with some modifications [63, 64]. C57BL/6 mice were immunized with 1.5 nmol ABD-iTEP-sMMP-pTRP2 or pTRP2 and 10 μg MPLA at each side of the tail base on day 0 and boosted on day 7. At day 17, splenocytes from naïve C57BL/6 mice were collected. The spelnocytes were splited into two populations: one was pulsed with 10 μM pTRP2 peptide at 37 °C for 2 h; the other one was not pulsed. The pulsed and non-pulsed cells were then labeled with 5 μM (CFSEhigh) and 0.5 μM (CFSElow) CFSE, respectively, at 37 °C for 20 min. Next, the pulsed and non-pulsed cells were mixed at 1:1 ratio. Last, 1×107 of the mixed cells were intravenously injected into one naïve and two groups of immunized mice. At 16 h after injection, mice were sacrificed and splenocytes were collected from these mice. The splenocytes were then analyzed by flow cytometry until 1×104 CFSElow cells were counted. The number of CFSEhigh and CFSElow cells among the analyzed splenocytes was determined by FlowJo software. The pTRP2-specific killing (%) was calculated using the following equation.
4.17. Statistical analysis
Two-tailed, unpaired Student’s t test and one-way analysis of variance (ANOVA) with the Bonferroni post-test were applied to do statistical analysis of the data. A significant difference was considered when P < 0.05.
Supplementary Material
Acknowledgements
We appreciate Kenneth Rock (University of Massachusetts) and Nilabh Shastri (University of California, Berkeley) for providing us the DC2.4 cell line and B3Z cell line, receptively. We thank Xiaomin Wang and Hu Dai for their assistance in mice study. We acknowledge Michael Kay and Debbie Eckert for helping us with the SPR assay. We also recognize the Core Facility of University of Utah for serving us with flow cytometry and IVIS imaging. This work was primarily supported by the University of Utah Start-up Fund, a Huntsman Cancer Institute Pilot Grant [Grant number 170301], and partially by a NIH grant [Grant number R21EB024083] to Mingnan Chen.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability
The raw/processed data required to reproduce these findings are available to download from [https://data.mendeley.com/datasets/b2cx5n27pv/1].
Competing interests
The authors declare no competing financial interests.
References
- [1].Gilbert SC, T-cell-inducing vaccines - what’s the future, Immunology 135(1) (2012) 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Appay V, Douek DC, Price DA, CD8+ T cell efficacy in vaccination and disease, Nat Med 14(6) (2008) 623–8. [DOI] [PubMed] [Google Scholar]
- [3].Nagata T, Koide Y, Induction of Specific CD8 T Cells against Intracellular Bacteria by CD8 T-Cell-Oriented Immunization Approaches, J Biomed Biotechnol 2010 (2010) 764542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Dermime S, Armstrong A, Hawkins RE, Stern PL, Cancer vaccines and immunotherapy, Br Med Bull 62 (2002) 149–62. [DOI] [PubMed] [Google Scholar]
- [5].Saslow D, Castle PE, Cox JT, Davey DD, Einstein MH, Ferris DG, Goldie SJ, Harper DM, Kinney W, Moscicki AB, Noller KL, Wheeler CM, Ades T, Andrews KS, Doroshenk MK, Kahn KG, Schmidt C, Shafey O, Smith RA, Partridge EE, Gynecologic Cancer Advisory G, Garcia F, American Cancer Society Guideline for human papillomavirus (HPV) vaccine use to prevent cervical cancer and its precursors, CA Cancer J Clin 57(1) (2007) 7–28. [DOI] [PubMed] [Google Scholar]
- [6].Centers for Disease C, Recommendation of the Immunization Practices Advisory Committee (ACIP). Inactivated hepatitis B virus vaccine, MMWR Morb Mortal Wkly Rep 31(24) (1982) 317–22, 327–8. [PubMed] [Google Scholar]
- [7].Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF, Redfern CH, Ferrari AC, Dreicer R, Sims RB, Xu Y, Frohlich MW, Schellhammer PF Sipuleucel-T Immunotherapy for Castration-Resistant Prostate Cancer, New Engl J Med 363(5) (2010) 411–422. [DOI] [PubMed] [Google Scholar]
- [8].Dallal RM, Lotze MT, The dendritic cell and human cancer vaccines, Curr Opin Immunol 12(5) (2000) 583–8. [DOI] [PubMed] [Google Scholar]
- [9].Brossart P, Wirths S, Stuhler G, Reichardt VL, Kanz L, Brugger W, Induction of cytotoxic T-lymphocyte responses in vivo after vaccinations with peptide-pulsed dendritic cells, Blood 96(9) (2000) 3102–8. [PubMed] [Google Scholar]
- [10].Yang HG, Hu BL, Xiao L, Wang P, Dendritic cell-directed lentivector vaccine induces antigen-specific immune responses against murine melanoma, Cancer Gene Ther 18(5) (2011) 370–80. [DOI] [PubMed] [Google Scholar]
- [11].O’Neill D, Bhardwaj N, Generation of autologous peptide- and protein-pulsed dendritic cells for patient-specific immunotherapy, Methods Mol Med 109 (2005) 97–112. [PubMed] [Google Scholar]
- [12].Prins RM, Wang X, Soto H, Young E, Lisiero DN, Fong B, Everson R, Yong WH, Lai A, Li G, Cloughesy TF, Liau LM, Comparison of glioma-associated antigen peptide-loaded versus autologous tumor lysate-loaded dendritic cell vaccination in malignant glioma patients, J Immunother 36(2) (2013) 152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Salcedo M, Bercovici N, Taylor R, Vereecken P, Massicard S, Duriau D, Vernel-Pauillac F, Boyer A, Baron-Bodo V, Mallard E, Bartholeyns J, Goxe B, Latour N, Leroy S, Prigent D, Martiat P, Sales F, Laporte M, Bruyns C, Romet-Lemonne JL, Abastado JP, Lehmann F, Velu T, Vaccination of melanoma patients using dendritic cells loaded with an allogeneic tumor cell lysate, Cancer Immunol Immunother 55(7) (2006) 819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sabado RL, Bhardwaj N, Dendritic cell immunotherapy, Ann N Y Acad Sci 1284 (2013) 31–45. [DOI] [PubMed] [Google Scholar]
- [15].Palucka K, Banchereau J, Dendritic-cell-based therapeutic cancer vaccines, Immunity 39(1) (2013) 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Maecker HT, Ghanekar SA, Suni MA, He XS, Picker LJ, Maino VC, Factors affecting the efficiency of CD8+ T cell cross-priming with exogenous antigens, J Immunol 166(12) (2001) 7268–75. [DOI] [PubMed] [Google Scholar]
- [17].Otahal P, Hutchinson SC, Mylin LM, Tevethia MJ, Tevethia SS, Schell TD, Inefficient cross-presentation limits the CD8+ T cell response to a subdominant tumor antigen epitope, J Immunol 175(2) (2005) 700–12. [DOI] [PubMed] [Google Scholar]
- [18].Tel J, Aarntzen EH, Baba T, Schreibelt G, Schulte BM, Benitez-Ribas D, Boerman OC, Croockewit S, Oyen WJ, van Rossum M, Winkels G, Coulie PG, Punt CJ, Figdor CG, de Vries IJ, Natural human plasmacytoid dendritic cells induce antigen-specific T-cell responses in melanoma patients, Cancer Res 73(3) (2013) 1063–75. [DOI] [PubMed] [Google Scholar]
- [19].Anguille S, Smits EL, Lion E, van Tendeloo VF, Berneman ZN, Clinical use of dendritic cells for cancer therapy, Lancet Oncol 15(7) (2014) E257–E267. [DOI] [PubMed] [Google Scholar]
- [20].Garg AD, Vandenberk L, Koks C, Verschuere T, Boon L, Van Gool SW, Agostinis P, Dendritic cell vaccines based on immunogenic cell death elicit danger signals and T cell-driven rejection of high-grade glioma, Sci Transl Med 8(328) (2016) 328ra27. [DOI] [PubMed] [Google Scholar]
- [21].Sabado RL, Bhardwaj N, Cancer immunotherapy: dendritic-cell vaccines on the move, Nature 519(7543) (2015) 300–1. [DOI] [PubMed] [Google Scholar]
- [22].Chambers JD, Neumann PJ, Listening to Provenge - What a Costly Cancer Treatment Says about Future Medicare Policy, New Engl J Med 364(18) (2011) 1687–1689. [DOI] [PubMed] [Google Scholar]
- [23].Anassi E, Ndefo UA, Sipuleucel-T (provenge) injection: the first immunotherapy agent (vaccine) for hormone-refractory prostate cancer, P T 36(4) (2011) 197–202. [PMC free article] [PubMed] [Google Scholar]
- [24].Liu Y, Xiao L, Joo KI, Hu B, Fang J, Wang P, In situ modulation of dendritic cells by injectable thermosensitive hydrogels for cancer vaccines in mice, Biomacromolecules 15(10) (2014) 3836–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zehn D, Cohen CJ, Reiter Y, Walden P, Efficiency of peptide presentation by dendritic cells compared with other cell types: implications for cross-priming, Int Immunol 18(12) (2006) 1647–54. [DOI] [PubMed] [Google Scholar]
- [26].Dong S, Wang P, Zhao P, Chen M, Direct Loading of iTEP-Delivered CTL Epitope onto MHC Class I Complexes on the Dendritic Cell Surface, Mol Pharm 14(10) (2017) 3312–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Randolph GJ, Angeli V, Swartz MA, Dendritic-cell trafficking to lymph nodes through lymphatic vessels, Nat Rev Immunol 5(8) (2005) 617–28. [DOI] [PubMed] [Google Scholar]
- [28].Martin-Fontecha A, Lanzavecchia A, Sallusto F, Dendritic cell migration to peripheral lymph nodes, Handb Exp Pharmacol (188) (2009) 31–49. [DOI] [PubMed] [Google Scholar]
- [29].Harrell MI, Iritani BM, Ruddell A, Lymph node mapping in the mouse, J Immunol Methods 332(1–2) (2008) 170–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Davies-Venn CA, Angermiller B, Wilganowski N, Ghosh P, Harvey BR, Wu G, Kwon S, Aldrich MB, Sevick-Muraca EM, Albumin-binding domain conjugate for near-infrared fluorescence lymphatic imaging, Mol Imaging Biol 14(3) (2012) 301–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang Y, Lang L, Huang P, Wang Z, Jacobson O, Kiesewetter DO, Ali IU, Teng G, Niu G, Chen X, In vivo albumin labeling and lymphatic imaging, Proc Natl Acad Sci U S A 112(1) (2015) 208–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu H, Moynihan KD, Zheng Y, Szeto GL, Li AV, Huang B, Van Egeren DS, Park C, Irvine DJ, Structure-based programming of lymph-node targeting in molecular vaccines, Nature 507(7493) (2014) 519–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhu G, Lynn GM, Jacobson O, Chen K, Liu Y, Zhang H, Ma Y, Zhang F, Tian R, Ni Q, Cheng S, Wang Z, Lu N, Yung BC, Wang Z, Lang L, Fu X, Jin A, Weiss ID, Vishwasrao H, Niu G, Shroff H, Klinman DM, Seder RA, Chen X, Albumin/vaccine nanocomplexes that assemble in vivo for combination cancer immunotherapy, Nat Commun 8(1) (2017) 1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Levy OE, Jodka CM, Ren SS, Mamedova L, Sharma A, Samant M, D’Souza LJ, Soares CJ, Yuskin DR, Jin LJ, Parkes DG, Tatarkiewicz K, Ghosh SS, Novel exenatide analogs with peptidic albumin binding domains: potent anti-diabetic agents with extended duration of action, PLoS One 9(2) (2014) e87704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Li R, Yang H, Jia D, Nie Q, Cai H, Fan Q, Wan L, Li L, Lu X, Fusion to an albumin-binding domain with a high affinity for albumin extends the circulatory half-life and enhances the in vivo antitumor effects of human TRAIL, J Control Release 228 (2016) 96–106. [DOI] [PubMed] [Google Scholar]
- [36].Stromberg P, Berglund M, Ekholm C, Su C, Sterky C, Edlund PO, Berghard L, Wiken M, Selen G, Nyhem CS, Development of Affibody (R) C5 inhibitors for versatile and efficient therapeutic targeting of the terminal complement pathway, Mol Immunol 61(2) (2014) 256–256. [Google Scholar]
- [37].Wang P, Zhao P, Dong S, Xu T, He X, Chen M, An Albumin-binding Polypeptide Both Targets Cytotoxic T Lymphocyte Vaccines to Lymph Nodes and Boosts Vaccine Presentation by Dendritic Cells, Theranostics 8(1) (2018) 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Phuphanich S, Wheeler CJ, Rudnick JD, Mazer M, Wang H, Nuno MA, Richardson JE, Fan X, Ji J, Chu RM, Bender JG, Hawkins ES, Patil CG, Black KL, Yu JS, Phase I trial of a multi-epitope-pulsed dendritic cell vaccine for patients with newly diagnosed glioblastoma, Cancer Immunol Immunother 62(1) (2013) 125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Rosenberg SA, Yang JC, Restifo NP, Cancer immunotherapy: moving beyond current vaccines, Nat Med 10(9) (2004) 909–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cho S, Dong S, Parent K, Chen M, Immune-tolerant elastin-like polypeptides (iTEPs) and their application as CTL vaccine carriers, Journal of drug targeting 24(4) (2016) 328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hu J, Zhang X, Nothnick WB, Spencer TE, Matrix metalloproteinases and their tissue inhibitors in the developing neonatal mouse uterus, Biol Reprod 71(5) (2004) 1598–604. [DOI] [PubMed] [Google Scholar]
- [42].Karttunen J, Sanderson S, Shastri N, Detection of rare antigen-presenting cells by the lacZ T-cell activation assay suggests an expression cloning strategy for T-cell antigens, Proc Natl Acad Sci U S A 89(13) (1992) 6020–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ghanekar SA, Nomura LE, Suni MA, Picker LJ, Maecker HT, Maino VC, Gamma Interferon Expression in CD8(+) T Cells Is a Marker for Circulating Cytotoxic T Lymphocytes That Recognize an HLA A2-Restricted Epitope of Human Cytomegalovirus Phosphoprotein pp65, Clinical and Diagnostic Laboratory Immunology 8(3) (2001) 628–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].den Haan JM, Lehar SM, Bevan MJ, CD8(+) but not CD8(−) dendritic cells cross-prime cytotoxic T cells in vivo, J Exp Med 192(12) (2000) 1685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lin ML, Zhan Y, Proietto AI, Prato S, Wu L, Heath WR, Villadangos JA, Lew AM, Selective suicide of cross-presenting CD8+ dendritic cells by cytochrome c injection shows functional heterogeneity within this subset, Proc Natl Acad Sci U S A 105(8) (2008) 3029–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Slingluff CL Jr., The present and future of peptide vaccines for cancer: single or multiple, long or short, alone or in combination?, Cancer J 17(5) (2011) 343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Li W, Joshi MD, Singhania S, Ramsey KH, Murthy AK, Peptide Vaccine: Progress and Challenges, Vaccines (Basel) 2(3) (2014) 515–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Casella CR, Mitchell TC, Putting endotoxin to work for us: monophosphoryl lipid A as a safe and effective vaccine adjuvant, Cell Mol Life Sci 65(20) (2008) 3231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Madan RA, Gulley JL, Sipuleucel-T: harbinger of a new age of therapeutics for prostate cancer, Expert Rev Vaccines 10(2) (2011) 141–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hammerstrom AE, Cauley DH, Atkinson BJ, Sharma P, Cancer immunotherapy: sipuleucel-T and beyond, Pharmacotherapy 31(8) (2011) 813–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wagner CS, Grotzke JE, Cresswell P, Intracellular events regulating cross-presentation, Front Immunol 3 (2012) 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Xu Z, Ramishetti S, Tseng YC, Guo S, Wang Y, Huang L, Multifunctional nanoparticles co-delivering Trp2 peptide and CpG adjuvant induce potent cytotoxic T-lymphocyte response against melanoma and its lung metastasis, J Control Release 172(1) (2013) 259–65. [DOI] [PubMed] [Google Scholar]
- [53].Keller S, Wilson JT, Patilea GI, Kern HB, Convertine AJ, Stayton PS, Neutral polymer micelle carriers with pH-responsive, endosome-releasing activity modulate antigen trafficking to enhance CD8(+) T cell responses, J Control Release 191 (2014) 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yan S, Gu W, Zhang B, Rolfe BE, Xu ZP, High adjuvant activity of layered double hydroxide nanoparticles and nanosheets in anti-tumour vaccine formulations, Dalton Trans 47(9) (2018) 2956–2964. [DOI] [PubMed] [Google Scholar]
- [55].Van Kaer L, Ashton-Rickardt PG, Ploegh HL, Tonegawa S, TAP1 mutant mice are deficient in antigen presentation, surface class I molecules, and CD4–8+ T cells, Cell 71(7) (1992) 1205–14. [DOI] [PubMed] [Google Scholar]
- [56].Kretzer NM, Theisen DJ, Tussiwand R, Briseno CG, Grajales-Reyes GE, Wu X, Durai V, Albring J, Bagadia P, Murphy TL, Murphy KM, RAB43 facilitates cross-presentation of cell-associated antigens by CD8alpha+ dendritic cells, J Exp Med 213(13) (2016) 2871–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Dong S, Xu T, Wang P, Zhao P, Chen M, Engineering of a self-adjuvanted iTEP-delivered CTL vaccine, Acta Pharmacol Sin 38(6) (2017) 914–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Liu S, Tobias R, McClure S, Styba G, Shi Q, Jackowski G, Removal of endotoxin from recombinant protein preparations, Clin Biochem 30(6) (1997) 455–63. [DOI] [PubMed] [Google Scholar]
- [59].Madaan A, Verma R, Singh AT, Jain SK, Jaggi M, A stepwise procedure for isolation of murine bone marrow and generation of dendritic cells, Journal of Biological Methods 1(1) (2014) e1. [Google Scholar]
- [60].Frankowski H, Gu Y-H, Heo JH, Milner R, del Zoppo GJ, Use of Gel Zymography to Examine Matrix Metalloproteinase (Gelatinase) Expression in Brain Tissue or in Primary Glial Cultures, Methods in molecular biology (Clifton, N.J.) 814 (2012) 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis, Nat Methods 9(7) (2012) 671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Markey KA, Gartlan KH, Kuns RD, MacDonald KP, Hill GR, Imaging the immunological synapse between dendritic cells and T cells, J Immunol Methods 423 (2015) 40–4. [DOI] [PubMed] [Google Scholar]
- [63].Schliehe C, Redaelli C, Engelhardt S, Fehlings M, Mueller M, van Rooijen N, Thiry M, Hildner K, Weller H, Groettrup M, CD8-dendritic cells and macrophages cross-present poly(D,L-lactate-co-glycolate) acid microsphere-encapsulated antigen in vivo, J Immunol 187(5) (2011) 2112–21. [DOI] [PubMed] [Google Scholar]
- [64].Xu ZH, Ramishetti S, Tseng YC, Guo ST, Wang YH, Huang L, Multifunctional nanoparticles co-delivering Trp2 peptide and CpG adjuvant induce potent cytotoxic T-lymphocyte response against melanoma and its lung metastasis, Journal of Controlled Release 172(1) (2013) 259–265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.