Abstract
The F element of the Drosophila karyotype (the fourth chromosome in Drosophila melanogaster) is often referred to as the “dot chromosome” because of its appearance in a metaphase chromosome spread. This chromosome is distinct from other Drosophila autosomes in possessing both a high level of repetitious sequences (in particular, remnants of transposable elements) and a gene density similar to that found in the other chromosome arms, ∼80 genes distributed throughout its 1.3-Mb “long arm.” The dot chromosome is notorious for its lack of recombination and is often neglected as a consequence. This and other features suggest that the F element is packaged as heterochromatin throughout. F element genes have distinct characteristics (e.g., low codon bias, and larger size due both to larger introns and an increased number of exons), but exhibit expression levels comparable to genes found in euchromatin. Mapping experiments show the presence of appropriate chromatin modifications for the formation of DNaseI hypersensitive sites and transcript initiation at the 5′ ends of active genes, but, in most cases, high levels of heterochromatin proteins are observed over the body of these genes. These various features raise many interesting questions about the relationships of chromatin structures with gene and chromosome function. The apparent evolution of the F element as an autosome from an ancestral sex chromosome also raises intriguing questions. The findings argue that the F element is a unique chromosome that occupies its own space in the nucleus. Further study of the F element should provide new insights into chromosome structure and function.
Keywords: fourth chromosome, F element, heterochromatin, HP1a, chromosome evolution, FlyBook
THE dot chromosome, or F element, of Drosophila melanogaster is unique in that it has many characteristics of heterochromatin, yet maintains a typical euchromatic gene density in its 1.3-Mb long arm. The basic karyotype of Dipterans consists of six distinct genomic elements, named A through F by Muller (1940). Five of the six Muller elements are large telocentric chromosomes or metacentric chromosome arms, while the sixth element, the F element, is typically represented by a small, dot-like chromosome (Figure 1). The five large Muller elements in D. melanogaster represent the X chromosome (A), chromosome 2 (2L = B; 2R = C), and chromosome 3 (3L = D; 3R = E), while chromosome 4 corresponds to the small F element (Muller 1940). Despite its dot-like appearance, two short arms can be discerned in mitotic chromosome spreads for chromosome 4 (Hochman 1976), only one of which is amplified in polytene chromosomes (Figure 1). The discussion here focuses on this right arm of chromosome 4 of D. melanogaster—and the homologous regions in other species—as the remainder of the chromosome consists of highly repetitive sequences that are largely uncharacterized. (For details on the terminology used, see Box 1.)
Figure 1.
D. melanogaster chromosome 4 is enriched for HP1a and POF. Immunohistochemical analysis of polytene chromosomes from the third instar larval salivary gland shows the genomic distribution of HP1a and POF. (A) D. melanogaster karyotype indicating the chromosome configuration and the six Muller elements A–F. Regions of constitutive heterochromatin are shown in dark grey. (B) Phase contrast image of a polytene chromosome spread from the salivary glands of a third instar larva (D. melanogaster). The chromosome arms have gone through ∼10 rounds of endoreduplication, while the pericentric heterochromatin is underreplicated and fuses in a common chromocenter. The rectangle marks the region containing chromosome 4 enlarged in (C–F). (C) Close-up of the phase contrast image. (D) Close-up showing staining with the monoclonal antibody C1A9 against HP1a (secondary goat anti-mouse-Alexa488 antibody); the pericentric heterochromatin and chromosome 4 are stained. (D) Close-up showing staining with the antibody MO459 against POF (secondary goat anti-rabbit-Alexa594 antibody); only chromosome 4 is stained. (F) Close-up merged image showing HP1a and POF signals superimposed. HP1a=green; POF=red. The chromosome squashing and staining protocol used is that described by Stephens and colleagues (Stephens et al. 2004).
Box 1. Glossary.
Chromosome 4
Used in reference to the fourth chromosome of D. melanogaster (see Figure 1). In D. melanogaster, chromosome 4 is the smallest chromosome and it appears as a “dot” in metaphase spreads due to its small size.
F element
In the genus Drosophila, the karyotype is conserved and there are typically five rod-shaped elements, as well as a small, dot-shaped chromosome. The gene content of these six elements is relatively constant across species; most of the same genes (∼90%) are found together on a given element in all Drosophila. However, there are a significant number of fusion events between these six basic elements within Drosophila, resulting in chromosome numbering systems that do not carry across species in many cases. Thus, the Muller labeling system, where each rod-shaped element is identified by a letter (A–F), is used, with D. melanogaster chromosome 4 receiving the F designation (see Figure 1). The F element does not appear as a dot in all species, but consistently identifies the chromosomal element containing the same group of genes (homologous to the D. melanogaster chromosome 4) in all Drosophila species.
Dot chromosome
An alternative name for the F element based on chromosome appearance. Due to its small nature and appearance as a dot in a metaphase spread, the term dot chromosome was applied to the D. melanogaster chromosome 4 and its homologous chromosomes in many other Drosophila species. However, the F element is not always a dot, as shown most strikingly in the case of D. ananassae.
Genomic domain
Portion of the genome that is characterized by a specific set of functional characteristics. Examples of genomic domains are regions of the genome under the control of the Polycomb system (Polycomb domains) or topologically associated domains (TADs) defined based on Hi-C data. The F element is considered a distinct genomic domain due to its high repeat content throughout the chromosome arm that contains the genes, its association with biochemical marks of heterochromatin across this region, and its association with POF, characteristics that together distinguish it from other regions of the genome.
Since the early days of Drosophila research, the dot chromosome has been a subject of interest due to its unusual size. Since then, results from genetic analyses, genomic studies, and biochemical investigations have revealed the dot chromosome to be unique, having a mixture of characteristics of euchromatin and of constitutive heterochromatin. The dot chromosome stands apart from the other Muller elements due to differences in sequence composition, biochemical make-up, and evolutionary history. These differences lead to a variety of emergent properties that distinguish the F element from the rest of the genome and broaden our view of functional chromosome organization. Here, we review the collective data supporting the unique status of the F element in Dipterans, and suggest how this chromosome might inform our thinking about the role of chromatin structure in gene function and the evolution of eukaryotic genomes.
The Dot Chromosome has a Very Low Incidence of Recombination, but Higher than Average Levels of Inversion
One of the first observations hinting at the unique nature of the dot chromosome came from genetic studies, as early fly geneticists quickly noticed that recombination levels on chromosome 4 were unusual. For example, in 1951, Sturtevant begins his article presenting a genetic map for the D. melanogaster chromosome 4 by stating that “Under ordinary conditions there is so little crossing over in the fourth chromosome of Drosophila melanogaster that the usual method of constructing a map is not practicable” (Sturtevant 1951). Since then, this lack of recombination for chromosome 4 in wild-type animals has been confirmed by several laboratories. For example, Sandler and Szauter examined ∼30,000 mitoses without finding evidence of recombination (Sandler and Szauter 1978), and McMahan and colleagues investigated 1,285,000 progeny from a reporter assay and found no evidence of recombination at the 102D site (McMahan et al. 2013). More recently, Hatkevich and colleagues examined 3112 progeny for recombination between ci and sv on chromosome 4 without recovering any recombinants, while a parallel experiment looking for recombination in a stretch of pericentric heterochromatin on chromosome 2L revealed eight recombination events among 7399 animals (Hartmann and Sekelsky 2017; Hatkevich et al. 2017). Thus, despite significant efforts, no recombination events on chromosome 4 of D. melanogaster have been observed under standard laboratory conditions in wild-type animals, illustrating the unusual nature of the dot chromosome.
Evidence for a lack, or at least a lower level, of recombination on the F element has also been collected in additional Drosophila species. Chino and Kikkawa (1933) observe some recombination on the D. virilis F element, between 0 and 1% depending on rearing temperature, but this recombination level is much lower than what is observed on the other Muller elements, in this study 22–54% for the B element (Chino and Kikkawa 1933). More recent analyses of genome sequence data reveal that, like the D. melanogaster chromosome 4, the F elements in D. virilis, D. erecta, D. mojavensis, D. grimshawi, and D. ananassae show evidence of low recombination; genes on these F elements show larger size, more introns, and less codon bias than genes in other genomic regions (Leung et al. 2010, 2015, 2017). These characteristics are exaggerated in D. ananassae, where the gene-containing region of the F element has expanded to at least 18.7 Mb in size (Schaeffer et al. 2008; Leung et al. 2017). Lower levels of codon bias were also reported for F element genes in the additional Drosophila species sequenced by the Drosophila 12 Genomes Consortium (Drosophila 12 Genomes Consortium et al. 2007; Vicario et al. 2007). These findings indicate that selection is less effective on the F element in general, implying that the lack of recombination is a general feature of F elements in the Drosophilids. Presumably as a consequence, F element genes show lower levels of polymorphism than the genome average (Berry et al. 1991; Arguello et al. 2010; Campos et al. 2014).
Studies exploring translocations of chromosome 4 sequences in D. melanogaster, as well as studies exploiting “natural experiments” where the F element has fused with another Muller element in a given species, have shed additional light on this lack of recombination. In D. melanogaster, recombination can occur when chromosome 4 sequences are translocated to other chromosome arms. For example, in a reciprocal X chromosome–chromosome 4 translocation, recombination was observed within the translocated chromosome 4 sequences between ey and sv (Osborne 1998). In contrast, no recombination was observed in the reciprocal translocation stock, which contains distal X chromosome sequences (∼2.7 Mb) attached to the chromosome 4 centromere (Osborne 1998). In D. willistoni and D. insularis, the F element is fused to the E element, with the F element genes located proximal to the centromere (Figure 4). Powell and colleagues have shown that in these fused F elements, linkage disequilibrium and recombination rates are similar to other genomic regions. There is a gradient in codon usage that likely reflects a gradient in recombination from the most centromere-proximal genes of the F element to the genes close to the fusion point with the E element; the steepness of the gradient suggests that an evolutionary equilibrium has not been reached in the ∼12 MY since the fusion occurred (Powell et al. 2011). These studies indicate that the unique lack of recombination observed for F elements can be impacted by rearrangements that presumably impact nuclear organization.
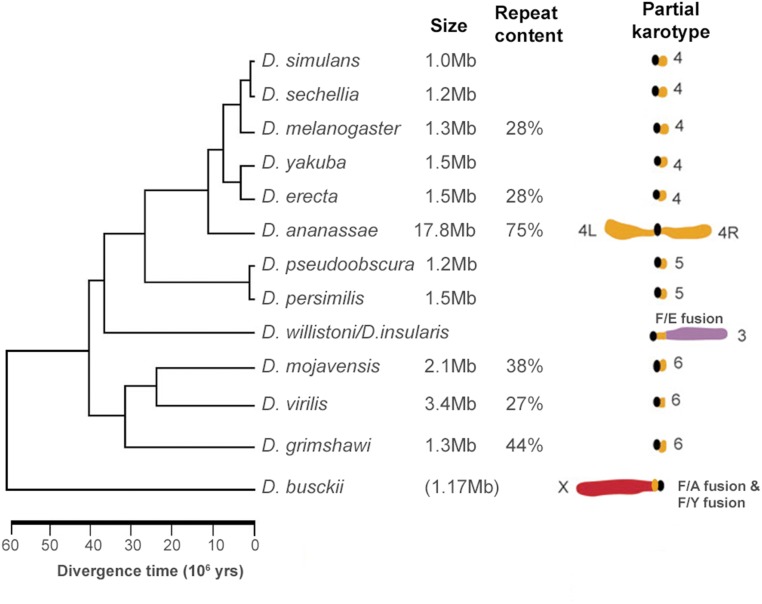
Figure 4.
Dot chromosome evolution within the Drosophilids illustrates the changeable nature of the chromosome, which nonetheless maintains common characteristics. Phylogenetic tree and chromosome diagram modified from Schaeffer and colleagues (Schaeffer et al. 2008) using additional information from Song et al. (2011) and Zhou and Bachtrog (2015). Chromosome size estimates of the F element arms (from the most proximal to most distal gene, not including the pericentric heterochromatin or telomeres) are from Schaeffer et al. (2008) and Zhou and Bachtrog (2015). Repeat content estimates for the F element gene-containing regions are derived from Leung et al. (2010, 2015, 2017). In the partial karyotype, fusions of the F element (orange) to other chromosomes are marked; in D. busckii, the F element is fused to the sex chromosomes (red), i.e., one copy of the F element fused to the Y chromosome (F/Y fusion), the other copy fused to the X chromosome, which is the A element in this species (F/A fusion). In D. willistoni, the F element is fused with the E element (purple).
One possibility that has been raised is the hypothesis that the lack of recombination is due to the “centromere effect,” a model based on the observation that recombination rates are low adjacent to the centromere for any chromosome (Beadle 1932). Recent research on the Bloom syndrome helicase (BLM) has revealed that recombination can occur on the D. melanogaster chromosome 4 in Blm mutants, where the centromere effect is absent (Hatkevich et al. 2017). As Blm mutants lose the general centromere effect (observed for all chromosomes), this supports suspicions that the lack of recombination on chromosome 4 is tied to that phenomenon in some way in D. melanogaster [recently reviewed in detail in Hartmann and Sekelsky (2017)]. But whether this implies a critical shift in chromatin structure, a change in nuclear compartmentalization, or other change is not known.
However, hypotheses that suggest that the lack of recombination is due solely to the F element’s small size, and hence the proximity of the gene-rich region to the centromere, appear to be ruled out by the results from D. ananassae. Despite the large size of the D. ananassae F element, these F element genes consistently show lower codon bias than genes on the D. ananassae D element. D. ananassae F element genes also show less optimal codon usage (as measured by the codon adaptation index) than their D. melanogaster orthologs, indicating a consistent lack of recombination despite the increasing distance from the centromere (Leung et al. 2017). Some further consideration, for example a unique biochemistry of DNA packaging or some aspect of nuclear organization, is needed to explain the low recombination rate on the F element. An intriguing question is whether the D. ananassae F element would respond to Blm mutations in the same way as reported for D. melanogaster (see above).
The F element also provides a contrast in the rates of inversion. While the rates of recombination are very low, the rates of inversion (as estimated by the size of syntenic blocks) are higher on the F element than the genome average. The syntenic block sizes of the D. mojavensis F element are ∼2.6× smaller than the genome average; for D. grimshawi, the comparable number is ∼2.3× smaller than the genome average (Leung et al. 2015). As syntenic block sizes are correlated with the rates of inversion, the results indicate that the F elements have a higher rate of inversions than the genomes as a whole. This finding argues (as does the normal levels of gene expression, discussed below) that the dot chromosome is not simply inaccessible to the multiprotein complexes used for processes such as recombination, but that a more complex model is needed to explain this feature.
The Dot Chromosome has Heterochromatic Characteristics
A salient characteristic of the dot chromosome that was noted early on is its heterochromatic nature. Initially, this assessment was based on genetic analyses and cytological studies of salivary gland polytene chromosomes, with biochemical and genomics analyses later confirming the finding. In 1935, Bridges reported that D. melanogaster chromosome 4 was regularly part of the heterochromatic chromocenter in polytene chromosome spreads from salivary glands (Bridges 1935) (see Figure 1). In 1947, L. V. Morgan noted that the visible eye phenotype imparted by spa, a recessive allele of sv located on chromosome 4, is dependent on the amount of heterochromatin present in the animals (Morgan 1947). This finding was interpreted as indicating that sv resides in a heterochromatic environment, given the contemporary studies of position-effect variegation (PEV, see Box 2). PEV had been linked clearly to heterochromatin formation, and variations in the amount of heterochromatin in the genome were known to impact phenotypic readouts. These examples illustrate that based on cytology and genetic analyses, early Drosophila researchers had classified the dot chromosome in D. melanogaster as heterochromatic.
Box 2. Position-Effect Variegation.
Chromosome rearrangements or transposition events that place euchromatic genes in cis to a breakpoint in heterochromatin can result in gene silencing [reviewed by Elgin and Reuter (2013)]. For genes that exhibit cell-autonomous expression, it can be seen that the silencing is mosaic, i.e., the gene is silenced in some, but not all, of the cells in which it is normally expressed, and so is described as “ever sporting” or variegating (Muller and Altenburg 1930). Extensive investigation has shown that this silencing is due to the spread of heterochromatin packaging in a stochastic fashion. Because the mutation has not altered the genes per se, but only their position in the genome, this phenotype is referred to as position-effect variegation. Expression of a variegating gene is impacted by the amount of heterochromatin in the genotype, presumably because of the titration of a fixed amount of key proteins required for heterochromatin formation.
As the Drosophila research toolkit expanded, additional findings supported this classification of the dot chromosome as heterochromatic. In situ hybridization studies revealed relatively high levels of repeated sequences in the polytenized portion of the D. melanogaster dot chromosome [for example see Strobel et al. (1979)], a characteristic of other classical heterochromatic regions such as centromeres and telomeres. The presence of high levels of repeats on chromosome 4, particularly remnants of transposable elements (TEs), was subsequently confirmed by whole-genome sequencing data (Adams et al. 2000; Drosophila 12 Genomes Consortium et al. 2007) (Figure 2). In D. melanogaster, the repeat level of the right arm of chromosome 4, at ∼30%, is lower than the repeat content of ∼70% seen in pericentric heterochromatin, but is significantly higher than the < 10% repeats seen in typical euchromatic regions of the genome (Drosophila 12 Genomes Consortium et al. 2007; Leung et al. 2010). While the exact repeat content is species-specific, high levels of repeated elements have been reported for F elements in other species (Drosophila 12 Genomes Consortium et al. 2007; Leung et al. 2010, 2015, 2017). Further, these repeats, particularly the TE remnants, are distributed within and between genes in a pattern similar to that seen in mammalian genomes, something not seen in the euchromatic long chromosome arms of Drosophila genomes. In D. melanogaster, the 1360 element (a DNA transposon remnant) plays an important, but not exclusive, role in maintaining the heterochromatic state of the F element (Sun et al. 2004; Riddle et al. 2008). Other TEs are more prominent in other Drosophila species; for example, LTR and LINE retrotransposons have played a prominent role in the expansion of the F element in D. ananassae. The DINE-1 element, first described in D. melanogaster (Locke et al. 1999a), is a prominent miniature inverted-repeat TE found in all Drosophila species examined (Yang and Barbash 2008). It is largely restricted to the F element and pericentric heterochromatin, and could play a role in driving heterochromatin formation (Leung et al. 2015). When antibodies to proteins characteristic of heterochromatin (e.g., HP1a and histone 3 modified by methylation at lysine 9) became available, dot chromosomes were confirmed to show high levels of these biochemical hallmarks of heterochromatin as well (James and Elgin 1986; James et al. 1989) (see Figure 1 and below for further discussion). The results from studies across species [e.g., Leung et al. (2015)] indicate that the heterochromatic nature of the dot chromosome is a conserved feature in the Drosophilids.
Figure 2.
Map illustrating the interspersed position-effect variegation-inducing (heterochromatic) and permissive domains on D. melanogaster chromosome 4. Hsp70-white reporter insertion sites are shown above the chromosome 4 map [diagram based on Riddle et al. (2008) with modifications]. Insertion sites that confer a red eye phenotype are indicated by red triangles, while insertion sites that confer a variegating eye phenotype are marked by dotted triangles. Above the map, two examples each of variegating and red eye phenotypes are shown. Beneath the chromosome map, the locations of transposable elements (TEs) and of genes are shown [FlyBase FB2018_02 (Gramates et al. 2017)]. The bottom track shows the distribution of the nine chromatin states (as defined by Kharchenko and colleagues) across chromosome 4 from BG3 cells (Kharchenko et al. 2011). State 1, enriched in H3K4me3 and other marks of active transcription, is in red; state 6, enriched in H3K27me3 and associated with Polycomb, is in dark gray; and states 7 and 8, enriched in H3K9me2/3 and associated with HP1a (heterochromatin), are in dark blue and light blue, respectively. ID, identifier.
An additional feature of the dot chromosome that supports its classification as heterochromatic was discovered in analyses of replication timing. A generally accepted attribute of heterochromatin, well documented in flies, is its tendency to replicate late in S phase (Lima-de-Faria 1959a,b; Spofford 1976; Allshire and Madhani 2018). In detailed studies of replication timing in D. melanogaster, chromosome 4 was found to generally replicate late in S phase, along with other domains of heterochromatin (Schübeler et al. 2002). Interestingly, a study that decreased heterochromatin integrity by RNA interference knockdown of HP1a, a protein essential for heterochromatin formation, found that replication timing of chromosome 4 shifted to a later stage in S phase than that seen under wild-type conditions. This result for chromosome 4 contrasts with the behavior of centromeric DNA, which is replicated earlier in S phase when heterochromatin is impaired (Schwaiger et al. 2010). These findings point to some shared features between heterochromatin in general and chromosome 4, but also some characteristics that set chromosome 4 apart from other heterochromatic domains.
This idea is also supported by genetic studies looking for an impact of chromosome 4 dosage (number of copies) or looking at the impact of various mutations in chromosomal proteins on gene expression. These studies show that reporters that reside in chromosome 4 exhibit different responses to several modifiers of PEV compared to reporters that reside in pericentric heterochromatin (Haynes et al. 2007; Brower-Toland et al. 2009; Phalke et al. 2009), thereby arguing for a unique set of requirements to assemble the heterochromatin of chromosome 4. For example, Su(var)3-9 is a suppressor of variegation for reporters in the pericentric heterochromatin, but a weak enhancer of variegation for a reporter in the banded arm of chromosome 4 (Brower-Toland et al. 2009). The observation that an additional copy of chromosome 4 acts as a suppressor of variegation for a reporter on chromosome 4, while an extra copy of the pericentric region alone does not, argues that there are one or more proteins unique to the long arm of chromosome 4 that are being titrated away (Haynes et al. 2007). Thus the F element, while clearly dependent on HP1a and H3K9me2/3, appears to have a unique heterochromatin composition.
The F Element is Not a Uniform Chromatin Domain
While the data discussed above provide ample evidence that the F element as a whole exhibits unusual characteristics compared to the other Muller elements, generally those associated with heterochromatin, there are several lines of evidence that demonstrate that the F element is not a uniform chromatin domain. The first observation suggesting nonuniformity again comes from the study of polytene chromosomes, which demonstrate that the dot chromosome, like the other chromosome arms, is banded, indicating the presence of both highly condensed and less condensed chromatin regions (Painter 1934) (Figure 1). Two distinct chromatin types on D. melanogaster chromosome 4 were also detected by studies of PEV. When a transgene reporter containing the white gene driven by an hsp70 promoter (hsp70-white) is inserted randomly throughout the genome by P-element transposition, insertions in regions of euchromatin result in animals with a red eye phenotype (full expression), while insertions in regions of heterochromatin result in animals with a variegating phenotype (i.e., silencing in some of the cells) (Wallrath and Elgin 1995). Interestingly, insertions of both types were recovered from chromosome 4 (Sun et al. 2000) (Figure 2), albeit sites allowing full expression were relatively rare. Most insertion sites, including most of those located in the body of a functional gene, resulted in a variegating phenotype, reflecting the heterochromatic nature of the surrounding chromatin. These results suggest that most of the chromosome 4 genes are not sequestered by boundaries or otherwise protected from heterochromatin formation, but can function in this environment. At the same time, the results also indicate that chromosome 4 is not entirely heterochromatic, as there are a few regions (four have been documented) permissive for the expression of the hsp70-white reporter.
The frequency of permissive sites initially appeared low, given that the genome sequence demonstrates that there are ∼80 genes within the long arm of D. melanogaster chromosome 4, in agreement with earlier estimates (Hochman 1976; Adams et al. 2000) (Figure 2). Thus, analysis of the genome sequence confirmed that the 1.3-Mb portion of chromosome 4 that is polytenized in D. melanogaster has an overall gene density similar to that of the other chromosomes, despite its significantly higher repeat density (Adams et al. 2000; Leung et al. 2010). The presence of a similar number of genes in the context of an increased repeat density has been confirmed for a variety of other Drosophila species (e.g., in D. erecta, D. ananassae, D. virilis, and D. mojavensis). In some cases, the size of the F element is expanded greatly by an influx of TEs, likely to enhance heterochromatin formation, but these shifts are tolerated by the F element genes (Drosophila 12 Genomes Consortium et al. 2007; Leung et al. 2015, 2017).
Comparison of the DNA sequences around the locations of D. melanogaster chromosome 4 reporter insertions leading to a variegating eye phenotype (indicative of a heterochromatin environment) with those leading to a red eye phenotype (indicative of a permissive environment) did not reveal any major differences in local repeat or gene density (Sun et al. 2004; Riddle et al. 2008). Variegating reporters were not found to be in regions with relatively high repeat density; instead, the study by Sun and colleagues found that 11 of 18 variegating transgene reporters on chromosome 4 were located within transcribed genes. Initial studies suggested that a particular repetitive element, the DNA transposon remnant 1360, might be a target for heterochromatin formation on chromosome 4 (Sun et al. 2004), a hypothesis that is supported by subsequent analyses (Haynes et al. 2006; Sentmanat and Elgin 2012). However, the distribution of 1360 within chromosome 4 argues that it cannot be the only target. Subsequent studies have demonstrated that other repetitive elements besides 1360 are required to explain the distribution of silencing and permissive domains on chromosome 4 as read out by the PEV reporters available at this time (Riddle et al. 2008). Further analysis using a P-element construct containing 1360 as well as the reporter demonstrated that proximity to a heterochromatic mass (most often the pericentric heterochromatin or the telomere-associated sequences) is required to obtain a PEV phenotype dependent on 1360 (Haynes et al. 2006; Sentmanat and Elgin 2012; Huisinga et al. 2016). A nearby mass of HP1a might facilitate heterochromatin spreading from the target 1360 to the adjacent reporter. This idea is supported by translocation studies: when a chromosome 4 segment containing a PEV reporter is translocated to another chromosome such that it is positioned distant from the chromocenter in the nucleus, PEV is suppressed and expression from the reporter increases (Cryderman et al. 1999); one also sees an increase in the rate of recombination (see above). While at least a fraction of HP1a is dynamic within the nucleus (Cheutin et al. 2003), its predominant association with the chromocenter might explain the impact of chromocenter proximity on PEV. Together, these findings illustrate the complexity of chromosome 4 packaging and suggest a constant competition between targeted silencing of repeats and targeted activation of the genes, resulting in closely interspersed chromatin types. Proximity to the chromocenter promotes heterochromatin formation and silencing, presumably by promoting heterochromatin spreading, but this effect can clearly be overcome to allow the expression of chromosome 4 genes, suggesting a dynamic equilibrium among chromatin types (Eissenberg and Elgin 2014).
Biochemical Studies Reveal the Complex Chromatin Makeup of Chromosome 4
Histone modifications
Analyses of the chromosome 4 chromatin structure in D. melanogaster using immunohistochemical staining of polytene chromosomes revealed that at this coarse level, chromosome 4 is strongly associated with biochemical signatures of heterochromatin. Chromosome 4 is enriched for the heterochromatin proteins HP1a (James and Elgin 1986; James et al. 1989) (Figure 1 and Figure 3) and HP2 (Shaffer et al. 2002), and it contains high levels of histone H3 methylated at lysine 9 (H3K9me2 and H3K9me3, Figure 3) (Jacobs et al. 2001; Cowell et al. 2002; Schotta et al. 2002); it also shows low levels of histone acetylation [for example H4K8ac (Haynes et al. 2004)], which typically marks transcriptionally active regions. Further studies revealed that two H3K9 histone methyltransferases are involved in the modification of chromosome 4, SU(VAR)3-9 and EGG with its cofactor WDE (Schotta et al. 2002; Stabell et al. 2006; Clough et al. 2007; Seum et al. 2007; Tzeng et al. 2007; Brower-Toland et al. 2009; Koch et al. 2009). For all of these marks, enrichment along chromosome 4 is not uniform, supporting the presence of multiple chromatin types.
Figure 3.
Genes on chromosome 4 are enriched for chromatin marks typical of heterochromatin irrespective of their expression status. (A) Metagene profiles of expressed genes residing in the pericentric heterochromatin (left) or on chromosome 4 of D. melanogaster (right), illustrating their enrichment for HP1a, H3K9me2, H3K9me3, and H3K36me3 in BG3 cells. In contrast to pericentric genes, chromosome 4 genes have higher levels of the heterochromatic marks HP1a, H3K9me2, and H3K9me3 across the body of the genes compared to intergenic regions. The metagene profile averages the 2-kb regions upstream and downstream of the gene, along with gene spans that have been scaled to 3 kb in size. x-axis: position along the metagene with 0 indicating the transcription start site; y-axis: enrichment relative to input [figure panels modified from Riddle et al. (2011)]. (B) Browser view illustrating that both an expressed gene (Ephrin) and a silent gene (CG1909) on chromosome 4 show similar levels of enrichment for heterochromatic marks, although there is a shift in the ratio of H3K9me2 to H3K9me3, with higher levels of H3K9me2 associated with silencing. x-axis: position along the chromosome; y-axis for chromatin immunoprecipitation panels: enrichment relative to input (University of California Santa Cruz Genome Browser; Kent et al. 2002). RefGenes, reference genes.
Additional studies mapping chromatin marks genome-wide by chromatin immunoprecipitation followed by microarray or next-generation sequencing analysis, primarily by the modENCODE (model organism ENCyclopedia Of DNA Elements) project, have provided further insights into the chromatin composition of D. melanogaster chromosome 4. One anticipates that chromatin marks associated with constitutive heterochromatin would be stable, while marks associated with a gene’s activity state would change over developmental time, and this prediction is supported by the data. These modENCODE studies revealed that the marks typically associated with heterochromatin (H3K9 methylation, HP1a, etc.) are not restricted to the repeats on chromosome 4 [Figure 3; for examples see Riddle et al. (2011, 2012)]. Rather, the heterochromatic marks are found over the body of most chromosome 4 genes, although there are differences in the level of enrichment compared to repeats. This finding is consistent across the cell types and developmental stages examined. Some specific regions lack heterochromatin marks altogether (Riddle et al. 2011, 2012; Figueiredo et al. 2012). In particular, the histone marks associated with transcription start sites (TSSs) are conserved between active chromosome 4 genes and active genes in other genomic compartments, as is the presence of DNaseI hypersensitive sites (DHSs) at active TSSs (Kharchenko et al. 2011). Careful analyses show that, while the bodies of transcribed genes tend to be enriched for heterochromatic marks, the TSSs of these genes are depleted for these same marks (Figure 3) (Riddle et al. 2011, 2012; Figueiredo et al. 2012). This lack of HP1a and H3K9 methylation at the TSSs of active chromosome 4 genes may be necessary to provide access for the transcription machinery in an otherwise refractory chromatin environment. Accordingly, one sees enrichment for RNA polymerase II (RPII) at the TSS and enrichment of H3K4me3 immediately downstream. These findings suggest that genes on chromosome 4 have adapted to function in a mostly heterochromatic environment by creating access at the TSS. Identification of the features of genome organization that make this arrangement possible is an area of active investigation. One can hypothesize a novel “pioneer” transcription factor (TF) that specifically destabilizes the nucleosomes at the TSSs of fourth chromosome genes, but no such factor has been identified to date. Alternatively, chromosome 4 genes might use the same TFs found in euchromatin, but have a higher local concentration of TF-binding sites (anchoring activation signals) to counter the silencing signals associated with the TEs, present in abundance. However, in general, the histone modification and chromatin protein-enrichment patterns seen on chromosome 4 do not resemble the patterns seen in the euchromatic chromosome arms of D. melanogaster. The most common epigenomic profile, based on the modENCODE 9-state model, is one where the 5′ end of the active gene is in state 1 (enriched for H3K4me3), while the body of the gene is in states 7 or 8 (enriched in H3K9me2/3). Inactive fourth chromosome genes are generally packaged throughout in states 7 or 8 (Kharchenko et al. 2011). These findings illustrate the unique chromatin structure of genes on the D. melanogaster chromosome 4.
The exception to these general conclusions, evident on examination of the genome-wide enrichment profiles generated by modENCODE (Kharchenko et al. 2011), is the presence of a small group of genes packaged with H3K27me3, indicating regulation by the Polycomb (PC) group and Trithorax (TRX) group proteins (state 6; see map from BG3 cells, Figure 2). The PC/TRX system is a highly conserved gene regulatory system essential for development, most famously for regulation of the bithorax complex genes (Lewis 1978). Genes under the control of the PC/TRX system are generally associated with PC when they are transcriptionally inactive and with TRX when they are transcriptionally active (Schuettengruber et al. 2017) (see Box 3 for the distinction between PC chromatin and facultative heterochromatin). In flies, genomic regions enriched for HP1a and H3K9 methylation are typically devoid of PC and its associated H3K27 methylation mark (Nestorov et al. 2013), making these two types of chromatin-based silencing distinct. Based on the available data, there are at least seven genes on chromosome 4 of D. melanogaster that are under the control of the PC/TRX system (ey, toy, zfh2, sv, ci, Sox102F, and fd102C) (Schwartz et al. 2006; Riddle et al. 2011, 2012). These regions correspond to the permissive insertion sites seen in the PEV screens using the hsp70-white P-element reporter discussed above; insertions into these PC (state 6) domains led to the recovery of fully expressed, red-eyed reporter lines (Riddle et al. 2012). The genes naturally found in these regions do not exhibit 5′ DHSs when packaged as heterochromatin (Kharchenko et al. 2011; states 7 and 8) in S2 cells, but do exhibit DHSs when packaged with PC (Kharchenko et al. 2011; state 6). This finding suggests that chromatin in state 6 (packaged with H3K27me3) is permissive for DHS formation, while chromatin in states 7 or 8 (packaged with H3K9me2/3) is not. The ability to generate DHSs may well determine the reporter response, resulting in full red expression when the reporter is inserted into a state 6 domain, but blocking access, resulting in PEV, when the reporter is inserted into a state 7/8 domain. These differences could be explained by differences in nucleosome stability, presumably reflecting contributions of both activating and silencing histone modifications (Eissenberg and Elgin 2014).
Box 3. Facultative Heterochromatin.
Chromatin domains that are heterochromatic in all cell types are referred to as constitutive heterochromatin, while those domains that appear heterochromatic in some cell types but not others are referred to as facultative heterochromatin. Some cell linages, notably the development of red blood cells in birds, show increasing accumulation of heterochromatin in the nucleus as the cell differentiates to a specialized state that uses only a few genes. Facultative heterochromatin formation has not been extensively studied using modern tools, but presumably reflects the accumulation of H3K9me2/3-dominated domains in the euchromatic long arms of the chromosomes; for an example, see the contrast between S2 (embryonic) cells and Bg3 (neuronal) cells in figures 2, S6, and S7 in Kharchenko et al. (2011). A second chromatin-based silencing system involved in cell type-specific gene regulation is defined by the Polycomb complex [reviewed by Grossniklaus and Paro (2014)]. This system also depends on a histone modification, H3K27me3. Because of these similarities, some literature has referred to Polycomb-associated domains as facultative heterochromatin. However, Polycomb-induced silencing is strictly limited to a small number of genes identified by Polycomb Response Elements. Because this mechanism cannot play the general role originally identified for facultative heterochromatin, the use of the term in discussions of Polycomb-induced silencing is inappropriate and will be avoided here.
Nonhistone chromosomal proteins
A further unique feature of the dot chromosome is the presence of a chromatin protein that specifically associates with this chromosome (Figure 1). Painting of fourth (Pof) was identified as an interactor of Zeste, a DNA-binding protein associated with nucleosome remodeling at the 5′ end of genes. POF was singled out due to its unusual localization pattern; in Drosophila polytene chromosome spreads, it is associated predominantly with chromosome 4 (Figure 1) (Larsson et al. 2001). POF targets the genes on chromosome 4, as do HP1a and EGG, but it does not appear to impact repeats (Lundberg et al. 2013b). Analysis of mutant strains for these various chromatin proteins has revealed complex interactions among them as well as interdependencies. While EGG has been identified as the main H3K9 histone methyltransferase acting on chromosome 4 (Clough et al. 2007; Seum et al. 2007; Tzeng et al. 2007; Brower-Toland et al. 2009), SU(VAR)3-9 is able to maintain a basal level of H3K9 methylation in the absence of EGG, acting mainly at repeats (Figueiredo et al. 2012; Riddle et al. 2012). Both POF and HP1a appear to be required for proper transcriptional regulation of genes on chromosome 4, and depletion of either protein results in lower expression for most fourth chromosome genes (Figueiredo et al. 2012; Riddle et al. 2012; Lundberg et al. 2013b). Despite its dominant role in heterochromatin formation, associated with silencing, HP1a has been observed to have a positive impact on gene expression in several other test systems (Piacentini and Pimpinelli 2010; Cryderman et al. 2011; Kwon and Workman 2011). For example, enhancement of transcript elongation by HP1a has been reported for the heat-shock loci at their endogenous sites in the euchromatic arms (Piacentini et al. 2003). On the fourth chromosome, HP1a and H3K9me3 levels are highest over the body of active genes (Figure 3). Under normal circumstances, one would anticipate that HP1a and H3K9me2/3 would work together to cross-link dinucleosomes (Machida et al. 2018), making transcription more difficult. Whether that is the case here, and how such a structure would be displaced to allow transcription, is unknown. Conversely, the dot chromosome as a whole, and the genes in particular, have a low melting temperature (Riddle et al. 2012; Leung et al. 2015), which might facilitate transcript elongation. In addition, the association with POF is expected to facilitate transcription (see below). The specialized chromatin structure of chromosome 4 genes impacts RPII dynamics, leading to lower levels of RPII pausing (Johansson et al. 2012; Riddle et al. 2012), which can also be seen as a lower half-life of paused RPII on chromosome 4 (Shao and Zeitlinger 2017). Together, the detailed analysis of chromatin marks reveals that chromatin packaging in chromosome 4 is complex, and clearly distinct from both that seen in the euchromatic arms and that found in the pericentric heterochromatin.
The Drosophila Dot Chromosome May be Derived From an Ancestral X Chromosome
Given the unique features of the dot chromosome, including its high repeat content, distinct chromatin structure, and lack of recombination, the question of its origin has attracted substantial attention. The earliest evidence that the dot chromosome might be linked to sex determination and the sex chromosomes comes from studies by Calvin Bridges in the 1920s investigating D. melanogaster intersex flies. In Drosophila, sex is determined by the ratio of X chromosomes to autosomes (Bridges 1921). Normally, females have an X to autosome ratio of 1 (two X chromosomes and two copies of each autosome), while males have an X to autosome ratio of 0.5 (one X chromosome and two copies of each autosome). Animals with two X chromosomes and three sets of autosomes (ratio of 2:3) develop into an intersex fly, exhibiting both male and female characteristics (Bridges 1921). When an extra copy of chromosome 4 is included in an intersex fly’s genome, the flies tend to exhibit more female characteristics, which led Bridges to conclude that the “fourth chromosome has a net female tendency, similar to that of the X” chromosome [Bridges (1925); see also Bridges (1921) and Ashburner et al. (2005)].
A second line of evidence linking the dot chromosome to the female sex chromosome comes from the work on POF. Given POF’s unique association with chromosome 4, as well as the links between the dot chromosome and the X chromosome in genetic studies [reviewed in Ashburner et al. (2005)], Larsson and colleagues have pointed out that the behavior of POF, a chromosome-wide mark, is similar to that of the male-specific lethal (MSL) complex, which is targeted specifically to the male X chromosome to achieve dosage compensation (Lucchesi and Kuroda 2015). Like the MSL complex, which upregulates gene expression on the male X chromosome (Keller and Akhtar 2015; Lucchesi and Kuroda 2015; Birchler 2016), the presence of POF positively correlates with transcription of most genes on chromosome 4: high levels of POF are associated with high levels of expression, and loss of POF leads to decreased levels of gene expression (Johansson et al. 2007, 2012; Lundberg et al. 2013b). Like the MSL complex, POF binding to chromosome 4 also involves RNA, although the exact mechanism is unclear (Johansson et al. 2012). Binding of POF to its two binding sites observed on the X chromosome is dependent on roX1/roX2, the two noncoding RNAs that are integral parts of the Drosophila MSL complex required for dosage compensation (Lundberg et al. 2013a). Conversely, in the absence of the roX RNAs, the MSL complex is recruited to chromosome 4 and pericentric heterochromatin at a higher level, apparently based on an affinity for repeat-enriched regions; this capacity to bind to repeats has been suggested to be an ancient but still intrinsic property of the MSL complex (Figueiredo et al. 2014). The similarities between POF and MSL suggest a parallel evolution, possibly with common antecedents. Finally, in males of D. busckii, POF specifically stains the entire X chromosome, which is a dot–X fusion(a fusion of the F and A Muller elements) (Larsson et al. 2001). Taken together, the results from work on POF point to a potential biochemical link between regulation of the dot chromosome and the regulation of the X chromosomes (the only two chromosomes with chromosome-wide recognition), presumably rooted in past history.
Detailed analyses of the evolution of the dot chromosome using DNA sequence data, looking both within and beyond the Drosophila genus, provide strong support for a connection between the dot and the present X chromosome (F and A elements, respectively). The collection of genes found within each Muller element has remained largely the same during the evolution of the Diptera (∼200 MY) (Vicoso and Bachtrog 2015; Sved et al. 2016). For example, ∼95% of the genes have remained on the same Muller element across 12 Drosophila species (Bhutkar et al. 2008). Similarly, analysis of the Queensland fruit fly Bactrocera tryoni (which diverged from the Drosophilids ∼60–70 MYA) shows that ∼90% of the genes have remained on the same Muller element between the Drosophila and Bactrocera genomes. However, this analysis also shows that 57 out of the 63 D. melanogaster F element genes that could be placed in the B. tryoni assembly are located on the B. tryoni X chromosome (Sved et al. 2016). Similarly, of the 59 X-linked genes found in the Australian sheep blowfly Lucilia cuprina, 49 are located on the D. melanogaster F element (Davis et al. 2018).
Consistent with these observations, a previous study by Vicoso and Bachtrog suggests that the Drosophila F element was derived from an ancestral X chromosome (Vicoso and Bachtrog 2013). Their investigations focused on the evolution of the sex chromosomes in the Drosophilids (Vicoso and Bachtrog 2013), including the black soldier fly Hermetia illucens (Stratiomyidae, a basal Brachycera), the olive fruitfly B. oleae (Tephritidae), the gray fleshfly Sarcophaga bullata (Sarcophagidae), and the zoophilic fruitfly Phortica variegata (Steganinae, a sister clade to Drosophila within Drosophilidae), as well as the basal Drosophila species D. busckii. Males and females from each species were sequenced using the Illumina sequencing platform and the reads assembled into scaffolds. The reads from the male and female samples were then mapped back to these scaffolds, which enabled the identification of the female sex chromosomes based on their underrepresentation in the genome sequence derived from males in comparison to that from females. This read coverage analysis shows that the Muller A element is the sex chromosome for the species within Drosophilidae (i.e., D. melanogaster, D. busckii, and P. variegata). In contrast, the read coverage analysis shows that the Muller A element is an autosome in the more distant outgroups (S. bullata, B. oleae, and H. illucens), and that the F element is the female sex chromosome in these species. This study also reveals that the fusion of chromosome 4 to the X chromosome (F to A) in D. busckii [where one sees F+A chromosome staining by POF in males (Larsson et al. 2001)], appears to be a derived feature (Vicoso and Bachtrog 2013). Given that the F element is an autosome within the Drosophilidae and a sex chromosome in the outgroup, Vicoso and Bachtrog postulated that the F element was originally a sex chromosome that has reverted back to an autosome in the Drosophila genus. How the F element within the genus Drosophila acquired its heterochromatic features, and whether and/or how those features might be linked to the ability of the chromosome to make such a transition, remains unresolved.
The Evolutionary History of the F Element Within the Genus Drosophila Sheds Some Light on its Unique Features
Evolutionary studies of the F element within the Drosophila genus provide some insights into its unique biology (Figure 4). As noted above, within Drosophila, the F element is a small autosome in most species. However, there are exceptions: In D. willistoni, the F element is fused to the E element (chromosome 3R in D. melanogaster) (Clayton and Wheeler 1975; Powell et al. 2011; Pita et al. 2014). In D. ananassae, the F element has expanded into a large metacentric chromosome of ≥ 18.7 Mb (Schaeffer et al. 2008; Leung et al. 2017; Davis et al. 2018). Several other species [such as D. takahashii (W. Leung, personal communication)] show a two–fourfold expansion. In D. pseudoobscura, the F element is fused to an ancestral Y chromosome (Larracuente and Clark 2014), and in D. busckii, the F element is fused to the X and Y chromosomes, restoring its ancestral status as a sex chromosome (Figure 4) (Zhou and Bachtrog 2015). In addition, several Hawaiian Drosophila species have rod-shaped or metacentric derivatives of the F element (Craddock et al. 2016). These findings demonstrate that the F element, which can be a dot chromosome, is remarkably tolerant to changes in its chromosome configuration and size, perhaps because the genes are already adapted to function within a heterochromatic environment. With this collection of chromosomal configurations and distinct evolutionary trajectories, Drosophila researchers have a rich resource to characterize key features that might be necessary and/or sufficient to generate the dot chromosome’s unique properties.
As noted above, genome sequence analyses have revealed that the F element in D. melanogaster is enriched for various repeated elements (Miklos et al. 1988; Locke et al. 1999b; Bartolomé et al. 2002; Hoskins et al. 2002; Kaminker et al. 2002; Slawson et al. 2006; Leung et al. 2010, 2015, 2017), which likely contributes to its unique chromatin features. Repeated sequences on the F element, primarily remnants of TEs, have been investigated in a variety of species and they significantly contribute to the evolution of the F element. For example, detailed analyses of genome sequences from D. melanogaster, D. virilis, D. erecta, D. grimshawi, and D. mojavensis have revealed that the transposon density in all of these species is consistently higher on the F element, ranging from 25 to 50%, than in a comparable euchromatin reference region at the base of the D element, which ranges from 3 to 11% (Leung et al. 2015). Despite a consistently high repeat content, the contribution of various types of repeats to the overall repeat content of the F differs among species. Taking transposon types as an example, the fraction of LINE (long interspersed nuclear element), LTR transposons, DINE-1, and DNA transposons is variable. While in D. mojavensis DINE-1 elements and their remnants make up the largest fraction of TEs, their contribution to the total amount of transposons on the F element in D. grimshawi is negligible relative to the contribution of LTR, LINE, and DNA transposons (Leung et al. 2015). These findings illustrate the fact that despite the high repeat content of F elements being a common characteristic, it is achieved in different ways in different species. Together, the data suggest multiple TE expansion events, with different species experiencing different events of this type.
Repeated elements also contribute to the evolutionary history of the F element. This point is well illustrated by the case of D. ananassae, noted above, where the dot chromosome has evolved into a large metacentric chromosome (Leung et al. 2017). In-depth analysis of the D. ananassae genomic sequence data reveals that the increase in the size of the arms of the F element is largely due to an increase in TE density (Leung et al. 2017). The D. ananassae F element repeat density is at least double that of D. melanogaster, a minimum of 56.4%–74.5% vs. 14.4%–29.5% depending on the analysis method used. In particular, the density of LINE and LTR transposons has increased significantly in the D. ananassae lineage. Interestingly, the additional transposons are not simply part of the intergenic space, or found in the regions close to the centromere and telomere. Of the 64 D. ananassae F element genes analyzed in this study, 59 genes (92%) show larger total intron size than their D. melanogaster orthologs, primarily due to the increased presence of TEs; this change does not appear to impact patterns or levels of expression (Leung et al. 2017). As noted above, these genes exhibit low codon bias, and this does not change with the distance from the centromere, indicating that the lack of recombination noted for the smaller dot chromosomes likely persists across the whole of these larger chromosome arms. The D. ananassae F element illustrates the remarkable ability of this domain to tolerate TE amplification; similar tolerance may be an important feature of eukaryotic genome evolution.
Past studies have also indicated that Wolbachia (an endosymbiont of Drosophila that invades the germ cells) is integrated into multiple strains of D. ananassae via lateral DNA transfer (Choi et al. 2015) and might contribute to genome expansion. Fluorescence in situ hybridization of mitotic chromosomes has shown that Wolbachia sequences are integrated into the region near the centromere of the D. ananassae F element (Klasson et al. 2014). However, Wolbachia sequences do not appear to have contributed substantially to the expansion of the chromosome arms (Leung and Elgin 2018).
Fusion of the F element to other chromosomal elements also alters its evolutionary trajectory. Support for this finding comes from studies of species such as D. willistoni, D. busckii, and D. pseudoobscura. In D. willistoni, the F element was fused to Muller’s E element, one of the large autosomal arms (Clayton and Wheeler 1975; Powell et al. 2011; Pita et al. 2014). When linkage disequilibrium, mutation rates, and recombination rates are compared between the F–E fusion chromosome in D. willistoni and the other chromosomes, no significant differences are found, in contrast to what is typically seen for the D. melanogaster F element (Powell et al. 2011). Equilibration of recombination rates between two recently joined distinct domains is underway in D. busckii, where the dot chromosome is fused to the X and Y chromosomes, creating a neo-X and a neo-Y chromosome (Zhou and Bachtrog 2015); the opposite example, an old Y chromosome fusing to the dot chromosome, has also been observed (Chang and Larracuente 2017). In D. busckii, the F element portion of the neo-Y chromosome is undergoing rapid changes leading to degeneration, with many more single-nucleotide polymorphisms, and insertions and deletions being detected in the neo-Y than the neo-X (Zhou and Bachtrog 2015). These findings suggest that while the F element is unique in many ways (low codon bias, low recombination rate, etc.), these characteristics are lost over time upon large-scale changes in chromosomal conformations. How these changes relate to changes in sequence composition remains an area of investigation.
While the above examples illustrate that the F element within Drosophila has undergone a number of changes, its gene content has been surprisingly constant. It has been suggested that the F element, as well as the other Muller elements, has been stable for the ∼200 MY of Dipteran evolution (Vicoso and Bachtrog 2015). Despite this constancy, several genes have been identified and studied that have moved between Muller’s elements. A 2015 study identified 12 genes (“wanderer genes”) of ∼80 that have moved on or off the D. melanogaster, D. erecta, D. mojavensis, and D. grimshawi F elements (Leung et al. 2015). This level of movement is similar to that seen for the other chromosomes, where ∼95% of the genes are found on the same Muller element across the 12 Drosophila species (Bhutkar et al. 2008). Movement of these wanderer genes is in both directions, on and off the F element. Most of the arrivals to the D. mojavensis and D. grimshawi F elements occur at a single “hotspot” of unknown significance (Leung et al. 2015). Thus, while the F element has been surprisingly constant in its genic content, individual genes can move on or off, most likely through transposition, at similar rates as on other chromosome arms (Drosophila 12 Genomes Consortium et al. 2007).
Conclusion and Outlook
As the data reviewed here demonstrate, the Drosophila dot chromosome has been the subject of extensive research since the earliest beginnings of Drosophila biology, and it has continued to fascinate biologists due to its intriguing and unique characteristics. While many of these characteristics have been known for close to a century, only recently, with the advances in high-throughput sequencing and other genomic techniques, have the origins of these various unique characteristics become clearer. Analyses of the F element DNA sequence has revealed why it exhibits a mix of heterochromatic and euchromatic characteristics: genes and repeats are interspersed, leading to broad regions in which silencing signals prevail, while in small regions at the 5′ ends of genes, activating signals dominate. Comparisons of the F element sequences between species have revealed its evolutionary history and supported earlier suggestions that the dot chromosome might have been derived from a sex chromosome, thereby providing a model for the origins of POF, as a potential remnant of an ancestral dosage-compensation or chromosome-marking complex. Thus, after decades of research, we are finally developing some understanding of why the F element is such a unique genomic domain.
While tremendous progress has been made, several questions remain unresolved. One set of questions concerns the genes that reside on the F element. How have these genes adapted to a chromatin environment that is dominated by silencing signals? What processes control the expression of these genes? Several issues need to be considered. How are DHSs (and a switch to appropriate histone modification marks for TSSs) established in this heterochromatin milieu at the TSSs of active F element genes? Is there a specific pioneer TF or other features unique to the F element, or simply a sufficient collection of positive TF-binding sites to compete successfully with the high density of silencing marks? Why are two H3K9 histone methyltransferases recruited to the dot chromosome, one [SU(VAR)3-9] apparently to the repetitious sequences, as anticipated, and another (EGG) to the body of transcribed genes? What are the recruitment mechanisms? It seems likely that the recruitment of SU(VAR)3-9 to the repetitious sequences (TEs and their remnants) uses the same cues as elsewhere in the genome; recent studies indicate that the piwi-interacting RNA system evolved to direct heterochromatin formation to help silence TEs (Castel and Martienssen 2013). But understanding both the recruitment and the role of EGG is a greater challenge. EGG appears to be required for the recruitment and stabilization of POF, and POF and HP1a exhibit mutual dependency (Riddle et al. 2012). Both POF and HP1a have been implicated in promoting transcript elongation. Thus, one suspects that HP1a functions very differently in complexes associated with fourth chromosome genes (with POF) as opposed to complexes associated with fourth chromosome repeats (without POF).
A second set of questions is raised by the low recombination rates of the F element. How is it that the F element resists recombination, while other chromosome events such as inversion and single-gene transposition both occur at the usual, or higher than usual, rates? Simple “occlusion” models will clearly not suffice, nor is a centromere effect per se sufficient, given the results with D. ananassae. The increase in recombination seen in Blm mutants (Hatkevich et al. 2017) has been interpreted in terms of a loss of the centromere effect, as noted above. Are Blm mutants defective in their heterochromatin and is that the limiting factor in determining recombination rates? What is the role of heterochromatin and of repeats in the centromere effect? It might be profitable to think in terms of recent models of heterochromatin based on phase separation (Larson et al. 2017; Strom et al. 2017), considering the possibility that the F element occupies a distinct nuclear compartment that for some reason excludes a critical component required for recombination. Alternatively, it is worth recalling that recombination does not occur in male flies. Some common features might be involved, albeit the F element is considered a feminizing chromosome, and recombination rates are normal or a bit higher on the X chromosome compared to the autosomes in D. melanogaster.
A third set of questions centers around the evolution of the F element, particularly issues associated with reverting from a sex chromosome to an autosome. Several potential pathways have been delineated by Vicoso and Bachtrog (2013), but how do these pathways play out at the level of chromatin structure? Sex chromosomes are marked to attract the dosage-compensation complex, specifically to double expression from the chromosome in the male. The dosage-compensation mechanism operating on this chromosome would need to be negated across the whole chromosome. One might argue for the spreading of pericentric heterochromatin to reduce the expression from the new autosome, providing a rationale for why such a reversion has only been seen for the small F element. Given the ability of heterochromatin formation to silence TEs, this state of affairs could make the F element susceptible to retaining repetitious elements, given the ability to silence these. Expression of the genes then becomes a balancing act between the positive effects from residual dosage compensation and the negative effects from high repeat density.
Answering these and other remaining questions will require innovative genetic manipulations and the application of new techniques, which have not been available in the past. Given the resourcefulness of Drosophila biologists, it is unlikely that these questions will remain unanswered for long.
Acknowledgments
We thank W. Leung for assistance with Figure 2, and our colleagues in our laboratories and the Drosophila community for useful discussions on these issues. Work by the authors is supported by National Science Foundation (NSF) grants MCB-1517266 and IUSE-1431407, and National Institutes of Health grant 1RO1 GM-117340, to S.C.R.E.; and by NSF grant MCB-1552586 to N.C.R.
Footnotes
Communicating editor: S. Celniker
Literature Cited
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., et al. , 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185–2195. 10.1126/science.287.5461.2185 [DOI] [PubMed] [Google Scholar]
- Allshire R. C., Madhani H. D., 2018. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19: 229–244. 10.1038/nrm.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello J. R., Zhang Y., Kado T., Fan C., Zhao R., et al. , 2010. Recombination yet inefficient selection along the Drosophila melanogaster subgroup’s fourth chromosome. Mol. Biol. Evol. 27: 848–861. 10.1093/molbev/msp291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Golic K. G., Hawley R. S., 2005. Drosophila: A Laboratory Handbook. Cold Spring Harbor Press, Cold Spring Harbor, NY. [Google Scholar]
- Bartolomé C., Maside X., Charlesworth B., 2002. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol. Biol. Evol. 19: 926–937. 10.1093/oxfordjournals.molbev.a004150 [DOI] [PubMed] [Google Scholar]
- Beadle G. W., 1932. A possible influence of the spindle fibre on crossing-over in Drosophila. Proc. Natl. Acad. Sci. USA 18: 160–165. 10.1073/pnas.18.2.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry A. J., Ajioka J. W., Kreitman M., 1991. Lack of polymorphism on the Drosophila fourth chromosome resulting from selection. Genetics 129: 1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutkar A., Schaeffer S. W., Russo S. M., Xu M., Smith T. F., et al. , 2008. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics 179: 1657–1680. 10.1534/genetics.107.086108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J. A., 2016. Parallel universes for models of X chromosome dosage compensation in Drosophila: a review. Cytogenet. Genome Res. 148: 52–67. 10.1159/000445924 [DOI] [PubMed] [Google Scholar]
- Bridges C. B., 1921. Triploid intersexes in Drosophila melanogaster. Science 54: 252–254. 10.1126/science.54.1394.252 [DOI] [PubMed] [Google Scholar]
- Bridges C. B., 1925. Sex in relation to chromosomes and genes. Am. Nat. 59: 127–137. 10.1086/280023 [DOI] [Google Scholar]
- Bridges C. B., 1935. SALIVARY CHROMOSOME MAPS: with a key to the banding of the chromosomes of Drosophila melanogaster. J. Hered. 26: 60–64. 10.1093/oxfordjournals.jhered.a104022 [DOI] [Google Scholar]
- Brower-Toland B., Riddle N. C., Jiang H., Huisinga K. L., Elgin S. C., 2009. Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics 181: 1303–1319. 10.1534/genetics.108.100271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos J. L., Halligan D. L., Haddrill P. R., Charlesworth B., 2014. The relation between recombination rate and patterns of molecular evolution and variation in Drosophila melanogaster. Mol. Biol. Evol. 31: 1010–1028. 10.1093/molbev/msu056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castel S. E., Martienssen R. A., 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14: 100–112. 10.1038/nrg3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. H., Larracuente A. M., 2017. Genomic changes following the reversal of a Y chromosome to an autosome in Drosophila pseudoobscura. Evolution 71: 1285–1296. 10.1111/evo.13229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T., McNairn A. J., Jenuwein T., Gilbert D. M., Singh P. B., et al. , 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299: 721–725. 10.1126/science.1078572 [DOI] [PubMed] [Google Scholar]
- Chino M., Kikkawa H., 1933. Mutants and crossing over in the dot-like chromosome of DROSOPHILA VIRILIS. Genetics 18: 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. Y., Bubnell J. E., Aquadro C. F., 2015. Population genomics of infectious and integrated Wolbachia pipientis genomes in Drosophila ananassae. Genome Biol. Evol. 7:2362–82. 10.1093/gbe/evv158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton F. E., Wheeler M. R., 1975. A catalog of Drosophila metaphase chromosome configurations, pp. 471–512 in Handbook of Genetics, edited by King R. C. Plenum Press, New York. [Google Scholar]
- Clough E., Moon W., Wang S., Smith K., Hazelrigg T., 2007. Histone methylation is required for oogenesis in Drosophila. Development 134: 157–165. 10.1242/dev.02698 [DOI] [PubMed] [Google Scholar]
- Cowell I. G., Aucott R., Mahadevaiah S. K., Burgoyne P. S., Huskisson N., et al. , 2002. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma 111: 22–36. 10.1007/s00412-002-0182-8 [DOI] [PubMed] [Google Scholar]
- Craddock E. M., Gall J. G., Jonas M., 2016. Hawaiian Drosophila genomes: size variation and evolutionary expansions. Genetica 144: 107–124. 10.1007/s10709-016-9882-5 [DOI] [PubMed] [Google Scholar]
- Cryderman D. E., Morris E. J., Biessmann H., Elgin S. C., Wallrath L. L., 1999. Silencing at Drosophila telomeres: nuclear organization and chromatin structure play critical roles. EMBO J. 18: 3724–3735. 10.1093/emboj/18.13.3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryderman D. E., Vitalini M. W., Wallrath L. L., 2011. Heterochromatin protein 1a is required for an open chromatin structure. Transcription 2: 95–99. 10.4161/trns.2.2.14687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R. J., E. J. Belikoff, E. H. Scholl, F. Li, and M. J. Scott, 2018 no blokes is essential for male viability and X chromosome gene expression in the Australian sheep blowfly. Curr. Biol. 28: 1987–1992.e3. DOI: 10.1016/j.cub.2018.05.005 [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium . Clark A. G., Eisen M. B., Smith D. R., Bergman C. M., et al. , 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., Elgin S. C., 2014. HP1a: a structural chromosomal protein regulating transcription. Trends Genet. 30: 103–110. 10.1016/j.tig.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin S. C., Reuter G., 2013. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb. Perspect. Biol. 5: a017780 10.1101/cshperspect.a017780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo M. L., Philip P., Stenberg P., Larsson J., 2012. HP1a recruitment to promoters is independent of H3K9 methylation in Drosophila melanogaster. PLoS Genet. 8: e1003061 10.1371/journal.pgen.1003061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo M. L., Kim M., Philip P., Allgardsson A., Stenberg P., et al. , 2014. Non-coding roX RNAs prevent the binding of the MSL-complex to heterochromatic regions. PLoS Genet. 10: e1004865 10.1371/journal.pgen.1004865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramates L. S., Marygold S. J., Santos G. D., Urbano J. M., Antonazzo G., et al. , 2017. FlyBase at 25: looking to the future. Nucleic Acids Res. 45: D663–D671. 10.1093/nar/gkw1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U., Paro R., 2014. Transcriptional silencing by polycomb-group proteins. Cold Spring Harb. Perspect. Biol. 6: a019331 10.1101/cshperspect.a019331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann M. A., Sekelsky J., 2017. The absence of crossovers on chromosome 4 in Drosophila melanogaster: imperfection or interesting exception? Fly (Austin) 11: 253–259. 10.1080/19336934.2017.132118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatkevich T., Kohl K. P., McMahan S., Hartmann M. A., Williams A. M., et al. , 2017. Bloom syndrome helicase promotes meiotic crossover patterning and homolog disjunction. Curr. Biol. 27: 96–102. 10.1016/j.cub.2016.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes K. A., Leibovitch B. A., Rangwala S. H., Craig C., Elgin S. C., 2004. Analyzing heterochromatin formation using chromosome 4 of Drosophila melanogaster. Cold Spring Harb. Symp. Quant. Biol. 69: 267–272. 10.1101/sqb.2004.69.267 [DOI] [PubMed] [Google Scholar]
- Haynes K. A., Caudy A. A., Collins L., Elgin S. C., 2006. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Curr. Biol. 16: 2222–2227. 10.1016/j.cub.2006.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes K. A., Gracheva E., Elgin S. C., 2007. A distinct type of heterochromatin within Drosophila melanogaster chromosome 4. Genetics 175: 1539–1542. 10.1534/genetics.106.066407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochman B., 1976. The fourth chromosome of Drosophila melanogaster, pp. 903–928 in The Genetics and Biology of Drosophila, edited by Ashburner M., Novitski E. Academic Press, London. [Google Scholar]
- Hoskins R. A., Smith C. D., Carlson J. W., Carvalho A. B., Halpern A., et al. , 2002. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 3: RESEARCH0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga K. L., Riddle N. C., Leung W., Shimonovich S., McDaniel S., et al. , 2016. Targeting of P-element reporters to heterochromatic domains by transposable element 1360 in Drosophila melanogaster. Genetics 202: 565–582. 10.1534/genetics.115.183228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S. A., Taverna S. D., Zhang Y., Briggs S. D., Li J., et al. , 2001. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 20: 5232–5241. 10.1093/emboj/20.18.5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T. C., Elgin S. C., 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 6: 3862–3872. 10.1128/MCB.6.11.3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James T. C., Eissenberg J. C., Craig C., Dietrich V., Hobson A., et al. , 1989. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 50: 170–180. [PubMed] [Google Scholar]
- Johansson A. M., Stenberg P., Pettersson F., Larsson J., 2007. POF and HP1 bind expressed exons, suggesting a balancing mechanism for gene regulation. PLoS Genet. 3: e209 10.1371/journal.pgen.0030209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A. M., Stenberg P., Allgardsson A., Larsson J., 2012. POF regulates the expression of genes on the fourth chromosome in Drosophila melanogaster by binding to nascent RNA. Mol. Cell. Biol. 32: 2121–2134. 10.1128/MCB.06622-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminker J. S., Bergman C. M., Kronmiller B., Carlson J., Svirskas R., et al. , 2002. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3: RESEARCH0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C. I., Akhtar A., 2015. The MSL complex: juggling RNA-protein interactions for dosage compensation and beyond. Curr. Opin. Genet. Dev. 31: 1–11. 10.1016/j.gde.2015.03.007 [DOI] [PubMed] [Google Scholar]
- Kent W. J., Sugnet C. W., Furey T. S., Roskin K. M., Pringle T. H., et al. , 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., Minoda A., Riddle N. C., et al. , 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485. 10.1038/nature09725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L., Kumar N., Bromley R., Sieber K., Flowers M., et al. , 2014. Extensive duplication of the Wolbachia DNA in chromosome four of Drosophila ananassae. BMC Genomics 15: 1097 10.1186/1471-2164-15-1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C. M., Honemann-Capito M., Egger-Adam D., Wodarz A., 2009. Windei, the Drosophila homolog of mAM/MCAF1, is an essential cofactor of the H3K9 methyl transferase dSETDB1/Eggless in germ line development. PLoS Genet. 5: e1000644 10.1371/journal.pgen.1000644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S. H., Workman J. L., 2011. The changing faces of HP1: from heterochromatin formation and gene silencing to euchromatic gene expression: HP1 acts as a positive regulator of transcription. BioEssays 33: 280–289. 10.1002/bies.201000138 [DOI] [PubMed] [Google Scholar]
- Larracuente A. M., Clark A. G., 2014. Recent selection on the Y-to-dot translocation in Drosophila pseudoobscura. Mol. Biol. Evol. 31: 846–856. 10.1093/molbev/msu002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson, A. G., D. Elnatan, M. M. Keenen, M. J. Trnka, J. B. Johnston et al., 2017 Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547: 236–240. 10.1038/nature22822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J., Chen J. D., Rasheva V., Rasmuson-Lestander A., Pirrotta V., 2001. Painting of fourth, a chromosome-specific protein in Drosophila. Proc. Natl. Acad. Sci. USA 98: 6273–6278. 10.1073/pnas.111581298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W., Elgin S. C. R., 2018. Response to the letter to the editor by Dunning Hotopp and Klasson. G3 (Bethesda) 8: 375 10.1534/g3.117.300379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W., Shaffer C. D., Cordonnier T., Wong J., Itano M. S., et al. , 2010. Evolution of a distinct genomic domain in Drosophila: comparative analysis of the dot chromosome in Drosophila melanogaster and Drosophila virilis. Genetics 185: 1519–1534. 10.1534/genetics.110.116129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W., Shaffer C. D., Reed L. K., Smith S. T., Barshop W., et al. , 2015. Drosophila muller f elements maintain a distinct set of genomic properties over 40 million years of evolution. G3 (Bethesda) 5: 719–740. 10.1534/g3.114.015966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W., Shaffer C. D., Chen E. J., Quisenberry T. J., Ko K., et al. , 2017. Retrotransposons are the major contributors to the expansion of the Drosophila ananassae Muller F element. G3 (Bethesda) 7: 2439–2460. 10.1534/g3.117.040907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B., 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- Lima-de-Faria A., 1959a. Incorporation of tritiated thymidine into meiotic chromosomes. Science 130: 503–504. 10.1126/science.130.3374.503 [DOI] [PubMed] [Google Scholar]
- Lima-de-Faria A., 1959b. Differential uptake of tritiated thymidine into hetero- and euchromatin in Melanoplus and Secale. J. Biophys. Biochem. Cytol. 6: 457–466. 10.1083/jcb.6.3.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke J., Howard L. T., Aippersbach N., Podemski L., Hodgetts R. B., 1999a. The characterization of DINE-1, a short, interspersed repetitive element present on chromosome and in the centric heterochromatin of Drosophila melanogaster. Chromosoma 108: 356–366. 10.1007/s004120050387 [DOI] [PubMed] [Google Scholar]
- Locke J., Podemski L., Roy K., Pilgrim D., Hodgetts R., 1999b. Analysis of two cosmid clones from chromosome 4 of Drosophila melanogaster reveals two new genes amid an unusual arrangement of repeated sequences. Genome Res. 9: 137–149. [PMC free article] [PubMed] [Google Scholar]
- Lucchesi J. C., Kuroda M. I., 2015. Dosage compensation in Drosophila. Cold Spring Harb. Perspect. Biol. 7: a019398 10.1101/cshperspect.a019398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg L. E., Kim M., Johansson A. M., Faucillion M. L., Josupeit R., et al. , 2013a. Targeting of painting of fourth to roX1 and roX2 proximal sites suggests evolutionary links between dosage compensation and the regulation of the fourth chromosome in Drosophila melanogaster. G3 (Bethesda) 3: 1325–1334. 10.1534/g3.113.006866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg L. E., Stenberg P., Larsson J., 2013b. HP1a, Su(var)3–9, SETDB1 and POF stimulate or repress gene expression depending on genomic position, gene length and expression pattern in Drosophila melanogaster. Nucleic Acids Res. 41: 4481–4494. 10.1093/nar/gkt158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida S., Takizawa Y., Ishimaru M., Sugita Y., Sekine S., et al. , 2018. Structural basis of heterochromatin formation by human HP1. Mol. Cell 69: 385–397.e8. 10.1016/j.molcel.2017.12.011 [DOI] [PubMed] [Google Scholar]
- McMahan S., Kohl K. P., Sekelsky J., 2013. Variation in meiotic recombination frequencies between allelic transgenes inserted at different sites in the Drosophila melanogaster genome. G3 (Bethesda) 3: 1419–1427. 10.1534/g3.113.006411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos G. L., Yamamoto M. T., Davies J., Pirrotta V., 1988. Microcloning reveals a high frequency of repetitive sequences characteristic of chromosome 4 and the beta-heterochromatin of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 85: 2051–2055. 10.1073/pnas.85.7.2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan L. V., 1947. A variable phenotype associated with the fourth chromosome of DROSOPHILA MELANOGASTER and affected by heterochromatin. Genetics 32: 200–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1940. Bearings of the Drosophila work on systematics, pp. 185–268 in The New Systematics, edited by Huxley J. Clarendon, Oxford. [Google Scholar]
- Muller H. J., Altenburg E., 1930. The frequency of translocations produced by X-rays in Drosophila. Genetics 15: 283–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestorov P., Tardat M., Peters A. H., 2013. H3K9/HP1 and Polycomb: two key epigenetic silencing pathways for gene regulation and embryo development. Curr. Top. Dev. Biol. 104: 243–291. 10.1016/B978-0-12-416027-9.00008-5 [DOI] [PubMed] [Google Scholar]
- Osborne, J. D., 1998 Crossing Over in a T(1;4) Translocation in Drosophila melanogaster. M.Sc. Thesis, University of Alberta, Edmonton. [Google Scholar]
- Painter T. S., 1934. A new method for the study of chromosome aberrations and the plotting of chromosome maps in Drosophila Melanogaster. Genetics 19: 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phalke S., Nickel O., Walluscheck D., Hortig F., Onorati M. C., et al. , 2009. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat. Genet. 41: 696–702. 10.1038/ng.360 [DOI] [PubMed] [Google Scholar]
- Piacentini L., Pimpinelli S., 2010. Positive regulation of euchromatic gene expression by HP1. Fly (Austin) 4: 299–301. 10.4161/fly.4.4.13261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini L., Fanti L., Berloco M., Perrini B., Pimpinelli S., 2003. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 161: 707–714. 10.1083/jcb.200303012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pita S., Panzera Y., Lucia da Silva Valente V., de Melo Z., Garcia C., et al. , 2014. Cytogenetic mapping of the Muller F element genes in Drosophila willistoni group. Genetica 142: 397–403. 10.1007/s10709-014-9784-3 [DOI] [PubMed] [Google Scholar]
- Powell J. R., Dion K., Papaceit M., Aguade M., Vicario S., et al. , 2011. Nonrecombining genes in a recombination environment: the Drosophila “dot” chromosome. Mol. Biol. Evol. 28: 825–833. 10.1093/molbev/msq258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle N. C., Leung W., Haynes K. A., Granok H., Wuller J., et al. , 2008. An investigation of heterochromatin domains on the fourth chromosome of Drosophila melanogaster. Genetics 178: 1177–1191. 10.1534/genetics.107.081828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle N. C., Minoda A., Kharchenko P. V., Alekseyenko A. A., Schwartz Y. B., et al. , 2011. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 21: 147–163. 10.1101/gr.110098.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle N. C., Jung Y. L., Gu T., Alekseyenko A. A., Asker D., et al. , 2012. Enrichment of HP1a on Drosophila chromosome 4 genes creates an alternate chromatin structure critical for regulation in this heterochromatic domain. PLoS Genet. 8: e1002954 10.1371/journal.pgen.1002954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler L., Szauter P., 1978. The effect of recombination-defective meiotic mutants on fourth-chromosome crossing over in Drosophila melanogaster. Genetics 90: 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer S. W., Bhutkar A., McAllister B. F., Matsuda M., Matzkin L. M., et al. , 2008. Polytene chromosomal maps of 11 Drosophila species: the order of genomic scaffolds inferred from genetic and physical maps. Genetics 179: 1601–1655. 10.1534/genetics.107.086074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G., Ebert A., Krauss V., Fischer A., Hoffmann J., et al. , 2002. Central role of Drosophila SU(VAR)3–9 in histone H3–K9 methylation and heterochromatic gene silencing. EMBO J. 21: 1121–1131. 10.1093/emboj/21.5.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D., Scalzo D., Kooperberg C., van Steensel B., Delrow J., et al. , 2002. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat. Genet. 32: 438–442. 10.1038/ng1005 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Bourbon H. M., Di Croce L., Cavalli G., 2017. Genome regulation by polycomb and trithorax: 70 years and counting. Cell 171: 34–57. 10.1016/j.cell.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Schwaiger M., Kohler H., Oakeley E. J., Stadler M. B., Schubeler D., 2010. Heterochromatin protein 1 (HP1) modulates replication timing of the Drosophila genome. Genome Res. 20: 771–780. 10.1101/gr.101790.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B., Kahn T. G., Nix D. A., Li X. Y., Bourgon R., et al. , 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38: 700–705. 10.1038/ng1817 [DOI] [PubMed] [Google Scholar]
- Sentmanat M. F., Elgin S. C., 2012. Ectopic assembly of heterochromatin in Drosophila melanogaster triggered by transposable elements. Proc. Natl. Acad. Sci. USA 109: 14104–14109. 10.1073/pnas.1207036109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seum C., Reo E., Peng H., Rauscher F. J., III, Spierer P., et al. , 2007. Drosophila SETDB1 is required for chromosome 4 silencing. PLoS Genet. 3: e76 10.1371/journal.pgen.0030076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer C. D., Stephens G. E., Thompson B. A., Funches L., Bernat J. A., et al. , 2002. Heterochromatin protein 2 (HP2), a partner of HP1 in Drosophila heterochromatin. Proc. Natl. Acad. Sci. USA 99: 14332–14337. 10.1073/pnas.212458899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W., Zeitlinger J., 2017. Paused RNA polymerase II inhibits new transcriptional initiation. Nat. Genet. 49: 1045–1051. 10.1038/ng.3867 [DOI] [PubMed] [Google Scholar]
- Slawson E. E., Shaffer C. D., Malone C. D., Leung W., Kellmann E., et al. , 2006. Comparison of dot chromosome sequences from D. melanogaster and D. virilis reveals an enrichment of DNA transposon sequences in heterochromatic domains. Genome Biol. 7: R15 10.1186/gb-2006-7-2-r15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X., Goicoechea J. L., Ammiraju J. S., Luo M., He R., et al. , 2011. The 19 genomes of Drosophila: a BAC library resource for genus-wide and genome-scale comparative evolutionary research. Genetics 187: 1023–1030. 10.1534/genetics.111.126540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spofford J. B., 1976. Position-effect variegation in Drosophila, pp. 955–1018 in The Genetics and Biology of Drosophila. Academic Press, New York. [Google Scholar]
- Stabell M., Bjorkmo M., Aalen R. B., Lambertsson A., 2006. The Drosophila SET domain encoding gene dEset is essential for proper development. Hereditas 143: 177–188. 10.1111/j.2006.0018-0661.01970.x [DOI] [PubMed] [Google Scholar]
- Stephens G. E., Craig C. A., Li Y., Wallrath L. L., Elgin S. C., 2004. Immunofluorescent staining of polytene chromosomes: exploiting genetic tools. Methods Enzymol. 376: 372–393. 10.1016/S0076-6879(03)76025-X [DOI] [PubMed] [Google Scholar]
- Strobel E., Dunsmuir P., Rubin G. M., 1979. Polymorphisms in the chromosomal locations of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell 17: 429–439. 10.1016/0092-8674(79)90169-7 [DOI] [PubMed] [Google Scholar]
- Strom A. R., Emelyanov A. V., Mir M., Fyodorov D. V., Darzacq X., et al. , 2017. Phase separation drives heterochromatin domain formation. Nature 547: 241–245. 10.1038/nature22989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant A. H., 1951. A map of the fourth chromosome of Drosophila melanogaster, based on crossing over in triploid females. Proc. Natl. Acad. Sci. USA 37: 405–407. 10.1073/pnas.37.7.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F. L., Cuaycong M. H., Craig C. A., Wallrath L. L., Locke J., et al. , 2000. The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc. Natl. Acad. Sci. USA 97: 5340–5345. 10.1073/pnas.090530797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F. L., Haynes K., Simpson C. L., Lee S. D., Collins L., et al. , 2004. cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol. Cell. Biol. 24: 8210–8220. 10.1128/MCB.24.18.8210-8220.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sved J. A., Chen Y., Shearman D., Frommer M., Gilchrist A. S., et al. , 2016. Extraordinary conservation of entire chromosomes in insects over long evolutionary periods. Evolution 70: 229–234. 10.1111/evo.12831 [DOI] [PubMed] [Google Scholar]
- Tzeng T. Y., Lee C. H., Chan L. W., Shen C. K., 2007. Epigenetic regulation of the Drosophila chromosome 4 by the histone H3K9 methyltransferase dSETDB1. Proc. Natl. Acad. Sci. USA 104: 12691–12696. 10.1073/pnas.0705534104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicario S., Moriyama E. N., Powell J. R., 2007. Codon usage in twelve species of Drosophila. BMC Evol. Biol. 7: 226 10.1186/1471-2148-7-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B., Bachtrog D., 2013. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature 499: 332–335. 10.1038/nature12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B., Bachtrog D., 2015. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 13: e1002078 10.1371/journal.pbio.1002078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath L. L., Elgin S. C., 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 9: 1263–1277. 10.1101/gad.9.10.1263 [DOI] [PubMed] [Google Scholar]
- Yang H. P., Barbash D. A., 2008. Abundant and species-specific DINE-1 transposable elements in 12 Drosophila genomes. Genome Biol. 9: R39 10.1186/gb-2008-9-2-r39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Bachtrog D., 2015. Ancestral chromatin configuration constrains chromatin evolution on differentiating sex chromosomes in Drosophila. PLoS Genet. 11: e1005331 10.1371/journal.pgen.1005331 [DOI] [PMC free article] [PubMed] [Google Scholar]