Abstract
Dense core vesicles (DCVs) can transmit signals by releasing neuropeptides from specialized synaptic regions called active zones. DCVs reach the active zone by motorized transport through a long axon. A reverse motor frequently interrupts progress by taking DCVs in the opposite direction. “Guided transport” refers to the mechanism by which outward movements ultimately dominate to bring DCVs to the synaptic region. After guided transport, DCVs alter their interactions with motors and enter a “captured” state. The mechanisms of guided transport and capture of DCVs are unknown. Here, we discovered two proteins that contribute to both processes in Caenorhabditis elegans. SAD kinase and a novel conserved protein we named Sentryn are the first proteins found to promote DCV capture. By imaging DCVs moving in various regions of single identified neurons in living animals, we found that DCV guided transport and capture are linked through SAD kinase, Sentryn, and Liprin-α. These proteins act together to regulate DCV motorized transport in a region-specific manner. Between the cell body and the synaptic region, they promote forward transport. In the synaptic region, where all three proteins are highly enriched at active zones, they promote DCV pausing by inhibiting transport in both directions. These three proteins appear to be part of a special subset of active zone-enriched proteins because other active zone proteins do not share their unique functions.
Keywords: dense core vesicle, Sentryn, SAD kinase, Liprin, axonal transport
NEURONS are perhaps the most complex of all cells. Their complexity arises from the need to send and receive signals from long processes that extend from the cell body. These processes include one or more dendrites, which receive signals, and a single axon. Part of the axon is specialized to form synapses. This part, the synaptic region, transmits signals to other neurons or muscle cells through the regulated fusion of signaling vesicles with the plasma membrane. Here, we report the discovery of two proteins that have important roles in guiding the motorized transport of one kind of signaling vesicle to the synaptic region and then ensuring that the vesicles halt their transport and become captured in the synaptic region.
Two types of signaling vesicles are found in the synaptic region: synaptic vesicles (SVs) and dense core vesicles (DCVs) (Richmond and Broadie 2002; Sudhof 2004). SVs cluster around a small site known as the active zone (Sudhof 2012), from which they focally release small molecule neurotransmitters in response to electrical signals in the axon. Neurotransmitter release activates postsynaptic receptors that align with the presynaptic active zones. Neuropeptide containing DCVs are often present in lower numbers in the synaptic region, with one study finding ∼7% as many DCVs as SVs at worm motor neuron synapses (Sossin and Scheller 1991; Levitan 2008; Hoover et al. 2014). DCV-mediated neuropeptide signaling is important because it can influence and coordinate the activities of neuronal circuits (Liu et al. 2007; Hu et al. 2011; Bhattacharya et al. 2014; Bhattacharya and Francis 2015; Choi et al. 2015; Chen et al. 2016; Lim et al. 2016; Banerjee et al. 2017) or generally modulate the responsiveness of the presynaptic and postsynaptic cells (Kupfermann 1991; Hu et al. 2015).
DCVs are enriched at synapses relative to the short interaxonal regions between synapses (Wong et al. 2012; Hoover et al. 2014). However, within synapses, the DCV distribution appears random or near-random, not clustered around active zones like SVs. Furthermore, docked DCVs are largely excluded from the active zone, where SVs dock and fuse (Weimer et al. 2006; Hammarlund et al. 2008; Hoover et al. 2014). If docked DCVs represent the only fusion locations, the apparent distributed signaling of DCVs within synapses contrasts sharply with the highly focal fusion of SVs at active zones.
The long distance axonal transport of SVs and DCVs requires a sophisticated cargo transport system. This system uses a network of microtubule tracks and motors that exhibit intrinsic directionality. The microtubules have a plus and a minus end, and there are dedicated plus- and minus-end directed motors. The microtubules in axons are almost uniformly oriented, with their plus-ends pointing outward into the axon (Burton and Paige 1981; Heidemann et al. 1981; Baas and Lin 2011). Both SVs and DCVs use the same motors. The plus-end directed (forward) motor KIF1A moves SVs and DCVs from the cell soma to the synaptic region (Hall and Hedgecock 1991; Pack-Chung et al. 2007; Edwards et al. 2015b), while the minus-end directed (reverse) motor dynein moves them in the opposite direction (Ou et al. 2010; Goodwin et al. 2012; Wong et al. 2012; Cavolo et al. 2015; Edwards et al. 2015b). A recent study in flies found that conventional Kinesin also contributes to the guided forward transport of DCVs (Bhattacharya et al. 2014). During transport from the soma to the synaptic region, both the forward and reverse motors act on the same vesicles, causing them to reverse direction multiple times en route to the synaptic region (Edwards et al. 2015b). Although the significance of this bidirectional transport is unknown, its existence necessitates a mechanism for ensuring the ultimate forward progress of vesicles. In other words, the forward motor(s) must outcompete dynein to ensure that optimal levels of SVs and DCVs accumulate in the synaptic region. We refer to this as “guided” axonal transport, or, simply, guided transport.
Adding complexity, neurons must also have a mechanism to enable SVs and DCVs to enter a captured state in the synaptic region. The mechanism by which DCV capture occurs is unknown. Although one possibility is a physical anchoring mechanism, this has not been proven to be an essential part of DCV capture. Although physical anchors may contribute to vesicle immobilization, entering a true captured state may require a mechanism that inhibits, blocks, or equalizes the actions of both motors.
There has been significant progress in identifying the proteins that contribute to the guided transport of SVs in axons, and to their capture in the synaptic region. These studies revealed that three active zone-enriched proteins, SAD-1 (SAD kinase), SYD-2 (Liprin-α), and SYD-1, affect the axonal transport of SVs (Miller et al. 2005; Wagner et al. 2009; Zheng et al. 2014; Edwards et al. 2015b). The involvement of active zone-enriched proteins in axonal transport was surprising because these proteins seem to affect cargo transport at sites far removed from their sites of enrichment. When acting in an axonal transport context, SAD-1, SYD-2, and SYD-1 promote the forward progress of cargos toward the synaptic region, and, thus, prevent their accumulation at microtubule minus ends in cell somas and dendrites (Miller et al. 2005; Edwards et al. 2015a,b). Earlier studies found that these three active zone-enriched proteins (SAD-1, SYD-2, and SYD-1) also contribute to SV cluster assembly in the synaptic region (Zhen and Jin 1999; Crump et al. 2001; Dai et al. 2006; Patel et al. 2006). Subsequent studies suggested that the role of these proteins in SV cluster assembly can be more sharply defined as a role in capturing SVs into clusters (Stigloher et al. 2011; Wu et al. 2013; Edwards et al. 2015b). Thus, by virtue of the common involvement of SAD kinase, SYD-2 (Liprin-α), and SYD-1 in both the guided transport of SVs and the capture of SVs in clusters, these two processes appear to be linked for SVs.
In contrast to SVs, much less is known about the molecular mechanisms that ensure the guided transport and capture of DCVs. Previous studies found that DCVs, like SVs, use SYD-2 (Liprin-α) to ensure that DCVs accumulate in the synaptic region of axons (Goodwin and Juo 2013). In the absence of SYD-2, there are significantly lower levels of DCVs in axons (Goodwin and Juo 2013) and significantly higher levels of DCVs in cell somas and dendrites (Goodwin and Juo 2013; Edwards et al. 2015b). However, it is unknown if SYD-2 contributes to DCV capture in the synaptic region, or, alternatively, if the lower axonal levels of DCVs in syd-2 mutant axons are simply caused by decreased outward transport of DCVs. In fact, not a single protein with DCV capturing activity has been identified to date.
In the current study, we used the model organism C. elegans to show that DCVs, despite lacking an obvious close association with the active zone like SVs, do in fact use the same specialized subset of active-zone-enriched proteins as SVs to ensure their guided transport and capture. This study revealed the first proteins found to promote the capture of DCVs. Further underscoring the fundamental nature and novelty of these findings, one of the DCV guided transport and capture proteins, which we named Sentryn, is conserved in all animals, and is described here for the first time.
Materials and Methods
C. elegans culture and strains
Worm culture and manipulation essentially followed previously described methods (Brenner 1974; Sulston and Hodgkin 1988; Stiernagle 2006). Briefly, culture media was modified NGM (referred to as NGM-LOB) (Hoover et al. 2014). Prior studies defined the culture plate types “spread plates,” “streak plates,” “locomotion plates,” “24-well plates,” and “96-well solid media culture plates” (Miller et al. 1999; Edwards et al. 2008, 2015b). The wild type strain was N2 except when the strain CB4856 was used for mapping the ce793 mutation and the insertion sites of integrated transgenes. Nonwild-type strains that were used in this study are listed in Supplemental Material, Table S1. The relevant mutations present in the worm strains, along with the methods we used for genotyping them in crosses, are listed Table S2 in File S1. Some strains contained one or more transgenic arrays. Transgenic arrays are listed in Table S3 in File S1 along with their plasmid contents, injection concentrations, and genomic form (extrachromosomal or integrated).
Forward genetic screen for altered DCV localization
We mutagenized 9000 L4s of the strain KG4247 ceIs201 with 27.6 mM EMS in M9 (Sulston and Hodgkin 1988) supplemented with OP-50 bacteria for 4 hr at 20°. Subsequent manipulation and growth procedures to produce F2 grandprogeny of mutagenized animals were essentially the same as we reported for previous genetic screens (Edwards et al. 2015a). At 20-min intervals, we pipetted 50 μl (∼12 worms) of L4-stage F2s from a stirring suspension into each of 18 wells of a 96-well Mat-Tek glass bottom plates (P96G-1.5-F-F; MatTek, Ashland, MA). Before animal distribution, each well was preloaded with 50 μl of 300 μM levamisole (L-9756; Sigma) in water per well. We screened animals in each well for decreased INS-22-Venus fluorescence in the dorsal cord axons, and increased INS-22-Venus fluorescence in the ventral cord somas. At the end of each 20-min screening session, after noting wells containing mutants, we pipetted the contents of each mutant-bearing well onto a predried streak plate using a Pasteur pipet, rinsed the well with 100 μl of M9, and then, immediately after the liquid dried in, clonally distributed the animals to a 96-well solid-medium culture plate (see section C. elegans culture and strains). After 4 days at 20°, we used a sterile toothpick to pick around six L4-stage animals from each well into Mat-Tek wells containing 150 μM Levamisole. After rescreening on the inverted microscope, we noted wells with 100% mutant phenotype and used the corresponding well on the 96-well plate to score behavioral and other phenotypes and to set up stocks. We repeated this screen for three cycles of a week each, for a total of 23,328 F2s. This computes to ∼4.67-fold genomic coverage for an average size gene (Greenwald and Horvitz 1980). However, taking into account an experimentally determined 23% loss of animals during recovery of the mutants, the actual fold-coverage was ∼3.6.
Complementation testing and mapping of new mutations
We determined that ce784 and ce776 are allelic using a complementation test similar to previously described methods (Edwards et al. 2015a). ce776 and ce784 were not further mapped, but were subjected to whole genome sequencing (see below). ce793 was roughly mapped relative to left arm, center, and right arm markers on X using methods similar to those previously described (Edwards et al. 2015a). By this method, we mapped ce793 to a region of X between −11.1 and 2.13 cM.
Whole genome sequencing and identification of phenotype-causing mutation
We produced genomic DNA and libraries for whole genome sequencing as described (Edwards et al. 2015a). Whole genome sequencing and analysis was performed as described (Edwards et al. 2015a). We identified eight missense mutations in the mapped interval, one of which, known only by its clone name C16E9.2, contained an early stop codon. To confirm that loss of function mutations in this gene are associated with the observed DCV trafficking phenotypes, we obtained the independently isolated alleles ok2996 and ok2975 from the Caenorhabditis Stock Center, crossed them into the ceIs201 background containing fluorescence DCVs, and quantitatively assayed the phenotypes. After this confirmation of ce793 as the phenotype-causing mutation isolated in the screen, we then did all further experiments, including rescue experiments, using either ok2996 or ok2975.
Targeted knockout of rimb-1 by CRISPR
We inserted the sequence GC TAG C TAA A TGA after codon 16 of the rimb-1 gene (out of 1276 codons total). The underlined sequence is a NheI site for snip-PCR screening (Paix et al. 2014). The insert contains three stop codons, each in a different reading frame (Paix et al. 2014). We used the oligonucleotide-templated coconversion strategy (Arribere et al. 2014), using dpy-10(cn64) as a coconversion marker, and screened for conversion by PCR followed by restriction digest.
We cloned the Cas9 target sequence CATGCCATAGGAGGATGCGG into the pJP118 gRNA expression cassette as previously described for cloning targeting sequences into pRB1017 (Arribere et al. 2014). pJP118 is a modified version of the published pRB1017 plasmid (Arribere et al. 2014). It contains a modified sgRNA (F+E), with an extended Cas9 binding structure, and removes a potential Pol III terminator by an A–U basepair flip. The oligo template contained 50 bases of homology on each side of this insertion for 113 bases total. The oligo was ordered from Sigma at the 0.2 μmole scale with PAGE purification. The injection mixture was pDD162 (Cas9 plasmid; 50 ng/μl) (Dickinson et al. 2013), pJA58 (dpy-10 gRNA plasmid; 25 ng/μl) (Arribere et al. 2014), KG#843 (rimb-1 gRNA plasmid; 25 ng/μl), and the dpy-10(cn64) and rimb-1(ce828) oligo templates (500 and 2400 nM, respectively). We injected 36 wild type animals with this mixture and cloned 48 F1 rollers, 47 of which yielded progeny. Of the 47 rollers, 34 (72%) showed successful edits, as indicated by NheI cleavage of a PCR product containing the insertion site.
DNA constructs
The plasmids used in this study, along with sources and/or key construction details, are listed in Table S4 in File S1. For Gibson Assembly plasmid construction using NEBuilder, we followed the manufacturer’s instructions, with the following options and modifications: (1) we designed primers with 30 bp of overlap, with all of the overlap on one of the two fragments to be combined; (2) we used 15 ng/μl as a starting template concentration for all PCR reactions, rather than the much lower concentration recommended by the manufacturer; (3) we performed the optional DpnI digestions, gel-purified the PCR fragments, and quantified them with a Nanodrop; (4) the assembly reaction contained 200 ng of the vector fragment and a threefold molar excess of each remaining fragment; (5) each reaction was performed in 10 μl total volume, starting with 5 μl frozen aliquots of 2× NEBuilder master mix; and (6) we directly transformed 2 μl of the reaction using electroporation. In all constructs involving the cloning of PCR fragments, we sequenced the inserts, and used clones containing no mutations in the fragment of interest to make the final stock.
Production and integration of transgenes
We prepared plasmids for microinjection using the Qiagen Tip-20 system according to the manufacturer’s instructions, except that we added a 0.1 M potassium acetate/two volumes of ethanol precipitation step after resuspending the isopropanol-precipitated pellet. We produced transgenic strains bearing extrachromosomal arrays by the method of Mello et al. (1991). For the strn-1 rescue experiments, the host was KG4786 strn-1(ok2975); ceIs201. For all other injection experiments, N2 was the host. We used pBluescript carrier DNA to bring the final concentration of DNA in each injection mixture to 165–175 ng/μl. We integrated transgenes into the genome using 9100 Rads of gamma rays and screening for 100% transmittance of the cotransformation marker as described (Reynolds et al. 2005; Charlie et al. 2006). We mapped the insertion site of ceIs308 by crossing the integrant through CB4856, reisolating and cloning homozygous animals in the F2 generation, and using the resulting mapping lines to map the integration sites relative to SNPs as described (Schade et al. 2005). Table S3 in File S1 lists all the transgenes in this study, their DNA contents, and the injection concentration of each DNA.
Strain constructions
We constructed strains containing multiple mutations or transgenes using standard genetic methods (Edwards et al. 2015a,b). After making a strain composed of two or more mutations, or one or more mutations plus an integrated transgenic array insertion or an extrachromosomal array, we confirmed the homozygosity of each mutation using the genotyping methods in Table S2.
High-pressure freezing electron microscopy
Worms were prepared using high-pressure freeze (HPF) fixation as described previously (Weimer 2006). Briefly, young adult hermaphrodites were placed in specimen chambers filled with Escherichia coli and frozen at −180° and high pressure using a Leica SPF HPM 100. Freeze substitution was then performed on the frozen samples using a Leica Reichert AFS. Samples were held at −90° for 107 hr with 0.1% tannic acid and 2% OsO4 in anhydrous acetone. The temperature was increased at 5°/hr to −20°, kept at −20° for 14 hr, and increased by 10°/hr to 20°. After fixation, samples were infiltrated with 50% Epon/acetone for 4 hr, 90% Epon/acetone for 18 hr, and 100% Epon for 5 hr. Finally, samples were embedded in Epon and incubated for 48 hr at 65°. All specimens were prepared in the same fixation and subsequently blinded for genotype. Ultrathin (40 nm) serial sections were cut using a Leica Ultracut 6, and collected on formvar-covered, carbon-coated copper grids (EMS, FCF2010-Cu). Poststaining was performed using 2.5% aqueous uranyl acetate for 4 min, followed by Reynolds lead citrate for 2 min. Images were acquired starting at the anterior reflex of the gonad using a Jeol JEM-1220 transmission electron microscope operating at 80 kV. Micrographs were collected using a Gatan digital camera at a magnification of 100,000×. Images were taken of cross-sections of the dorsal cord of three animals for each strain. Cholinergic synapses were identified based on established morphological criteria (White et al. 1986). The presynaptic density of cholinergic synapses generally orients at a ∼45° angle to its postsynaptic targets (muscle), and often forms a dyadic synapse with another nerve process and muscle. A synapse was defined as a set of serial sections containing a dense projection (DP) and two flanking sections from both sides without DPs. DCVs were identified as spherical structures with a dark gray center and an average diameter of ∼40 nm. Consecutive sections were aligned, so it was clear when a DCV appear to span more than one section based on its location in the image. DCVs that appeared to span more than one section were counted only in the section in which they were most pronounced—darkest color and clearest membranes. Images were analyzed using NIH ImageJ software and macros provided by the Jorgensen laboratory. Serial images were aligned using TrakEM2 before annotating organelle structures in ImageJ. Analysis of these structures was performed using MATLAB scripts written by the Jorgensen laboratory and Ricardo Fleury.
Light level imaging
File S1 describes the methods used for growth and mounting of strains for imaging, image acquisition and processing, quantitative image analysis, special imaging methods, immunostaining of formaldehyde-fixed animals, and time lapse microscopy.
Quantification and statistical analysis
We performed all statistical comparisons using the unpaired t-test, Welch corrected (for comparisons between two selected groups) or ANOVA followed by the Tukey Kramer post-test (for comparisons involving three or more groups) using Graphpad Instat 3 (Graphpad Software). All statistical parameters, including the exact value of n, what n represents, and what error bars represent are reported in the figure legends, or in the text for data presented in text form.
Data availability
Strains and plasmids are available upon request from the corresponding author, from the Caenorhabditis Genetics Center, or from AddGene. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental files available at FigShare. File S1 contains Figures S1–S8 and Tables S1–S4, Supplemental Materials and Methods and Supplemental Material References. File S2 contains detailed EM DCV counts broken down by strain, worm, and synapse. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6965684.
Results
SAD kinase and Sentryn mutants isolated in a genetic screen for DCV transport defects
To identify proteins that contribute to the axonal transport and/or synaptic capture of DCVs, we performed a forward genetic screen for mutants with both lower DCV levels in their axons and higher DCV levels in their cell somas (Figure 1A). To mark DCVs, we used a genomically integrated transgene that expresses a fluorescent DCV cargo in cholinergic motor neurons. Synapses in these neurons use SVs to release acetylcholine (ACh) onto muscles, but they also corelease neuropeptides from DCVs. We screened ∼23,300 grandprogeny of EMS-mutagenized animals, which is ∼3.6-fold genomic coverage for knocking out an average size gene (see Materials and Methods).
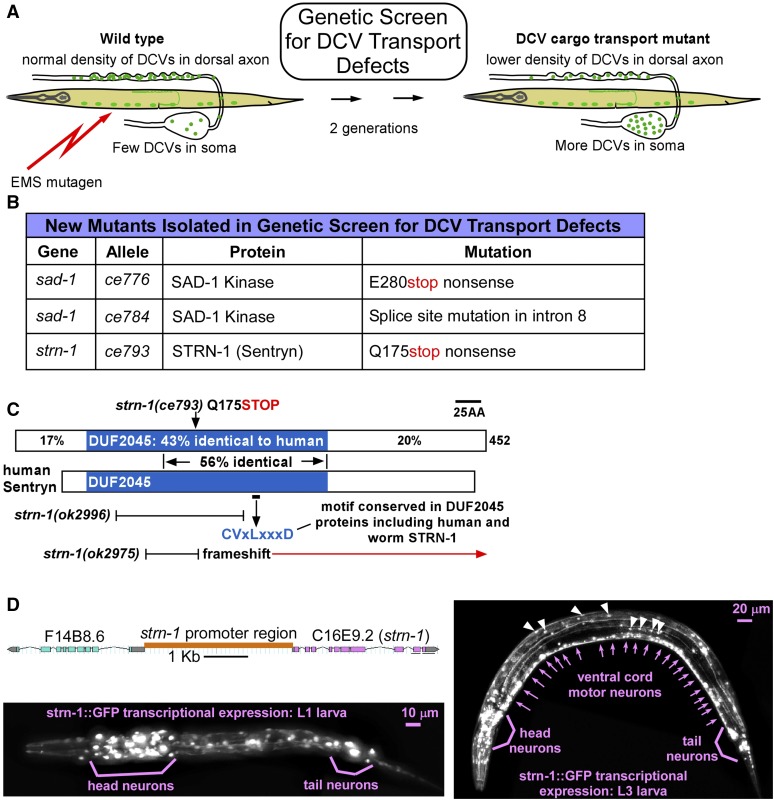
Figure 1.
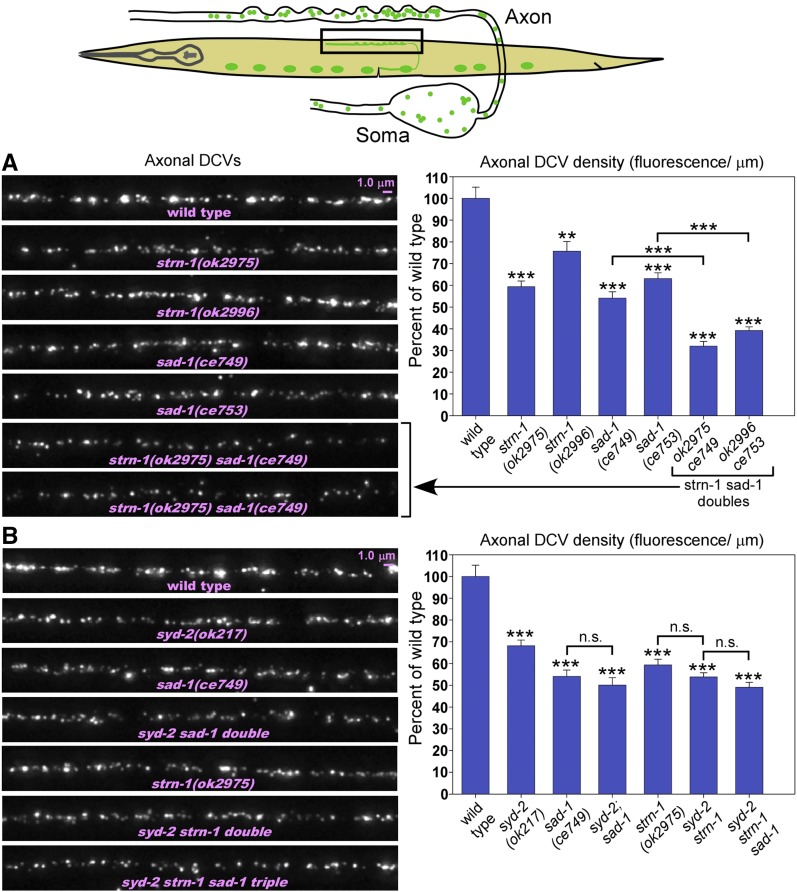
Forward genetic screen for DCV transport mutants identifies SAD kinase and a novel, conserved neuronal protein. (A) Drawings illustrate the forward genetic screen. The screen used EMS to mutagenize wild type animals carrying the integrated transgene ceIs201, which coexpresses the neuropeptide INS-22-Venus (to mark dense core vesicles) and mCherry (as an expression control) in ventral cord cholinergic motor neurons. We screened the F2 grandprogeny of mutagenized animals on 96-well glass bottom Mat-Tek plates using an inverted microscope and selected animals with lower densities of DCVs in their axons and/or higher densities in somas. (B) Summary of mutants isolated in the genetic screen. (C) Comparison of human and worm Sentryn proteins and mutation locations. Sentryn’s human ortholog is similar in length. Percent identities between corresponding regions of the human and worm proteins are indicated. The conserved motif “CVLxxxD” is found in both human and worm Sentryn in a large region that is 56% identical between the two proteins. Three allele lesions disrupting the conserved DUF2045 region (domain of unknown function) are shown. The two proteins were aligned using the Align X module of Vector NTI. (D) Scale drawing of strn-1 promoter region and images of larvae expressing GFP from the strn-1 promoter in the ceEx461 extrachromosomal transgenic array. Sentryn is highly expressed throughout the nervous system. White arrowheads denote two hypodermal nuclei (lateral) and two muscle nuclei (medial) that also express the transcriptional reporter.
We investigated four mutants from this screen in detail. We previously reported that one mutant contains a temperature-sensitive mutation in UNC-104 (KIF1A), a plus-end (forward) motor for SVs and DCVs (Edwards et al. 2015b). Complementation tests, mapping, and whole-genome sequencing revealed that two of the remaining mutants had loss-of-function mutations in SAD-1 (SAD kinase), and one mutant had a loss-of-function mutation in a novel protein with no known functional domains (Figure 1B).
We named the novel protein STRN-1 (Sentryn) as a metaphor for its function (see Discussion). C. elegans Sentryn is predicted to be a medium-size cytosolic protein of ∼53 kDa. It appears to be conserved in all animals. Human Sentryn is slightly smaller. Human and worm Sentryn share 29% overall amino acid identity, 43% identity over a domain of unknown function (DUF) that covers about half of each protein, and 56% identity in a segment covering about two-thirds of the DUF (Figure 1C).
To determine the cell types in which Sentryn is expressed, we made transgenic animals that expressed a strn-1 (C16E9.2) transcriptional reporter driving GFP. The GFP reporter was highly expressed throughout the nervous system, but was also expressed at lower levels in at least some other cell types, including some hypodermal and muscle cells (Figure 1D). Although additional regulatory elements could contribute to Sentryn’s expression, our findings agree with a prior microarray profiling study, which found that transcripts of Sentryn (then referred to as C16E9.2) were enriched pan-neuronally in embryos and larvae (Von Stetina et al. 2007).
Sentryn and SAD kinase act cell-autonomously to control the distribution of DCVs in cholinergic motor neurons
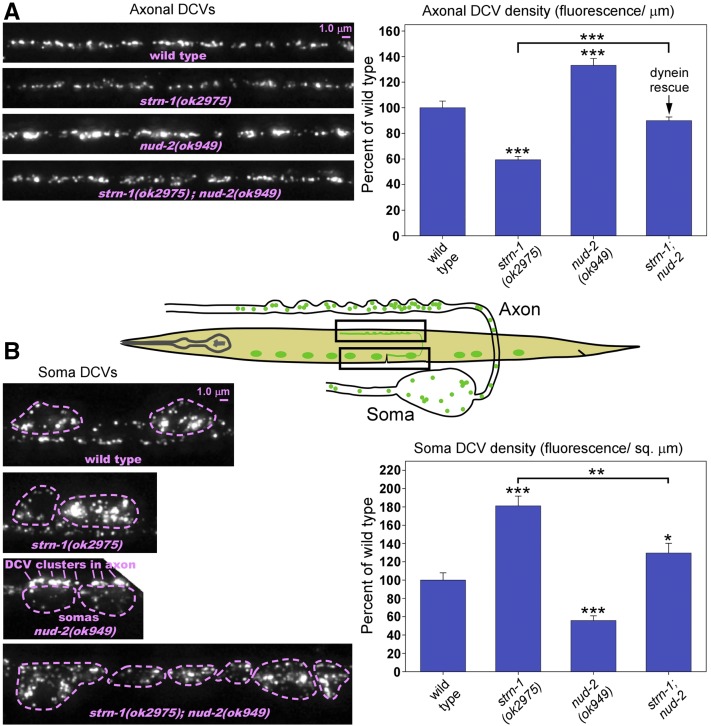
Quantitative imaging of fluorescently tagged DCVs in cholinergic motor neurons revealed that knocking out either Sentryn or SAD kinase strongly affected the distribution of DCVs. For the strn-1 mutant quantitative experiments, we used the previously isolated null deletion allele strn-1(ok2975). In strn-1(ok2975) mutant axons, the DCV density was reduced to 60% of wild type, while DCV density in mutant somas was increased to ∼180% of wild type (Figure 2, A and B). Transgenic expression of a wild type strn-1 cDNA in cholinergic motor neurons fully rescued the axonal phenotype, showing that the decreased axonal DCV density results from the absence of Sentryn in these neurons (Figure 2A). The soma phenotype was also significantly rescued, although not to wild-type levels (Figure 2B). Multiple, independently isolated alleles of strn-1 showed a similar severity of the soma phenotype (Figure 2B). This suggests that the soma phenotype results from absence of Sentryn rather than second-site mutations, and that the transgenic rescue strain may not have had optimal expression timing or levels.
Figure 2.
Sentryn acts cell-autonomously to control the distribution of DCVs in ventral cord cholinergic motor neurons. (A and B) Rectangles in drawing indicate regions imaged. Representative images and quantification of DCV fluorescence in axons and somas in the indicated genotypes. The DCV marker INS-22-Venus (a tagged neuropeptide) is expressed from the integrated transgene ceIs201. Images are identically scaled. Dashed lines in (B) outline cell somas. Graph data are means and SE from 14–15 animals each. Unmarked bars are not significantly different from wild type. *, **, and *** indicate P-values that are <0.05, <0.01, or <0.001, respectively. Asterisks that are not above relationship bars compare the indicated bar to wild type. See Figure S1 for analogous sad-1 null mutant data. See Figure S2 for expression controls, which showed no significant difference from wild type.
We obtained similar results in quantitative analyses of a sad-1 null mutant. For sad-1 mutant experiments, we used the previously isolated putative null alleles sad-1(ce749) or sad-1(ce753) (Edwards et al. 2015a). A sad-1 null mutant was not significantly different from the strn-1 null mutant in its axonal and cell soma DCV levels (55 and 180%, respectively), nor in the extent to which a sad-1 cDNA expressed in cholinergic motor neurons rescued each of these phenotypes (Figure S1, A and B). Similar to the strn-1 null, the significant, but incomplete, rescue of the soma phenotype likely results from suboptimal transgene expression timing or levels. The independently isolated sad-1(ce753) allele also showed a build-up of DCVs in its soma that was not significantly different from sad-1(ce749) (Figure S5).
Sentryn’s function in ensuring optimal levels of DCVs in axons appears to have been conserved from worms to humans because the expression of a human Sentryn cDNA in worm cholinergic motor neurons fully rescued axonal DCV levels in the Sentryn null mutant (Figure 2A). The mutant rescued with human Sentryn had a small but significant hypermorphic phenotype of increased DCV levels in axons (Figure 2A). However, the human Sentryn cDNA was unable to rescue the cell soma phenotype (Figure 2B). Similar to the strn-1 and sad-1 rescue experiments with the worm cDNAs, this is consistent with the idea that the functions of Sentryn and SAD kinase in somas are sensitive to precise protein levels and/or timing of expression.
In summary, the above results show that Sentryn and SAD kinase each function cell autonomously to ensure an optimal distribution of DCVs in neurons. The absence of either protein results in strong cell biological effects, in which DCV levels are reduced by almost half in the synaptic regions of axons, and increased almost twofold in somas.
The altered distribution of DCVs in mutants lacking Sentryn or SAD kinase occurs in a dynein-dependent manner
The results of a previous study suggested that SAD kinase can alter the interactions of SVs with the SV motorized transport system (Edwards et al. 2015b). Based on those earlier findings, and the shared phenotypes of strn-1 and sad-1 mutants, we hypothesized that the high soma/low axon DCV ratio in strn-1 and sad-1 mutants results, in part, from a disruption in the balance of forward and reverse transport (i.e., the guided transport) of DCVs in axons. This disruption may ultimately cause dynein-mediated (reverse) transport of DCVs to prevail relative to forward transport, thus leading to a build-up of DCVs in cell somas and contributing to lower DCV levels in axons.
Consistent with this hypothesis, reducing the function of dynein in neurons significantly rescued the disrupted DCV distribution in mutants lacking Sentryn or SAD kinase. As dynein null mutants are sterile lethal in C. elegans, we used a null mutation in the dynein regulator NUD-2 for this experiment. A prior study found that the deletion allele nud-2(ok949) strongly reduces dynein motor activity on SVs in C. elegans motor neurons to an extent indistinguishable from strong loss-of-function mutations in DHC-1 (dynein heavy chain) (Ou et al. 2010). Reducing dynein function with this mutation restored wild-type DCV levels to strn-1 null mutant axons (Figure 3A), and significantly lowered cell soma DCV accumulation (Figure 3B). We found similar results for sad-1; nud-2 double mutants (Figure S3, A and B).
Figure 3.
Altered distribution of DCVs in mutants lacking Sentryn occurs in a dynein-dependent manner. (A and B) Rectangles in drawing indicate regions imaged. Representative images and quantification of DCV fluorescence in axons (A) and somas (B) in the indicated genotypes. The DCV marker INS-22-Venus (a tagged neuropeptide) is expressed from the integrated transgene ceIs201. Images are identically scaled. Dashed lines in (B) outline cell somas. Graph data are means and SE from 14–15 animals each. Bars without asterisks are not significantly different from wild type. *, **, and *** indicate P-values that are <0.05, <0.01, or <0.001, respectively. Asterisks that are not above relationship bars compare the indicated bar to wild type. See Figure S2 for soluble mCherry expression controls, which showed no difference from wild type. See Figure S3 for analogous sad-1 null mutant data.
The above results are consistent with the hypothesis that Sentryn and SAD kinase normally reduce the impact of the dynein motor on DCVs in axons by affecting the balance of plus and minus-end directed transport. Theoretically, this could occur by inhibiting dynein, by promoting the activity of one or more plus-end motors, or both.
Sentryn and SAD kinase promote anterograde DCV transport in the axon initial segment
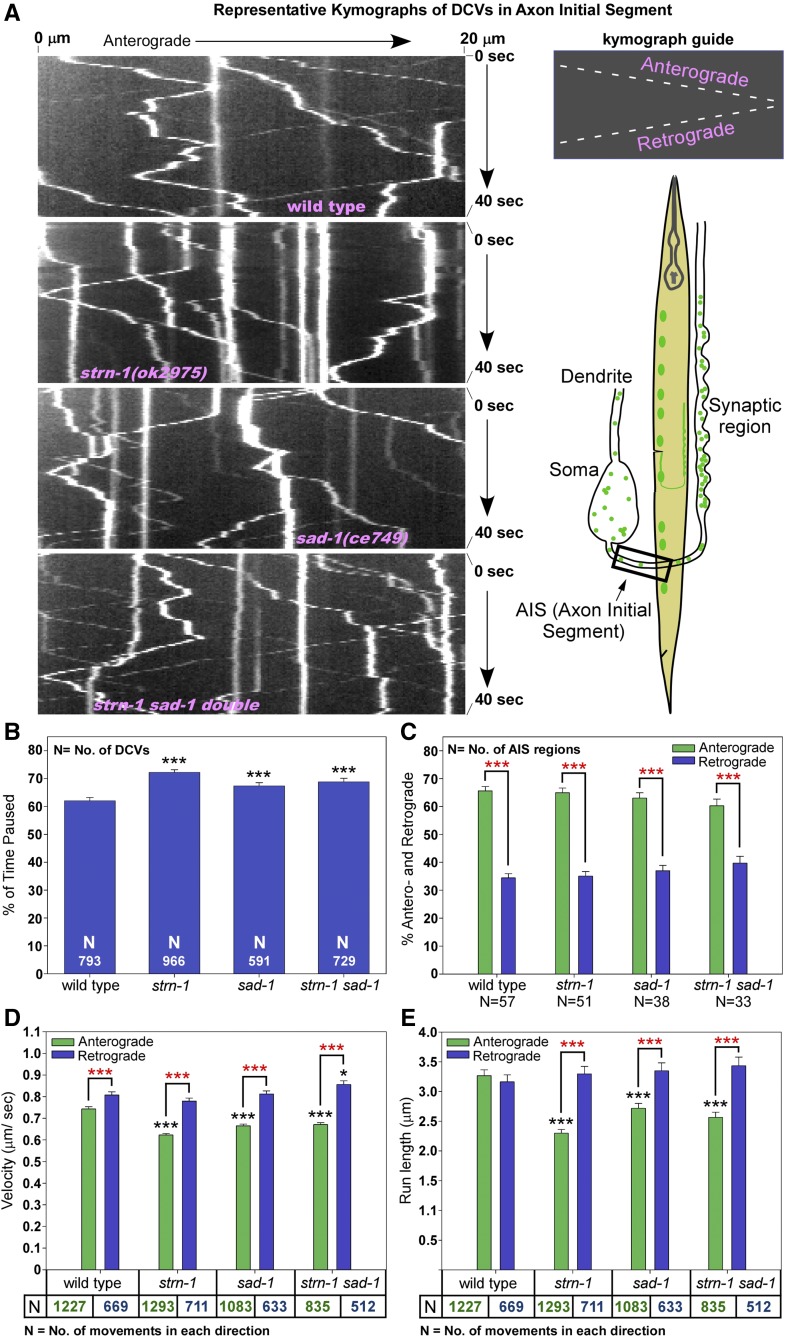
The results thus far do not elucidate whether Sentryn and SAD kinase act primarily during DCV guided transport, or later in ensuring that captured DCVs do not leave the synaptic region via motors, or in both processes. Defects in either process would ultimately alter the steady-state distribution of DCVs between the soma and the synaptic region. To determine if Sentryn and SAD kinase can influence the guided transport of DCVs in the axonal region near the cell soma (i.e., far removed from the synaptic region), we analyzed the steady-state levels, as well as the transport, of DCVs in the axon initial segment of wild type and the mutants.
The axon initial segment in C. elegans cholinergic motor neurons is >60 μm away from the synaptic region. In the axon initial segment, sad-1 and strn-1 null mutants accumulated DCVs at levels that were ∼180% of wild type (Figure S4A). This defect occurred in multiple independent alleles of each gene and could be rescued cell-autonomously in a strn-1 null mutant (Figure S4A). Similar to the cell soma build-up in the same mutants, the phenotype was not exacerbated in a strn-1 sad-1 double mutant, indicating that both mutations disrupt the same process or pathway in this context. However, unlike the cell soma and synaptic region defects, the axon initial segment phenotype could not be rescued by strongly reducing the activity of dynein using a nud-2 null mutation (Figure S4B).
To directly test if the build-up of DCVs in the axon initial segment correlates with one or more guided transport defects, we collected time lapse images of fluorescently tagged DCVs moving in wild type animals and in mutants lacking Sentryn and/or SAD kinase. In total, we analyzed ∼2000 DCV movements per strain, and a similar number of DCV pauses from ∼6000 images per strain. We analyzed run lengths and velocities for both anterograde (forward) and retrograde (reverse) movements, as well as the percent of time each DCV spent in a paused state. Figure 4A shows representative kymographs for each strain. In wild type, DCVs spent ∼40% of their time moving and ∼60% of their time in a paused state (Figure 4B). Mutants lacking Sentryn and/or SAD kinase showed a highly significant increase (∼16%) in the percentage of time each DCV spent in a paused state (Figure 4B).
Figure 4.
Reduced anterograde motor activity in the axon initial segment in mutants lacking Sentryn or SAD-1. (A) Representative kymographs of INS-22-Venus–loaded DCVs in the axon initial segment of DA and DB-type cholinergic motor neurons. Worm drawing indicates region imaged. INS-22-Venus is expressed from the integrated transgene ceIs201. (B–E) Graphs plotting various indicated parameters extracted from the time lapse analyses. For (C) the percent of movements in each direction is based on the combined movements of all DCVs per time lapse recording rather than the combined movements of each DCV in the recording. Error bars are SEM. Bars without asterisks are not significantly different from wild type. *, **, and *** indicate P-values that are <0.05, <0.01, or <0.001, respectively. Black asterisks compare the marked bar to the wild type value. Red asterisks compare the indicated two bars in a group. Unmarked bars are not significantly different from wild type or the other group member. Total recorded time lapse time (minutes:seconds in order of strains as shown): 42:45, 38:15, 24:45, and 28:30.
All strains, including wild type, showed bidirectional DCV movements, with similar percentages of DCVs moving in the dominant, anterograde direction (∼60–65%; Figure 4C). DCV movements in mutants lacking Sentryn and/or SAD kinase had a similar retrograde velocity and retrograde run length as wild type (Figure 4, D and E). However, DCV movements in the mutants lacking Sentryn and/or SAD kinase had lower anterograde velocities and anterograde run lengths than wild type (∼24% lower). This effect was highly significant (Figure 4, D and E). The fact that anterograde run length and anterograde velocity were similarly reduced suggests that the reduction in anterograde velocity contributes to the reduction in anterograde run length, although other factors may contribute to the anterograde run-length reduction since it seemed proportionately greater. These results demonstrate that Sentryn and SAD kinase normally help one or more forward motors outcompete dynein during bidirectional transport of DCVs in this region, and that they also act to prevent DCVs from persisting in a paused state in this region. The combined effects of the pausing and forward transport defects are likely additive and could be sufficient to explain the buildup of DCVs in this region in the mutants. In addition, the effect on anterograde motor velocity suggests that Sentryn and SAD kinase can directly or indirectly stimulate anterograde motor activity.
Overall, the major relevant point of the above findings is that Sentryn and SAD kinase can exert their effects on guided transport occurring at a site that is far-removed from the synaptic region, and thus far-removed from the sites of DCV capture. A simple explanation may be that Sentryn and SAD kinase act locally in the axon initial segment to modulate DCV guided transport. Relevant to this point, Liprin-α, which is highly enriched at synapses and acts in the same system as Sentryn and SAD kinase, has both synaptic and nonsynaptic sites-of-action in neurons (Wyszynski et al. 2002; Ko et al. 2003a). Alternatively, Sentryn and SAD kinase may exert a long distance effect transmitted by motors.
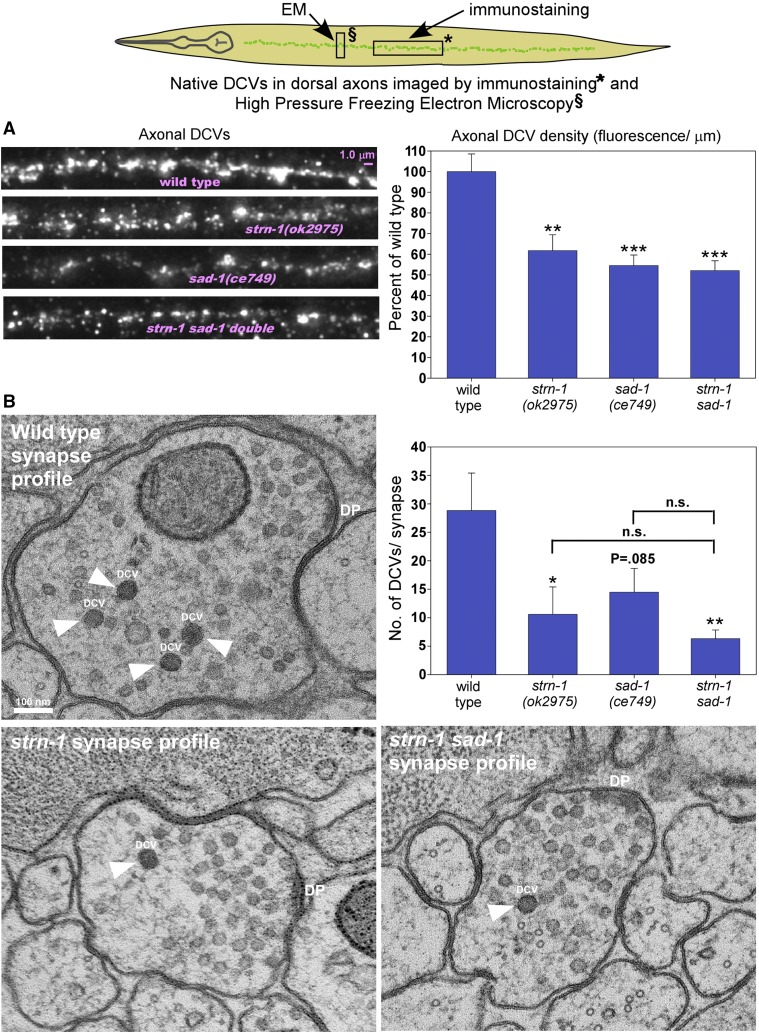
Two other complementary approaches demonstrate reduced DCVs in axons and synapses of mutants lacking Sentryn and/or SAD kinase
To confirm that our transgenic model accurately represents native DCVs with endogenous cargos, we first immunostained nontransgenic animals with an antibody against the DCV cargo EGL-21 (Carboxypeptidase E), which is a neuropeptide processing enzyme that is known to be copackaged with neuropeptides in DCVs (Fricker 1988; Cool et al. 1997; Rindler 1998) and has previously been used to image DCVs in C. elegans (Hoover et al. 2014). We imaged the dorsal nerve cord, which contains only neuronal processes, mostly axons, and is devoid of cell somas. The results showed that DCV densities were reduced to ∼60 and 55% of wild type in strn-1 and sad-1 null mutants, respectively, which closely matched the transgenic results (Figure 5A). strn-1 sad-1 double mutants had axonal DCV densities that were not significantly different from either single mutant, suggesting that, in most of these axons, Sentryn and SAD kinase function in the same process or pathway. We were unable to reliably visualize immunostained motor neuron somas in any of the strains.
Figure 5.
Two other complementary approaches demonstrating DCV distribution defects in mutants lacking Sentryn and/or SAD kinase. Rectangles in drawing show regions imaged in (A) (immunostaining) and (B) (HPF-EM). (A) Representative images and quantification of native EGL-21 (Carboxypeptidase E) immunostaining in dorsal cord axons of animals with the indicated genotypes. Graph data are means and SE from 18 animals each. ** and *** indicate P-values that are <0.01 or <0.001, respectively. (B) Representative single-section EM images and quantification of cholinergic motor neuron synapses visualized by HPF EM. Graph data are means and SE of total DCVs from 17, 12, 17, and 16 synapses reconstructed from 220, 115, 150, and 134 40-nm sections from a total of three animals each for wild type, strn-1, sad-1, and strn-1 sad-1 double mutants, respectively. See File S2 for DCV counts from each synapse in each animal. * and ** indicate P-values that are <0.05 or <0.01, respectively. Labels in the representative images are as follows: White arrowheads and “DCV” (dense core vesicle), DP (dense projection). Note that each single 40-nm section shown in the representative images is only 7–10% of a synapse.
To determine if the reductions in axonal DCVs were occurring at synapses, we quantified native DCVs in cholinergic motor neuron synapses using HPF electron microscopy (EM). In all, we sampled an average of 15 synapses each for wild type and strains lacking Sentryn and/or SAD kinase. The analysis was performed in a genotype-blinded manner. The results closely matched the results we obtained by imaging transgenically tagged DCVs in axons, where we had no knowledge of the precise localization of the DCVs relative to synapses. Wild type synapses, reconstructed from an average of 13 thin (40 nm) sections that spanned each synapse, had an average of 29 DCVs (Figure 5B). DCVs at strn-1 and sad-1 null mutant synapses were reduced to 38 and 48% of wild type, respectively. Due to the small sample size and high variability of DCV numbers between synapses, this was only statistically significant for the strn-1 mutant, but the sad-1 mutant value was close to significant (Figure 5B). Native DCVs at strn-1 sad-1 double mutant synapses appeared further reduced, to just 22% of wild type. Although this was highly significant when compared to wild type, its levels were not statistically different from the levels of either single mutant for this sample size (Figure 5B).
When comparing the immunofluorescence and EM results, note that axonal DCVs, especially in the strn-1 sad-1 double, appear more strongly reduced in the EM study than in the immuofluorescence study. Note that the EM study imaged only cholinergic motor axons, while the immunofluorescence study imaged all axons and all processes in the dorsal nerve cord, including cholinergic axons, GABAergic axons and postsynaptic processes, and axons from the large peptidergic RID process that largely lacks active zones (Lim et al. 2016). These differences indicate that noncholinergic and/or solely peptidergic neurons may have different requirements for Sentryn and/or SAD kinase than cholinergic motor neurons.
From the combined immunostaining and EM results we conclude that our transgenic model accurately represents native DCVs with endogenous cargos, and that the reductions in axonal DCVs, at least in the case of the strn-1 sad-1 double mutant in the EM analysis, are occurring at synapses. Furthermore, the EM results hint at a possible synthetic interaction between the strn-1 and sad-1 mutations at cholinergic synapses, because axonal DCV levels in the double were more significantly different from wild type than either single mutant and were reduced to about half of each single mutant. However, the EM data alone lacks the statistical power to conclude this.
Sentryn and SAD kinase act in the same DCV transport/capture pathway as Liprin-α
A prior study found that SAD kinase acts in the same pathway as Liprin-α (SYD-2) for SV cluster assembly (Patel et al. 2006). Similarly, we previously showed that SAD kinase and Liprin-α act in the same pathway for organelle transport and SV cluster stability (Edwards et al. 2015a,b). Having established the validity of the transgenic model using immunostaining and EM, we returned to its greater quantitative and statistical power to investigate the functional relationship between Sentryn, SAD kinase, and Liprin-α in DCV transport and capture. To do this, we constructed all possible combinations of double mutants and the triple mutant. Two different combinations of strn-1 sad-1 double mutants showed a synthetic interaction in which the double was significantly worse than either single mutant. Axonal DCV densities in these doubles were reduced from about half of wild type in each single mutant, to about one-third of wild type in the doubles (Figure 6A). However, the cell soma DCV levels in the doubles were about the same as each single mutant (Figure S5). This suggests that Sentryn and SAD kinase have at least one nonoverlapping function in ensuring wild type levels of captured DCVs in axons. However, we did not observe this effect in a syd-2 (Liprin-α) null background. The syd-2 strn-1 sad-1 triple was not significantly different from either the syd-2 strn-1 or syd-2sad-1 doubles for the low axonal DCV density phenotype (Figure 6B). Furthermore, the syd-2 strn-1 and syd-2sad-1 doubles were not significantly different from the strn-1 and sad-1 singles (Figure 6B). We observed no additive effects for the cell soma phenotype for any combination of mutants (Figure S5).
Figure 6.
In axons, Sentryn and SAD kinase act in the same DCV transport/capture pathway as the active-zone-enriched protein SYD-2 (Liprin-α). (A and B) Rectangle in drawing shows region imaged. Representative images and quantification of DCV fluorescence in axons in the indicated genotypes. The DCV marker INS-22-Venus (a tagged neuropeptide) is expressed from the integrated transgene ceIs201. Graph data are means and SE from 14–16 animals each. n.s., **, and *** indicate P-values that are not significant, <0.01, or <0.001, respectively. Asterisks not above relationship bars compare the indicated bar to wild type. See Figure S5 for the corresponding data for cell soma DCV density and the soluble mCherry expression controls, the latter of which showed no significant difference from wild type.
The above data suggest that Sentryn, SAD kinase, and Liprin-α largely function in the same process or pathway in controlling the distribution of DCVs between the cell soma and capture sites in the axons, although Sentryn and SAD kinase each have at least one nonoverlapping function in determining DCV levels in cholinergic motor neuron axons.
Although the data show that, for the most part, Liprin-α is not able to fully exert its functions without Sentryn or SAD kinase, we did find at least one exception to this rule. Sentryn is not required for the neuron to maintain wild type levels of DCVs in its dendrite (Figure S6), whereas dendrites lacking either SAD kinase or (SYD-2) Liprin-α have strong accumulations of DCVs in dendrites compared to wild type (Figure S6 and previously reported for syd-2 mutants) (Goodwin and Juo 2013; Edwards et al. 2015b). Thus, both SAD kinase and Liprin-α appear able to function without Sentryn in restricting DCV accumulation in dendrites.
SAD kinase and Liprin-α are a specialized subset of active zone-enriched proteins
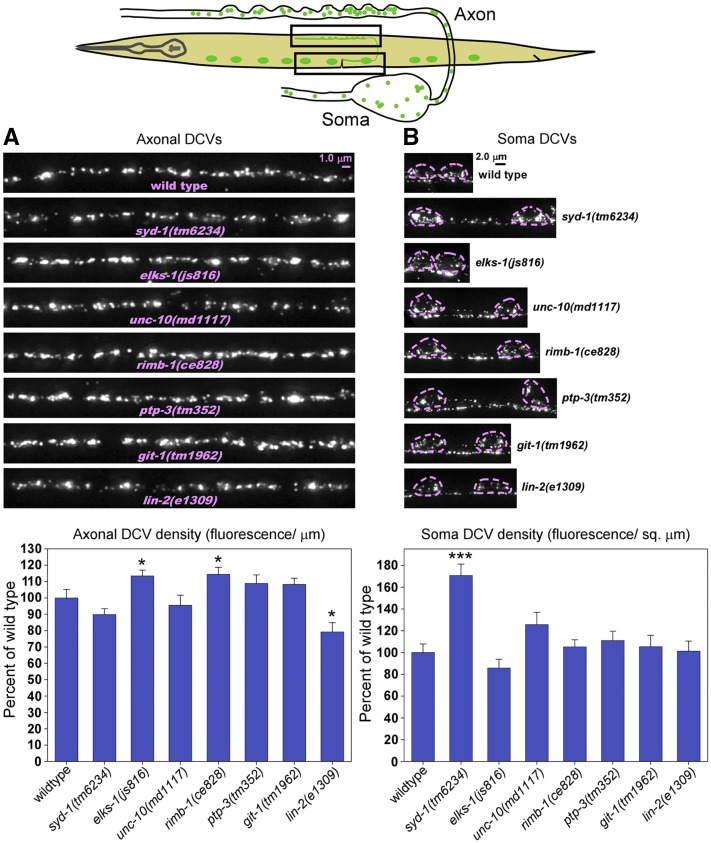
Liprin-α is highly enriched near active zones (Ackley et al. 2005; Yeh et al. 2005; Weimer et al. 2006; Fouquet et al. 2009), which are the specialized presynaptic regions around which SVs are clustered and where SV exocytosis occurs. Similarly, SAD kinase is also enriched at active zones (Inoue et al. 2006). To test whether other active-zone-enriched proteins have roles in DCV transport/capture, we quantified the axonal and cell soma DCV levels of null mutants for seven other active-zone-enriched proteins.
None of the mutants tested showed the unique combination of low axonal/high cell soma DCV density that we observed in syd-2, sad-1, and strn-1 mutants. However, one of the mutants, the syd-1 null, had high cell soma DCV levels, comparable to syd-2, sad-1, and strn-1 mutants. Its axonal DCV levels were lower than wild type, but not quite significantly so (Figure 7, A and B). Thus, although SYD-1 may be equally as important as Liprin-α, SAD kinase, and Sentryn for preventing the build-up of DCVs in cell somas, its role in ensuring optimal transport and/or capture of DCVs is redundant or significantly less essential.
Figure 7.
Null mutations in most other active-zone-enriched proteins do not significantly impact DCV transport and/or capture in cholinergic motor neurons. (A and B) Rectangles in drawing indicate regions imaged. Representative images and quantification of DCV fluorescence in axons and somas in the indicated genotypes. The DCV marker INS-22-Venus (a tagged neuropeptide) is expressed from the integrated transgene ceIs201. Images are identically scaled. Dashed lines in (B) outline cell somas. Graph data are means and SE from 13–15 animals each. Unmarked bars are not significantly different from wild type. * and *** indicate P-values that are <0.05 or <0.001, respectively, and compare the indicated bar to wild type. See Figure S7 for soluble mCherry expression controls, which showed no difference from wild type. The C. elegans gene names and the common names of each protein are as follows, along with one or more references demonstrating that the protein is enriched at active zones: SYD-1 (SYD-1) (Owald et al. 2010), ELKS-1 (ELKS/ERC/Bruchpilot) (Ohtsuka et al. 2002; Wang et al. 2002; Deken et al. 2005; Wagh et al. 2006), UNC-10 (RIM: Rab3 interacting molecule) (Wang et al. 1997; Koushika et al. 2001), RIMB-1 (RIM-binding protein) (Wang et al. 2000; Liu et al. 2011), PTP-3 (LAR: Leukocyte-common antigen related) (Ackley et al. 2005), GIT-1 (GIT: G protein coupled receptor kinase 2 interacting protein) (Kim et al. 2003), LIN-2 (CASK: name ontology unknown) (Olsen et al. 2005).
One of the mutants, the lin-2 (CASK) null, had axonal DCV levels that were slightly, but significantly, lower than wild type (∼79% of wild type); however, its cell soma levels were not significantly different (Figure 7, A and B). Due to the weakness of this lin-2 DCV phenotype, we did not investigate it further.
DCVs are captured to a specialized region of the axon
The results presented earlier show that mutants lacking Sentryn and/or SAD kinase have defects in the guided transport of DCVs. Although guided transport defects could contribute to the high soma/low synaptic region distribution of DCVs in Sentryn and SAD kinase mutants, defects in DCV capture could also cause, or contribute to, this altered distribution. The reason a capture defect could contribute is because the synaptic region of C. elegans cholinergic motor neuron axons is restricted to a short segment in the middle of the axon that is flanked by long proximal and distal asynaptic regions (White et al. 1986). Therefore, a failure to capture DCVs could result in lower DCV levels in the synaptic region as one or more forward motors carry uncaptured DCVs beyond the synaptic region, and/or as dynein competes to carry uncaptured DCVs back toward the soma. Indeed, the motorized loss of DCVs from boutons (synaptic sites) after capture failure in wild type animals has been demonstrated for DCVs in Drosophila motor neurons (Wong et al. 2012; Bulgari et al. 2014).
DCV capture defects in cholinergic motor neurons could theoretically be observed by quantifying the accumulation of DCVs in the distal asynaptic region, which would occur if the motorized transport of DCVs failed to halt in the synaptic region. However, before assaying for uncaptured DCVs in this region it was important to test whether DCVs are even captured to the synaptic region of C. elegans motor neuron axons as opposed to being uniformly distributed in those axons. To test this, we analyzed the distribution of DCVs in a single motor neuron: DA9. The axon in DA9 consists of the following segments: a commissure that leads from the cell soma to the synaptic region, a 100 μm synaptic region composed of ∼40 SV clusters that are often associated with boutons (swellings) in the axon, and a 400 μm long, distal asynaptic region. SVs are tightly captured in the synaptic region and, with the exception of a few escapers, do not populate the long asynaptic region (Wu et al. 2013; Edwards et al. 2015b).
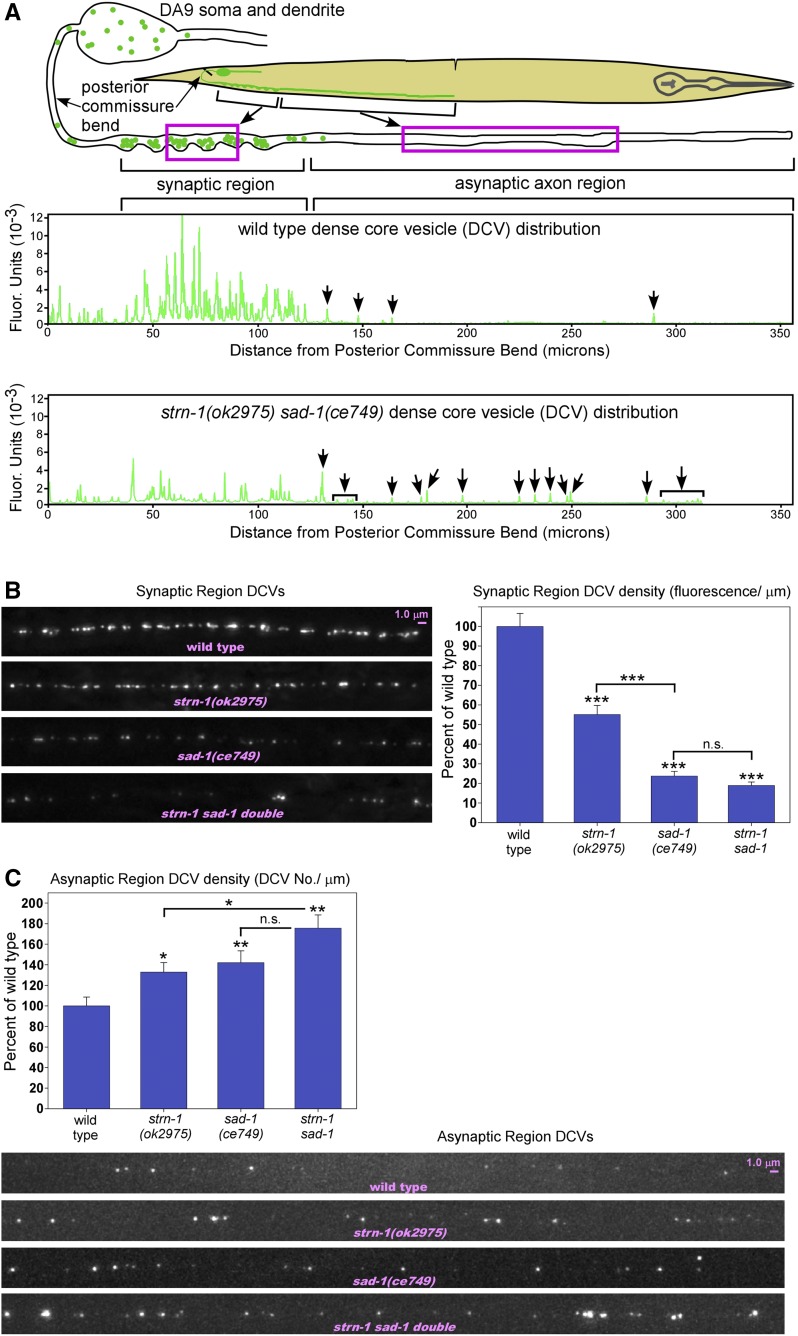
Using a new integrated transgene that tags DCVs in DA9, we visualized DCVs at high resolution along the entire ∼0.5 mm length of the DA9 axon in wild type and used overlapping images from a large field-of-view camera to reconstruct composite plots of fluorescence intensity along the axon. The wild type plot in Figure 8A (the top plot) shows a representative fluorescence distribution plot chosen from 15 sets of overlapping images. These data demonstrate that DCVs, like SVs, are highly enriched in the synaptic region and are largely excluded from the distal asynaptic region, consistent with a region-specific capture mechanism for DCVs in this axon.
Figure 8.
Sentryn and SAD kinase inhibit the plus-end accumulation of DCVs in distal asynaptic region to keep them captured in the synaptic region. (A) Rectangles in drawing indicate regions of the DA9 motor neuron that were imaged. Graphs plot fluorescence intensity of the DCV marker INS-22-Emerald as a function of distance from the posterior commissure bend. High resolution images acquired with a large field-of-view Flash 4.0 camera were used to reconstruct the dorsal axon from multiple images. Arrows indicate DCVs in the asynaptic region. In wildtype, DCVs are tightly captured in a small region of the axon and do not significantly accumulate in the long asynaptic region, whereas, in strn-1 sad-1 double mutants, DCVs do accumulate in this region (arrows). INS-22-Emerald is expressed from the genomically integrated transgene array ceIs308. See Figure S8 for mCherry expression controls showing that the observed differences are not caused by changes in expression of the transgene. (B and C) Representative images and quantification of DCV fluorescence/μm in the synaptic region (B) or DCV number/μm in the asynaptic region (C) in the indicated genotypes. Note that only the center part of each region was imaged as indicated in the drawing in (A). The DCV marker INS-22-Emerald is expressed from the integrated transgene array ceIs308. Graph data are means and SE from 14–15 animals each. n.s., *, **, and *** indicate P-values that are not significant, <0.05, <0.01, or <0.001, respectively.
Sentryn and SAD kinase are DCV capture proteins
We next used this new transgene to determine the extent to which Sentryn and SAD kinase are responsible for capturing DCVs in the DA9 synaptic region. To do this we first quantified DCVs in the synaptic and asynaptic regions in mutants lacking these proteins. Because the asynaptic region we quantified is distal to the synaptic region, a capture defect would cause DCVs to accumulate in this region.
DCVs in the synaptic region were reduced to ∼50% of wild type in strn-1 null mutants, while synaptic region DCVs in sad-1 mutants and in the strn-1 sad-1 double were more strongly reduced to ∼20% of wild type (Figure 8B). In the asynaptic region, DCV density (number/μm) was significantly increased over wild type in the mutants lacking Sentryn and/or SAD kinase (Figure 8C). Even though strn-1 sad-1 double mutants had ∼fivefold fewer DCVs than wild type in their synaptic regions, thus biasing the mutants against random DCVs escaping capture, strn-1 sad-1 double mutants had almost twice as many uncaptured DCVs as wild type in their long asynaptic region (Figure 8, A and C). We obtained similar results in a quantitative analysis of syd-2(ok217) null mutants (synaptic region DCV density reduced to 17.5 ± 1.5% of wild type, P < 0.0001; asynaptic region DCV density increased to 136.1 ± 9.5% of wild type, P = 0.0094; N = 14 animals).
The above results are consistent with a defect in capture, but they do not rule out the possibility that DCVs are accumulating in the distal asynaptic region by a process other than active transport, such as diffusion. If true, this could still be considered a capture defect (for example, a defect in tethering that could lead to diffusion of DCVs away from the synaptic region). However, our data thus far suggest that Sentryn and SAD-1 exert their effects on guided transport by regulating the motorized DCV transport system, and we thus hypothesized that capture failure in mutants lacking Sentryn and/or SAD-1 would be related to motorized transport misregulation.
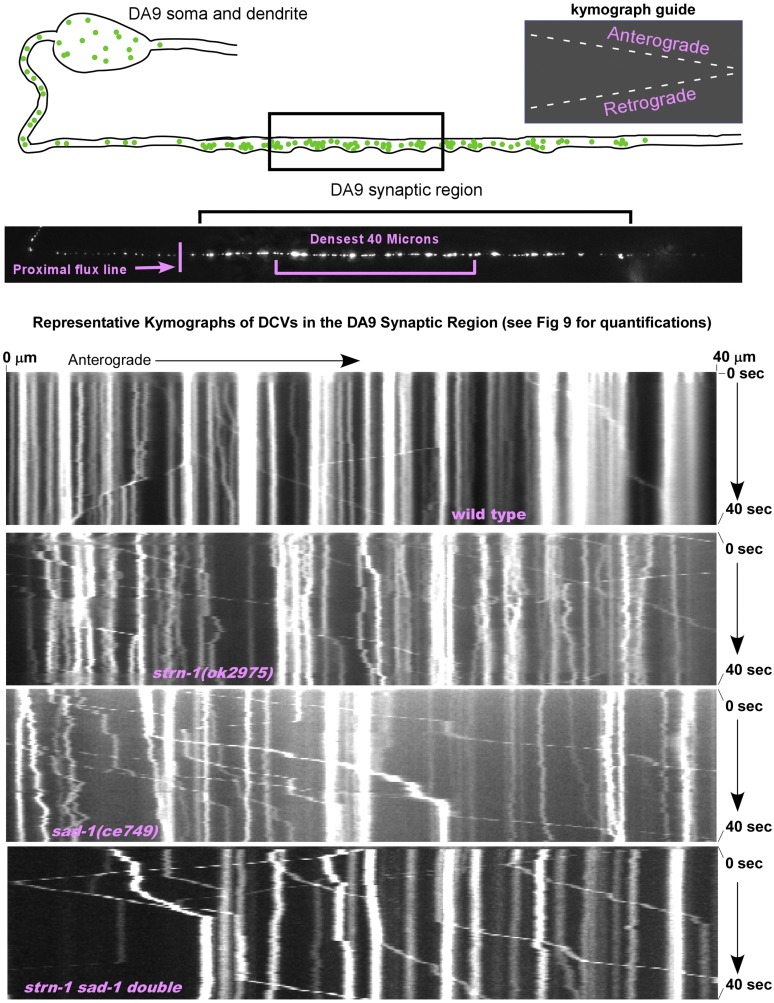
If DCV capture has a significant motor regulation component, and if mutants lacking Sentryn and/or SAD kinase have a defect in capturing DCVs, it should be detectable as an overall increase in DCV active transport in the synaptic region. To test this, we recorded time lapse movies of DCVs moving in the interior of the DA9 neuron synaptic region in wild type and mutants lacking Sentryn and/or SAD kinase. We then analyzed the DCV movements using kymographs (Figure 9A and Figure 10).
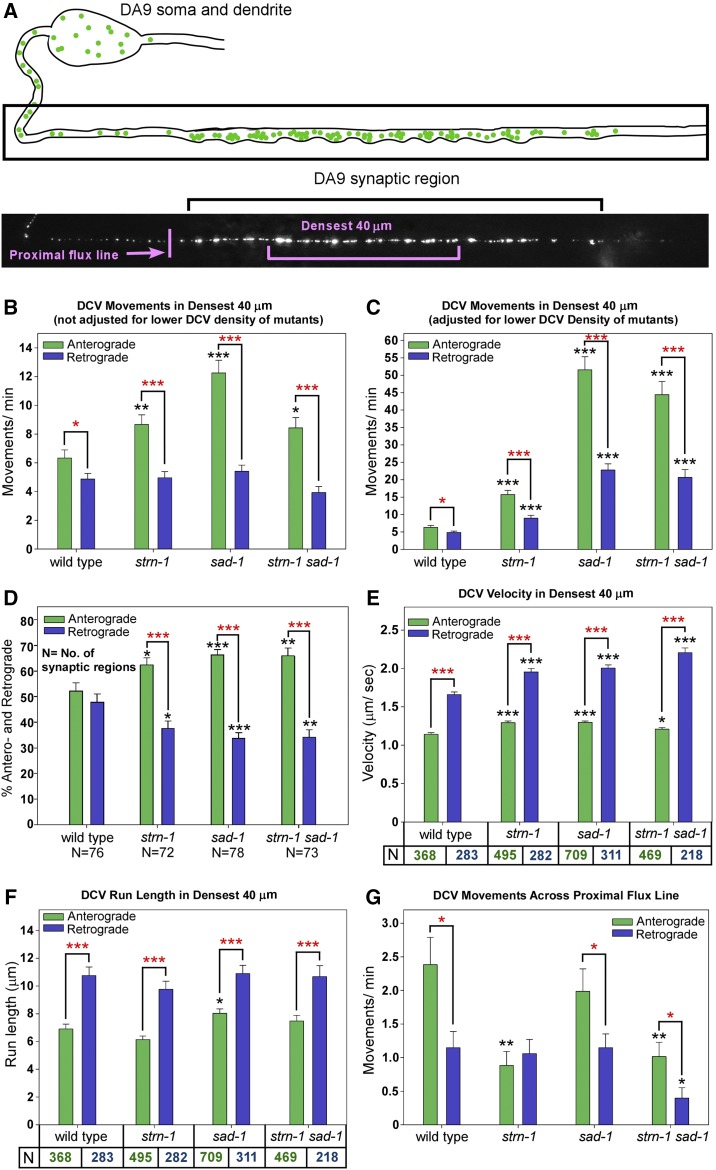
Figure 9.
Impaired DCV capture in mutants lacking Sentryn and/or SAD-1. (A) Drawing and representative image of the wildtype DA9 motor neuron synaptic region. Noted are the “Densest 40 μm” and the “Proximal flux line” used for this experiment. The DCV marker INS-22-Emerald is expressed from the integrated transgene array ceIs308. (B and C) Quantification of anterograde and retrograde DCV movements in the densest 40 μm of the synaptic region in the indicated genotypes. Both the absolute number of movements (B) and the number of movements normalized for DCV density (C) are shown. Normalization used the data from Figure 8B, which quantified DCV density in the densest 40 μm of the same strains. Data are from the following numbers of animals, in the same left-right order as the figures: 77, 76, 24, and 27. Each time lapse lasted for 45 sec. Error bars are SEM. *, **, and *** indicate P-values that are <0.05, <0.01, or <0.001, respectively. Black asterisks compare the marked bar to the wild type value. Red asterisks compare the indicated two bars in a group. Unmarked bars are not significantly different from wild type or the other group member. See Figure 10 for representative kymographs showing DCV movements in all four strains. See (E and F) for Ns. (D) The percentage of movements in each direction is based on the combined movements of all DCVs per time lapse recording rather than the combined movements of each DCV in the recording. The indicated Ns are the number of time lapse recordings with at least one DCV movement in the synaptic region. See (B and C) for statistical annotations. (E and F) Mean DCV velocity and run length of DCVs in the densest 40 μm of the synaptic region. N is the number of movements analyzed in each direction and is also the number of movements analyzed in (B and C). See (B and C) for statistical annotations. (G) Mean DVCs fluxing in each direction across a line just proximal to the start of the synaptic region. N = 30 animals = 30 time lapses for each strain. Each time lapse lasted 45 sec. See (B and C) for statistical annotations.
Figure 10.
Representative kymographs of DCVs in the DA9 synaptic region. Representative kymographs of INS-22-Emerald-loaded DCVs in the synaptic region of the DA9 cholinergic motor neuron. Rectangle in drawing indicates region imaged. INS-22-Emerald is expressed from the integrated transgene ceIs308. See Figure 9 for the quantifications of various parameters of DCV movements in this region. Note that the three mutant strains have fewer DCVs in this region, but they tend to be more active (less captured) than wild-type on a per vesicle basis and tend to move more often in an anterograde direction.
Based on our EM data showing an average of 29 DCVs per cholinergic motor neuron SV cluster (Figure 5B) and based on precise counts of SV clusters in the 100 μm DA9 synaptic region averaging 40.59 ± 1.33 (N = 17 animals), the 40 μm densest region should contain a minimum of 471 DCVs. We found that 11 puncta moved by active transport in either direction minute in wild type (Figure 9B and Figure 10). Assuming each punctum represents a single DCV, this estimate characterizes the captured state in cholinergic motor neurons as 2.3% of DCVs moving per minute.
Despite having 50 and 23% as many DCVs in this region as wild type, mutants lacking Sentryn and/or SAD kinase had significantly higher anterograde DCV movements/min than wild type, but their retrograde movements/min were similar to wild type (Figure 9B and Figure 10). However, after normalizing to the wild type density, all three mutants had highly significant increases in DCV movements/min/vesicle in both directions (Figure 9C). These density-normalized fold-increases for anterograde movements/min ranged from ∼threefold in the strn-1 mutant to ∼9- to 10-fold in the sad-1 mutant and the strn-1 sad-1 double. The latter two strains were not significantly different from each other in anterograde or retrograde movements per minute per vesicle.
The results showed that, even though the increased activity was highly significant for both directions of movement, the mean proportion of movements in the anterograde direction for mutants lacking Sentryn and/or SAD kinase was significantly higher than in wild type, and the mean proportion of movements in the retrograde direction was significantly lower (Figure 9D and Figure 10). About two-thirds of the DCV movements in the mutants occurred in the anterograde direction, whereas in wild type about half of the movements occurred in each direction (Figure 9D and Figure 10). We note that this combination of both a higher level of motorized DCV movements (i.e., uncaptured DCVs) coupled with unbalanced anterograde transport could contribute to the decreased levels of DCVs in the synaptic region in the mutants, and to the accumulation of DCVs in the distal asynaptic region, which is anterograde to the synaptic region. This contrasts with motor forces acting on DCVs during transport (i.e., in the axon initial segment, Figure 4) and at the proximal synaptic region boundary (see below), which would tend to cause DCVs to move toward and accumulate in the soma and dendrite in mutants lacking Sentryn and/or SAD kinase.
DCVs in the mutants had significantly higher anterograde and retrograde velocities than wild type, with increases ranging from ∼6 to 34%, depending on the mutant and the direction of movement (Figure 9E). We cannot rule out that DCV collisions/crowding contributed to these relatively small velocity changes since wild type has more DCVs than the mutants. However, the DCV movements in the synaptic region of wild type were quite processive, with run lengths averaging 11 and 7 μm per movement in the retrograde and anterograde directions, respectively (Figure 9F). These distances spanned multiple mini-clusters averaging ∼1/4th of the target region. However, although the mutants exhibited many-fold more DCV movements than wild type on a per vesicle basis, their DCV run lengths were not significantly different from wild type in either direction, with the exception of the sad-1 single mutant, which had a slightly (∼16%), but significantly, longer anterograde run length (Figure 9F).
We next sought to determine whether increased DCVs entering the synaptic region might contribute to the increased proportion of uncaptured DCVs in the synaptic region of the mutants. For example, the moving DCVs in the mutants may represent DCVs that have recently entered the synaptic region but have not yet dissociated from their microtubule tracks at the time of observation. To test this, we measured the flux of DCVs in both directions across a line immediately proximal to the start of the synaptic region. Wild type DCVs entered the synaptic region at a rate of 2.4/min, with significantly fewer DCVs moving in the opposite direction (1.1/min; Figure 9G). In sad-1 mutants, the number of DCVs fluxing across the proximal synaptic region boundary in both directions was not significantly different from wild type. strn-1 mutants had a similar number of DCVs as wild type fluxing away from the synaptic region. However, strn-1 mutants had significantly fewer DCVs moving toward the synaptic region, such that the number entering and exiting was not significantly different. The strn-1 sad-1 double showed significantly fewer DCVs moving across the proximal boundary in both directions (Figure 9F). Thus, the increased DCV movements per minute in the synaptic region of the mutants cannot be explained by a greater flux of DCVs into the synaptic region.
When analyzing reverse (retrograde) flux at the boundary of the synaptic region, it is important to remember that mutants lacking STRN-1, SAD-1, or both proteins have 50, 23, and 19% as many DCVs in their DA9 synaptic region as wild type, respectively (Figure 8B). Thus, their driving forces for retrograde flux are effectively reduced by these percentages, and their actual per vesicle tendencies to move retrogradely across the boundary line are estimated to be around two, four, and five times higher than actually observed, respectively (the inverse of those percentages). By this reasoning, DCVs at the proximal synaptic region boundary in mutants lacking Sentryn and/or SAD kinase could have tendencies for retrograde movement that equal or exceed their tendencies for anterograde movements. If this is true, then a major barrier preventing even greater retrograde depletion of DCVs from the synaptic region due to unbalanced transport at the proximal synaptic region boundary in the mutants could be the equilibrium reached when DCVs are lost from the synaptic region (anterogradely and retrogradely) and the driving (mass action) force from the synaptic region is reduced.
In summary of this section, the results support the hypothesis that DCV capture is closely coupled to regulation of the active transport system (i.e., motors and/or microtubule tracks) by Sentryn and SAD kinase. DCVs in the synaptic region of mutants lacking Sentryn and/or SAD kinase have many-fold more movements per minute per DCV than wild type. Within the synaptic region, the increased active transport activity occurs in both directions, but it is disproportionately increased in the anterograde direction. These results cannot be explained by an increase in new DCVs entering the synaptic region in the mutants.
Discussion
In this study, we used a forward genetic screen to discover roles for SAD kinase and Sentryn in DCV guided axonal transport and synaptic capture. SAD-1 was first discovered in C. elegans over 17 years ago based, in part, on the diffuse localization of SVs in sad-1 mutant axons (Crump et al. 2001). Subsequent studies focused on the role of SAD-1 role in regulating the distribution of SVs and the assembly of SV clusters in the synaptic region (Patel et al. 2006; Hung et al. 2007; Kim et al. 2008, 2010; Klassen et al. 2010; Chia et al. 2012). Part of the significance of the current study is that it is the first to demonstrate a role for SAD kinase in the guided transport and synaptic capture of DCVs.
Adding significance, the current study is also the first to report a function for a novel protein that is conserved in all animals. We chose the name Sentryn for the new protein. Based on “sentry,” the name is a metaphor for its role in “standing guard” over captured vesicles and “protecting” them from counterproductive motor activity, both during guided transport and after capture.
In addition to discovering the first proteins with roles in promoting DCV capture (Sentryn, SAD kinase, and Liprin-α), three significant conceptual advances came out of this study. First, we showed that the seemingly distinct functions of the guided transport of DCVs and the synaptic capture of DCVs are connected, with the identical group of molecules acting in both processes to regulate active transport. An important implication of this finding is that these same proteins must also contribute to the critical transition between transport and capture. Second, we showed that Sentryn, SAD kinase, and Liprin-α can all act as part of the same system in both contexts (guided transport and capture), although SAD kinase and Sentryn may have one or more nonoverlapping functions in some contexts. And third, we showed that the dual functions of DCV guided transport and DCV capture arise from a distinct group of active-zone-enriched proteins, because most active-zone-enriched proteins do not share these functions. Below, we expand on these concepts and explain their significance.
Sentryn, SAD kinase, and Liprin-α contribute to capturing DCVs in a specialized region of the axon
This study shows that DCVs can be captured to a specialized nonterminal region of an axon. The region with the captured DCVs precisely coincides with the region where SVs are captured into clusters in the same neuron (Edwards et al. 2015b). Prior studies in Drosophila have demonstrated that DCVs have specific capture mechanisms involving the regulation of DCV active transport under various activity states in the terminal boutons of motor neurons (Shakiryanova et al. 2006; Wong et al. 2012; Bulgari et al. 2014, 2017; Cavolo et al. 2016). However, it was unknown whether DCV capture mechanisms were, like SV capture mechanisms, sufficiently robust to restrict DCVs to a nonterminal bouton region, or whether DCVs would be relatively evenly dispersed throughout the axon. The implication of our analysis of the DA9 motor neuron is that DCVs, like SVs, have a robust capture mechanism that keeps them concentrated at synapses regardless of where those synapses are located (i.e., DCVs do not just move to the terminal region of the axon by default).
A link between DCV transport and synaptic capture: shared molecules regulating motorized transport at two different locations
Studies of Drosophila DCVs moving in the synaptic region provided the first observations of DCVs being captured in neurons in vivo (Shakiryanova et al. 2006; Wong et al. 2012). These and other studies also provided the first clues suggesting that the capture mechanism can be altered for capturing DCVs under different physiological conditions (Cavolo et al. 2016; Bulgari et al. 2017). However, prior to the current study, no proteins that promote DCV capture had been identified, although Drosophila Huntingtin was recently identified as an inhibitor of DCV capture (Bulgari et al. 2017).
A significant finding of this study is that DCV guided transport and synaptic capture appear to be linked through a common set of molecules (Sentryn, SAD kinase, and Liprin-α). One common function that links guided transport and capture is the regulation of DCV motorized transport. Our results do not distinguish between regulation by direct effects on motor activity vs. regulation by modifications of the microtubule tracks on which the motors run. However, relevant to this, we note that past studies have found direct interactions between Liprin-α and the KIF1A motor (Shin et al. 2003; Wagner et al. 2009) as well as between Liprin-α and conventional Kinesin (Miller et al. 2005). This suggests that direct motor regulation may at least contribute to Liprin’s functions. This study does not rule out the possibility that other motors in addition to KIF1A are relevant for DCV anterograde transport. Indeed, a recent study found that, in Drosophila, conventional Kinesin functions together with UNC-104 (KIF1A) after DCVs enter the axon and begin guided transport (Lim et al. 2017).
Although our results show that both guided transport and capture are connected through a common set of molecules acting together in the same neuron at the same time, the relationship of SAD-1/Sentryn/Liprin-α to the motorized transport system must drastically change as DCVs switch from between guided transport and capture. During guided transport, Sentryn, SAD kinase, and Liprin-α must ensure the dominance of outward DCV transport. However, after DCVs reach the synaptic region and begin the transition to the captured state, these three proteins must suppress unbalanced plus- and minus-end movements to prevent most DCVs from moving past the synaptic region or returning to the soma. Our Sentryn mutant data provide an example of this transition. In the axon initial segment, Sentryn promotes escape from this region by reducing pausing and stimulating the velocity and run length of anterograde movements (Figure 4, D and E). This promotes guided transport to the synaptic region. However, in the synaptic region, Sentryn now inhibits transport in both directions, such that any movements that do occur are balanced and do not cause DCVs to permanently exit the synaptic region (Figure 9, B and C). Thus, in addition to suppressing DCV movements in both directions in the synaptic region, Sentryn and SAD kinase ensure balanced anterograde and retrograde movements for any DCVs that escape capture. This favors the recapture of DCVs in the synaptic region.
One possible explanation for these seemingly opposite actions of a single group of proteins in different regions of the axon is that the focally high concentration of Liprin-α and SAD kinase at active zone regions, as well as Sentryn (Edwards et al. 2018) may alter their interactions with DCVs in this region and trigger the transport-to-capture transition. This may occur as DCVs pass near active zones during transport. Indeed, although Sentryn appears enriched at active zones, it also appears at lower concentrations throughout the synaptic region (Edwards et al. 2018). This opens the possibility that DCVs need not contact the active zone to become captured if they can interact with lower concentrations of Sentryn, and perhaps other capture proteins peripheral to the active zone.
Capture may not be an immediate and strong effect. Indeed, direct observations of DCVs being captured in living animals have shown that DCVs often shoot by proximal boutons before becoming captured in more distal boutons (Wong et al. 2012). Sometimes DCVs even reverse course and exit the synaptic region before circulating back and becoming captured (Wong et al. 2012). Additionally, neuronal activity can alter capture dynamics (Shakiryanova et al. 2006; Cavolo et al. 2016).
A previous study reported that DCVs and SVs in Drosophila motor neurons accumulate in a distal asynaptic region in Liprin-α mutants (Li et al. 2014). Interestingly, that study also found that what appeared to be “floating” fragments of active zones also accumulated along with the vesicles. Since SVs are known to physically associate with active zones (Stigloher et al. 2011; Kittelmann et al. 2013), we find this intriguing and consistent with the idea that, in mutants lacking active-transport-inhibiting capture proteins, motors may rip vesicles plus attached or tethered active zone components from synaptic sites and carry the complexes to asynaptic regions.
The meaning of “capture”
It is important to note that, in this context, “capture” need not involve physical restraints such as tethers. Indeed our results are at least equally consistent with the concept that capture results from motor regulation or from a combination of motor regulation and physical tethering. Our finding that Sentryn and SAD kinase can regulate motorized transport in both the axon initial segment and the synaptic region, coupled with our finding that Sentryn and SAD kinase contribute to DCV capture, make it reasonable to suggest that regulation of motorized transport contributes to capture. An alternative, but more complex, explanation is that Sentryn and SAD kinase function as motorized transport regulators in one context (i.e., in the axon initial segment) and promote physical tethers in another (i.e., in the synaptic region).
If tethering mediates capture, then the most obvious anchor for tethering is the DP of the active zone, from which fine filaments can be seen connecting nearby SVs in a network (Stigloher et al. 2011). One potential problem with this “active zone tethering capture hypothesis” is that two recent studies found that dual elimination of either RIM plus RIM-binding protein, or ELKS plus RIM, led to disassembly of the active zone/DP and eliminated tethering and priming/docking of SVs (Acuna et al. 2016; Wang et al. 2016). However, in those double mutants, wild-type numbers of SVs were still captured at synapses (Acuna et al. 2016; Wang et al. 2016). Importantly, however, the presynaptic localization of Liprin-α was not affected (Wang et al. 2016). This result, combined with the current study, supports the hypothesis that Liprin-α (and, by extension, Sentryn and SAD kinase that act in the same system) can mediate SV capture even in the near-absence of an active zone and its associated clustering and tethering properties.
It is also possible that tethering contributes to capture by preventing diffusion of DCVs after motor inhibition, or that tethering represents a second capture mechanism that operates in a specific region of the synapse. A previous study found evidence that at least some DCVs appear connected to fine-filament tethers (Stigloher et al. 2011). However, it is unclear whether DCV tethers would be sufficient to protect against the forces exerted by motors. Future studies may benefit from a thoughtful consideration that the captured state may involve a significant transport regulation component.
The mechanism by which DCVs ultimately disengage from the motorized transport system to become captured is unknown. A disengagement pathway may be triggered when a DCV passes through a subregion that exceeds a threshold concentration of unsaturated capture proteins. Our data suggest that disengagement involves a transition from anterograde-dominated transport (during guided transport through the axon) to a reversible paused state (capture in the synaptic region) with rare reversions to balanced bidirectional transport (within the synaptic region), as suggested by the data in Figure 4 and Figure 9.
A specialized set of active-zone enriched proteins with dual functions in DCV guided transport and synaptic capture
The connection we established between a subset of active-zone-enriched proteins and DCV capture is significant because no previous study has found a special relationship between DCVs and active zones. Prior studies of active zones have focused on their specificity in promoting the capture and docking of SVs (Stigloher et al. 2011; Sudhof 2012; Kittelmann et al. 2013; Wu et al. 2013; Edwards et al. 2015b, 2018). Active-zone-enriched proteins have not, to our knowledge, been shown to regulate the capture of DCVs. Indeed, unlike SVs, docked DCVs analyzed by EM are largely excluded from the region near the active zone, which is mostly occupied by SVs, and the DCV distribution within synapses appears random or near-random (Weimer et al. 2006; Hammarlund et al. 2008; Hoover et al. 2014).
Among the 12 known active-zone-enriched proteins in C. elegans, SAD kinase and Liprin-α appear to form a special subset with roles in DCV guided transport and synaptic capture. We can now add Sentryn to this small subset, since it also appears to be an active zone enriched protein (Edwards et al. 2018). It was reasonable for us to test other active zone proteins for roles in DCV guided transport and synaptic capture. Indeed, six of the seven active-zone-enriched proteins we tested have been shown to interact directly or indirectly with Liprin-α (SYD-2) in C. elegans and/or other animals: SYD-1 (Chia et al. 2012), ELKS (Ko et al. 2003b; Dai et al. 2006; Kittelmann et al. 2013), RIM (Schoch et al. 2002), CASK (Olsen et al. 2005), LAR (Serra-Pages et al. 1998), and GIT (Kim et al. 2003).
This study did not test two other known active-zone-enriched proteins in C. elegans: UNC-13 and the recently reported Clarinet protein (Xuan et al. 2017). A prior study found that unc-13 near-null mutants have increased DCV levels in C. elegans motor neuron axons (Sieburth et al. 2007), which is the opposite phenotype from what we observed in strn-1, sad-1, and syd-2 mutants. Although the role of UNC-13 in DCV exocytosis (Sieburth et al. 2007) could mask any effects on the transport/capture of DCVs, there is currently no evidence that UNC-13 promotes the transport or capture of DCVs or SVs.
It is important to note that “active-zone-enriched” does not mean that the proteins exclusively localize to, or exclusively function at, the active zone. In their guided transport role, SAD kinase, Sentryn, and Liprin-α can affect the motorized transport of vesicles and organelles at sites far removed from active zones, such as in dendrites, the boundaries between somas and dendrites or axons, and the axon initial segment (this study) (Goodwin and Juo 2013; Edwards et al. 2015a,b, 2018). In immunostaining studies of mammalian cultured neurons (both at the light level and the EM level using multiple independent polyclonal and monoclonal antibodies), Liprin-α localization is described as “partially synaptic,” and there is extensive somatodendritic staining, especially near the postsynaptic density and postsynaptic membrane where Liprin-α functions in AMPA receptor targeting (Wyszynski et al. 2002; Ko et al. 2003a). In C. elegans, SYD-2 (Liprin-α) is broadly expressed, even in non-neuronal cells (Stefanakis et al. 2015).
Sentryn interacting proteins?
In a separate study, we showed that Sentryn is required to produce normal length DPs (the structure at the center of the active zone) (Edwards et al. 2018), similar to what was previously shown for syd-2 (Liprin-α) null mutants (Kittelmann et al. 2013). Sentryn is completely dependent on SYD-2 for its localization to active zones, but not for its transport to the synaptic region (Edwards et al. 2018). Thus, at active zones, Sentryn may physically interact with SYD-2, or with a SYD-2-dependent complex. However, this interaction, if present, may be transient or unstable because Sentryn has eluded discovery by conventional biochemical approaches for over 25 years despite intensive investigation of SVs, DCVs, and active zone proteins by many laboratories.
Acknowledgments
We thank Edwin Levitan for providing a plasmid containing Emerald, and Joshua Arribere and Andrew Fire for providing the gRNA plasmid pJP118. Many critical strains in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440) and by Shohei Mitani as part of the Japanese National Bioresource Project. This work was supported by a grant from the National Institute of General Medical Sciences of the NIH (R01GM080765 to K.G.M.) and by a grant from the Oklahoma Center for the Advancement of Science and Technology (HR14-003 to K.G.M.). This work made use of the BioCryo facility of Northwestern University’s NUANCE Center, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-1542205); the MRSEC program (NSF DMR-1720139) at the Materials Research Center; the International Institute for Nanotechnology (IIN); and the State of Illinois, through the IIN. It also made use of the CryoCluster equipment, which has received support from the Major Research Instrumentation (MRI) program (NSF DMR-1229693).
Note added in proof: See Edwards et al. 2018 (pp 947–968) in this issue for a related work.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6965684.
Communicating editor: M. Johnston
Literature Cited
- Ackley B. D., Harrington R. J., Hudson M. L., Williams L., Kenyon C. J., et al. , 2005. The two isoforms of the Caenorhabditis elegans leukocyte-common antigen related receptor tyrosine phosphatase PTP-3 function independently in axon guidance and synapse formation. J. Neurosci. 25: 7517–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acuna C., Liu X., Sudhof T. C., 2016. How to make an active zone: unexpected universal functional redundancy between RIMs and RIM-BPs. Neuron 91: 792–807. 10.1016/j.neuron.2016.07.042 [DOI] [PubMed] [Google Scholar]
- Arribere J. A., Bell R. T., Fu B. X., Artiles K. L., Hartman P. S., et al. , 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198: 837–846. 10.1534/genetics.114.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas P. W., Lin S., 2011. Hooks and comets: the story of microtubule polarity orientation in the neuron. Dev. Neurobiol. 71: 403–418. 10.1002/dneu.20818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee N., Bhattacharya R., Gorczyca M., Collins K. M., Francis M. M., 2017. Local neuropeptide signaling modulates serotonergic transmission to shape the temporal organization of C. elegans egg-laying behavior. PLoS Genet. 13: e1006697 10.1371/journal.pgen.1006697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya R., Francis M. M., 2015. In the proper context: neuropeptide regulation of behavioral transitions during food searching. Worm 4: e1062971 10.1080/21624054.2015.1062971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya R., Touroutine D., Barbagallo B., Climer J., Lambert C. M., et al. , 2014. A conserved dopamine-cholecystokinin signaling pathway shapes context-dependent Caenorhabditis elegans behavior. PLoS Genet. 10: e1004584 10.1371/journal.pgen.1004584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of C. elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgari D., Zhou C., Hewes R. S., Deitcher D. L., Levitan E. S., 2014. Vesicle capture, not delivery, scales up neuropeptide storage in neuroendocrine terminals. Proc. Natl. Acad. Sci. USA 111: 3597–3601. 10.1073/pnas.1322170111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgari D., Deitcher D. L., Levitan E. S., 2017. Loss of Huntingtin stimulates capture of retrograde dense-core vesicles to increase synaptic neuropeptide stores. Eur. J. Cell Biol. 96: 402–406. 10.1016/j.ejcb.2017.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton P. R., Paige J. L., 1981. Polarity of axoplasmic microtubules in the olfactory nerve of the frog. Proc. Natl. Acad. Sci. USA 78: 3269–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavolo S. L., Zhou C., Ketcham S. A., Suzuki M. M., Ukalovic K., et al. , 2015. Mycalolide B dissociates dynactin and abolishes retrograde axonal transport of dense-core vesicles. Mol. Biol. Cell 26: 2664–2672. 10.1091/mbc.E14-11-1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavolo S. L., Bulgari D., Deitcher D. L., Levitan E. S., 2016. Activity induces Fmr1-sensitive synaptic capture of anterograde circulating neuropeptide vesicles. J. Neurosci. 36: 11781–11787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlie N. K., Schade M. A., Thomure A. M., Miller K. G., 2006. Presynaptic UNC-31 (CAPS) is required to activate the G alpha(s) pathway of the Caenorhabditis elegans synaptic signaling network. Genetics 172: 943–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Taylor K. P., Hall Q., Kaplan J. M., 2016. The neuropeptides FLP-2 and PDF-1 act in concert to arouse Caenorhabditis elegans locomotion. Genetics 204: 1151–1159. 10.1534/genetics.116.192898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia P. H., Patel M. R., Shen K., 2012. NAB-1 instructs synapse assembly by linking adhesion molecules and F-actin to active zone proteins. Nat. Neurosci. 15: 234–242. 10.1038/nn.2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Taylor K. P., Chatzigeorgiou M., Hu Z., Schafer W. R., et al. , 2015. Sensory neurons arouse C. elegans locomotion via both glutamate and neuropeptide release. PLoS Genet. 11: e1005359 10.1371/journal.pgen.1005359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cool D. R., Normant E., Shen F., Chen H. C., Pannell L., et al. , 1997. Carboxypeptidase E is a regulated secretory pathway sorting receptor: genetic obliteration leads to endocrine disorders in Cpe(fat) mice. Cell 88: 73–83. [DOI] [PubMed] [Google Scholar]
- Crump J. G., Zhen M., Jin Y., Bargmann C. I., 2001. The SAD-1 kinase regulates presynaptic vesicle clustering and axon termination. Neuron 29: 115–129. [DOI] [PubMed] [Google Scholar]
- Dai Y., Taru H., Deken S. L., Grill B., Ackley B., et al. , 2006. SYD-2 liprin-alpha organizes presynaptic active zone formation through ELKS. Nat. Neurosci. 9: 1479–1487. [DOI] [PubMed] [Google Scholar]
- Deken S. L., Vincent R., Hadwiger G., Liu Q., Wang Z. W., et al. , 2005. Redundant localization mechanisms of RIM and ELKS in Caenorhabditis elegans. J. Neurosci. 25: 5975–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. L., Charlie N. K., Milfort M. C., Brown B. S., Gravlin C. N., et al. , 2008. A novel molecular solution for ultraviolet light detection in Caenorhabditis elegans. PLoS Biol. 6: e198 10.1371/journal.pbio.0060198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. L., Morrison L. M., Yorks R. M., Hoover C. M., Boominathan S., et al. , 2015a. UNC-16 (JIP3) acts through synapse-assembly proteins to inhibit the active transport of cell soma organelles to Caenorhabditis elegans motor neuron axons. Genetics 201: 117–141. 10.1534/genetics.115.177345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. L., Yorks R. M., Morrison L. M., Hoover C. M., Miller K. G., 2015b. Synapse-assembly proteins maintain synaptic vesicle cluster stability and regulate synaptic vesicle transport in Caenorhabditis elegans. Genetics 201: 91–116. 10.1534/genetics.115.177337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S. L., Morrison L. M., Manning L., Stec N., Richmond J. E., et al. , 2018. Sentryn acts with a subset of active zone proteins to optimize the localization of synaptic vesicles in Caenorhabditis elegans. Genetics 210: 947–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouquet W., Owald D., Wichmann C., Mertel S., Depner H., et al. , 2009. Maturation of active zone assembly by Drosophila Bruchpilot. J. Cell Biol. 186: 129–145. 10.1083/jcb.200812150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker L. D., 1988. Carboxypeptidase E. Annu. Rev. Physiol. 50: 309–321. [DOI] [PubMed] [Google Scholar]
- Goodwin P. R., Juo P., 2013. The scaffolding protein SYD-2/liprin-alpha regulates the mobility and polarized distribution of dense-core vesicles in C. elegans motor neurons. PLoS One 8: e54763 10.1371/journal.pone.0054763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin P. R., Sasaki J. M., Juo P., 2012. Cyclin-dependent kinase 5 regulates the polarized trafficking of neuropeptide-containing dense-core vesicles in Caenorhabditis elegans motor neurons. J. Neurosci. 32: 8158–8172. 10.1523/JNEUROSCI.0251-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald I. S., Horvitz H. R., 1980. unc-93 (e1500): a behavioral mutant of Caenorhabditis elegans that defines a gene with a wild type null phenotype. Genetics 96: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. H., Hedgecock E. M., 1991. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell 65: 837–847. [DOI] [PubMed] [Google Scholar]
- Hammarlund M., Watanabe S., Schuske K., Jorgensen E. M., 2008. CAPS and syntaxin dock dense core vesicles to the plasma membrane in neurons. J. Cell Biol. 180: 483–491. 10.1083/jcb.200708018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann S. R., Landers J. M., Hamborg M. A., 1981. Polarity orientation of axonal microtubules. J. Cell Biol. 91: 661–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover C. M., Edwards S. L., Yu S. C., Kittelmann M., Richmond J. E., et al. , 2014. A novel CaM kinase II pathway controls the location of neuropeptide release from Caenorhabditis elegans motor neurons. Genetics 196: 745–765. 10.1534/genetics.113.158568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Pym E. C., Babu K., Vashlishan Murray A. B., Kaplan J. M., 2011. A neuropeptide-mediated stretch response links muscle contraction to changes in neurotransmitter release. Neuron 71: 92–102. 10.1016/j.neuron.2011.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Vashlishan-Murray A. B., Kaplan J. M., 2015. NLP-12 engages different UNC-13 proteins to potentiate tonic and evoked release. J. Neurosci. 35: 1038–1042. 10.1523/JNEUROSCI.2825-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung W., Hwang C., Po M. D., Zhen M., 2007. Neuronal polarity is regulated by a direct interaction between a scaffolding protein, Neurabin, and a presynaptic SAD-1 kinase in Caenorhabditis elegans. Development 134: 237–249. [DOI] [PubMed] [Google Scholar]
- Inoue E., Mochida S., Takagi H., Higa S., Deguchi-Tawarada M., et al. , 2006. SAD: a presynaptic kinase associated with synaptic vesicles and the active zone cytomatrix that regulates neurotransmitter release. Neuron 50: 261–275. [DOI] [PubMed] [Google Scholar]
- Kim J. S., Lilley B. N., Zhang C., Shokat K. M., Sanes J. R., et al. , 2008. A chemical-genetic strategy reveals distinct temporal requirements for SAD-1 kinase in neuronal polarization and synapse formation. Neural Dev. 3: 23 10.1186/1749-8104-3-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Hung W., Narbonne P., Roy R., Zhen M., 2010. C. elegans STRADalpha and SAD cooperatively regulate neuronal polarity and synaptic organization. Development 137: 93–102. 10.1242/dev.041459 [DOI] [PubMed] [Google Scholar]
- Kim S., Ko J., Shin H., Lee J. R., Lim C., et al. , 2003. The GIT family of proteins forms multimers and associates with the presynaptic cytomatrix protein Piccolo. J. Biol. Chem. 278: 6291–6300. [DOI] [PubMed] [Google Scholar]
- Kittelmann M., Hegermann J., Goncharov A., Taru H., Ellisman M. H., et al. , 2013. Liprin-alpha/SYD-2 determines the size of dense projections in presynaptic active zones in C. elegans. J. Cell Biol. 203: 849–863. 10.1083/jcb.201302022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen M. P., Wu Y. E., Maeder C. I., Nakae I., Cueva J. G., et al. , 2010. An Arf-like small G protein, ARL-8, promotes the axonal transport of presynaptic cargoes by suppressing vesicle aggregation. Neuron 66: 710–723. 10.1016/j.neuron.2010.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J., Kim S., Valtschanoff J. G., Shin H., Lee J. R., et al. , 2003a. Interaction between liprin-alpha and GIT1 is required for AMPA receptor targeting. J. Neurosci. 23: 1667–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J., Na M., Kim S., Lee J. R., Kim E., 2003b. Interaction of the ERC family of RIM-binding proteins with the liprin-alpha family of multidomain proteins. J. Biol. Chem. 278: 42377–42385. [DOI] [PubMed] [Google Scholar]
- Koushika S. P., Richmond J. E., Hadwiger G., Weimer R. M., Jorgensen E. M., et al. , 2001. A post-docking role for active zone protein Rim. Nat. Neurosci. 4: 997–1005. 10.1038/nn732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfermann I., 1991. Functional studies of cotransmission. Physiol. Rev. 71: 683–732. [DOI] [PubMed] [Google Scholar]
- Levitan E. S., 2008. Signaling for vesicle mobilization and synaptic plasticity. Mol. Neurobiol. 37: 39–43. 10.1007/s12035-008-8014-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Tian X., Zhu M., Bulgari D., Bohme M. A., et al. , 2014. Drosophila Syd-1, liprin-alpha, and protein phosphatase 2A B’ subunit Wrd function in a linear pathway to prevent ectopic accumulation of synaptic materials in distal axons. J. Neurosci. 34: 8474–8487. 10.1523/JNEUROSCI.0409-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A., Rechtsteiner A., Saxton W. M., 2017. Two kinesins drive anterograde neuropeptide transport. Mol. Biol. Cell 28: 3542–3553. 10.1091/mbc.E16-12-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M. A., Chitturi J., Laskova V., Meng J., Findeis D., et al. , 2016. Neuroendocrine modulation sustains the C. elegans forward motor state. eLife 5: pii: e19887 [corrigenda: eLife 6 (2017)] 10.7554/eLife.19887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K. S., Siebert M., Mertel S., Knoche E., Wegener S., et al. , 2011. RIM-binding protein, a central part of the active zone, is essential for neurotransmitter release. Science 334: 1565–1569. 10.1126/science.1212991 [DOI] [PubMed] [Google Scholar]
- Liu T., Kim K., Li C., Barr M. M., 2007. FMRFamide-like neuropeptides and mechanosensory touch receptor neurons regulate male sexual turning behavior in Caenorhabditis elegans. J. Neurosci. 27: 7174–7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10: 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. E., DeProto J., Kaufmann N., Patel B. N., Duckworth A., et al. , 2005. Direct observation demonstrates that liprin-alpha is required for trafficking of synaptic vesicles. Curr. Biol. 15: 684–689. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Emerson M. D., Rand J. B., 1999. Goalpha and diacylglycerol kinase negatively regulate the Gqalpha pathway in C. elegans. Neuron 24: 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka T., Takao-Rikitsu E., Inoue E., Inoue M., Takeuchi M., et al. , 2002. Cast: a novel protein of the cytomatrix at the active zone of synapses that forms a ternary complex with RIM1 and munc13–1. J. Cell Biol. 158: 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen O., Moore K. A., Fukata M., Kazuta T., Trinidad J. C., et al. , 2005. Neurotransmitter release regulated by a MALS-liprin-alpha presynaptic complex. J. Cell Biol. 170: 1127–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou C. Y., Poon V. Y., Maeder C. I., Watanabe S., Lehrman E. K., et al. , 2010. Two cyclin-dependent kinase pathways are essential for polarized trafficking of presynaptic components. Cell 141: 846–858. 10.1016/j.cell.2010.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owald D., Fouquet W., Schmidt M., Wichmann C., Mertel S., et al. , 2010. A Syd-1 homologue regulates pre- and postsynaptic maturation in Drosophila. J. Cell Biol. 188: 565–579. 10.1083/jcb.200908055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack-Chung E., Kurshan P. T., Dickman D. K., Schwarz T. L., 2007. A Drosophila kinesin required for synaptic bouton formation and synaptic vesicle transport. Nat. Neurosci. 10: 980–989. [DOI] [PubMed] [Google Scholar]
- Paix A., Wang Y., Smith H. E., Lee C. Y., Calidas D., et al. , 2014. Scalable and versatile genome editing using linear DNAs with microhomology to Cas9 sites in Caenorhabditis elegans. Genetics 198: 1347–1356. 10.1534/genetics.114.170423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M. R., Lehrman E. K., Poon V. Y., Crump J. G., Zhen M., et al. , 2006. Hierarchical assembly of presynaptic components in defined C. elegans synapses. Nat. Neurosci. 9: 1488–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N. K., Schade M. A., Miller K. G., 2005. Convergent, RIC-8-dependent Galpha signaling pathways in the Caenorhabditis elegans synaptic signaling network. Genetics 169: 651–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond J. E., Broadie K. S., 2002. The synaptic vesicle cycle: exocytosis and endocytosis in Drosophila and C. elegans. Curr. Opin. Neurobiol. 12: 499–507. [DOI] [PubMed] [Google Scholar]
- Rindler M. J., 1998. Carboxypeptidase E, a peripheral membrane protein implicated in the targeting of hormones to secretory granules, co-aggregates with granule content proteins at acidic pH. J. Biol. Chem. 273: 31180–31185. [DOI] [PubMed] [Google Scholar]
- Schade M. A., Reynolds N. K., Dollins C. M., Miller K. G., 2005. Mutations that rescue the paralysis of Caenorhabditis elegans ric-8 (synembryn) mutants activate the G alpha(s) pathway and define a third major branch of the synaptic signaling network. Genetics 169: 631–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch S., Castillo P. E., Jo T., Mukherjee K., Geppert M., et al. , 2002. RIM1alpha forms a protein scaffold for regulating neurotransmitter release at the active zone. Nature 415: 321–326. [DOI] [PubMed] [Google Scholar]
- Serra-Pages C., Medley Q. G., Tang M., Hart A., Streuli M., 1998. Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J. Biol. Chem. 273: 15611–15620. [DOI] [PubMed] [Google Scholar]
- Shakiryanova D., Tully A., Levitan E. S., 2006. Activity-dependent synaptic capture of transiting peptidergic vesicles. Nat. Neurosci. 9: 896–900. [DOI] [PubMed] [Google Scholar]
- Shin H., Wyszynski M., Huh K. H., Valtschanoff J. G., Lee J. R., et al. , 2003. Association of the kinesin motor KIF1A with the multimodular protein liprin-alpha. J. Biol. Chem. 278: 11393–11401. [DOI] [PubMed] [Google Scholar]
- Sieburth D., Madison J. M., Kaplan J. M., 2007. PKC-1 regulates secretion of neuropeptides. Nat. Neurosci. 10: 49–57. [DOI] [PubMed] [Google Scholar]
- Sossin W. S., Scheller R. H., 1991. Biosynthesis and sorting of neuropeptides. Curr. Opin. Neurobiol. 1: 79–83. [DOI] [PubMed] [Google Scholar]
- Stefanakis N., Carrera I., Hobert O., 2015. Regulatory logic of Pan-neuronal gene expression in C. elegans. Neuron 87: 733–750. 10.1016/j.neuron.2015.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T., 2006. Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.101.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stigloher C., Zhan H., Zhen M., Richmond J., Bessereau J. L., 2011. The presynaptic dense projection of the Caenorhabditis elegans cholinergic neuromuscular junction localizes synaptic vesicles at the active zone through SYD-2/liprin and UNC-10/RIM-dependent interactions. J. Neurosci. 31: 4388–4396. 10.1523/JNEUROSCI.6164-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof T. C., 2004. The synaptic vesicle cycle. Annu. Rev. Neurosci. 27: 509–547. [DOI] [PubMed] [Google Scholar]
- Sudhof T. C., 2012. The presynaptic active zone. Neuron 75: 11–25. 10.1016/j.neuron.2012.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J., Hodgkin J., 1988. Methods, pp. 596–597 in The Nematode Caenorhabditis elegans, edited by Wood W. B. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Von Stetina S. E., Watson J. D., Fox R. M., Olszewski K. L., Spencer W. C., et al. , 2007. Cell-specific microarray profiling experiments reveal a comprehensive picture of gene expression in the C. elegans nervous system. Genome Biol. 8: R135 10.1186/gb-2007-8-7-r135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh D. A., Rasse T. M., Asan E., Hofbauer A., Schwenkert I., et al. , 2006. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron 49: 833–844. [DOI] [PubMed] [Google Scholar]
- Wagner O. I., Esposito A., Kohler B., Chen C. W., Shen C. P., et al. , 2009. Synaptic scaffolding protein SYD-2 clusters and activates kinesin-3 UNC-104 in C. elegans. Proc. Natl. Acad. Sci. USA 106: 19605–19610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. S., Held R. G., Wong M. Y., Liu C., Karakhanyan A., et al. , 2016. Fusion competent synaptic vesicles persist upon active zone disruption and loss of vesicle docking. Neuron 91: 777–791. 10.1016/j.neuron.2016.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Okamoto M., Schmitz F., Hofmann K., Sudhof T. C., 1997. Rim is a putative Rab3 effector in regulating synaptic-vesicle fusion. Nature 388: 593–598. [DOI] [PubMed] [Google Scholar]
- Wang Y., Sugita S., Sudhof T. C., 2000. The RIM/NIM family of neuronal C2 domain proteins. Interactions with Rab3 and a new class of Src homology 3 domain proteins. J. Biol. Chem. 275: 20033–20044. [DOI] [PubMed] [Google Scholar]
- Wang Y., Liu X., Biederer T., Sudhof T. C., 2002. A family of RIM-binding proteins regulated by alternative splicing: implications for the genesis of synaptic active zones. Proc. Natl. Acad. Sci. USA 99: 14464–14469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer R. M., 2006. Preservation of C. elegans tissue via high-pressure freezing and freeze-substitution for ultrastructural analysis and immunocytochemistry. Methods Mol. Biol. 351: 203–221. [DOI] [PubMed] [Google Scholar]
- Weimer R. M., Gracheva E. O., Meyrignac O., Miller K. G., Richmond J. E., et al. , 2006. UNC-13 and UNC-10/rim localize synaptic vesicles to specific membrane domains. J. Neurosci. 26: 8040–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N., Brenner S., 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314: 1–340. [DOI] [PubMed] [Google Scholar]
- Wong M. Y., Zhou C., Shakiryanova D., Lloyd T. E., Deitcher D. L., et al. , 2012. Neuropeptide delivery to synapses by long-range vesicle circulation and sporadic capture. Cell 148: 1029–1038. 10.1016/j.cell.2011.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. E., Huo L., Maeder C. I., Feng W., Shen K., 2013. The balance between capture and dissociation of presynaptic proteins controls the spatial distribution of synapses. Neuron 78: 994–1011. 10.1016/j.neuron.2013.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski M., Kim E., Dunah A. W., Passafaro M., Valtschanoff J. G., et al. , 2002. Interaction between GRIP and liprin-alpha/SYD2 is required for AMPA receptor targeting. Neuron 34: 39–52. [DOI] [PubMed] [Google Scholar]
- Xuan Z., Manning L., Nelson J., Richmond J. E., Colon-Ramos D. A., et al. , 2017. Clarinet (CLA-1), a novel active zone protein required for synaptic vesicle clustering and release. eLife 6: e29276 10.7554/eLife.29276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Kawano T., Weimer R. M., Bessereau J. L., Zhen M., 2005. Identification of genes involved in synaptogenesis using a fluorescent active zone marker in Caenorhabditis elegans. J. Neurosci. 25: 3833–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen M., Jin Y., 1999. The liprin protein SYD-2 regulates the differentiation of presynaptic termini in C. elegans. Nature 401: 371–375. [DOI] [PubMed] [Google Scholar]
- Zheng Q., Ahlawat S., Schaefer A., Mahoney T., Koushika S. P., et al. , 2014. The vesicle protein SAM-4 regulates the processivity of synaptic vesicle transport. PLoS Genet. 10: e1004644 10.1371/journal.pgen.1004644 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request from the corresponding author, from the Caenorhabditis Genetics Center, or from AddGene. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures, and tables. Supplemental files available at FigShare. File S1 contains Figures S1–S8 and Tables S1–S4, Supplemental Materials and Methods and Supplemental Material References. File S2 contains detailed EM DCV counts broken down by strain, worm, and synapse. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6965684.