As histone lysine methylation “readers”, chromo domain-containing proteins can provide a link between the chromatin landscape and downstream gene regulation. Saltzman et al. define new roles for two uncharacterized Caenorhabditis elegans chromo domain proteins...
Keywords: C. elegans, CEC, chromodomain proteins, gene silencing, H3K27me3, histone methyl-lysine readers, mortal germline, Polycomb proteins, PRC2, worm
Abstract
Chromatin modifications, including methylation of histone H3 at lysine 27 (H3K27me) by the Polycomb group proteins, play a broadly conserved role in the maintenance of cell fate. Diverse chromatin organization modifier (chromo) domain proteins act as “readers” of histone methylation states. However, understanding the functional relationships among chromo domains and their roles in the inheritance of gene expression patterns remains challenging. Here, we identify two chromo-domain proteins, CEC-1 and CEC-6, as potential readers of H3K27me in Caenorhabditis elegans, where they have divergent expression patterns and contribute to distinct phenotypes. Both cec-1 and cec-6 genetically interact with another chromo-domain gene, cec-3, a reader of H3K9 methylation. Combined loss of cec-1 and cec-3 leads to developmental defects in the adult that result in decreased fitness. Furthermore, loss of cec-6 and cec-3 surprisingly leads to a progressive loss of fertility across generations, a “mortal germline” phenotype. Our results provide evidence of functional compensation between H3K27me and H3K9me heterochromatin pathways, and show that histone methylation readers contribute to both somatic development and transgenerational fitness.
CHROMATIN regulation is essential for the proper establishment and maintenance of cell identity. The Polycomb group proteins are evolutionarily conserved chromatin regulators that form multiprotein complexes involved in maintaining gene repression. Although the Polycomb group proteins were initially identified in Drosophila for their role in body patterning, they have since also proved important for the control of many genes in development, differentiation, and cell proliferation pathways across species (Di Croce and Helin 2013; Simon and Kingston 2013). In addition, disruption of Polycomb regulation is associated with several cancers (Pasini and Di Croce 2016). Gene silencing by Polycomb proteins can be maintained during development, and intriguing studies further support a role for transgenerational inheritance of Polycomb-repressed chromatin states (Gaydos et al. 2014; Ciabrelli et al. 2017; Zenk et al. 2017). However, the mechanisms governing the inheritance of silencing through cell division are not fully understood, and the requirements for this inheritance and its evolutionary conservation remain to be fully elucidated (Campos et al. 2014; Steffen and Ringrose 2014).
Gene silencing by Polycomb group proteins is mediated in part through the di/trimethylation of histone H3 at lysine (K) 27 (H3K27me) by Polycomb Repressive Complex 2 (PRC2). In the model roundworm Caenorhabditis elegans, the PRC2-related genes [maternal effect sterile (mes)-2, mes-3, and mes-6] controlling histone H3K27me deposition are required maternally for germline development (Capowski et al. 1991; Bender et al. 2004), and restrict cell fate plasticity in the germline (Patel et al. 2012) and early embryo (Yuzyuk et al. 2009). In addition, both PRC2/mes and H3K27me3 can be transgenerationally inherited (Gaydos et al. 2014). In the soma, mutation of mes genes leads to subtle effects on anteroposterior patterning (Ross and Zarkower 2003). Thus, in contrast to the essential role of PRC2/mes in the germline, the functions of H3K27me in C. elegans postembryonic development are poorly understood.
In flies and mammals, the H3K27me3 modification serves as a recognition site for the specialized chromatin organization modifier (chromo)-domain subunit of canonical PRC1. This chromo-domain protein is encoded by Polycomb in Drosophila or one of several Chromobox (CBX) homolog genes in mammals (CBX2, CBX4, and CBX6–8), which have distinct functions in pluripotent and differentiated cell types (Laugesen and Helin 2014). Although two C. elegans PRC1 component homologs (Karakuzu et al. 2009) and several more distantly related PRC1-like genes (Zhang et al. 2003; Yang et al. 2007) have been characterized, to date, no C. elegans chromo-domain proteins that recognize H3K27me have been identified. As such, the relationship of PRC1-related genes to H3K27me and the existence of a PRC1-like complex in C. elegans have remained in question. Characterization of H3K27me-readers would therefore provide new insight into the evolutionary plasticity of Polycomb silencing mechanisms, and their roles in germline maintenance and somatic development.
Drosophila Polycomb and mammalian CBX proteins are part of a larger group of methyl-lysine reader chromo-domain proteins with roles in chromatin and transcriptional regulation (Eissenberg 2012). Two C. elegans chromo-domain (cec) genes (Aasland and Stewart 1995; Agostoni et al. 1996), cec-3 (also known as eap-1) (Greer et al. 2014) and cec-4 (Gonzalez-Sandoval et al. 2015), as well as the chromo domain-containing genes heterochromatin protein 1(HP1)-like 1 (hpl-1) and hpl-2 (Schott et al. 2006), recognize histone H3K9 methylation, a mark associated with heterochromatin. These genes have both common and distinct roles in cell fate and fertility. For example, both cec-3 and hpl-2 play a role in neuronal subtype-specific gene expression (Zheng et al. 2013), and cec-3 affects the transgenerational fertility of a histone H3K4me1/2-demethylase mutant (Katz et al. 2009; Nottke et al. 2011; Greer et al. 2014). However, the C. elegans genome encodes several additional chromo-domain proteins whose methyl-lysine-binding specificity remains entirely unknown (Supplemental Material, Figure S1). These genes are probable candidates to test for interaction with H3K27me, and their characterization has great potential to reveal both conserved and novel functions associated with these versatile readers of chromatin states.
In this study, we set out to identify H3K27me readers in C. elegans. We used a candidate biochemical interaction approach to characterize two C. elegans chromo-domain proteins, CEC-1 and CEC-6, which directly recognize H3K27me. By constructing strains to examine their expression patterns, loss-of-function phenotypes, and genetic interactions, we find that CEC-1 and CEC-6 contribute to somatic and germline functions, respectively. Taking advantage of the stereotyped development and short generation time of C. elegans, our work indicates that multiple chromo-domain proteins contribute to developmental robustness and germline immortality over generations in this organism. Together, our findings expand the functions of H3K27me readers, and uncover new evidence for functional overlap between readers of H3K27me and H3K9me heterochromatic histone modifications.
Materials and Methods
Worm maintenance
Strains were maintained on nematode growth medium (NGM) agar with Escherichia coli OP50-1 as a food source as described (Stiernagle 2006) at 22° unless specified otherwise. The N2 (Bristol) strain was used as wild-type (Brenner 1974). Strains and alleles used are listed in Table S1.
Generation of transgenic animals
Single-guide RNA design for clustered regularly interspaced short palindromic repeats/Cas9-mediated genome editing:
For gene deletions, one or two single-guide (sg)RNAs were designed to target in the gene body. For GFP or HA epitope tag knock-ins, one or two sgRNAs were designed to target near the insertion site. To select sgRNAs, potential target sites with a 3′GG (Farboud and Meyer 2015) in the protospacer were identified, and those with the best score and minimal predicted off-target sites were used (Ran et al. 2013; Heigwer et al. 2014). The sgRNAs were cloned by Phusion site-directed mutagenesis (Thermo Fisher Scientific) of plasmid pU6::unc-119_sgRNA (# 46169; Addgene plasmid) (Friedland et al. 2013).
Clustered regularly interspaced short palindromic repeats/Cas9-mediated gene deletions:
Strains harboring gene deletions were generated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9-mediated homologous recombination (HR) using an excisable selection cassette, as described (Norris et al. 2015). Briefly, HR repair templates were designed to replace the target gene (from start to penultimate codon) with a floxed selection cassette. The upstream and downstream homology arms (∼1 kb) were cloned into the SacII and NotI sites, respectively, of pPmyo-2::GFP-Prps-27::NeoR-loxP (Norris et al. 2015) using the Gibson isothermal assembly (Gibson et al. 2009). The HR and sgRNA plasmids were microinjected with Peft-3(eef-1A.1)::cas9-SV40_NLS::tbb-2 3′UTR (#46168; Addgene plasmid) and co-injection markers as described (Norris et al. 2015). Transgenic animals were selected from progeny of injected animals as described (Norris et al. 2015), and verified by PCR across the upstream and downstream insertion junctions. Strains were backcrossed twice to N2. The selection cassette was then excised by injection of plasmid pDD104 (Peft-3::Cre, #47551; Addgene plasmid) as described (Dickinson et al. 2013). Excision of the selection cassette was verified by PCR and strains were again backcrossed twice to N2. The deletion alleles were confirmed by PCR from worm lysates and direct Sanger sequencing of the amplified bands. Additional backcrossing prior to mortal germline assays was also performed as described in the text (Figure 5B and Figure 6C).
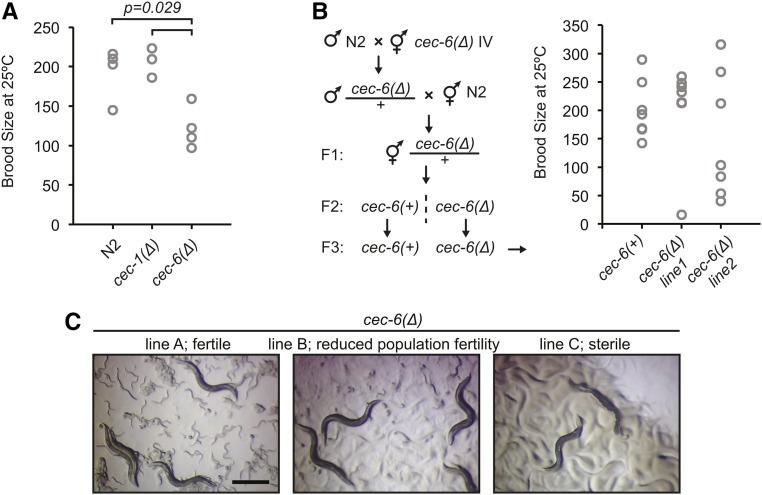
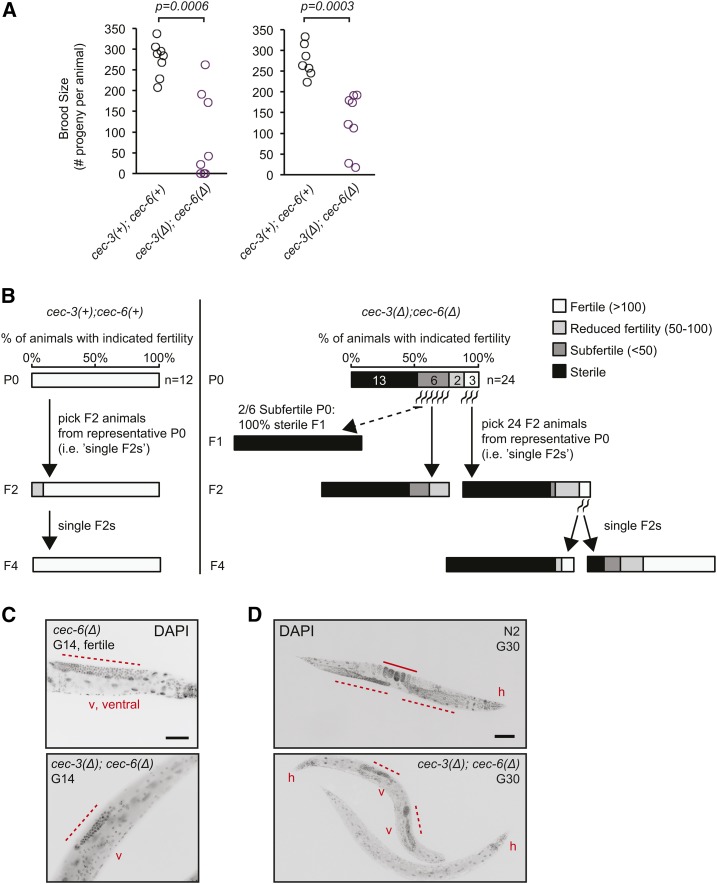
Figure 5.
Variability in fertility of cec-6Δ animals. (A) Brood size per animal for worms of indicated genotypes. The distribution of the cec-6Δ animals is shifted relative to the N2 and cec-1Δ animals (Wilcoxon rank-sum test, one-sided). The distributions of N2 and cec-1Δ brood sizes were not significantly different. (B) Brood size per animal after outcrossing to N2 as indicated in the scheme. Differences in distributions were not significant (Wilcoxon rank-sum test, two-sided). (C) Stereoscope images of growth plates 6 days after six L1 worms were plated. Images represent lines at various stages of population fertility. When lines became sterile, most animals had a dark appearance with a clear area where embryos are normally located (Capowski et al. 1991; Katz et al. 2009) (right panel). Bar, 0.5 mm.
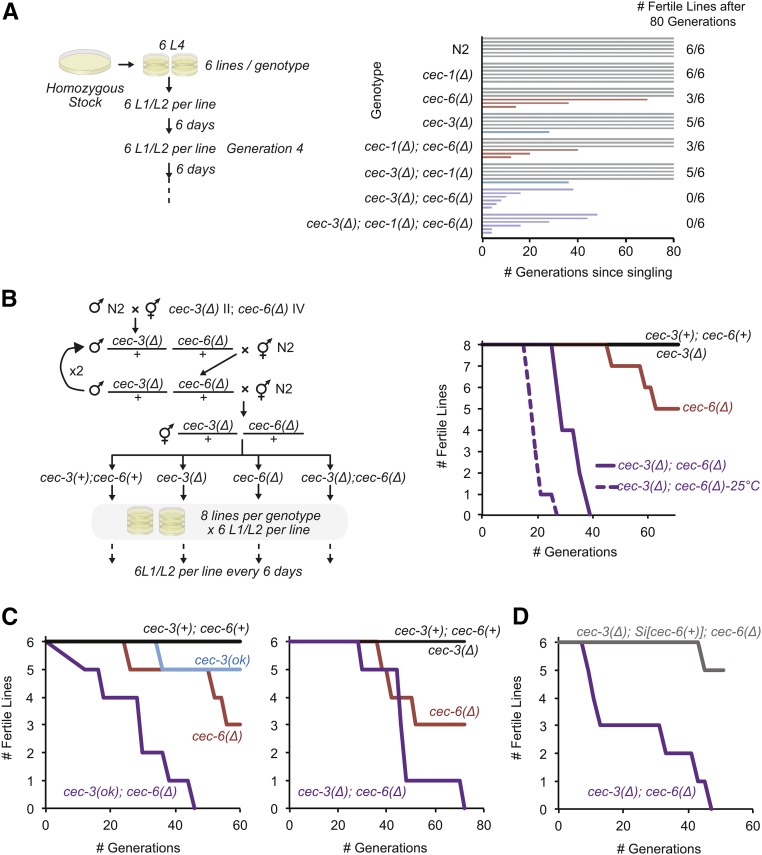
Figure 6.
Germline immortality is compromised in cec-3Δ;cec-6Δ animals. (A and B) Mortal germline assays starting from homozygous mutant (A) or freshly outcrossed (B) strains. In (B), strains harboring cec-3Δ and cec-6Δ deletions were backcrossed to N2 (wild-type) worms four times consecutively, while maintaining both cec-3 and cec-6 in a heterozygous state until the final generation. All assays were performed at 22° except as indicated [dotted line in (B)]. (C) Independent biological replicates of mortal germline assays. Animals with indicated alleles were outcrossed five times to N2 before the start of the assays. Experiments in the left and right panels were conducted using independent alleles of cec-3 [left panel: cec-3(ok3432), from the C elegans knockout consortium; right panel: cec-3Δ, this study]. (D) The mortal germline of cec-3Δ;cec-6Δ animals is rescued by a cec-6(+) single-copy transgene inserted using Minimal Mos1 (see text).
CRISPR/Cas9-mediated epitope tag knock-ins:
Insertion of epitope tags in-frame with gene coding sequences was performed as described with (Norris et al. 2015) or without (Tzur et al. 2013) insertion of an excisable selection cassette. Briefly, for insertion of 3xHA at the cec-1 gene, an HR donor vector coding for an N-terminal in-frame 3xHA tag [codon-optimized sequence (Ooi et al. 2010)] and flanking ∼1-kb homology arms was constructed. Injection and identification of F1 animals expressing the co-injection markers was performed as described (Tzur et al. 2013). Animals carrying the insertion were identified by single-worm PCR. The insertion was verified by direct Sanger sequencing of the amplified bands. For insertion of a GFP::3xHA C-terminal tag into cec-6 or cec-1, the excisable selection cassette described above for gene deletions was used. An HR donor plasmid was constructed by Gibson isothermal assembly (Gibson et al. 2009) containing ∼1-kb homology arms flanking an in-frame C-terminal tag encoding GFP::3xHA. The CRE-excisable Pmyo-2::GFP-Prps-27::NeoR-loxP selection cassette was cloned into the second intron of GFP in the HR donor construct. Worm microinjection, selection, and CRE excision was performed as described above.
Minimal Mos1 transgene insertion:
Minimal Mos1 (miniMos) transgenesis with G418 selection was performed as described (Frøkjær-Jensen et al. 2014). For the cec-1 rescue construct, the region including the cec-1 promoter and gene (5.3 kb including 2896/463 bp up/downstream of the gene body, LGIII: 8,423,444–8,428,728; ce11) was PCR-amplified from C. elegans genomic DNA. For the cec-6 rescue construct, the promoter region upstream of the cec-6-containing operon (CEOP4168, LGIV: 5,537,933–5,538,489; ce11) and the gene (4.4 kb including the 3′-UTR, LGIV: 5,544,860–5,549,302; ce11) were PCR-amplified from genomic DNA. Fragments were cloned into SpeI/StuI-digested pCFJ910 (pMinimos-NeoR-MCS, #44481; Addgene plasmid) using Gibson isothermal assembly (Gibson et al. 2009). The cec-1(+) and cec-6(+) miniMos transgenes were injected into cec-3Δ;cec-1Δ or N2 animals, respectively. Plasmids were microinjected along with the transposase vector pCFJ601 (Peft-3:Mos1, #34874; Addgene plasmid) and co-injection markers as described (Frøkjær-Jensen et al. 2014). Transgenic animals were selected, and transgene insertion sites of individual lines were mapped by restriction enzyme digestion and inverse PCR of genomic DNA essentially as described (Frøkjær-Jensen et al. 2014).
Protein sequence alignment
Protein sequences were obtained from the National Center for Biotechnology Information (NCBI Resource Coordinators 2017) and chromo domains were aligned with T-Coffee (Tree-based Consistency Objective Function for alignment Evaluation) using default settings (Notredame et al. 2000; Di Tommaso et al. 2011). The alignments were visualized using the ESpript server (Robert and Gouet 2014) (http://espript.ibcp.fr). Numbering of chromo domains is relative to the following protein sequence accessions: CeCec-1, NP_498862.1; CeCec-6, NP_500828.1; DmPc, NP_524199.1; HsCBX2, NP_005180.1; HsCBX4, NP_003646.2; HsCBX6, NP_055107.3; HsCBX7, NP_783640.1; and HsCBX8, NP_065700.1.
Recombinant protein purification
cDNAs were reverse-transcribed and PCR-amplified from C. elegans total RNA, and cloned into a pGEX-2T (GE Healthcare)-based vector modified to allow PreScission Protease (GE Healthcare) cleavage of the N-terminal GST and to encode a C-terminal 6xHis epitope. Construct sequences were verified by plasmid Sanger sequencing (Macrogen). Proteins were expressed in E. coli BL21(DE3)pLysS (Invitrogen, Carlsbad, CA). Following sonication, GST/6xHis fusion proteins were affinity-purified in two steps from the soluble fraction. First, proteins were bound to Glutathione Sepharose 4B (GE Healthcare), washed, and eluted by PreScission Protease cleavage. The eluted 6xHis fusion proteins were then purified on Talon metal affinity resin (Clontech), washed, eluted with imidazole, dialyzed into storage buffer (25 mM HEPES pH 7.5, 100 mM KCl, 0.2 mM EDTA, 1 mM DTT, and 20% (v/v) glycerol), aliquoted, and snap-frozen. Protein concentration was determined using the Bradford assay (Bio-Rad, Hercules, CA).
Histone peptide array assays
Protein–peptide interaction studies used MODified Histone Peptide Arrays (Active Motif) according to the manufacturer’s recommendations with minor modifications. Arrays were washed in phosphate-buffered saline (PBS) containing 0.05% (v/v) Tween-20 (PBST) (Sigma [Sigma Chemical], St. Louis, MO), blocked for 1 hr in 5% (w/v) skimmed milk powder/PBST, washed in PBST, and washed in 3% BSA/PBST. Next, 6xHis-tagged proteins were bound overnight at 4° in binding buffer [PBST with 0.45% (w/v) BSA, 0.5 mM EDTA, 0.1 mM DTT, and 10% (v/v) glycerol] at the indicated final concentrations (CEC-1, 125 nM; CEC-6, 150 nM; and CEC-3 200 nM). Arrays were washed three times with PBST, incubated with an anti-His(Cterm)-HRP-conjugated antibody (Invitrogen) at 1:3500 in 5% milk/PBST, washed three times with PBST, incubated with ECL reagents (Perkin-Elmer [Perkin-Elmer Cetus], Norwalk, CT), and exposed to film. Digital film scans were quantified with ArrayAnalyze software (Active Motif). A normalized relative signal was calculated for each array spot from the raw intensity value: (spot intensity-background)/(max spot intensity-background), where the background signal was defined as the 60th percentile of the signal intensity from the negative control array spots. Spots with intensities lower than the background spots were assigned a relative signal of zero.
Synchronous worm growth
Embryos were isolated from gravid adults using alkaline hypochlorite (“bleaching”) as described (Stiernagle 2006). After three washes in M9 buffer, embryos were allowed to hatch overnight in the absence of food. The arrested L1s were washed once in M9 buffer and plated on NGM agar with OP50-1.
Western blots of staged animals
Worms were synchronized by bleaching as described above and grown in liquid culture in S-medium (Stiernagle 2006) with OP50-1 to the indicated stages. Mixed-stage embryos were obtained by bleaching a culture grown to gravid adulthood. Worms or embryos were washed three times in M9 buffer and once in 0.1 M NaCl with 0.01% (v/v) Triton X-100. Samples were snap-frozen in liquid nitrogen in 0.1 M NaCl/0.01% Triton X-100 containing protease inhibitors (Complete Mini, EDTA-free; Roche). An equal volume of 2× lysis buffer (100 mM HEPES-Na pH 7.5, 300 mM NaCl, 2 mM EDTA, 0.2% sodium deoxycholate, 0.2% sodium N-laurylsarcosine, 2% Triton X-100, and 1 mM DTT) was added to samples thawed on ice, followed by sonication (QSonica) and removal of insoluble material by centrifugation. Protein concentration was determined by Bradford assay (Bio-Rad). Lysates were resolved on a 4–12% Bis-Tris NuPage gel (Invitrogen), transferred to nitrocellulose, and probed with an anti-HA tag antibody (rabbit monoclonal C29F4, 3724; Cell Signaling Technology), or with anti-histone H3 (rabbit polyclonal, ab1791; Abcam) or anti-β-tubulin (mouse monoclonal TUB2.1, T4026; Sigma) antibodies as loading controls.
RNA isolation and RT-PCR
Worms were synchronized by bleaching as described above and grown to the young adult stage. Worms were collected in M9 buffer containing 0.01% Triton X-100 and washed two to three times. Trizol (Invitrogen) was added to the worm pellet and three cycles of snap-freezing, thawing, and vortexing (1 min) were performed to assist in homogenization. RNA isolation was then performed according to the manufacturer’s instructions. To digest any remaining DNA, the RNA was treated with Turbo DNAse (Invitrogen) and purified on an RNA clean-and-concentrator-5 column (Zymo Research). RT-PCR was performed according to the manufacturer’s instructions (OneStep RT-PCR kit; QIAGEN, Valencia, CA) in a 10-μl reaction using 5–10 ng of total RNA and with addition of RiboLock RNase inhibitor (Thermo Fisher Scientific). Reaction products were resolved on a 2% agarose/TBE gel stained with ethidium bromide. Primers flanked exon–exon junctions, and a lack of DNA contamination was further confirmed by the absence of a band when the RT step was omitted.
Immunofluorescence
For whole-adult worm staining, animals were prepared for immunofluorescence as described (Bettinger et al. 1996) with incubation in fixation buffer for 2 hr on ice. Antibody incubations were performed in PBS containing 1% (w/v) BSA, 0.5% (v/v) Triton X-100, and 1mM EDTA (Sigma). For other stages, anti-HA immunofluorescence was performed essentially as described (Crittenden and Kimble 2009) with minor modifications, and PBS was used in place of Tris-buffered saline throughout. For L1 immunofluorescence, embryos were isolated by bleaching gravid adults and allowed to hatch overnight in M9 buffer. L1s were then fed on E. coli OP50-1 for 7–8 hr and washed in M9 buffer. L1s were processed using the whole-mount freeze-cracking method (Crittenden and Kimble 2009) with methanol/acetone fixation (15 and 5 min at −20°, respectively). For staining of dissected germlines, the tissue extrusion method (Crittenden and Kimble 2009) was used with fixation in 1% formaldehyde. For staining of embryos, gravid animals were dissected on slides and permeabilized by freeze-cracking (Crittenden and Kimble 2009). Fixation was in methanol and acetone (2–10 and 5 min at −20°, respectively), or in methanol (1 min at −20°), followed by 4% formaldehyde on ice (Greer et al. 2014). For RAD-51 staining of germlines, day 1 adults were dissected and fixed in 1% formaldehyde for 5 min, permeabilized by freeze-cracking, and incubated in −20° methanol for 1 min. Blocking and antibody incubations were performed in 1% BSA in PBS or PBS containing 0.1% (v/v) Tween-20 (Sigma). Washes were performed in PBS or PBS containing 0.1% (v/v) Tween-20 (Sigma).
Primary antibody incubation was performed overnight at the following concentrations: anti-HA (1:200–1:500, rabbit monoclonal C29F4, 3724; Cell Signaling Technology), anti-HA (1:200, mouse monoclonal H3663; Sigma), anti-RNA polymerase II C-terminal domain repeat (1:500, mouse monoclonal 8WG16, ab817; Abcam), anti-RNA polymerase II Ser5P CTD repeat (1:1000, mouse monoclonal 4H8, 05–623; Millipore, Bedford, MA), anti-PGL-1 [1:10, mouse monoclonal K76; Developmental Studies Hybridoma Bank (Strome and Wood 1983)], and anti-RAD-51 (1:500–1:1000, rabbit polyclonal, a kind gift from M. Zetka, McGill University). Secondary antibody incubation was performed for 2–4 hr using goat anti-rabbit-Alexa555 (1:500–1:1000; Molecular Probes, Eugene, OR) or goat anti-mouse Alexa488 (1:500–1:250; Molecular Probes or Jackson ImmunoResearch). Samples were mounted in Vectashield media containing DAPI (Vector Laboratories, Burlingame, CA). To control for antibody specificity, slides omitting primary antibodies, and samples containing control worms without a HA tag, were processed in parallel. Images were captured using an ORCA-ER-1394 CCD camera (Hamamatsu) and Volocity Software (Perkin-Elmer) on a Nikon (Garden City, NY) Eclipse 90i microscope using a 20×/0.75, 40×/1.3, or 60×/1.4 oil immersion objective (Nikon), or on a Zeiss ([Carl Zeiss], Thornwood, NY) Axio Imager Z1 using a 40×/0.75 air objective. Maximum intensity z-projections of images taken through the sample using a motorized stage are shown, with the exception of images for CEC-1 in the head (Figure 2C), and CEC-6 in embryos and the germline (Figure 2B, left lower panels, Figure 2E, middle panels), for which single z-plane images are shown.
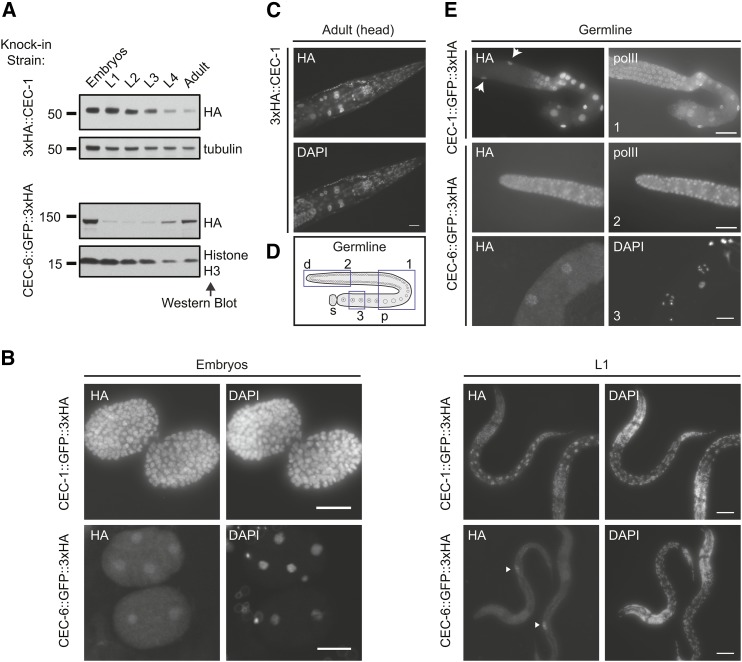
Figure 2.
Distinct expression patterns of C. elegans chromo-domain proteins CEC-1 and CEC-6. (A) Relative expression levels of CEC-1 and CEC-6 across developmental stages. The 3xHA (hemagglutinin) epitope was inserted in-frame at the endogenous locus using clustered regularly interspaced short palindromic repeats/Cas9 genome editing (see also Figure S3C). L1–L4, larval stages 1–4. (B) Localization of CEC-1 and CEC-6 by anti-HA tag immunofluorescence in embryos and L1 animals. The arrowheads [(B), lower right] indicate the primordial germ cells (see also Figure S3D). (C) Broad nuclear expression of CEC-1 in the adult head. (D) Schematic of an isolated germline arm indicating three regions depicted in (E). The distal germline is a syncytium, with germ cells surrounding a central canal. d, distal; p, proximal; and s, spermatheca. (E) Expression of CEC-1 and CEC-6 in dissected germlines. The arrows (top panel) indicate nuclei of the somatic gonad cells. Germlines in (C) were costained with an antibody detecting the RNA polymerase C-terminal domain (polII) to control for antibody penetration.
Scoring adult gross morphology phenotype
Worms were synchronized by bleaching as described above and the arrested L1s were washed once in M9 buffer, counted, and plated on NGM agar with OP50-1 (250–400 animals per plate). Animals were scored after 54–58 hr. Alternatively, a synchronous population was obtained by allowing 10–15 gravid adults to lay embryos on NGM agar with OP50-1 for 2 hr. The adults were removed and the progeny were scored after 66–68 hr of development. At these timepoints, ∼99% of wild-type animals are gravid adults. The other category (Figure 3B) represents ∼0–5% of animals (see graph) and includes, if present, developmentally delayed (larval) animals, males, sterile adults, and ruptured animals. The number of worms scored for each biological replicate was: N2: 278, 890, 689, 656, and 741; cec-1Δ: 265, 839, 742, 639, and 627; cec-1(ok1005): 318 and 695; cec-3Δ: 283, 720, 939, 751, and 870; cec-3(ok3432): 270 and 371; cec-3(Δ);cec-1(Δ): 597, 360, 513, 393, and 412; cec-3(ok3432);cec-1(ok1005): 310 and 558; Si[cec-1(+)];cec-3(Δ);cec-1(Δ): 563, 699, 462, and 866; cec-6(Δ): 638; cec-1(Δ) cec-6(Δ): 646, 640, 273, and 539; and cec-3(Δ);cec-6(Δ): 792, 607, 330, and 690.
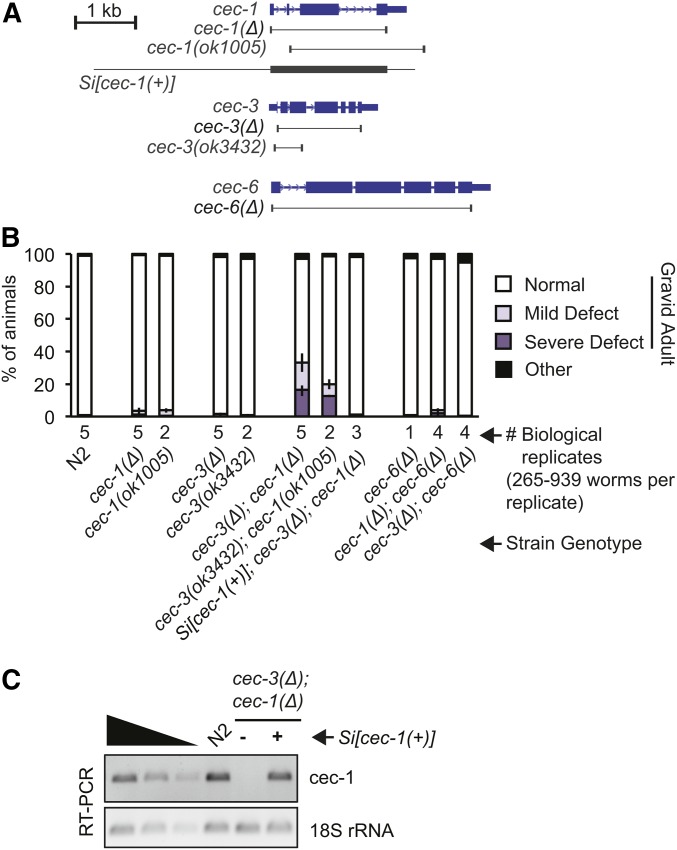
Figure 3.
Loss of cec-1 and cec-3 leads to adult-onset defects with partial penetrance. (A) Schematic of alleles and transgenes used in this study (see also Figure S5). The borders of deletion alleles are indicated below the gene diagrams. The genomic fragment used for the Minimal Mos1 (miniMos) transgene rescue (Si[cec-1(+)]) is indicated. (B) Quantification of abnormal gross morphology phenotypes in synchronized worm populations of indicated genotypes grown to gravid adulthood. Error bars represent the range of proportions of abnormal worms from all biological replicates scored. For images of abnormal adult animals, see Figure 4A. (C) The miniMos cec-1 rescue transgene (Si[cec-1(+)]) mRNA is expressed at similar levels to endogenous cec-1, as assayed by RT-PCR.
Competition assay
Competition assays were performed essentially as described (Norris et al. 2017). Worms were semisynchronized by allowing five gravid adults from well-fed plates to lay embryos for 3 hr and then removing the adults. Two days later, an equal number of L4 animals from each test strain and the cec-1(Pmyo2::GFP+)-marked control strain were picked to a fresh plate. After 5 days, the proportion of GFP- vs. GFP+ worms was determined. Worms were immobilized by incubation at 4° for several hours and counted on a fluorescence stereoscope (Zeiss). In some instances, the growth plate was chunked onto a fresh unseeded NGM agar plate for counting. Three to six fields containing a total of 141–513 animals were counted per plate. Three or four biological replicates were performed for each strain, with three plates per strain per replicate. The total number of animals counted per strain per biological replicate was: N2: 1170, 745, 732, and 646; cec-1Δ: 761, 705, and 680; cec-3Δ: 595, 657, and 580; cec-3(Δ);cec-1(Δ): 638, 601, 535, and 652; and Si[cec-1(+)];cec-3(Δ);cec-1(Δ): 563, 649, and 575. The average relative fitness (RF) for each biological replicate was calculated as follows: (#GFP(−)/#Total)/0.5, where 0.5 is the expected value for equal fitness between the GFP(+) and GFP(−) strains (Norris et al. 2017).
Life span assay
Worms were grown synchronously as described above using either a synchronized egg lay or bleaching. At the L4 stage, worms were picked onto fresh plates to begin the assay: 35 worms per plate on three plates per strain (biological replicate 1) or 30 worms per plate on two plates per strain (biological replicate 2). Worms were transferred to fresh plates daily until the end of egg laying. Worms were scored for every 1–2 days by gently tapping with an eyelash or platinum wire. Worms that died by vulval bursting or “bagging”, or that crawled off the plate, were censored in the analysis. Survival curves and life span statistics were calculated using OASIS 2 software (Han et al. 2016).
Brood size and fertility assays
To determine brood size per animal, L4 hermaphrodites were “singled” and allowed to develop into adults. After the onset of egg laying, the mother was moved to a new plate daily until egg laying ceased. Hatched progeny were counted the day after the mother was removed or 2 days later for assays at 15°. Differences in the distributions of brood sizes were assessed using the Exact Dynamic Programming Solution of the Wilcoxon rank-sum test (Marx et al. 2016). The calculated P-values are shown for all pairwise comparisons where P < 0.05.
To estimate and follow the fertility of a larger number of animals over several generations, single worms were picked to plates (as L4s or as L1s), and the number of F1 progeny was counted on the third or fourth day of adulthood, when F1s laid on the first 2 days of adulthood were L4/adults. Worms were scored as “fertile” if they had > 100 F1 progeny, or as “reduced fertility” (50–100 F1) or “subfertile” (< 50 F1). Worms were scored as sterile if no live progeny were observed, and these plates were checked again for progeny after an additional 2 days.
Mortal germline assay
The mortal germline assay was performed essentially as described (Ahmed and Hodgkin 2000). Briefly, six to eight lines were established for each genotype. For each line, six L1/L2 worms were picked to a fresh growth plate every 6–7 days (∼2 generations) at 22°. For the assay at 25°, animals were transferred every 5 days (Figure 6B, dotted line only), and for the assay at 15° animals were transferred every 10–12 days (Figure 8). Lines were scored as sterile when no live progeny were produced. For assays in Figure 6, B and C, strains were freshly outcrossed to N2 four or five times, and the rehomozygosed worms were considered to be the “first” generation.
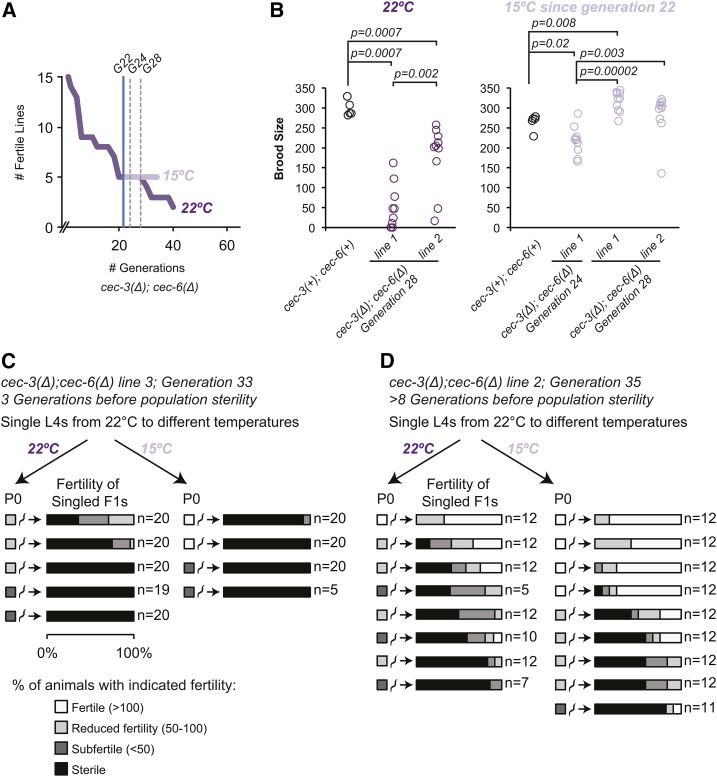
Figure 8.
Effect of temperature downshift on late-generation cec-3Δ;cec-6Δ animals. (A) A mortal germline assay was initiated using 15 lines at 22°. When five fertile lines remained [generation (G) 22, blue vertical line), lines were passaged to both 22 and 15° and mrt assays were continued in parallel at both temperatures. The break on the x-axis indicates that the generation count was restarted when these 15 lines were initiated from fertile plates from the mortal germline assay presented in Figure 6C (right panel). (B) Brood size assays were conducted on animals selected from plates at each temperature two and six generations (G24 and G28, dotted gray vertical lines) after the temperature shift. The P-values are shown for all pairwise comparisons where P < 0.05 (two-sided Wilcoxon rank-sum test). (C and D) L4 animals from the 22° mrt assay (A) were singled to the indicated temperatures. Their fertility was scored as indicated by the color scale and described in Materials and Methods. Approximately 12–20 F1 animals were singled per line and their fertility was also scored. Assays were initiated at the generation number indicated in the figure.
Bright field microscopy and DAPI staining
Bright field images of worms on growth plates were captured using a Focus microscope camera (Exo Labs) mounted on a Leica M60 stereoscope. In Figure 4A, worms were anesthetized with 30 mM sodium azide. For whole-animal DAPI staining, worms were fixed in methanol or ethanol, and mounted on microscope slides in Vectashield media containing DAPI (Vector Laboratories). Images were captured with an ORCA-ER-1394 CCD camera (Hamamatsu) on a Nikon Eclipse 90i or Zeiss Axio Imager Z1 microscope using Volocity Software (Perkin-Elmer), or a Nikon TE-2000E inverted microscope using OpenLab (Perkin-Elmer).
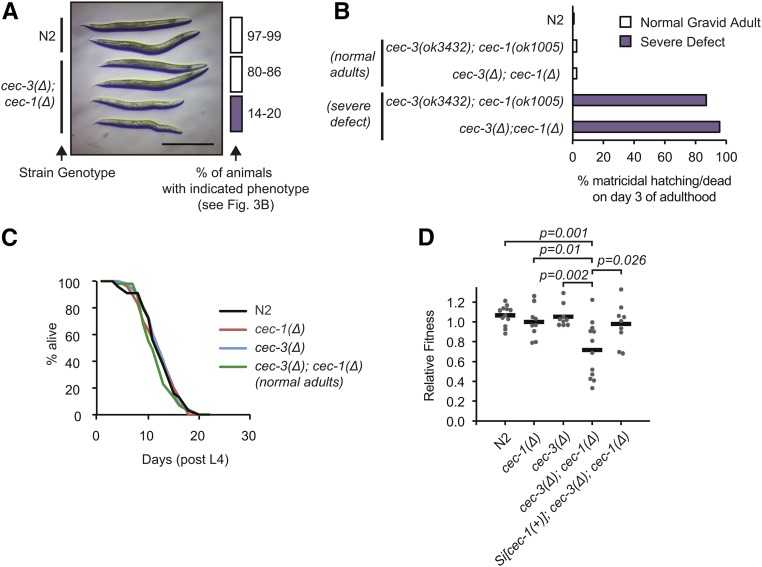
Figure 4.
Synthetic fitness defect in cec-3Δ;cec-1Δ adult animals. (A) Stereoscope image of wild-type (N2) and age-matched cec-3Δ;cec-1Δ normal or severely abnormal gravid animals as quantified in Figure 3B. Bar, 0.5 mm. (B) Nearly all severely abnormal cec-3Δ;cec-1Δ adults do not survive past day 3 of adulthood. (C) The life span of phenotypically normal cec-3Δ;cec-1Δ animals is not significantly different from wild-type or single-mutant animals. Number of animals scored: N2, 55; cec-1Δ, 53; cec-3Δ, 58; and cec-3Δ;cec-1Δ, 55. (D) Phenotype of cec-3Δ;cec-1Δ animals in a competitive fitness assay. Animals of the indicated genotypes were grown in competition with a GFP-marked strain, cec-1(Pmyo-2::GFP+). The P-values indicate a two-sample Student’s t-test.
Data availability
All strains and plasmids are available upon request. Strains and alleles are listed in Table S1. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7045148.
Results
Identification of C. elegans H3K27me-binding chromo-domain proteins
To identify C. elegans proteins that directly recognize histone H3K27me, we used the Basic Local Alignment Search Tool (Altschul et al. 1990) to search for genes encoding chromo domains with similarity to known H3K27me3 readers, Drosophila Polycomb (Pc), and the related human CBX family (Fischle et al. 2003; Min et al. 2003; Kaustov et al. 2011), and which share the single-domain architecture of Pc/CBX. We focus on the C. elegans cec genes (Aasland and Stewart 1995; Agostoni et al. 1996), cec-1 and cec-6, which contain chromo domains with 69 (37/53) and 53% (29/54) amino acid similarity to the human CBX2 chromo domain, respectively (Figure 1A).
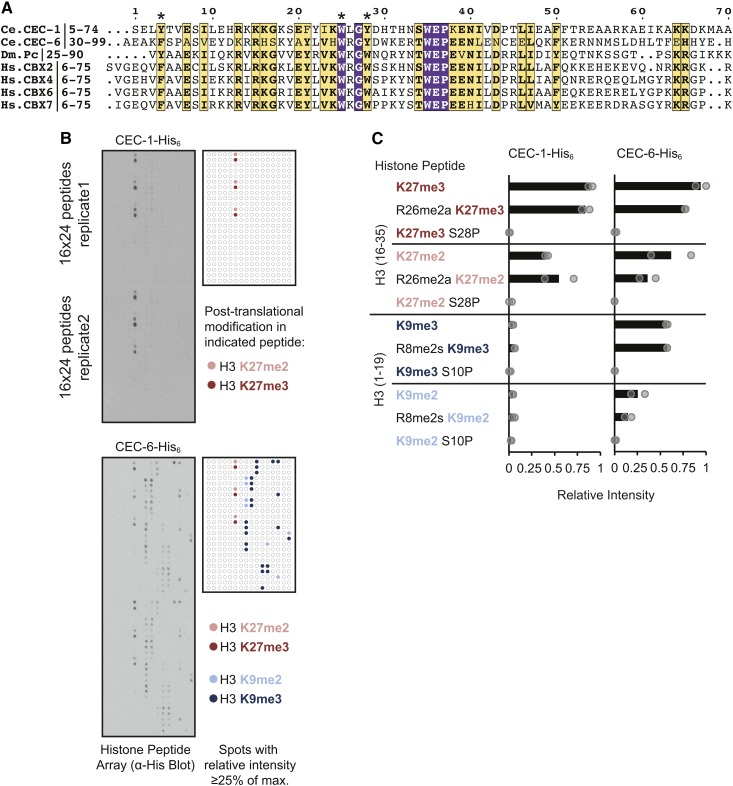
Figure 1.
Binding specificity of C. elegans chromo-domain proteins CEC-1 and CEC-6 for histone H3 peptides with tri- or dimethylation at lysine (K) 27 or K9 (H3K27me/H3K9me). (A) Multiple sequence alignment showing amino acid similarity of the chromo domains of CEC-1 and CEC-6 to those of Drosophila polycomb (Dm_Pc) and the human Chromobox (Hs_CBX) proteins. Conserved aromatic residues involved in methyl-lysine binding (Fischle et al. 2003; Min et al. 2003) are indicated by asterisks. Residue coloring: purple boxes, amino acids identical in all seven sequences; yellow boxes and bold text, amino acids with similar properties in all seven sequences; yellow boxes, bold and normal text, identical amino acids in six of seven sequences. (B) Binding of recombinant His6-tagged proteins to histone peptide arrays detected by HRPase-conjugated anti-His antibody and chemiluminescence. Arrays contain 384 peptide spots in duplicates of 19mer histone peptides bearing single or multiple post-translational modifications. The map highlights all spots with a relative intensity ≥ 0.25, where the array spot with the greatest intensity is set to 1, as described in the Materials and Methods. Colors show that the indicated spots contain H3K27me3/2 or H3K9me3/2. (C) Quantification of peptide array signals showing relative binding of CEC-1 or CEC-6 to H3K27me3/2 and H3K9me3/2 in combination with other peptide modifications. R8/26me2s/a refers to symmetric or asymmetric arginine dimethylation. Relative intensities of individual peptide spots are indicated by circles and the average is shown as a bar. For additional quantification, see also Figure S2 and Table S2.
To determine the methyl-lysine-binding specificity of CEC-1 and CEC-6, we used a histone peptide array (Figure 1B and Figure S2) (Bock et al. 2011). The array contained 384 peptides in duplicate, representing eight regions of histones H3, H4, H2A, and H2B with single and combinations of post-translational modifications (Table S2). The arrays were probed with recombinant, full-length 6xHis-tagged proteins, which revealed that the most highly-bound peptide spots for both CEC-1 and CEC-6 contained trimethylated histone H3-Lys27 (H3K27me3). Both CEC-1 and CEC-6 also showed binding to H3K27me2, with on average 55% intensity compared to the H3K27me3 signals. In addition, CEC-6 recognized H3K9me3 (on average ∼70% of H3K27me3 signal) and H3K9me2 (∼48% of H3K9me3 signal) peptides. In contrast, CEC-1 showed less cross-reactivity to H3K9me-peptides (on average ∼6% of H3K27me3 signal). There was no detectable binding of CEC-1 or CEC-6 to unmodified histone peptides, or to peptides tri- or dimethylated at other residues (Figure 1B, Figure S2, and Table S2). In contrast to CEC-1 and CEC-6, the chromo-domain protein CEC-3 recognized H3K9me but not H3K27me peptides, in agreement with a previous study (Greer et al. 2014) and in support of the specificity of our assay (Figure S2). The binding of CEC-1 or CEC-6 to H3K27me3/2 or K9me3/2 peptides was abrogated by phosphorylation of the neighboring serine residue, S28P or S10P (Figure 1C), a switch that plays a role in the chromatin targeting of other chromo-domain proteins (Fischle et al. 2005; Hirota et al. 2005; Gehani et al. 2010). Additionally, as seen here for CEC-6, the chromo domains of the Polycomb-related mammalian CBX family also recognize both H3K27me3 and H3K9me3 in vitro (Bernstein et al. 2006; Kaustov et al. 2011). These binding assays suggest that CEC-1 and CEC-6 are novel readers of H3K27 methylation in C. elegans.
To examine the developmental expression patterns of cec-1 and cec-6, we generated epitope-tagged knock-in strains using CRISPR/Cas9 and examined published gene expression profiles [RNA-Seq (RNA-sequencing) data]. Western blots of tagged proteins from staged worm populations indicated that both CEC-1 and CEC-6 are present in embryos (Figure 2, A and B and Figure S3). While CEC-1 is also expressed throughout development, CEC-6 levels were low until the L4 and adult stages, when the number of germ cells dramatically increases and gametes are produced (Kimble and Crittenden 2007). The temporal expression patterns of CEC-1 and CEC-6 proteins agreed well with published RNA-Seq data (Boeck et al. 2016), and the germline expression of cec-6 was supported by its enrichment in dissected adult germlines (Boeck et al. 2016) and in germline cells from L2 animals in single-cell RNA-Seq (Cao et al. 2017) (Figure S3). Using immunofluorescence, we found that CEC-1 was nuclear and broadly expressed in the embryonic, larval, and adult animals (Figure 2, B and C). Although CEC-1 was suggested to be soma-specific (Agostoni et al. 1996), its transcript was detected in the oogenic germline by RNA-Seq (Ortiz et al. 2014; Boeck et al. 2016). Consistent with these findings, while we do not observe CEC-1 in the distal germline, it is detected in proximal germ cells and oocytes (Figure 2, D and E and Figure S4), albeit at a lower level than somatic cells. Although CEC-6 was enriched in the nucleus in early embryos and the adult germline, it was also detected outside the nucleus, particularly in the primordial germ cells in L1 animals (Figure 2 and Figure S3). While we cannot rule out that interference from the tag contributes to this localization pattern, cytoplasmic localization of chromatin factors, including histone methyltransferases, has been reported (Loyola et al. 2009; Towbin et al. 2012; Bodega et al. 2017). In addition, the detection of CEC-6 protein in oocytes and early embryos suggests that it may be maternally deposited. However, this prediction remains to be verified by genetic analysis. Together, the distinct expression patterns of cec-1 and cec-6 suggest that, although they have overlapping histone methylation-binding profiles, they are likely to contribute to distinct functions.
cec-1 and cec-3 contribute to robust development
To investigate the roles of these chromo-domain proteins in C. elegans development, we generated deletion mutants using CRISPR/Cas9, referred to below as cec-1Δ and cec-6Δ, and obtained deletion mutants from the C. elegans knockout consortium (C. elegans Deletion Mutant Consortium 2012) (see Materials and Methods, Figure 3A, Figure S5, and Table S1). Under typical growth conditions, the mutant strains appeared superficially normal and fertile, with growth rates similar to wild-type animals (Figure 3B and data not shown). These results suggest that, although they may play roles in specific biological processes not examined here, cec-1 and cec-6 are individually dispensable for gross somatic development.
To examine potential functional relationships among the related cec genes, we mated single-mutant strains to construct pair-wise double mutants of cec-1, cec-6, and cec-3, which recognizes H3K9 methylation (Greer et al. 2014). These strains were grown synchronously and observed for differences in development. Surprisingly, we found that cec-3Δ;cec-1Δ double mutants displayed abnormalities that became apparent during the reproductive phase of adulthood. At this time, 95–99% of wild-type or single-mutant animals were embryo-containing (gravid) adults that appeared relatively uniform in size (Figure 3B). In contrast, ∼16% (range 14–20%) of cec-3Δ;cec-1Δ mutants were noticeably thinner and shorter than wild-type animals (“severe”; Figure 3B and Figure 4A). This severely affected subset of cec-3Δ;cec-1Δ gravid adults also had a scrawny or frail appearance. An additional 17% (11–23%) of cec-3Δ;cec-1Δ animals were less severely affected, thin-looking gravid adults (“mild,” Figure 3B). Worms with independent deletion alleles of cec-1 and cec-3 (cec-3(ok3432);cec-1(ok1005); Figure 3A) displayed these developmental abnormalities at a similar frequency, whereas other cec mutant combinations did not (Figure 3B). To confirm the specificity of this phenotype, we introduced a cec-1 genomic transgene using miniMos transposon insertion (Frøkjær-Jensen et al. 2014) (Si[cec-1(+)], Figure 3A) into cec-3Δ;cec-1Δ mutants. This transgene was expressed at comparable levels to endogenous cec-1 (Figure 3C) and was able to rescue the adult developmental abnormalities of cec-3Δ;cec-1Δ mutants.
To examine the heritability of the cec-3Δ;cec-1Δ phenotype, severely affected animals or those with wild-type morphology were picked to separate plates. Severely affected animals had progeny with wild-type adult morphology and vice versa (data not shown). In addition, the proportion of affected animals in the population did not change substantially among assays repeated several months apart (Figure 3B). Thus, we did not find evidence for heritability of the abnormal morphology phenotype. Overall, the results above suggest a genetic interaction and functional synergy between cec-1 and cec-3, genes encoding readers of H3K27me and H3K9me, respectively, that is manifest at adulthood and affects the robustness of somatic development.
Life span and fitness of cec-3Δ;cec-1Δ animals
To further characterize the severely affected cec-3Δ;cec-1Δ animals, we examined their survival as adults. On the third day of adulthood, when nearly all N2, cec-1Δ, cec-3Δ, and morphologically normal cec-3Δ;cec-1Δ double-mutant adults are healthy, the survival of severely affected cec-3Δ;cec-1Δ animals was compromised. Eighty-seven percent (n = 126) or 95% (n = 123) of severely affected cec-3(ok3432);cec-1(ok1005) or cec-3Δ;cec-1Δ animals, respectively, were dead, mostly by hatching of embryos inside the mother (also known as bagging or “matricidal hatching”) (Figure 4B). In contrast, the life span of morphologically normal, isogenic cec-3Δ;cec-1Δ adults was not significantly different from N2, or from the cec-1Δ and cec-3Δ single-mutant animals (Figure 4C and Figure S5B). Overall, combined loss of cec-3 and cec-1 affected the adult survival and therefore reproductive capacity of a subset (14–20%) of animals.
Given the partially penetrant phenotype observed in cec-3Δ;cec-1Δ mutants, we sought to quantify whether loss of both chromo-domain genes would lead to a fitness defect using a competitive growth assay (Norris et al. 2017) (Figure 4D). In contrast to our estimation of the penetrance of developmental defects in the scoring assays above, the fitness assay enables a quantitative and unbiased measurement of the effect of the genetic interaction between cec-1Δ and cec-3Δ at the population level. Wild-type (N2), and single- and double-mutant worms were grown in competition with a cec-1 deletion mutant, in which the cec-1 coding region was replaced with a marker expressing GFP in the pharynx [cec-1(Δ::Pmyo-2::GFP)]. The relative fitness (RF) of strains was assessed by counting GFP-positive vs. non-GFP-positive animals after the food on the plate was consumed. As expected, the fitness of N2, cec-1Δ, or cec-3Δ single-mutant animals was similar to cec-1(Δ::Pmyo-2::GFP) (average RF = 1.1, 1.0, and 1.1, respectively; Figure 4D). In contrast, cec-3Δ;cec-1Δ mutants were less fit (RF = 0.72) and fitness was rescued in strains expressing a cec-1(+) transgene (Si[cec-1(+)];cec-3Δ;cec-1Δ; RF = 0.98). These results provide further evidence of an aggravating genetic interaction between cec-1 and cec-3. Despite the partial penetrance of the cec-3Δ;cec-1Δ developmental phenotype, the effect on survival and reproduction translates into a competitive disadvantage of the double-mutant strain.
Chromo-domain proteins affect germline maintenance across generations
To determine the role of chromo-domain proteins in germline maintenance, we examined the fertility of our mutant strains. Chromatin regulation pathways are important for C. elegans germline function (Kelly 2014) and mutation of the genes responsible for histone H3K27 methylation results in a maternal effect sterility (mes) phenotype (Capowski et al. 1991; Bender et al. 2004). We found that cec-6Δ, but not cec-1Δ, animals had a reduced brood size at 25° (Figure 5A). However, after two backcrosses to N2, cec-6Δ animals had variable brood sizes, with some animals showing broods similar to wild-type (Figure 5B). In addition, individual lines passaged from small numbers of cec-6Δ animals showed variable population fertility. Three representative growth plates display a range of fertility (Figure 5C). Whereas most lines had qualitatively normal fertility, some lines had reduced fertility, evident by the lack of food consumption 1 week after six animals were placed on the plate (Figure 5C, middle), and other lines became sterile, giving rise to no live progeny (Figure 5C, right). These data suggested that there may be a progressive multigenerational fertility defect.
To directly address this possibility, a mortal germline assay (Ahmed and Hodgkin 2000) was carried out using single-, double-, and triple-mutant strains (Figure 6). Six lines per genotype were each initiated from six L4-stage animals randomly selected from populations passaged for many generations as homozygotes of the indicated genotypes (Figure 6A). Strikingly, cec-3Δ;cec-6Δ lines started to become sterile at generation 4 and all six lines were sterile by generation 38. The cec-6Δ single-mutant animals had an intermediate phenotype, with three of six lines becoming sterile during the 80-generation time period, whereas cec-3Δ single-mutant worms remained mostly fertile (five of six lines), with one line becoming sterile after 28 generations (Figure 6A). In contrast, all wild-type (N2) and cec-1Δ lines remained fertile (6/6 lines) throughout the 80 generations monitored. The fertility of double- and triple-mutant combinations with cec-1Δ, cec-3Δ;cec-1Δ, cec-1Δ;cec-6Δ, and cec-3Δ;cec-1Δ;cec-6Δ were similar to cec-3Δ, cec-6Δ, and cec-3Δ;cec-6Δ, respectively. These results indicate that loss of cec-1 does not aggravate the fertility phenotype of cec-3Δ;cec-6Δ animals.
To more specifically address the timing and contributions of cec-6 and cec-3 to the mortal germline (mrt) phenotype of the cec-3Δ;cec-6Δ double mutant, cec-3Δ;cec-6Δ animals were backcrossed to N2 four times while maintaining both alleles as heterozygotes (cec-3Δ/+;cec-6Δ/+). Homozygous wild-type and mutant animals were isolated from the progeny of these crosses, and the mrt assay was repeated (Figure 6B). Consistent with the results above, eight out of eight cec-3Δ;cec-6Δ lines became sterile between generations 27 and 39. This phenotype was exacerbated at high temperature (25°), where eight out of eight lines became sterile 12 generations earlier than at 22° (between generation 17 and 27; Figure 6B). The cec-3Δ lines remained fertile through 70 generations. However, as observed in the initial experiment, a subset of cec-6Δ lines (three out of eight) became sterile at later generations (47–63). The mrt phenotype of cec-3Δ;cec-6Δ and cec-3(ok3432);cec-6Δ animals was also supported in independent biological replicates (Figure 6C). To confirm the role of cec-6 in the mrt phenotype, we introduced a cec-6 rescue transgene into N2 animals using miniMos transposon insertion (Frøkjær-Jensen et al. 2014) (Si[cec-6(+)]) and mated them with freshly backcrossed cec-3Δ;cec-6Δ animals. Whereas the cec-3Δ;cec-6Δ lines became sterile, the fertility of cec-3Δ;Si[cec-6(+)];cec-6Δ lines was rescued to a degree similar to cec-3Δ single mutants (five out of six lines fertile after 50 generations; Figure 6D). These genetic interactions demonstrate that cec-6 and cec-3 both play important roles in germline maintenance, and that mutating both genes amplifies the germline mortality phenotype.
To examine the variability of fertility among animals approaching population sterility, brood size assays were performed on animals selected at random from late-generation plates in the mrt assay (Figure 7A). Animals wild-type for cec-3 and cec-6 [cec-3(+);cec-6(+)] had consistently large broods (median 284 F1 progeny, range 207–337). In contrast, cec-3Δ;cec-6Δ animals had variable fertility (Figure 7A). Three of 16 animals assayed were sterile, four of 16 were subfertile (<50 progeny), and the remaining nine had brood sizes from 113 to 262 (median 179). Thus, late-generation cec-3Δ;cec-6Δ animals exhibit a range of fertility defects.
Figure 7.
Germline and fertility defects in individual late-generation cec-3Δ;cec-6Δ animals. (A) Fertility defects of late-generation cec-3Δ;cec-6Δ animals. Individual worms were randomly selected for brood size assays 14 (left) or 10 (right) generations (G) before population sterility, corresponding to G58 and G62 from the mortal germline assay in Figure 6C, right. The P-values represent a two-sided Wilcoxon rank-sum test. (B) Transgenerational fertility of late-generation single animals during the mortal germline assay. For cec-3Δ;cec-6Δ animals (right panel), F2 progeny were analyzed from P0 animals with varying levels of fertility (G66 from the mortal germline assay in Figure 6C, right). All assays were performed at 22°. (C and D) Germline morphology in fertile, sterile, or near-sterile DAPI-stained fixed animals of the indicated genotypes. Animals in (C and D) are from the mortal germline assays shown in Figure 6, A and B, respectively, at the indicated generations. Bar, (C) 50 μm; (D) 100 μm. v, ventral; h, head. Dotted lines indicate the distal germline and the solid line in (C) indicates embryos in the uterus. For additional images, see Figure S6.
Transgenerational fertility and germline defects in cec-3Δ;cec-6Δ mrt animals
To examine the dynamics and heritability of fertility defects in cec-3Δ;cec-6Δ animals, individual worm lineages were followed across several generations. Single animals from late generations in the mrt assay were picked to plates (singled) and their fertility scored (Figure 7B). In the initial generation (P0), 54% (13/24) of the randomly selected cec-3Δ;cec-6Δ P0 animals were sterile, while the remaining 11 animals had variable fertility. To determine how the fertility of individual P0 animals was related to the fertility of their progeny, we then followed the fertility of F1 or F2 animals from these P0 animals. Among six subfertile P0 animals (defined as animals with <50 F1 progeny, Figure 7B), two animals gave rise to only sterile F1 progeny. The other four subfertile P0 animals gave rise to a mixture of both sterile and fertile F1 animals. When F2 progeny were analyzed from a subfertile P0 animal, they had a range of fertility defects and ∼65% were sterile.
In parallel, the F2 progeny of a representative fertile cec-3Δ;cec-6Δ P0 were analyzed (Figure 7B, rightmost). These progeny also had a range of fertility defects, including two animals with near-wild-type fertility. When F2 progeny from these two fertile animals were examined, the distribution of fertility was distinct. For one animal, 85% of examined F2s were sterile, while for the second animal, only 13% were sterile and the majority were fertile. Therefore, as was the case in the brood size assays above (Figure 7A), there was variability in the fertility level of individual animals both within and across generations. However, an overall pattern emerged in which, as subfertile animals arose in the population, they initiated lineages with transgenerational fertility defects. These lineages varied in the number of generations until complete sterility of progeny was reached, which may underlie the gradual progression of population-level sterility.
To examine germline morphology in the sterile animals, worms were fixed and nuclei visualized by DNA staining with DAPI (Figure 7, C and D and Figure S6). In fertile animals, the nuclei of well-proliferated distal germlines (Figure 7, C and D, left) and embryos in utero (Figure 7C, left) are easily visualized. In many sterile cec-3Δ;cec-6Δ animals examined, embryos were not present, and the germlines were generally small and irregular (Figure 7, C and D) or hardly discernible (Figure 7D; bottom animal). A similar phenotype was observed in 10/10 sterile animals in an independent experiment (Figure S6). Thus, loss of cec-6 and cec-3 leads to abnormal germline development, including disorganized germlines with drastically reduced numbers of germ cells compared to wild-type animals.
The mrt phenotype of cec-3Δ;cec-6Δ animals is partially reversible by low temperature
To determine if the mrt phenotype is reversible, we shifted mrt assay animals from 22 to 15° and then followed fertility at each temperature in parallel (Figure 8A). In addition, brood sizes were compared among animals at each temperature at the second and sixth generations after the temperature shift (Figure 8B). These assays indicated that shifting animals from 22 to 15° for several generations at least partially restores fertility. However, given the number of generations to sterility at 22° and the longer intergenerational time at 15°, we could not distinguish between a complete reversal of the mrt phenotype and a much longer delay in reaching sterility at 15°.
To determine how a temperature shift would affect the fertility of individual late-generation cec-3Δ;cec-6Δ animals, we also transferred individual animals as L4s from 22° to either 22 or 15°, and assessed their fertility and the fertility of their progeny at each temperature (Figure 8, C and D). Consistent with the results in Figure 7, most progeny of subfertile animals were subfertile or sterile. We found that the fertility of F1 animals from P0 mothers raised at 22° until L4 and then transferred was similar at both 22 and 15° (Figure 8, C and D). These results suggest that rescue of fertility by transfer to 15° requires several generations of growth at the lower temperature.
Previous work has found that germline defects in met-2set-25 mutants lacking H3K9me may be the result of extensive DNA damage, which can be monitored by ectopic foci of the DNA repair protein RAD-51 in the mitotic germline (Zeller et al. 2016). To determine whether such genetic changes may contribute to the mrt phenotype of cec-3Δ;cec-6Δ animals, dissected germlines were stained with an anti-RAD-51 antibody (Figure S7). However, we did not see evidence for increased RAD-51 foci in these animals. Together with the temperature shift analysis, these data are consistent with a partially reversible mechanism for germline mortality in cec-3Δ;cec-6Δ animals.
Discussion
Chromo-domain proteins and other histone post-translational modification readers can act as effectors of chromatin states. Identifying and characterizing the binding specificity of these reader proteins is therefore a crucial step toward understanding the roles of histone modifications in development and epigenetic inheritance. We have identified two C. elegans chromo domains that directly recognize PRC2-dependent histone methylation (H3K27me2/3) in vitro. In addition, we uncovered surprising functional interactions that indicate a key role for multiple heterochromatin readers in both somatic development and in the maintenance of germline immortality across generations.
Our work has unveiled new potential heterochromatin reader proteins in a widely studied model organism. It is also noteworthy that there are several additional C. elegans chromo-domain proteins with unknown binding specificity or functions (Figure S1). Future analysis of these additional candidate chromatin readers will be important to understand how they genetically interact with CEC-1, CEC-6, and CEC-3, and whether they affect the severity of the developmental and fertility phenotypes we have described. In addition, based on our in vitro-binding data, and by analogy to similar chromo-domain proteins such as Polycomb and mammalian CBX proteins, we propose that CEC-1 and CEC-6 associate with H3K27me-marked chromatin in vivo. It will be of interest in future studies to fully characterize the genomic binding patterns of CEC-1 and CEC-6, and to understand the extent to which these and other potential chromatin readers may regulate common or distinct genomic targets.
Histone H3K27me3 and H3K9me2 are considered hallmarks of facultative heterochromatin, and H3K9me3 a hallmark of constitutive heterochromatin (Nestorov et al. 2013). Although these repressive chromatin pathways are generally considered mechanistically distinct, our finding of a genetic interaction between cec-1 and cec-3, which have divergent binding specificity for H3K27me3/2 and H3K9me, respectively, suggests a compensatory interaction between these pathways during development. In support of this interplay, H3K9 methylation can partially compensate for loss of H3K27 methylation to repress the X chromosome in the C. elegans male germline (Gaydos et al. 2014). In addition to the unexpected synthetic interaction between two readers of constitutive vs. facultative heterochromatin marks, the incomplete penetrance of developmental defects in cec-3Δ;cec-1Δ animals is intriguing. Mutation or loss of chromo-domain proteins that recognize H3K9me3 or H3K27me3 can lead to redistribution of H3K27 methylation to constitutive heterochromatin in several model systems (Basenko et al. 2015; Dumesic et al. 2015; Jamieson et al. 2016), and loss of H3K9me3 from heterochromatic loci in human cells (Tchasovnikarova et al. 2015). One model that may account for the incomplete penetrance in cec-3;cec-1 mutants is a stochastic loss of transcriptional silencing, analogous to position-effect variegation, leading to variations in gene expression levels and phenotypic differences among isogenic individuals (Raj et al. 2010).
Genomic redistribution of chromatin modifications may also play a role in the synthetic mrt phenotype of cec-3;cec-6 mutant animals. Studies of the mes genes (Capowski et al. 1991; Xu et al. 2001; Bender et al. 2004) have revealed the importance of balance between the opposing machineries of H3K27 and H3K36 methylation in ensuring proper germline gene expression (Gaydos et al. 2012). In addition, genes with opposing enzymatic effects on histone methylation, such as the H3K4me2/3 methyltransferase set-2 (Xiao et al. 2011) and the H3K4me2-demethylase spr-5 (Katz et al. 2009; Kerr et al. 2014), can lead to a mrt phenotype. However, in contrast to other chromatin modifiers with mrt phenotypes (Andersen and Horvitz 2007; Simonet et al. 2007; Katz et al. 2009; Li and Kelly 2011; Xiao et al. 2011; Kerr et al. 2014; Robert et al. 2014; Weiser et al. 2017), cec-6 and cec-3 do not encode domains with enzymatic activities, such as a histone methyltransferase or demethylase. Therefore, they may influence the genomic distribution, rather than global levels, of histone modifications. Binding of the chromo-domain protein at a locus may influence the assembly or spreading of heterochromatin. Alternatively, in a manner similar to other chromo-domain proteins in Drosophila and mammals (Di Croce and Helin 2013; Simon and Kingston 2013), cec-6 and cec-3 (Greer et al. 2014) may interact with enzymatic chromatin modification complexes. Assays such as chromatin immunoprecipitation sequencing in the germlines of cec-6;cec-3 mutants will be important to further understand how these chromo-domain proteins regulate chromatin states and contribute to germline maintenance.
In addition to chromatin regulation, C. elegans germline immortality is also compromised in mutants affecting small RNA (Buckley et al. 2012; Sakaguchi et al. 2014; Simon et al. 2014) and genome stability (Ahmed and Hodgkin 2000; Chin and Villeneuve 2001; Hofmann et al. 2002; Grabowski et al. 2005; Ahmed 2006; Yanowitz 2008; Meier et al. 2009) pathways. There is also mechanistic overlap between these mechanisms and chromatin regulation, as mrt phenotypes of chromatin regulators can also involve small RNA pathways (Lev et al. 2017; Weiser et al. 2017) and impact DNA repair (Nottke et al. 2011; Zeller et al. 2016). Furthermore, genes regulating histone H3K27 and H3K9 methylation are required for the inheritance of piwi-interacting RNA-mediated silencing (Ashe et al. 2012; Shirayama et al. 2012) and silencing of repetitive transgenes (Kelly and Fire 1998; Klosin et al. 2017). It is noteworthy that that cec-3 and cec-6 were identified in an RNA interference screen to play a role in cosuppression of a repetitive transgene in the germline (Robert et al. 2005). These results suggest that small RNA silencing pathways and derepression of endogenous repetitive sequences, which has been observed in some mrt strains (Sakaguchi et al. 2014; Simon et al. 2014), may play a role in the mrt phenotype of cec-3Δ;cec-6Δ animals. Our temperature downshift and rad-51-staining experiments suggest that reversible mechanisms, rather than irreversible genetic changes such as extensive DNA damage, are likely important in the cec-3Δ;cec-6Δ mrt phenotype. However, as described above, chromatin regulators can impact multiple pathways. Additional genetic interaction studies may help to reveal the extent to which reversible mechanisms and genetic changes may contribute in parallel to the mrt phenotype.
The potential roles of histone methylation and the contribution of chromatin readers to transgenerational inheritance are poorly understood. Our finding that a genetic interaction between cec-3 and cec-6 leads to synthetic progressive sterility provides new evidence for chromo-domain proteins as important factors in transgenerational inheritance. In addition, chromatin regulation is thought to play a role in phenotypic variation or noise (Whitelaw et al. 2010; Pujadas and Feinberg 2012). We speculate that the effect on C. elegans developmental robustness observed upon deletion of cec-1 and cec-3 may provide a new tool to understand the role of chromo-domain proteins in this phenomenon. Overall, our findings reveal new roles for two heterochromatin readers, cec-1 and cec-6, as part of a network of chromatin factors regulating the reproductive and developmental fitness of an organism.
Acknowledgments
We thank S. F. Pinter (University of Connecticut), J. A. Calarco, J. M. Claycomb, and J. A. Mitchell (University of Toronto) for critical feedback on the manuscript; J. A. Calarco and A. D. Norris (Harvard University) for plasmids and assistance with CRISPR genome editing; D. Ray (T. Hughes laboratory, University of Toronto), B. Ardehali and S. Kundu [Kingston laboratory, Massachusetts General Hospital (MGH)], and C. Phillips (Ruvkun laboratory, MGH) for helpful advice, discussions, and reagents; A. E. E. Bruce (University of Toronto) for sharing equipment; and M. Zetka (McGill) for the rad-51 antibody. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD-010440). This work was supported by NIH grant R01 GM-090278 to J.T.L. J.T.L. is an Investigator of the Howard Hughes Medical Institute. A.L.S. gratefully acknowledges funding from the Canadian Institute for Health Research and Banting Postdoctoral fellowships, the National Research Council of Canada H. L. Holmes Award for Post-Doctoral Studies, and the MGH Executive Committee on Research.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7045148.
Communicating editor: D. Greenstein
Literature Cited
- Aasland R., Stewart A. F., 1995. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 23: 3168–3173. 10.1093/nar/23.16.3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostoni E., Albertson D., Wittmann C., Hill F., Tobler H., et al. , 1996. cec-1, a soma-specific chromobox-containing gene in C. elegans. Dev. Biol. 178: 316–326. 10.1006/dbio.1996.0221 [DOI] [PubMed] [Google Scholar]
- Ahmed S., 2006. Uncoupling of pathways that promote postmitotic life span and apoptosis from replicative immortality of Caenorhabditis elegans germ cells. Aging Cell 5: 559–563. 10.1111/j.1474-9726.2006.00244.x [DOI] [PubMed] [Google Scholar]
- Ahmed S., Hodgkin J., 2000. MRT-2 checkpoint protein is required for germline immortality and telomere replication in C. elegans. Nature 403: 159–164. 10.1038/35003120 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Andersen E. C., Horvitz H. R., 2007. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134: 2991–2999. 10.1242/dev.009373 [DOI] [PubMed] [Google Scholar]
- Ashe A., Sapetschnig A., Weick E. M., Mitchell J., Bagijn M. P., et al. , 2012. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 150: 88–99. 10.1016/j.cell.2012.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basenko E. Y., Sasaki T., Ji L., Prybol C. J., Burckhardt R. M., et al. , 2015. Genome-wide redistribution of H3K27me3 is linked to genotoxic stress and defective growth. Proc. Natl. Acad. Sci. USA 112: E6339–E6348. 10.1073/pnas.1511377112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender L. B., Cao R., Zhang Y., Strome S., 2004. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr. Biol. 14: 1639–1643. 10.1016/j.cub.2004.08.062 [DOI] [PubMed] [Google Scholar]
- Bernstein E., Duncan E. M., Masui O., Gil J., Heard E., et al. , 2006. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 26: 2560–2569. 10.1128/MCB.26.7.2560-2569.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettinger J. C., Lee K., Rougvie A. E., 1996. Stage-specific accumulation of the terminal differentiation factor LIN-29 during Caenorhabditis elegans development. Development 122: 2517–2527. [DOI] [PubMed] [Google Scholar]
- Bock I., Kudithipudi S., Tamas R., Kungulovski G., Dhayalan A., et al. , 2011. Application of Celluspots peptide arrays for the analysis of the binding specificity of epigenetic reading domains to modified histone tails. BMC Biochem. 12: 48 10.1186/1471-2091-12-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodega B., Marasca F., Ranzani V., Cherubini A., Della Valle F., et al. , 2017. A cytosolic Ezh1 isoform modulates a PRC2-Ezh1 epigenetic adaptive response in postmitotic cells. Nat. Struct. Mol. Biol. 24: 444–452. 10.1038/nsmb.3392 [DOI] [PubMed] [Google Scholar]
- Boeck M. E., Huynh C., Gevirtzman L., Thompson O. A., Wang G., et al. , 2016. The time-resolved transcriptome of C. elegans. Genome Res. 26: 1441–1450. 10.1101/gr.202663.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley B. A., Burkhart K. B., Gu S. G., Spracklin G., Kershner A., et al. , 2012. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 489: 447–451. 10.1038/nature11352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos E. I., Stafford J. M., Reinberg D., 2014. Epigenetic inheritance: histone bookmarks across generations. Trends Cell Biol. 24: 664–674. 10.1016/j.tcb.2014.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Packer J. S., Ramani V., Cusanovich D. A., Huynh C., et al. , 2017. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357: 661–667. 10.1126/science.aam8940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capowski E. E., Martin P., Garvin C., Strome S., 1991. Identification of grandchildless loci whose products are required for normal germ-line development in the nematode Caenorhabditis elegans. Genetics 129: 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Deletion Mutant Consortium , 2012. Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda) 2: 1415–1425. 10.1534/g3.112.003830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin G. M., Villeneuve A. M., 2001. C. elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G(2) DNA damage checkpoint. Genes Dev. 15: 522–534. 10.1101/gad.864101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciabrelli F., Comoglio F., Fellous S., Bonev B., Ninova M., et al. , 2017. Stable Polycomb-dependent transgenerational inheritance of chromatin states in Drosophila. Nat. Genet. 49: 876–886. 10.1038/ng.3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden S., Kimble J., 2009. Preparation and immunolabeling of Caenorhabditis elegans. Cold Spring Harb. Protoc. 2009: pdb.prot5216. [DOI] [PubMed] [Google Scholar]
- Dickinson D. J., Ward J. D., Reiner D. J., Goldstein B., 2013. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 10: 1028–1034. 10.1038/nmeth.2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Croce L., Helin K., 2013. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 20: 1147–1155. 10.1038/nsmb.2669 [DOI] [PubMed] [Google Scholar]
- Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., et al. , 2011. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39: W13–W17. 10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumesic P. A., Homer C. M., Moresco J. J., Pack L. R., Shanle E. K., et al. , 2015. Product binding enforces the genomic specificity of a yeast polycomb repressive complex. Cell 160: 204–218. 10.1016/j.cell.2014.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg J. C., 2012. Structural biology of the chromodomain: form and function. Gene 496: 69–78. 10.1016/j.gene.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Farboud B., Meyer B. J., 2015. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics 199: 959–971. 10.1534/genetics.115.175166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Wang Y., Jacobs S. A., Kim Y., Allis C. D., et al. , 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17: 1870–1881. 10.1101/gad.1110503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Tseng B. S., Dormann H. L., Ueberheide B. M., Garcia B. A., et al. , 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438: 1116–1122. 10.1038/nature04219 [DOI] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiacovo M. P., Church G. M., et al. , 2013. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods 10: 741–743. 10.1038/nmeth.2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Sarov M., Taylor J., Flibotte S., et al. , 2014. Random and targeted transgene insertion in Caenorhabditis elegans using a modified Mos1 transposon. Nat. Methods 11: 529–534. 10.1038/nmeth.2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos L. J., Rechtsteiner A., Egelhofer T. A., Carroll C. R., Strome S., 2012. Antagonism between MES-4 and Polycomb repressive complex 2 promotes appropriate gene expression in C. elegans germ cells. Cell Rep. 2: 1169–1177. 10.1016/j.celrep.2012.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos L. J., Wang W., Strome S., 2014. Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science 345: 1515–1518. 10.1126/science.1255023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehani S. S., Agrawal-Singh S., Dietrich N., Christophersen N. S., Helin K., et al. , 2010. Polycomb group protein displacement and gene activation through MSK-dependent H3K27me3S28 phosphorylation. Mol. Cell 39: 886–900. 10.1016/j.molcel.2010.08.020 [DOI] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A., III, et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sandoval A., Towbin B. D., Kalck V., Cabianca D. S., Gaidatzis D., et al. , 2015. Perinuclear anchoring of H3K9-Methylated chromatin stabilizes induced cell fate in C. elegans embryos. Cell 163: 1333–1347. 10.1016/j.cell.2015.10.066 [DOI] [PubMed] [Google Scholar]
- Grabowski M. M., Svrzikapa N., Tissenbaum H. A., 2005. Bloom syndrome ortholog HIM-6 maintains genomic stability in C. elegans. Mech. Ageing Dev. 126: 1314–1321. 10.1016/j.mad.2005.08.005 [DOI] [PubMed] [Google Scholar]
- Greer E. L., Beese-Sims S. E., Brookes E., Spadafora R., Zhu Y., et al. , 2014. A histone methylation network regulates transgenerational epigenetic memory in C. elegans. Cell Rep. 7: 113–126. 10.1016/j.celrep.2014.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. K., Lee D., Lee H., Kim D., Son H. G., et al. , 2016. OASIS 2: online application for survival analysis 2 with features for the analysis of maximal lifespan and healthspan in aging research. Oncotarget 7: 56147–56152. 10.18632/oncotarget.11269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F., Kerr G., Boutros M., 2014. E-CRISP: fast CRISPR target site identification. Nat. Methods 11: 122–123. 10.1038/nmeth.2812 [DOI] [PubMed] [Google Scholar]
- Hirota T., Lipp J. J., Toh B. H., Peters J. M., 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438: 1176–1180. 10.1038/nature04254 [DOI] [PubMed] [Google Scholar]
- Hofmann E. R., Milstein S., Boulton S. J., Ye M., Hofmann J. J., et al. , 2002. Caenorhabditis elegans HUS-1 is a DNA damage checkpoint protein required for genome stability and EGL-1-mediated apoptosis. Curr. Biol. 12: 1908–1918. 10.1016/S0960-9822(02)01262-9 [DOI] [PubMed] [Google Scholar]
- Jamieson K., Wiles E. T., McNaught K. J., Sidoli S., Leggett N., et al. , 2016. Loss of HP1 causes depletion of H3K27me3 from facultative heterochromatin and gain of H3K27me2 at constitutive heterochromatin. Genome Res. 26: 97–107. 10.1101/gr.194555.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakuzu O., Wang D. P., Cameron S., 2009. MIG-32 and SPAT-3A are PRC1 homologs that control neuronal migration in Caenorhabditis elegans. Development 136: 943–953. 10.1242/dev.029363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz D. J., Edwards T. M., Reinke V., Kelly W. G., 2009. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137: 308–320. 10.1016/j.cell.2009.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaustov L., Ouyang H., Amaya M., Lemak A., Nady N., et al. , 2011. Recognition and specificity determinants of the human cbx chromodomains. J. Biol. Chem. 286: 521–529. 10.1074/jbc.M110.191411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., 2014. Transgenerational epigenetics in the germline cycle of Caenorhabditis elegans. Epigenetics Chromatin 7: 6 10.1186/1756-8935-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Fire A., 1998. Chromatin silencing and the maintenance of a functional germline in Caenorhabditis elegans. Development 125: 2451–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr S. C., Ruppersburg C. C., Francis J. W., Katz D. J., 2014. SPR-5 and MET-2 function cooperatively to reestablish an epigenetic ground state during passage through the germ line. Proc. Natl. Acad. Sci. USA 111: 9509–9514. 10.1073/pnas.1321843111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., Crittenden S. L., 2007. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 23: 405–433. 10.1146/annurev.cellbio.23.090506.123326 [DOI] [PubMed] [Google Scholar]
- Klosin A., Casas E., Hidalgo-Carcedo C., Vavouri T., Lehner B., 2017. Transgenerational transmission of environmental information in C. elegans. Science 356: 320–323. 10.1126/science.aah6412 [DOI] [PubMed] [Google Scholar]
- Laugesen A., Helin K., 2014. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell 14: 735–751. 10.1016/j.stem.2014.05.006 [DOI] [PubMed] [Google Scholar]
- Lev I., Seroussi U., Gingold H., Bril R., Anava S., et al. , 2017. MET-2-dependent H3K9 methylation suppresses transgenerational small RNA inheritance. Curr. Biol. 27: 1138–1147. 10.1016/j.cub.2017.03.008 [DOI] [PubMed] [Google Scholar]
- Li T., Kelly W. G., 2011. A role for Set1/MLL-related components in epigenetic regulation of the Caenorhabditis elegans germ line. PLoS Genet. 7: e1001349 10.1371/journal.pgen.1001349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyola A., Tagami H., Bonaldi T., Roche D., Quivy J. P., et al. , 2009. The HP1alpha-CAF1-SetDB1-containing complex provides H3K9me1 for Suv39-mediated K9me3 in pericentric heterochromatin. EMBO Rep. 10: 769–775. 10.1038/embor.2009.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx A., Backes C., Meese E., Lenhof H. P., Keller A., 2016. EDISON-WMW: exact dynamic programing solution of the Wilcoxon—Mann—Whitney test. Genomics Proteomics Bioinformatics 14: 55–61. 10.1016/j.gpb.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier B., Barber L. J., Liu Y., Shtessel L., Boulton S. J., et al. , 2009. The MRT-1 nuclease is required for DNA crosslink repair and telomerase activity in vivo in Caenorhabditis elegans. EMBO J. 28: 3549–3563. 10.1038/emboj.2009.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J., Zhang Y., Xu R. M., 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 17: 1823–1828. 10.1101/gad.269603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators , 2017. Database resources of the national center for biotechnology information. Nucleic Acids Res. 45: D12–D17. 10.1093/nar/gkw1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestorov P., Tardat M., Peters A. H., 2013. H3K9/HP1 and Polycomb: two key epigenetic silencing pathways for gene regulation and embryo development. Curr. Top. Dev. Biol. 104: 243–291. 10.1016/B978-0-12-416027-9.00008-5 [DOI] [PubMed] [Google Scholar]
- Norris A. D., Kim H. M., Colaiacovo M. P., Calarco J. A., 2015. Efficient genome editing in Caenorhabditis elegans with a toolkit of dual-Marker selection cassettes. Genetics 201: 449–458. 10.1534/genetics.115.180679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris A. D., Gracida X., Calarco J. A., 2017. CRISPR-mediated genetic interaction profiling identifies RNA binding proteins controlling metazoan fitness. Elife 6: e28129 10.7554/eLife.28129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notredame C., Higgins D. G., Heringa J., 2000. T-coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302: 205–217. 10.1006/jmbi.2000.4042 [DOI] [PubMed] [Google Scholar]
- Nottke A. C., Beese-Sims S. E., Pantalena L. F., Reinke V., Shi Y., et al. , 2011. SPR-5 is a histone H3K4 demethylase with a role in meiotic double-strand break repair. Proc. Natl. Acad. Sci. USA 108: 12805–12810. 10.1073/pnas.1102298108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi S. L., Henikoff J. G., Henikoff S., 2010. A native chromatin purification system for epigenomic profiling in Caenorhabditis elegans. Nucleic Acids Res. 38: e26 10.1093/nar/gkp1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M. A., Noble D., Sorokin E. P., Kimble J., 2014. A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 (Bethesda) 4: 1765–1772. 10.1534/g3.114.012351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D., Di Croce L., 2016. Emerging roles for Polycomb proteins in cancer. Curr. Opin. Genet. Dev. 36: 50–58. 10.1016/j.gde.2016.03.013 [DOI] [PubMed] [Google Scholar]
- Patel T., Tursun B., Rahe D. P., Hobert O., 2012. Removal of Polycomb repressive complex 2 makes C. elegans germ cells susceptible to direct conversion into specific somatic cell types. Cell Rep. 2: 1178–1186. 10.1016/j.celrep.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujadas E., Feinberg A. P., 2012. Regulated noise in the epigenetic landscape of development and disease. Cell 148: 1123–1131. 10.1016/j.cell.2012.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A., Rifkin S. A., Andersen E., van Oudenaarden A., 2010. Variability in gene expression underlies incomplete penetrance. Nature 463: 913–918. 10.1038/nature08781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., et al. , 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8: 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V. J., Sijen T., van Wolfswinkel J., Plasterk R. H., 2005. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 19: 782–787. 10.1101/gad.332305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert V. J., Mercier M. G., Bedet C., Janczarski S., Merlet J., et al. , 2014. The SET-2/SET1 histone H3K4 methyltransferase maintains pluripotency in the Caenorhabditis elegans germline. Cell Rep. 9: 443–450. 10.1016/j.celrep.2014.09.018 [DOI] [PubMed] [Google Scholar]
- Robert X., Gouet P., 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42: W320–W324. 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. M., Zarkower D., 2003. Polycomb group regulation of Hox gene expression in C. elegans. Dev. Cell 4: 891–901. 10.1016/S1534-5807(03)00135-7 [DOI] [PubMed] [Google Scholar]
- Sakaguchi A., Sarkies P., Simon M., Doebley A. L., Goldstein L. D., et al. , 2014. Caenorhabditis elegans RSD-2 and RSD-6 promote germ cell immortality by maintaining small interfering RNA populations. Proc. Natl. Acad. Sci. USA 111: E4323–E4331. 10.1073/pnas.1406131111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott S., Coustham V., Simonet T., Bedet C., Palladino F., 2006. Unique and redundant functions of C. elegans HP1 proteins in post-embryonic development. Dev. Biol. 298: 176–187. 10.1016/j.ydbio.2006.06.039 [DOI] [PubMed] [Google Scholar]
- Shirayama M., Seth M., Lee H. C., Gu W., Ishidate T., et al. , 2012. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 150: 65–77. 10.1016/j.cell.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J. A., Kingston R. E., 2013. Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49: 808–824. 10.1016/j.molcel.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M., Sarkies P., Ikegami K., Doebley A. L., Goldstein L. D., et al. , 2014. Reduced insulin/IGF-1 signaling restores germ cell immortality to Caenorhabditis elegans Piwi mutants. Cell Rep. 7: 762–773. 10.1016/j.celrep.2014.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet T., Dulermo R., Schott S., Palladino F., 2007. Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev. Biol. 312: 367–383. 10.1016/j.ydbio.2007.09.035 [DOI] [PubMed] [Google Scholar]
- Steffen P. A., Ringrose L., 2014. What are memories made of? How Polycomb and Trithorax proteins mediate epigenetic memory. Nat. Rev. Mol. Cell Biol. 15: 340–356. 10.1038/nrm3789 [DOI] [PubMed] [Google Scholar]
- Stiernagle, T., 2006 Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.101.1, http//www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Strome S., Wood W. B., 1983. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35: 15–25. 10.1016/0092-8674(83)90203-9 [DOI] [PubMed] [Google Scholar]
- Tchasovnikarova I. A., Timms R. T., Matheson N. J., Wals K., Antrobus R., et al. , 2015. GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 348: 1481–1485. 10.1126/science.aaa7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin B. D., Gonzalez-Aguilera C., Sack R., Gaidatzis D., Kalck V., et al. , 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150: 934–947. 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]
- Tzur Y. B., Friedland A. E., Nadarajan S., Church G. M., Calarco J. A., et al. , 2013. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics 195: 1181–1185. 10.1534/genetics.113.156075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser N. E., Yang D. X., Feng S., Kalinava N., Brown K. C., et al. , 2017. MORC-1 integrates nuclear RNAi and transgenerational chromatin architecture to promote germline immortality. Dev. Cell 41: 408–423.e7. 10.1016/j.devcel.2017.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw N. C., Chong S., Whitelaw E., 2010. Tuning in to noise: epigenetics and intangible variation. Dev. Cell 19: 649–650. 10.1016/j.devcel.2010.11.001 [DOI] [PubMed] [Google Scholar]
- Xiao Y., Bedet C., Robert V. J., Simonet T., Dunkelbarger S., et al. , 2011. Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proc. Natl. Acad. Sci. USA 108: 8305–8310. 10.1073/pnas.1019290108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Fong Y., Strome S., 2001. The Caenorhabditis elegans maternal-effect sterile proteins, MES-2, MES-3, and MES-6, are associated in a complex in embryos. Proc. Natl. Acad. Sci. USA 98: 5061–5066. 10.1073/pnas.081016198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Sun Y., Luo X., Zhang Y., Chen Y., et al. , 2007. Polycomb-like genes are necessary for specification of dopaminergic and serotonergic neurons in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 104: 852–857. 10.1073/pnas.0610261104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanowitz J. L., 2008. Genome integrity is regulated by the Caenorhabditis elegans Rad51D homolog rfs-1. Genetics 179: 249–262. 10.1534/genetics.107.076877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzyuk T., Fakhouri T. H., Kiefer J., Mango S. E., 2009. The polycomb complex protein mes-2/E(z) promotes the transition from developmental plasticity to differentiation in C. elegans embryos. Dev. Cell 16: 699–710. 10.1016/j.devcel.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller P., Padeken J., van Schendel R., Kalck V., Tijsterman M., et al. , 2016. Histone H3K9 methylation is dispensable for Caenorhabditis elegans development but suppresses RNA:DNA hybrid-associated repeat instability. Nat. Genet. 48: 1385–1395. 10.1038/ng.3672 [DOI] [PubMed] [Google Scholar]
- Zenk F., Loeser E., Schiavo R., Kilpert F., Bogdanovic O., et al. , 2017. Germ line-inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science 357: 212–216. 10.1126/science.aam5339 [DOI] [PubMed] [Google Scholar]
- Zhang H., Azevedo R. B., Lints R., Doyle C., Teng Y., et al. , 2003. Global regulation of Hox gene expression in C. elegans by a SAM domain protein. Dev. Cell 4: 903–915. 10.1016/S1534-5807(03)00136-9 [DOI] [PubMed] [Google Scholar]
- Zheng C., Karimzadegan S., Chiang V., Chalfie M., 2013. Histone methylation restrains the expression of subtype-specific genes during terminal neuronal differentiation in Caenorhabditis elegans. PLoS Genet. 9: e1004017 10.1371/journal.pgen.1004017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All strains and plasmids are available upon request. Strains and alleles are listed in Table S1. Supplemental material available at Figshare: https://doi.org/10.6084/m9.figshare.7045148.