Abstract
Polyploidy has played a pivotal and recurring role in angiosperm evolution. Allotetraploids arise from hybridization between species and possess duplicated gene copies (homeologs) that serve redundant roles immediately after polyploidization. Although polyploidization is a major contributor to plant evolution, it remains poorly understood. We describe an analytical approach for assessing homeolog-specific expression that begins with de novo assembly of parental transcriptomes and effectively (i) reduces redundancy in de novo assemblies, (ii) identifies putative orthologs, (iii) isolates common regions between orthologs, and (iv) assesses homeolog-specific expression using a robust Bayesian Poisson-Gamma model to account for sequence bias when mapping polyploid reads back to parental references. Using this novel methodology, we examine differential homeolog contributions to the transcriptome in the recently formed allopolyploids Tragopogon mirus and T. miscellus (Compositae). Notably, we assess a larger Tragopogon gene set than previous studies of this system. Using carefully identified orthologous regions and filtering biased orthologs, we find in both allopolyploids largely balanced expression with no strong parental bias. These new methods can be used to examine homeolog expression in any tetrapolyploid system without requiring a reference genome.
Keywords: RNA-Seq, allopolyploid, nonmodel, homeolog-specific expression, transcriptome
POLYPLOIDY has played a major role in angiosperm evolution and has received considerable attention for nearly a century (Müntzing 1936; Darlington 1937; Clausen et al. 1945; Stebbins 1947), with all angiosperms now known to be of ancient polyploid ancestry (Doyle et al. 2008; Soltis et al. 2008; Jiao et al. 2011; Amborella Genome Project 2013). Polyploidization provides an immediate doubling of genetic material and results in increased biodiversity, instant speciation and genetic robustness as seen in both heterosis and the masking of deleterious recessive mutations (Levin 2002; Madlung 2013). These features of genome doubling are evident in a diverse range of angiosperms that have been extensively examined from crop species such as potatoes (Spooner et al. 2014), sugar cane (Jannoo et al. 2007), tobacco (Deng et al. 2012), cotton (Wright et al. 1998), rice (Paterson et al. 2004), and maize (Schnable et al. 2009) to the model organism Arabidopsis (Vision et al. 2000) and many evolutionary model plants, including polyploid species in Tragopogon (Soltis et al. 2004), Spartina (Chelaifa et al. 2010), Senecio (Abbott and Lowe 2004), and Glycine (Doyle et al. 2004; reviewed in Adams and Wendel 2005b).
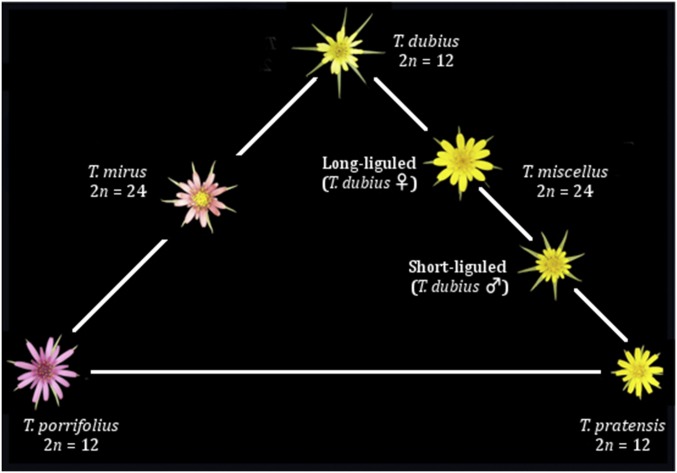
The flowering plant genus Tragopogon (Compositae) provides a textbook example of recent, naturally occurring allopolyploid speciation (reviewed in Soltis et al. 2012). The allotetraploids T. mirus and T. miscellus (Ownbey 1950; Soltis et al. 2004) have formed recently (∼80 years ago) (Soltis et al. 2004; Mavrodiev et al. 2008) and repeatedly in North America (Soltis et al. 1995; reviewed in Soltis et al. (2004), Symonds et al. (2010); reviewed in Soltis et al. 2012), and provide the opportunity to examine incipient genome evolution after natural polyploid formation (Figure 1).
Figure 1.
Tragopogon Triangle. Triangle depicting the relationships between diploid and polyploid Tragopogon species. Diploid individuals occupy the corners of the triangle with polyploids resting between their corresponding diploid progenitors. The polyploid T. mirus is formed with a paternal T. dubius and maternal T. porrifolius. In this study, we are using the short-liguled T. miscellus, which is formed by paternal T. dubius and maternal T. pratensis. There are no naturally occurring polyploids between T. porrifolius and T. pratensis, although these two species do form diploid hybrids in nature.
Advances in RNA sequencing technologies and de novo assembly provide opportunities to examine gene content, gene expression, and genome dynamics within nonmodel organisms without available reference genome sequences (Ozsolak and Milos 2011). These advances have enabled preliminary studies into biased homeolog expression in Tragopogon (Buggs et al. 2010a, 2011) as well as other polyploid plant systems (reviewed in Grover et al. 2012; Yoo et al. 2014; Wendel et al. 2018) such as maize (Springer and Stupar 2007), strawberry (Schaart et al. 2005) and cotton (Adams and Wendel 2005a; Liu and Adams 2007). Examining the expression patterns of genes within allopolyploids requires accurate assignment of transcript reads to homeologous gene copies. The software packages Hylite (Duchemin et al. 2015) and Polycat (Page et al. 2013) have been developed to achieve this. However, Hylite requires a reference genome sequence of a third species sufficiently closely related to the original diploid parents that the allopolyploid RNA-Seq reads will align to it, while Polycat requires a set of SNPs to be identified a priori between a set of extant diploid relatives. Neither of these methods solely exploits RNA-Seq data, which is often the most cost-effective means of obtaining genomic information from the majority of uncharacterized, nonmodel allopolyploid species.
De novo transcriptome assembly is error-prone, and assembling polyploidy transcriptomes compounds these challenges. There have been multiple attempts to develop assemblers that can generate haplotype assemblies from species of varying ploidal levels, but optimal solutions are computationally intensive even for diploid genomes (Das and Vikalo 2015). Assembly accuracy is further compromised by the number of duplicates of each subgenome, the degree of similarity among inparalogs (paralogs within a subgenome), outparalogs (paralogs between subgenomes), homeologs, and, in the case of transcriptome assembly, multiple copies of varying isoforms (reviewed in Martin and Wang 2011). Attempts have been made to limit these difficulties by creating multispecies assemblies containing parents and polyploids, which is justified on the premise that homeologs in the polyploid demonstrate more similarity to the diploid transcripts from which they derive than to one another (Flagel et al. 2012). However, this depends on a good understanding of the genome evolution for each species involved.
Resolving sequence differences between homeologs is crucial both for quantification of homeolog expression and for understanding the biology of polyploid species (Krasileva et al. 2013). Krasileva et al. (2013) demonstrated that more fractionated transcripts occur upon directly assembling a recently duplicated polyploid genome compared to assembly of the corresponding diploid progenitors, and similarities between the subgenomes result in substantial numbers of chimeric homeologs. Thus, failure to assess carefully the quality of a transcriptome assembly prior to using it as a reference for short-read alignment to evaluate expression will result in a significant number of reads that either fail to map or mismap due to deficiencies in the ability of sequencers to distinguish between homeologs (Krasileva et al. 2013). Instead, assembly of parental transcriptomes followed by detection of orthologs between the assemblies offers a simplified approach to assessing homeolog-specific expression (HSE) in the polyploid transcriptome where parental assemblies serve as polyploid transcriptome references and parental orthologs represent polyploid homeologs (Krasileva et al. 2013; Soltis et al. 2013).
Here, we describe a new analytical approach to examine biased homeolog expression from RNA-Seq data. The methodology (i) reduces redundancy in de novo assemblies, (ii) identifies putative orthologs between diploid assemblies, (iii) isolates common orthologous regions between orthologs, and (iv) assesses HSE using a Bayesian Poisson-Gamma model to account for sequence bias when mapping polyploid reads back to parental references.
We then apply our methodology to RNA-Seq data from T. mirus and T. miscellus and their corresponding diploid progenitors (T. dubius and T. porrifolius, and T. dubius and T. pratensis) to assess changes in HSE in the early generations following allopolyploidization. We focus on HSE where expression may be biased toward one parentally derived homeolog relative to the other, and examine the relative contribution of each homeolog to the total expression of those loci. We additionally examine additive expression, where the polyploid expression is the arithmetic mean of its parents and further distinguish between additive expression occurring when parental expression levels are either the same (i.e., not differentially expressed) or different (differentially expressed). Additivity assesses the total expression of homeologous pairs in the polyploid, whereas HSE examines the relative contribution of each homeolog to the total expression of those loci.
Materials and Methods
Sample processing
Leaf tissue was collected from the leaf tissue of 6-week-old plants grown from seed in controlled growth chambers under uniform conditions; RNA was extracted using a modified CTAB method, as described in Tate et al. (2006). A MicroPoly(A) Purist kit was used to purify mRNAs from 100 μg of total RNA. Three individuals of each species were sampled: the diploids T. porrifolius, T. dubius, and T. pratensis and the allotetraploids T. mirus and T. miscellus. The seed material originated from natural populations: collections 2674-4 (Oakesdale, WA) for T. dubius, 2878-2 (Pullman-2, WA) for T. porrifolius, 2893-26 (Garfield, WA) for T. pratensis, 2880-19 (Pullman-2, WA) for T. mirus, and 2894-2 (Garfield, WA) for T. miscellus. Herbarium vouchers for all of these collections are at FLAS. RNA-Seq samples were barcoded and processed using the Illumina TruSeq kit.
Sequencing and assembly
Sequencing was performed at the University of Florida’s Interdisciplinary Center for Biotechnology Research. We obtained paired-end 100-bp reads from three individuals of each species using Illumina HiSeq 2000. Samples were multiplexed and run across three lanes to account for possible lane effects (Auer and Doerge 2010). Approximately 245, 265, and 310 million read pairs were obtained from the diploids T. porrifolius, T. dubius, and T. pratensis, respectively (Supplemental Material, Table S1), and ∼144 and 145 million read pairs were acquired for T. mirus and T. miscellus, respectively. Adapters were removed using CutAdapt [v1.7.1, -O = 5] (Martin 2011), and the first 10 bp were trimmed from reads using Trimmomatic [v0.32, HEADCROP:10, LEADING:3 TRAILING:3, SLIDINGWINDOW:4:15, MINLEN:60] (Bolger et al. 2014). Reads were assembled jointly for all individuals within each species with Trinity de novo assembler [v2013-02-25] (Grabherr et al. 2011) using default parameters with normalization at a maximum coverage of 50 and a minimum coverage of 2. Assembly was performed for reads both with and without head cropping, resulting in two assemblies per species that were subsequently concatenated and subjected to redundancy removal to maximize assembly continuity (Table S2).
Minimizing redundant isoforms
We generated a pipeline to minimize redundant isoforms within our assemblies (Figure S1). To avoid the generation of chimeric contigs, we opted to cluster contigs within assemblies. Clusters were established by performing a self-WU-BLAST blastn [v2.0] (W. Gish, personal communication) for each assembly. In this manner, contigs that were highly similar (90% identity, P-value ≤1E−100 and alignment length >90% of the subject length) were placed into a single cluster, and then each cluster was processed individually by CAP3 under less stringent parameters [v2012-07-05, -o 25, -p 80] (Huang and Madan 1999). CAP3 clustering resulted in significantly reduced transcriptome complexity (Table S3). A subsequent self-BLAST revealed that, while redundancy was reduced, CAP3 largely failed to sufficiently collapse heterozygous or fragmented transcripts. We further processed these assemblies by reclustering contigs using WU-BLAST blastn [95% identity, P-value ≤1E−100 and alignment length >90% of the subject length] and performing a multiple sequence alignment on each cluster using MAFFT [v7.127,–adjustdirection,–clustalout,–preservecase] (Katoh and Standley 2013). A consensus sequence was subsequently generated for each multiple sequence alignment using the Align package within Biopython [v1.65] where the predominant nucleotide for each position had to be present >50% of the time or it was considered ambiguous (replaced with N) (Cock et al. 2009).
Trinotate annotation and gene ontology enrichment
Each assembly was annotated using Trinotate [v3.0.1] (Grabherr et al. 2011), which wrapped TransDecoder [3.0.0] (Grabherr et al. 2011), TMHMM [v2.0–short] (Krogh et al. 2001), RNAmmer [v3.0.1] (Lagesen et al. 2007), hmmscan [from HMMER v3.1b1–domtblout] (Finn et al. 2011; Punta et al. 2012), and BLAST [BLASTX and BLASTP using TransDecoder peptides] in conjunction with the Swiss-Prot database [-max_target_seqs 1 -evalue 1e-5] (Altschul et al. 1990). Annotations were generated in XLS format with accompanying gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms (Ashburner et al. 2000; Kanehisa et al. 2011). GO enrichment and depletion were tested using the GO terms identified by Trinotate and the GOSeq (Young et al. 2012) pipeline included with Trinity (Grabherr et al. 2011). The additive and HSE datasets were tested for GO enrichment using the set of GO terms determined from the Trinotate annotation with respect to the T. dubius assembly. The backgrounds included those loci that both possessed identifiable GO terms and were used in assessing additivity or HSE.
Reciprocal best-hit orthologs
Pairwise ortholog calling was performed between the T. dubius assembly and both T. pratensis and T. porrifolius using a reciprocal best-hit approach (Moreno-Hagelsieb and Latimer 2008). Sequences subjected to ortholog calling were required to meet minimum standards of similarity set at 90% identity, an E-value of 1E−100, 1 kb minimum contig length and an alignment length of 90% of either the subject or query total length, which is a more robust requirement than other methods that depend upon WU-BLAST reciprocal best-hits (Li et al. 2003; W. Gish, personal communication). This stringent definition is expected to reduce the number of paralog mismatches in the identification of orthologs.
Common orthologous regions
Common orthologous regions (COREs) were identified for each orthologous pair from the local alignment provided by WU-BLAST (W. Gish, personal communication). BED files for the CORES were generated using the BLAST alignments that were used to identify orthologs and a custom python script (Files S7–S10). These BED files were used to identify homologous regions between the orthologous pairs. In this manner, reads from the polyploids had comparable references for each parent. We required that the homologous region be 90% sequence-identical, and have 90% of the individual species contig (with minimum contig length set at 1 kb) included in the pair.
Poisson-Gamma model
Adapter-trimmed and quality filtered reads used in the assembly process above were aligned to complete references from both parents independently using Bowtie [v0.12.9, -m 1, -v 3] (Langmead et al. 2009) and Last [v531, -l 25] (Frith et al. 2010; Graze et al. 2012; Munger et al. 2014). Individualized BED files containing CORE coordinates for each ortholog set were used to filter SAM files, one from each parent. Alignments were compared using a custom script initially developed in PERL (Graze et al. 2012) and updated in python for this project (https://github.com/McIntyre-Lab/mcscript/blob/master/sam_compare.py), and reads mapping in the COREs were classified as mapping equally well to both parental orthologs, or as mapping better to one of the two parents based on edit distance.
A Poisson-Gamma model (León-Novelo et al. 2014) that can use a fixed parameter (q) was used to identify and correct for mapping biases to the CORE reference regions. Mean theta, the real proportion of reads from the paternal allele, was estimated for each species after mapping to the corresponding references using three separate estimations where the prior (q) was set at 0.4, 0.5, and 0.6, to account for potential bias toward T. dubius, no bias and bias toward the alternative diploid parent, respectively. The ortholog was considered to have homeologous expression bias if the credible interval did not overlap 0.5 for all priors. This approach has been shown to conservatively control type I error (Fear et al. 2016). For diploid read alignment, orthologous regions where reads were predominantly mapping to the “wrong” parent reference were filtered out, and a set of nonbiased contigs was identified for each diploid. The sets of contigs from each diploid were compared to generate an overlapping set of orthologs with low read-mapping bias. This same approach was then used to determine HSE using mapped reads from the polyploids to nonbiased orthologous COREs.
Differential expression analysis
Counts were taken from reads mapped to both of the COREs between orthologous pairs. Thus, counts are taken from highly similar regions of the orthologs to account for differences in length between orthologous pairs resulting from incompletely reconstructed orthologs. The resulting CORE count matrix was filtered by removing loci with <10 counts per million based on the average library size. The voom function (Law et al. 2014) in the R package limma (Ritchie et al. 2015) was used to estimate the mean-variance relationship of the log2 counts to add mean-variance weights before calculating test statistics using the empirical Bayes analysis pipeline. Differentially expressed (DE) loci were identified between diploid parents and filtered based on the Benjamini and Hochberg false discovery rate (FDR) correction at 0.05 FDR (Benjamini and Hochberg 1995).
Putative homeolog silencing and loss
The reads for each polyploid individual were mapped to the corresponding diploid contig references. COREs that had reads mapped to one reference, but not the other, and demonstrated HSE were identified as putative silencing or loss events for polyploid individuals. Loci demonstrating putative loss/silencing were tabulated and subjected to a reciprocal best-hit blastn, with at least 95% identity, a P-value ≤1E−10 and an alignment length ≥150 bases against 114 previously identified Tragopogon genes demonstrating loss (Buggs et al. 2010a, 2012).
Data availability
Sequenced reads are available in the NCBI Sequence Read Archive under SRP026656. Scripts used in this study are available at https://github.com/BBarbazukLab/papers/. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6207317.
Results and Discussion
Assembly and redundancy removal
Diploid reference transcriptomes were assembled using the Trinity de novo assembler (Grabherr et al. 2011) and cleaned of redundant sequences (Figure S1, Files S11–S13, and Tables S2 and S3), and orthologs were identified using WU-BLAST (Materials and Methods). The parental assemblies also serve as polyploid transcriptome references, and parental ortholog pairs represent polyploid homeologs (Table S3) (Krasileva et al. 2013; Soltis et al. 2013).
Ortholog detection, CORE identification, and quality checks
We identified 15,587 orthologs between T. dubius and T. porrifolius and 15,493 orthologs between T. dubius and T. pratensis using a reciprocal best-hit approach (Moreno-Hagelsieb and Latimer 2008). A CORE was delimited to account for sequence length bias between orthologous pairs (Figure S2), which is likely to occur due to incomplete assembly and would ultimately result in mapping and expression biases. Length difference between orthologous COREs was a maximum of 16 bp as a result of the inherent properties of the BLAST algorithm parameters (Figures S3 and S4). The difference between CORE percent GC content was generally ≤2% (Figures S5 and S6); the COREs were 90% sequence-identical and contained ≥90% of the shortest ortholog in the pair. These CORE metrics lend further support to the identification of highly similar reference regions between orthologous pairs. We then verified the orthologs by mapping reads originating from one parent to all parental genomes. If reads originating in one parent mapped preferentially to an ortholog from a different parent, that ortholog was removed from further analysis.
HSE was assessed using the Poisson-Gamma model and the unfiltered COREs generated from the two diploid ortholog sets (León-Novelo et al. 2014). From the 15,587 orthologs identified between T. dubius and T. porrifolius, 11,513 orthologous pairs exhibited low mapping bias for T. dubius reads and 11,223 for T. porrifolius. Similarly, of the 15,493 T. dubius and T. pratensis orthologous pairs, 11,502 exhibited low mapping bias for T. dubius reads and 10,194 for T. pratensis. Notably, the sets of unbiased orthologous pairs identified for the T. dubius and T. porrifolius loci is relatively equal (∼11,000 each), whereas the T. dubius and T. pratensis orthologous pairs differ by >1000 loci. This is noteworthy because, if biased loci were not filtered and HSE in the polyploids was purely random, we would expect relatively equal HSE within T. mirus and a bias toward higher T. pratensis-derived homeologs in T. miscellus. This finding underscores the importance of assessing read mapping bias prior to identifying HSE. We then isolated the overlapping orthologous pairs between T. dubius and T. porrifolius that did not exhibit mapping bias for parental reads, resulting in 8064 orthologous pairs with low read-mapping bias for the T. mirus analysis, and similarly identified 7202 orthologous pairs for the T. miscellus analysis.
Homeolog-specific expression
Differences in homeolog expression can arise from variation in cis- or trans- regulatory sequences (Wittkopp et al. 2004; Williams et al. 2007) and epistatic effects (Graze et al. 2012), and are expected to be observed after polyploidization (Buggs et al. 2010a; Koh et al. 2010; De Smet and Van de Peer 2012; reviewed in Yoo et al. 2014). Sequence polymorphisms and redundancy often affect read mapping, which hinders accurate measurement of allele-specific expression and the degree of allelic imbalance (León-Novelo et al. 2014). Additional mapping issues include biases between references due to sequence ambiguity, incomplete or missing transcripts, chimeric transcripts, misidentified orthologs and structural differences (Degner et al. 2009; León-Novelo et al. 2014). To account for potential reference sequence biases, we utilized a Bayesian Poisson-Gamma model to identify biased ortholog references and subsequently determine HSE based on the remaining set of unbiased orthologs (León-Novelo et al. 2014).
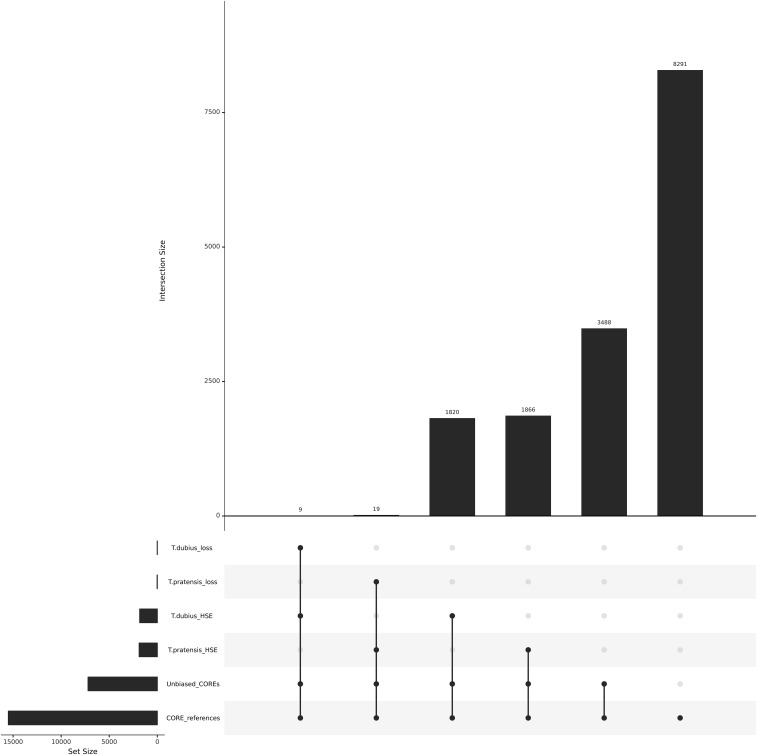
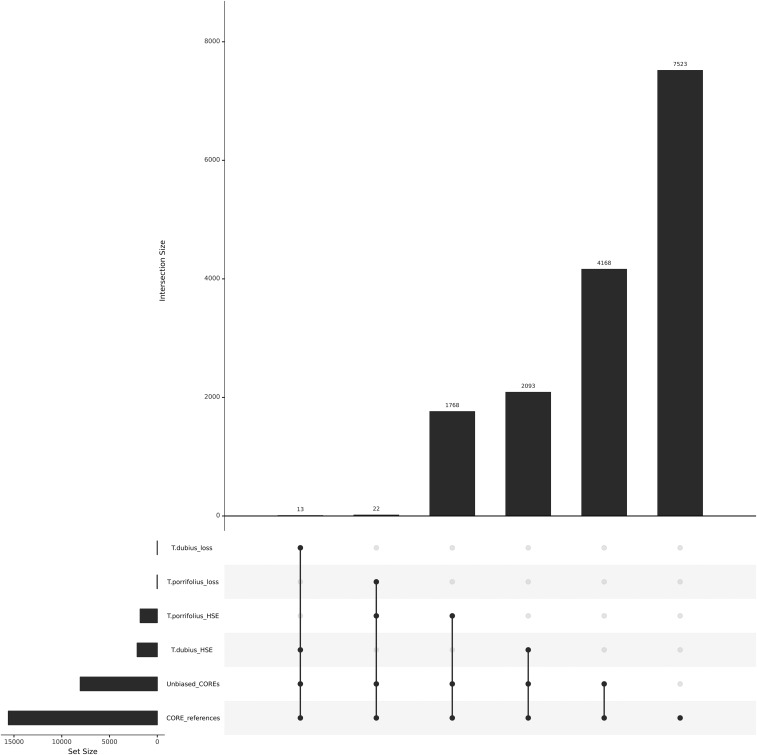
In a previous study examining HSE in T. miscellus, homeolog expression levels showed a slight bias toward higher T. dubius expression over T. pratensis (Buggs et al. 2010a). Application of our COREs and rigorous exclusion of alignment bias determined that HSE in T. miscellus was equally distributed between both parental homeologs, with 1820 contigs biased toward T. dubius and 1866 contigs biased toward T. pratensis when parental expression is ignored (Figure 2). Similarly, HSE in T. mirus was approximately equal, with a potentially small expression bias toward T. dubius (2093) compared to T. porrifolius (1768), resulting in slightly higher T. dubius expression at those loci regardless of whether parental expression was equal or not (Figure 3).
Figure 2.
Tragopgon miscellus homeolog-specific expression and putative loss. CORE references represent all of the COREs for homeologous pairs (15,493). Unbiased COREs are those references that do not biasedly map reads from the alternate parent (7202) of which 3488 loci do not exhibit HSE. Tragopogon pratensis HSE represents the loci demonstrating HSE toward the T. pratensis homeolog (1885) and T. dubius toward the corresponding paternal homeolog (1829). The loss events represent putative silencing or loss and are a subset of the HSE analysis.
Figure 3.
Tragopogon mirus homeolog-specific expression and putative loss. CORE references represent all of the COREs for homeologous pairs (15,587). Unbiased COREs are those references that do not biasedly map reads from the alternate parent (8064) of which 4168 loci do not exhibit HSE. Tragopogon porrifolius HSE represents the loci demonstrating HSE toward the T. porrifolius homeolog (1790) and T. dubius toward the corresponding paternal homeolog (2106). The loss events represent putative silencing or loss and are a subset of the HSE analysis.
We also examined HSE in light of parental expression patterns where parental expression may be the same or different. Differences in parental expression were tested using voom (Law et al. 2014). After filtering for minimum read depth, the reduced contig sets had final sizes of 10,677 for T. dubius-T. porrifolius orthologs and 10,248 for the T. dubius-T. pratensis set. For T. dubius and T. pratensis, 5176 of 10,248 loci were differentially expressed, and 5800 of 10,677 loci were differentially expressed for T. dubius and T. porrifolius. Expression in T. mirus showed a slight bias toward T. dubius homeolog expression (Table 1), which agrees with previous studies examining far fewer loci (Kovarik et al. 2005; Buggs et al. 2010a). Expression of parental homeologs in T. miscellus was equal when parental expression was different but showed a slight bias toward T. pratensis when parental expression levels were the same.
Table 1. Homeolog-specific expression.
| T. dubius | T. porrifolius | T. pratensis | |
|---|---|---|---|
| Parents same | |||
| T. mirus | 715 | 645 | — |
| T. miscellus | 680 | — | 749 |
| Parents different | |||
| T. mirus | 1325 | 1065 | — |
| T. miscellus | 1081 | — | 1059 |
Counts represent total number of loci demonstrating expression bias toward a particular parental homeolog. Homeolog expression biases are examined in light of loci expression levels being the same in the diploid parents and loci expression levels being different in the diploid parents.
These results represent a substantial (100× or more) increase in gene sample size compared to previous expression studies in Tragopogon (Tate et al. 2006; Buggs et al. 2010a, 2011; Koh et al. 2010). Importantly, the differences in expression between our work and previous studies are actually minor, and may be due to differences in the scale of the analyses, the stringent requirements for orthology, stochasticity (Buggs et al. 2011, 2012), direction of the cross, or tissue analyzed (Buggs et al. 2009, 2010b). As orthologs were stringently defined, the relatively balanced HSE may also be a product of examining more conserved orthologs. As such, HSE may become increasingly imbalanced as sequence identity between orthologs decreases. It may also be that differences in the number of loci demonstrating HSE, which is only 325 of the ∼8000 loci analyzed for T. mirus, are within a predictable range of variation such that independent polyploidization events result in relatively balanced HSE with some independently formed lineages being moderately biased.
Homeolog expression has been a subject of interest in other allopolyploid plants (reviewed in Grover et al. 2012; Yoo et al. 2014; Wendel et al. 2018). In the allotetraploid Arabidopsis suecica, which formed from maternal Arabidopsis thaliana and paternal Arabidopsis arenosa, HSE favors expression of the A. arenosa subgenome (Wang et al. 2006a; Chang et al. 2010; Shi et al. 2012). In one study using Gossypium hirsutum and 40 genes, homeologs demonstrated extreme biases that were organ-specific (Adams et al. 2003), but, in another study examining >13,000 genes in allotetraploid Gossypium tomentosum and two cultivars of G. hirsutum, HSE was balanced (Rambani et al. 2014). However, the degree of HSE in G. hirsutum has also been shown to be unbalanced, with bias toward the D-genome based on 1383 homeologs (Flagel et al. 2008) or toward the A-genome based on a revised Cotton Gene Index version 7 containing 55,673 unique sequences (Yang et al. 2006). Parental bias has also shown tissue-specific biases in synthetics, where HSE varied in both degree and direction (Adams et al. 2004). HSE may also vary between natural and synthetic allopolyploids where natural allopolyploids have shown balanced HSE and synthetic are biased toward the A-genome in Gossypium (Yoo et al. 2013). In the same study, the degree of HSE was additionally shown to increase with increasing time since polyploidization (Yoo et al. 2013). Genome expression biases also appear to occur in Senecio cambrensis, where bias was reproduced in natural and synthetic allopolyploids (Hegarty et al. 2012). Overall, there seems to be little consensus across allopolyploid plant species. However, it tentatively appears that biased expression patterns may at least be reproducible within certain clades and stochastic within others (reviewed in Yoo et al. 2014).
Additively expressed loci
Most loci displayed expression levels in the polyploids consistent with additivity, with ∼64% of the T. miscellus loci demonstrating additivity and ∼76% of the T. mirus DE loci demonstrating additivity when parental expression is different (Table 2). These results are consistent with studies in hybrid maize (Schadt et al. 2003; Guo et al. 2006; Stupar and Springer 2006) with synthetic allotetraploid Arabidopsis (Wang et al. 2006b) and synthetic allohexaploid Brassica (Zhao et al. 2013). Expression patterns observed after polyploidization are, at least in part, a product of genome doubling and hybridization (reviewed in Yoo et al. 2014). However, the degree to which each contributes to these expression changes is unclear (reviewed in Soltis et al. 2016b). Patterns of nonadditive expression in cotton have been proposed to be a long-term evolutionary response, where older polyploids exhibit more nonadditive expression (reviewed in Yoo et al. 2014). Early studies in allopolyploid Tragopogon also demonstrated additive enzyme phenotypes in T. miscellus (Roose and Gottlieb 1980). Proteome additivity for T. mirus occurred for 408 of 476 proteins (Koh et al. 2012). However, it is still not clear to what degree the transcriptome accurately portrays the proteome (reviewed in Vogel and Marcotte 2012; Soltis et al. 2016a). Ecological data suggest intermediate habitat occupation for both allotetraploids compared to their parents (Sauber 2000; Soltis et al. 2004), suggesting some degree of phenotypic additivity. However, most morphological phenotypes in T. mirus and T. miscellus are only intermediate in terms of recombining dominant traits from each diploid (Ownbey 1950). Interestingly, dominant phenotypes may be explained by dosage imbalance, where interacting partners in the same pathway may be coordinately regulated to ameliorate imbalances (Veitia 2004).
Table 2. Test for additivity in polyploid expression.
| Not Additive | Consistent with Additive | |
|---|---|---|
| T. mirus | ||
| Parents same | 716 | 4625a |
| Parents different | 1080 | 3494b |
| T. miscellus | ||
| Parents same | 1258 | 3426a |
| Parents different | 1785 | 3210b |
Counts represent loci where parental expression is not significantly different or is significantly different and polyploid expression is either additive or nonadditive in the polyploid individuals of independent origin in T. mirus or T. miscellus.
These loci are not strictly additive as polyploid expression could deviate from midparent expression and yet be consistent with additive when parental expression is the same.
These loci have power issues because the hybrid mean expression falls within the diploid mean expression levels.
GO enrichment
We found no significant enrichment or depletion in additive, nonadditive, HSE or non-HSE sets. However, if only Arabidopsis annotations are utilized, there is enrichment for transferases and binding proteins. This result may suggest that limiting GO enrichment analyses to single-species orthology calls may bias identification of highly conserved loci and skew enrichment results. Thus, it is crucial that GO assignment methods utilize more comprehensive databases such as the nonredundant database, TrEMBL, or SwissProt. Paterson and colleagues (2006) found that shared protein functional domains were more useful for identifying gene retention patterns than broad GO categories in Arabidopsis, Oryza, Saccharomyces, and Tetraodon (Paterson et al. 2006). Similarly, GO enrichment alone may not be the most appropriate method to identify conserved patterns of expression such as HSE after polyploidization. Other models, such as domain-based (Addou et al. 2009) or network-based (Veitia 2004; Chang et al. 2010) models, may better describe the processes governing expression change in newly formed polyploids.
Homeolog silencing and potential loss
Biased fractionation and silencing have been observed in Arabidopsis suecica (Wang et al. 2006b; Chang et al. 2010), Brassica rapa (Wang et al. 2011; Cheng et al. 2012; Woodhouse et al. 2014) and Zea mays (Schnable et al. 2011; reviewed in Yoo et al. 2014), and may result from epigenetic changes (reviewed in Wendel et al. 2018), and be influenced by stoichiometric constraints in accord with the gene dosage balance hypothesis (reviewed in Freeling 2009; Wendel et al. 2018). We cannot differentiate between physical loss of a homeolog or silencing using mapped RNA-Seq reads because both phenomena result in the absence of aligned reads. Notably, these potential silencing/loss events were few in both T. miscellus (Figure 2) and T. mirus (Figure 3). As there are so few putative events detected, it is difficult to speculate on potential homeolog-specific biases in loss. However, it tentatively appears that silencing/loss may occur more frequently in the maternally derived homeologs (i.e., T. pratensis and T. porrifolius). This is contrary to previous, targeted studies where T. miscellus exhibits preferential loss of T. dubius (Tate et al. 2006; Buggs et al. 2009, 2012; Soltis et al. 2009) and T. mirus shows preferential silencing (Buggs et al. 2010b) or loss (Koh et al. 2010) of the T. dubius homeolog based upon 13 and 30 genes, respectively. However, data from a larger set of anonymous loci assayed via the Sequenom approach show greater silencing of the maternal homeolog in T. mirus (the T. porrifolius homeolog) and of the T. dubius homeolog—regardless of maternal or paternal ancestry—in T. miscellus (Jordon-Thaden et al., personal communication). We reiterate that the scale of the current study is larger than previous analyses, and loss events are only putative loss events as they are derived from lack of expression alone rather than in conjunction with direct assessment of DNA. These putatively lost loci were additionally compared to 114 loci, some of which previously demonstrated loss in Tragopogon (Buggs et al. 2010a, 2012). We identified 111 of the 114 loci in the T. dubius assembly, 103 in T. pratensis and 106 in T. porrifolius. However, none of the loci potentially demonstrating loss here overlapped with those from the previous studies. In fact, most of the loci (∼80 of 114), which previously showed loss, were identified within our ortholog sets and expressed. This lack of overlap for loci demonstrating homeolog silencing/loss may support that the processes governing gene expression may be largely stochastic and prone to differ between individuals (Buggs et al. 2009, 2010b, 2011). Indeed, every individual surveyed in Jordon-Thaden et al. (personal communication) differs in its profile of lost loci. Alternatively, it may reflect our more stringently applied requirements for ortholog definition, sequence alignment, correction of mapping bias, and statistically robust expression assessment that may have been an unrecognized shortcoming of previous studies.
Final remarks
Here, we present a robust methodology for assessing homeolog-specific expression in polyploids from RNA-Seq data and employ it to elucidate the large-scale changes in gene expression that have occurred in the recently formed (∼40 generations ago) Tragopogon allopolyploids. Our methodology reduces redundancy in de novo assemblies, identifies putative orthologs between assemblies and isolates common orthologous regions between ortholog pairs for the purpose of identifying unbiased references to assess changes in HSE. Notably, this study assesses significantly more contigs than previous studies in Tragopogon (Tate et al. 2006, 2009; Buggs et al. 2009, 2010b, 2012; Koh et al. 2010) and represents a notable increase in both scale and confidence of HSE assessment in Tragopogon. While our results confirm earlier observations that T. mirus homeolog expression is slightly biased toward T. dubius (Kovarik et al. 2005; Buggs et al. 2010a), we observe equal HSE toward T. pratensis and T. dubius homeologs in T. miscellus, which contradicts earlier small-scale studies (Tate et al. 2006; Buggs et al. 2011). Additionally, the relatively balanced HSE seen in Tragopogon contradicts studies in other allopolyploid species (e.g., A. suecica, Gossypium, or S. cambrensis). Because our analysis is based on a large sample size of well-defined orthologous genes that have been robustly filtered for bias, these differences between our analysis and previous studies of homeolog expression may largely be a product of methodological rigor.
Acknowledgments
We thank Oleksandr Moskalenko for rewriting sam_compare from PERL to CPython (https://github.com/McIntyre-Lab/mcscript/blob/master/sam_compare.py). The High-Performance Computing Center of the University of Florida (UF) provided computational resources and support. This work was supported by the UF Office of Research, UF Department of Biology, National Science Foundation (NSF) grant DEB-1146065, and National Institutes of Health (NIH) grant GMS102227. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NSF or NIH. The authors declare no conflict of interest.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6207317.
Communicating editor: J. Birchler
Literature Cited
- Abbott R. J., Lowe A. J., 2004. Origins, establishment and evolution of new polyploid species: Senecio cambrensis and S. eboracensis in the British Isles. Biol. J. Linn. Soc. Lond. 82: 467–474. 10.1111/j.1095-8312.2004.00333.x [DOI] [Google Scholar]
- Adams K. L., Wendel J. F., 2005a Allele-specific, bidirectional silencing of an alcohol dehydrogenase gene in different organs of interspecific diploid cotton hybrids. Genetics 171: 2139–2142. 10.1534/genetics.105.047357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams K. L., Wendel J. F., 2005b Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141. 10.1016/j.pbi.2005.01.001 [DOI] [PubMed] [Google Scholar]
- Adams K. L., Cronn R., Percifield R., Wendel J. F., 2003. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100: 4649–4654. 10.1073/pnas.0630618100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams K. L., Percifield R., Wendel J. F., 2004. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 168: 2217–2226. 10.1534/genetics.104.033522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addou S., Rentzsch R., Lee D., Orengo C. A., 2009. Domain-based and family-specific sequence identity thresholds increase the levels of reliable protein function transfer. J. Mol. Biol. 387: 416–430. 10.1016/j.jmb.2008.12.045 [DOI] [PubMed] [Google Scholar]
- Amborella Genome Project , 2013. The Amborella genome and the evolution of flowering plants. Science 342: 1241089 10.1126/science.1241089 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., et al. , 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer P. L., Doerge R., 2010. Statistical design and analysis of RNA sequencing data. Genetics 185: 405–416. 10.1534/genetics.110.114983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300. [Google Scholar]
- Bolger A. M., Lohse M., Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggs R., Doust A., Tate J., Koh J., Soltis K., et al. , 2009. Gene loss and silencing in Tragopogon miscellus (Asteraceae): comparison of natural and synthetic allotetraploids. Heredity 103: 73–81. 10.1038/hdy.2009.24 [DOI] [PubMed] [Google Scholar]
- Buggs R. J., Chamala S., Wu W., Gao L., May G. D., et al. , 2010a Characterization of duplicate gene evolution in the recent natural allopolyploid Tragopogon miscellus by next-generation sequencing and Sequenom iPLEX MassARRAY genotyping. Mol. Ecol. 19: 132–146. 10.1111/j.1365-294X.2009.04469.x [DOI] [PubMed] [Google Scholar]
- Buggs R. J., Elliott N. M., Zhang L., Koh J., Viccini L. F., et al. , 2010b Tissue‐specific silencing of homoeologs in natural populations of the recent allopolyploid Tragopogon mirus. New Phytol. 186: 175–183. 10.1111/j.1469-8137.2010.03205.x [DOI] [PubMed] [Google Scholar]
- Buggs R. J., Zhang L., Miles N., Tate J. A., Gao L., et al. , 2011. Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr. Biol. 21: 551–556. 10.1016/j.cub.2011.02.016 [DOI] [PubMed] [Google Scholar]
- Buggs R. J., Chamala S., Wu W., Tate J. A., Schnable P. S., et al. , 2012. Rapid, repeated, and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Curr. Biol. 22: 248–252. 10.1016/j.cub.2011.12.027 [DOI] [PubMed] [Google Scholar]
- Chang P. L., Dilkes B. P., McMahon M., Comai L., Nuzhdin S. V., 2010. Homoeolog-specific retention and use in allotetraploid Arabidopsis suecica depends on parent of origin and network partners. Genome Biol. 11: R125 10.1186/gb-2010-11-12-r125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelaifa H., Monnier A., Ainouche M., 2010. Transcriptomic changes following recent natural hybridization and allopolyploidy in the salt marsh species Spartina× townsendii and Spartina anglica (Poaceae). New Phytol. 186: 161–174. 10.1111/j.1469-8137.2010.03179.x [DOI] [PubMed] [Google Scholar]
- Cheng F., Wu J., Fang L., Sun S., Liu B., et al. , 2012. Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One 7: e36442 10.1371/journal.pone.0036442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen J., Keck D. D., Hiesey W. M., 1945. Experimental Studies on the Nature of Species. II. Plant Evolution Through Amphiploidy, with Examples From the Madiinae. Carnegie Inst., Washington. Publ. 564. [Google Scholar]
- Cock P. J., Antao T., Chang J. T., Chapman B. A., Cox C. J., et al. , 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25: 1422–1423. 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlington C. D., 1937. Recent Advances in Cytology. P. Blakiston’s Son and Co., Philadelphia. [Google Scholar]
- Das S., Vikalo H., 2015. SDhaP: haplotype assembly for diploids and polyploids via semi-definite programming. BMC Genomics 16: 260 10.1186/s12864-015-1408-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degner J. F., Marioni J. C., Pai A. A., Pickrell J. K., Nkadori E., et al. , 2009. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics 25: 3207–3212. 10.1093/bioinformatics/btp579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng B., Du W., Liu C., Sun W., Tian S., et al. , 2012. Antioxidant response to drought, cold and nutrient stress in two ploidy levels of tobacco plants: low resource requirement confers polytolerance in polyploids? Plant Growth Regul. 66: 37–47. 10.1007/s10725-011-9626-6 [DOI] [Google Scholar]
- De Smet R., Van de Peer Y., 2012. Redundancy and rewiring of genetic networks following genome-wide duplication events. Curr. Opin. Plant Biol. 15: 168–176. 10.1016/j.pbi.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L., Rauscher J. T., Brown A., 2004. Evolution of the perennial soybean polyploid complex (Glycine subgenus Glycine): a study of contrasts. Biol. J. Linn. Soc. Lond. 82: 583–597. 10.1111/j.1095-8312.2004.00343.x [DOI] [Google Scholar]
- Doyle J. J., Flagel L. E., Paterson A. H., Rapp R. A., Soltis D. E., et al. , 2008. Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42: 443–461. 10.1146/annurev.genet.42.110807.091524 [DOI] [PubMed] [Google Scholar]
- Duchemin W., Dupont P.-Y., Campbell M. A., Ganley A. R., Cox M. P., 2015. HyLiTE: accurate and flexible analysis of gene expression in hybrid and allopolyploid species. BMC Bioinformatics 16: 8 10.1186/s12859-014-0433-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fear J. M., Leon-Novelo L. G., Morse A. M., Gerken A. R., Van Lehman K., et al. , 2016. Buffering of genetic regulatory networks in Drosophila melanogaster. Genetics 203: 1177–1190. 10.1534/genetics.116.188797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Clements J., Eddy S. R., 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39: W29–W37. 10.1093/nar/gkr367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L., Udall J., Nettleton D., Wendel J., 2008. Duplicate gene expression in allopolyploid Gossypium reveals two temporally distinct phases of expression evolution. BMC Biol. 6: 16 10.1186/1741-7007-6-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel L. E., Wendel J. F., Udall J. A., 2012. Duplicate gene evolution, homoeologous recombination, and transcriptome characterization in allopolyploid cotton. BMC Genomics 13: 302 10.1186/1471-2164-13-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., 2009. Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 60: 433–453. 10.1146/annurev.arplant.043008.092122 [DOI] [PubMed] [Google Scholar]
- Frith M. C., Wan R., Horton P., 2010. Incorporating sequence quality data into alignment improves DNA read mapping. Nucleic Acids Res. 38: e100 10.1093/nar/gkq010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., et al. , 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graze R., Novelo L., Amin V., Fear J., Casella G., et al. , 2012. Allelic imbalance in Drosophila hybrid heads: exons, isoforms, and evolution. Mol. Biol. Evol. 29: 1521–1532. 10.1093/molbev/msr318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover C., Gallagher J., Szadkowski E., Yoo M., Flagel L., et al. , 2012. Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 196: 966–971. 10.1111/j.1469-8137.2012.04365.x [DOI] [PubMed] [Google Scholar]
- Guo M., Rupe M. A., Yang X., Crasta O., Zinselmeier C., et al. , 2006. Genome-wide transcript analysis of maize hybrids: allelic additive gene expression and yield heterosis. Theor. Appl. Genet. 113: 831–845. 10.1007/s00122-006-0335-x [DOI] [PubMed] [Google Scholar]
- Hegarty M. J., Abbott R. J., Hiscock S. J., 2012. Allopolyploid speciation in action: the origins and evolution of Senecio cambrensis, pp. 245–270 in Polyploidy and Genome Evolution. Springer, New York. [Google Scholar]
- Huang X., Madan A., 1999. CAP3: a DNA sequence assembly program. Genome Res. 9: 868–877. 10.1101/gr.9.9.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannoo N., Grivet L., Chantret N., Garsmeur O., Glaszmann J. C., et al. , 2007. Orthologous comparison in a gene‐rich region among grasses reveals stability in the sugarcane polyploid genome. Plant J. 50: 574–585. 10.1111/j.1365-313X.2007.03082.x [DOI] [PubMed] [Google Scholar]
- Jiao Y., Wickett N. J., Ayyampalayam S., Chanderbali A. S., Landherr L., et al. , 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. 10.1038/nature09916 [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M., 2011. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40: D109–D114. 10.1093/nar/gkr988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh J., Soltis P. S., Soltis D. E., 2010. Homeolog loss and expression changes in natural populations of the recently and repeatedly formed allotetraploid Tragopogon mirus (Asteraceae). BMC Genomics 11: 97 10.1186/1471-2164-11-97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh J., Chen S., Zhu N., Yu F., Soltis P. S., et al. , 2012. Comparative proteomics of the recently and recurrently formed natural allopolyploid Tragopogon mirus (Asteraceae) and its parents. New Phytol. 196: 292–305. 10.1111/j.1469-8137.2012.04251.x [DOI] [PubMed] [Google Scholar]
- Kovarik A., Pires J. C., Leitch A. R., Lim K. Y., Sherwood A., et al. , 2005. Rapid concerted evolution of nuclear ribosomal DNA in two Tragopogon allopolyploids of recent and recurrent origin. Genetics 169: 931–944. 10.1534/genetics.104.032839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva K. V., Buffalo V., Bailey P., Pearce S., Ayling S., et al. , 2013. Separating homeologs by phasing in the tetraploid wheat transcriptome. Genome Biol. 14: R66 10.1186/gb-2013-14-6-r66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Larsson B., Von Heijne G., Sonnhammer E. L., 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580. 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- Lagesen K., Hallin P., Rødland E., Stærfeldt H., Rognes T., et al. , 2007. RNammer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35: 3100–3108. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C. W., Chen Y., Shi W., Smyth G. K., 2014. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15: R29 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- León-Novelo L. G., McIntyre L. M., Fear J. M., Graze R. M., 2014. A flexible Bayesian method for detecting allelic imbalance in RNA-seq data. BMC Genomics 15: 920 10.1186/1471-2164-15-920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. A., 2002. The Role of Chromosomal Change in Plant Evolution. Oxford University Press, Oxford. [Google Scholar]
- Li L., Stoeckert C. J., Roos D. S., 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13: 2178–2189. 10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Adams K. L., 2007. Expression partitioning between genes duplicated by polyploidy under abiotic stress and during organ development. Curr. Biol. 17: 1669–1674. 10.1016/j.cub.2007.08.030 [DOI] [PubMed] [Google Scholar]
- Madlung A., 2013. Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity 110: 99–104. 10.1038/hdy.2012.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. A., Wang Z., 2011. Next-generation transcriptome assembly. Nat. Rev. Genet. 12: 671–682. 10.1038/nrg3068 [DOI] [PubMed] [Google Scholar]
- Martin M., 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. Journal 17: 10–12. [Google Scholar]
- Mavrodiev E. V., Soltis P. S., Soltis D. E., 2008. Putative parentage of six Old World polyploids in Tragopogon L.(Asteraceae: Scorzonerinae) based on ITS, ETS, and plastid sequence data. Taxon 57: 1215. [Google Scholar]
- Moreno-Hagelsieb G., Latimer K., 2008. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 24: 319–324. 10.1093/bioinformatics/btm585 [DOI] [PubMed] [Google Scholar]
- Munger S. C., Raghupathy N., Choi K., Simons A. K., Gatti D. M., et al. , 2014. RNA-Seq alignment to individualized genomes improves transcript abundance estimates in multiparent populations. Genetics 198: 59–73. 10.1534/genetics.114.165886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müntzing A., 1936. The evolutionary significance of autopolyploidy. Hereditas 21: 363–378. 10.1111/j.1601-5223.1936.tb03204.x [DOI] [Google Scholar]
- Ownbey M., 1950. Natural hybridization and amphiploidy in the genus Tragopogon. Am. J. Bot. 37: 487–499. 10.1002/j.1537-2197.1950.tb11033.x [DOI] [Google Scholar]
- Ozsolak F., Milos P. M., 2011. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 12: 87–98. 10.1038/nrg2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page J. T., Gingle A. R., Udall J. A., 2013. PolyCat: a resource for genome categorization of sequencing reads from allopolyploid organisms. G3 (Bethesda) 3: 517–525. 10.1534/g3.112.005298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A., Bowers J., Chapman B., 2004. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 101: 9903–9908. 10.1073/pnas.0307901101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Chapman B. A., Kissinger J. C., Bowers J. E., Feltus F. A., et al. , 2006. Many gene and domain families have convergent fates following independent whole-genome duplication events in Arabidopsis, Oryza, Saccharomyces and Tetraodon. Trends Genet. 22: 597–602. 10.1016/j.tig.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., et al. , 2012. The Pfam protein families database. Nucleic Acids Res. 40: D290–D301. 10.1093/nar/gkr1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambani A., Page J. T., Udall J. A., 2014. Polyploidy and the petal transcriptome of Gossypium. BMC Plant Biol. 14: 3 10.1186/1471-2229-14-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., et al. , 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose M., Gottlieb L., 1980. Biochemical properties and level of expression of alcohol dehydrogenases in the allotetraploid plant Tragopogon miscellus and its diploid progenitors. Biochem. Genet. 18: 1065–1085. 10.1007/BF00484339 [DOI] [PubMed] [Google Scholar]
- Sauber, K. L., 2000 Habitat differentiation between derivative tetraploid and progenitor diploid species of Tragopogon. Masters of Science thesis, Washington State University, Pullman, WA. [Google Scholar]
- Schaart J. G., Mehli L., Schouten H. J., 2005. Quantification of allele‐specific expression of a gene encoding strawberry polygalacturonase‐inhibiting protein (PGIP) using PyrosequencingTM. Plant J. 41: 493–500. 10.1111/j.1365-313X.2004.02299.x [DOI] [PubMed] [Google Scholar]
- Schadt E. E., Monks S. A., Drake T. A., Lusis A. J., Che N., et al. , 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422: 297–302. 10.1038/nature01434 [DOI] [PubMed] [Google Scholar]
- Schnable J. C., Springer N. M., Freeling M., 2011. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074. 10.1073/pnas.1101368108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., et al. , 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. 10.1126/science.1178534 [DOI] [PubMed] [Google Scholar]
- Shi X., Ng D. W., Zhang C., Comai L., Ye W., et al. , 2012. Cis-and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat. Commun. 3: 950 10.1038/ncomms1954 [DOI] [PubMed] [Google Scholar]
- Soltis D., Buggs R., Barbazuk W., Schnable P., Soltis P., 2009. On the origins of species: does evolution repeat itself in polyploid populations of independent origin? in Cold Spring Harbor Symposia on Quantitative Biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed] [Google Scholar]
- Soltis D. E., Soltis P. S., Pires J. C., Kovarik A., Tate J. A., et al. , 2004. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biol. J. Linn. Soc. Lond. 82: 485–501. 10.1111/j.1095-8312.2004.00335.x [DOI] [Google Scholar]
- Soltis D. E., Albert V. A., Leebens-Mack J., Palmer J. D., Wing R. A., et al. , 2008. The Amborella genome: an evolutionary reference for plant biology. Genome Biol. 9: 402 10.1186/gb-2008-9-3-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D. E., Mavrodiev E. V., Meyers S. C., Severns P. M., Zhang L., et al. , 2012. Additional origins of Ownbey’s Tragopogon mirus. Bot. J. Linn. Soc. 169: 297–311. 10.1111/j.1095-8339.2012.01244.x [DOI] [Google Scholar]
- Soltis D. E., Gitzendanner M. A., Stull G., Chester M., Chanderbali A., et al. , 2013. The potential of genomics in plant systematics. Taxon 62: 886–898. 10.12705/625.13 [DOI] [Google Scholar]
- Soltis D. E., Misra B. B., Shan S., Chen S., Soltis P. S., 2016a Polyploidy and the proteome. Biochim. Biophys. Acta 1864: 896–907. 10.1016/j.bbapap.2016.03.010 [DOI] [PubMed] [Google Scholar]
- Soltis D. E., Visger C. J., Marchant D. B., Soltis P. S., 2016b Polyploidy: pitfalls and paths to a paradigm. Am. J. Bot. 103: 1146–1166. 10.3732/ajb.1500501 [DOI] [PubMed] [Google Scholar]
- Soltis P. S., Plunkett G. M., Novak S. J., Soltis D. E., 1995. Genetic variation in Tragopogon species: additional origins of the allotetraploids T. mirus and T. miscellus (Compositae). Am. J. Bot. 82: 1329–1341. 10.1002/j.1537-2197.1995.tb12666.x [DOI] [Google Scholar]
- Spooner D. M., Ghislain M., Simon R., Jansky S. H., Gavrilenko T., 2014. Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. Bot. Rev. 80: 283–383. 10.1007/s12229-014-9146-y [DOI] [Google Scholar]
- Springer N. M., Stupar R. M., 2007. Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. The Plant Cell Online 19: 2391–2402. 10.1105/tpc.107.052258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins G., 1947. Types of polyploids: their classification and significance. Adv. Genet. 1: 403–429. [DOI] [PubMed] [Google Scholar]
- Stupar R. M., Springer N. M., 2006. Cis-transcriptional variation in maize inbred lines B73 and Mo17 leads to additive expression patterns in the F1 hybrid. Genetics 173: 2199–2210. 10.1534/genetics.106.060699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds V. V., Soltis P. S., Soltis D. E., 2010. Dynamics of polyploid formation in Tragopogon (Asteraceae): recurrent formation, gene flow, and population structure. Evolution 64: 1984–2003. 10.1111/j.1558-5646.2010.00978.x [DOI] [PubMed] [Google Scholar]
- Tate J. A., Ni Z., Scheen A.-C., Koh J., Gilbert C. A., et al. , 2006. Evolution and expression of homeologous loci in Tragopogon miscellus (Asteraceae), a recent and reciprocally formed allopolyploid. Genetics 173: 1599–1611. 10.1534/genetics.106.057646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate J. A., Symonds V. V., Doust A. N., Buggs R. J., Mavrodiev E., et al. , 2009. Synthetic polyploids of Tragopogon miscellus and T. mirus (Asteraceae): 60 years after Ownbey’s discovery. Am. J. Bot. 96: 979–988. 10.3732/ajb.0800299 [DOI] [PubMed] [Google Scholar]
- Veitia R. A., 2004. Gene dosage balance in cellular pathways: implications for dominance and gene duplicability. Genetics 168: 569–574. 10.1534/genetics.104.029785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision T. J., Brown D. G., Tanksley S. D., 2000. The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117. 10.1126/science.290.5499.2114 [DOI] [PubMed] [Google Scholar]
- Vogel C., Marcotte E. M., 2012. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13: 227–232. 10.1038/nrg3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H.-S., Chen Z. J., 2006a Nonadditive regulation of FRI and FLC loci mediates flowering-time variation in Arabidopsis allopolyploids. Genetics 173: 965–974. 10.1534/genetics.106.056580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Tian L., Lee H.-S., Wei N. E., Jiang H., et al. , 2006b Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517. 10.1534/genetics.105.047894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang H., Wang J., Sun R., Wu J., et al. , 2011. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039. 10.1038/ng.919 [DOI] [PubMed] [Google Scholar]
- Wendel J. F., Lisch D., Hu G., Mason A. S., 2018. The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr. Opin. Genet. Dev. 49: 1–7. 10.1016/j.gde.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Williams R. B., Chan E. K., Cowley M. J., Little P. F., 2007. The influence of genetic variation on gene expression. Genome Res. 17: 1707–1716. 10.1101/gr.6981507 [DOI] [PubMed] [Google Scholar]
- Wittkopp P. J., Haerum B. K., Clark A. G., 2004. Evolutionary changes in cis and trans gene regulation. Nature 430: 85–88. 10.1038/nature02698 [DOI] [PubMed] [Google Scholar]
- Woodhouse M. R., Cheng F., Pires J. C., Lisch D., Freeling M., et al. , 2014. Origin, inheritance, and gene regulatory consequences of genome dominance in polyploids. Proc. Natl. Acad. Sci. USA 111: 5283–5288 (erratum: Proc. Natl. Acad. Sci. USA 111: 6527) 10.1073/pnas.1402475111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. J., Thaxton P. M., El-Zik K. M., Paterson A. H., 1998. D-subgenome bias of Xcm resistance genes in tetraploid Gossypium (cotton) suggests that polyploid formation has created novel avenues for evolution. Genetics 149: 1987–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Cheung F., Lee J. J., Ha M., Wei N. E., et al. , 2006. Accumulation of genome‐specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 47: 761–775. 10.1111/j.1365-313X.2006.02829.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo M., Szadkowski E., Wendel J., 2013. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 110: 171–180. 10.1038/hdy.2012.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo M.-J., Liu X., Pires J. C., Soltis P. S., Soltis D. E., 2014. Nonadditive gene expression in polyploids. Annu. Rev. Genet. 48: 485–517. 10.1146/annurev-genet-120213-092159 [DOI] [PubMed] [Google Scholar]
- Young, M. D., M. J. Wakefield, G. K. Smyth, and A. Oshlack, 2012 goseq: gene ontology testing for RNA-seq datasets. R Bioconductor. Available at: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.366.9571&rep=rep1&type=pdf.
- Zhao Q., Zou J., Meng J., Mei S., Wang J., 2013. Tracing the transcriptomic changes in synthetic trigenomic allohexaploids of Brassica using an RNA-Seq approach. PLoS One 8: e68883 10.1371/journal.pone.0068883 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequenced reads are available in the NCBI Sequence Read Archive under SRP026656. Scripts used in this study are available at https://github.com/BBarbazukLab/papers/. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6207317.