Abstract
Evolutionary dynamics driven out of equilibrium by growth, expansion, or adaptation often generate a characteristically skewed distribution of descendant numbers: the earliest, the most advanced, or the fittest ancestors have exceptionally large number of descendants, which Luria and Delbrück called “jackpot” events. Here, I show that recurrent jackpot events generate a deterministic median bias favoring majority alleles, which is akin to positive frequency-dependent selection (proportional to the log ratio of the frequencies of mutant and wild-type alleles). This fictitious selection force results from the fact that majority alleles tend to sample deeper into the tail of the descendant distribution. The flip side of this sampling effect is the rare occurrence of large frequency hikes in favor of minority alleles, which ensures that the allele frequency dynamics remains neutral in expectation, unless genuine selection is present. The resulting picture of a selection-like bias compensated by rare big jumps allows for an intuitive understanding of allele frequency trajectories and enables the exact calculation of transition densities for a range of important scenarios, including population-size variations and different forms of natural selection. As a general signature of evolution by rare events, fictitious selection hampers the establishment of new beneficial mutations, counteracts balancing selection, and confounds methods to infer selection from data over limited timescales.
Keywords: coalescent, dynamics of adaptation, fixation probability, jackpot events, multiple mergers, selection
ONE of the virtues of mathematizing Darwin’s theory of evolution is that one obtains quantitative predictions for the dynamics of allele frequencies that can be tested with increasing rigor as experimental techniques, sequencing methods, and computational power advance. The Wright–Fisher model is arguably the simplest null model of how allele frequencies change across time (Fisher 1930). It states that a new generation of alleles is formed by random sampling (with replacement) from the current generation. In the limit of large populations, this Wright–Fisher sampling generates a stochastic dynamics akin to Brownian motion with a frequency-dependent diffusivity (Crow and Kimura 1970). Nowadays, the limiting diffusion process without selection is often replaced by the corresponding backward-in-time coalescent process (Kingman 1982), which has opened many ways for statistical inference from neutral genetic diversities. However, forward-in-time approaches are still unrivaled in their ability to include the effects of natural selection. As such, the Wright–Fisher model has been instrumental for shaping the intuition of generations of population geneticists about the basic dynamics of neutral and selected variants. Transition densities derived from the Wright–Fisher model also find tangible application in scans for selection in time-series data (Coop and Griffiths 2004; Bollback et al. 2008; Illingworth et al. 2011).
The Wright–Fisher model is remarkably versatile as it can be adjusted to many scenarios by the use of effective model parameters: an effective population size, an effective mutation rate, an effective generation time, and effective selection coefficients. But, crucially, these reparameterizations cannot account for extremely skewed offspring distributions. A large variance in offspring number seems common among species who need to balance a high mortality early in life with large numbers of offspring (Eldon and Wakeley 2006). This applies, for instance, to the Atlantic cod or the Pacific oyster, which can lay millions of eggs (May 1967; Oosthuizen and Daan 1974; Li and Hedgecock 1998). Yet, the overall number of reported high fecundity species is small and it is challenging to characterize the tails of their offspring distributions. However, in the microbial and viral world, skewed clone size distributions are common and they have a characteristic tail produced by the combination of exponential growth and recurrent mutations. This was highlighted 75 years ago by Luria and Delbrück (1943) who noticed that mutations that occur early in an exponential growth process will produce an exceptionally large number of descendants. The distribution of such mutational “jackpot” events has a particular power-law tail in well-mixed populations (Yule 1925), as is briefly explained in Figure 2A. The simplest models of continual evolution (Neher 2013) and related models of traveling waves (van Saarloos 2003) can be viewed, on a coarse-grained level, as repeatedly sampling from this jackpot distribution. Although the number of draws and the resampling timescale depends on the model, the descendant distribution always has the characteristic tail of the Luria–Delbrück distribution. It is, by now, well established that the ensuing genealogies are described by a particular multiple-merger coalescent (Schweinsberg 2003, 2017a,b; Brunet et al. 2007; Brunet and Derrida 2013; Desai et al. 2013; Neher and Hallatschek 2013), first identified by Bolthausen and Sznitman (1998).
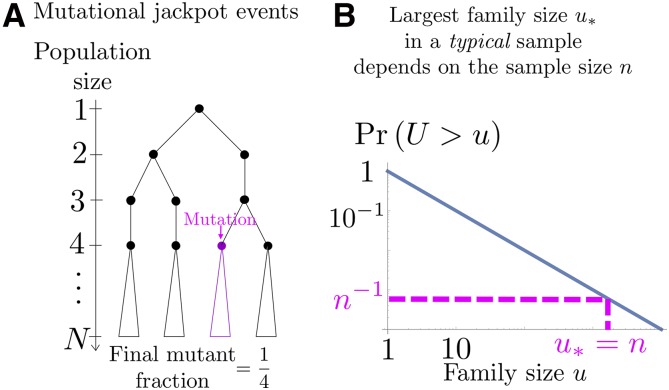
Figure 2.
(A) Illustration of mutational jackpot events, studied by Luria and Delbrück (1943). Consider the growth of a well-mixed population of microbes starting from a single cell (ignore death). A mutation that occurs in the jth cell division will have a final frequency of about ( in the illustration) and thus a clone size of Hence, the probability for a mutation to reach an even larger clone size U is equal to the probability that the mutation occurs prior to the nth cell division. The probability density to acquire a clone size u therefore exhibits a broad power-law tail (Our argument ignores the discreteness of the cell division events, which however does not change the asymptotic behavior.) (B) The blue line indicates the probability that the clone size U of a mutation is larger than u. The largest offspring number in a sample of n jackpot events should typically be of order n because the probability of sampling an even larger event is (dashed lines). A typical n sample, therefore, has a mean offspring number of order ln which is obtained upon truncating the offspring distribution at It turns out that this effect generates a selection-like bias favoring majority alleles, which is compensated by rare sampling events that favor minority alleles.
Other dynamical mechanisms can give rise to jackpot events as well. For instance, mutations that arise at the edge of spatially expanding populations can rise to high frequencies just by chance as a result of gene surfing (Fusco et al. 2016). When stationary bacterial populations are suddenly supplied with fresh media, jackpot events can arise from cells that leave dormancy anomalously early (Wright and Vetsigian 2018). In sexual populations, completely neutral mutations can rise to large frequencies by chance if they are closely linked to sites of strong selection (genetic “draft”) (Durrett and Schweinsberg 2005; Neher 2013). The resulting clone size distributions in general differ from the Luria–Delbrück distribution, but they too have power-law tails with diverging mean and variance.
One approach to account for these jackpot events is to consider an effective model where, in each generation, the population is sampled from an effective offspring number distribution with a broad, power-law tail. While such extensions of the Wright–Fisher diffusion process to skewed offspring distributions have been formally constructed (Donnelly and Kurtz 1999; Pitman 1999; Bertoin and Le Gall 2003; Berestycki 2009; Griffiths 2014), also including selection and mutations (Etheridge et al. 2010; Der et al. 2012; Foucart 2013; Der and Plotkin 2014; Baake et al. 2016), we still lack explicit finite time predictions for the probability distribution of allele frequency trajectories. My goal here is to fill this gap for the particular case of the Luria–Delbrück jackpot distribution, by characterizing the allele frequency process with and without selection in such a way that it can be generalized, intuitively understood, and integrated in time.
Sampling Allele Frequencies Across Generations
Our starting point is a simple model for the change of the frequency of an allele as a population passes from generation to generation. It is useful (although not necessary) to think of the model as emulating a population where each individual contributes a very large random number of seeds to a common seed pool. Due to competition for a finite resource, only a small fraction of these seeds survives to become the adults of the new generation. The production of seeds and the subsequent down-sampling are two separate steps of the model, illustrated in Figure 1. A similar seed-pool metaphor is often used to motivate the Wright–Fisher model, with each individual contributing the exact same number of seeds (see, e.g., Otto and Day 2011). In general, however, we want to assume that the individual contributions to the seed pool are random variables (independent and identically distributed) drawn from a given offspring distribution.
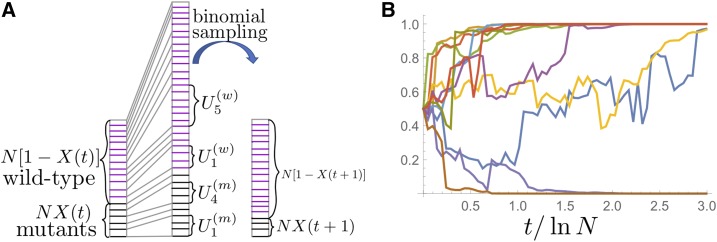
Figure 1.
(A) Illustration of how our model resamples allele frequencies from generation to generation. From a given frequency of mutants in generation t, the frequency of the next generation is produced in two steps. First, each individual gets an offspring number drawn from a given offspring distribution. This step generates random numbers for the mutant family sizes, and for the wild-type family sizes (a couple of large family sizes are indicated in the illustration). After this first step, the total population size will strongly deviate from the starting size N (middle column is much higher than the left column). Thus, we binomially sample with replacement from this intermediate population exactly N individuals (right column has the same size as left column). The resulting fraction of mutants is the mutant frequency in generation (B) A total of 10 sample paths are shown for a population size and an offspring distribution with density All sample paths start with the same initial frequency 0.5. The time axis is measured in units of which turns out to be the coalescence timescale (for large N).
To define the model mathematically, let be the frequency of one type, the “mutants,” within a population of size at the discrete generation t. (The population size is generally allowed to change from generation to generation.) The mutant frequency in generation is produced from the frequency in generation t, as follows: first, each individual gets to draw a statistical weight U from a given probability density function (nonzero only for and the same for both mutants and wild types). This generates random variables for the mutants and random variables for the wild types. In the seed-pool picture, one can interpret and as the statistical weight of the contribution of mutant i and wild-type j to the seed pool, respectively. The new discrete mutant number is obtained in a second step by binomially sampling times with the success probability
| (1) |
which depends on the sums and One can think of M and W as representing the total weight of the mutant and wild-type seeds in the seed pool. After the subsampling step, one finally ends up with new frequencies and of mutants and wild types, respectively. By construction, the final mutant and wild-type numbers are discrete, irrespective of whether the individual contributions to the seed pool are discrete variables. We therefore allow the weights and to be drawn, in general, from a continuous distribution.
Note that the deviation of the new mutant frequency from the simple fraction in Equation 1, is of order For most of this work, we are interested in large enough N and a broad enough offspring distribution such that the binomial sampling error, which represents classical random genetic drift, is negligible compared to the fluctuations induced by sampling from the offspring distribution. Also note that, because all random variables are independently drawn from the same distribution, there is no expected bias in the mutant frequency: The expected allele frequency in generation t is constant and equal to the starting allele frequency [ is a “martingale”].
If has finite mean and variance, binomial sampling does matter and the above population model generates allele frequencies that are described by Wright–Fisher diffusion in the large N limit, which has genealogies described by the Kingman coalescent (Schweinsberg 2003). (Note that the Wright–Fisher model is obtained if the offspring distribution is a Δ function, so that all weights are identical in the first step of our reproduction model. More generally, our population model belongs to the class of Cannings models.)
But what are the characteristic features of the forward allele-frequency process for offspring distributions so broad that even the mean diverges? As I show in the next sections using numerical exploration and heuristic arguments, this leads to a sampling-induced bias for alleles that are in the majority, an apparent rich-get-richer effect. I will then describe and illustrate phenomena driven by this effect, including biased time series, a high-frequency uptick in site frequency spectra (SFS), and a low probability of fixation of beneficial mutations. In the technical section of this article, I discuss a suitable large-N scaling limit of in which the stochastic dynamics for the particular case of the Luria–Delbrück distribution can be fully predicted.
Data availability
The author states that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Simulations
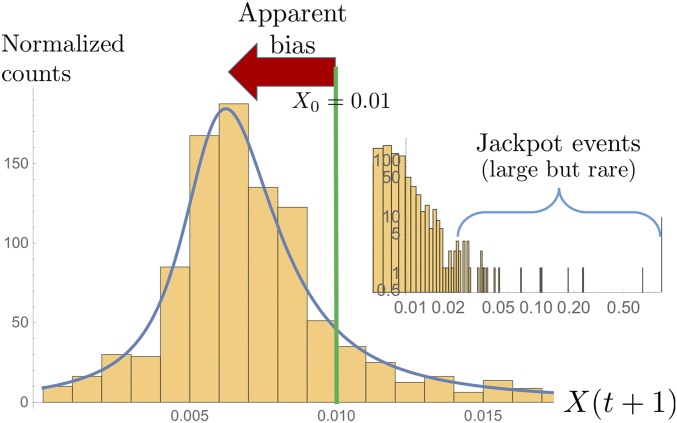
Since offspring numbers affect the way reproduction cycles scatter allele frequencies, we first explore numerically how an allele frequency changes in a single generation when family sizes are drawn from the density which arises in the Luria–Delbrück case (Figure 2). For a low allele frequency in a given generation, Figure 3 shows a histogram of 800 next-generation frequencies As one might have guessed, the distribution is peaked; the more peaked, the larger the size of the population (increasing the population size generally suppresses the scatter). Importantly, however, the bulk of the distribution, including its peak and the median, is shifted downward relative to the starting frequency.
Figure 3.
Histogram obtained from resampling 800 times a new allele frequency given that The population size is Notice the apparent shift to lower frequencies of the bulk of the histogram compared to the initial frequency (green line). The blue line shows the asymptotic resampling distribution (Equation E8). The inset shows the same histogram on a double-logarithmic scale and reveals that, rarely, the resampled frequency reaches very large values, up to 0.71 in the present case. Those jackpot events turn out to balance the apparent bias (←) such that neutrality is preserved.
One might wonder how this apparent bias can be consistent with overall neutrality—the mean of the distribution must be unbiased, by the construction of our model. The mean of the histogram in Figure 3 is indeed close to X, much closer than the median. The difference between median and mean results from the extreme skew of the histogram. The few largest frequencies in the sample are visible in the inset and contribute substantially to the value of the mean, but they only contribute marginally to the median. The median, therefore, gives a better idea of the “typical” behavior upon resampling, which is dominated by the peak of the resampling distribution.
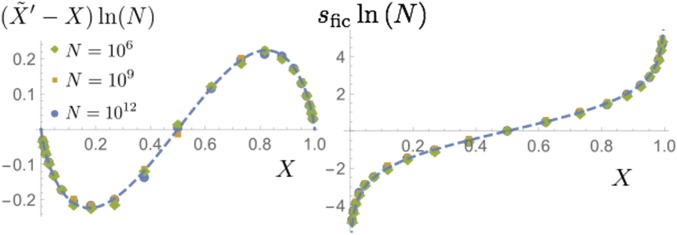
The direction and magnitude of the median bias depends on the starting frequency. Choosing a range of starting frequencies shows that the median bias always favors the majority type: the frequency increases (decreases) for Thus, the majority allele tends to become even more abundant under resampling. Moreover, the magnitude of the bias is of order of the inverse of the logarithm of the population size, as the data collapse in Figure 4 suggests. The remaining variation follows the theoretical dashed line, which is derived below.
Figure 4.
Left: The displacement of the median of next-generation frequencies from the starting frequency X, scaled by is plotted as a function of starting frequency. The medians were obtained for three different population sizes (see legend) from samples each of size The chosen scaling ensures that the data for different population sizes nearly collapse on the dashed line, which represents the function The median shift is negative for and positive for thus favoring the majority allele, and its overall magnitude is of order . Right: This apparent bias can be turned into an effective selection coefficient upon dividing by which closely follows
In summary, the mean frequency in a new generation is (by construction) unbiased, but its unbiased value relies on sampling rare events of large frequency change. Most of the time, generational resampling tends to favor the majority type, reflected in a bias of the median, proportional to the inverse of the logarithm of the population size. Natural selection represents a genuine bias, which produces a generational frequency change where s is the selective difference between the two alleles in question. Inspired by this relation, we divide the median bias by the product to obtain an apparent selection coefficient. The resulting effective selection coefficient vanishes at a frequency of 50%, but diverges logarithmically toward fixation and extinction, see the right-hand side of Figure 4.
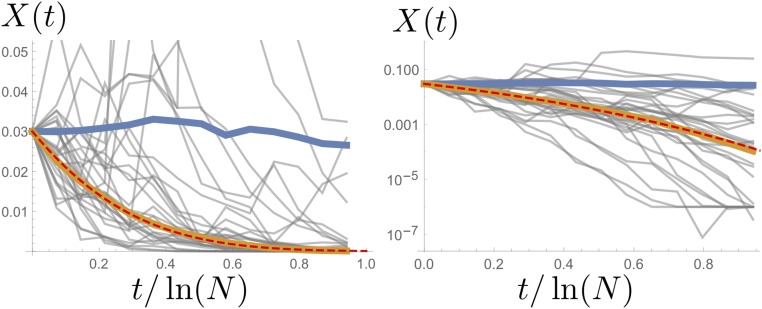
Even though the bias of the median frequency might be small in a single generation, it can accumulate over a chain of many generations and generate a strong discrepancy between typical allele frequency trajectories and the expected ones. This can be seen in Figure 5, which shows both the median and the mean of many allele frequency trajectories, all starting at the same initial frequency. The mean tends to stay at the initial frequency, as it must by construction, even though marked fluctuations (due to rare events) are visible. The median, on the other hand, follows a smoothed, biased curve toward extinction for starting frequencies <50%. Importantly, the median trajectory closely follows the dashed-line trajectories, which are generated by the apparent frequency-dependent selection identified above upon ignoring any source of stochasticity.
Figure 5.
For a population size of and initial allele frequency 1000 allele frequency trajectories were simulated, 30 of which are shown in gray. The trajectory of the mean (blue) is consistent with the constant expectation, although considerable fluctuations are visible even in such large samples. By contrast, the median (yellow) closely follows the smooth dashed line, which is the behavior expected under frequency-dependent selection with selection coefficient (Figure 4) (ignoring any source of random number fluctuations). The plot on the right is the semilogarithmic version of the one on the left.
These numerical results call for an explanation of the difference between typical and mean frequency dynamics in terms of the tail of the offspring-number distribution, and raise the question of whether that difference has any real consequences for observable population genetic data and the fate of selected variants. Does the apparent bias act similarly to real frequency-dependent selection? In this case, it should hamper the invasion of new alleles until they become sufficiently common, thereby suppressing their fixation probabilities.
Resampling typically favors the majority type
It is useful to start our analysis with purely heuristic arguments to see that, for broad enough offspring distributions, population resampling typically favors the majority type. These arguments provide an intuitive basis for the phenomena I discuss and derive further below.
Suppose an allele is currently at frequency X, and we would like to estimate the frequency after resampling. According to our reproduction rules stated above, we need to estimate the total number of offspring of both mutants, M, and wild types, W, which represent sums of many random offspring numbers (if N is large). Such estimates are challenging for skewed offspring distributions, especially when the mean depends on the largest families that occur in a sample. Yet, a mean offspring number of a typical sample of n offspring numbers can be estimated by the truncated expectation
| (2) |
With high probability, the mean offspring number of a sample will be close to this typical value (Appendix A). The cutoff of the integral represents the largest offspring number in a typical n-sample, illustrated in Figure 2B, which can be estimated by the extremal criterion (Krapivsky et al. 2010)
| (3) |
Note that the typical value becomes essentially equal to the expectation if the integral in Equation 2 is not sensitive to the upper bound. The key phenomena discussed in the article, however, rely on being sufficiently fat tailed so that is dependent on the sample size n. This occurs when decays like or more slowly so that the mean offspring number diverges. Throughout most of this article, I will in fact focus on the particularly interesting marginal case of which as mentioned arises in population models that combine stochastic jumps with exponential growth. In this Luria–Delbrück case,
| (4) |
| (5) |
| (6) |
Hence, the mean offspring number of a typical sample from a mutant population currently at frequency X can be estimated by
| (7) |
and likewise for the wild-type population. The frequency dependence of this expression is the crux of our study, as it implies that more abundant mutants (larger X) typically behave as if they have higher fitness (larger offspring numbers).
Assuming an initial allele frequency X, the typical frequency after resampling can then be estimated by
| (8) |
The typical discrepancy between the resampled frequency from the original frequency X simplifies in the limit to a deterministic advection velocity
| (9) |
which can be interpreted as a traditional selection term with a frequency-dependent selection coefficient:
| (10) |
Akin to positive frequency-dependent selection, this term tends to increase the frequency of the majority type and results from the fact that the majority type is able to sample deeper into the tail of the offspring number distribution. This explains the magnitude and functional form of the apparent bias detected in our numerical experiments of Figure 4. Since this bias acts like genuine selection term, yet is of purely probabilistic origin, I will call it fictitious selection.
As mentioned above, overall neutrality of our generation model demands that the mutant frequency has to stay constant on average, in contrast to what I argued should typically happen. It turns out that the ignored atypical events ensure neutrality: a detailed analysis of the resampling distribution (Appendix E and Appendix F) shows that neutrality holds overall because rare big events rescue the minority type. Both effects, a nearly deterministic advection toward the majority type and compensating rare jumps in favor of the minority type, can be appreciated from the histogram in Figure 3, which shows resampled frequencies conditional on an initial frequency For large N, a pronounced peak appears below at a scale consistent with the selection coefficient determined above, see Figure 3. Yet, the histogram still has some support at very large frequencies, which correspond to the just-mentioned compensating jumps.
Consequences
I will now describe tangible phenomena driven by fictitious selection and the compensating rare jumps. The mathematical description of these phenomena follows from an analytically solvable mathematical framework described below.
Trajectories:
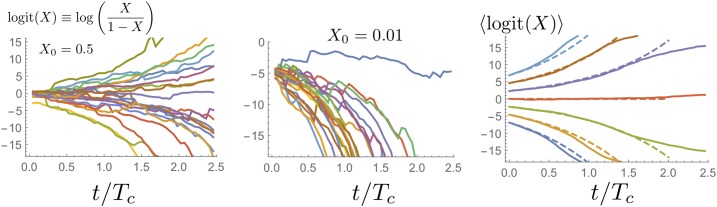
If we ignore the effect of rare jumps, one would expect a characteristic allele frequency trajectory to move toward fixation or extinction, following the fictitious selection force. The resulting deterministic trajectory is most easily described in terms of the logit, which obeys
| (11) |
It turns out that the expectation of the logit indeed follows these dynamics,
| (12) |
as the simulations in Figure 6 demonstrate. Thus, while neutrality requires the expectation of allele frequencies to remain constant, one finds that the expectation of the log ratio of frequencies moves away from zero and, moreover, exponentially fast. This peculiar behavior reflects the fact that a log-transformed stochastic variable is much less sensitive to rare, big jumps, which are needed to compensate for the fictitious selection force. In frequency space, the trajectories resemble nearly deterministic trajectory pieces glued together by rare compensating jumps.
Figure 6.
Frequency trajectories for the log ratio of mutant frequency and wild-type frequency, also called the logit frequency. The population size was which sets the coalescence time Trajectories were chosen to start at in the left panel and in the middle panel, respectively. The panel on the right shows the average over 100 sample paths of the log ratio (solid lines) for different starting frequencies. The dashed lines are the theoretical expectation in the continuum limit, which simply is an exponential function in logit space (Equation 12).
The dynamics of the logit expectation in Equation 12 implies that it typically takes a time of order generations to change the allele frequency by an order of one, and generations to reach one of the absorbing boundaries, which have a logit frequency of order Both timescales are known from the associated Bolthausen–Sznitman coalescent: is the time (in generations) to coalesce a random sample of two lineages, and is the timescale to coalesce all lineages in a population of constant size N (Pitman 1999; Berestycki 2009).
Fictitious selection can be most easily detected at the high-frequency end of the SFS. As with regular selection, the SFS near fixation solely depends on the advection term, SFS as as if allele frequencies are only advected and jumps are negligible. This leads to an uptick at high frequencies (Neher and Hallatschek 2013) that has the same form as the SFS of a selected allele with frequency-dependent selection coefficient The excess of common alleles relative to intermediate frequencies results from the advection term slowing down as allele frequencies increase toward fixation.
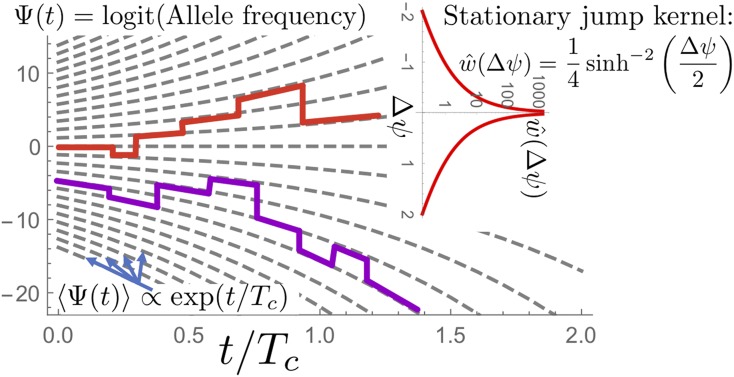
If one includes both deterministic advection and stochastic jumps, one finds that the logit of the frequency, continuously switches between exponentially diverging deterministic trajectories via jumps drawn from a jump kernel, which is a power law for small jump distances but exponentially decaying for large jumps. The ensuing stochastic process is illustrated in Figure 7.
Figure 7.
Illustration of the limiting allele-frequency process, which is most easily described for the log ratio of the frequency of mutants and wild types, Trajectories in this logit space on average follow exponentially diverging paths (gray lines). Switching between these paths occurs at a rate which depends (only) on the distance in logit space. Small jumps happen frequently, in fact as but impactful jumps of order 1 or larger roughly take a coalescence time to occur. The advection force drives the ultimate fixation or extinction of alleles, which takes a time of order
Interestingly, the jump rate between two positions in logit space only depends on their distance (the kernel is stationary). This makes it possible to compute exactly, for a general time-dependent population size, the probability density that a trajectory will move from logit position to ψ in a time period t. This transition density is given by
| (13) |
in terms of a rescaled time variable The scale factor appearing in this time conversion represents the coalescence rate of two lineages at time t. For a constant population size, the time conversion simplifies to where is the mean coalescence time of two lineages. From the transition density in Equation 13 [which for constant population size also arises in a simple model of microbial adaptation (Desai et al. 2013)], it is possible to characterize a number of interesting statistics, including sojourn times and the SFS (Kosheleva and Desai 2013; Neher and Hallatschek 2013), and directly confirm the duality with the Bolthausen–Sznitman coalescent (Appendix C).
Selection:
Selection modifies the above dynamics by introducing a true bias (no jumps to leading order). A mutation with a frequency-independent fitness effect s leads to
| (14) |
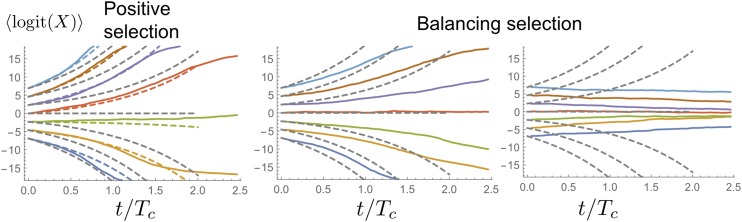
where a constant coalescence rate is assumed. From this expression, we can see that genuine selection indeed competes with fictitious selection: the logit will, on average, increase with time only if is >0, otherwise it will decay over time. Mean sample paths are shown in Figure 8.
Figure 8.
Frequency trajectories for positive (left) and balancing selection (middle, right) for The positive selection coefficient was chosen to be In the middle and right panels, balancing selection coefficient was chosen below and above threshold , respectively. The dashed lines correspond to the neutral expectation.
In fact, the detailed analysis below shows that the stochastic dynamics of a selected allele starting at frequency is identical to the dynamics of a neutral allele properly shifted in logit space. The exact statement is
| (15) |
in terms of a transition density of a selected allele with selective advantage s. Since the fixation probability of a neutral allele at frequency X simply is X, this mapping implies
| (16) |
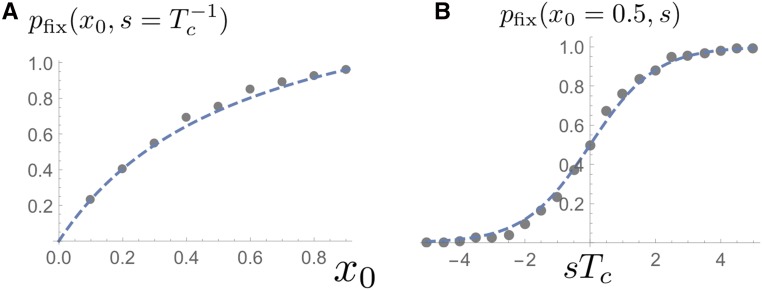
If the selected allele is initially rare, we have So, a selective advantage does increase the odds of fixation exponentially. But, because for a single mutant, the fixation probability of newly arising beneficial mutations is very small, unless The comparison of theory and simulations in Figure 9 confirm these predictions for the fixation probability.
Figure 9.
The probability of fixation as a function of frequency (left; ) and as a function of scaled selection coefficient (right; ). Each gray dot was obtained by collecting 100 fixation or extinction events for a population size of The dashed lines are the theoretical predictions in Equation 16.
Finally, since the effective bias tries to push allele frequencies toward fixation, one may ask what happens in the presence of balancing selection opposing fictitious selection. So, let us consider an allele under balancing selection modeled by in the sense of a Taylor expansion in logit space. The first moment in logit space now satisfies
| (17) |
The most important feature of this expression is a threshold phenomenon at For balancing selection will not be able to maintain diversity. For both alleles will be maintained in an equilibrium between balancing selection and the fluctuations induced by sampling large family sizes. The variance of the equilibrium distribution diverges as a approaches
Limiting stochastic process
I now show how the above phenomena follow from a detailed mathematical analysis of the asymptotic allele frequency dynamics as the population size tends to infinity. The resulting process belongs to the class of Λ-Fleming–Viot processes, which are dual to multiple-merger coalescents in a similar way as Wright–Fisher diffusion is dual to the Kingman coalescent. Although Λ-Fleming–Viot processes (Donnelly and Kurtz 1999; Pitman 1999; Bertoin and Le Gall 2003; Berestycki 2009) and extensions involving selection and mutations (Etheridge et al. 2010; Der et al. 2012; Foucart 2013; Baake et al. 2016) have been extensively studied, closed-form predictions for the transition density of allele frequency trajectories are still lacking. It turns out that, upon formulating the process in terms of a jump-drift process, such closed-form predictions can be obtained for the particular case of the Luria–Delbrück offspring-number distribution, which generates a Λ-Fleming–Viot process dual to the Bolthausen–Sznitman coalescent. Although somewhat technical in nature, the analysis will elucidate how fluctuations, the sampling-induced bias, and an actual bias (natural selection) combine to control the fate of alleles, pointing to further theoretical directions that could be explored. Readers not interested in mathematical details may jump right to the Discussion section.
The larger the population size N, the more deterministic is the resampling of allele frequencies, and the slower the ensuing stochastic process. Hence, to obtain an interesting time-continuous stochastic process, it is natural to slow down the progression of time. It turns out that a well-behaved, time-continuous, Markov process is obtained in terms of the time variable τ with differential upon sending Since proves to be the coalescence rate for two lineages, one can say that we measure time in units of the inverse coalescence rate. For a constant population size, one simply has where is the mean coalescence time of two lineages.
Any such time-continuous (sufficiently well-behaved) Markov process is defined by an advection velocity, diffusion coefficient, and jump kernel (Gardiner 1985). To state this triplet, we define to be the probability density to sample a frequency if we start with a frequency In our new units of time, the rate of jumps from to follows from a scaling limit of
| (18) |
The advection velocity and diffusion coefficient are defined by the rate of change in mean and variance,
| (19) |
| (20) |
The next task is to evaluate these limits for the resampling distribution of our population model with broad offspring numbers. Recall that, for the Wright–Fisher model, the resampling distribution is a binomial distribution, giving rise to a finite diffusivity while both jump kernel and advection velocity vanish. Up to a change in (coalescent) timescale, the same limiting results are obtained for many other neutral population models with narrow offspring-number distributions, as a consequence of the central limit theorem. In the case of broad offspring numbers, one must use a generalization of the central limit theorem (Gnedenko and Kolmogorov 1954), which in our case reveals a close connection between our resampling distribution and the so-called Landau distribution (Landau 1944). From the asymptotic analysis in Appendix E, one concludes
| (21) |
| (22) |
| (23) |
This triplet has several interesting features. First, the vanishing diffusivity contrasts with the defining feature of Wright–Fisher diffusion, which has in properly scaled time units. So, the classical picture of diffusing allele frequencies does not apply to the present case, which is characterized by jumps encoded in the kernel and the advection velocity —both of them vanish in the neutral Wright–Fisher diffusion (the inclusion of natural selection will be discussed further below).
The form of the jump kernel may not be surprising as it can be obtained by just thinking about the effect of individual jackpot events drawn from the offspring distribution: the denominator comes from the offspring distribution and the numerator represents the probability that the jackpot occurs on one allele multiplied by a normalizing factor. [Suppose the mutants are currently at frequency . If a jackpot of size arises in the wild-type subpopulation, the new allele frequency will be (in the large population size limit). The probability that a jackpot changes the allele frequency from to is given by the probability that the jackpot arises in the wild-type subpopulation multiplied by the probability that the jackpot is larger than . Thus, or which also results from the analogous argument for jackpots that arise on the mutant background.]
The advection term, however, is perhaps unusual, as I have argued in the heuristic part of this study: It emerges from the fact that mutants can typically sample deeper in the tail of the offspring number distribution if they are in the majority (and vice versa). The advection term consequently biases frequencies away from and, consequently, acts like positive frequency-dependent selection, with an effective selective coefficient in our scaled units of time. In Appendix D, I provide an alternative, but somewhat technical, way of deriving the advection term using the so-called Λ-Fleming–Viot generator, which in its standard form does not reveal the advection term.
All aspects of the ensuing stochastic process are encoded in the probability density that a neutral allele evolves from initial frequency to a frequency x within the time period τ. This transition density satisfies the differential equation
| (24) |
in terms of the frequency-dependent advection velocity and jump kernel given in Equation 21 ( denotes the Cauchy principle value). Equations of the type of Equation 24 are sometimes called differential Chapman–Kolmogorov equations (Gardiner 1985), which we will adopt in the following.
We have the important consistency check that the entire dynamics is neutral [ is a martingale]: multiplying Equation 24 by x and integrating yields an equation for the first moment
| (25) |
The integrals on the right-hand side can be performed easily in the limit so that the transition density becomes a Δ function
| (26) |
| (27) |
| (28) |
So we see that the advection term is necessary to balance the fact that the symmetric jump kernel has a different extent for lowering the frequency than for increasing the frequency, unless the starting frequency is precisely at Neutrality can be used to rationalize the advection term in Equation 21 if one happens to know the jump kernel.
The analysis of moments can be pushed further. The initial rates of change of higher moments directly yield the coalescence rates associated with the genealogical process. As shown in Appendix C, these rates are precisely the ones of the Bolthausen–Sznitman coalescent, confirming the duality between the process and the Bolthausen–Sznitman coalescent.
But, as with many processes that involve broad-tailed jump distributions, moments say little about the typical behavior of frequencies, which depends on all moments. Fortunately, the analysis massively simplifies if we describe the dynamics in terms of the log ratio of the frequency of both alleles, also called the logit of x. The corresponding transition density again satisfies a differential Chapman–Kolmogorov equation
| (29) |
in terms of a transformed advection velocity being simply linear in ψ, and the transformed jump kernel
| (30) |
which depends on the jump distance
Thus, the stochastic process has a simple description in logit space,
| (31) |
where it consists of linear deterministic advection combined with a pure jump process as illustrated in Figure 7. The jumps are drawn from a stationary kernel (Equation 30), which has the property that small jumps occur at a power-law rate diverging as and big jumps are exponentially suppressed,
Because the logit ψ runs from to and the jump kernel is symmetric with respect to the jump displacement the jump displacement has to vanish on average, This implies that the expectation of the random variable is controlled just by the fictitious selection force,
| (32) |
as was anticipated in Equation 14.
Moreover, because the jump kernel only depends on the jump distance, and not the jump start or end point separately, we can use a Fourier transform to solve Equation 29: this converts the integral on the right-hand side into a simple product, as shown in Appendix B. The final result for the transition density is
| (33) |
in logit space, and
| (34) |
in frequency space. Note that the form of the transition density in Equation 34 is the same as the one found for a particular model of microbial adaptation (Desai et al. 2013; Kosheleva and Desai 2013) if one replaces with where integer k denotes a discrete fitness class and the quantity is related to the largest fitness class q typically occupied. With this substitution, all findings for the dynamics of neutral mutations from these studies carry over to the present population model, including the SFS and sojourn times, if the population size does not vary with time.
Genuine selection:
The allele-frequency process admits a number of natural extensions. For instance, the differential Chapman–Kolmogorov equation can be modified to include fluctuations in the offspring-number distribution, mutations, and classical genetic drift; or it can be turned into a backward equation, which allows the discussion of (certain) first-hitting-time problems. Most importantly, we can now include selection which is notoriously hard to include in coalescence processes.
The most obvious example for a nonneutral scenario is the rise of unconditionally beneficial or deleterious mutations in a population with skewed offspring-number distributions. In traveling-wave models, this scenario arises effectively when a mutation occurs that (slightly) changes the wave speed. Range expansions, for instance, are accelerated by the fixation of mutations that increase the linear growth rate, the dispersal rate, or by mutations that broaden the dispersal kernel (Hallatschek and Fisher 2014). In models of adaptation, the rate of adaptation can be increased through mutations that increase the mutation rate (by mutator alleles) or the frequency of beneficial mutations (potentiating mutations). The analysis below also lends itself to a discussion of balancing selection, which could model ecological interactions or some generic fitness-landscape roughness.
A selective difference between mutants and wild types modifies (to leading order) the allele-frequency dynamics in the same way as it affects Wright–Fisher diffusion, namely as a part of the advection velocity (Etheridge et al. 2010; Griffiths 2014)
| (35) |
Here, I have introduced the selective difference which is not necessarily a small quantity as it represents the action of selection accumulated over generations, or the coalescence time of two lineages.
In logit space, including selection leads to the simple change
| (36) |
showing that positive/negative selection is competing with the fictitious selection term, if the mutant is in the minority/majority.
First, consider the case where selection is not frequency dependent, constant. In this case, we can perform a simple shift in logit position to map the transition density for the nonneutral dynamics onto the neutral one:
| (37) |
In frequency space, we have
| (38) |
Equivalently, we can say that the stochastic variable
| (39) |
is a martingale. Remarkably, this implies that the fixation probability of a selected allele at frequency is the same as the fixation probability of a neutral allele at initial frequency which is simply equal to We have thus proved the result Equation 16 for the fixation probability.
We can further account for a simple form of balancing selection,
| (40) |
where the term acts as a restoring force trying to push the frequency to the frequency value (in the Results section, I used the unscaled variable ). The terms and may be viewed as the first two terms of a Taylor expansion of the selection term in logit space.
The mean logit of the allele frequency now obeys
| (41) |
The deterministic dynamics of the mean has the fixed point but it is repelling if and attractive for Thus, unless balancing selection is strong enough, we still have a runaway effect: diversity cannot be maintained in our model, although the gradual loss of diversity now proceeds at a slower pace [fixation times are amplified by a factor ].
The Fourier transform of the associated transition density can be obtained as above via the method of characteristics, yielding
| (42) |
but no simple analytical form of the Fourier back transform seems to exist for general α.
The stationary distribution for the attractive case, is given by
| (43) |
where I used the short-hand The Fourier back integral can be evaluated numerically. While a closed form does not seem to exist, one can show that the distribution approaches a normal distribution with SD as
Discussion
I described a number of phenomena driven by an apparent bias for majority alleles in populations with a strongly skewed offspring-number distribution (with a cutoff-dependent mean offspring number). The majority allele is typically at an advantage compared to the minority allele because it samples more often, and thus deeper, into the tail of the offspring distribution. This leads to a larger apparent fitness of the majority type if the offspring number distribution has a diverging mean. The word typical is important here because the neutrality of the process is restored by untypical events by which the minority type hikes up in frequency—while typically the rich get richer, the poor can occasionally turn the tide.
Heuristic arguments suggest a typical bias in favor of the majority compensated by rare, but large, jumps in favor of the minority to be a general signature of offspring distributions with diverging means. Here, I have focused on the marginal case where the mean of the offspring distribution diverges logarithmically. As I have briefly reviewed in Figure 2, such a broad, effective offspring distribution emerges in population models that combine homogeneous growth and stochastic mutations as first highlighted by the seminal work on spontaneous mutations by Luria and Delbrück (1943). The corresponding sampling-induced bias for the majority type was found to take the form of a selection term, with a strength proportional to the log ratio of the frequencies of both alleles and inversely proportional to the logarithm of the population size. Since this term looks and acts like (frequency-dependent) selection but does not result from phenotypic differences, I have termed this force fictitious selection.
Biased time series and comparison with data
Despite overall neutrality, allele-frequency trajectories typically look biased (Figure 6), especially if only short time series are available that do not sample the rare compensating jumps. Hence, the possibility of a sampling-induced bias should be considered in attempts to infer selection from allele-frequency trajectories.
In light of these biases, one may wonder how long time series should be to sample enough compensating jumps, such that neutral dynamics could be distinguished from a truly biased one. A satisfying answer to this question is beyond the scope of this article but may relate to the following estimates. The rate of jumps of varying sizes is encoded in the jump kernel, Equation 21. So, in the continuum limit, the rate of jumps is infinite but, importantly, most of them are tiny. The jumps that matter in the sense that they balance fictitious selection in Equation 26, Equation 27, and Equation 28 are of order of the current frequency X or larger, assuming Those occur at a finite rate: at a given frequency X, one can expect the first balancing jumps to occur after a time of order of the coalescence time But it takes much longer, of order to have sampled enough long jumps to establish full neutrality. Turned around, one could say that a time trace hovering about a frequency X typically exhibits a bias characterized by an apparent selection coefficient of order assuming the duration t obeys In application it might be informative to study trajectories over time windows of varying length directly (Bollback et al. 2008; Illingworth et al. 2011) or indirectly by integrating information across genomic length scales (Weissman and Hallatschek 2017). A timescale-dependent selection strength could then serve as an indicator for broad offspring numbers.
Note that the problem of apparent timescale-dependent biases is absent in Wright–Fisher diffusion: if complete information about a time trace of a Wright–Fisher diffusion process is given, one can decide unequivocally on neutrality, no matter how long the time trace is. Of course, deviations from the continuum limit and incomplete information will complicate the decision of whether a process is truly neutral, but these problems affect both Wright–Fisher diffusion as well as the allele frequency dynamics generated by broad offspring-number distributions.
Time series of allele frequencies are increasingly generated for natural populations, which frequently go through range expansions and also show signatures of adaptation. As the resolution in time and frequency improves, these data sets may ultimately reveal the footprints of fictitious selection. Yet, currently the best type of data is high-resolution barcoding trajectories determined from experimental evolution studies (Levy et al. 2015). Those data span several orders in magnitude so that it should be possible to detect the typical bias when the rate of change of the logit is plotted vs. the logit itself—on average, one should obtain straight lines. Other observables can be used to directly probe the jump kernel. For instance, the median squared frequency deviation from the deterministic trajectory is predicted to grow quadratically as a function of time. This behavior would be a clear departure from a linear behavior that Wright–Fisher diffusion implies.
Effective parameters
Even though our initially discrete population model was characterized by two parameters, an effective population size and an effective generation time we found that the time-continuous allele frequency obtained for large only depended on the product which sets the coalescence time of two lineages. This is true at least when the effective population size is time independent. In general, the allele-frequency process depends on the coalescence rate of two lineages, which can be a time-dependent quantity. So, if one wants to study the likelihood of some observed allele-frequency trajectories under the neutral allele-frequency process I have described, one merely needs to scale time by the correct coalescence rate.
Logit space picture
Mathematically, the neutral allele-frequency process is best described in logit space: the logit of frequency X follows exponentially diverging trajectories except for jumps drawn from a symmetrical and stationary jump kernel, as illustrated in Figure 7. The ensuing stochastic process can be described by a jump-advection process [dual to the Bolthausen–Sznitman coalescent (Bolthausen and Sznitman 1998)] and, via a Fourier transform, integrated in time to obtain exact expressions for the neutral transition density (Equation 13).
Accounting for selection
Studying allele frequencies in logit space has the advantage that it enables the analysis of natural selection, which is notoriously hard in the backward picture of the coalescent [although not impossible (Krone and Neuhauser 1997; Neuhauser and Krone 1997)]. Remarkably, I found that, in logit space, allele frequencies under constant selection pressure simply follow a shifted version of the neutral dynamics (Equation 15). This allowed, for instance, the exact calculation of fixation probabilities (Equation 16). Other forms of frequency-dependent selection can also be analyzed to leading order in an expansion in logit space (e.g., balancing, disruptive, or fluctuating selection).
Impact of selection
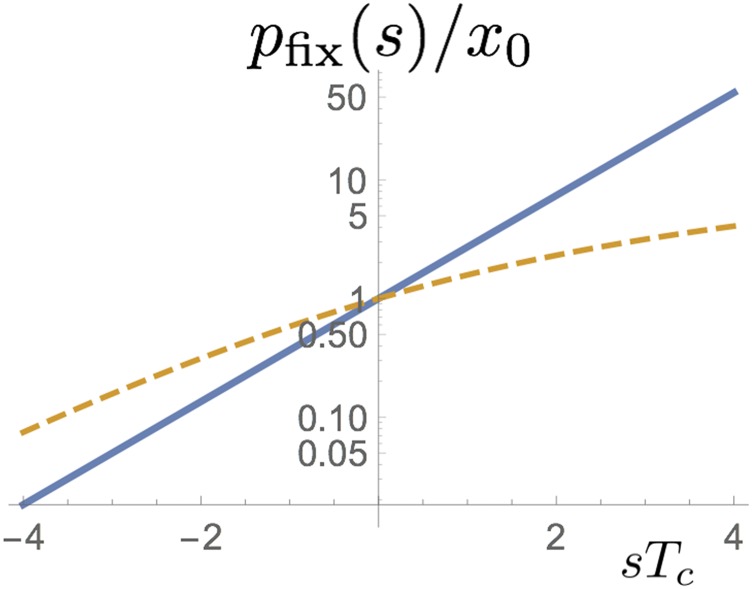
The dynamics under selection turns out to be strongly shaped by the competition between genuine and fictitious selection. For a beneficial mutation to reach fixation, allele frequencies need to become large enough by chance for the frequency-dependent fictitious selection to succumb to constant positive selection. This causes a low probability of fixation compared to a Wright–Fisher model with the same population size. By contrast, if one chooses to compare with a Wright–Fisher model that has the same coalescence time (or variance effective population size), the fixation probability with jackpot events is larger for all values of see Figure 10. Such a comparison makes sense if is known for a given population, say from its pairwise nucleotide diversity, and one wants to explore allele-frequency dynamics under different effective offspring distributions. The observation that increases with broader offspring numbers for fixed agrees qualitatively with the numerical results of Der et al. (2012). Note, however, that this statement relies on fixing If, instead, one fixes the census population sizes, fixation probabilities always go down with broader offspring numbers, simply because success becomes more a matter of luck (of drawing large family sizes) than of fitness.
Figure 10.
The probability of fixation of an allele initially at frequency of as a function of in our model with Luria–Delbrück jackpot events (solid) and Wright–Fisher diffusion (dashed).
I have also considered the case of balancing selection, to see under which conditions it may be able to tame the disruptive force of fictitious selection. One finds an interesting threshold phenomenon. Only if balancing selection is strong enough will both alleles coexist. In the case of coexistence, the allele-frequency distribution is in general off-centered with respect to the fitness optimum. Since fictitious selection scales as one can also conclude that polymorphisms are stably maintained at a given strength of balancing selection if the population size is large enough.
Mapping to models of adaptation and other types of noisy traveling waves
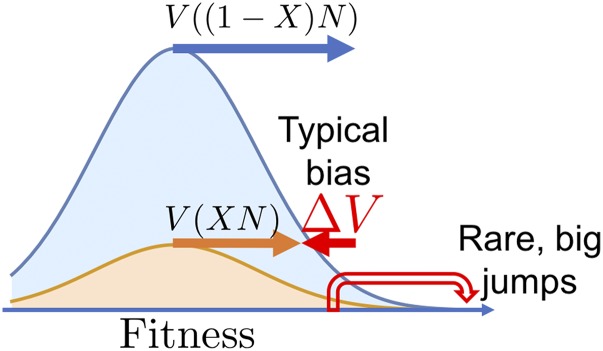
Even though some marine species have a surprisingly skewed offspring-number distribution (Eldon and Wakeley 2006), it is unlikely to be of the type with logarithmically diverging mean, on which I have focused in the present article. But our analysis serves as a coarse-grained picture of population models from which such an effective offspring-number distribution emerge. Large effective offspring numbers arise in population models that generate traveling waves: the most advanced individuals in the tip of a traveling wave generate an exponentially larger number of descendants than individuals in the bulk of the populations. A classic example are Fisher–Kolmogorov waves, which have been used to describe range expansions of species or the spread of epidemics. Recent evolutionary theory has repeatedly shown how similar waves arise in models of rampant adaptation in both asexual and sexual populations, where a traveling bell-like shape describes the gradual increase in fitness, as illustrated in Figure 11 (Neher 2013; Neher et al. 2013; Weissman and Hallatschek 2014).
Figure 11.
The combination of a typical bias against the minority allele and rare compensating jumps can be appreciated directly in models of adaptation. A subpopulation of mutants at frequency will typically fall behind the wild-type population because of a speed differential between wild type and mutants resulting from the population-size dependence of the speed of adaptation. Neutrality is restored by rare leap-frog events by which highly fit mutants overtake the most-fit wild types.
In all of these models of traveling waves [more precisely “pulled” waves (van Saarloos 2003)], the effective descendant number follows the characteristic jackpot distribution, when integrated over an appropriately chosen intermediate timescale. For instance, in traveling waves of the Fisher–Kolmogorov type, this characteristic time is of the order microscopic generations, which represents the time lineages need to diffusively mix within the wave tip consisting of individuals (K, r, and D are the carrying capacity, growth rate, and diffusivity, respectively). Hence, resampling from the offspring-number distribution occurs once every microscopic generations, setting the effective generation time in our discrete population model. This implies that the coalescence time of Fisher–Kolomogorov waves should scale as which indeed is by now well established (Brunet et al. 2007).
The mapping to traveling waves provides another intuitive interpretation of the fictitious selection force, as illustrated in Figure 11 using the examples of traveling fitness waves as they arise in models of rampant adaptation. The key point is that a subpopulation of a neutral mutation at low frequency X approximately resembles a traveling wave with lower population size and, usually, moves with a correspondingly lower speed compared to the total population. In all known pulled waves (those that generate descendant distributions of the Luria–Delbrück type), the speed differential asymptotically approaches as the population size is increased, where the function decays slowly (logarithmically) with population size. The speed differential between subpopulation and the entire population will lead to a continual reduction in the frequency of the subpopulation in the tip of the wave, described by a fictitious selection term of the type identified in this study. Neutrality is preserved only by rare jumps whereby individuals in the tip of the subpopulation move anomalously far ahead. These rare jumps also control the diffusion constant of noisy traveling waves (Brunet 2016) and presumably in other types of pulled waves as well.
In this traveling-wave picture, it also becomes clear how genuine selection for mutants arises: suppose a population entirely consisting of mutants has a wave speed relation compared to the wave speed of the wild type. In a situation where mutants are at frequency X, and wild type at frequency one will then have the speed differential Range expansions, for instance, are accelerated by the fixation of mutations that increase the linear growth rate, the dispersal rate, or by mutations that broaden the dispersal kernel (Hallatschek and Fisher 2014). In models of adaptation, the rate of adaptation can be increased through mutations that increase the mutation rate (by mutator alleles) or the frequency of beneficial mutations (potentiating mutations).
Potential significance of our results on balancing selection
The results on balancing selection may be useful to get some intuition on the coexistence of two eco-types in an overall adapting population, as has been repeatedly found in evolution experiments even with deliberately simple environments. The stable maintenance of a polymorphism requires balancing selection to be strong enough to overcome the fictitious selection force. A balancing selection term may also serve as a simple way to model some generic roughness of the fitness landscape. Imagine, for instance, a high-dimensional fitness landscape where continual adaptation requires one to move along fitness plateaus, or even across valleys, rather than always following the steepest uphill direction. In such a landscape, following the steepest uphill direction will generate large frequencies on the short run. On longer timescales, however, these lineages will have a harder time to extend their uphill paths, which slows their speed of adaptation. This leads to a negative frequency-dependent term, which in a Taylor-expansion sense could be captured by the term in Equation 40. This, of course, is just a hypothesis and should be backed up by evolution simulations in a high-dimensional fitness landscape.
Link to disordered systems
Since the Bolthausen–Sznitman coalescent was first identified in spin glass models, one may wonder what the above forward-in-time process implies for these statistical mechanics problems. The significance can be appreciated at least in very simple mean-field models of spin glasses or polymers in random media, which can be mapped onto traveling waves (Derrida and Spohn 1988). Increasing time corresponds to increasing the length of the spin chains and of the polymer, respectively. The precise mapping from chain length to time depends on whether the number of relevant chain conformations stays constant or increases (possibly exponentially) with chain length. The average of the logit variable, maps (up to prefactors) onto a difference in disorder-averaged free energy between two parts of phase space. The runaway of this average, (Equation 14), then represents the phenomenon of ergodicity breaking: an ever-increasing free energy barrier between phase-space regions as the system tends to the thermodynamic limit.
Acknowledgments
I thank Jason Schweinsberg, Eric Brunet, and Benjamin H. Good for in-depth discussions and a critical reading of the manuscript. I also thank the Kavli Institute for Theoretical Physics in Santa Barbara for providing an inspiring environment, which allowed me to finalize this work. Research reported in this publication was supported by a National Science Foundation Career Award (#1555330) and by a Simons Investigator Award from the Simons Foundation (#327934).
Appendix A: Brief Note On “Typicality”
The notion of typicality arises formally when one applies the central limit theorem to the logarithm of the probability of drawing a given sample This log probability is narrowly distributed about the most-likely probability if the sample size is large enough. The typical set are all those samples [ many, where is the Shannon entropy of U] that have probability close to that most-likely one. The outcome of a randomly drawn sample almost certainly belongs to this typical set as one increases the sample size n, which in our case scales with the population size N. More details on the definition of typicality can be found, e.g., in section 4.4 of MacKay (2003). I use to denote the expectation of a random variable Z. The notation is used for the mean of the random variable P in a typical sample of size n, as defined in Equation 2 for the case of offspring numbers.
Appendix B: The Transition Density
As I have pointed in the main text, the jump kernel for an offspring-number distribution is stationary in logit space, such that the jump rate from ψ to only depends on their difference, This pleasant feature enables the use of a Fourier transform to convert convolutions involving the jump kernel into a simple product. I will use this strategy here to solve the differential Chapman–Kolmogorov equation Equation 29 to obtain the probability density that a trajectory moves from to ψ in the time period τ.
In terms of the Fourier transform
| (B1) |
Equation 29 takes the form
| (B2) |
where I have introduced
| (B3) |
We apply the method of characteristics to solve this linear partial differential equation: Introduce and and rewrite Equation B2 as
| (B4) |
which is easily solved by
| (B5) |
| (B6) |
| (B7) |
| (B8) |
Since we need to choose the initial condition and obtain
| (B9) |
| (B10) |
A Fourier back transform yields the propagator in Equation 13.
Appendix C: Duality
In the main text, we have seen that the rate of change of the first moment of the frequency of a neutral mutation vanishes for the forward-in-time process defined by Equation 21 and Equation 24, as required by the neutrality of the process. I will now analyze the rate of change of the higher moments: they are characteristic of the ensuing genealogical process, allowing us to confirm the duality of the process and the Bolthausen–Sznitman coalescent.
Multiplying Equation 24 by and integrating yields
| (C1) |
| (C2) |
In going from the first to the second line, I used and, in the first term, integration by parts. The remaining integral has an expression in terms of a power series in
| (C3) |
where the coefficients are given by
| (C4) |
Nonincidentally, represents precisely the rate at which a given set of lineages in a sample of lineages coalesce in the Bolthausen–Sznitman coalescent.
In fact, relation Equation C3 shows directly that our process describes the evolution of a subpopulation forward in time, whose ancestral lineages coalesce according to the Bolthausen–Sznitman coalescent backward in time. This remarkable duality relation can be intuitively understood as follows. Suppose we sample at random n individuals at time The probability that all sampled individuals are mutants is given by On the other hand, we can imagine tracing the lineages by backward in time. We then have in the likely case that no coalescence occurred in if two lineages coalesced, if three lineages coalesced, and so on (multiple coalescence events can be ignored as in the Bolthausen–Sznitman coalescent). Using the probability that k lineages coalesce, the expected rate of change can thus be represented as a power series in which yields Equation C3.
Appendix D: Backward Equation and Link to Λ-Fleming–Viot Generator
The generator of the differential Chapman–Kolmogorov equation Equation 24 for the transition density acts on the variables characterizing the allele frequencies in the final state. An equivalent backward equation can be obtained using the adjoined generator. In terms of the kernel this backward equation reads
| (D1) |
This backward equation can be derived from the identity in the limit The operator appearing on the right-hand side is the adjoined operator to the one on the right-hand side of the forward equation Equation 24.
Our process is dual to the Bolthausen–Sznitman coalescent and, as such, belongs to the larger class of so-called Λ-Fleming–Viot processes. The generator of the above backward equation is in the literature on Λ-Fleming–Viot processes usually presented in a somewhat different form, in which the drift term is less evident. In our notation, the standard formulation reads (see, e.g., Etheridge et al. 2010; Griffiths 2014)
| (D2) |
It is not immediately evident that the generators on the right-hand sides of both equations are identical, but they in fact are. I show how Equation D1 emerges from Equation D2. First note that the integral on the right-hand side of Equation D2 can be rewritten as
| (D3) |
| (D4) |
| (D5) |
which can be split into two parts:
| (D6) |
with
| (D7) |
| (D8) |
Next, we change the integration variables. In A, we use running from to 1. In B, we use running from 0 to These substitutions yield
| (D9) |
| (D10) |
Obviously, adding both terms yields a single integral running from to Moreover, the last term can be split off as an advection term if the remaining integral is interpreted in terms of the Cauchy principle value (PV),
| (D11) |
| (D12) |
establishing
Appendix E: Resampling Distribution
Here, I determine the distribution that describes how allele frequencies change from one generation to the next in the large N limit, in which we can ignore the binomial sampling error.
The resampling distribution is fully characterized by the probability of the event (the allele frequency in generation is y) given (the allele frequency x in generation t). We can write this probability as
where the average is taken over the distributions of the statistical weights M of mutants and W of wild types, respectively, given the initial frequency x. The main challenge of our analysis stems from the fact that we are looking for the distribution of the ratio rather than M itself. M is a sum of many independent random variables, which approaches well-known limiting distributions that depend on the tail of the offspring-number distribution. In our case, where M follows the Landau distribution (see below). However, M itself is not properly normalized and, in our case, has a diverging mean and variance. The frequency is instead well behaved in that it has a finite variance and finite mean equal to starting frequency x. Its limiting distribution is to our knowledge not known, but can be related to the Landau distribution as we now show.
Before we compute the resampling distribution, it is convenient to express the Dirac Δ function in terms of its Fourier transform:
| (E1) |
| (E2) |
| (E3) |
| (E4) |
where we substituted and introduced which is (up to a constant) the reverse cumulative distribution.
Since M and W are independently drawn, the average factorizes and we can write as
| (E5) |
where I introduced the characteristic function
of the distribution of M and used the fact that the distribution of W can be obtained from the distribution of X by replacing which implies Once we have figured out the average for large N, can be determined by integration.
But obtaining the characteristic function is standard: recall that M is the sum of random variables each distributed according to a power law According to the generalized central limit theorem of Gnedenko and Kolmogorov (1954), the distribution of M must thus tend for large N to a scaled version of the so-called Landau distribution, which is the Lévy α-stable distribution with stability and skewness parameters both equal to one. Equivalently, the characteristic function of M must for large N approach the one of the Landau distribution up to a stretching factor,
| (E6) |
A heuristic derivation of the characteristic function of the Landau distribution is provided in Appendix F. It is noteworthy that the probability density corresponding to has a peak at and the peak has a width of order So, as we increase the population size, the peak becomes increasingly sharper relative to its position. This behavior is responsible for the asymptotic vanishing of the diffusion coefficient (see below).
Inserting in Equation E5 leads to
| (E7) |
where I substituted
Since for and the integral of the imaginary part of the integrand vanishes, we can rewrite this integral as
| (E8) |
The remaining integral can only be evaluated numerically, as shown in Figure 3.
Large N Limit
In the limit we can simplify the last integral in Equation E8 further: unless y is very close to x, the integrand is rapidly oscillating because of the first term in the argument of the sine function multipling a factor of These oscillations cutoff the integration at σ values of order which yields integral values of order The integral becomes nonvanishing in the large N limit only when x is very close to y. To leading order, we can therefore replace y by x in the subdominant terms inside the argument of the sine function, leading to
| (E9) |
The remaining integral reduces to an elementary expression in terms of an inverse tangent, which to leading order in is given by
| (E10) |
in terms of the shift
| (E11) |
The probability distribution of the increments reads
| (E12) |
| (E13) |
and approaches in the limit a stretched and shifted Cauchy distribution
| (E14) |
where is restricted to the interval and is the scale factor
| (E15) |
It is now straightforward to evaluate the limits in Equation 18, Equation 19, and Equation 20 for the jump kernel , advection speed and diffusivity D, respectively,
| (E16) |
| (E17) |
| (E18) |
The second and third limits follow from being ever more sharply peaked at as
The triplet controls the allele-frequency dynamics in the continuous time limit, as discussed in the main text.
Appendix F: Heuristic Calculation of the Laplace Transform of the Landau Distribution
Here, I provide a purely heuristic calculation of the Laplace transform of the sum
| (F1) |
| (F2) |
| (F3) |
| (F4) |
| (F5) |
Here, the variable stands for the offspring number of the ith individual. In the first line, I integrate over the offspring number distribution for In the third line, I replaced the upper integration interval by knowing that larger P-values are cut off by the decaying exponential. In the fourth and fifth line, I assumed Note that the resulting Equation F5 is a stretched version of the Laplace transform of the Landau distribution. The characteristic function of U follows from an analytic continuation
Footnotes
Communicating editor: J. Wakeley
Literature Cited
- Baake E., Lenz U., Wakolbinger A., 2016. The common ancestor type distribution of a lambda-wright-fisher process with selection and mutation. Electron. Commun. Probab. 21: 1–16. [Google Scholar]
- Berestycki N., 2009. Recent progress in coalescent theory. Ensaios Matematicos 16: 1–193. [Google Scholar]
- Bertoin J., Le Gall J.-F., 2003. Stochastic flows associated to coalescent processes. Probab. Theory Relat. Fields 126: 261–288. 10.1007/s00440-003-0264-4 [DOI] [Google Scholar]
- Bollback J. P., York T. L., Nielsen R., 2008. Estimation of 2Nes from temporal allele frequency data. Genetics 179: 497–502. }{\\}{ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolthausen E., Sznitman A.-S., 1998. On ruelle’s probability cascades and an abstract cavity method. Commun. Math. Phys. 197: 247–276. 10.1007/s002200050450 [DOI] [Google Scholar]
- Brunet, É., 2016 Some aspects of the Fisher-KPP equation and the branching Brownian motion. Statistical Mechanics [cond-mat.stat-mech]. UPMC.
- Brunet É., Derrida B., 2013. Genealogies in simple models of evolution. J. Stat. Mech. 2013: P01006 10.1088/1742-5468/2013/01/P01006 [DOI] [Google Scholar]
- Brunet É., Derrida B., Mueller A. H., Munier S., 2007. Effect of selection on ancestry: an exactly soluble case and its phenomenological generalization. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 76: 041104 10.1103/PhysRevE.76.041104 [DOI] [PubMed] [Google Scholar]
- Coop G., Griffiths R. C., 2004. Ancestral inference on gene trees under selection. Theor. Popul. Biol. 66: 219–232. 10.1016/j.tpb.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Crow J. F., Kimura M., 1970. An Introduction to Population Genetics Theory. Burgess Publishing Company, Minneapolis. [Google Scholar]
- Der R., Plotkin J. B., 2014. The equilibrium allele frequency distribution for a population with reproductive skew. Genetics 196: 1199–1216. 10.1534/genetics.114.161422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Der R., Epstein C., Plotkin J. B., 2012. Dynamics of neutral and selected alleles when the offspring distribution is skewed. Genetics 191: 1331–1344. 10.1534/genetics.112.140038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrida B., Spohn H., 1988. Polymers on disordered trees, spin glasses, and traveling waves. J. Stat. Phys. 51: 817–840. 10.1007/BF01014886 [DOI] [Google Scholar]
- Desai M. M., Walczak A. M., Fisher D. S., 2013. Genetic diversity and the structure of genealogies in rapidly adapting populations. Genetics 193: 565–585. 10.1534/genetics.112.147157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly P., Kurtz T. G., 1999. Particle representations for measure-valued population models. Ann. Probab. 27: 166–205. 10.1214/aop/1022677258 [DOI] [Google Scholar]
- Durrett R., Schweinsberg J., 2005. A coalescent model for the effect of advantageous mutations on the genealogy of a population. Stochastic Process. Appl. 115: 1628–1657. 10.1016/j.spa.2005.04.009 [DOI] [Google Scholar]
- Eldon B., Wakeley J., 2006. Coalescent processes when the distribution of offspring number among individuals is highly skewed. Genetics 172: 2621–2633. 10.1534/genetics.105.052175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge A. M., Griffiths R. C., Taylor J. E., 2010. A coalescent dual process in a moran model with genic selection, and the lambda coalescent limit. Theor. Popul. Biol. 78: 77–92. 10.1016/j.tpb.2010.05.004 [DOI] [PubMed] [Google Scholar]
- Fisher R., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford. }{\\}{ [Google Scholar]
- Foucart C., 2013. The impact of selection in the lambda-wright-fisher model. Electron. Commun. Probab. 18: 72 [corrigenda: Electron. Commun. Probab. 19: 15 (2014)]. [Google Scholar]
- Fusco D., Gralka M., Kayser J., Anderson A., Hallatschek O., 2016. Excess of mutational jackpot events in expanding populations revealed by spatial luria–delbrück experiments. Nat. Commun. 7: 12760 10.1038/ncomms12760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner C., 1985. Stochastic Methods (Springer Series in Synergetics). Springer-Verlag, Berlin. [Google Scholar]
- Gnedenko B., Kolmogorov A., 1954. Independent Random Variables. Addison-Wesley, Cambridge, MA. [Google Scholar]
- Griffiths R. C., 2014. The λ-fleming-viot process and a connection with wright-fisher diffusion. Adv. Appl. Probab. 46: 1009–1035. 10.1239/aap/1418396241 [DOI] [Google Scholar]
- Hallatschek O., Fisher D. S., 2014. Acceleration of evolutionary spread by long-range dispersal. Proc. Natl. Acad. Sci. USA 111: E4911–E4919. 10.1073/pnas.1404663111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth C. J., Parts L., Schiffels S., Liti G., Mustonen V., 2011. Quantifying selection acting on a complex trait using allele frequency time series data. Mol. Biol. Evol. 29: 1187–1197. 10.1093/molbev/msr289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingman J. F. C., 1982. The coalescent. Stochastic Process. Appl. 13: 235–248. 10.1016/0304-4149(82)90011-4 [DOI] [Google Scholar]
- Kosheleva K., Desai M. M., 2013. The dynamics of genetic draft in rapidly adapting populations. Genetics 195: 1007–1025. 10.1534/genetics.113.156430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapivsky P. L., Redner S., Ben-Naim E., 2010. A Kinetic View of Statistical Physics. Cambridge University Press, Cambridge, UK: 10.1017/CBO9780511780516 [DOI] [Google Scholar]
- Krone S. M., Neuhauser C., 1997. Ancestral processes with selection. Theor. Popul. Biol. 51: 210–237. 10.1006/tpbi.1997.1299 [DOI] [PubMed] [Google Scholar]
- Landau L. D., 1944. On the energy loss of fast particles by ionization. J. Phys. 8: 201–205. [Google Scholar]
- Levy S. F., Blundell J. R., Venkataram S., Petrov D. A., Fisher D. S., et al. , 2015. Quantitative evolutionary dynamics using high-resolution lineage tracking. Nature 519: 181–186. 10.1038/nature14279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Hedgecock D., 1998. Genetic heterogeneity, detected by pcr-sscp, among samples of larval pacific oysters (crassostrea gigas) supports the hypothesis of large variance in reproductive success. Can. J. Fish. Aquat. Sci. 55: 1025–1033. 10.1139/f97-312 [DOI] [Google Scholar]
- Luria S. E., Delbrück M., 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28: 491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKay D. J., 2003. Information Theory, Inference and Learning Algorithms. Cambridge University Press, Cambridge, UK. [Google Scholar]
- May A., 1967. Fecundity of atlantic cod. J. Fish. Res. Board Can. 24: 1531–1551. 10.1139/f67-127 [DOI] [Google Scholar]
- Neher R. A., 2013. Genetic draft, selective interference, and population genetics of rapid adaptation. Annu. Rev. Ecol. Evol. Syst. 44: 195–215. 10.1146/annurev-ecolsys-110512-135920 [DOI] [Google Scholar]
- Neher R. A., Hallatschek O., 2013. Genealogies of rapidly adapting populations. Proc. Natl. Acad. Sci. USA 110: 437–442. 10.1073/pnas.1213113110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neher R. A., Kessinger T. A., Shraiman B. I., 2013. Coalescence and genetic diversity in sexual populations under selection. Proc. Natl. Acad. Sci. USA 110: 15836–15841. 10.1073/pnas.1309697110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauser C., Krone S. M., 1997. The genealogy of samples in models with selection. Genetics 145: 519–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosthuizen E., Daan N., 1974. Egg fecundity and maturity of north sea cod, gadus morhua. Neth. J. Sea Res. 8: 378–397. 10.1016/0077-7579(74)90006-4 [DOI] [Google Scholar]
- Otto S. P., Day T., 2011. A Biologist’s Guide to Mathematical Modeling in Ecology and Evolution. Princeton University Press, Princeton, NJ. [Google Scholar]
- Pitman J., 1999. Coalescents with multiple collisions. Ann. Probab. 27: 1870–1902. 10.1214/aop/1022874819 [DOI] [Google Scholar]
- Schweinsberg J., 2003. Coalescent processes obtained from supercritical galton–watson processes. Stochastic Process. Appl. 106: 107–139. 10.1016/S0304-4149(03)00028-0 [DOI] [Google Scholar]
- Schweinsberg J., 2017a Rigorous results for a population model with selection i: evolution of the fitness distribution. Electron. J. Probab. 22: 37. [Google Scholar]
- Schweinsberg J., 2017b Rigorous results for a population model with selection ii: genealogy of the population. Electron. J. Probab. 22: 38. [Google Scholar]
- van Saarloos W., 2003. Front propagation into unstable states. Phys. Rep. 386: 29–222. 10.1016/j.physrep.2003.08.001 [DOI] [Google Scholar]
- Weissman D. B., Hallatschek O., 2014. The rate of adaptation in large sexual populations with linear chromosomes. Genetics 196: 1167–1183. 10.1534/genetics.113.160705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D. B., Hallatschek O., 2017. Minimal-assumption inference from population-genomic data. eLife 6: e24836 10.7554/eLife.24836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E. S., Vetsigian K., 2018. Stochastic exits from dormancy give rise to heavy-tailed distributions of descendants in bacterial populations. bioRxiv 246629. DOI https//.org/10.1101/246629. [DOI] [PubMed] [Google Scholar]
- Yule G. U., 1925. A mathematical theory of evolution, based on the conclusions of Dr. JC Willis, FRS. Philos. Trans. R. Soc. Lond., B. 213: 21–87. 10.1098/rstb.1925.0002 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The author states that all data necessary for confirming the conclusions presented in the article are represented fully within the article.