Abstract
Titin (TTN) has multifunctional roles in sarcomere assembly, mechanosignaling transduction and muscle stiffness. TTN splicing generates variable protein sizes with different functions. Therefore, understanding TTN splicing is important to develop novel treatment for TTN-based diseases. The I-band TTN splicing regulated by RBM20 have been extensively studied. However, the Z- and M-band splicing and regulation remain poorly understood. Herein, we aimed to define the Z- and M-band splicing in striated muscles, and determined whether RBM20 regulates the Z- and M-band splicing. We discovered four new Z-band TTN splicing variants, and one of them dominates in mouse, rat, sheep and human heart. But only one form can be detected in frog and chicken hearts. In skeletal muscles, three new Z repeats (Zr) were detected, and Zr4–6 exclusion dominates in the fast muscles, whereas Zr4 skipping dominates in the slow muscle. No developmental changes were detected in the Z-band. In the M-band, two new variants were discovered with alternative 3’ splice site in exon363 (Mex5) and alternative 5’ splice site in intron362. However, only sheep heart expresses two new variants rather than other species. Skeletal muscles express three M-band variants with altered ratios of Mex5 inclusion to Mex5 exclusion. Lastly, we revealed that RBM20 does not regulate the Z- and M-band splicing in the heart, but does in skeletal muscles. Taken together, we characterized the Z- and M-band splicing and provided the first evidence of the role of RBM20 in the Z- and M-band TTN splicing.
Keywords: TTN, RBM20, Z-BAND, M-BAND, ALTERNATIVE SPLICING, HEART DISEASE
Introduction
TTN is the third most abundant protein in vertebrate striated muscles (cardiac and skeletal muscles) (Labeit et al., 1997). It spans each half sarcomere from the Z-band to the M-band located in the center of the sarcomere (Furst et al., 1988; Labeit et al., 1995), with its carboxy (C)-terminal cross-linking to another C-terminal in the M-band, and its amino (N)-terminal attaching another N-terminal from adjacent sarcomere in the Z-band (Trinick et al., 1999; Obermann et al., 1996; Gregorio et al., 1998). This arrangement allows TTN molecules to form a continuous system along the myofibril and enables it to function as a molecular blueprint for maintenance of sarcomere integrity and precise assembly of the regulatory, contractile, and structural sarcomere proteins (Tskhovrebova et al., 2003; Granzier et al., 2004; Freiburg et al., 1996). TTN, the largest protein found in cardiac and skeletal muscle is increasingly regarded as one of the important molecular origins of cardiomyopathy and heart failure (Linke et al., 2014; Yin et al., 2015), and plays a critical role in elastic recoil of the cardiac myocytes and contributes to diastolic function during the left ventricular filling phase due to its elasticity (Granzier et al., 2004; Herman et al., 2012; LeWinter et al., 2014; Taylor et al., 2011). Previous studies have indicated that TTN elasticity is highly variable which is largely attributed to the ratio changes of TTN isoforms as a result of alternative splicing (Makarenko et al., 2004; Guo et al., 2018).
TTN has two major classes of isoforms in cardiac muscle, N2B and N2BA, and one class of isoform known as N2A with variable sizes in skeletal muscle as a result of alternative splicing (Labeit et al., 1995; Freiburg et al., 2000; Cazorla et al., 2000; Bang et al., 2001). These isoforms are mainly generated from alternative splicing of the I-band exons encoding the middle immunoglobulin (Ig) region and PEVK (proline (P), glutamate (E), valine (V), and lysine (K) residues) region (Zhu et al., 2017; Greaser et al., 2008; Guo et al., 2012; 2013; Li et al., 2012; 2013). The major splicing pathways in the middle Ig domain have been well characterized (Zhu et al., 2017; Rexiati et al., 2018). Recently, a splicing factor called RNA binding motif 20 (RBM20) has been identified that regulates TTN middle Ig exon splicing (Guo et al., 2012; Li et al., 2013; Methawasin et al., 2014). Furthermore, in addition to the TTN middle Ig and PEVK domains, the TTN Z-band and M-band also underwent alternative splicing. In the Z-band, the first 28 TTN exons encode nine Z-band domains (Z1-Z9) with large inter-domain insertions, and there are seven alternatively spliced repeats between Z3 and Z4 termed Zr1–7. These seven repeats are encoded by seven exons from exon 8 to exon 14. Alternative splicing of these seven exons leads to variation in the differential expression of Z-repeats from four to seven Z repeats. Current studies in the human heart and skeletal muscles showed that in the adult heart, all seven repeats are included, but Zr4–6 is skipped in the fetal heart (Guo et al., 2010; 2018; Gautel et al., 1996; Linke et al., 2008; Sorimachi et al., 1997). In the slow muscle soleus, two variants have been identified, with one excluding Zr4–6 and the other skipping Zr6 (Fig. S1A) (Guo et al., 2018; Gautel et al., 1996; Linke et al., 2008; Sorimachi et al., 1997). The M-band consists of only six exons from 359 to 364, which are also termed M-band exons 1 to 6 (Mex1 to Mex6). Interestingly, only Mex5 was found an alternatively used exon. It is not frequently spliced in cardiac muscle, but is in skeletal muscle (Fig. S1B) (Guo et al., 2010; Granzier et al., 2002; Kolmerer et al., 1996). However, the splicing pattern of the Z-band and M-band has not been well characterized. Especially, it is completely unknown whether the recently identified splicing factor RBM20 regulates the splicing of the Z-band and M-band domain. In this study, we characterized the splicing pattern of the Z-band and M-band exons across developmental stages, species and muscle types, and examined whether RBM20 is a major regulator in the splicing events of the Z-band and M-band using a Rbm20 knockout (KO) rat model in cardiac muscle as well as in skeletal muscle.
Methods and Materials
Experimental animals and tissue samples
We studied wild type (WT) and homozygous Rbm20 KO rats. The KO rats were derived from a previously described spontaneous mutant (Makarenko et al., 2004; Guo et al., 2012). Rats were crosses of Sprague-Dawley (SD) X Brown Norway (BN). All strains were originally obtained from Harlan Sprague Dawley, Indianapolis, IN. Mice and rats were maintained on standard rodent chow. All animals were maintained in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All procedures pertaining to animals were approved by the Institutional Animal Use and Care Committee of the University of Wyoming. Sheep heart and skeletal muscle samples were obtained from the Center for Fetal Programming at University of Wyoming. Chicken heart tissues were obtained from Dr. Paul Mozdziak at North Carolina State University and frog heart tissues were provided by Dr. Dan Levy at the University of Wyoming. The RNA sample of human left ventricular tissue from a de-identified donor heart was obtained from Dr. Ying Ge at University of Wisconsin-Madison with approval from the Institutional Review Board. Tissue samples with developmental stages were collected from fetal (late gestation) and adult animals, and all other tissues used in this study were from adult animals. All tissue samples were snap-frozen in liquid nitrogen and stored at −800C.
Z- and M-band genomic sequence analysis of multiple species
TTN gene sequences of different species were obtained through gene database of either University of California Santa Cruz (UCSC, http://genome.ucsc.edu/) or Ensembl (http://www.ensembl.org/index.html) genome browsers or National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) nucleotide database. The investigated species were human (Homo sapiens, GRCh38), mouse (Mus musculus, GRCm38), rat (Rattus norvegicus, Rnor_6), chicken (Gallus gallus, Gallus_gallus-5), frog (Xenopus laevis, Xenopus_laevis_v2), and sheep (Ovis aries, Oar v4.0). The sequences of around Z-band and M-band splicing region were retrieved by basic local alignment search tool (BLAST). All assembled Z-band and M-band sequences were manually edited and checked for consistency. Multiple sequence alignments were processed with ClustalX (Jeanmougin et al., 1998).
RT-PCR and DNA gel electrophoresis
Total RNAs from human, sheep, mouse, rat, chicken and frog heart samples were isolated with Trizol (Invitrogen, USA) and further treated with RQ1 RNase-free DNase (Promega, USA) to remove genomic DNA contamination. Reverse transcriptase (RT) reactions were carried out using ImProm-II Reverse Transcription System (Promega, USA) with random primers. Standard RT-PCR reactions were carried out using the C1000 Thermal Cycler (BIO-RAD, USA). Primer pairs for amplification of the Z- and M-band splicing sequence transcripts were listed in the supplement table 1 (Table S1). PCR products were analyzed with DNA agarose gel electrophoresis. DNA gel was stained with ethidium bromide and visualized under UV light. The gel images were captured with ChemiDoc Imaging System (Bio-Rad). DNA band density was quantified with NIH ImageJ (Schneider et al., 2012).
DNA sequencing
From the gel electrophoresis, the bands of interest were excised with blade and purified with Wizard SV gel and PCR clean-up system (Promega). Purified DNA bands were sent out and sequenced to confirm their identity with service provided by Eurofins Genomics.
Statistics
GraphPad prism software was used for statistical analysis. Results were expressed as means ± SEM. Three replicates were used in each group. The significance threshold was set at P<0.05
Results
Z-band TTN splicing pattern in cardiac muscle of multiple species
The Z-band TTN splicing occurs in the Z repeat (Zr) region from Zr1 to 7. Previous studies determined the splicing pattern of the Z repeats in human heart, and indicated that three splicing variants are present in the human heart with fully expressed seven Z repeats, four repeats with Zr4–6 skipping and six repeats with Zr6 exclusion (Fig. S1A). Here, we examined the Z-band splicing pattern in multiple species including the frog, chicken, mouse, rat, sheep as well as human using primers spanning Zr1–7, Zr1–3, Zr3–7, Zr3–5 and Zr4–6. Using primer Zr1–7 that covers all seven Z repeats, we found that multiple splicing variants exist in mouse, rat, sheep and human, but only one major splicing variant was observed in frog and chicken (Fig. 1A). Sequencing results indicated that the major splicing form in frogs and chicken is four repeats with Zr4–6 skipping (V5) and six repeats with Zr4 skipping (V2) respectively (Fig. 1D). In other species, we designed the primers covering Zr1–3 and Zr3–7, and performed PCR in human, sheep and rat. The results showed that no splicing occurs in first three repeats (Zr1–3), but does in last four repeats (Zr3–7) (Fig. 1B), which is consistent with previous findings (Guo et al., 2010; 2018; Gautel et al., 1996; Linke et al., 2008; Sorimachi et al., 1997). To further test which repeats are spliced in and out in the last four repeats, we performed RT-PCR using primers spanning Zr3–5 and Zr4–6, and observed multiple bands in DNA agarose gel, indicating alternative splicing occurs from repeats 3 to 5 as well as from 4 to 6 (Fig. 1C). All major bands (labeled with V plus numbers) were sequenced and the sequencing results demonstrate that the splicing patterns in mouse, rat, sheep and human from upper band to lower band in Fig. 1A are seven repeats (V1), six repeats with Zr4 skipping (V2), six repeats with Zr5 skipping (V3), five repeats with Zr4–5 skipping (V4), and four repeats with Zr4–6 skipping (V5) (Fig. 1D). From these results, it can be seen that the major splicing variants in each species are different. For example, only six repeats and four repeats were observed in the chicken and frog respectively; seven repeats and six repeats with Zr4 skipping are predominantly expressed in the mouse; six repeats with Zr4 skipping is predominantly expressed in the rat and sheep, and a nearly equal level of seven repeats, six repeats with Zr4 skipping, five repeats with Zr4–5 skipping and four repeats with Zr4–6 skipping are present in human heart (Fig. 1A). Most importantly, three new Z-band splicing variants (six repeats with Zr4 skipping (V2), six repeats with Zr5 skipping (V3), and five repeats with Zr4–5 skipping (V4)) were identified in this study by comparing to previous reports (Guo et al., 2010; Gautel et al., 1996; Linke et al., 2008; Sorimachi et al., 1997).
Figure 1.
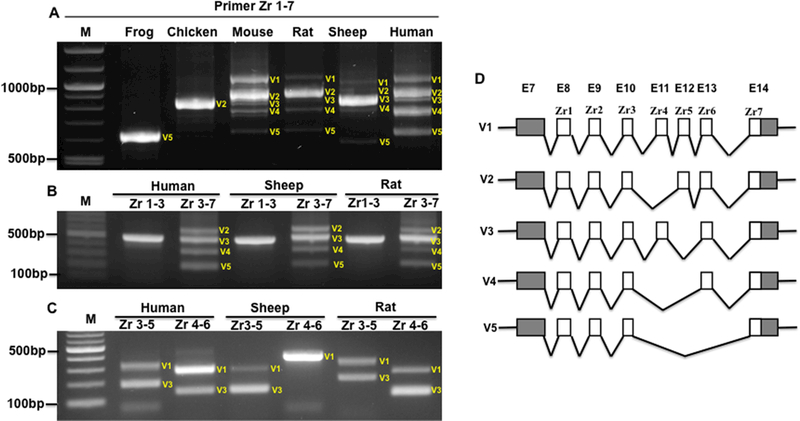
Z-band splicing in the adult heart across multiple species. A, PCR products with primers spanning Zr1 to 7 across species; B, PCR products with primers spanning Zr1 to 3, and Zr3 to 7 in human, sheep and rat heart; C, PCR products with primers spanning Zr3 to 5, and Zr4 to 6 in human, sheep and rat heart; D, Schematic diagram of splicing pattern of Z repeats based on PCR products and sequencing results across species; Zr, Z repeats; M, 1kb DNA marker; E, exon. White box, alternative exons; Grey box, exon; V, variant.
Z-band TTN splicing pattern in different skeletal muscle types
Next, we determined the splicing pattern of the Z-band in distinct muscle types in rat. RT-PCR was performed in the longissimus muscle (LM), tibialis anterior (TA), extensor digitorum longus (EDL), gastrocnemius (GAS) and soleus (SOL) using primers spanning Zr1–7. The agarose gel analysis detected three splicing variants (Fig. 2A). To identify which Z repeats are frequently spliced, we employed primers spanning Zr1–3 and Zr3–7 to amplify GAS and SOL. The results indicated that the repeats Zr1–3 does not undergo splicing, whereas splicing only occurs in fast muscle type GAS, but not in the slow muscle type SOL with primer Zr3–7 (Fig. 2B). Sequencing data showed that these three splicing variants are made up of six repeats with Zr4 skipping (V2), five repeats with Zr4–5 skipping (V4) and four repeats with Zr4–6 (V5) skipping (Fig. 2C). In fast or mixed muscle types such as LM, TA, EDL and GAS, the four repeats with Zr4–6 skipping is the major splicing variant, whereas in slow muscle SOL, the six repeats with Zr4 skipping is the dominant variant (Fig. 2A and2B). Interestingly, we did not observe the splicing pattern of six Z repeats with Zr6 skipping in the slow muscle SOL as reported previously (Gautel et al., 1996; Sorimachi et al., 1997), which could be due to the undetectable level of this splicing form.
Figure 2.
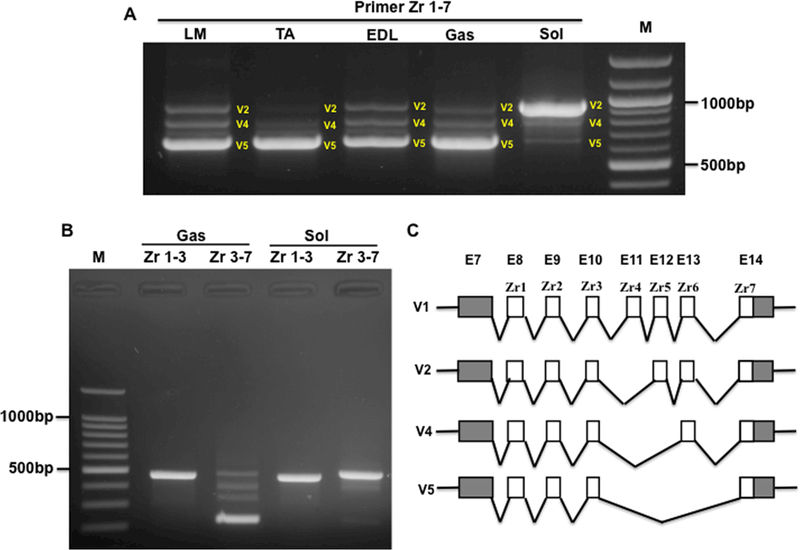
Z-Band splicing in different type of skeletal muscles in adult rats. A, PCR products with primer spanning Zr1–7 in different type of skeletal muscles; B, PCR products with primer spanning Zr1–3 and Zr 3–7 in Gas and Sol muscles; C, Schematic diagram of Z-band splicing pattern in skeletal muscles; Zr, Z repeats; M, 1kb DNA marker; E, exon; LM, Longissimus Muscle; TA, Tibialis Anterior; EDL, Extensor Digitorum Longus; Gas, Gastrocnemius; Sol, Soleus; White box, Z repeat; Grey box, exon; V, Variant.
M-band TTN splicing pattern in cardiac and skeletal muscles of multiple species
The M-band is the end of the TTN C-terminal located in the middle of the sarcomere. It is composed of only six exons from exon 359 to 364 or Mex1 to Mex6. Previous observations in the rabbit heart and skeletal muscles showed that exon Mex5 is predominantly included in heart muscle, and co-expression of Mex5+ (Mex5 inclusion) to Mex5- (Mex5 exclusion or skipping) is observed in rabbit skeletal muscle (Fig. S1B). Here, we examined the M-band TTN splicing pattern in the heart of other species including the mouse, rat, sheep and human using the primer spanning exon362 (Mex4) to 364 (Mex6). The PCR results indicated that only one DNA gel band can be observed in mouse, rat and human heart tissue, but three bands were shown in sheep heart muscle (Fig. 3A). The sequencing results revealed that the observed band in mouse, rat and human heart includes Mex5 (Fig. 3B), which is consistent with previous finding in rabbit heart (Kolmerer et al., 1996). However, in sheep heart, in addition to the intense upper band with Mex5+, we observed two additional bands. The middle band contains Mex5 with 195bp skipping due to alternative 3’ splice site (Fig. 3B). The lower band has Mex5- with 21bp inclusion of intron 362 resulting from alternative 5’ splice site (Fig. 3B). We then examined the M-band TTN splicing pattern in different rat skeletal muscle types (SOL, GAS, EDL, TA, LM) using the same primer as used in the heart. In the fast and mixed muscle types, the splicing pattern is exactly the same as in the sheep heart (Fig. 3A and3C). In the slow muscle type SOL, the splicing pattern of the mouse, rat and human heart is the same as in rabbit which predominantly expresses Mex5+ (Fig. 3A and3C). The equal or lower ratios of Mex5+ to Mex5- in skeletal muscles as reported previously (Kolmerer et al., 1996) only occur in fast or mixed muscle types, but not in the slow muscle type. The slow muscle SOL expresses 100% of Mex5+ (Fig. 3D).
Figure 3.
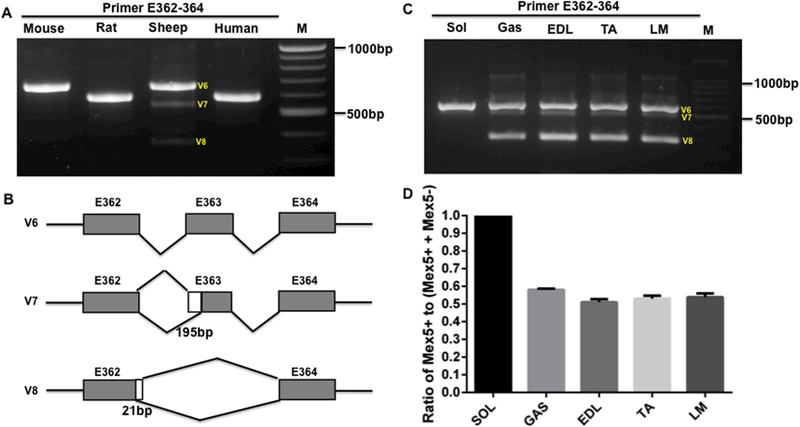
M-line splicing in the adult heart across species and in different type of skeletal muscles. A, PCR products with primer spanning E362–364 in heart muscle of mouse, rat, sheep and human; B, Schematic diagram of M-band splicing pattern in heart and skeletal muscles; C, PCR products with primer spanning E362–364 in different type of skeletal muscles in rat; D, Percentage of Mex5+ to (Mex5+ + Mex5-); M, 1kb DNA marker; E, exon; LM, Longissimus Muscle; TA, Tibialis Anterior; EDL, Extensor Digitorum Longus; Gas, Gastrocnemius; Sol, Soleus; White box, Alternative 5’ donor sites; Grey box, exon; bp, base pair; V, variant. Mean ± SEM (n=3).
Z-band TTN splicing in cardiac and skeletal muscle with development in rat and sheep
Previous study in human heart during development showed that in the adult heart, all seven repeats are included, whereas in the fetal heart, the splicing variant of skipped Zr4–6 is expressed (Fig. S1A) (Sorimachi et al., 1997). However, changes in the splicing pattern of the Z-band in skeletal muscle during development have not been studied. Here, we tested the Z-band splicing in fetal and adult heart and skeletal muscle of the rat and sheep using primers Zr1–7, Zr3–5 and Zr4–6. The PCR results indicated that the splicing patterns of the Z-band in fetal and adult heart muscle show no differences in both rat and sheep (Fig. 4A, 4B and 4C). We did not observe increased expression of four Z repeats with Zr4–6 skipping in fetal heart as previously reported (Sorimachi et al., 1997). Further, we examined the splicing pattern of the Z-band in fetal and adult skeletal muscle semitendinosus (ST) and longissimus Muscle (LM) of sheep. The results revealed that no difference was found in both muscle types with development in sheep (Fig. 4D, 4E and 4F). In sheep skeletal muscle, three splicing variants of the Z-band were observed using primer spanning full length of the seven Z repeats (Zr1–7) (Fig. 4D). Sequencing of the three splicing variants showed that they are variants of seven Z-repeats (V1), five Z repeats with Zr5–6 (V9) skipping, and four repeats with Zr4–6 skipping (V4) (Fig. 4G). These results suggest a new splicing pattern with Zr5–6 (V9) skipping appears in sheep skeletal muscle.
Figure 4.
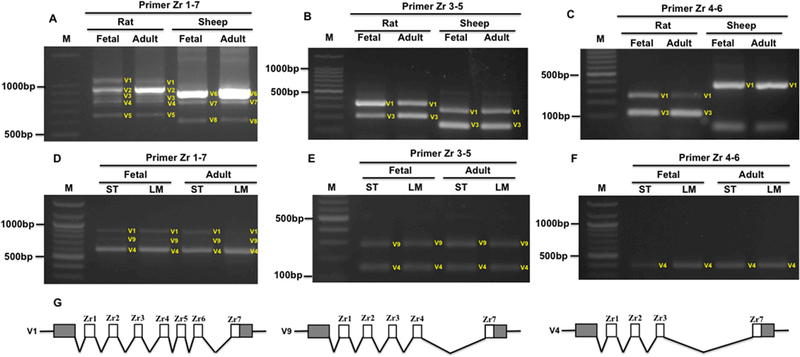
Z-band splicing in heart and skeletal muscles during development. A-C, PCR products with primerspanning Zr1–7, Zr3–5 and Zr4–6 in heart muscle of rat and sheep during development; D-F, PCR products with primer spanning Zr1–7, Zr3–5 and Zr4–6 in skeletal muscle of sheep during development; G, Schematic diagram of Z-band splicing pattern in skeletal muscles of sheep; M, 1kb DNA marker; E, exon; Zr, Z repeats; ST, Semitendinosus; LM, Longissimus Muscle; White box, Alternative exons; Grey box, exon. V, variant.
M-band TTN splicing in cardiac and skeletal muscle with development in rat and sheep
In previous study on developmental splicing changes of the M-band exons, alternative splicing of Mex5 was not observed in the heart during mouse development (Kolmerer et al., 1996). There were no previous reports on M-band TTN splicing pattern in skeletal muscle. In this study, we examined the developmental splicing changes of the M-band in fetal and adult heart muscles of rat and sheep with RT-PCR using primer spanning Mex4 to 6. The results indicated that there are no changes in fetal, day 1 and adult heart in rat as well as in fetal and adult sheep heart (Fig. 5A). In rat heart, the primary splicing variant of the M-band is the inclusion of Mex5, but in sheep heart, three splicing variants are all present with highest expression level of Mex5+ (Fig. 5A and5C). We then studied the splicing pattern of the M-band in sheep skeletal muscles during development. The PCR results revealed that three splicing variants are expressed in both fetal and adult ST muscle. The larger splicing variant with Mex5+ (upper band) has almost equal level to the smallest splicing form with Mex5- (lower band), and the splicing variant with partial Mex5+ (middle band) due to alternative 3’ splice site has lower expression level, which accounts for approximately 27% of total splicing variants (Fig. 5B and5D). Interestingly, the fetal and adult LM muscle also expresses three splicing vairants, but the lower band dominates in adult than in fetal LM muscle, and the upper band is significantly reduced during development. There is no significant difference observed in middle band during development (Fig. 5B and5D).
Figure 5.
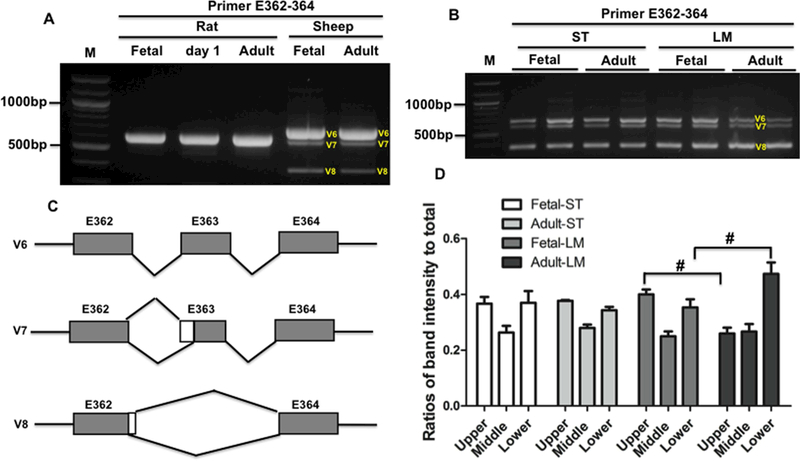
M-band titin splicing in heart and skeletal muscles during development. A, PCR products with primer spanning E362–364 in heart muscle of rat and sheep during development; B, PCR products with primer spanning E362–364 in skeletal muscle of sheep during development; C, Schematic diagram of Mband splicing pattern in heart and skeletal muscles during development; D, Ratios of each splicing form to total splicing forms, and lower band is significantly higher and upper band is significantly reduced in adult LM; M, 1kb DNA marker; E, exon; ST, Semitendinosus; LM, Longissimus Muscle; V, varaiant. Mean ± SEM (n=3). #, P<0.05.
RBM20 regulation of the Z- and M-band TTN splicing in heart and skeletal muscle
RBM20 is considered as a muscle specific splicing factor serving as a major regulator of TTN gene splicing (Guo et al., 2012). Splicing regulation of RBM20 in TTN splicing has been validated only in TTN middle Ig and PEVK region (Guo et al., 2012; 2013; Li et al., 2012; 2013; Rexiati et al., 2018). It is unknown whether RBM20 also regulates the Z-band and M-band TTN splicing. To determine the role of RBM20 in the regulation of the Z-band and M-band TTN splicing in cardiac and skeletal muscles, we employed a spontaneous Rbm20 KO rat model to examine the splicing pattern between WT and KO rats. Western blot results indicated that RBM20 is completely depleted in heart and different skeletal muscles (Fig. 6A). In heart muscle, the splicing pattern of the Z repeats shows no differences between WT and KO when using primers spanning Zr1–3, Zr3–7 and Zr1–7 (Fig. 6B). In skeletal muscle, we tested different muscle types including TA, EDL, SOL, GAS and LM. The PCR results revealed that no splicing difference was observed in these muscles between WT and KO using primer Zr1–3 (Fig. 6C). However, differences were observed in distinct skeletal muscle types using primers Zr3–7 and Zr1–7 (Fig. 6D and6E). We found a small PCR product were present in all skeletal muscle types of Rbm20 KO, but in slow muscle SOL, only part of larger variant became smaller, which is different from fast and mixed muscle types (Fig. 6D). We cloned and sequenced the small PCR product in Fig. 6D and6E, and found that the smaller band represents two Z repeats with Zr2–6 skipping (Fig. 6F). These results revealed that RBM20 regulates the Z-band TTN splicing in skeletal muscle but not in heart muscle. Furthermore, the results indicate that the role of RBM20 in the regulation of the Z-band TTN splicing slightly differs between slow and fast skeletal muscles.
Figure 6.
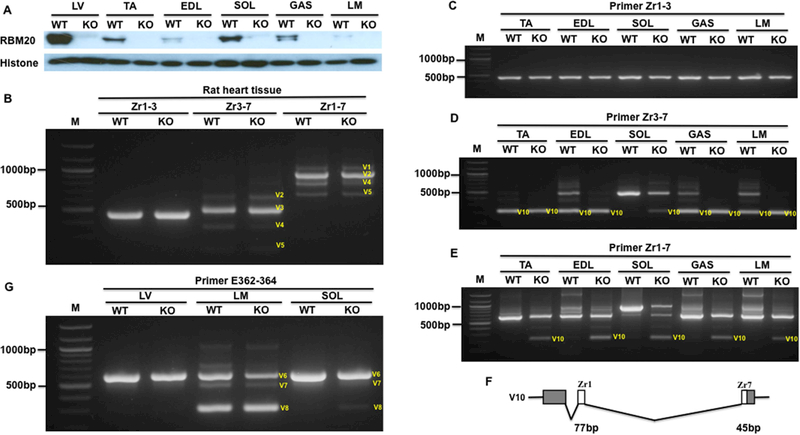
RBM20 regulation of the Z-band and M-band splicing in adult heart and skeletal muscles of RBM20 KO rats. A, RBM20 expression in heart and skeletal muscle of WT and KO rats; B, PCR products with primer spanning Zr1–7, Zr1–3 and Zr3–7 in heart muscle of WT and KO rat; C-E, PCR products with primer spanning Zr1–3, Zr3–7 and Zr1–7 in skeletal muscles of WT and KO rat; F, Schematic diagram of a new Z-band splicing form in skeletal muscles of KO rats; G, PCR products with primer spanning E362–364 in heart and skeletal muscles of WT and KO rat; M, 1kb DNA marker; E, exon; Zr, Z repeat; TA, Tibialis Anterior; EDL, Extensor Digitorum Longus; Gas, Gastrocnemius; Sol, Soleus; LM, Longissimus Muscle; LV, Left ventricle; WT, Wilt type; KO, RBM20 knockout; V, Variant; Histone, protein loading control.
Next, we determined the role of RBM20 in the regulation of the M-band TTN splicing in cardiac and skeletal muscle types. The results revealed that no splicing difference occurs in heart muscle between WT and KO, and also no differences were observed in fast skeletal muscle LM between WT and KO. However, in slow muscle type SOL, smaller splicing forms appear in Rbm20 KO rats. Even though the expression level is low, they are detectable (Fig. 6G). These results suggest that RBM20 has no impact on the M-band splicing in heart muscle and fast skeletal muscle, but a slight effect on the splicing of the M-band in the slow muscle type.
Discussion
Mechanical stresses play a central role in the regulation of physiological function in the striated muscles (Linke, 2008). Dysregulation of mechanical force transduction can lead to chronic muscle diseases such as hypertrophic (HCM) or dilated cardiomyopathy (DCM) (Linke, 2008). The molecular mechanisms by which muscle structures discern physical loads and transduce them into biochemical signals to change gene expression and modify cellular structure and function, are incompletely understood (Heineke et al., 2006). In the mechanotransduction network of the striated muscles, external force signals are transmitted from the extracellular matrix to the muscle cell cytoskeleton to generate forces from the opposite direction of a sarcomere. This bidirectional force transduction is mediated by highly specialized nodal points of mechanosensation: the Z-bands and the M-bands (Lange et al., 2006; Pyle et al., 2004; Frank et al., 2006; Agarkova et al., 2005).
TTN, as a myofibrillar backbone, links the Z-band to the M-band, and it is conceivable that TTN plays a central role in the mechanical stress response. Z-band TTN is encoded by the first 28 exons and consists of nine Ig domains (Z1-Z9). The Ig domain Z1-Z4 is an integral segment of the Z-band, whereas the Z5-Z9 reaches beyond the Z-band edge but is still tightly associated with the thin filament actin (Trombitas et al., 1997). The TTN Z-band interacts with numerous proteins that are associated with TTN assembly, mechanosignaling, and hypertrophic signaling (Heineke et al., 2006; Kontrogianni-Konstantopoulos et al., 2003; Makarenko et al., 2004; Knoll et al., 2002). Particularly, the scaffold protein α-actinin interacts with the seven 45-amino acid repeats located between Z3 to Z4 known as “TTN Z-repeats” (exons 8–14) (Granzier et al., 2004; Lange et al., 2006). The Z repeats undergo alternative splicing and could generate a variable number of Z repeat combinations which may change the binding sites of the scaffold protein α-actinin, and thus influence the mechanical stability of the sarcomere structure. However, the splicing pattern of Z repeats has not been well defined. In our comprehensive analysis of the splicing patterns of Z repeats across species, muscle types and during development, we revealed that in addition to reported splicing patterns including all seven repeats (V1), four repeats with Zr4–6 skipping (V5), and six repeats with Zr6 skipping (not found in this study) (Sorimachi et al., 1997), another four new splicing patterns were discovered. These four new Z repeats are six repeats with Zr4 skipping (V2), six repeats with Zr5 skipping (V3), five repeats with Zr4–5 skipping (V4) and five repeats with Zr5–6 skipping (V9). Five splicing forms of seven repeats, six repeats with Zr4 skipping, six repeats with Zr5 skipping, five repeats with Zr4–5 skipping, and four repeats with Zr4–6 skipping are all expressed in mouse, rat, sheep and human heart with highest level of six repeats with Zr4 skipping. However, in frog and chicken heart, only one major splicing form was observed with four repeats with Zr4–6 skipping and six repeats with Zr4 skipping respectively. Three new splicing forms of six repeats with Zr4 skipping (V2), five repeats with Zr4–5 skipping (V4), and five repeats with Zr5–6 skipping (V9) were discovered in skeletal muscles.
The M-band TTN is encoded by last six exons of TTN gene from exon 359 (Mex1) to 364 (Mex6). The M-band plays a crucial role in thick filament assembly. Deletion of the entire M-band TTN in cardiac muscle prevented sarcomere formation (Weinert et al., 2006). Deletion of the M-band Mex 1 and 2 in a conditional knockout mouse caused sarcomere disassembly in both skeletal and cardiac muscle and early death (Gotthardt et al., 2003). In particular, Mex1 encodes a TTN-kinase domain that is centrally involved in myocyte stress signaling, but the downstream effectors remains incompletely defined (Lange et al., 2005; Grater et al., 2005). Further, the unique sequence in TTN M-band encoded by Mex5 interacts with the protease calpain-3/p94 (Ojima et al., 2005; Duguez et al., 2006). Interestingly, data from a previous study indicated that Mex5 is an alternatively used exon that can be excluded in both cardiac and skeletal muscles (Kolmerer et al., 1996). The detailed alternative usage of Mex5 across species, muscle types and during development is not clear. In our current study, we comprehensively examined the splicing pattern of the M-band TTN in mouse, rat, sheep and human cardiac and skeletal muscle and during development. Most importantly, we found two new splicing patterns with alternative 3’ splice site in Mex5 (V7) and alternative 3’ splice site in intron 362 (V8). In the heart muscle of mouse, rat and human, only one splicing form with Mex5+ was observed, while, in sheep heart, we revealed that three splicing forms including two new splicing variants and one variant with Mex5+ were expressed with high expression level of the variant containing Mex5. In skeletal muscle, we observed that slow muscle type predominantly expresses a splicing variant with Mex5+ similar to heart muscle in mouse, rat and human, whereas, in fast and mixed muscle types, three splicing forms were observed which is similar to the splicing pattern in sheep heart. But the expression level of the smaller splicing forms (middle and lower bands) in skeletal muscle is increased. We also observed the changes of the M-band splicing pattern in skeletal muscle during development.
Our last major goal was to address whether the major TTN splicing factor RBM20 also regulates the alternative splicing of the Z-band and the M-band TTN exons in addition to TTN middle Ig and PEVK domains. Using the Rbm20 KO rat model, we revealed that RBM20 neither regulates Z-band TTN splicing nor M-band TTN splicing in heart muscle, but it regulates Z-band and M-band TTN splicing in skeletal muscle. RBM20 deficiency results in a smaller splicing form in the skeletal muscle Z-band TTN. Sequencing of the small form revealed another new splicing form with Zr2–6 skipping (V10). In the M-band, RBM20 deficiency leads to slight changes in slow muscle where two smaller variants appear, but no changes in fast muscle types. Our western blot results with antibody against RBM20 demonstrated that RBM20 expression levels differ in different muscle type. Whether differential splicing forms in different muscle types are associated with variable RBM20 levels remains unclear.
In summary, our results provide the first evidence that the Z repeats have five new splicing forms in cardiac muscle and/or skeletal muscle, and the M-band has two additional splicing patterns that are present in sheep heart as well as in skeletal muscles of other species. Interestingly, we did not observe the splicing form with only Mex5 skipping as reported previously (Kolmerer et al., 1996). Most importantly, we first confirmed the role of RBM20 in the regulation of TTN splicing in the Z-band and the M-band domains. Although the functional role of these new splicing forms found in our study remains poorly understood, these splicing forms could be associated with sarcomere assembly, mechanosignaling and protein quality control. However, the functional role of these splicing changes merits further studies.
Supplementary Material
Acknowledgments:
This work was supported by the National Institute of Health/National Institute of General Medical Sciences [NIGMSP20GM103432]; the BGIA from the American Heart Association [16BGIA27790136 to WG]; the USDA National Institute of Food and Agriculture [Hatch project 1009266 to WG]; NICHD HD070096 (SPF and PWN). Ying Ge would like to acknowledge NIH R01 HL109810 and HL096971. We gratefully thank Dan Levy at the University of Wyoming to provide frog heart tissues, and David M. Hanna for the animal care and laboratory management. We thank the Chinese Scholarship Council (CSC, Beijing, China) to support Zhilong Chen. We also thank Drs. Paul Mozdziak and Peter W. Nathanielsz for their critical reading and corrections on grammatical errors.
Grant sponsor: National Institute of General Medical Sciences; Grant number: NIGMSP20GM103432; Grant sponsor: American Heart Association; Grant number: 16BGIA27790136; Grant sponsor: the USDA National Institute of Food and Agriculture; Hatch; Grant number: 1009266; Grant sponsor: NICHD Grant number: HD070096. Grant sponsor: NIH; Grant number: HL109810 and HL096971.
Footnotes
Conflict of interest: The authors declare that they have no conflicts of interest with the contents of this article.
References
- Agarkova I, and Perriard JC 2005. The M-band: an elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 15: 477–485 [DOI] [PubMed] [Google Scholar]
- Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, and Labeit S 2001. The complete gene sequence of titin, expression of an unusual ≈ 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 89: 1065–1072 [DOI] [PubMed] [Google Scholar]
- Cazorla O, Freiburg A, Helmes M, Centner T, McNabb M, Wu Y, Trombitás K, Labeit S, and Granzier H 2000. Differential expression of cardiac titin isoforms and modulation of cellular stiffness. Circ. Res. 86: 59–67 [DOI] [PubMed] [Google Scholar]
- Duguez S, Bartoli M, and Richard I 2006. Calpain-3: a key regulator of the sarcomere? FEBS J. 273: 3427–3436 [DOI] [PubMed] [Google Scholar]
- Frank D, Kuhn C, Katus HA, and Frey N 2006. The sarcomeric Z-disc: a nodal point in signalling and disease. J. Mol. Med. 84: 446–468 [DOI] [PubMed] [Google Scholar]
- Freiburg A, Trombitas K, Hell W, Cazorla O, Fougerousse F, Centner T, Kolmerer B, Witt C, Beckmann JS, Gregorio CC, Granzier H, and Labeit S 2000. Series of exon-skipping events in the elastic spring region of titin as the structural basis for myofibrillar elastic diversity. Circ. Res. 86: 1114–1121 [DOI] [PubMed] [Google Scholar]
- Freiburg A, and Gautel M 1996. A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur. J. Biochem. 235: 317–323 [DOI] [PubMed] [Google Scholar]
- Fürst DO, Osborn M, Nave R, and Weber K 1988. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z-line extends close to the M line. J. Cell Biol. 106: 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautel M, Goulding D, Bullard B, Weber K, and Fürst, D. O. 1996. The central Z-disk region of titin is assembled from a novel repeat in variable copy numbers. J. Cell Sci. 109: 2747–2754 [DOI] [PubMed] [Google Scholar]
- Gotthardt M, Hammer RE, Hubner N, Monti J, Witt CC, McNabb M, Richardson JA, Granzier H, Labeit S, and Herz J 2003. Conditional expression of mutant M-line titins results in cardiomyopathy with altered sarcomere structure. J. Biol. Chem. 278: 6059–6065 [DOI] [PubMed] [Google Scholar]
- Granzier H, and Labeit S 2002. Cardiac titin: an adjustable multi-functional spring. J. Physiol. 541:335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzier HL, and Labeit S 2004. The giant protein titin: a major player in myocardial mechanics, signaling, and disease. Circ. Res. 94: 284–295 [DOI] [PubMed] [Google Scholar]
- Grater F, Shen J, Jiang H, Gautel M, and Grubmuller H 2005. Mechanically induced titin kinase activation studied by force-probe molecular dynamics simulations. Biophys. J. 88: 790–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaser ML, Warren CM, Esbona K, Guo W, Duan Y, Parrish AM, Krzesinski PR, Norman HS, Dunning S, Fitzsimons DP, and Moss RL 2008. Mutation that dramatically alters rat titin isoform expression and cardiomyocyte passive tension. J. Mol. Cell. Cardiol. 44: 983–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio CC, Trombitás K, Centner T, Kolmerer B, Stier G, Kunke K, Suzuki K, Obermayr F, Herrmann B, Granzier H, Sorimachi H, and Labeit S 1998. The NH2 terminus of titin spans the Z-disc: its interaction with a novel 19-kD ligand (T-cap) is required for sarcomeric integrity. J. Cell Biol.143:1013–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Bharmal SJ, Esbona K, and Greaser ML 2010. Titin diversity--alternative splicing gone wild. J. Biomed. Biotechnol. 2010: 753675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Pleitner JM, Saupe KW, and Greaser ML 2013. Pathophysiological defects and transcriptional profiling in the RBM20−/− rat model. PLoS One 8: e84281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V, Vakeel P, Klaassen S, Gerull B, Thierfelder L, Regitz-Zagrosek V, Hacker TA, Saupe KW, Dec GW, Ellinor PT, MacRae CA, Spallek B, Fischer R, Perrot A, Özcelik C, Saar K, Hubner N, and Gotthardt M 2012. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat. Med. 18: 766–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, and Sun M 2018. RBM20, a potential target for treatment of cardiomyopathy via titin isoform switching. Biophys. Rev. 10:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineke J, and Molkentin JD 2006. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 7: 589–600 [DOI] [PubMed] [Google Scholar]
- Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJ, Cook SA, Mestroni L, Seidman JG, and Seidman CE 2012. Truncations of titin causing dilated cardiomyopathy. N. Engl. J. Med. 366:619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, and Gibson TJ 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23: 403–405 [DOI] [PubMed] [Google Scholar]
- Knöll R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang ML, Hayashi T, Shiga N, Yasukawa H, Schaper W, McKenna W, Yokoyama M, Schork NJ, Omens JH, McCulloch AD, Kimura A, Gregorio CC, Poller W, Schaper J, Schultheiss HP, and Chien KR 2002. The cardiac mechanical stretch sensor machinery involves a Z-disc complex that is defective in a subset of human dilated cardiomyopathy. Cell 111: 943–955 [DOI] [PubMed] [Google Scholar]
- Kolmerer B, Olivieri N, Witt CC, Herrmann BG, and Labeit S 1996. Genomic organization of M line titin and its tissue-specific expression in two distinct isoforms. J. Mol. Biol. 256: 556–563 [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Jones EM, Van Rossum DB, and Bloch RJ 2003. Obscurin is a ligand for small ankyrin-1 in skeletal muscle. Mol. Biol. Cell 14: 1138–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeit S, Kolmerer B, and Linke WA 1997. The giant protein titin: emerging roles in physiology and pathophysiology. Circ. Res. 80: 290–294 [DOI] [PubMed] [Google Scholar]
- Labeit S, and Kolmerer B 1995. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science 270: 293–296 [DOI] [PubMed] [Google Scholar]
- Lange S, Ehler E, and Gautel M 2006. From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 16: 11–18 [DOI] [PubMed] [Google Scholar]
- Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, Gunnarsson LG, Hughes SM, Marchand S, Sejersen T, Richard I, Edström L, Ehler E, Udd B, and Gautel M 2005. The kinase domain of titin controls muscle gene expression and protein turnover. Science 308: 1599–1603 [DOI] [PubMed] [Google Scholar]
- LeWinter MM, and Granzier HL 2014. Cardiac titin and heart disease. J. Cardiovasc. Pharmacol. 63: 207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Guo W, Schmitt BM, and Greaser ML 2012. Comprehensive analysis of titin protein isoform and alternative splicing in normal andmutant rats. J. Cell. Biochem. 113: 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Guo W, Dewey CN, and Greaser ML 2013. Rbm20 regulates titin alternative splicing as a splicing repressor. Nucleic Acids Res. 41: 2659–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke WA 2008. Sense and stretchability: the role of titin and titin-associated proteins in myocardial stress-sensing and mechanical dysfunction. Cardiovasc. Res. 77: 637–648 [DOI] [PubMed] [Google Scholar]
- Linke WA, and Hamdani N 2014. Gigantic business: titin properties and function through thick and thin. Circ. Res. 114: 1052–1068 [DOI] [PubMed] [Google Scholar]
- Makarenko I, Opitz CA, Leake MC, Neagoe C, Kulke M, Gwathmey JK, del Monte F, Hajjar RJ, and Linke WA 2004. Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts. Circ. Res. 95: 708–716 [DOI] [PubMed] [Google Scholar]
- Mestroni L 2011. Genetic variation in titin in arrhythmogenic right ventricular cardiomyopathy-overlap syndromes. Circulation 124: 876–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methawasin M, Hutchinson KR, Lee EJ, Smith JE, Saripalli C, Hidalgo CG, Ottenheijm CA, and Granzier H 2014. Experimentally increasing titin compliance in a novel mouse model attenuates the Frank-Starling mechanism but has a beneficial effect on diastole. Circulation 129: 1924–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann WM, Gautel M, Steiner F, van der Ven PF, Weber K, and Fürst DO 1996. The structure of the sarcomeric M band: localization of defined domains of myomesin, M-protein, and the 250-kD carboxy-terminal region of titin by immunoelectron microscopy. J. Cell Biol. 134: 1441–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojima K, Ono Y, Hata S, Koyama S, Doi N, and Sorimachi H 2005. Possible functions of p94 in connectin-mediated signaling pathways in skeletal muscle cells. J. Muscle Res. Cell Motil. 26: 409–417 [DOI] [PubMed] [Google Scholar]
- Pyle WG, and Solaro RJ 2004. At the crossroads of myocardial signaling: the role of Z-discs in intracellular signaling and cardiac function. Circ. Res. 94: 296–305 [DOI] [PubMed] [Google Scholar]
- Rexiati M, Sun M, and Guo W 2018. Muscle-Specific Mis-Splicing and Heart Disease Exemplified by RBM20. Genes (Basel) 9: E18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorimachi H, Freiburg A, Kolmerer B, Ishiura S, Stier G, Gregorio CC, Labeit D, Linke WA, Suzuki K, and Labeit S 1997. Tissue-specific expression and alpha-actinin binding properties of the Z-disc titin: implications for the nature of vertebrate Z-discs. J. Mol. Biol. 270: 688–695 [DOI] [PubMed] [Google Scholar]
- Taylor M, Graw S, Sinagra G, Barnes C, Slavov D, Brun F, Pinamonti B, Salcedo EE, Sauer W, Pyxaras S, Anderson B, Simon B, Bogomolovas J, Labeit S, Granzier H, and Makarenko I, Opitz CA, Leake MC, Neagoe C, Kulke M, Gwathmey JK, del Monte F, Hajjar RJ, and Linke WA. 2004. Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts. Circ. Res. 95: 708–716 [DOI] [PubMed] [Google Scholar]
- Trinick J, and Tskhovrebova L 1999. Titin: a molecular control freak. Trends Cell Biol. 9: 377–380 [DOI] [PubMed] [Google Scholar]
- Trombitas K, and Granzier H 1997. Actin removal from cardiac myocytes shows that near Z-line titin attaches to actin while under tension. Am. J. Physiol. 273: C662–C670 [DOI] [PubMed] [Google Scholar]
- Tskhovrebova L, and Trinick J 2003. Titin: properties and family relationships. Nat. Rev. Mol. Cell Biol. 4: 679–689 [DOI] [PubMed] [Google Scholar]
- Weinert S, Bergmann N, Luo X, Erdmann B, and Gotthardt M 2006. M-line-deficient titin causes cardiac lethality through impaired maturation of the sarcomere. J. Cell Biol. 173: 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Ren J, and Guo W 2015. Sarcomeric protein isoform transitions in cardiac muscle: a journey to heart failure. Biochim. Biophys. Acta. 1852: 47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Chen Z, and Guo W 2017. Pre-mRNA mis-splicing of sarcomeric genes in heart failure. Biochim. Biophys. Acta. 1863: 2056–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


