Abstract
We measured and compared heritability estimates for measures of functional brain connectivity extracted using the Enhancing Neuroimaging Genetics through Meta‐Analysis (ENIGMA) rsfMRI analysis pipeline in two cohorts: the genetics of brain structure (GOBS) cohort and the HCP (the Human Connectome Project) cohort. These two cohorts were assessed using conventional (GOBS) and advanced (HCP) rsfMRI protocols, offering a test case for harmonization of rsfMRI phenotypes, and to determine measures that show consistent heritability for in‐depth genome‐wide analysis. The GOBS cohort consisted of 334 Mexican‐American individuals (124M/210F, average age = 47.9 ± 13.2 years) from 29 extended pedigrees (average family size = 9 people; range 5–32). The GOBS rsfMRI data was collected using a 7.5‐min acquisition sequence (spatial resolution = 1.72 × 1.72 × 3 mm3). The HCP cohort consisted of 518 twins and family members (240M/278F; average age = 28.7 ± 3.7 years). rsfMRI data was collected using 28.8‐min sequence (spatial resolution = 2 × 2 × 2 mm3). We used the single‐modality ENIGMA rsfMRI preprocessing pipeline to estimate heritability values for measures from eight major functional networks, using (1) seed‐based connectivity and (2) dual regression approaches. We observed significant heritability (h 2 = 0.2–0.4, p < .05) for functional connections from seven networks across both cohorts, with a significant positive correlation between heritability estimates across two cohorts. The similarity in heritability estimates for resting state connectivity measurements suggests that the additive genetic contribution to functional connectivity is robustly detectable across populations and imaging acquisition parameters. The overarching genetic influence, and means to consistently detect it, provides an opportunity to define a common genetic search space for future gene discovery studies.
Keywords: functional connectivity, heritable, seed‐based connectivity
1. INTRODUCTION
Resting state functional MRI (rsfMRI) connectivity analysis takes advantage of temporal coherence in low‐frequency blood oxygenation level‐dependent fluctuations, when the brain is not engaged in a goal‐directed activity (Fox & Raichle, 2007; Smith et al., 2009). This activity is synchronized across interconnected brain regions and may be used to link them into functional networks (Biswal, Yetkin, Haughton, & Hyde, 1995; Fox & Raichle, 2007; Margulies et al., 2007; Smith et al., 2009). Functional networks identified during the resting state are also detectable when the brain is engaged in cognitive activities. Functional networks may also be derived through independent component analysis of brain activation maps, such as these stored in the BrainMap, the largest database (over 100,000) of functional imaging studies (Smith et al., 2009). These functional networks are even stable enough to be detectable to some extent when a person is asleep, in a coma, or under anesthesia, and are present in infants and in nonhuman primates (Chen et al., 2017; Mitra et al., 2017; Wey et al., 2014; Wu et al., 2016). The stability and reproducible structure of functional networks suggests that they are a fundamental property of the brain, making it of interest to what extent individual variance depends on genetics versus other factors. We examined and compared heritability estimates of resting state phenotypes in two independent genetically informed cohorts collected using two familial study designs (family and twin‐siblings), though the datasets were collected 10 years apart—using a conventional rsfMRI and state‐of‐the‐art connectivity protocols.
Genetic analyses of rsfMRI phenotypes can be challenging due to limitations in statistical power and methodological differences in the available analysis approaches. One way to improve statistical power and consistency of the outcomes is by pooling data from multiple cohorts. As has been noted for other phenotypes from structure MRI and diffusion MRI, very large, highly powered, and representative samples are difficult to collect at a single site (Ioannidis, 2014; Jahanshad et al., 2013), although there are some notable exceptions, for example, the UK Biobank (Elliott et al., 2017). Multisite studies can collect larger and more representative samples but require pre‐processing steps to address site‐specific sources of methodological variance. The ENIGMA consortium developed an rsfMRI analysis pipeline to perform consistent analysis and extraction of resting state connectivity measures across data collected using diverse protocols (Adhikari et al., 2017). Here, we used this pipeline to (i) replicate findings in the first report that demonstrated significant additive genetic contribution to the inter‐subject variance in the default mode network using the original (N = 334) genetics of brain structure (GOBS) cohort (Glahn et al., 2010); (ii) extend this analysis to other resting state networks and (iii) replicate these findings in an independent dataset from the Human Connectome Project (HCP; Van Essen et al., 2013). The motivation for this work is to show that genetic effects on resting state connectivity measures are robust enough to serve as targets for cross‐site discovery of genetic variants that affect cerebral connectivity. A key milestone in this quest is to determine resting state connectivity measures that are reliable and consistently heritable regardless of the population and/or data collection protocols.
The ENIGMA rsfMRI pipeline differs from other pipelines in two respects. Many fMRI analysis pipelines require a structural T1‐weighted (T1w) brain MRI scan to regress out signal measured in the cerebrospinal fluid (CSF) and cerebral white matter and for registration of all data to a common atlas space (Jenkinson, Beckmann, Behrens, Woolrich, & Smith, 2012; Smith et al., 2004). In the spirit of ENIGMA, this pipeline is a single‐modality and uses a deformable template created from 1,100 individual images provided by ENIGMA sites to incorporate and compensate for shape distortions common to fMRI images (Adhikari et al., 2017). This alleviates potential pitfalls of site‐to‐site variance in the quality of T1w data and coregistration biases that may influence extraction of rsfMRI phenotypes. Clearly both approaches are possible, and the current approach offers a reasonable and feasible approach to test and apply across sites.
Another notable difference is that the ENIGMA rsfMRI pipeline uses a noise reduction technique based on the Marchenko–Pastur Principal Component Analysis (MP‐PCA) to improve signal‐to‐noise ratio (SNR). The MP‐PCA approach takes advantage of the redundancy in the time‐series data (such as that in fMRI and/or diffusion tensor imaging) to quantify the contribution of the thermal noise to the voxel‐wise signal. MP‐PCA‐based denoising reduces signal fluctuations to increase the temporal SNR (tSNR) without altering the spatial resolution of the data (Adhikari et al., 2017). The thermal noise‐selective is based on data redundancy in the PCA domain (Veraart, Novikov, et al., 2016b). The bulk of the PCA eigenvalues arise due to noise and can be approximated by the universal Marchenko–Pastur distribution. This Marchenko–Pastur parameterization allows us to estimate the noise level in a local neighborhood based on the singular value decomposition of a matrix combining neighborhood voxels (Veraart, Fieremans, & Novikov, 2016a). Regression of these components enhances SNR by suppression of the noise contribution to the signal.
The advantages of improving SNR using an MP‐PCA approach has already been demonstrated for homogenization of multi‐site analyses of diffusion MRI data (Kochunov et al., 2018). In addition, the spatial map of the thermal noise serves as an important quality assurance measurement (Kochunov et al., 2018). Thermal noise contribution to the image is expected to be regionally uniform and any regional variations in the thermal noise can indicate nonlinearity in the RF‐receiving system or other scanner problems (Kochunov et al., 2018).
Additive genetic effects on brain metrics can be detected and estimated by modeling the degree of relatedness across individuals when studying the variance in a database of brain scans. First, we computed heritability measures from two commonly used familial study designs: GOBS subjects were recruited from an extended pedigree, and HCP participants were recruited from a twin/siblings registry. To examine some alternative metrics, we used two complementary measures of functional connectivity commonly used in brain mapping research: region‐based (or “seed”‐based) analysis, and dual regression. In the first approach, the connectivity strength is estimated based on the degree of covariance in the signal measured from two or more regions. In the second approach, the “group‐wise‐average” trend is regressed from the data, yielding a statistical inference on how individual subjects differ from the group average. To reduce the biases inherent in having to extract resting state networks from different cohorts, we used the pattern derived through the meta‐analysis of functional activations stored in the BrainMap database (Smith et al., 2009). Overall, similarity in the heritability measurements across two diverse cohorts is helpful to offer further evidence for the suitability of the ENIGMA rsfMRI protocol and connectivity measures for more in‐depth multisite genetic analyses of cerebral functional connectivity.
2. METHODS AND MATERIALS
2.1. Study subjects and imaging protocols
We performed analyses in two independently collected rsfMRI datasets: GOBS and HCP, detailed below.
2.1.1. GOBS and function study
This cohort consisted of 334 Mexican‐American individuals (124M/210F, age: 47.9 ± 13.2 years) from 29 extended pedigrees (average family size, 9 people; range 5–32) who participated in the GOBS and Function study (Glahn et al., 2010). Individuals were selected from the community as part of a large family epidemiological study focused on the San Antonio, TX region. Individuals with a history of neurological illness, stroke, or other major neurological events were excluded, as were people with brain MRI contraindications. All participants provided written informed consent on forms approved by the institutional review board at the University of Texas Health Science Center San Antonio (UTHSCSA).
All imaging was performed at the Research Imaging Institute, UTHSCSA, on a Siemens 3 T Trio scanner, using a multichannel phased array head coil. Whole‐brain, resting‐state functional imaging was performed using a gradient‐echo echo planar imaging (EPI) sequence sensitive to the BOLD effect. The acquisition parameters were: repetition and echo times TR/TE = 3,000/30 ms, spatial resolution = 1.72 × 1.72 × 3 mm3, flip angle = 90°. The resting‐state protocol included 43 slices acquired parallel to the sagittal plane containing the anterior and posterior commissures; the total scan time was 7.5 min.
2.1.2. Human connectome project
Human connectome project (HCP) cohort consisted of 518 participants (240M/278F, average age 28.7 ± 3.7) from the HCP dataset, released in March 2017. Participants were recruited from the Missouri Family and Twin Registry (Van Essen et al., 2013). All HCP participants were from young adult sibships of average size 3–4 that included a monozygotic (MZ) or dizygotic (DZ) twin pair and (where available) their nontwin siblings. Out of 518 participants, 128 were MZ twins and 89 were DZ twins. The zygosity of the same‐sex twins was verified based on the genetic screening. Subjects ranged in age from 22 to 37 years. This age range corresponds to a period after neurodevelopment is largely completed and before the typical age of onset of neurodegenerative changes. The inclusion and exclusion criteria are detailed elsewhere (Van Essen et al., 2013). The HCP subjects are healthy young adults within a restricted age range and free from major psychiatric or neurological illnesses (Edens, Glowinski, Pergadia, Lessov‐Schlaggar, & Bucholz, 2010; Sartor et al., 2011). All subjects provided written informed consent on forms approved by the Institutional Review Board of Washington University in St. Louis.
All HCP subjects are scanned on a customized Siemens 3 T “Connectome Skyra” scanner at Washington University in St. Louis, using a standard 32‐channel Siemens receive head coil. RsfMRI data consisted of two runs in one session. Within a session, oblique axial acquisitions alternated between phase encoding in a right‐to‐left direction in one run and phase encoding in a left‐to‐right direction in the other run. Resting state images were collected using a gradient‐echo echo planar imaging (EPI) sequence. The acquisition parameters were: TR/TE = 720 and 33.1 ms, flip angle = 52°, field of view (FOV) = 208 × 180 mm, matrix = 104 × 90, 2.0 mm isotropic voxels, 72 axial slices, multiband factor = 8; scan time was 28.8 min.
2.2. Functional image analysis
2.2.1. ENIGMA resting state analysis
ENIGMA developed a single‐modality resting state analysis pipeline (Adhikari et al., 2017) by implementing it in the Analysis of Functional NeuroImages (AFNI) software (Cox, 1996) (Figure 1). It includes the application of principal components analysis (PCA)‐based denoising (Veraart, Fieremans,et al., 2016a; Veraart, Novikov, et al., 2016b). The denoising is the first step in this analysis pipeline to improve SNR and tSNR properties of the time series data (Adhikari et al., 2017), without losing image spatial resolution, and avoids introducing of additional partial volume effects that complicate further analyses. The MP‐PCA approach does not alter the resting state network activation patterns, whereas spatial smoothing using a Gaussian kernel leads to partial voxel averaging, spreading the activations across gray and white matter regions and removing smaller nodes. Finally, the noise‐maps produced by the MP‐PCA approach provide valuable information for quality control—deviations from the expected uniform pattern of thermal noise, or one that slowly varies in space, may indicate problems with the coil or other scanner hardware.
Figure 1.
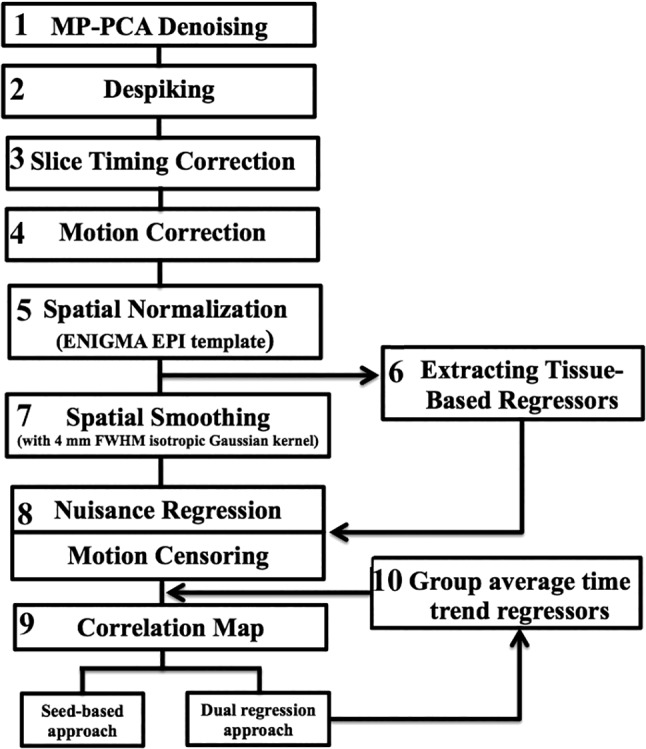
Flowchart of ENIGMA rsfMRI analysis pipeline
In the next step, supplementary data, if provided, is used to correct spatial distortions associated with long‐TE gradient echo imaging. Two available corrections are the gradient‐echo “fieldmap” or the reversed‐gradient approach. Here, we implemented the spatial distortion corrections in the HCP dataset but distortion correction data was not collected in GOBS dataset. In the next step, a transformation is computed registering the base volume to the ENIGMA EPI template that was derived from 1,100 datasets collected across 22 sites (Adhikari et al., 2017) to develop a spatial template and spatial atlas. This atlas has a dual purpose: it is used for regression of the global signal, but it also offers a common anatomical spatial reference frame. Next, correction for head motion is performed by registering each functional volume to the volume with the minimum outlier fraction (suggesting it has little motion), where each transformation is concatenated with the transformation to standard space, to avoid unnecessary interpolation. Nuisance variables such as the linear trend, 6 motion parameters (3 rotational and 3 translational directions), their 6 temporal derivatives (rate of change in rotational and translational motion), and time courses from the local white matter and cerebrospinal fluid (CSF) from lateral ventricles are modeled using multiple linear regression analysis, which were then removed as regressors of no interest. Time points with excessive motion (>0.2 mm), estimated as the magnitude of displacement from one time point to the next, including neighboring time points and outlier voxels fraction (>0.1) are censored from statistical analysis. Images were spatially normalized to the ENIGMA EPI template in Montreal Neurological Institute (MNI) standard space for group analysis.
2.3. Functional connectivity analysis
Resting state network templates were defined based on the probabilistic regions of interest (ROIs) from 20‐component analysis of the BrainMap activation database and resting fMRI dataset (Smith et al., 2009). We defined the binary masks of the resting state template regions from auditory network (AN), attention network (AttN), default mode network (DMN), executive control network (ECN), fronto‐parietal network (FPN), salience network (SN), sensorimotor network (SMN), and visual network (VN) (Figure 2). The colored regions in Figure 2 represent the “seeds” for the functional connectivity analysis. Mean time series were extracted from the seed regions of each network and connectivity maps corresponding to each seed region were obtained by accessing correlations along the time series for different regions. Next, Fisher's r‐to‐z transformations were applied to obtain a normal distribution. We calculated seed‐based functional connectivity values between seed regions in each network and performed heritability calculation. Furthermore, we performed dual regression analysis for these all network template ROIs, and calculated the functional connectivity measures and hence heritability estimates on both datasets. (The same GOBS dataset was used in a prior study by Glahn et al. [2010]).
Figure 2.
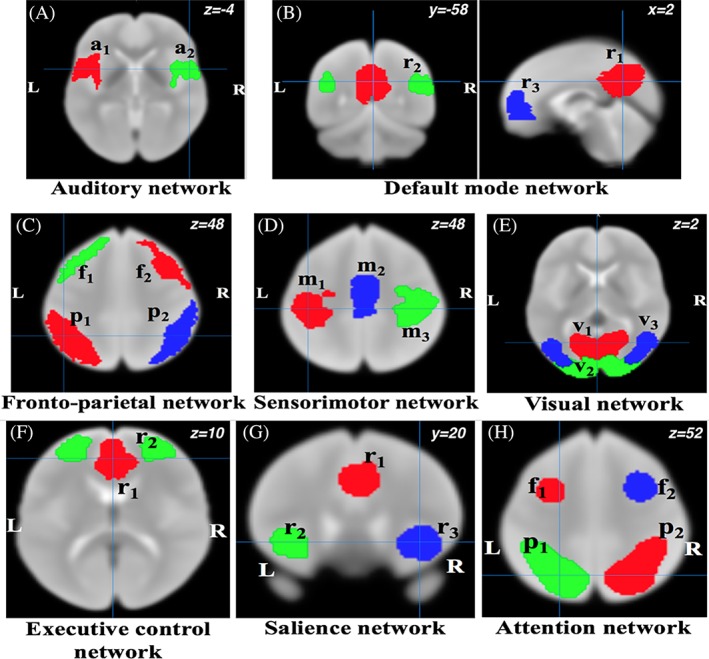
Resting state network template ROIs based on the BrainMap activation database (Smith et al., 2009). Here, L = left, R = right, in (a) a1/a2 = left/right primary and association auditory cortices, in (b) r1 = posterior cingulate/precuneus, r2 = bilateral temporal–parietal regions and, r3 = ventromedial frontal cortex, in (c) f1/f2 = left/right frontal area and p1/p2 = left/right parietal area, in (d) m1/m3 = left/right motor area and m2 = supplementary motor area, in (e) v1 = medial visual areas, v2 = occipital visual areas, and v3 = lateral visual areas, in (f) r1 = anterior cingulate cortex and r2 = bilateral medial frontal gyrus, in (g) r1 = anterior cingulate cortex and r2/r3 = left/right insula, in (h) f1/f2 = left/right middle frontal gyrus and p1/p2 = left/right superior parietal lobule [Color figure can be viewed at http://wileyonlinelibrary.com]
We developed a modification of the dual regression approaches as implemented in the FSL software (Beckmann, Mackay, Filippini, & Smith, 2009; Filippini et al., 2009). FSLs dual regression approach uses the spatial maps from the group‐average analysis to generate subject‐specific spatial maps, and extract subject‐specific time series. For each subject, the group‐average set of spatial maps is regressed into the subject's 4D space–time dataset. This results in a set of subject‐specific time series, one per group‐level spatial map. Next, those time series are regressed into the same 4D dataset, resulting in a set of subject‐specific spatial maps, one per group‐level spatial map.
We have used the spatial maps template of resting networks from independent component analysis (ICA)‐based analyses of the BrainMap database. Following the pre‐processing steps, we computed the average time series for the ROIs included in the given network template for each subject. We obtained the average time series from all subjects to represent the group's time‐trend for the corresponding network. We regressed out this trend from individual subject's data. This was performed by including the group average trend for a network as an additional regressor appended to the matrix. This was achieved by re‐running the step 10 (in Figure 1) because the group average trend is not expected to be orthogonal to the other nuisance regressors for any given subject (e.g., motion parameters). After regressing out the effects of the group average trend, we calculated correlation maps corresponding to each seed region for a given network, performed Fisher's r‐to‐z transformations and calculated FC values between seed regions in each network template before performing heritability calculation on these measures. The dual regression approach provides a statistical inference on how the individual subjects differ from the group average.
2.4. Genetic analyses
We estimated the degree of additive genetic variance (heritability) in the intersubject variability in the FC values. The first set of analyses was focused on replicating findings of significant heritability in the default mode network reported in the GOBS cohort (Glahn et al., 2010). The same subjects and the same genetic analyses were used to estimate the additive and environmental components of the phenotypic variance. Specifically, we used the variance components method, as implemented in the Sequential Oligogenic Linkage Analysis Routines (SOLAR) Eclipse software package (http://www.solar-eclipse-genetics.org) (Almasy & Blangero, 1998). SOLAR uses maximum likelihood variance decomposition methods, extensions of the strategy developed by Amos and colleagues (Amos, 1994). The covariance matrix Ω for a pedigree is given by: +I , where is the genetic variance due to the additive genetic factors, φ is the kinship matrix representing the pair‐wise kinship coefficients among all individuals, is the variance due to individual—unique environmental effects and measurement error, and I is an identity matrix (under the assumption that all environmental effects are uncorrelated among family members). Narrow sense heritability is defined as the fraction of phenotypic variance attributable to additive genetic factors,
The variance parameters are estimated by comparing the observed phenotypic covariance matrix with the covariance matrix predicted by kinship (Almasy & Blangero, 1998). Significance of the heritability estimate is tested by comparing the likelihood of the model in which is constrained to zero with that of a model in which is estimated. Twice the difference between the log‐likelihoods of these models yields a test statistic, which is asymptotically distributed as a 1/2:1/2 mixture of x 2 variables with 1‐of‐freedom and a point mass at zero (Beasley, Erickson, & Allison, 2009). Prior to the heritability estimation, phenotype values (FC measures) from each dataset were adjusted for covariates including sex, age, age2, age × sex interaction, and age2 × sex interaction. Inverse Gaussian transformation was also applied to ensure normality of the distribution. Outputs from SOLAR include the heritability estimate (h 2), the significance value (p), and the standard error for each trait (SE).
3. RESULTS
3.1. Seed‐based analysis
Heritability estimates of seed‐based connectivity phenotypes for both cohorts are summarized in Table 1. We replicated the significant heritability for default mode network (DMN) functional connectivity from the posterior cingulate/precuneus to bilateral temporal–parietal regions (h 2 = 34%, p = .014) and ventromedial frontal cortex (h 2 = 35%, p = .014) in the GOBS dataset. We further replicated significant heritability of these phenotypes, in the HCP dataset. Both datasets showed the significantly heritable functional connectivity between the fronto‐parietal network nodes. In case of the attention network, significant heritable functional connectivity was found between right middle frontal gyrus and superior parietal lobule, for both datasets. The right and left motor cortices are characterized by the significantly heritable functional connections for both datasets. Heritability estimates in these and other networks showed a similar pattern of genetic control in both the GOBS and the HCP with greater evidence for statistical significance (i.e., lower p‐values) observed in the HCP subjects.
Table 1.
Heritability estimates for measures derived from resting state networks (RSNs) from seed‐based analysis approach
| GOBS (N = 334) | HCP (N = 518) | ||||
|---|---|---|---|---|---|
| Network | Connectionsa | Heritabilityb | p‐value | Heritabilityb | p‐value |
| Auditory network (AN) | Auditory cortices L–Auditory cortices R | 0.12 (0.16) | .209 | 0.05 (0.09) | .275 |
| Auditory cortices R–Auditory cortices L | 0.05 (0.14) | .356 | 0.05 (0.08) | .260 | |
| Attention network (AttN) | Middle FG L–SPL L | 0.20 (0.12) | .031 | 0.10 (0.15) | .288 |
| SPL L–Middle FG L | 0.21 (0.12) | .024 | 0.08 (0.11) | .213 | |
| Middle FG R–SPL R | 0.32 (0.12) | .001 | 0.27 (0.14) | .018 | |
| SPL R–Middle FG R | 0.31 (0.12) | .002 | 0.32 (0.14) | .005 | |
| Default mode network (DMN) | PCC/precuneus–Bilateral temporal–parietal region | 0.34 (0.16) | .014 | 0.27 (0.09) | 1.0 × 10−7 |
| Bilateral temporal–parietal region–vmFC | 0 | .500 | 0.14 (0.09) | .008 | |
| vmFC–PCC/precuneus | 0.09 (0.15) | .276 | 0.15 (0.11) | .025 | |
| Bilateral temporal–parietal region–PCC/precuneus | 0.09 (0.13) | .244 | 0.27 (0.09) | 4.6 × 10−8 | |
| vmFC– Bilateral temporal–parietal region | 0 | .500 | 0.23 (0.12) | .002 | |
| PCC/precuneus–vmFC | 0.35 (0.17) | .014 | 0.09 (0.1) | .120 | |
| Executive control network (ECN) | ACC–Bilateral medial FG | 0.17 (0.14) | .088 | 0.17 (0.11) | .023 |
| Bilateral medial FG–ACC | 0.23 (0.14) | .034 | 0.23 (0.11) | .003 | |
| Fronto‐parietal network (FPN) | Frontal area L–Parietal area L | 0.14 (0.14) | .149 | 0.16 (0.11) | .019 |
| Parietal area L–Frontal area L | 0.13 (0.14) | .169 | 0.16 (0.11) | .018 | |
| Frontal area R–Parietal area R | 0.31 (0.15) | .016 | 0.19 (0.14) | .034 | |
| Parietal area R–Frontal area R | 0.29 (0.15) | .025 | 0.27 (0.14) | .042 | |
| Salience network (SN) | ACC–INS L | 0.20 (0.13) | .062 | 0.07 (0.09) | .121 |
| INS L–INS R | 0.24 (0.12) | .019 | 0.25 (0.11) | .002 | |
| INS R–ACC | 0 | .500 | 0.13 (0.08) | .005 | |
| INS L–ACC | 0.16 (0.12) | .084 | 0.20 (0.11) | .002 | |
| INS R–INS L | 0.18 (0.12) | .049 | 0.31 (0.12) | 3.8 × 10−4 | |
| ACC–INS R | 0 | .500 | 0.05 (0.06) | .142 | |
| Sensorimotor network (SMN) | Motor area L–SMA | 0.09 (0.14) | .255 | 0.29 (0.15) | .017 |
| SMA–Motor area R | 0 | .500 | 0.14 (0.12) | .113 | |
| Motor area R–Motor area L | 0.32 (0.20) | .041 | 0.27 (0.14) | .009 | |
| SMA–Motor area L | 0.06 (0.12) | .302 | 0 | .500 | |
| Motor area R–SMA | 0 | .500 | 0.15 (0.13) | .108 | |
| Motor area L–Motor area R | 0.32 (0.20) | .045 | 0.25 (0.13) | .008 | |
| Visual network (VN) | Medial visual areas–occipital visual areas | 0.21 (0.15) | .062 | 0.14 (0.09) | .021 |
| Occipital visual areas–lateral visual areas | 0.36 (0.14) | .004 | 0.17 (0.11) | .029 | |
| Lateral visual areas–Medial visual areas | 0.12 (0.14) | .168 | 0.03 (0.04) | .191 | |
| Occipital visual areas–medial visual areas | 0.32 (0.15) | .009 | 0.13 (0.09) | .042 | |
| Lateral visual areas–occipital visual areas | 0.13 (0.14) | .161 | 0.15 (0.09) | .040 | |
| Medial visual areas–Lateral visual areas | 0.17 (0.14) | .100 | 0.06 (0.05) | .060 | |
GOBS = genetic of brain structure and function study; HCP = human connectome project; L = left; R = right; FG = frontal gyrus; SPL = superior parietal lobule; PCC = posterior cingulate cortex; vmFC = ventromedial frontal cortex; ACC = anterior cingulate cortex; INS = insula; SMA = supplementary motor area.
Bolded figures are significant at 5% false discovery rate (FDR). Regions are based off of Figure 2.
Estimated heritability, h 2 (SE).
3.2. Dual regression analysis
Heritability estimates of dual‐regression‐based connectivity phenotypes for both cohorts are summarized in Table 2. Heritability estimates were similar for the connectivity values calculated by the seed‐based and dual regression approaches. For example, in the GOBS dataset, connectivity values for the DMN ROIs were significantly heritable from posterior cingulate/precuneus to bilateral temporal–parietal regions (h 2 = 30%, p = .014) and ventromedial frontal cortex (h 2 = 25%, p = .038) (Table 2). The connection from bilateral temporal–parietal regions to posterior cingulate/precuneus was likewise significantly heritable (h 2 = 26%, p = .035). The HCP dataset showed the greater evidence of statistical significance (heritability from posterior cingulate/precuneus to bilateral temporal–parietal regions; h 2 = 31%, p = 4.0 × 10−8 and from bilateral temporal–parietal regions to posterior cingulate/precuneus; h 2 = 30%, p = 3.8 × 10−8). Heritability estimates in other networks showed a similar pattern of genetic control in both datasets. We found some additional significantly heritable functional connections in the dual regression analysis compared with the seed based analysis approach for both datasets. For GOBS dataset, the mean heritability estimate for the regions that showed significantly heritable FC within DMN, using a dual regression approach implemented in the FSL software, was 0.35 (±0.06) (Glahn, et al., 2010). With a modification of this approach, the mean heritability estimate for the functional connections that showed significantly heritable FC measures was 0.27 (±0.03). The mean heritability value was 0.34 (±0.01) in the seed‐based analysis. For HCP dataset, mean heritability estimates were 0.22 (±0.08) for dual regression, and 0.21 (±0.06) for the seed‐based analysis.
Table 2.
Heritability estimates for measures derived from resting state networks (RSNs) from dual regression analysis approach
| GOBS (N = 334) | HCP (N = 518) | ||||
|---|---|---|---|---|---|
| Network | Connectionsa | Heritabilityb | p‐value | Heritabilityb | p‐value |
| Auditory network (AN) | Auditory cortices L–Auditory cortices R | 0.07 (0.13) | .479 | 0.07 (0.09) | .188 |
| Auditory cortices R–Auditory cortices L | 0 | . 500 | 0.06 (0.08) | .221 | |
|
Attention network (AttN) |
Middle FG L–SPL L | 0.25 (0.15) | .041 | 0.27 (0.15) | .031 |
| SPL L–Middle FG L | 0.25 (0.15) | .043 | 0.17 (0.13) | .091 | |
| Middle FG R–SPL R | 0.29 (0.15) | .029 | 0.31 (0.13) | .009 | |
| SPL R–Middle FG R | 0.36 (0.16) | .010 | 0.40 (0.12) | .002 | |
|
Default mode network (DMN) |
PCC/precuneus–Bilateral temporal–parietal region | 0.30 (0.16) | .014 | 0.31 (0.09) | 4.0×10−8 |
| Bilateral temporal–parietal region–vmFC | 0 | .500 | 0.15 (0.09) | .010 | |
| vmFC–PCC/precuneus | 0.09 (0.11) | .279 | 0.14 (0.10) | .038 | |
| Bilateral temporal–parietal region–PCC/precuneus | 0.26 (0.16) | .035 | 0.30 (0.09) | 3.8×10−8 | |
| vmFC– Bilateral temporal–parietal region | 0 | .500 | 0.20 (0.11) | .004 | |
| PCC/precuneus–vmFC | 0.25 (0.17) | .038 | 0.08 (0.09) | .131 | |
| Executive control network (ECN) | ACC–Bilateral medial FG | 0.26 (0.14) | .026 | 0.20 (0.12) | .016 |
| Bilateral medial FG–ACC | 0.26 (0.13) | .024 | 0.22 (0.10) | .009 | |
|
Fronto‐parietal network (FPN) |
Frontal area L–Parietal area L | 0.13 (0.14) | .159 | 0.17 (0.12) | .022 |
| Parietal area L–Frontal area L | 0.10 (0.15) | .235 | 0.15 (0.11) | .027 | |
| Frontal area R–Parietal area R | 0.33 (0.14) | .010 | 0.23 (0.14) | .041 | |
| Parietal area R–Frontal area R | 0.30 (0.16) | .027 | 0.30 (0.14) | .023 | |
|
Salience network (SN) |
ACC–INS L | 0.26 (0.15) | .047 | 0.07 (0.08) | .116 |
| INS L–INS R | 0.26 (0.16) | .043 | 0.26 (0.12) | .001 | |
| INS R–ACC | 0.07 (0.15) | .318 | 0.15 (0.09) | .004 | |
| INS L–ACC | 0.14 (0.14) | .160 | 0.20 (0.11) | .002 | |
| INS R–INS L | 0.25 (0.14) | .037 | 0.35 (0.12) | .001 | |
| ACC–INS R | 0 | .500 | 0.07 (0.06) | .099 | |
|
Sensorimotor network (SMN) |
Motor area L–SMA | 0.12 (0.14) | .179 | 0.30 (0.13) | .013 |
| SMA–Motor area R | 0.05 (0.18) | .397 | 0.24 (0.14) | .009 | |
| Motor area R–Motor area L | 0.37 (0.18) | .013 | 0.27 (0.13) | .008 | |
| SMA–Motor area L | 0.13 (0.13) | .151 | 0.26 (0.14) | .009 | |
| Motor area R–SMA | 0.06 (0.16) | .352 | 0.16 (0.14) | .088 | |
| Motor area L–Motor area R | 0.40 (0.16) | .012 | 0.13 (0.12) | .135 | |
|
Visual network (VN) |
Medial visual areas–occipital visual areas | 0.25 (0.15) | .044 | 0.24 (0.08) | .006 |
| Occipital visual areas–lateral visual areas | 0.35 (0.14) | .005 | 0.32 (0.11) | .001 | |
| Lateral visual areas–Medial visual areas | 0.14 (0.14) | .135 | 0.04 (0.05) | .194 | |
| Occipital visual areas–medial visual areas | 0.31 (0.14) | .012 | 0.22 (0.11) | .009 | |
| Lateral visual areas–occipital visual areas | 0.16 (0.14) | .121 | 0.15 (0.09) | .044 | |
| Medial visual areas–Lateral visual areas | 0.22 (0.13) | .050 | 0.12 (0.06) | .026 | |
GOBS = genetic of brain structure and function study; HCP = human connectome project; L = left; R = right; FG = frontal gyrus; SPL = superior parietal lobule; PCC = posterior cingulate cortex; vmFC = ventromedial frontal cortex; ACC = anterior cingulate cortex; INS = insula; SMA = supplementary motor area.
Bolded figures are significant at 5% FDR. Regions are based off of Figure 2.
Estimated heritability, h 2 (SE).
3.2.1. Comparison of GOBS and HCP heritability measurements
Heritability estimates for functional connectivity measures in the GOBS cohorts were plotted versus heritability estimates for these in the HCP cohorts for seed‐based analysis and dual regression analysis approaches (Figure 3). The lines represent the result of the linear correlation between two cohorts. There was a significant and positive correlation (r = .38, p = .023 for seed‐based analysis approach and r = .50, p = .002 for dual regression analysis approach) between heritability estimates for functional connectivity measurements in the GOBS and HCP cohorts.
Figure 3.
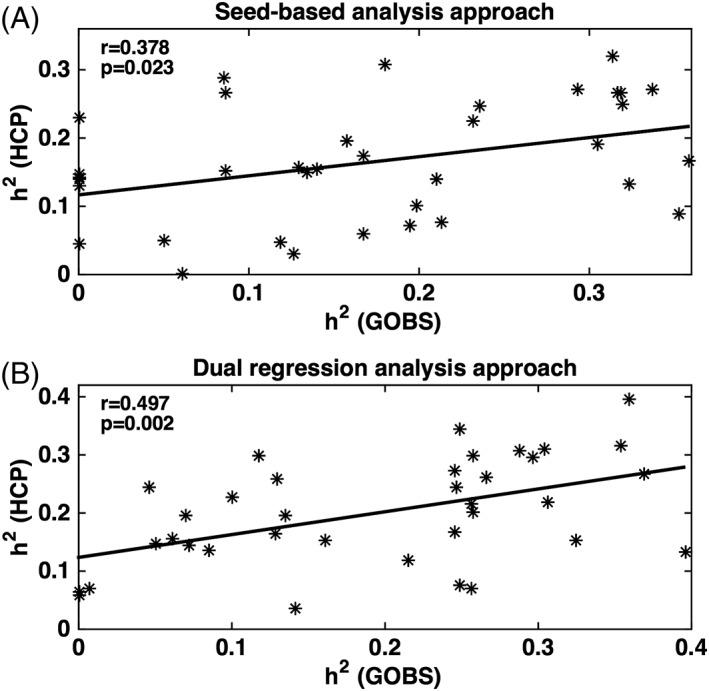
Heritability estimates for functional connectivity measured in the GOBS are presented as a scatter versus heritability estimates calculated in the HCP cohorts using seed‐based analysis approach (a) and dual regression analysis approach (b). The line represents the result of the linear correlation between two cohorts that reported a positive and significant correlation [r = .378, p = .023 for seed‐based approach (a) and r = .497, p = .002 for dual regression approach (b)]
3.2.2. Execution time
All analyses were carried out at the Washington University Center for High‐Performance Computing. Dual regression analysis of GOBS resting data took about 30 min per subject/network on a modern linux server node. Analysis of HCP data consisting of N = 2,400 fMRI volumes took about 6 hr per subject/network.
4. DISCUSSION
We performed heritability analyses of the connectivity phenotypes extracted from GOBS and HCP. We used two BigData developments: the ENIGMA rsfMRI pipeline and the spatial patterns of resting state networks derived from BrainMap database. We show that about approximately 31% of the variance in functional connectivity may be explained by additive genetic contribution. We performed these analyses in two datasets collected 10 years using a conventional and state‐of‐the‐art connectivity protocols. We observed good consistency in the magnitude of genetic influences on resting state connectivity. Building on prior work in individual cohorts, this experiment provides direct evidence that connectivity within the DMN and other intrinsic brain networks is influenced by genetic factors. Establishing heritability of resting state functional connectivity provides important information to support the use of these measures as phenotypes for genetic studies or functionally characterize genes influencing brain functions in health and illness. Showing significant heritability is the first step before these metrics they can be considered an intermediate phenotype or endophenotype for neurological or mental illnesses (Glahn et al., 2012). Overall, our study demonstrated robust estimates of additive genetic variability for functional connectivity measurements and showed a good agreement between two independent cohorts and with the previously reported values (Glahn et al., 2010).
We showed that functional connectivity measurements showed a consistent heritability pattern in two independent cohorts collected 10 years apart and using different recruitment and imaging protocols. Some discrepancies in heritability estimates and ACE decomposition are expected between cohorts as different as the ones chosen, and some proportion of the differences in heritability are likely to be real. For instance, the GOBS study recruited members of the extended families, aged 18–80 years; HCP collected data from twins and siblings in a narrow age range to minimize age‐related phenotypic variance and the age‐by‐genotype interaction (Batouli et al., 2013; Brouwer et al., 2012; Glahn et al., 2013). GOBS data was collected with the conventional rsfMRI protocol; HCP data was collected using a state‐of‐art temporal and spatial resolution enabled by advanced gradient system. Despite these differences, we still observed very similar heritability values and the pattern between two datasets. The findings are consistent with prior studies that showed similarities in heritability values and pattern between HCP and conventional datasets. For instance, the spatial pattern of heritability values for the fractional anisotropy of water diffusion calculated from aggregated ENIGMA datasets that included GOBS data was predictive of the heritability pattern in HCP data (Kochunov, Jahanshad, et al., 2015a). Likewise, there was an excellent agreement in the genetic relationship between processing speed and white matter integrity measured in HCP and members of Amish Connectome Project (Kochunov, Thompson, et al., 2015b). Overall, these data show that the resting state connectivity metrics are under a modest‐to‐moderate genetic control and its heritability is stable across populations and in terms of image acquisition, in legacy, and state‐of‐the art samples.
4.1. Relationship to other analysis approaches
The ENIGMA‐rsfMRI pipeline is uni‐modal and is based on a population‐based brain template. Prior works on rsfMRI analysis are generally multimodal and use functional and structural data (Calhoun et al., 2017). In such approaches, a spatial transformation of the functional data to the structural image for each subject is followed by nonlinear registration of the structural data to an anatomic template. The transformation field is then applied to the rsfMRI data for regressions of global connectivity signals and ROI analyses in a common anatomical frame. Arguably, a disadvantage of this approach is that spatial distortions in rsfMRI data may prevent consistent registration with the structural data unless corrected for. Calhoun et al. examined the unimodal and multimodal approaches in four datasets and registering functional data to a population template led to superior registration quality, lower inter‐subject variance, and higher statistical significance when compared with the multimodal approach (Calhoun, et al., 2017). This problem is likely to be encountered in ENIGMA studies that aggregate data collected over the past 20 years. The site‐to‐site variability in the quality of the T1w data and the variance in registration quality between T1w and rsfMRI images is likely be more prominent for ENIGMA studies and therefore poses a risk of influencing the results of the overall rsfMRI analysis. Likewise, we showed that the use of a deformable template improved registration for individual EPI images, including ventricular overlap, when compared with the standard ICBM‐152 template (Adhikari et al., 2017).
Our analysis pipeline incorporates the MP‐PCA denoising algorithm that improves SNR/tSNR properties of the time series data (Veraart, Fieremans, et al., 2016a; Veraart, Novikov, et al., 2016b), with no loss of spatial resolution of the image. MP‐PCA removes thermal noise artifacts that can have prominent site‐specific spatial patterns. In a multi‐site study of diffusion MRI findings in schizophrenia, the MP‐PCA approach improved the agreement in findings across three sites and improved the overall agreement with the previous meta‐analytic findings in this disorder (Kochunov et al., 2018). The MP‐PCA improvements were especially prominent in the HCP data. The HCP protocol pushed the limits of spatial and temporal resolution of the rsfMRI but the tSNR values in this dataset (~21) were less than the suggested minimum tSNR (30–40) (Murphy, Bodurka, & Bandettini, 2007). MP‐PCA approach improved the average tSNR value to 40 without introduction of spatial or temporal smoothing that may have partially negated the advantages of this protocol.
The ENIGMA rsfMRI pipeline is built using NIH‐supported software—AFNI—that is freely available to both noncommercial and commercial users. Free‐license software opens ENIGMA collaboration to commercial entities such as pharmacological companies. It is a unimodal analysis workflow designed for consistent retrospective analyses of state‐of‐the‐art and legacy data. The pipeline incorporates stringent quality assurance (QA) and quality control steps. It incorporates traditional QA measurements to detect and censor motion and other types artifacts that are detectable visually. It also uses novel analysis of the heterogeneity of the thermal noise within imaging volume to enable identification of more subtle artifacts such as time‐and‐space related variability in coil sensitivity profiles. Efforts are now ongoing to compare the performance of ENIGMA rsfMRI analysis pipeline across multiple cohorts with other rsfMRI analysis pipelines.
4.2. Limitations
Reduction of methodological differences between two cohorts through regression of thermal noise and harmonization analyses should lead to an overall decrease in the noise (random environment) component of the variance and improved agreement in heritability values. However, this may also lead to an elevation in the shared environmental variance if some or all family members tend to be scanned on the same day. The impact of common environmental factors was not evaluated here, in order to maintain the same design as in the prior study (Glahn et al., 2010). Moreover, there was a good agreement between heritability estimates in GOBS cohort where data collection for extended family members were separated in time and HCP (twin data were collected on the same day). This suggests that the effects of elevation in common environmental variance due to improvements in SNR and homogenization approaches are likely to be minor.
This study was aimed at replicating previous heritability findings using a novel resting state analysis workflow. Additional work will be needed to formally evaluate the differences in the heritability between two cohorts.
ACKNOWLEDGMENTS
Support was received from NIH grants U54EB020403, U01MH108148, 2R01EB015611, R01MH112180, R01DA027680, R01MH085646, and T32MH067533. We are grateful to Human Connectome Project and the Center for High‐Performance Computing at Washington University, University of Washington St. Louis for allowing the use of their computational facility for this project.
Adhikari BM, Jahanshad N, Shukla D, et al. Comparison of heritability estimates on resting state fMRI connectivity phenotypes using the ENIGMA analysis pipeline. Hum Brain Mapp. 2018;39:4893–4902. 10.1002/hbm.24331
Funding information NIH, Grant/Award Numbers: T32MH067533, R01MH085646, R01DA027680, R01MH112180, 2R01EB015611, U01MH108148, U54EB020403
REFERENCES
- Adhikari, B. M. , Jahanshad, N. , Shukla, D. K. , Turner, J. A. , Grotegerd, D. , Dannlowski, U. , … Kochunov, P. (2017). A resting state fMRI analysis pipeline for pooling inference across diverse cohorts: The ENIGMA rs‐fMRI Protocol. Brain Imaging and Behavior, under review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy, L. , & Blangero, J. (1998). Multipoint quantitative‐trait linkage analysis in general pedigrees. American Journal of Human Genetics, 62, 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos, C. I. (1994). Robust variance‐components approach for assessing genetic linkage in pedigrees. American Journal of Human Genetics, 54, 535–543. [PMC free article] [PubMed] [Google Scholar]
- Batouli, S. A. , Sachdev, P. S. , Wen, W. , Wright, M. J. , Ames, D. , & Trollor, J. N. (2013). Heritability of brain volumes in older adults: The older Australian twins study. Neurobiology of Aging, 35(937), e5–e18. [DOI] [PubMed] [Google Scholar]
- Beasley, T. M. , Erickson, S. , & Allison, D. B. (2009). Rank‐based inverse normal transformations are increasingly used, but are they merited? Behavior Genetics, 39, 580–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann, C. F. , Mackay, C. E. , Filippini, N. , & Smith, S. M. (2009). Group comparison of resting‐state FMRI data using multi‐subject ICA and dual regression. OHBM, 2009. NeuroImage, 47(Suppl 1), S148. [Google Scholar]
- Biswal, B. , Yetkin, F. Z. , Haughton, V. M. , & Hyde, J. S. (1995). Functional connectivity in the motor cortex of resting human brain using echo‐planar MRI. Magnetic Resonance in Medicine, 34, 537–541. [DOI] [PubMed] [Google Scholar]
- Brouwer, R. M. , Mandl, R. C. , Schnack, H. G. , van Soelen, I. L. , van Baal, G. C. , Peper, J. S. , … Hulshoff Pol, H. E. (2012). White matter development in early puberty: A longitudinal volumetric and diffusion tensor imaging twin study. PLoS One, 7, e32316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun, V. D. , Wager, T. D. , Krishnan, A. , Rosch, K. S. , Seymour, K. E. , Nebel, M. B. , … Kiehl, K. (2017). The impact of T1 versus EPI spatial normalization templates for fMRI data analyses. Human Brain Mapping, 38, 5331–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. M. , Yang, P. F. , Wang, F. , Mishra, A. , Shi, Z. , Wu, R. , … Gore, J. C. (2017). Biophysical and neural basis of resting state functional connectivity: Evidence from non‐human primates. Magnetic Resonance Imaging, 39, 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, R. W. (1996). AFNI: Software for analysis and visualization of functional magnetic resonance neuroimages. Computers and Biomedical Research, 29, 162–173. [DOI] [PubMed] [Google Scholar]
- Edens, E. L. , Glowinski, A. L. , Pergadia, M. L. , Lessov‐Schlaggar, C. N. , & Bucholz, K. K. (2010). Nicotine addiction in light smoking African American mothers. Journal of Addiction Medicine, 4, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, L. , Sharp, K. , Alfaro‐Almagro, F. , Douaud, G. , Miller, K. , Marchini, J. , & Smith, S. (2017) The genetic basis of human brain structure and function: 1,262 genome‐wide associations found from 3,144 GWAS of multimodal brain imaging phenotypes from 9,707 UK Biobank participants. biorXiv.
- Filippini, N. , MacIntosh, B. J. , Hough, M. G. , Goodwin, G. M. , Frisoni, G. B. , Smith, S. M. , … Mackay, C. E. (2009). Distinct patterns of brain activity in young carriers of the APOE‐ε4 allele. Proceedings of the National Academy of Sciences of the United States of America, 106, 7209–7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, M. D. , & Raichle, M. E. (2007). Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nature Reviews. Neuroscience, 8, 700–711. [DOI] [PubMed] [Google Scholar]
- Glahn, D. C. , Curran, J. E. , Winkler, A. M. , Carless, M. A. , Kent, J. W., Jr. , Charlesworth, J. C. , … Blangero, J. (2012). High dimensional Endophenotype ranking in the search for major depression risk genes. Biological Psychiatry, 71, 6–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn, D. C. , Kent, J. W., Jr. , Sprooten, E. , Diego, V. P. , Winkler, A. M. , Curran, J. E. , … Blangero, J. (2013). Genetic basis of neurocognitive decline and reduced white‐matter integrity in normal human brain aging. Proceedings of the National Academy of Sciences of the United States of America, 110, 19006–19011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn, D. C. , Winkler, A. M. , Kochunov, P. , Almasy, L. , Duggirala, R. , Carless, M. A. , … Blangero, J. (2010). Genetic control over the resting brain. Proceedings of the National Academy of Sciences of the United States of America, 107, 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis, J. P. A. (2014). How to make more published research true. PLoS Medicine, 11, e1001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad, N. , Kochunov, P. , Sprooten, E. , Mandl, R. C. , Nichols, T. E. , Almassy, L. , … Glahn, D. C. (2013). Multi‐site genetic analysis of diffusion images and voxelwise heritability analysis: A pilot project of the ENIGMA‐DTI working group. Neuroimage, 81, 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson, M. , Beckmann, C. F. , Behrens, T. E. , Woolrich, M. W. , & Smith, S. M. (2012). FSL. NeuroImage, 62, 782–790. [DOI] [PubMed] [Google Scholar]
- Kochunov, P. , Dickie, E. W. , Viviano, J. D. , Turner, J. , Kingsley, P. B. , Jahanshad, N. , … Voineskos, A. N. (2018). Integration of routine QA data into mega‐analysis may improve quality and sensitivity of multisite diffusion tensor imaging studies. Human Brain Mapping, 39, 1015–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov, P. , Jahanshad, N. , Marcus, D. , Winkler, A. , Sprooten, E. , Nichols, T. E. , … Van Essen, D. C. (2015a). Heritability of fractional anisotropy in human white matter: A comparison of human Connectome project and ENIGMA‐DTI data. NeuroImage, 111, 300–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov, P. , Thompson, P. M. , Winkler, A. , Morrissey, M. , Fu, M. , Coyle, T. R. , … Hong, L. E. (2015b). The common genetic influence over processing speed and white matter microstructure: Evidence from the old order Amish and human Connectome projects. NeuroImage, 125, 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies, D. S. , Clare Kellya, A. M. , Uddin, L. Q. , Biswal, B. B. , Xavier Castellanos, F. , & Milham, M. P. (2007). Mapping the functional connectivity of anterior cingulate cortex. NeuroImage, 37, 579–588. [DOI] [PubMed] [Google Scholar]
- Mitra, A. , Snyder, A. Z. , Tagliazucchi, E. , Laufs, H. , Elison, J. , Emerson, R. W. , … Raichle, M. (2017). Resting‐state fMRI in sleeping infants more closely resembles adult sleep than adult wakefulness. PLoS One, 12, e0188122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, K. , Bodurka, J. , & Bandettini, P. A. (2007). How long to scan? The relationship between fMRI temporal signal to noise and necessary scan duration. NeuroImage, 34, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor, C. E. , McCutcheon, V. V. , Pommer, N. E. , Nelson, E. C. , Grant, J. D. , Duncan, A. E. , … Heath, A. C. (2011). Common genetic and environmental contributions to post‐traumatic stress disorder and alcohol dependence in young women. Psychological Medicine, 41, 1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. M. , Fox, P. T. , Miller, K. L. , Glahn, D. C. , Fox, P. M. , Mackay, C. E. , … Beckmann, C. F. (2009). Correspondence of the brain's functional architecture during activation and rest. Proceedings of the National Academy of Sciences of the United States of America, 106, 13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. M. , Jenkinson, M. , Woolrich, M. W. , Beckmann, C. F. , Behrens, T. E. J. , Johansen‐Berg, H. , … Matthews, P. M. (2004). Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage, 23, S208–S219. [DOI] [PubMed] [Google Scholar]
- Van Essen, D. C. , Smith, S. M. , Barch, D. M. , Behrens, T. E. , Yacoub, E. , & Ugurbil, K. (2013). The WU‐Minn human Connectome project: An overview. NeuroImage, 80, 62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraart, J. , Fieremans, E. , & Novikov, D. S. (2016a). Diffusion MRI noise mapping using random matrix theory. Magnetic Resonance in Medicine, 76, 1582–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraart, J. , Novikov, D. S. , Christiaens, D. , Ades‐Aron, B. , Sijbers, J. , & Fieremans, E. (2016b). Denoising of diffusion MRI using random matrix theory. NeuroImage, 142, 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wey, H. Y. , Phillips, K. A. , McKay, D. R. , Laird, A. R. , Kochunov, P. , Davis, M. D. , … Fox, P. T. (2014). Multi‐region hemispheric specialization differentiates human from nonhuman primate brain function. Brain Structure & Function, 219, 2187–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T. L. , Wang, F. , Anderson, A. W. , Chen, L. M. , Ding, Z. , & Gore, J. C. (2016). Effects of anesthesia on resting state BOLD signals in white matter of non‐human primates. Magnetic Resonance Imaging, 34, 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]


