Abstract
Docosahexaenoic acid (DHA, 22:6) is an n-3 polyunsaturated fatty acid (n-3 PUFA) that influences immunological, metabolic, and neurological responses through complex mechanisms. One structural mechanism by which DHA exerts its biological effects is through its ability to modify the physical organization of plasma membrane signaling assemblies known as sphingomyelin/cholesterol (SM/chol)-enriched lipid rafts. Here we studied how DHA acyl chains esterified in the sn-2 position of phosphatidylcholine (PC) regulate the formation of raft and non-raft domains in mixtures with SM and chol on differing size scales. Coarse grained molecular dynamics simulations showed that 1-palmitoyl-2-docosahexaenoylphosphatylcholine (PDPC) enhances segregation into domains more than the monounsaturated control, 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC). Solid state 2H NMR and neutron scattering experiments provided direct experimental evidence that substituting PDPC for POPC increases the size of raft-like domains on the nanoscale. Confocal imaging of giant unilamellar vesicles with a non-raft fluorescent probe revealed that POPC had no influence on phase separation in the presence of SM/chol whereas PDPC drove strong domain segregation. Finally, monolayer compression studies suggest that PDPC increases lipid-lipid immiscibility in the presence of SM/chol compared to POPC. Collectively, the data across model systems provide compelling support for the emerging model that DHA acyl chains of PC lipids tune the size of lipid rafts, which has potential implications for signaling networks that rely on the compartmentalization of proteins within and outside of rafts.
Keywords: Lipid rafts, polyunsaturated fatty acids, biomimetic membranes, cholesterol, docosahexaenoic acid (DHA)
1. INTRODUCTION
Docosahexaenoic acid1 (DHA) is an n-3 polyunsaturated fatty acid (n-3 PUFA) that is generally consumed in low amounts in the western diet [1]. Pre-clinical and clinical studies suggest that dietary DHA supplementation has the potential to improve cardiovascular and inflammatory endpoints for a range of metabolic diseases [2–9]. Furthermore, DHA plays a critical role in neuronal development, which has implications for a range of neurodegenerative disorders [10]. Therefore, the clinical development of DHA for differing diseases requires a fundamental understanding of its underlying mechanism of action.
DHA influences cellular function through overlapping and complex mechanisms. These include DHA serving as a substrate for enzymes such as lipoxygenases to generate potent downstream metabolites, targeting of transcription factors to modify gene expression, and controlling cellular signaling [11] (and references within). Many studies demonstrate that DHA acyl chains, upon esterification into plasma membrane phospholipids, reorganize plasma membrane phospholipid-protein spatial distribution that is critical for downstream signaling networks and gene activation across cell types [12;13]. In particular, DHA acyl chains remodel the architecture of lipid raft domains, which are operationally defined as sphingolipid/cholesterol-enriched nanoscale assemblies that can coalesce into larger signaling platforms [13–17].
Studies of protein-free model membranes of well-defined composition have established that DHA strongly influences the molecular composition and formation of lipid raft-like domains enriched in sphingomyelin (SM) and cholesterol (chol) [12;18]. We previously reported that DHA-containing phosphatidylethanolamines (PE), due to the poor affinity of DHA for chol, laterally separated from lipid raft molecules in monolayer and bilayer models [19–21]. Strikingly, in subsequent NMR work we found that a DHA-containing phosphatidylcholine (PC) directly infiltrated raft-like domains [22]. This finding was in agreement with data from in vitro and mouse models showing that DHA incorporated directly into lipid raft-like domains [17;23–27]. However, what DHA does to raft organization is unclear – whether rafts become bigger or smaller remains controversial [23;28]. To address this issue, our focus here is on how DHA esterified to a phospholipid affects the size of rafts.
In this study, we investigated the influence that a heteroacid PC containing palmitic acid in the sn-1 position and DHA in the sn-2 position has on domains formed in mixtures with the lipid raft molecules SM and chol (Figure 1). By a combination of coarse grained molecular dynamics (CG MD) simulations, solid state 2H NMR, small angle neutron scattering (SANS) and imaging of giant unilamellar vesicles (GUVs), we observed the impact of 1-palmitoyl-2-docosahexaenoyl-phosphatidylcholine (16:0–22:6PC, PDPC) vs. 1-palmitoyl-2-oleoyl-phosphatidylcholine (16:0–18:1PC, POPC), as a monounsaturated control, on the size of raft-like and non-raft domains on the nanometer and micron scale. Complementary studies with monolayers were conducted to calculate the excess area per molecule and Gibbs free energy of lipid mixing.
Figure 1. Molecular structure of lipids.
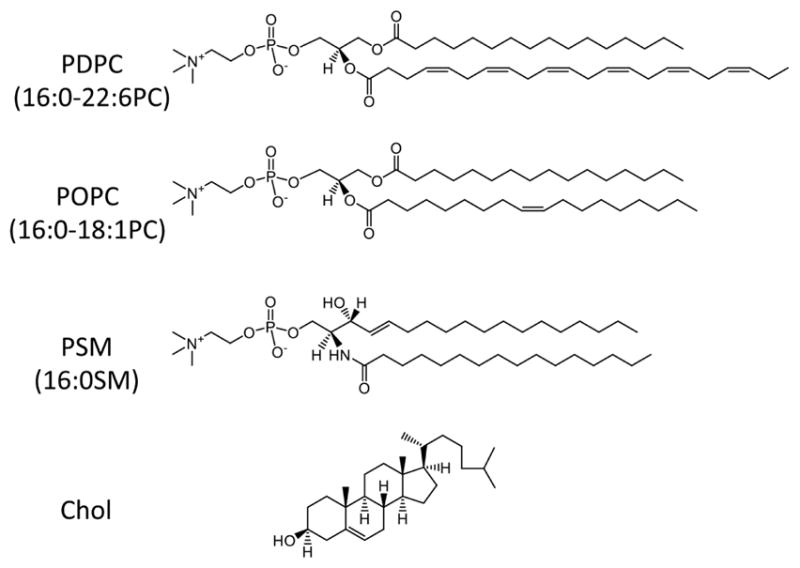
Detailed structures for the following lipids are illustrated: 1-palmitoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine (PDPC; 16:0–22:6PC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC; 16:0–18:1PC), palmitoyl-sphingomyelin (PSM; 16:0SM), cholesterol (Chol).
2. MATERIALS AND METHODS
2.1 MD simulations
The CG MD simulations of membrane bilayers were performed by the GROMACS program v4.6.5 [29] with the Martini CG force field (version 2.1) [30]. Followed by the standard setup protocols, the initial membrane-water system was constructed using the Insane python tool [31] with an initial size of 17.5 nm × 17.5 nm in the xy dimension (plane of the membrane) and 8 nm total length in the z direction (normal to the membrane). The simulated bilayers were composed of 352 unsaturated lipids (POPC or PDPC), 352 SM and 352 chol molecules (1:1:1 mol). There were 8544 CG waters and 94 Na+ and Cl− ions in the POPC/SM/Chol system, and 8254 CG waters and 91 Na+ and Cl− ions in the PDPC/SM/Chol system. The topology files used in the simulations were obtained from http://cgmartini.nl.
The initial systems were energy minimized and equilibrated for 10 ns with a 0.02 ps time step. After that, 30 μs production runs with 0.03 ps time steps were performed. All simulations were run in the NPT (isobaric-isothermal) ensemble with periodic boundary conditions. The van der Waals (vdW) interactions were cut off at 1.2 nm, the Lennard-Jones interactions were shifted to zero in the range of 0.9–1.2 nm, the Columbic interactions were smoothly shifted to zero from 0 to 1.2 nm and the relative dielectric constant was 15 as the default value used in the force field [30]. V-rescale heat baths [32] were coupled to both the bilayer and the solution separately with a temperature of 303 K and a coupling constant of 1 ps. The semi-isotropic Parrinello-Rahman pressure coupling scheme [33] was applied to the system to maintain the pressure at 1 bar with a coupling constant of 12 ps and a compressibility of 3×10−4 bar−1. A 10 ns time step was used to update the neighbor list of non-bonded interactions, and the corresponding cutoff was 1.4 nm. The simulations were run on the Karst high-throughput computing cluster Karst at Indiana University. Trajectory analysis and visualizations were done in VMD [34].
2.2 Solid state 2H NMR
[2H31]-N-palmitoyl-sphingomyelin (PSM-d31) was synthesized as previously described [35], while the sources for PDPC and POPC and for chol were Avanti Polar Lipids (Alabaster, AL) and Sigma Chemical (St. Louis, MO), respectively. Multilamellar dispersions of 50 wt% PDPC/PSM-d31/chol and POPC/PSM-d31/chol (1:1:1 mol) were prepared in 50 mM Tris (pH 7.5) buffer implementing precautions to minimize oxidation as described in earlier publications [22].
Solid state 2H NMR experiments were performed on a homebuilt NMR spectrometer operating at 46.0 MHz with a 7.05 T superconducting magnet (Oxford Instruments, Osney Mead, UK) [22]. A phase-alternated quadrupolar echo sequence (90°x-τ-90°y-aquire-delay) was used to eliminate spectral distortion due to the receiver recovery time [22]. Parameters were 90° pulse width = 3.7 μs; separation between pulses τ = 50 μs; delay between pulse sequences = 1.0 s; sweep width = ±100 kHz; and number of scans = 8,192. The 2H NMR spectra acquired are a superposition of powder patterns from all positions of isotopic labeling in the amide-linked chain of PSM-d31 [36]. Each powder pattern has a pair of most intense peaks split in frequency by Δvr that relates to an order parameter SCD for the C-2H bond in a chain segment according to a standard expression [37].
2.3 Small-angle neutron scattering
Large unilamellar vesicle (LUV) samples were prepared as previously described [38], with most steps performed in a glove box purged with argon to < 0.1% O2 (OXY-sen oxygen monitor, Alpha Omega Instruments). POPC and PDPC were obtained from Avanti Polar Lipids and chol from Nu Check Prep, Inc. (Elysian, MN). Briefly, lipid mixtures were prepared by transferring volumes of lipids and chol stocks in chloroform to a glass vial with a glass syringe. Organic solvent was removed with a nitrogen stream, followed by vacuum drying for > 3 h. Dry lipid films were hydrated to a concentration of 15 mg/ml, with a 29.0 % (v/v) D2O/H2O mixture preheated to 40 °C, and then vortexed to generate multilamellar vesicles (MLVs). The MLV suspension was incubated at 40 °C for 1 h, followed by 5 freeze/thaw cycles between −80 and 40 °C. LUVs were prepared using a miniextruder (Avanti Polar Lipids) assembled with a single 50 nm pore size polycarbonate filter, and heated to 40 °C. After extrusion, a 0.3 ml aliquot of the sample was loaded into a 1 mm path-length quartz banjo cell (Hellma USA, Plainview, NY). A separate 0.2 ml sample aliquot was mixed with 0.4 mL D2O, and the resulting 78% D2O sample (~ 5 mg/ml lipid concentration) was loaded into a 2 mm path-length banjo cell.
Small-angle neutron scattering (SANS) experiments were conducted at Oak Ridge National Laboratory (ORNL) using the CG-3 BioSANS instrument of the High Flux Isotope Reactor (HFIR). Banjo cells containing the LUV suspensions were mounted in a temperature-controlled cell holder with an accuracy of 1°C. Data were collected at a 14.5 m sample-to-detector distance (SDD) using 6 Å wavelength neutrons (FWHM 15%), resulting in a total usable scattering vector of 0.006 < q < 0.05 Å−1. Scattered neutrons were collected with a two-dimensional (1 × 1 m2) 3He position-sensitive detector (ORDELA, Inc., Oak Ridge, TN) with 192 × 192 pixels. The 2D data were reduced using the software package Mantid [39]. During reduction, data were corrected for detector pixel sensitivity, dark current, sample transmission, and background scattering from water. The one-dimensional scattering intensity I(q) [q = 4π sin(θ)/λ, where λ is the neutron wavelength and 2θ is the scattering angle relative to the incident beam] was obtained by radial averaging of the corrected 2D data.
Nanodomain sizes were determined by analysis of I(q) data using a coarse-grained Monte Carlo method described elsewhere [40]. Additional details are found in the Supporting Information.
2.4 Confocal imaging and analyses of giant unilamellar vesicles
GUVs were constructed by electroformation as previously described [41]. Briefly, PDPC/SM/chol (1:1:1) or POPC/SM/chol (1:1:1) were co-dissolved in the presence of 0.1 mol% of the non-raft probe Texas-Red DHPE. 5.0 μg of total lipid were spread onto the conductive side of an indium tin oxide coated glass slide. The lipid-coated slide was subjected to overnight vacuum pumping in the dark to remove excess solvent. GUV electroformation was performed at 55 °C, in the dark, using a 250 mM sucrose solution. Imaging was conducted with an Olympus FV1000 Confocal Microscope using a 60X 1.35NA oil immersion objective (Olympus, Waltham, MA). The Texas-Red DHPE probe was excited with an Argon laser as previously described. All acquired images were of GUV equatorial cross-sections. Analysis of lipid domains was conducted with NIH ImageJ software [41;42].
2.5 Langmuir monolayer compression studies
Pressure-area isotherms of PDPC/SM/chol (1:1:1) or POPC/SM/chol (1:1:1) were generated at 23°C [41;42]. Isotherms were acquired by spotting lipids on a degassed subphase of 10 mM sodium phosphate buffer (pH 7.4). Excess chloroform was allowed to evaporate for 10 min prior to monolayer compression. The surface pressure-area isotherms were used to calculate the surface elasticity modulus ( ):
| (Eq. 1) |
A represents the mean molecular area of the lipid mixture of interest at the indicated surface pressure (π) [43]. The ideal mean molecular area of multi-component mixed monolayers at a constant surface pressure (π) was calculated from the isotherms with:
| (Eq. 2) |
Xn represents the mol fraction of each individual component and An represents the mean molecular area of each component [43]. The excess area/molecule (Aex) was calculated at a given surface pressure (π) by:
| (Eq. 3) |
Here, (A12…n) represents the mean molecular area of the mixed monolayer of interest at a given surface pressure (π) [44–46]. A negative excess area/molecule indicates attractive forces and a positive excess area/molecule indicates repulsive forces [44]. The Gibbs free energy of mixing (ΔGmix) was calculated by the following relation to quantify lipid-lipid miscibility:
| (Eq. 4) |
ΔGex represents the excess Gibbs free energy of mixing and ΔGideal is the ideal Gibbs free energy of mixing. ΔGex was calculated from the pressure-area isotherms according to the following expression:
| (Eq. 5) |
An and Xn are the mean molecular area and mol fraction of each component, respectively, at a given surface pressure (π) [44]. ΔGideal was calculated from:
| (Eq. 6) |
R represents the ideal gas constant, and T represents the temperature in Kelvin [44]. All lipid mixtures were acquired multiple times to ensure reproducibility.
2.6 Statistical analyses
Statistical analyses for imaging and lipid-lipid mixing studies were conducted as previously described. We first ensured data sets displayed parametric distributions followed by two-tailed Student’s t test. For the NMR spectroscopy studies, a reproducibility of ±1–2% applies to the first moment that characterizes the spectral shape (29) obtained from multiple acquisitions in single 2H NMR experiment. P values <0.05 were considered significant.
3. RESULTS
3.1 CG MD simulations reveal PDPC enhances segregation into domains in raft-forming mixtures
We performed CG MD simulations comparing the evolution in lateral distribution of lipids in POPC/PSM/chol and PDPC/SM/chol bilayers (1:1:1 mol) at 30 °C. Top-view snapshots of the simulated monolayers (POPC- and PDPC-containing in upper and lower left panels, respectively) at the beginning (left column) and end (right column) of a 30 μs production run are presented in Figure 2. Color-coded circles represent the lateral location of the lipids, while chol is hidden. By visual inspection, it is apparent that the PDPC-containing mixture becomes more heterogeneous over time than the POPC-containing mixture. The grouping together of like lipids at the end of simulation is greater in PDPC/SM/chol (PDPC with PDPC (blue) and SM with SM (red) - Fig.2, lower right panel) than in POPC/SM/chol (POPC with POPC (green) and SM with SM (red) - Fig.2, upper right panel).
Figure 2. CG MD simulations of POPC and PDPC in the presence of SM/chol.
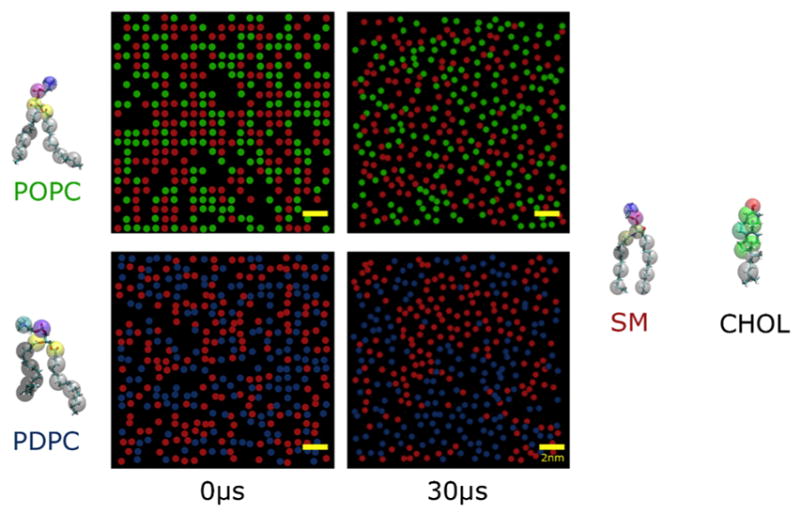
Top-view snapshots of monolayers of POPC/SM/chol (upper panel) and PDPC/SM/chol (lower panel) at the beginning (t = 0 μs, left column) and end (t = 30 μs, right column) of CG MD simulation production runs. Scale bar (yellow) is 2 nm. Color-coded circles indicate the lateral location of the phosphate head group on SM (red), POPC (green) and PDPC (blue). Chol is hidden to improve image clarity. The CG MD representation of the molecular structure of the lipids is shown to the side of the snapshots.
A quantitative measure of lipid clustering was achieved by counting nearest neighbors, which we define as lipid molecules within 1 nm of each other. The results are formatted as bar graphs of the number of nearest neighbor lipid [POPC (green bar), PDPC (blue bar), SM (red bar) and chol (white bar)] molecules around each POPC or PDPC and SM molecule on average over the entire duration of the simulation (Figure 3, upper panel). This analysis confirms that PDPC molecules segregate away from SM and chol in the PDPC/SM/chol bilayer, a segregation that is not apparent in the data for POPC in the POPC/SM/chol bilayer. PDPC has more PDPC (4.16 ± 0.23) and less SM (2.74 ± 0.23) and chol (3.30 ± 0.14) molecules for nearest neighbors, while SM has more SM (4.55 ± 0.35) and chol (4.49 ± 0.19) and less PDPC (2.74 ± 0.28) for nearest neighbors (Fig. 3, right upper panel). POPC has the same number of POPC (3.48 ± 0.13), SM (3.47 ± 0.13) and chol (3.71 ± 0.09) molecules for nearest neighbors as, within uncertainty, SM has SM (3.48 ± 0.13), POPC (3.47 ± 0.13) and chol (3.75 ± 0.09) molecules for nearest neighbors (Fig. 3, left upper panel).
Figure 3. Nearest neighbor and order parameter analyses of CG MD simulations.
Upper panel: Number of POPC (green bar), SM (red bar) and chol (white bar) molecules as nearest neighbors around POPC and SM in PDPC/SM/chol (left panel) and of PDPC (blue bar), SM (red bar) and chol (white bar) around PDPC and SM in PDPC/SM/chol (right). A nearest neighbor is defined as a lipid molecule that is within 1 nm of another lipid molecule, with the position of a lipid defined by its head group (phosphate bead for PC and SM, and polar OH bead for chol). Lower panel: Order parameters Smol for the second bead of the sn-1 chain in POPC (green bar) or PDPC (blue bar) and of the sphingosine chain in SM (red bar) in POPC/SM/chol (left panel) and PDPC/SM/chol (right panel) bilayers. Values are an average over the entire trajectory and the standard deviation is indicated.
Order parameters Smol for lipid chains were calculated to further examine the lateral redistribution in location produced by PDPC vs. POPC in the bilayers with SM and chol. Values of Smol for the second bead, in practice describing a time and ensemble average of the angular fluctuations undergone by a vector connecting the first and third beads, in the CG MD representation of the sn-1 chain in POPC (green bar) or PDPC (blue bar) and of the sphingosine chain in SM (red bar), are compared in Figure 3 (lower panel). Smol illustrates the definitive role that the high disorder conferred on PDPC by DHA plays in driving SM into domains. The difference in order parameter for PDPC (Smol = 0.549 ± 0.013) relative to SM (Smol = 0.717 ± 0.017) in PDPC/SM/chol (Fig. 3, right lower panel) substantially exceeds that for POPC (Smol = 0.694 ± 0.012) (Fig. 3, left lower panel) relative to SM (Smol = 0.753 ± 0.012) in POPC/SM/chol.
3.2 2H NMR spectroscopy shows PDPC increases the size of domains in raft-forming mixtures on the nanometer scale
Solid state 2H NMR spectra recorded for multilamellar dispersions of PDPC/PSM-d31/chol and POPC/PSM-d31/chol (1:1:1 mol) at 35 °C are plotted in Figure 4. Their shape is determined by the molecular organization of PSM-d31 in the mixed membranes. The contrast in shape illustrates that DHA increases domain size.
Figure 4. 2H NMR spectra for PDPC and POPC in the presence of SM/Chol.
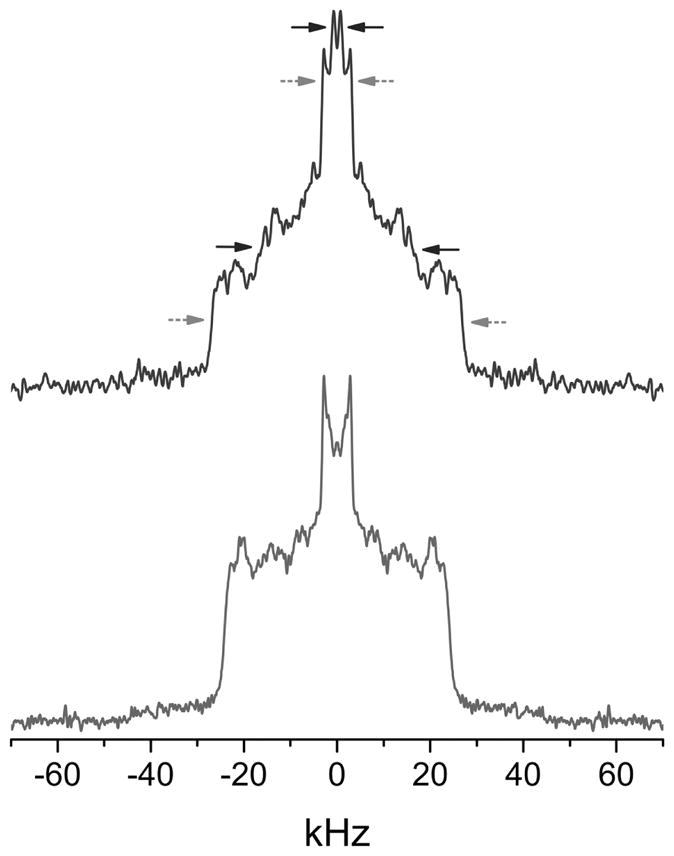
2H NMR spectra at 35 °C for PDPC/PSM-d31/chol (top) and POPC/PSM-d31/chol (1:1:1 mol) (bottom). The arrows designate pairs of signals assigned to the terminal methyl group (upper) and to methylene groups in the plateau region (lower) on PSM-d31 (upper) in more ordered SM-rich/chol-rich (grey dashed arrows - outer splitting) and more disordered PDPC-rich/chol-poor (black full arrows - inner splitting) domains for the PDPC/PSM-d31/chol mixture. The POPC/PSM-d31/chol spectrum lacks these features. Spectra are symmetrized about the central frequency to enhance signal/noise.
The 2H NMR spectrum for POPC/PSM-d31/chol (Fig. 4, bottom) has a shape that is characteristic of lipid bilayers in the liquid crystalline state [36]. It consists of a superposition of powder patterns from all segments along the perdeuterated palmitic chain of PSM-d31 in the mixture with POPC and chol. A plateau region of methylene groups with approximately constant order parameter (SCD ~ 0.38) in the upper portion is responsible for the sharp edges (±~24 kHz). Progressively more disordered methylene groups in the lower portion of the chain produce progressively narrower powder patterns, culminating in a pair of peaks (±~3 kHz) in the center of the spectrum due to the highly disordered terminal methyl group (SCD ~ 0.05). As thoroughly discussed in our recent study [47], we interpret the spectrum in terms of the presence of more ordered SM-rich and less ordered POPC-rich domains that are nanometer scale in size. The exchange of PSM-d31 between the domains, which is mediated by lateral diffusion, is fast because the domains are small (r < 45 nm) - the rate of exchange is faster than the difference in splitting of powder patterns for PSM-d31 in each domain - and a time averaged spectrum results [48;49].
Two spectral components assigned to PSM-d31 in more ordered SM-rich (raft like) and disordered PC-rich (non-raft) domains are resolved in the 2H NMR spectrum for PDPC/PSM-d31/chol (Fig. 4, top). Grey and red arrows indicate the edges of spectral components (±~27 and ±~16 kHz) assigned to the plateau region of order (SCD~ 0.42 and ~ 0.25) in the upper portion of the chain in PSM-d31 and to the peaks (±~3.2 and ±~1.0 kHz) assigned to terminal methyl group (SCD ~ 0.05 and ~ 0.02) of PSM-d31 in the respective domains. Separate components are observed because the domains are larger in the DHA-containing mixture. As a result, the rate of diffusion-mediated exchange of PSM-d31 in and out of them is too slow to produce time averaging [48;49]. The difference in the splitting (Δν) of the powder pattern for the spectral component assigned to each domain provides a lower limit to the lifetime (τ > (2πΔν)−1) for PSM-d31 in a domain from which, assuming a typical value reported for the coefficient of lateral diffusion (D~5 × 10−12m2/s) [50], a rough estimate of the corresponding domain size ( ) can be calculated (Table 1). The values for the size obtained in this way from the differential in splitting between domains for the terminal methyl and plateau regions are 28 and 12 nm, respectively. These values in the same range as estimates made previously on the basis of the splitting of signals resolved in 2H NMR spectra for deuterium labeled analogs of PDPC and chol, as well as PSM-d31, in PDPC/SM/chol mixtures (1:1:1 mol) [47]
Table 1. Estimates of domain size.
The estimates are based upon the quadrupolar splitting of the signals resolved for the terminal methyl groups and the methylene groups in the upper (plateau region) portion of the chain for PSM-d31 in ordered (raft-like) and disordered (non raft) domains in PDPC/PSM-d31/chol (1:1:1) at 35 °C.
| Ordered Δνr (kHz) | Disordered Δνr (kHz) | Δν (kHz) | τ (μs) | r (nm) | |
|---|---|---|---|---|---|
| Terminal methyl | 6.2 | 2.0 | 4.2 | 37.8 | 28 |
| Plateau | 54 | 32 | 22 | 7.2 | 12 |
3.3 Neutron scattering reveals a 2.5-fold increase in ordered domain size for PSM/Chol mixtures containing PDPC, compared to POPC
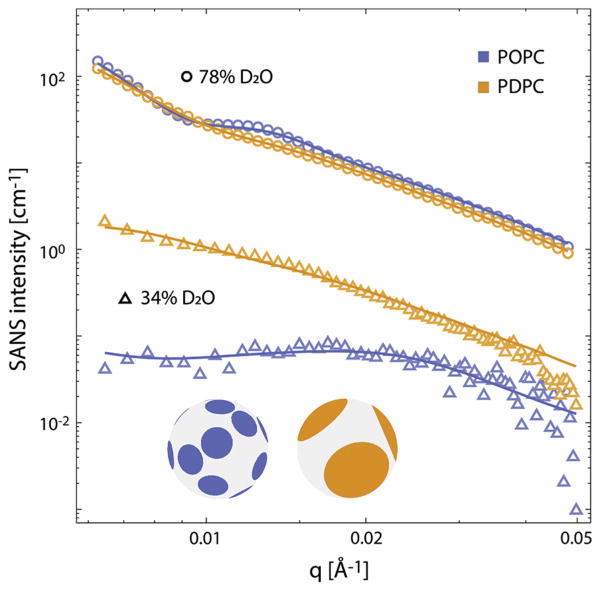
SANS is sensitive to lateral lipid inhomogeneities on nanometer length scales in mixtures of protiated and deuterated lipids - due to the large difference in coherent scattering lengths of hydrogen and its stable isotope deuterium [51]. It is an approach that has previously been applied to monitor the size of domains in mixtures of POPC and 1,2-dioleoylphosphatidylcholine (DOPC) with 1,2-distearoylphosphatidylcholine (DSPC) and cholesterol and of POPC or DOPC with SM and cholesterol [40;52]. Figure 5 shows SANS data for ~ 50 nm diameter vesicles composed of PSM-d31/POPC/chol (1:1:0.56 mol, blue symbols) and PSM-d31/PDPC/chol (1:1:0.56 mol, orange symbols). At 78% D2O, the scattering signal is dominated by the large contrast between the solvent and bilayer, and the SANS intensity can be adequately modeled with a conventional homogeneous bilayer form factor (solid lines) to recover the vesicle size distribution [53]. At 29% D2O, the solvent NSLD is approximately matched to the average bilayer NSLD, which dramatically attenuates the homogeneous component of the signal as evidenced by a nearly 100-fold reduction in intensity. Under these conditions, the NSLD contrast arises primarily from lateral segregation of the protiated low-melting lipid (i.e., POPC or PDPC) and chain-deuterated PSM-d31 between the raft and non-raft domains, and the scattering form factor depends strongly on the size, shape and spatial configuration of domains [54]. The stronger scattering and lack of a pronounced low-q peak are qualitatively consistent with more extensive lipid segregation and larger domains for the PDPC-containing mixture.
Figure 5. SANS data for POPC and PDPC in the presence of SM/Chol.
SANS data at 20 °C for vesicles composed of POPC/PSM-d31/chol (1:1:0.56 mol, blue symbols) or PDPC/PSM-d31/chol (1:1:0.56 mol, orange symbols), in 78% D2O solvent (open circles) or 29% D2O solvent (open triangles). Solid lines show fits to the data using either a homogeneous form factor for the 78% D2O data, or a laterally heterogeneous form factor for the 29% D2O data, as described in the text and Supporting Information. The inset shows schematic vesicle images corresponding to the average domain structure for the POPC-containing (blue) and PDPC-containing (orange) systems.
To gain further insight, we used a previously-described Monte Carlo method [40] to model the lateral structure in terms of circular liquid ordered (lo) domains randomly arranged in a continuous liquid disordered (ld) matrix, where the sole adjustable parameter was the domain radius. Consistent with the qualitative interpretation of the scattering data, modeling indicated that replacing POPC with PDPC results in a ~ 2.5-fold increase in the average domain radius, from 9 nm for the POPC-containing sample to 24 nm for the PDPC-containing sample (Supplemental Tables).
3.4 Imaging reveals PDPC, but not POPC, promotes phase separation in the presence of SM and chol
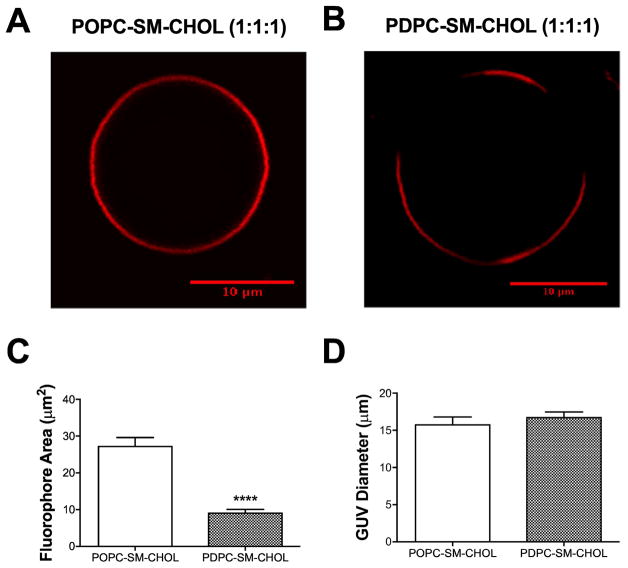
The next set of experiments tested the effect of PDPC on macroscopic phase separation. To directly image the formation of lipid domains on the scale of confocal microscopy, we constructed GUVs of POPC/SM/chol (1:1:1) (Fig. 6A) and PDPC/SM/chol (1:1:1) (Fig. 6B). The non-raft probe Texas-Red DHPE was employed for these studies. Note that kinetic studies were not conducted with GUV formation.
Figure 6. PDPC promotes phase separation in raft-containing GUVs.
Sample GUV image of (A) POPC/SM/Chol (1:1:1) and (B) PDPC/SM/Chol (1:1:1). GUVs were imaged with the non-raft fluorescent probe DHPE-Texas Red. (C) The average domain area and (D) GUV diameter for POPC and PDPC containing GUVs. Data are average ± SEM from 3 independent experiments. ****P<0.0001.
GUVs containing POPC showed no evidence of macroscopic phase separation (Fig. 6A). The incorporation of PDPC into SM/chol produced strong phase separation between domains (Fig. 6B). This contrast in behavior is reflected in the average area occupied by the non-raft fluorophore (Fig. 6C). In the PDPC/SM/chol vesicles, the fluorophore is presumably excluded from the raft domains promoted by PDPC and occupies a smaller area than in POPC/SM/chol where, because phase separation is not exhibited, the fluorophore is found throughout the perimeter of GUV. It should be noted that the differences in domain areas occupied by the Texas-Red DHPE probe were not due to differences in the diameter of the POPC and PDPC containing GUVs (Fig. 6D).
3.5 PDPC in the presence of SM/chol increases the excess area per molecule and lowers the Gibbs free energy of mixing relative to POPC/SM/chol
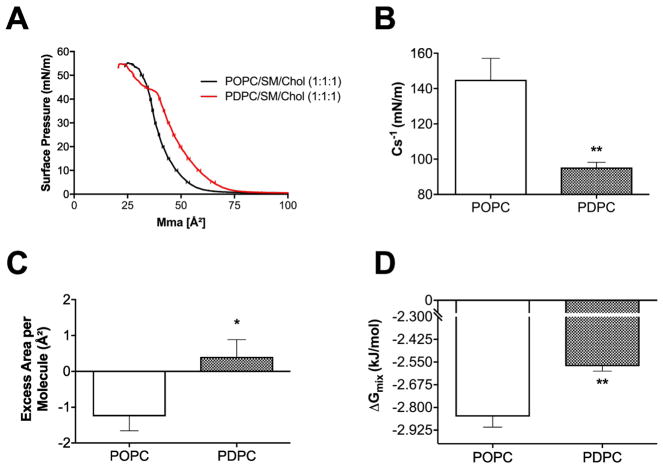
We performed experiments to explain how PDPC promoted the formation of lipid domains. Studies were specifically constructed to determine if PDPC influenced the excess area/molecule and Gibbs free energy of mixing in a manner that was distinct from POPC in the presence of SM/chol. To address this, pressure-area isotherms were generated for POPC/SM/chol (1:1:1) and PDPC/SM/chol (1:1:1) monolayers. All analyses were conducted at the physiologically relevant surface pressure of 30 mN/m.
Sample pressure area isotherms are presented in Fig. 7A. The inverse elasticity modulus (Cs−1) was significantly decreased (Fig. 7B) and the excess area per molecule was increased by ~ 1.5 Å2 (Fig. 7C) in the presence of PDPC relative to POPC containing mixtures. The Gibbs free energy of mixing was significantly lowered with PDPC relative to POPC-containing monolayers (Fig. 7D).
Figure 7. PDPC promotes lipid-lipid immiscibility in the presence of SM/Chol.
(A) Pressure area isotherms of POPC/SM/Chol (1:1:1) and PDPC/SM/Chol (1:1:1) generated at 23°C on a buffer subphase (pH=7.4). Pressure area isotherms were analyzed at 30 mN/m for the (B) inverse elasticity modulus Cs −1, (C) excess area per molecule and (D) Gibbs free energy of mixing. Data are average ± SEM from 5–6 independent experiments. *P<0.05, **P<0.01.
4. DISCUSSION
4.1. PDPC increases domain size
A major result from this study was that PDPC, compared to POPC, enhances the size of lipid rafts on the nanometer and micrometer size scales. The applied methods ranged from MD simulation to spectroscopies and imaging, and all directly and/or indirectly supported the notion that the DHA acyl chains of PC promote phase separation and enhance raft size. The rationale for using biomimetic membranes was to tightly control the lipid composition, which is difficult to do in cellular systems due to changes in lipid metabolism. We are able to present a unified picture of how substituting DHA (PDPC) for OA (POPC) affects the size of raft-like domains in mixtures with the same raft forming lipid molecules - SM and cholesterol.
Two spectral components that we assigned to PSM-d31 in raft-like (more ordered) and non-raft (less ordered) domains were resolved in the 2H NMR spectrum for PDPC/PSM-d31/chol (Fig. 4, top), while only a single component was resolved for PSM-d31 in the spectrum for POPC/PSM-d31/chol (Fig. 4, bottom). Our interpretation involves fast exchange between small domains in the POPC-containing mixture slowing when domains become larger in the PDPC-containing mixture on the nanoscale (Table 1). This interpretation is substantiated by the Monte Carlo modeling of SANS data for LUVs (Fig. 5) in terms of lo (raft-like) domains, surrounded by a continuous ld (non-raft) environment. Those data indicate an increase in domain size from 9 to 24 nm upon replacing POPC by PDPC in mixtures with PSM-d31 and chol (Table S1). On the microscale, confocal imaging of Texas-Red DHPE in GUVs detects the presence of lo domains in PDPC/SM/chol (Fig. 6B) that are not apparent in POPC/SM/chol (Fig. 6A).
The enhanced disorder associated with the multiple double bonds in DHA is the molecular origin for the formation of larger domains by PDPC. Our CG MD simulations illustrate this point. The order parameter Smol calculated for PDPC is relatively small and substantially reduced in comparison to SM, while the value of Smol for POPC is bigger and closer to that for SM (Fig. 3, lower panel). Visual inspection of snapshots (Fig. 2) and counts of nearest neighbor molecules (Fig. 3, upper panel) show that PDPC molecules separate away from SM and chol molecules more than POPC molecules. The high disorder of DHA chains also deters close packing with the ordered saturated chains of SM and rigid steroid moiety of chol [55], which is reflected in the poor miscibility seen for monolayers of PDPC/SM/chol compared to POPC/SM/chol (Fig. 7). An alternative explanation may be that PDPC is not influencing large-scale lipid-lipid miscibility in the monolayer model but is driving changes in cholesterol-induced condensation. This will require further investigation in future studies.
The results presented here add to the growing consensus from earlier work that DHA increases the size and stability of raft domains. Following a study of the effects of DHA on molecular organization in model membranes and intact plasma membranes (PM) from mammalian cells, it was concluded that disordered DHA-containing lipids enhance the stability of ordered rafts by increasing the difference in order between raft and the non-raft environments [56]. The study included CG MD simulations on PDPC or POPC in a bilayer with 1,2-dipamitoyl-phosphatidylcholine (16:0–16:0PC, DPPC) (serving as a proxy for SM) and chol that, like the current simulations, displayed an evolution with time into domains with the polyunsaturated phospholipid but not its monounsaturated counterpart. A switch from nano- to micro-scopic phase separation into lo and ld domains when 1-stearoyl-2-docosahexaenoyl-phosphatidylcholine (18:0–22:6PC, SDPC) replaced POPC in PC/SM (brain)/chol mixtures was reported on the basis of Förster resonance energy transfer (FRET) and fluorescence imaging microscopy (fluorescent dye C12:0 DiI) [57]. Lo/ld phase separation into domains in PC/SM/chol mixtures that become micron size, as opposed to nano size, with PDPC vs. POPC was observed by a combination of fluorescence imaging (fluorescent dye egg rhodamine PE), ESR and XRD [58].
Our results support an emerging view that the level of DHA incorporated into phospholipids in PM plays an important role in regulating the size of rafts. The molecular origin lies in the high disorder of the PUFA. To give context here, order parameters are ~10% lower when DHA replaces OA at the sn-2 position in PC with a saturated sn-1 chain [55]. This differential in order parameter is comparable to the reduction measured between DOPC that is another, albeit non-physiological, lipid known to promote phase coexistence [40], and POPC [59]. However, the mechanism by which DHA promotes the formation of larger domains has yet to be determined. There are two schools of thought. According to one scenario, DHA-rich non-raft regions are thinned by the increased cross-sectional area that accompanies greater disorder, causing rafts to grow to relieve the line tension associated with enhanced hydrophobic mismatch [40]. The alternative scenario links increased raft size to the greater differential in order between raft and non-raft domains [58;60]. We plan to address this issue in the future by a combination of 2H NMR, SANS and XRD methodologies that will enable us to non-invasively determine the composition, order, size and thickness of domains [40;47].
4.2 Limitations of the study
It is important to acknowledge limitations of the current study. One, the concentration of PDPC was very high. Our previous in vivo work in human and mouse intervention studies with fish oil show that DHA levels can increase several fold [23;61]; however, the levels of DHA-containing phospholipids do not reach an equimolar concentration relative to SM/Chol. Thus, future studies need to establish how lower concentrations of PDPC influence raft size.
Vulnerability to oxidation is a concern in all experiments on PUFA and has the potential to influence membrane architecture. Precautions were taken during sample preparation and data collection to minimize this problem. In the future, we aim to intentionally oxidize PDPC to determine how oxidization of DHA influences raft molecular organization. This is of high relevance since dietary intake of oxidized DHA, or oxidation in response to changes in cellular metabolism, would have a strong influence on cellular signaling.
4.3. Implications of a DHA-containing phospholipid to promote phase separation for immunological outcomes
The ability of PDPC to promote raft formation has strong implications for numerous biological systems. For simplicity, we focus on immunological studies given that our labs are currently studying how DHA, due to its immunomodulatory properties, can be used for improving chronic inflammation and simultaneously enhancing humoral immunity. We previously reported that dietary administration of DHA to mice and humans enhances B lymphocyte activation [25;61;62]. This was relevant for select clinical populations such as the obese that have impaired B cell activation and thereby poor responses to infections and vaccinations [63;64]. Mechanistically, we discovered that DHA enhanced the binding of the cholera toxin subunit B (which reports on GM1-enriched rafts) to the B lymphocyte plasma membrane [23]. The data from this manuscript now advance studies at the murine and human level by explaining how DHA may be enhancing lipid raft formation.
DHA, which incorporates into PEs and PCs of B lymphocytes, enhances phase separation and increases the size of lipid rafts, as measured with confocal and total internal reflection microscopy [23]. Thus, we speculate that enhanced raft formation allows for increased cellular signaling through metabolites generated from DHA and ultimately targeting of transcription factors and gene expression. The results from this study also have consequences for other immune cells such as helper T cells and the formation of the immunological synapse, which is a key regulator of inflammatory signaling [65]. Finally, we speculate that DHA-induced modification to raft size could have an effect on neurotransmission, which has implications for a variety of mental health disorders such as depression [66].
Supplementary Material
Highlights.
DHA regulates raft size in model membranes
DHA enhances phase segregation
DHA promotes unfavorable lipid-lipid interactions
Acknowledgments
We thank Helgi Ingolfsson for guidance in running CG MD simulations and Bob Bittman, now sadly deceased, for PSM-d31 (NMR work). Xubo Lin gave us advice on calculating order parameters in CG simulations. The work was funded, in part, by a grant from the National Institutes of Health to S.R.S. (NIH R01AT008375). F.A.H and J.K are supported through the Scientific User Facilities Division of the Department of Energy (DOE) Office of Science, sponsored by the Basic Energy Science (BES) Program, DOE Office of Science, under contract no. DEAC05-00OR22725. A portion of this research used resources at the High Flux Isotope Reactor, a DOE Office of Science User Facility operated by the Oak Ridge National Laboratory.
Footnotes
Abbreviations: docosahexaenoic acid (DHA), oleic acid (OA), omega-3 polyunsaturated fatty acid(s) (n-3 PUFA), 1-palmitoyl-2-docosahexaenoyl-phosphatidylcholine (PDPC), 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC), phosphatidylcholine (PC), sphingomyelin (SM)
AUTHOR CONTRIBUTIONS
S.R.S. analyzed data, wrote parts of the manuscript and takes responsibility for the content and analyses of GUV imaging and monolayer studies, E.R.P. conducted studies with monolayers, GUV imaging, and data analyses, D.J., conducted GUV imaging and data analyses, S.W.C. and X.L ran and analyzed CG MD simulations, and wrote parts of the manuscript. J.J.K. and A.T.C. performed NMR experiments, analyzed spectra and wrote parts of the manuscript. S.R.W. wrote parts of the manuscript and takes primary responsibility for NMR and CG MD simulation data. F.A.H. performed SANS experiments, analyzed the data, and takes primary responsibility for the SANS data. F.A.H. and J.K. wrote parts of the manuscript.
Conflict of interest
There are no conflict’s of interest and all authors have read and approved the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blasbalg TL, Hibbeln JR, Ramsden CE, Majchrzak SF, Rawlings RR. Changes in consumption of omega-3 and omega-6 fatty acids in the United States during the 20th century. Am J Clin Nutr. 2011;93:950–962. doi: 10.3945/ajcn.110.006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calder PC. Marine omega-3 fatty acids and inflammatory processes: Effects, mechanisms and clinical relevance. Biochim Biophys Acta. 2015;1851:469–484. doi: 10.1016/j.bbalip.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Siscovick DS, Barringer TA, Fretts AM, Wu JHY, Lichtenstein AH, Costello RB, Kris-Etherton PM, Jacobson TA, Engler MB, Alger HM, Appel LJ, Mozaffarian D. Omega-3 polyunsaturated fatty acid (fish oil) supplementation and the prevention of clinical cardiovascular disease. A science advisory from the American Heart Association. Circulation. 2017;135:e867–e884. doi: 10.1161/CIR.0000000000000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mozaffarian D, Wu JHY. (n-3) Fatty acids and cardiovascular health: Are effects of EPA and DHA shared or complementary? J Nutr. 2012;142:614S–625S. doi: 10.3945/jn.111.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asztalos IB, Gleason JA, Sever S, Gedik R, Asztalos BF, Horvath KV, Dansinger ML, Lamon-Fava S, Schaefer EJ. Effects of eicosapentaenoic acid and docosahexaenoic acid on cardiovascular disease risk factors: A randomized clinical trial. Metabolism. 2016;65:1636–1645. doi: 10.1016/j.metabol.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Laidlaw M, Cockerline CA, Rowe WJ. A randomized clinical trial to determine the efficacy of manufacturers recommended doses of omega-3 fatty acids from different sources in facilitating cardiovascular disease risk reduction. Lipids Health Dis. 2014;13:99–99. doi: 10.1186/1476-511X-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kromhout D, Giltay EJ, Geleijnse JM. n–3 Fatty acids and cardiovascular events after myocardial infarction. N Engl J Med. 2010;363:2015–2026. doi: 10.1056/NEJMoa1003603. [DOI] [PubMed] [Google Scholar]
- 8.de Luis D, Domingo JC, Izaola O, Casanueva FF, Bellido D, Sajoux I. Effect of DHA supplementation in a very low-calorie ketogenic diet in the treatment of obesity: A randomized clinical trial. Endocrine. 2016;54:111–122. doi: 10.1007/s12020-016-0964-z. [DOI] [PubMed] [Google Scholar]
- 9.Allaire J, Couture P, Leclerc M, Charest A, Marin J, Lépine M, Talbot D, Tchernof A, Lamarche B. A randomized, crossover, head-to-head comparison of eicosapentaenoic acid and docosahexaenoic acid supplementation to reduce inflammation markers in men and women: The Comparing EPA to DHA (ComparED) Study. Am J Clin Nutr. 2016;104:280–287. doi: 10.3945/ajcn.116.131896. [DOI] [PubMed] [Google Scholar]
- 10.Cao D, Kevala K, Kim J, Moon HS, Jun SB, Lovinger D, Kim HY. Docosahexaenoic acid promotes hippocampal neuronal development and synaptic function. J Neurochem. 2009;111:510–521. doi: 10.1111/j.1471-4159.2009.06335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calder PC. Mechanisms of action of (n-3) fatty acids. J Nutr. 2012;142:592S–599S. doi: 10.3945/jn.111.155259. [DOI] [PubMed] [Google Scholar]
- 12.Shaikh SR, Kinnun JJ, Leng X, Williams JA, Wassall SR. How polyunsaturated fatty acids modify molecular organization in membranes: Insight from NMR studies of model systems. Biochim Biophys Acta. 2015;1848:211–219. doi: 10.1016/j.bbamem.2014.04.020. [DOI] [PubMed] [Google Scholar]
- 13.Hou TY, Barhoumi R, Fan Y, Rivera GM, Hannoush RN, McMurray DN, Chapkin RS. n-3 Polyunsaturated fatty acids suppress CD4+ T cell proliferation by altering phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] organization. Biochim Biophys Acta. 2016;1858:85–96. doi: 10.1016/j.bbamem.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 15.Levental I, Veatch SL. The continuing mystery of lipid rafts. J Mol Biol. 2016;428:4749–4764. doi: 10.1016/j.jmb.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schley PD, Brindley DN, Field CJ. (n-3) PUFA Alter raft lipid composition and decrease epidermal growth factor receptor levels in lipid rafts of human breast cancer cells. J Nutr. 2007;137:548–553. doi: 10.1093/jn/137.3.548. [DOI] [PubMed] [Google Scholar]
- 17.Shaikh SR, Rockett BD, Salameh M, Carraway K. Docosahexaenoic acid modifies the clustering and size of lipid rafts and the lateral organization and surface expression of MHC class I of EL4 cells. J Nutr. 2009;139:1632–1639. doi: 10.3945/jn.109.108720. [DOI] [PubMed] [Google Scholar]
- 18.Shaikh SR, Wassall SR, Brown DA, Kosaraju R. n-3 Polyunsaturated fatty acids, lipid microclusters, and vitamin E. Curr Top Membr. 2015;75:209–231. doi: 10.1016/bs.ctm.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Shaikh SR, Cherezov V, Caffrey M, Soni SP, LoCascio D, Stillwell W, Wassall SR. Molecular organization of cholesterol in unsaturated phosphatidylethanolamines: X-ray diffraction and solid state 2H NMR reveal differences with phosphatidylcholines. J Am Chem Soc. 2006;128:5375–5383. doi: 10.1021/ja057949b. [DOI] [PubMed] [Google Scholar]
- 20.Soni SP, LoCascio DS, Liu Y, Williams JA, Bittman R, Stillwell W, Wassall SR. Docosahexaenoic acid enhances segregation of lipids between raft and nonraft domains: 2H-NMR study. Biophys J. 2008;95:203–214. doi: 10.1529/biophysj.107.123612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaikh SR, Dumaual AC, Castillo A, LoCascio D, Siddiqui RA, Stillwell W, Wassall SR. Oleic and docosahexaenoic acid differentially phase separate from lipid raft molecules: A comparative NMR, DSC, AFM, and detergent extraction study. Biophys J. 2004;87:1752–1766. doi: 10.1529/biophysj.104.044552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams JA, Batten SE, Harris M, Rockett BD, Shaikh SR, Stillwell W, Wassall SR. Docosahexaenoic and eicosapentaenoic acids segregate differently between raft and nonraft domains. Biophys J. 2012;103:228–237. doi: 10.1016/j.bpj.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rockett BD, Teague H, Harris M, Melton M, Williams J, Wassall SR, Shaikh SR. Fish oil increases raft size and membrane order of B cells accompanied by differential effects on function. J Lipid Res. 2012;53:674–685. doi: 10.1194/jlr.M021782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rockett BD, Franklin A, Harris M, Teague H, Rockett A, Shaikh SR. Membrane raft organization is more sensitive to disruption by (n-3) PUFA than nonraft organization in EL4 and B cells. J Nutr. 2011;141:1041–1048. doi: 10.3945/jn.111.138750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Teague H, Harris M, Fenton J, Lallemand P, Shewchuk B, Shaikh SR. Eicosapentaenoic and docosahexaenoic acid ethyl esters differentially enhance B-cell activity in murine obesity. J Lipid Res. 2014;55:1420–1433. doi: 10.1194/jlr.M049809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim W, Fan YY, Barhoumi R, Smith R, McMurray DN, Chapkin RS. n-3 Polyunsaturated fatty acids suppress the localization and activation of signaling proteins at the immunological synapse in murine CD4+ T cells by affecting lipid raft formation. J Immunol. 2008;181:6236–6243. doi: 10.4049/jimmunol.181.9.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan Y-Y, McMurray DN, Ly LH, Chapkin RS. Dietary (n-3) polyunsaturated fatty acids remodel mouse T-cell lipid rafts. J Nutr. 2003;133:1913–1920. doi: 10.1093/jn/133.6.1913. [DOI] [PubMed] [Google Scholar]
- 28.Fan Y-Y, Fuentes NR, Hou TY, Barhoumi R, Li XC, Deutz NEP, Engelen MPKJ, McMurray DN, Chapkin RS. Remodelling of primary human CD4+ T cell plasma membrane order by n-3 PUFA. Br J Nutr. 2018;119:163–175. doi: 10.1017/S0007114517003385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 30.Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J Phys Chem B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 31.Wassenaar TA, Ingólfsson HI, Böckmann RA, Tieleman DP, Marrink SJ. Computational lipidomics with insane: A versatile tool for generating custom membranes for molecular simulations. J Chem Theory Comput. 2015;11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- 32.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 33.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 34.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 35.Bittman R, Verbicky CA. Methanolysis of sphingomyelin: Toward an epimerization-free methodology for the preparation of d-erythro-sphingosylphosphocholine. J Lipid Res. 2000;41:2089–2093. [PubMed] [Google Scholar]
- 36.Davis JH. The description of membrane lipid conformation, order and dynamics by 2H-NMR. Biochim Biophys Acta. 1983;737:117–171. doi: 10.1016/0304-4157(83)90015-1. [DOI] [PubMed] [Google Scholar]
- 37.Seelig J. Deuterium magnetic resonance theory and application to lipid membranes. Q Rev Biophys. 1977;10:353–418. doi: 10.1017/s0033583500002948. [DOI] [PubMed] [Google Scholar]
- 38.Usery RD, Enoki TA, Wickramasinghe SP, Weiner MD, Tsai W-C, Kim MB, Wang S, Torng TL, Ackerman DG, Heberle FA, Katsaras J, Feigenson GW. Line tension controls liquid-disordered + liquid-ordered domain size transition in lipid bilayers. Biophys J. 2017;112:1431–1443. doi: 10.1016/j.bpj.2017.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arnold O, Bilheux JC, Borreguero JM, Buts A, Campbell SI, Chapon L, Doucet M, Draper N, Ferraz Leal R, Gigg MA, Lynch VE, Markvardsen A, Mikkelson DJ, Mikkelson RL, Miller R, Palmen K, Parker P, Passos G, Perring TG, Peterson PF, Ren S, Reuter MA, Savici AT, Taylor JW, Taylor RJ, Tolchenov R, Zhou W, Zikovsky J. Mantid—Data analysis and visualization package for neutron scattering and μ SR experiments. Nucl Instr Meth Phys Res A. 2014;764:156–166. [Google Scholar]
- 40.Heberle FA, Petruzielo RS, Pan J, Drazba P, Kučerka N, Standaert RF, Feigenson GW, Katsaras J. Bilayer thickness mismatch controls domain size in model membranes. J Am Chem Soc. 2013;135:6853–6859. doi: 10.1021/ja3113615. [DOI] [PubMed] [Google Scholar]
- 41.Pennington ER, Fix A, Sullivan EM, Brown DA, Kennedy A, Shaikh SR. Distinct membrane properties are differentially influenced by cardiolipin content and acyl chain composition in biomimetic membranes. Biochim Biophys Acta. 2017;1859:257–267. doi: 10.1016/j.bbamem.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan EM, Pennington ER, Sparagna GC, Torres MJ, Neufer PD, Harris M, Washington J, Anderson EJ, Zeczycki TN, Brown DA, Shaikh SR. Docosahexaenoic acid lowers cardiac mitochondrial enzyme activity by replacing linoleic acid in the phospholipidome. J Biol Chem. 2017 doi: 10.1074/jbc.M117.812834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shaikh SR, Dumaual AC, Jenski LJ, Stillwell W. Lipid phase separation in phospholipid bilayers and monolayers modeling the plasma membrane. Biochim Biophys Acta. 2001;1512:317–328. doi: 10.1016/s0005-2736(01)00335-2. [DOI] [PubMed] [Google Scholar]
- 44.Nichols-Smith S, The S, Kuhl TL. Thermodynamic and mechanical properties of model mitochondrial membranes. Biochim Biophys Acta. 2004;1663:82–88. doi: 10.1016/j.bbamem.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 45.Domènech Ò, Sanz F, Montero MT, Hernández-Borrell J. Thermodynamic and structural study of the main phospholipid components comprising the mitochondrial inner membrane. Biochim Biophys Acta. 2006;1758:213–221. doi: 10.1016/j.bbamem.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 46.Chou T, Chang C. Thermodynamic characteristics of mixed DPPC/DHDP monolayers on water and phosphate buffer subphases. Langmuir. 2000;16:3385–3390. [Google Scholar]
- 47.Kinnun JJ, Bittman R, Shaikh SR, Wassall SR. DHA modifies the size and composition of raftlike domains: A solid-state 2H NMR study. Biophys J. 2018;114:380–391. doi: 10.1016/j.bpj.2017.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thewalt JL. Essential insights into lipid membrane organization from essential fatty acids. Biophys J. 2018;114:254–255. doi: 10.1016/j.bpj.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bloom M, Thewalt J. Spectroscopic determination of lipid dynamics in membranes. Chem Phys Lipids. 1994;73:27–38. [Google Scholar]
- 50.Orädd G, Shahedi V, Lindblom G. Effect of sterol structure on the bending rigidity of lipid membranes: A 2H NMR transverse relaxation study. Biochim Biophys Acta. 2009;1788:1762–1771. doi: 10.1016/j.bbamem.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 51.Marquardt D, Heberle FA, Nickels JD, Pabst G, Katsaras J. On scattered waves and lipid domains: Detecting membrane rafts with X-rays and neutrons. Soft Matter. 2015;11:9055–9072. doi: 10.1039/c5sm01807b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petruzielo RS, Heberle FA, Drazba P, Katsaras J, Feigenson GW. Phase behavior and domain size in sphingomyelin-containing lipid bilayers. Biochim Biophys Acta. 2013;1828:1302–1313. doi: 10.1016/j.bbamem.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pencer J, Krueger S, Adams CP, Katsaras J. Method of separated form factors for polydisperse vesicles. J Appl Cryst. 2006;39:293–303. [Google Scholar]
- 54.Heberle FA, Anghel VNP, Katsaras J. Scattering from phase-separated vesicles. I. An analytical form factor for multiple static domains. J Appl Cryst. 2015;48:1391–1404. [Google Scholar]
- 55.Leng X, Kinnun JJ, Cavazos AT, Canner SW, Shaikh SR, Feller SE, Wassall SR. All n-3 PUFA are not the same: MD simulations reveal differences in membrane organization for EPA, DHA and DPA. Biochim Biophys Acta. 2018 doi: 10.1016/j.bbamem.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levental KR, Lorent JH, Lin X, Skinkle AD, Surma MA, Stockenbojer EA, Gorfe AA, Levental I. Polyunsaturated lipids regulate membrane domain stability by tuning membrane order. Biophys J. 2016;110:1800–1810. doi: 10.1016/j.bpj.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Konyakhina TM, Feigenson GW. Phase diagram of a polyunsaturated lipid mixture: Brain sphingomyelin/1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine/cholesterol. Biochim Biophys Acta. 2016;1858:153–161. doi: 10.1016/j.bbamem.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Georgieva R, Chachaty C, Hazarosova R, Tessier C, Nuss P, Momchilova A, Staneva G. Docosahexaenoic acid promotes micron scale liquid-ordered domains. A comparison study of docosahexaenoic versus oleic acid containing phosphatidylcholine in raft-like mixtures. Biochim Biophys Acta. 2015;1848:1424–1435. doi: 10.1016/j.bbamem.2015.02.027. [DOI] [PubMed] [Google Scholar]
- 59.Marquardt D, Williams JA, Kučerka N, Atkinson J, Wassall SR, Katsaras J, Harroun TA. Tocopherol activity correlates with its location in a membrane: A new perspective on the antioxidant vitamin E. J Am Chem Soc. 2013;135:7523–7533. doi: 10.1021/ja312665r. [DOI] [PubMed] [Google Scholar]
- 60.Lin X, Lorent JH, Skinkle AD, Levental KR, Waxham MN, Gorfe AA, Levental I. Domain stability in biomimetic membranes driven by lipid polyunsaturation. J Phys Chem B. 2016;120:11930–11941. doi: 10.1021/acs.jpcb.6b06815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guesdon W, Kosaraju R, Brophy P, Clark A, Dillingham S, Aziz S, Moyer F, Willson K, Dick JR, Patil SP, Balestrieri N, Armstrong M, Reisdroph N, Shaikh SR. Effects of fish oils on ex vivo B-cell responses of obese subjects upon BCR/TLR stimulation: A pilot study. J Nutr Biochem. 2018;53:72–80. doi: 10.1016/j.jnutbio.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teague H, Fhaner CJ, Harris M, Duriancik DM, Reid GE, Shaikh SR. n-3 PUFAs enhance the frequency of murine B-cell subsets and restore the impairment of antibody production to a T-independent antigen in obesity. J Lipid Res. 2013;54:3130–3138. doi: 10.1194/jlr.M042457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Green WD, Beck MA. Obesity impairs the adaptive immune response to influenza virus. Ann Am Thorac Soc. 2017;14:S406–S409. doi: 10.1513/AnnalsATS.201706-447AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kosaraju R, Guesdon W, Crouch MJ, Teague HL, Sullivan EM, Karlsson EA, Schultz-Cherry S, Gowdy K, Bridges LC, Reese LR, Neufer PD, Armstrong M, Reisdorph N, Milner JJ, Beck M, Shaikh SR. B cell activity is impaired in human and mouse obesity and is responsive to an essential fatty acid upon murine influenza infection. J Immunol. 2017;198:4738–4752. doi: 10.4049/jimmunol.1601031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hou TY, Barhoumi R, Fan Y-Y, Rivera GM, Hannoush RN, McMurray DN, Chapkin RS. n-3 Polyunsaturated fatty acids suppress CD4+ T cell proliferation by altering phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] organization. Biochim Biophys Acta. 2016;1858:85–96. doi: 10.1016/j.bbamem.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gananca L, Galfalvy HC, Oquendo MA, Hezghia A, Cooper T, Mann JJ, Sublette ME. Lipid correlates of antidepressant response to omega-3 polyunsaturated fatty acid supplementation: A pilot study. Prostaglandins Leukot Essent Fatty Acids. 2017;119:38–44. doi: 10.1016/j.plefa.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.