Abstract
Extensive and accurate chromatin remodeling is essential during primordial germ cell (PGC) development for the perpetuation of genetic information across generations. Here, we report that distal cis-regulatory elements (CREs) marked by DNase I-hypersensitive sites (DHSs) show temporally restricted activities during mouse and human PGC development. Using DHS maps as proxy, we accurately locate the genome-wide binding sites of pluripotency transcription factors in mouse PGCs. Unexpectedly, we found that mouse female meiotic recombination hotspots can be captured by DHSs, and for the first time, we identified 12,211 recombination hotspots in mouse female PGCs. In contrast to that of meiotic female PGCs, the chromatin of mitotic-arrested male PGCs is permissive through nuclear transcription factor Y (NFY) binding in the distal regulatory regions. Furthermore, we examined the evolutionary pressure on PGC CREs, and comparative genomic analysis revealed that mouse and human PGC CREs are evolutionarily conserved and show strong conservation across the vertebrate tree outside the mammals. Therefore, our results reveal unique, temporally accessible chromatin configurations during mouse and human PGC development.
Introduction
Primordial germ cells (PGCs), the precursors of sperm and eggs, are established from post-implantation epiblast cells during early embryogenesis in mammals, and then, they migrate to the developing gonads, where PGCs undergo meiosis and differentiate into gametes.1 Mouse PGCs are specified at around embryonic day 7.25 (~E7.25). They regain potential pluripotency at early stage and then initiate extensive epigenetic reprogramming to erase somatic epigenetic memories, including global DNA demethylation and reorganization of chromatin modifications.1,2 At ~E14.5, female and male PGCs enter meiosis and mitotic arrest, respectively.3,4 The trajectory of human PGC development is less clear than that of mouse PGCs. Recent studies have demonstrated that an individual embryo simultaneously contains several PGC subpopulations characterized by distinct transcriptional features,5,6 illustrating the asynchronous and heterogeneous properties of human PGCs.
Although significant progress has been made toward uncovering the regulatory mechanisms of PGC development, previous studies have primarily focused on DNA methylation patterns and transcription analysis.5–8 It remains to be determined how chromatin reconfigures and regulates transcription programs during mouse and human PGC development. Cells of a specific type or at a particular state possess a unique set of cis-regulatory elements (CREs), such as promoters, enhancers, insulators and locus control regions.9 CREs are known to regulate developmentally expressed genes and often exhibit spatially restricted activity patterns,10 but their temporally regulatory activities and global contributions during mouse and human PGC development remain poorly understood. Mouse early-stage PGCs have potential pluripotency by repressing a somatic mesodermal program and regaining a transcriptional network for pluripotency.2,11 However, genome-wide mapping of pluripotency transcription factor (TF) binding sites in PGCs has been impeded owing to the paucity of PGCs. Following the development of PGCs, female PGCs enter into meiosis at ~E14.5, while male PGCs undergo mitotic arrest at around the same stage.2 Meiosis and mitotic arrest are distinct developmental events, but the chromatin conformation difference between female and male PGCs is still unclear. Moreover, PGC programs are speculated to be evolutionarily conserved to ensure the proper transmission of genetic and epigenetic information across the generations.12,13 Although extensive studies have proved that changes at the DNA sequence level in regulatory elements underlie evolutionary differences,10,14,15 the evolutionary pressure on mouse and human PGC CREs remains enigmatic.
To address these issues, we developed a low-input DNase-seq method and applied it to mouse and human PGCs across multiple developmental stages. We show that distal CREs exhibit temporally restricted activities and mirror the trajectory of PGC development. Genome-wide binding sites of pluripotency TFs were accurately located in mouse PGCs. Moreover, female recombination hotspots could be captured by DNase I-hypersensitive sites (DHSs) in meiotic female PGCs, while the chromatin of mitotic-arrested male PGCs is permissive by nuclear transcription factor Y (NFY) binding in the distal regulatory regions. Finally, we examined the evolutionary pressure on mouse and human PGC CREs and uncovered their uniquely conserved features.
Results
Mapping accessible chromatin in mouse and human PGCs
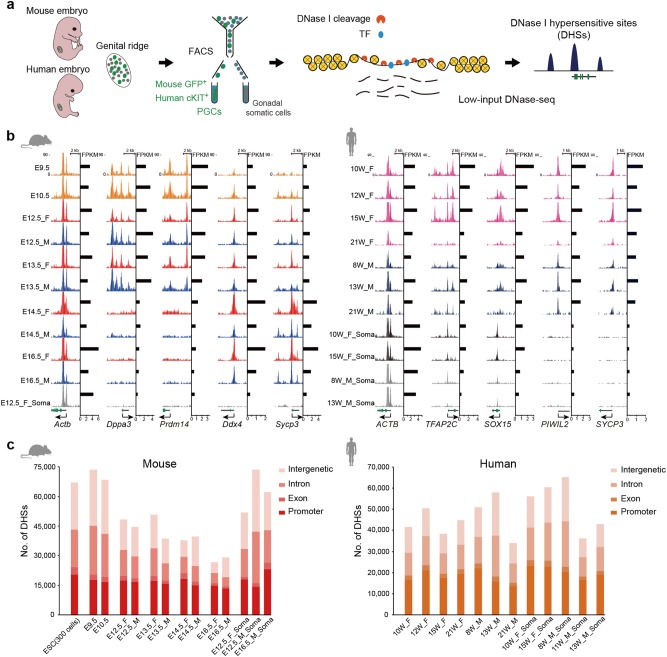
To obtain a comprehensive landscape of the accessible chromatin in mouse and human PGCs, we developed a low-input DNase-seq method that enables us to estimate data quality before sequencing (Supplementary information, Fig. S1a, b). We first verified that DHS maps generated from 300 or 3000 mouse embryonic stem cells (ESCs) recapitulated the results of the ENCODE project using 10–20 million ESCs (Supplementary information, Fig. S1c–f). Next, we screened PGCs using the fluorescence assisted cell sorting (FACS) method as previously described.5,16 For mouse, Oct4-GFP-positive PGCs from E9.5–E16.5 embryos were collected. For human, we enriched cKIT (also known as CD117)-positive PGCs from 8–21-week old embryos. In total, we determined DHSs in 20 mouse and 13 human PGC samples, 4 mouse and 10 human gonadal somatic cell samples, 2 mouse ESC samples and 2 mouse epiblast-like cell (EpiLC) samples (Fig. 1a; Supplementary information, Table S1).
Fig. 1.
Accessible chromatin landscapes in mouse and human PGCs. a Schematic of low-input DNase-seq for probing accessible chromatin in mouse and human PGCs and gonadal somatic cells. b Genome browser view of DNase signal enrichment at certain representative genes in mouse and human PGCs and gonadal somatic cells. Shown on the right are normalized DHS signals. c Bar graphs showing the number of DHSs from PGCs and gonadal somatic cells and their distribution across genomic features. E embryonic day, W week, F female PGCs, M male PGCs, Soma gonadal somatic cells
We conducted several analyses to validate the DNase-seq data in PGCs and gonadal somatic cells. First, we demonstrated that the replicates in each developmental stage had high reproducibility (Supplementary information, Fig. S2a). Thus, we combined the data and used the common DHSs from the two replicates with an additional stringent cutoff requirement of DNase signal FPKM ≥ 3. Next, we confirmed that genes with promoter DHSs were highly expressed compared to those without promoter DHSs (Supplementary information, Fig. S2b), which is consistent with previous studies in other cell types.9,17,18 Then, we focused on the chromatin accessibility levels around certain representative genes. The promoter regions of pluripotency marker genes, such as Dppa3 and Prdm14, were accessible only in mouse early-stage PGCs until E13.5, whereas germline-specific genes, such as Ddx4 and Sycp3, were highly accessible in mouse late-stage PGCs (Fig. 1b). In human, the chromatin around germline marker genes, such as PIWIL2 and SYCP3, were accessible only in PGCs but not in gonadal somatic cells (Fig. 1b). Finally, we examined the number and distribution of DHSs generated in this study. In contrast to promoter regions, where the quantity of DHSs was relatively constant, the number of distal DHSs was highly dynamic across developmental stages (Fig. 1c). This result is in line with the notion that distal regulatory elements show transient functions in defining cell identity during development.10,19,20 Taken together, these data suggest that our low-input DNase-seq approach could map accessible chromatin in mouse and human PGCs at high resolution.
Distal DHSs mirror the trajectory of PGC development
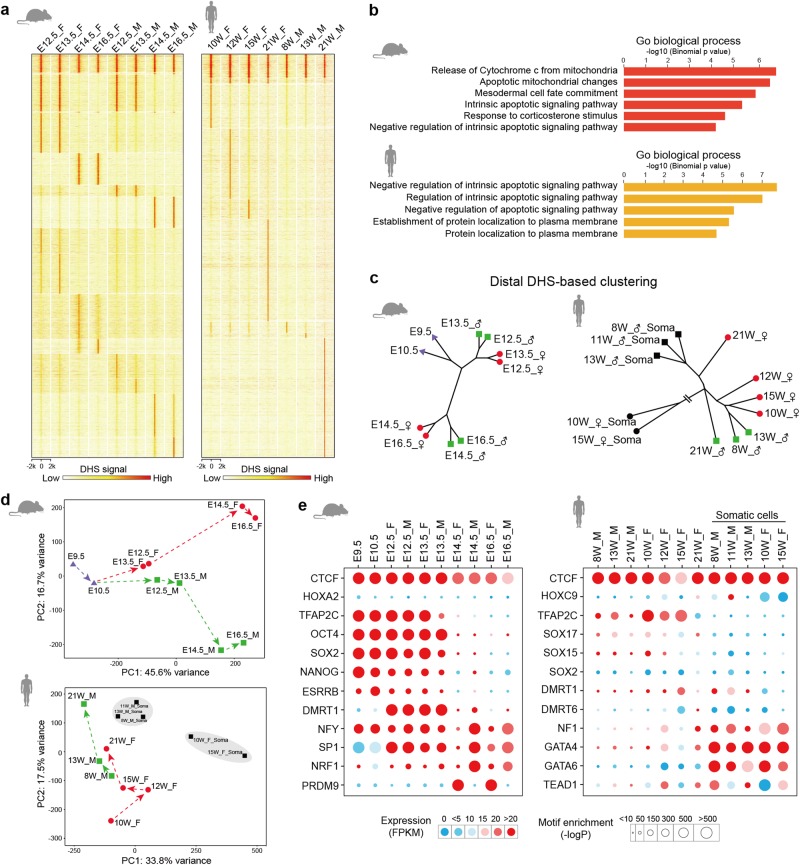
Accessible chromatin can mark CREs such as proximal promoters and distal enhancers.18,21 Our time-course profiling of DHS signals showed that the majority of promoters remained constantly accessible (Supplementary information, Fig. S2c), whereas most distal DHSs (away from TSS ± 1 kb) spanned merely a subset of the developmental stages examined (Fig. 2a). This result coincides with the differential distribution patterns of promoter and distal DHSs (Fig. 1c), illustrating the rapidly changing activities of distal regulatory elements during mouse and human PGC development. Functional annotation of the 2334 mouse and 2669 human distal DHSs showing constant activities revealed that these common DHSs were mostly enriched in the apoptosis signaling pathway (Fig. 2a, b; Supplementary information, Table S2). This result is also supported by transcription analysis performed in mouse and human PGCs,6,12,22 suggesting that the elimination of abnormally developed PGCs through apoptosis may be a general regulatory mechanism.
Fig. 2.
Distal DHSs exhibit temporally restricted activities during PGC development. a Heat maps show the dynamic changes of distal DHSs (away from TSS ± 1 kb) in mouse (n = 52,127) and human (n = 46,252) PGCs. Each row represents the DNase signal intensity at a DHS ± 2 kb. b Functional annotation of mouse common distal DHSs from E12.5-E16.5 PGCs (n = 2334) and human common distal DHSs from 8W-21W PGCs (n = 2669). c Clustering dendrogram of mouse and human DNase-seq samples based on DNase signals of distal DHSs. d Principal component analysis of distal DHS signals from mouse and human DNase-seq samples. e Representative TF motifs identified from mouse and human distal DHSs and the expression levels of related TFs
Next, we investigated whether the distal regulatory activities of each sample reflected spatiotemporal relationships by conducting a hierarchical clustering analysis of distal DHSs. We found that mouse early-stage PGCs (E9.5-E13.5) were clearly separated from those at late stages (E14.5-E16.5), and PGCs of the same sex from adjacent developmental stages were clustered together within each subgroup (Fig. 2c). Similar conclusions could also be reached in human samples (Fig. 2c). Principal component analyses (PCA) of distal DHSs further supported the grouping results (Fig. 2d). For mouse PGCs, the PC1 axis was dominated by differences of developmental stage, while the PC2 axis showed differences between meiotic female PGCs and mitotic-arrested male PGCs. For human samples, PGCs and gonadal somatic cells were separated along the PC1 axis, and the PC2 axis recapitulated the differential developmental paths of female and male PGCs. These results suggest that accessible chromatin marked by DHSs in distal regulatory regions can mirror the trajectory of PGC development precisely.
As DHSs are known to be hotspots for transcriptional regulator binding,23 we wondered whether distal DHSs possessed motifs for TFs that regulate PGC development. To this end, we conducted a TF binding motif enrichment analysis. We found that TF motifs were enriched in a highly stage- and sex-specific manner, and the motif enrichment coincided with the expression of their corresponding TFs (Fig. 2e; Supplementary information, Fig. S2d). For example, motifs for pluripotency TFs, such as the SOX family, were enriched from E9.5 to E13.5, while the PRDM9 (meiosis-specific methyltransferase) motif was highly enriched only in mouse E14.5 and E16.5 female PGCs. Similar observations were also obtained in the human DNase-seq samples. The motif for TFAP2C (required for PGC specification) was enriched only in PGCs, whereas GATA factor (cellular differentiation and organogenesis related) motifs were mostly enriched in human gonadal somatic cells. These results suggest that temporally restricted distal DHSs harbor binding motifs for TFs that play vital roles in defining PGC identity.
Identification of pluripotency transcription factor binding sites in mouse PGCs
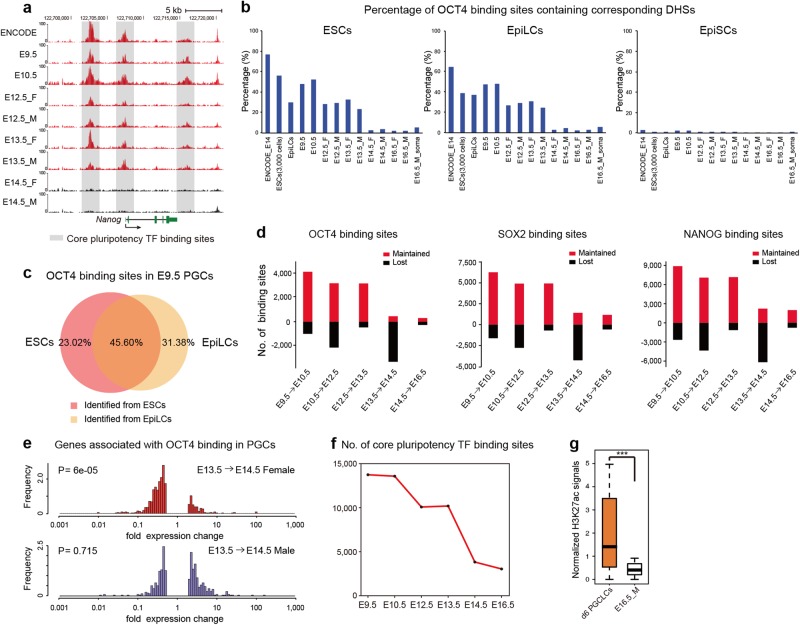
Mouse PGCs are known to have potential pluripotency.2,11 However, genome-wide mapping of pluripotency TF binding sites in PGCs has been impeded by the fact that cross-linking chromatin immunoprecipitation (X-ChIP)-based methods require millions of cells,24,25 yet only hundreds or thousands of early-stage PGCs could be collected at a time. Surprisingly, we observed that core pluripotency TF (OCT4, SOX2 and NANOG) binding sites in ESCs exactly overlapped the DHSs in mouse PGCs until E13.5 (Fig. 3a). This result is consistent with our previous finding that motifs for pluripotency TFs were highly enriched in mouse early-stage PGCs (Fig. 2e; Supplementary information, Fig. S2d). As TF binding to DNA in place of canonical nucleosomes results in markedly increased accessibility of the chromatin,21,26 it was possible to identify pluripotency TF binding sites in mouse PGCs by examining whether they contained corresponding PGC DHSs.
Fig. 3.
Identification of core pluripotency TF binding sites in mouse PGCs. a Genome browser view showing the DHS signal enrichment at core pluripotency TF binding sites around the Nanog loci in mouse ESCs and PGCs. b OCT4 binding sites from ESCs, EpiLCs and EpiSCs have different percentages of corresponding DHSs in ESCs, EpiLCs, PGCs and gonadal somatic cells. c Venn diagram shows the overlap of OCT4 binding sites (in E9.5 PGCs) identified from ESCs and those identified from EpiLCs. d Bar graphs illustrate the numbers of OCT4, SOX2 and NANOG binding sites that are maintained and lost between adjacent developmental stages in mouse PGCs. e The expression fold changes of genes associated with the lost OCT4 binding sites (from E13.5 to E14.5) in mouse female (upper) and male (lower) PGCs. f The number of core pluripotency TF binding sites identified in each stage of PGCs. g Box plot showing the H3K27ac signals around core pluripotency TF-bound regions in d6 (day 6) PGCLCs and E16.5 male PGCs. The difference is statistically significant by Wilcoxon rank sum test (***P < 0.001)
A similar overlap pattern with PGC DHSs was also observed for OCT4 binding sites in EpiLCs24 (Fig. 3b, c) but not in the more developmentally advanced epiblast stem cells (EpiSCs) (Fig. 3b), which represent a primed pluripotency state.27 Thus, we combined the OCT4 binding sites in ESCs and EpiLCs to represent the OCT4 occupancy pool. By examining which OCT4 binding sites contain corresponding PGC DHSs at a specific stage, we identified the OCT4 binding sites in mouse PGCs during development. We further compared the OCT4 binding sites in PGCs between adjacent developmental stages and found a dramatic decrease during the E13.5-to-E14.5 transition (Fig. 3d). The same dynamic pattern was also observed for SOX2 and NANOG binding sites identified in mouse PGCs (Fig. 3d). The rapid removal of core pluripotency TFs from their occupancy regions in E14.5 PGCs accords with the fact that female and male PGCs enter meiosis and mitotic arrest at this stage, respectively.1,2,4 Next, we correlated pluripotency TF binding sites with nearby genes and investigated whether the expression levels of the associated genes were coincident with this rapid loss process. Interestingly, genes associated with the binding sites that lost the occupancy at E14.5 were significantly downregulated only in females but not in males (Fig. 3e; Supplementary information, Fig. S3a). The possible explanation may be that the transcripts of the associated genes are not immediately degraded in mitosis-arrested male PGCs upon the removal of pluripotency TF occupancy. Collectively, these results demonstrate that key characteristics of PGCs during development could be directly and immediately reflected by the reconfiguration of accessible chromatin.
To obtain an overview of the dynamic patterns of core pluripotency TF binding sites during mouse PGC development, we combined the OCT4, SOX2 and NANOG binding sites identified in PGCs. There were two waves of decrease: a minor decrease in these binding sites was observed from E10.5 to E12.5, and the most significant decline occurred during the E13.5-to-E14.5 transition (Fig. 3f). Moreover, we further examined whether the removal of core pluripotency TFs from their binding sites was accompanied with changes in chromatin patterns by mapping the H3K27ac modifications of E16.5 male PGCs (Supplementary information, Table S1). Compared to the day 6 (d6) primordial germ cell-like cells (PGCLCs), which acquire epigenetic properties similar to those of PGCs at ~E9.5,12 the H3K27ac enrichment level around the core pluripotency TF binding regions dramatically decreased in E16.5 male PGCs (Fig. 3g). This result further confirmed the accuracy of our identification.
The binding sites of PRDM14, a PR domain-containing transcriptional regulator pivotal for PGC specification and maintenance,28 were also identified using similar methods (Supplementary information, Fig. S3b, c). Most PRDM14 binding sites in PGCs showed co-occupancy with core pluripotency TFs (Supplementary information, Fig. S3d). This result is consistent with its critical role in reacquisition of potential pluripotency in mouse PGCs.2,28 As loss of Prdm14 caused a decrease in Dppa3 expression and PGC death,28 a PRDM14 and core pluripotency TF colocalized region we identified may be responsible for Dppa3 activation in mouse PGCs (Supplementary information, Fig. S3e).
Female recombination hotspots can be captured by DHSs
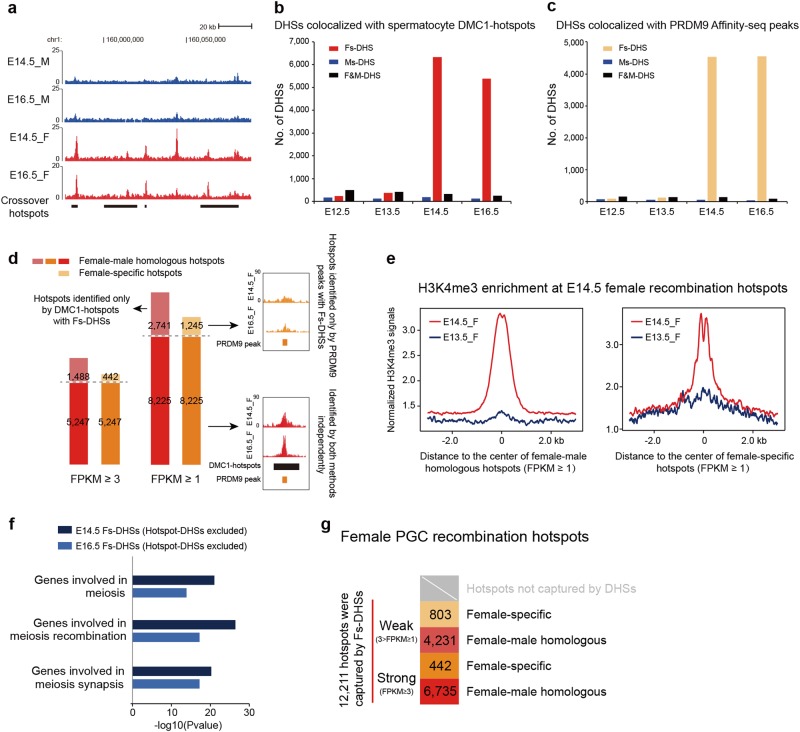
As mouse female PGCs enter into meiosis at ~E14.5, homologous recombination preferentially occurs in specialized sites called meiotic recombination hotspots.29 Male recombination hotspots could be determined by DMC1 (DNA meiotic recombinase 1) ChIP-seq (single-stranded DNA sequencing (SSDS)) in postnatal spermatocytes.30 However, female recombination hotspots are not known, due to the paucity of PGCs. The exact position of recombination hotspots depends on the activity of a histone H3 methyltransferase, PR domain-containing 9 (PRDM9),29,31–33 and our motif analysis indicates that PRDM9 signals are highly enriched only in E14.5 and E16.5 female PGCs (Fig. 2e). Moreover, four known crossover recombination hotspots identified in pedigrees34 were found to exactly overlap with E14.5 and E16.5 female PGC DHSs (Fig. 4a). Based on these observations, we asked whether recombination hotspots where double-stranded breaks (DSBs) occur tend to be friable and can be captured by DHSs.
Fig. 4.
Identification of female meiotic recombination hotspots using PGC Fs-DHSs. a Genome browser view of DHS signal enrichment at the positions of four known crossover recombination hotspots. b Bar plot shows the number of Fs-, Ms- and F&M-DHSs colocalized with spermatocyte DMC1-hotspots. c Bar plot shows the number of Fs-, Ms- and F&M-DHSs colocalized with PRDM9 Affinity-seq peaks. d Bar charts showing the number of female recombination hotspots identified by DMC1-hotspots colocalized with Fs-DHSs (FPKM ≥ 3, n = 6735; FPKM ≥ 1, n = 10,966) and those identified by PRDM9 Affinity-seq peaks colocalized with Fs-DHSs (FPKM ≥ 3, n = 5689; FPKM ≥ 1, n = 9470). Right insets show the genome browser view of DHS signal enrichment at a representative female-specific hotspot (top) and a representative female-male homologous hotspot (bottom). e The H3K4me3 signal density profiles of E13.5 and E14.5 female PGCs at E14.5 female recombination hotspots (left, female-male homologous hotspots; right, female-specific hotspots). f Functional annotation of E14.5 and E16.5 Fs-DHSs excluding hotspot-linked DHSs. g Model illustrating the number of female recombination hotspots captured by DHSs. Strong hotspots were defined as DHS signal with FPKM ≥ 3, and weak hotspots were defined as DHS signal with 3 > FPKM ≥ 1. Fs female PGC-specific, Ms male PGC-specific, F&M female-male PGC common
To address the above question, we used spermatocyte hotspots (denoted as DMC1-hotspots) determined in C57BL/6J (B6) mice as coordinates to examine whether female PGCs have DHSs at the same positions. After a genome-wide examination, we observed that 6330 E14.5 and 5386 E16.5 female PGC-specific DHSs (Fs-DHSs) were colocalized with DMC1-hotspots, whereas only 244 E12.5 and 374 E13.5 Fs-DHSs were colocalized with DMC1-hotspots (Fig. 4b). As meiotic recombination occurs only in female PGCs, male PGC-specific DHSs (Ms-DHSs) and female-male common DHSs (F&M-DHSs) were rarely colocalized with DMC1-hotspots (Fig. 4b). This result indicates that recombination hotspots could be marked only by the E14.5 and E16.5 Fs-DHSs. To further verify this result, we took advantage of DMC1-hotspots from strains having different Prdm9 alleles from B6. As expected, E14.5 and E16.5 Fs-DHSs were colocalized with DMC1-hotspots in a strain-specific manner (Supplementary information, Fig. S4a), and loss of Prdm9 led to aberrant overlap between Ms-DHSs and DMC1-hotspots (Supplementary information, Fig. S4b). Further examinations showed that the Fs-DHSs colocalized with DMC1-hotspots were enriched around the centers of DMC1-hotspots (Supplementary information, Fig. S4c), and the majority of these Fs-DHSs marked only one DMC1-hotspot (Supplementary information, Fig. S4d). These results indicate that the high overlap between Fs-DHSs and DMC1-hotspots was not random. Moreover, similar proportions of Fs-DHSs were colocalized with DMC1-hotspots when we retained DHSs with FPKM ≥ 3 or 1 (Supplementary information, Fig. S4e). Collectively, these results indicate that female recombination hotspots in PGCs tend to be DNase I-hypersensitive, and with spermatocyte DMC1-hotspots as coordinates, we identified the precise locations of female-male homologous recombination hotspots.
To rule out context dependency and gender bias caused by spermatocyte DMC1-hotspots, we took advantage of PRDM9 Affinity-seq, which could detect PRDM9 binding sites in vitro and quantitate its relative affinities.35 By using PRDM9 Affinity-seq peaks as unbiased recombination hotspot coordinates, we found that PRDM9 peaks had high overlap with E14.5 and E16.5 Fs-DHSs but not Ms-DHSs or F&M-DHSs (Fig. 4c). Strikingly, of the 5689 hotspots identified by PRDM9 peaks overlapping with Fs-DHSs, 5247 were previously identified by comparing DMC1-hotspots with Fs-DHSs (Fig. 4d; Supplementary information, Fig. S4f). When we retained DHSs with FPKM ≥ 1, 8255 female PGC hotspots could be detected by both methods independently (Fig. 4d). As in vitro Affinity-seq is sexually independent, Fs-DHSs overlapping with in vitro PRDM9 Affinity-seq peaks but not with spermatocyte DMC1-hotspots (442, FPKM ≥ 3; 1245, FPKM ≥ 1) were defined as hotspots unique to female PGCs (Fig. 4d). These results indicate that combining in vitro PRDM9 Affinity-seq with low-input DNase-seq could provide an alternative way to identify female recombination hotspots, which bypasses the difficulties in direct pull-down of DMC1 from the limited quantity of PGCs. Although both spermatocyte DMC1-hotspots and PRDM9 Affinity-seq peaks could be combined with Fs-DHSs to identify female hotspots and yielded consistent results (Fig. 4d; Supplementary information, Fig. S4g), the coordinates provided by PRDM9 Affinity-seq were not limited to determine female-male homologous hotspots but also female-specific hotspots. Furthermore, in vitro PRDM9 Affinity-seq can be easily performed in different mouse strains and even other species.
Since PRDM9 binds to recombination hotspots and adds methyl groups to H3 on nearby nucleosomes,29,31 we further mapped the H3K4me3 modifications of E14.5 female PGCs (Supplementary information, Table S1). The results revealed that H3K4me3 signals were highly enriched at female-male homologous and female-specific hotspots in E14.5 but not E13.5 female PGCs before they entered meiosis (Fig. 4e; Supplementary information, Fig. S4h). Moreover, functional annotation of E14.5 and E16.5 Fs-DHSs revealed that Fs-DHSs excluding hotspot-linked DHSs are enriched in meiosis-related regulatory functions (Fig. 4f; Supplementary information, Table S2), whereas DHSs marking recombination hotspots have no meiosis-related regulatory activities, which is consistent with their self-destructive nature during meiosis.36
We applied a similar analysis to human 21 W PGCs. The results showed that more Fs-DHSs overlapped with spermatocyte DMC1-hotspots (PRDM9A and PRDM9B)37 than Ms-DHSs (Supplementary information, Fig. S4i), and some representative motifs for human spermatocyte DMC1-hotspots were highly enriched in 21 W Fs-DHSs but not in Ms-DHSs (Supplementary information, Fig. S4j). However, due to the heterogeneity and long developmental period of human PGCs (Supplementary information, Fig. S4k),1,5,6 genome-wide identification of human female recombination hotspots using PGC DHSs requires further investigations.
Using spermatocyte DMC1-hotspots and in vitro PRDM9 Affinity-seq peaks as coordinates, for the first time, we identified 12,211 recombination hotspots in mouse female PGCs. Based on the strengths of the DHS signals, 6735 female-male homologous hotspots were recognized as strong hotspots (FPKM ≥ 3), while 4321 homologous hotspots were relatively weak (3 > FPKM ≥ 1); 442 and 803 hotspots were classified as strong and weak female-specific hotspots, respectively (Fig. 4g).
The chromatin of mitotic-arrested male PGCs is permissive through NFY binding
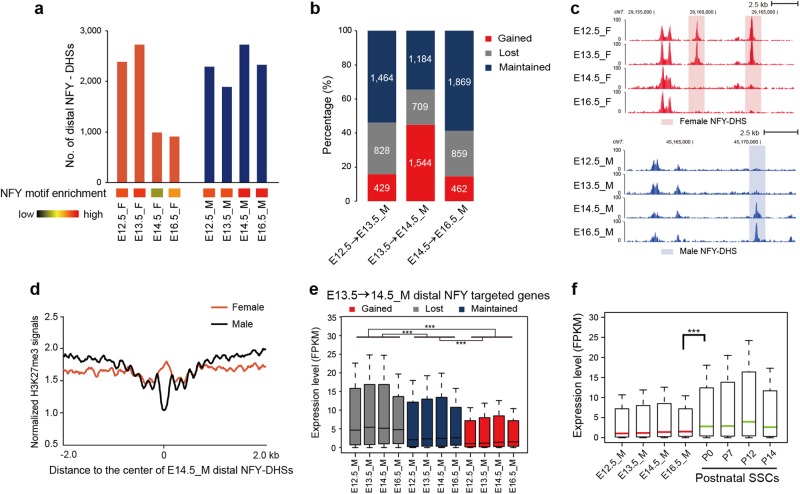
During mouse fetal development, E14.5 female PGCs initiate meiosis, whereas male PGCs undergo mitotic arrest and remain quiescent for the remaining embryonic period.2 Although late-stage (E14.5-E16.5) female and male PGCs are characterized by distinct developmental signatures, the numbers of DHSs in female and male PGCs were equivalent (Fig. 1c). Since thousands of female DHSs were identified to mark female recombination hotspots, an excess of DHSs in quiescent male PGCs raised the possibility that distinct chromatin configurations may be present in mitotic-arrested male PGCs.
Interestingly, we observed that motif signals for the TF NFY in E14.5 and E16.5 male PGCs were significantly higher than those in female PGCs (Fig. 2e). NFY is known as a nucleosome-like TF capable of promoting chromatin accessibility through its sequence-specific (CCAAT) DNA binding.38,39 To determine whether NFY may participate in shaping the chromatin conformation of mitotic-arrested male PGCs, we used the motif analysis tool HOMER40 to identify NFY binding sites as NFY-DHSs in PGCs (Fig. 5a). The number of NFY-DHSs was relatively stable in the promoter regions of E12.5-E16.5 female and male PGCs (Supplementary information, Fig. S5a, b). In contrast, the number of distal NFY-DHSs precipitously declined in female PGCs but increased in male PGCs from E13.5 to E14.5 (Fig. 5a–c). Mouse PGCs are known to undergo extensive epigenetic reprogramming during development, including reduction of H3K9me2 and elevation of H3K27me3 modifications.1,2,12 To understand whether histone modifications may participate in this process, we mapped the H3K27me3 modifications in E14.5 PGCs (Supplementary information, Table S1). The results revealed that H3K27me3 were highly enriched at distal NFY binding sites in female PGCs (Fig. 5d), which is consistent with the fact that female PGCs lost many distal NFY-DHSs at E14.5 (Fig. 5a). These results indicate that the chromatin could be opened through NFY binding in distal regulatory regions even though late-stage male PGCs were arrested in mitosis.
Fig. 5.
Annotation of distal NFY-DHSs in mouse mitotic-arrested male PGCs. a Bar charts showing the number of NFY-DHSs at distal regions (away from TSS ± 1 kb) in mouse female and male PGCs. The bottom heat maps show the corresponding NFY motif enrichment at distal DHSs. b Bar charts representing the percentages of gained, lost and maintained distal NFY-DHSs in male PGCs during each developmental stage transition. c Genome browser view of representative distal NFY-DHSs that were lost in E14.5 female PGCs and gained in E14.5 male PGCs. d Profiles of H3K27me3 signal density at distal NFY-DHSs and their surrounding regions in E14.5 male and female PGCs. e Expression levels of genes associated with E14.5 gained, lost and maintained distal NFY-DHSs in male PGCs. The difference is statistically significant by Wilcoxon rank sum test (***P < 0.001). f The expression of genes associated with male E14.5-gained distal NFY-DHSs in PGCs and postnatal spermatogonial stem cells (SSCs). The difference is statistically significant by Wilcoxon rank sum test (***P < 0.001)
To further uncover the regulatory activity of NFY-DHSs in PGCs, we examined the expression levels of genes associated with NFY-DHSs. With NFY occupancy in promoter regions, the expression levels of genes were significantly higher than those without promoter NFY-DHSs across all stages in male PGCs (Supplementary information, Fig. S5c), which is in line with the notion that NFY acts as a transcriptional activator via its promoter-proximal binding.41,42 However, genes associated with E14.5-gained distal NFY-DHSs in male PGCs were repressed compared to those associated with the lost or maintained distal NFY-DHSs (Fig. 5e). Consistently, the promoter regions of these repressed genes showed relatively low DHS signals (Supplementary information, Fig. S5d) and high H3K27me3 enrichment (Supplementary information, Fig. S5e). Notably, such a unique expression pattern of distal NFY-DHS associated genes could be observed only during the E13.5-to-E14.5 transition in male PGCs (Fig. 5e; Supplementary information, Fig. S5f). Further examination revealed that these repressed genes associated with E14.5-gained distal NFY-DHSs were subsequently activated in postnatal spermatogonial stem cells (SSCs)43 (Fig. 5f), and they were enriched in RAS signaling pathways that play crucial roles in SSC self-renewal and maintenance44–46 (Supplementary information, Fig. S5g and Table S2). Together, these findings suggested that the chromatin of mitotic-arrested male PGCs is permissive through NFY binding in distal regulatory regions and ready for reconfiguration in subsequent developmental stages.
Evolutionary pressure on PGC CREs
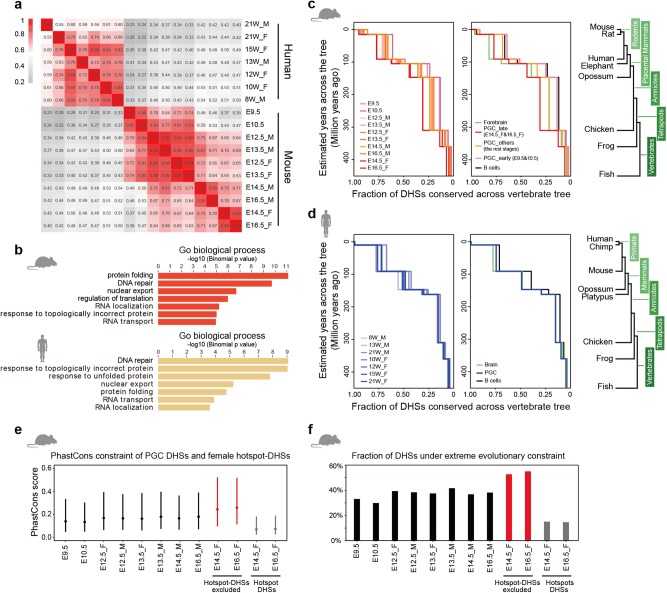
The developmental programs of germline are speculated to be evolutionarily conserved because they transmit genetic and epigenetic information across generations and maintain the continuance of the species.2,47 Our time-course profiling of accessible chromatin in mouse and human PGCs with consistent methodology provides an opportunity to examine the evolutionary pressure on PGC CREs using comparative genomic measurements. We first demonstrated that DHS signals from mouse and human PGC orthologous regions were well correlated between stages in each species (Fig. 6a), and PCA of DHSs in orthologous regions recapitulated the differential developmental paths of mouse and human PGCs (Supplementary information, Fig. S6a). To obtain a more accurate depiction of the evolutionary conservation between species, we further compared the DHS signals of the 59,754 mouse and human PGC orthologous regions, and 40,571 of them exhibited comparable chromatin accessibility levels. We asked whether orthologous regions showing similar DHS signals possessed conserved regulatory activities in mouse and human PGCs by allocating nearby genes to these orthologous DHSs. The functional annotations of associated genes in mouse and human PGCs were both enriched for fundamental functions (Fig. 6b; Supplementary information, Table S2). Notably, DNA repair was highly enriched in both species (Fig. 6b), indicating that DNA damage response is an evolutionarily conserved mechanism to preserve the genomic integrity of mouse and human PGCs. The high conservation of the 40,571 PGC orthologous sites was further confirmed by their high phastCons48 scores and numerous overlaps with the most highly constrained phastCons elements (Supplementary information, Fig. S6b, c).
Fig. 6.
Evolutionary pressures on cis-regulatory activities in mouse and human PGCs. a Spearman correlation matrix of DHS signals at orthologous sites in mouse and human PGCs. b Functional annotation of orthologous DHSs that exhibit comparable chromatin accessibility levels in mouse and human PGCs (n = 1458). c Plots showing cumulative proportion of mouse PGC DHSs conserved across the vertebrate tree (shown on far right) as defined by sequence homology. The left panel shows evolutionary differences among mouse PGC DHSs across developmental stages and between sexes. The right panel displays the average evolutionary conservation levels in mouse PGCs, forebrain and B cells. Mouse PGCs were divided into three categories based on their different evolutionary conservation levels. d Similar analysis to (c), performed in human PGCs, brain and B cells. e PhastCons constraint of mouse PGC DHSs and female hotspot-linked DHSs. For E14.5 and E16.5 female PGCs, DHSs representing recombination hotspots were excluded. f The fractions of mouse PGC DHSs that were under strong vertebrate constraint (score > 300)
Distal CREs marked by DHSs were demonstrated to be highly diverse during mouse and human PGC development (Fig. 2a), evolutionary differences of PGC CREs were expected across stages or between genders within species. Based on the evolutionary distances reflected in the vertebrate tree,10,18,49 we first verified that our evolutionary analysis of CREs from mouse forebrain, liver and heart was consistent with previous findings based on P300 binding49 or H3K27ac enrichment10 (Supplementary information, Fig. S6d). Strikingly, unlike these three tissues, in which the evolutionary conservation levels of CREs gradually decline during embryogenesis, the conservation of mouse PGC CREs exhibited a contradictory tendency during fetal development. In particular, the CREs of late-stage female PGCs (E14.5_F and E16.5_F) showed the maximum evolutionary conservation levels, whereas the CREs of early-stage PGCs (E9.5 and E10.5) were the least conserved, and intermediate CRE conservation levels were observed in the rest of the stages examined (Fig. 6c). We speculated that, as mouse PGCs gradually developed into gametes, they became increasingly evolutionarily conserved to ensure the proper delivery of genetic information. However, the evolutionary conservation of human PGC CREs was relatively irregular (Fig. 6d). Analysis of ENCODE DNase-seq data from human tissues such as brain and heart spanning a longer time course also showed disordered states (Supplementary information, Fig. S6e), which may be explained by the heterogeneity of human embryogenesis.5,6
Next, we concentrated on the evolutionary differences across tissues and cell types within species. Interestingly, we found that CREs in mouse E14.5 and E16.5 female PGCs were more evolutionarily conserved than those in forebrain outside the placental mammals (Fig. 6c). Similarly, CREs in human PGCs are more conserved than those in brain outside the mammals (Fig. 6d). The possible explanation may be that PGCs, as ancestors of gametes, are generally more conserved on a longer time scale, while brain became highly conserved to maintain its complex functions and structures within the mammal clade. Since thousands of E14.5 and E16.5 female DHSs were found to mark female recombination hotspots (Fig. 4g), we speculated that the high evolutionary conservation of late-stage female PGCs (E14.5_F and E16.5_F) was attributable to meiotic regulatory elements marked by DHSs but not hotspot-linked DHSs. Furthermore, meiosis occurs in all sexually reproducing single-celled and multicellular eukaryotes,50 whereas recombination hotspots are short-lived and self-destructive during meiosis.36 To verify our assumption, we examined the phastCons constraint of female recombination hotspots captured by DHSs. We found that the conservation levels of female recombination hotspot-linked DHSs were even lower than those of the least conserved E9.5 and E10.5 PGC CREs (Fig. 6e, f). Consistently, male spermatocyte recombination hotspots had very low evolutionary conservation, irrespective of mouse strain, compared to Prdm9–/– B6, which had aberrant hotspots in highly conserved promoter regions29 (Supplementary information, Fig. S6f). All these evidence indicates that the strong conservation of mouse late-stage female PGCs results from meiotic regulatory activities marked by DHSs but not from recombination hotspot-linked DHSs.
Discussion
We present time-course profiles of accessible chromatin in mouse and human PGCs at a high resolution. The DHS maps obtained from PGCs across multiple stages of the mammalian lifespan provide insights into chromatin conformation and cell fate regulation during PGC development. Distal CREs, such as enhancers, insulators and locus control regions, have long been found to associate with developmentally expressed genes. Here, we demonstrate that distal CREs marked by DHSs in PGCs show temporal activity patterns, highlighting the rapidly changing activities of distal regulatory elements during mouse and human PGC development.
We believe that DHS maps could provide a scaffold, on which combining data from TF ChIP-seq, histone modification experiments or other approaches enables accurate annotation of accessible chromatin during PGC development. Using core pluripotency TF binding sites from ESCs and EpiLCs as an occupancy pool, we accurately identified the binding sites of pluripotency TFs in mouse PGCs. We demonstrated that these binding sites had two waves of decrease during mouse PGC development: a minor decrease was observed from E10.5 to E12.5, and the most significant decline occurred during the E13.5-to-E14.5 transition (Fig. 3f). Following PGC development, late-stage female PGCs enter meiosis. Meiotic recombination in the male occurs in spermatocytes, and the recombination hotspots of that process can be determined by DMC1 ChIP-seq.29,30 However, the same method could be hardly applied to the female, owing to the limited cell number of PGCs. One of our most surprising results is that mouse female recombination hotspots can be captured by DHSs, and identification of hotspots in female PGCs could be achieved by combining low-input DNase-seq with in vitro PRDM9 Affinity-seq, bypassing direct pull-down of DMC1. Low-input DNase-seq could map DHSs using a small number of cells at a high resolution, and in vitro PRDM9 Affinity-seq only requires genomic DNA independent of cell type;35 therefore, combining low-input DNase-seq with PRDM9 Affinity-seq is most likely the only applicable solution to identify female recombination hotspots in human and other species. In contrast to meiotic female PGCs, male PGCs enter into mitotic arrest at around the same stage and stay quiescent for the remaining embryonic period.2 We prove that NFY may act as a bifunctional TF in mitotic-arrested male PGCs depending on its occupancy position in mitotic-arrested male PGCs. NFY-DHSs can pre-open the chromatin of mitotic-arrested male PGCs in the distal regulatory regions, leaving them ready for reconfiguration in subsequent developmental stages.
In sexually reproducing organisms, germ cells provide the enduring link between the generations, transmitting the genetic and epigenetic information required to construct a new organism. The evolutionary pressures on mouse and human PGC cis-regulatory activities reveal that evolutionary differences exist between species, across tissues and among developmental stages, consistent with previous studies in other models.10,49,51 Distinct evolutionary features of mouse late-stage female PGC CREs and hotspot-linked DHSs suggest that evolutionary examinations should not remain limited to the cellular scale but also be evaluated across genomic features within cells.
Due to the scarcity of PGCs, X-ChIP-based methods for mapping the genomic occupancy of TFs, such as pluripotency TFs and the meiotic recombination protein DMC1, have been impeded until now. Our accurate annotation of PGC accessible chromatin not only unveiled the chromatin landscapes in mouse and human PGCs, but also provided an alternative way to overcome these limitations. In particular, combining low-input DNase-seq with other approaches will help to advance many aspects of chromatin characterization.
Materials and methods
No statistical methods were used to predetermine sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment.
All animal maintenance and experimental procedures were performed in accordance with the University of Health Guide for the Care and Use of Laboratory Animals and were approved by the Biological Research Ethics Committee of Tongji University. Mouse were housed under a 12-h light/dark cycle under pathogen-free conditions at 22 ± 2 ℃ and fed with free access to standard mouse chow and tap water. The donors in this study were pregnant women who underwent medical termination of pregnancy. All of the patients signed informed consents and voluntarily donated the fetal tissues for this study. The experiments conducted in this study were approved by the Shanghai First Maternity and Infant Hospital.
Mouse and human PGC collection
Male mice homozygous for the Oct4-EGFP transgene in the C57BL/6J background were crossed with wild-type C57BL/6J females. When a copulatory plug was evident around noon, that time was counted as E0.5. PGCs were isolated from E9.5, E10.5, E12.5, E13.5, E14.5, and E16.5 embryos. Male and female samples were collected separately starting from E12.5, as gonads can be distinguished morphologically from E12.5 based on the presence or absence of testicular cords. Gonads were treated with 0.25% trypsin-EDTA for 10 min at 37 °C, and then, trypsin was inactivated by addition of 15% fetal bovine serum (FBS)/DMEM. Cells were filtered through 40 µm cell strainers to obtain a single-cell suspension for FACS (FACSAria II SORP, BD Bioscience).
Human PGCs and gonadal somatic cells were isolated as previously described.5 In addition to isolating CD117 (BD PharMingen #555714, clone YB5.B8)-positive PGCs, we also collected CD117-negative cells as gonadal somatic cells by FACS.
Cell culture
The R1 ESCs were cultured on mitomycin-C-treated MEFs in ESC medium containing DMEM supplemented with 15% (v/v) FBS, 1 mM L-glutamine, 0.1 mM mercaptoethanol, 1% nonessential amino acid stock, penicillin/streptomycin, nucleosides and 1000 U/mL LIF. EpiLCs were induced by plating 1.0 × 105 ESCs on a 12-well plate coated with human plasma fibronectin (16.7 mg/mL) in N2B27 medium containing activin A (20 ng/mL), bFGF (12 ng/mL), and KSR (1%).
Low-input DNase-seq
FACS-sorted cells were first washed in 0.5% bovine serum albumin in phosphate-buffered saline (BSA-PBS, Sigma) solution to avoid potential contamination. After washing, fresh cells were collected in 1.5 mL tubes and resuspended in 45 μL lysis buffer (40 mM Tris-HCl, pH 7.5, 10 mM NaCl, 6 mM MgCl2, 6 mM CaCl2, 0.1% NP40) and then incubated on ice for 5 min to release the nuclei. DNase I (10,000 U/mL, Roche, 04716728001) was added to a final concentration of 30 U/mL and incubated at 37 °C for exactly 5 min. DNase I concentrations to achieve optimal smearing sizes may differ for each cell line and therefore should be determined empirically for each cell type. After DNase I digestion, the reaction was terminated immediately by adding 50 μL Stop Buffer (40 mM Tris-HCl, pH 7.5, 10 mM NaCl, 0.5% SDS, 10 mM EDTA) containing 1 μL Proteinase K (20 mg/mL, Life Technologies). After incubation at 55 °C for 1 h, DNA was purified by extraction with phenol-chloroform and precipitated by ethanol in the presence of glycogen (20 mg/mL, Roche, 10901393001) for >3 h at –20 °C.
Sequencing libraries were generated using the KAPA Hyper Prep Kit for the Illumina platform (kk8504), following the manufacturer’s instructions. To assess library complexity, we chose agarose gel electrophoresis instead of SPRI bead-based methods for size selection. Because DHSs are not binary, only the smearing pattern samples under appropriate digestion were ideal for generating high-quality DNase data. DNA fragments of 150–300 bp were extracted from the agarose gel for a second PCR amplification. The DNA was eluted in 15 μL of elution buffer (2.5 mM Tris, pH 7.6, 0.05 mM EDTA) and quantified by a Qubit dsDNA HS assay kit (ThermoFisher Scientific, Q32854) and Agilent High Sensitivity assay kit (Agilent Technologies). The libraries were sequenced on an Illumina HiSeq 2500 with a single-end 50 bp protocol.
ULI-NChIP-seq
The ULI-NChIP procedure was performed as previously described.52 Approximately 3000 PGCs were used per reaction. One microgram of histone H3K4me3 antibody (Cell Signaling Technology, #9727), histone H3K27me3 antibody (Diagenode, C15410069) or histone H3K27ac antibody (Active Motif, #39133) was used for each immunoprecipitation reaction. The sequence libraries were generated using the KAPA Hyper Prep Kit according to the manufacturer’s instructions. The libraries were sequenced on an Illumina HiSeq 2500 with a single-end 50 bp protocol at Berry Genomics Co., Ltd.
DNase-seq data analysis
Sequencing reads were first trimmed of low-quality and adaptor sequences using cutadapt (v1.12)53 and then mapped to the mouse genome (mm10) or human genome (hg38) using bowtie (v1.2)54 with the parameters: -v 1 -m 1 --mm --no-unal. Uniquely mapped reads were kept for further analysis using SAMtools (v1.6).55 The DHS peaks were identified based on the above retained alignment results by the tool Hotspot (v4)56 with FDR < 0.01. The DHS peaks from biological replicates were merged using ‘bedtools merge’ from bedtools (v2.27.0)57 and only the overlapping peaks were retained. The signals of the DHS peaks were calculated and normalized as FPKM (fragments per kilobase per million uniquely mapped fragments) using ‘multiBamSummary --centerReads’ from deepTools (2.5.4),58 then divided by the uniquely mapped fragments and the length of the peak. To identify more reliable DHS peaks, we further filtered out peaks with RPKM < 3. The retained peaks were used for the analysis in this study.
RNA-seq data analysis
RNA-seq data of mouse PGCs were obtained from published works (see Supplementary information, Table S3). Sequencing reads were first trimmed similarly to those in the DNase-seq data analysis and then mapped to the annotated mouse transcripts (UCSC mm10) using Hisat2 (v2.1.0)59 with the parameters: --dta-cufflinks --no-unal. Mapped reads were subsequently assembled into transcripts guided by the UCSC gtf annotation files using Cuffdiff (v2.2.1).60 The expression level of each gene was quantified with normalized FPKM. For human PGC RNA-seq data, we directly used the gene expression levels provided by the published works.5,6
Histone modification ChIP-seq data analysis
Sequencing reads were trimmed and then mapped to the mouse genome (mm10) similarly to those in the DNase-seq data analysis. Uniquely mapped reads were kept for further analysis using SAMtools (v1.6).55 Genome-wide histone modification signals were calculated using a 25-bp window and normalized to the uniquely mapped fragments using ‘bam Coverage’ from deepTools (2.5.4).58 The distribution of a histone modification signal relative to the center of a set of DHS peaks was calculated as follows: the distances to the peak center were binned in 10-bp intervals. The sum of the histone modification signals in the same bin was normalized by the number of DHS peaks.
TF ChIP-seq data analysis and peak calling
TF ChIP-seq data were taken from published works (see Supplementary information, Table S3). For three samples (OCT4 for mouse ESC and EpiLC, and PRDM9 for mouse in vitro experiments), sequencing reads were trimmed and then mapped to the mouse genome (mm10) similarly to those in the DNase-seq data analysis. Uniquely mapped reads were retained for further analysis using SAMtools (v1.6).55 MACS2 (v2.1.1)61 was used to perform peak calling with P-value < 0.01. For other samples (OCT4 for mouse EpiSC, SOX2 and NANOG for mouse ESC, PRDM14 for mouse PGC, and DMC1 for different mouse strains), we directly used the peaks provided by the published works.
Motif enrichment and analysis in DHS peaks
Taking the coordinates of DHS peaks in BED format as an input file, the tool Homer (v4.9)40 was applied to identify the enriched motifs in the peaks with the parameters: -size 150 -mask. The enrichment of the identified motifs was plotted in heatmaps. Additionally, we scanned each DHS peak using the tool Homer with the parameters: -size 150 -mask -find NFY_motif in the NFY-DHSs analysis. Note that “NFY_motif” is the motif matrix of the transcription factor NFY provided in Homer. Thus, the DHS peaks were categorized into two groups: containing or not containing the NFY motif.
Functional annotation of DHS peaks
To predict the function of dynamic DHS during PGC development, the tool GREAT62 was applied to annotate the potential functions of the following DHS peak sets: distal peaks common in male and female mouse PGCs from E12.5 to E16.5; distal peaks common in male and female human PGCs from 8 to 21 W; E14.5 and E16.5 female-specific DHS peaks that did not overlap hotspots; orthologous regions with similar DHS signals in both mouse and human. Significant enrichment was defined as a Bonferroni-corrected binomial P-value < 0.05. KEGG pathway analysis for male distal DHS peaks (away from TSS ± 1 kb) containing the NFY motif at E14.5 but not at E13.5 was performed with DAVID (P-value < 0.05).63
Dynamic analysis of DHS peaks
We compared DHS peaks between two adjacent stages and then classified them into three states by ‘bedtools intersect’ from bedtools. Here, “gained” means that the DHS peak existed in the next stage but was not detected in the previous stage, while “lost” is the reverse situation, and “maintained” means that the DHS peak existed in both stages with at least 30% overlap.
Conservation estimation of DHS peaks throughout vertebrate evolution
Orthologous DHS peaks between species were identified as described previously.10,18 In brief, pairwise genome alignments (“over” chain files) were downloaded from the UCSC Genome Browser database between mouse (mm10) and human (hg19) to the following genomes: mouse (mm10), rat (rn6), human (hg38), elephant (loxAfr3), opossum (monDom5), chicken (galGal5), frog (xenTro3), zebrafish (danRer11), chimp (panTro5) and platypus (ornAna1). Based on these chain files, DHS peaks were mapped between species using the liftOver tool with -minMatch = 0.5 as the homology cutoff. A DHS peak was two-way orthologous if the two-way mappings satisfied the following: mapping from query to target species (e.g., mouse → human) was unique, and the reciprocal mapping from target species back to query (e.g., human → mouse) was also unique. Only such two-way orthologous DHS peaks were retained for further evolution-related analysis.
The DHS signals in the two-way orthologous DHS peaks between mouse and human were recalculated in mouse and human, respectively. The pairwise Spearman correlations between the signals in these DHS peaks were calculated. We further compared the DHS signals in the two-way orthologous regions. If the mouse DHS signals in an orthologous region were 3 times higher than those of human, we identified that DHS as a mouse-specific peak. Human-specific DHS peaks in orthologous regions were defined using the same standard. The remaining orthologous regions that had comparable DHS signals (≤3-fold in either) were termed mouse and human conserved sites.
To examine the evolutionary history of DHS peaks, we defined the fraction of DHS peaks conserved throughout vertebrate evolution as their degree of conservation. A DHS peak was conserved if it was two-way orthologous. Then, we overlaid the estimated divergence times of species provided by the published studies64–66 to estimate the proportion of DHS peaks conserved throughout vertebrate evolution.
DNase-seq data for different mouse and human tissues and cell lines were downloaded from the ENCODE project (see Supplementary information, Table S3). The DHS peaks provided by the ENCODE project were directly used in this study. Similarly, two-way orthologous DHS peaks were identified and retained for the evolutionary analysis described above.
The per-base phastCons48 scores and phastCons elements for multiple alignments of vertebrate species were downloaded from the UCSC Genome Browser database. To assess conservation, we computed the fractions of phastCons elements that overlapped mouse- and human-specific DHS peaks and orthologous DHSs between mouse and human, respectively, and calculated the exactly phastCons scores on these regions. We also do the similar analysis on PGC DHSs, female hotspot DHSs and other mouse male recombination hotspots.
Data and software availability
The accession number for all sequencing data (DNase-seq and ChIP-seq) reported in this paper is GEO: GSE109770. You can also visit our processed data without downloading any files: http://wukong.tongji.edu.cn/new_home/sjshen/PGC_CR_data_availability.html.
All analyses were performed based on custom shell, python and R code, and available upon request.
Electronic supplementary material
Acknowledgements
We are grateful to our colleagues in the laboratory for their assistance with the experiments and in the preparation of this manuscript. This work was primarily supported by the National Key R&D Program of China (2016YFA0100400) and the National Natural Science Foundation of China (31721003). This work was also supported by the Ministry of Science and Technology of China (2017YFA0102602 and 2015CB964800), the National Natural Science Foundation of China (81630035, 31771419, 31871446, 31871448 and 81501250), the Shanghai Rising-Star Program (17QA1402700), the Shanghai Chenguang Program (16CG17 and 16CG19), the Shanghai Municipal Medical and Health Discipline Construction Projects (2017ZZ02015), and the Shanghai Municipal Commission of Health and Family Planning (201540364 and 20164037).
Author contributions
J.L. and S.G. designed the experiments; J.L. conducted the experiments with assistance from J.C., W.L., X.L., B.W., J.Y., L.W., M.W., L.G. and H.W.; J.L., S.S. and C.J. analyzed the data with assistance from Q.Z. and X.C.; J.L., S.S., C.J. and S.G. wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Cizhong Jiang, Email: czjiang@tongji.edu.cn.
Shaorong Gao, Email: gaoshaorong@tongji.edu.cn.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0096-5.
References
- 1.Tang WW, Kobayashi T, Irie N, Dietmann S, Surani MA. Specification and epigenetic programming of the human germ line. Nat. Rev. Genet. 2016;17:585–600. doi: 10.1038/nrg.2016.88. [DOI] [PubMed] [Google Scholar]
- 2.Saitou M, Yamaji M. Primordial germ cells in mice. Cold Spring Harb. Perspect. Biol. 2012;4:pii: a008375. doi: 10.1101/cshperspect.a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spiller C, Wilhelm D, Koopman P. Cell cycle analysis of fetal germ cells during sex differentiation in mice. Biol. Cell. 2009;101:587–598. doi: 10.1042/BC20090021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Western PS, Miles DC, van den Bergen JA, Burton M, Sinclair AH. Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells. 2008;26:339–347. doi: 10.1634/stemcells.2007-0622. [DOI] [PubMed] [Google Scholar]
- 5.Guo F, et al. The transcriptome and DNA methylome landscapes of human primordial germ cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 6.Li L, et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell. Stem. Cell. 2017;20:858–873. doi: 10.1016/j.stem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Seisenberger S, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kobayashi H, et al. High-resolution DNA methylome analysis of primordial germ cells identifies gender-specific reprogramming in mice. Genome Res. 2013;23:616–627. doi: 10.1101/gr.148023.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu F, et al. Establishing chromatin regulatory landscape during mouse preimplantation development. Cell. 2016;165:1375–1388. doi: 10.1016/j.cell.2016.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nord AS, et al. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leitch HG, Smith A. The mammalian germline as a pluripotency cycle. Development. 2013;140:2495–2501. doi: 10.1242/dev.091603. [DOI] [PubMed] [Google Scholar]
- 12.Kurimoto K, et al. Quantitative dynamics of chromatin remodeling during germ cell specification from mouse embryonic stem cells. Cell. Stem. Cell. 2015;16:517–532. doi: 10.1016/j.stem.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Hajkova P, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016;167:1170–1187. doi: 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villar D, et al. Enhancer evolution across 20 mammalian species. Cell. 2015;160:554–566. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesch BJ, Dokshin GA, Young RA, McCarrey JR, Page DC. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc. . Natl. Acad. Sci. U. S. A. 2013;110:16061–16066. doi: 10.1073/pnas.1315204110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vierstra J, et al. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science. 2014;346:1007–1012. doi: 10.1126/science.1246426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soucie EL, et al. Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Science. 2016;351:aad5510. doi: 10.1126/science.aad5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lara-Astiaso D, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345:943–949. doi: 10.1126/science.1256271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neph S, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Runyan C, et al. Steel factor controls midline cell death of primordial germ cells and is essential for their normal proliferation and migration. Development. 2006;133:4861–4869. doi: 10.1242/dev.02688. [DOI] [PubMed] [Google Scholar]
- 23.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buecker C, et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell. Stem. Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li M, Belmonte JC. Ground rules of the pluripotency gene regulatory network. Nat. Rev. Genet. 2017;18:180–191. doi: 10.1038/nrg.2016.156. [DOI] [PubMed] [Google Scholar]
- 26.He HH, et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat. Methods. 2014;11:73–78. doi: 10.1038/nmeth.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kojima Y, et al. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell. Stem. Cell. 2014;14:107–120. doi: 10.1016/j.stem.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 28.Yamaji M, et al. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat. Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- 29.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khil PP, Smagulova F, Brick KM, Camerini-Otero RD, Petukhova GV. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res. 2012;22:957–965. doi: 10.1101/gr.130583.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smagulova F, et al. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baudat F, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies B, et al. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature. 2016;530:171–176. doi: 10.1038/nature16931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelmenson PM, et al. A torrid zone on mouse chromosome 1 containing a cluster of recombinational hotspots. Genetics. 2005;169:833–841. doi: 10.1534/genetics.104.035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walker M, et al. Affinity-seq detects genome-wide PRDM9 binding sites and reveals the impact of prior chromatin modifications on mammalian recombination hotspot usage. Epigenetics Chromatin. 2015;8:31. doi: 10.1186/s13072-015-0024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai IJ, Burt A, Koufopanou V. Conservation of recombination hotspots in yeast. Proc. . Natl. Acad. Sci. U. S. A. 2010;107:7847–7852. doi: 10.1073/pnas.0908774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pratto F, et al. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014;346:1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nardone V, Chaves-Sanjuan A, Nardini M. Structural determinants for NF-Y/DNA interaction at the CCAAT box. Biochim. Biophys. Acta. 2017;1860:571–580. doi: 10.1016/j.bbagrm.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Nardini M, et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell. 2013;152:132–143. doi: 10.1016/j.cell.2012.11.047. [DOI] [PubMed] [Google Scholar]
- 40.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oldfield AJ, et al. Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol. Cell. 2014;55:708–722. doi: 10.1016/j.molcel.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bungartz G, Land H, Scadden DT, Emerson SG. NF-Y is necessary for hematopoietic stem cell proliferation and survival. Blood. 2012;119:1380–1389. doi: 10.1182/blood-2011-06-359406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammoud SS, et al. Transcription and imprinting dynamics in developing postnatal male germline stem cells. Genes Dev. 2015;29:2312–2324. doi: 10.1101/gad.261925.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanatsu-Shinohara M, Shinohara T. Spermatogonial stem cell self-renewal and development. Annu. Rev. Cell. Dev. Biol. 2013;29:163–187. doi: 10.1146/annurev-cellbio-101512-122353. [DOI] [PubMed] [Google Scholar]
- 45.Meng X, et al. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 46.He Z, et al. Gdnf upregulates c-Fos transcription via the Ras/Erk1/2 pathway to promote mouse spermatogonial stem cell proliferation. Stem Cells. 2008;26:266–278. doi: 10.1634/stemcells.2007-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whittle CA, Extavour CG. Causes and evolutionary consequences of primordial germ-cell specification mode in metazoans. Proc. . Natl. Acad. Sci. U. S. A. 2017;114:5784–5791. doi: 10.1073/pnas.1610600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blow MJ, et al. ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 2010;42:806–810. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 51.Cotney J, et al. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell. 2013;154:185–196. doi: 10.1016/j.cell.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537:558–562. doi: 10.1038/nature19362. [DOI] [PubMed] [Google Scholar]
- 53.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 54.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, et al. The Sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.John S, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramirez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 64.Hedges SB. The origin and evolution of model organisms. Nat. Rev. Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 65.Murphy WJ, Pringle TH, Crider TA, Springer MS, Miller W. Using genomic data to unravel the root of the placental mammal phylogeny. Genome Res. 2007;17:413–421. doi: 10.1101/gr.5918807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bininda-Emonds OR, et al. The delayed rise of present-day mammals. Nature. 2007;446:507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.