Abstract
Oryza officinalis is an accessible alien donor for genetic improvement of rice. Comparison across a representative panel of Oryza species showed that the wild O. officinalis and cultivated O. sativa ssp. japonica have similar cold tolerance potentials. The possibility that either distinct or similar genetic mechanisms are involved in the low temperature responses of each species was addressed by comparing their transcriptional networks. General similarities were supported by shared transcriptomic signatures indicative of equivalent metabolic, hormonal, and defense status. However, O. officinalis has maintained an elaborate cold-responsive brassinosteroid-regulated BES1-network that appeared to have been fragmented in O. sativa. BES1-network is potentially important for integrating growth-related responses with physiological adjustments and defenses through the protection of photosynthetic machinery and maintenance of stomatal aperture, oxidative defenses, and osmotic adjustment. Equivalent physiological processes are functional in O. sativa but their genetic mechanisms are under the direct control of ABA-dependent, DREB-dependent and/or oxidative-mediated networks uncoupled to BES1. While O. officinalis and O. sativa represent long periods of speciation and domestication, their comparable cold tolerance potentials involve equivalent physiological processes but distinct genetic networks. BES1-network represents a novel attribute of O. officinalis with potential applications in diversifying or complementing other mechanisms in the cultivated germplasm.
Introduction
The domesticated rice (Oryza sativa L.) represents only a fraction of the total genetic potential of the genus1–3. Geographic distribution, developmental and physiological variation across cultivars and wild species reflect a gradient of ecological adaptation that were optimized during speciation and domestication4. Taxonomic sub-division in the genus includes the sativa, officinalis, ridleyi, and granulata species complexes, and sections ridleyanae Tateoka and brachyantha B.R. Lu. Under these sub-divisions are two cultivated and 22 wild species, defined by six diploid (2n = 24; AA, BB, CC, EE, FF, GG) and four allotetraploid (4n = 48; BBCC, CCDD, HHJJ, KKLL) genomes5,6. The sativa-complex includes the temperate (japonica) and tropical (indica) Asian rice O. sativa, African rice O. glaberrima, and their AA-genome progenitors O. rufipogon, O. nivara, and O. barthii, respectively. Harnessing the potential of this gene pool to widen the genetic base of cultivars has been successful2.
The officinalis-complex is the largest group that includes the genomes BB (O. punctata), CC (O. officinalis, O. rhizomatis, O. eichingeri) and EE (O. australiensis), and their allotetraploid combinations BBCC (O. minuta, O. punctata) and CCDD (O. latifolia, O. alta, O. grandiglumis). O. officinalis is a highly valued member of this complex because of its pivotal role in multiple tetraploidization events7. Habitat distribution indicate that O. officinalis is a rich reservoir of adaptive traits for the enhancement of cultivars8–12. Successful efforts for agronomic trait introgression from wild Oryza to cultivars involved either a diploid CC-genome (O. officinalis) or allotetraploid CC-combination (O. minuta, O. latifolia) as donors13–15. This shows that CC-genome is a more accessible alien donor for the diversification of the genetic base of cultivars16.
While the Asian cultivated rice is generally very cold-sensitive, a number of temperate japonica cultivars have been used as donors of tolerance-associated traits17,18. Based on population distribution and eco-geographic niches, bioclimatic models placed O. officinalis in the middle range of stress tolerance potentials relative to 21 other Oryza species4. However, despite the predictions of the bioclimatic models, the true potentials of the wild Oryza germplasm as a source of novel mechanisms for cold tolerance that may not exist in temperate japonica have not been critically examined.
In this study, the cold stress transcriptional network of the CC-genome reference O. officinalis accession IRGC100896 was reconstructed and compared with the corresponding network in the AA-genome reference O. sativa ssp. japonica cv. Nipponbare. The IRGC100896 was chosen as reference O. officinalis because it is widely used as an alien donor in rice breeding16,19. The major goal was to reveal shared and contrasting regulatory network signatures across the two reference genotypes with comparable cold tolerance potentials in order to understand the significance of such networks in context of possible complementation effects in recombinants. The genetic mechanism revealed in this study is an important first step for understanding the finer details behind the hidden potentials of the Oryza CC-genome for diversifying the genetic mechanisms that exist in cultivars.
Results
Cold tolerance potential of IRGC100896 and Nipponbare
Guided by the bioclimatic models of Atwell et al.6, a panel of wild accessions used as alien donors in IRRI’s breeding program was compared for sensitivity to low temperature (4 °C). Evaluation was based on standardized metrics that included IRRI’s Standard Evaluation Score (SES) for plant recovery, and plant injury measurements through the cellular electrolyte leakage index (ELI)18. Overall, the relative ranking of accessions revealed by SES and ELI was generally consistent with the proposed ranking in the bioclimatic model. The IRGC100896 and Nipponbare were more similar to each other in terms of cold tolerance, both having significantly lower ELI and SES compared to the highly sensitive check IR64 (O. sativa ssp. indica) and other diploid wild accessions (Fig. 1; Supplementary Figure S1).
Figure 1.

Comparison of plant injury by tissue electrolyte leakage index (ELI) and plant recovery expressed as standard evaluation score (SES) across a panel of wild and cultivated Oryza. Recovery is expressed in a scale of 0 (0%) to 10 (100%). ELI is the ratio of induced electrolyte leakage between control and stress conditions, with ELI >1.0 indicating significant injury (p < 0.05, n = 6). Cultivars: Osj = sativa-japonica, Nipponbare; Osi = sativa-indica, IR64; AA-genome: Or = rufipogon, Ol = longistaminata; CC-genome: Oo = officinalis; Oe = O. eichingeri; Or = rhizomatis; EE-genome: Oa = australiensis; FF-genome: Ob = brachyantha; CCDD-genome: Op = punctata.
General features of CC and AA cold stress transcriptomes
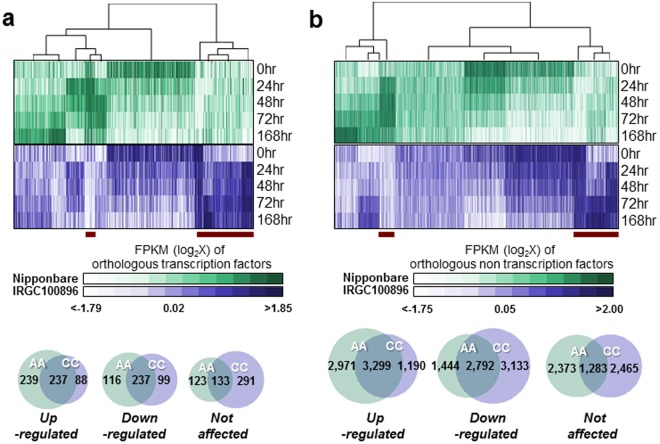
A series of RNA-Seq libraries (DDBJ-DRA006704) was constructed to investigate whether distinct mechanisms are involved in the expression of similar cold tolerance potential in IRGC100896 and Nipponbare (Supplementary Table S2). RNA-Seq libraries of Nipponbare detected 1,168 unique transcription factor transcripts and 20,699 unique non-transcription factor transcripts across expression profiles, which represent 88% and 87% of the total transcripts for each category in the Nipponbare reference. These results are generally consistent with previously reported microarray-based transcriptome datasets20–22. RNA-Seq libraries of IRGC100896 detected 1,107 unique transcription factor transcripts and 17,403 unique non-transcription factor transcripts across expression profiles, representing 83% and 73% of total transcripts for each category in the reference (Fig. 2; Supplementary Figure S3).
Figure 2.
General trends in the cold stress transcriptomes of O. officinalis (IRGC100896; purple) and O. sativa ssp. japonica (Nipponbare; green). Mapped reads in each RNA-Seq library were normalized to allow direct comparison of relative transcript abundance across orthologous loci. (a) Top panel shows the temporal expression of 943 orthologous pairs of transcription factors in IRGC100896 and Nipponbare. Profiles that are common between the two genotypes and those unique to one genotype (red bars) are indicated. Venn diagram (bottom) summarizes the proportion of orthologous transcription factors with either similar or unique profile across genotypes. (b) Expression patterns across 14,162 pairs of orthologous non-transcription factors in IRGC100896 and Nipponbare presented in similar context as in (a).
Overall, expression of orthologous genes did not follow a perfect one-to-one trend across the IRGC100896 and Nipponbare transcriptomes, which indicated that both overlapping and unique sets of genes are involved in each genotype’s genetic networks (Fig. 2; Supplementary Figure S3). Among the transcripts expressed from orthologous genes, 476, 211, and 256 represent transcription factors that were upregulated, downregulated, and not affected by cold, respectively in Nipponbare. In IRGC100896, 325, 194, and 424 transcription factors were upregulated, downregulated and not affected by cold, respectively. Only 50% of cold-upregulated transcription factors in Nipponbare were also upregulated in IRGC100896, while only 73% of cold-upregulated transcription factors in IRGC100896 were also upregulated in Nipponbare. Similar reciprocal and non-reciprocal patterns were observed for downregulated genes.
Metabolic status inferred from cold stress transcriptome signatures
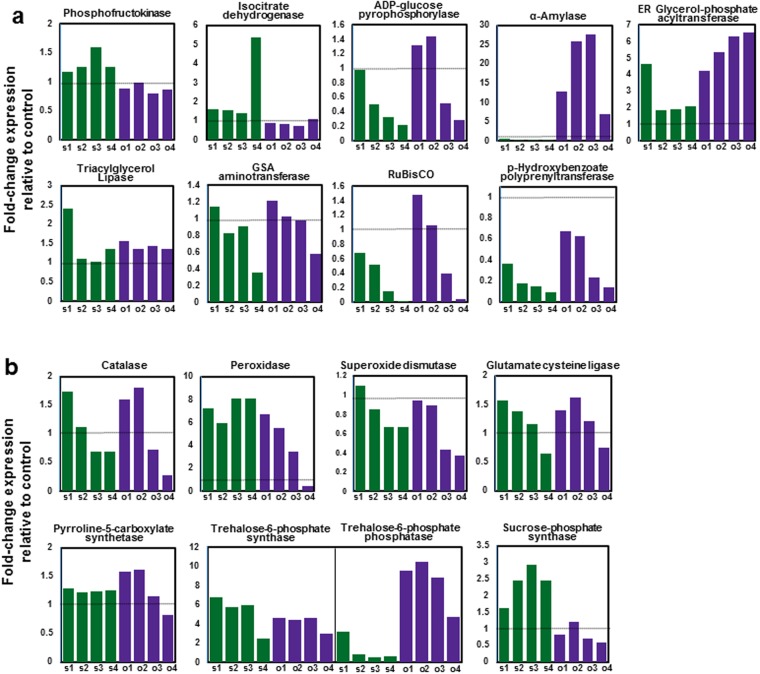
To compare the physiological status of IRGC100896 and Nipponbare under cold stress, KaPPA-View transcript maps were assembled for the primary metabolic pathways (Fig. 3a; Supplementary Figures S4–S8). Glycolysis and TCA cycle were not drastically different across genotypes based on the expression of their respective rate-limiting enzymes phosphofructokinase (EC.2.7.1.90) and isocitrate dehydrogenase (EC.1.1.1.42). Starch biosynthetic and catabolic pathways were assessed through the expression of ADP-glucose pyrophosphorylase (E.C.2.7.7.27) and α-Amylase (E.C.3.2.1.1), respectively. Opposite trends for biosynthesis and catabolism suggest less perturbation or penalty in IRGC100896. Based on the expression of glycerol-phosphate acyltransferase (E.C.2.3.1.15), triacylglyceride biosynthesis appeared to be differentially affected by cold, being highly upregulated in IRGC100896. Triacylglyceride catabolism as reported by triacylglycerol lipase (E.C.3.1.1.3) expression was not very different. Based on the expression of the enzyme p-hydroxybenzoate polyprenyltransferase (E.C.2.5.1.39) for ubiquinone and ATP biosynthesis, it appeared that the energy status of IRGC100896 and Nipponbare were similarly impaired under cold stress. As expected, photosynthesis was generally impaired in both genotypes as indicated by the downregulation of the rate-limiting biosynthetic enzyme glutamate-1-semialdehyde aminotransferase (E.C.5.4.3.8). Reduced activity of Calvin cycle as suggested by downregulation of RuBisCo (E.C.4.1.1.39) was consistent with the downregulation of chlorophyll biosynthesis (Fig. 3a; Supplementary Figures S9, S10).
Figure 3.
Transcriptome signatures representing the primary metabolic, radical scavenging, and osmotic adjustment status of IRGC100896 (purple) and Nipponbare (green). The s1/o1, s2/o2, s3/o3, and s4/o4 notations represent the transcript fold-change in Nipponbare (s) and IRGC100896 (o) at 24, 48, 72 and 144 hr, respectively. (a) Expression of rate-limiting enzymes phosphofructokinase (glycolysis), isocitrate dehydrogenase (TCA cycle), ADP-glucose phosphorylase (starch biosynthesis), α-amylase (starch catabolism), glycerol-phosphate acyltransferase (triacylglyceride biosynthesis), triacylglycerol lipase (triacylglyceride catabolism), GSA aminotransferase (chlorophyll biosynthesis), RuBisCO (photosynthesis/Calvin cycle), and p-hydroxybenzoate polyprenyltransferase (ubiquinone biosynthesis). KaPPA-View transcript maps in Supplementary Figures S4–S10. (b) Differential expression of radical scavenging (catalase, peroxidase, superoxide dismutase, glutathione synthesis), trehalose biosynthetic (trehalose-6-phosphate synthase, trehalose-6-phospate phosphatase), proline biosynthetic (pyrroline-5-carboxylate synthetase), and sucrose biosynthetic (sucrose-phosphate synthase) genes in IRGC100896 (purple) and Nipponbare (green). KaPPA-View maps are in Supplementary Figures S11–S12.
Primary defense capacity by radical scavenging (peroxidases, catalases, superoxide dismutases, glutathione-SH), and osmotic adjustment (sucrose, trehalose, proline) were also compared (Fig. 3b; Supplementary Figures S11, S12). Catalase (E.C.1.11.1.6; Os03g0131200) and peroxidase (E.C.1.11.1.7; Os02g0240300) were upregulated in both genotypes, particularly during the early periods of stress, while superoxide dismutase (E.C.1.15.1.1; Os05g0323900) was not drastically affected. Glutathione biosynthesis as indicated by the expression of glutamate cysteine ligase (E.C.6.3.2.2; Os07g0462000, Os05g0129000) was similar to the patterns of catalase and peroxidase, suggesting that the mechanisms of radical scavenging are similar in IRGC100896 and Nipponbare. Trehalose biosynthesis as indicated by the expression of trehalose-6-phosphate synthase (E.C.2.4.1.15) and trehalose-6-phosphate phosphatase (E.C.3.1.3.12) appeared to be higher IRGC100896. Increased expression of pyrroline-5-carboxylate synthetase (E.C.2.7.2.11 1.2.1.41; Os05g0455500) suggests an enhanced proline biosynthesis in both genotypes. Sucrose biosynthesis as indicated by sucrose-phosphate synthase (E.C.2.7.2.11; E.C.1.2.1.41) was upregulated in Nipponbare but downregulated in IRGC100896.
Hormonal status inferred from transcriptome signatures
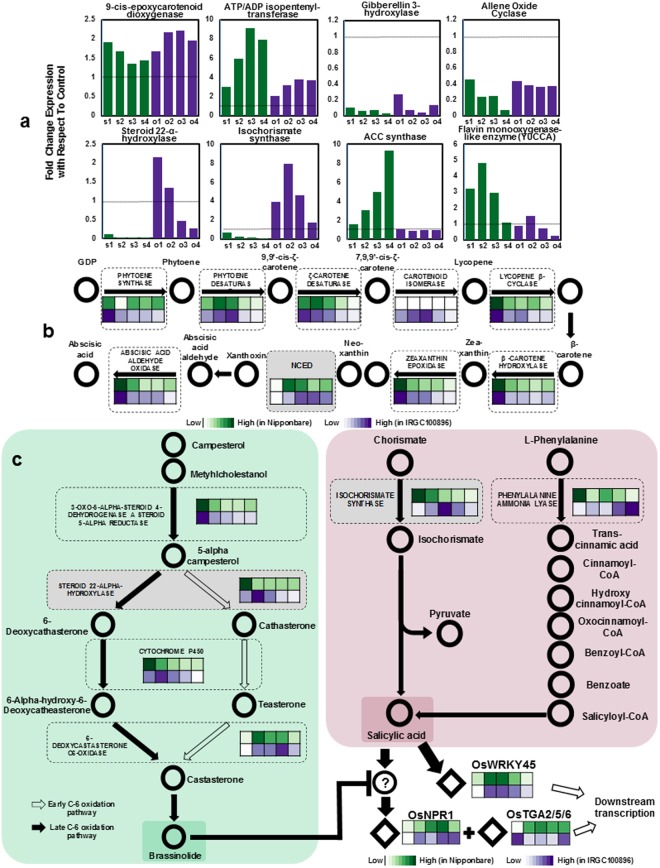
Stress and growth-related responses are integrated by hormonal signals. To compare the relative activities of hormone biosynthetic pathways across IRGC100896 and Nipponbare, KaPPA-View pathway maps were established for abscisic acid (ABA), gibberellic acid (GA), auxin (IAA) cytokinin (ZT), brassinosteroid (BL), salicylic acid (SA), ethylene (C2H4), and jasmonic acid (JA). Patterns in GA, ZT, and JA pathways were very similar across the two genotypes as indicated by the expression of their rate-limiting enzymes (Fig. 4a, b; Supplementary Figures S13–S15). Based on the expression of 9-cis-epoxycarotenoid dioxygenase (NCED; EC 1.13.11.51) and ATP/ADP isopentenyl transferase (E.C.2.5.1.8), ABA and ZT pathways appeared to be similarly enhanced by cold in both genotypes. GA and JA pathways were similarly downregulated as indicated by rate-limiting enzymes GA3-hydroxylase (E.C.1.14.11.15) and allene oxide cyclase (E.C.5.3.99.6). Downregulation of these pathways correlate well with the observed impairment of plant growth under cold stress.
Figure 4.
Transcriptome signatures representing the hormone biosynthetic status in IRGC100896 (purple) and Nipponbare (green). (a) Expression of rate-limiting enzymes 9-cis-epoxycarotenoid dioxygenase or NCED (ABA), ATP/ADP isopentenyl transferase (ZT biosynthesis), gibberellin 3-hydroxylase (GA) and allene oxide cyclase (JA), steroid 22-α-hydroxylase (BL), isochorismate synthase (SA), ACC synthase (C2H4) and tryptamine monooxygenase (IAA). The s1/o1, s2/o2, s3/o3, and s4/o4 notations represent the transcript fold-change in Nipponbare (s) and IRGC100896 (o) at 24, 48, 72 and 144 hr, respectively. (b) KaPPA-View transcript maps showing upregulation of ABA biosynthesis in both IRGC100896 (purple) and Nipponbare (green). (c) Parallel upregulation of BL and SA biosynthesis in IRGC100896 (purple) but not in Nipponbare (green), and cross-talks between BL and SA pathways through OsNPR1, OsTGA2/5/6 and OsWRKY45. Rate-limiting steps are highlighted in the grey box. Maps for other hormones are in Supplementary Figures S13–S17.
BL, SA, C2H4, and IAA pathways had the most significant contrasts between IRGC100896 and Nipponbare, based on the expression of their rate-limiting enzymes steroid 22-alpha-hydroxylase (E.C.1.14.13.-), isochorismate synthase (E.C.5.4.4.2), ACC synthase (E.C.4.4.1.14), and YUCCA flavin monooxygenase (E.C.1.14.13.8), respectively (Fig. 4c; Supplementary Figures S16–S17). The most notable contrast was the upregulation of BL and SA pathways in IRGC100896 but downregulation in Nipponbare. C2H4 and IAA pathways were upregulated in Nipponbare but downregulated in IRGC100896, consistent with the observed senescence and differential patterns in chlorophyll biosynthesis and Rubisco activity.
Transcriptional consequences of BL biosynthetic signatures
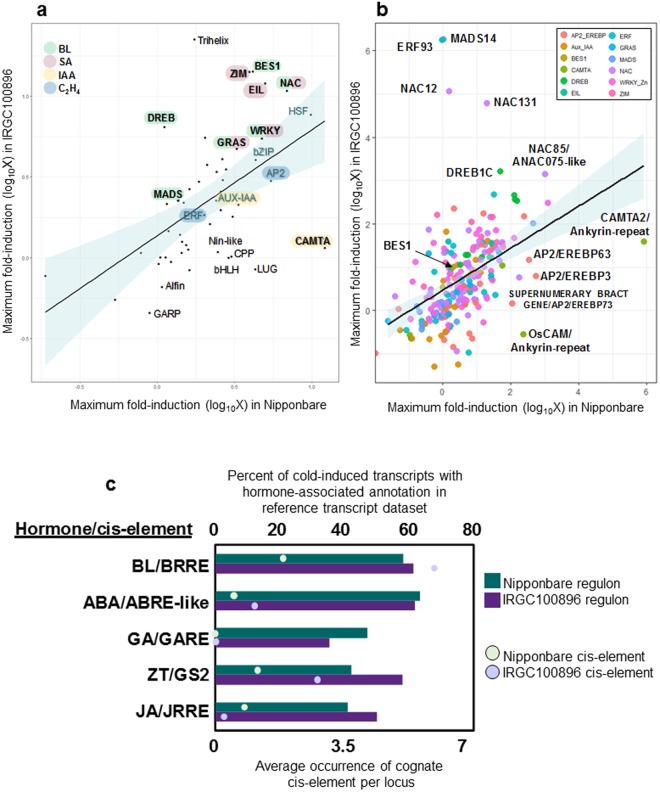
Analysis of the upstream sequences of genes that were cold-responsive in IRGC100896 identified the brassinosteroid response elements (BRRE) among the most significantly enriched motifs (Supplementary Figure S18). To establish a network of genes associated with the enhanced BL biosynthesis in IRGC100896, transcription factors that were cold-upregulated in IRGC100896 but not in Nipponbare were identified. It was assumed that such group of transcription factors are likely to include the direct targets of enhanced BL biosynthesis in IRGC100896, and their transcriptional networks can be reconstructed by capturing other co-expressed genes.
Patterns of transcription factor expression are shown in Fig. 5a and b with expression in Nipponbare in x-axis and IRGC100896 in y-axis (Supplementary Figure S19). Upregulation of bZIP transcription factors associated with ABA-responses is an example of a common signature of IRGC100896 and Nipponbare, mirroring the enhanced expression of ABA biosynthetic genes in both genotypes (Fig. 4; Supplementary Figures S13–S15). On the other hand, transcription factors that were most prominently upregulated only in IRGC100896 include BES1 (Os07g0580500), NAC (Os12g0123700), WRKY (Os05g0322900), and DREB2B (Os02g0752800) (Fig. 5a,b). BES1 is a well-known regulator of BL-mediated transcription, while NAC, WRKY, and DREB2B are known downstream targets of BES1. BL signaling is known to cross-talk with SA signaling through NAC, EIL, WRKY, GRAS, and ZIM transcription factors, all of which were upregulated in IRGC100896. Upregulation of these transcription factors was consistent with parallel upregulation of genes in the BL and SA biosynthetic pathways. Transcription factors associated with IAA (Aux/IAA, CAMTA), and C2H4 (AP2/ERF, bHLH) signaling were upregulated only in Nipponbare, mirroring the general trends in IAA and C2H4 biosynthetic pathways (Fig. 4; Supplementary Figures S13–S15).
Figure 5.
Shared or unique network signatures across IRGC100896 and Nipponbare. (a) Transcription factor families that were similarly or uniquely expressed across IRGC100896 and Nipponbare. BES1, NAC, WRKY, DREB, GRAS and MADS are BL-associated transcription factors that were upregulated in IRGC100896 but not in Nipponbare. WRKY, NAC and ZIM are involved in cross-talks between BL and SA that were upregulated in IRGC100896 but not in Nipponbare. (b) Non-differentially expressed and differentially expressed transcription factors across IRGC100896 and Nipponbare. (c) Cold-upregulated genes associated hormone response in RiceXPro were examined for BRRE (BL), ABRE-like (ABA), GARE (GA), GS2 (ZT), and JRRE (JA) enrichment along the −1,500 to +500 regions. Enrichment of each class of cis-elements within a cluster of co-regulated genes are shown by the colored dots across the bar graph.
To establish a network of BL-regulated genes in IRGC100896, the cold-induced portion of the transcriptome was further searched for genes with BL-associated functional annotation. The enrichment of cis-elements associated with known hormone-responsive genes in Nipponbare was also investigated across each co-expressed cluster (Fig. 5c). While the number of annotated BL-regulated genes that were also induced by cold was very similar in IRGC100896 and Nipponbare, the IRGC100896 cluster was more significantly enriched with brassinosteroid-response elements (BRRE). Co-regulated clusters for ABA, GA, CYT, and JA, which were very similar in composition in IRGC100896 and Nipponbare, also had very similar enrichments of associated cis-elements ABRE, GARE, AuxRE, GS2, JRRE, respectively. These trends suggest that the cold stress genetic networks mediated by ABA, GA, CYT, and JA are shared features of IRGC100896 and Nipponbare, while the genetic networks mediated by BL with BES1 as its core, and its possible interaction with SA signaling represents a unique feature of IRGC100896.
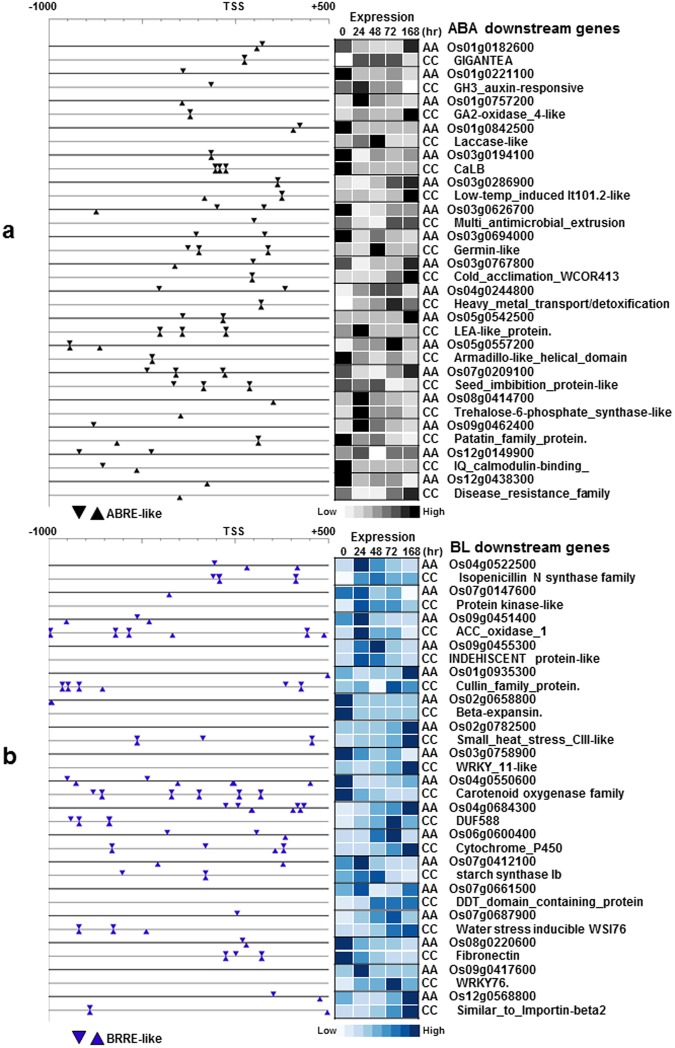
Figure 6 shows the gene-by-gene analysis of cis-element spatial distribution across a subset of co-regulated genes in the ABA network that is common between IRGC100896 and Nipponbare, and BL network that is unique to IRGC100896. Patterns of cis-element enrichment across these contrasting networks were consistent with the trends in Fig. 5c, suggesting that BL-regulated gene expression is an important feature of O. officinalis genetic mechanism that is deemphasized in O. sativa ssp. japonica.
Figure 6.
Distribution of critical cis-element motifs (−1,000 to +500) among genes associated with ABA-network (a) and those associated with BES1 network (b). Cis-element maps show similar enrichment of ABRE-like motifs across the co-regulated clusters in both IRGC100896 and Nipponbare. Cis-element maps for BRRE show an enrichment bias towards the BES1-associated genes in IRGC100896.
Cold stress mediated BES1-network
The BES1-network was assembled to facilitate the interpretation of the biological significance of the major contrasts in the genetic mechanisms of IRGC100896 and Nipponbare. A subset of BRRE-enriched genes that were also upregulated by cold in IRGC100896 was established (Supplementary Figure S20). For each transcript, upstream sequences of their genomic loci were extracted from the unpublished draft of O. officinalis W0002 genome and Nipponbare reference. Final dataset included 140 pairs of orthologous genes across O. officinalis and O. sativa, with either high (8 to 10) or very high (≥10) densities of BRRE/BRRE-like motifs (Supplementary Table S21).
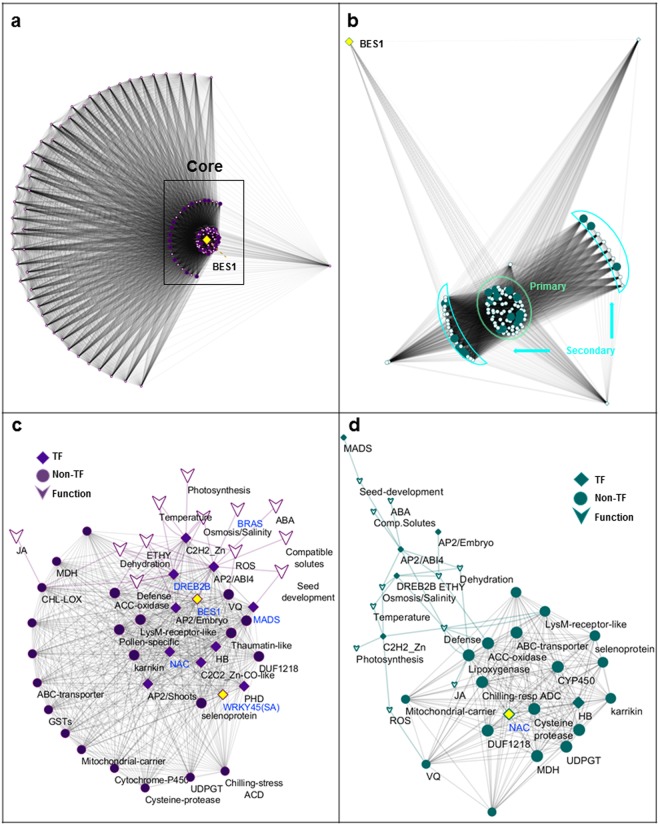
Figure 7 shows the organization of cold-regulated BES1-network in IRGC100896 and Nipponbare, and their putative biochemical and biological functions. The 140-pairs of orthologous genes in the network were all upregulated by cold in IRGC100896 but not necessarily in Nipponbare. In the larger network for IRGC100896 that included all 140 genes (Fig. 7a), BES1 occupies the core, consistent with the fact that all 140 genes were significantly enriched with BRRE. The primary, secondary and tertiary co-expressed genes assembled in an organized fashion around the putative master regulator BES1. In contrast, the larger network consisting of all 140 orthologs in Nipponbare appeared fragmented into three sub-networks that were at best only loosely linked to the central hub BES1 (Fig. 7b). Patterns in the larger network of Nipponbare were consistent with the fact that BRRE was significantly depleted among those 140 genes.
Figure 7.
BES1-networks in IRGC100896 and Nipponbare based on co-expression and cis-element enrichment. Gene ontology enrichments are shown. (a) Larger BES1-network in IRGC100896 consisting of 140 most significantly co-expressed genes. Primary, secondary, and tertiary components are shown with BES1 (yellow diamond) at the core. (b) Larger network in Nipponbare with linkages among 140 genes that were orthologous to the tightly co-expressed genes in IRGC100896. BES1 (yellow diamond) is not co-expressed with the network components, indicating network fragmentation and uncoupling to BL signaling. (c) Primary BES1-regulon in IRGC100896 comprised of 34 core network genes, and their associated biological and biochemical functions. Transcription factors downstream to BES1 (DERB2B, NAC, WRKY45) and associated with SA signaling are shown with direct connection to BL biosynthesis (BRAS). (d) Model of BES1-independent core regulon consisting of 20 genes that were uncoupled to BES1 in Nipponbare. The central hub is NAC transcription factor downstream to BES1 in the IRGC100896 core regulon but uncoupled to BL biosynthesis (BRAS).
Zoomed-in views of IRGC100896 and Nipponbare networks highlighting the core regulons are shown in Fig. 7c,d. In IRGC100896, the core regulon is comprised of a much larger subset of 34 tightly co-expressed genes that are directly connected to and organized around BES1. The significance of this core network is further strengthened by the fact that known BES1-target genes (DREB2B/Os02g0752800, NAC/Os12g0123700, WRKY45/Os05g0322900, and MADS-box/Os02g0170300) were indeed tightly co-expressed with BES1, and captured within its core regulon. These genes are also known regulators of SA signaling, consistent with the observed parallel upregulation of BL and SA biosynthetic pathways.
The biochemical and biological functions associated with the core BES1-regulon in IRGC100896 included brassinosteroid biosynthesis (BRAS), responses to dehydration, salinity and cold, responses to ABA, regulation of embryo maturation, seed dormancy, and germination, radical scavenging and oxidative defenses, osmotic adjustment, transport regulation, responses to pathogens, and photosynthesis (Fig. 7c; Supplementary Table S21). These functions are directly relevant to physiological adjustments and defenses21,22.
The core regulon of Nipponbare was a stark contrast to the core regulon of IRGC100896 for two reasons. First, it was comprised of much fewer genes (total = 20) that formed a relatively dispersed network uncoupled to BES1 but directly linked to NAC transcription factor (Os12g0123700) that is downstream to BES1 in the IRGC100896 network. Second, other transcription factors downstream to BES1 in the IRGC100896 network (DREB2B/Os02g0752800, MADS-box protein/Os02g0170300) were in fact not co-expressed in Nipponbare (Fig. 7d). The putative biochemical and biological functions of the NAC-regulated core regulon of Nipponbare include responses to dehydration, salinity, cold, and ABA, regulation of embryo maturation and seed dormancy, radical scavenging and oxidative defenses, osmotic adjustment, transport regulation, responses to pathogens, and photosynthesis. These are generally similar to the predicted outcomes of the more elaborate BES1-network in IRGC100896.
Previous analysis of the Nipponbare cold stress transcriptional network highlighted the primary roles of oxidative-mediated, ABA-dependent, and DREB-mediated regulons for early defenses20–22. While BRRE was a unique signature of cold-regulated genes in IRGC100896, the enrichment of cis-elements associated with ROS-mediated (Myb2-box, as1/ocs/TGA-like), ABA-dependent (ABRE, ABRE-like, ABRE-ACGT, hex-3), and DREB-mediated (DRE/CRT) regulons were very similar in IRGC100896 and Nipponbare (Fig. 8). These results highlight the importance of BES1-network as a defining feature of O. officinalis cold stress genetic mechanism.
Figure 8.
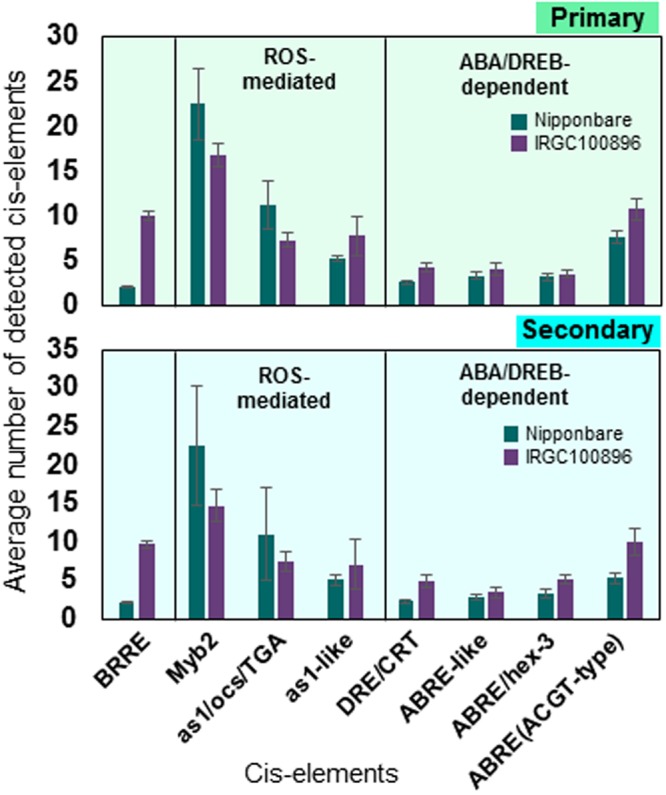
Enrichment of cis-elements associated with ROS-mediated (Myb2-box, as1/ocs/TGA-like), ABA-dependent (ABRE, ABRE-like, ABRE-ACGT, hex-3), and DREB-mediated (DRE/CRT) regulons across all genes in the BES1-network of IRGC100896, and across all genes in BES1-independent network of Nipponbare. Patterns of enrichment for these cis-elements across the members of the primary/core and secondary regulons were very similar in IRGC100896 and Nipponbare.
Discussion
Sequence variation showed that the CC-genome is more similar to AA-genome than to any other genomes in the genus Oryza5,9,23–29. Understanding the implications of such variation in context of regulatory networks is important for harnessing novel physiological traits in breeding. Analysis of habitat and geographic distribution in the genus Oryza as described in bioclimatic models indicated that temperature and moisture constraints are key determinants of species ecological boundaries. O. officinalis was placed in the middle of the spectrum of stress tolerance variation4,24. Improving cold tolerance has always been a major goal of rice breeding, yet there is no clear consensus as to which Oryza species are the most suitable alien genetic donors. The comparability of the cold tolerance potentials of IRGC100896 and Nipponbare20–22, led to two important questions: Which of the two genotypes is a better donor for cold-sensitive tropical indica cultivars? Is cold tolerance in IRGC100896 and Nipponbare due to conserved genetic mechanisms, or is it a similar outcome of distinct genetic mechanisms? It is important to consider a previously established theory that phenotype alone may not always be an adequate indicator of the true genetic potential of a donor in breeding25–28. The possibility exists that the underlying genetic mechanisms may be distinct between two individuals without an obvious phenotypic contrast like in the case of the cold tolerance of O. sativa and O. officinalis. Complementation and epistasis may create novel effects when the positive attributes of each parent are combined by recombination.
This study represents the very first in-depth examination of the cold stress genetic mechanism of O. officinalis, and its uniqueness relative to the mechanism in O. sativa ssp. japonica. Because an annotated CC-genome reference is still underway, the IRGC100896 transcriptome was examined in this study with specific focus on genes with clear orthologs in Nipponbare21,22. Several major trends became apparent. First, transcriptome signatures showed similar trends in primary metabolic pathways and primary defenses such as radical scavenging and osmotic adjustment. In other words, no drastic differences in transcriptome signatures were evident to support major physiological differences between IRGC100896 and Nipponbare. It appeared that the baseline mechanisms for physiological adjustments and defenses are somehow conserved across the two representatives from the AA and CC genome groups.
Second, gene expression associated with most hormone biosynthetic pathways were also not significantly different between IRGC100896 and Nipponbare. The notion that ABA-regulatory mechanisms could fully explain stress tolerance variation across genotypes above and beyond the baseline defenses is questionable17. Similar patterns in ABA biosynthetic genes across IRGC100896 and Nipponbare were mirrored by similar ABA transcriptional networks as defined by the patterns in ABRE enrichment and expression of cognate activators ABFs/bZIP/ABI4. This was also echoed by the conserved patterns for ABA-independent regulon involving the CRT/DRE-DREB and as1/ocs-bZIP/TGA across IRGC100896 and Nipponbare20,21,29. ABA-mediated regulon along with ABA-independent (CRT/DRE-DREB) and oxidative-mediated (as1/ocs-bZIP/TGA) regulons are clearly shared features of the genetic mechanisms of IRGC100896 and Nipponbare.
Third, BL and SA biosynthetic pathways were uniquely upregulated in IRGC100896. BL-mediated network and its possible cross-talk with SA-signaling appeared to be an important feature of O. officinalis mechanism that was extensively fragmented in O. sativa ssp. japonica. BL is primarily involved in vascular bundle development, photomorphogenesis, stem and leaf angle and elongation, tillering, and germination30,31. It is also implicated with the maintenance of photosynthesis through the enhancement of PSII efficiency, intercellular CO2 concentration, stomatal regulation, antioxidant defense system, and osmotic adjustment32–35.
Perception of BL by BRI1 protein leads to the accumulation of dephosphorylated BES1 transcription factor in the nucleus, and consequent transcriptional changes facilitated by BES1 binding to BRRE or E-box elements in target genes36–38. BES1 activates DREB, WRKY, NAC, and GRAS transcription factors that control different sub-regulons implicated with stomatal regulation, antioxidant defense, osmotic adjustment, intracellular CO2 maintenance, and photosynthetic efficiency39–42. In IRGC100896 but not in Nipponbare, BES1-network genes were tightly co-upregulated with other major regulators known to be downstream to BES1 (i.e., DREB, WRKY, NAC, GRAS). These genes have important roles in maintaining photosynthesis, radical scavenging, and osmotic adjustment.
Downstream targets of SA-mediated defenses in rice include OsNPR1 and its interacting partners OsWRKY45 and OsTGA2/5/6, all of which were prominent in the BL-mediated network of IRGC100896. BL modulates these transcription factors through an unknown regulator, while C2H4 has been suggested to antagonize the effects of SA and BL on these transcription factors43,44. These connections further strengthen the biological significance of the coupling of BL and SA biosynthesis in IRGC100896.
Earlier studies in Nipponbare showed that the mechanisms associated with early defenses to cold involve three sequentially expressing regulons17,20,21,45. First is the oxidative-mediated regulon involving bZIP-TGA-type transcription factors binding to the as1/ocs-like cis-elements. Second is ABA-dependent regulon that involves ABF, bZIP or Myb transcription factors, controlling their target genes through the ABRE cis-elements. Third is ABA-independent regulon that involves DREB/CBF transcription factors, controlling their target genes through the CRT/DRE cis-elements. These regulons contribute cumulatively to physiological adjustments and defenses through the enhancement of radical scavenging, repair of oxidative injuries, stomatal regulation, and osmotic adjustment. In IRGC100896, these processes appeared to be controlled by BES1 through a different set of regulators and effectors.
Analysis of the larger BES1 network (140 genes) indicated its extensive overlap with oxidative-mediated, ABA-dependent, and DREB/CBF-mediated regulons, all of which were active in both IRG1008096 and Nipponbare. This implies that in IRGC100896, the mechanisms for radical defense, stomatal regulation and osmotic adjustment are likely due to the synergy of all three regulatory pathways. In Nipponbare, similar physiological mechanisms appeared to be due mainly to oxidative-mediated, ABA-dependent, and DREB/CBF-mediated networks without BES1 mechanism, hence BES1 may be providing an extra layer of control for integrating defenses with growth in O. officinalis. This novel fine-tuning mechanism appeared fragmented in O. sativa ssp. japonica, perhaps as a consequence of domestication. BES1-network may also be an offshoot of a prominent role of BL in regulating growth under extreme environments where O. officinalis is well adapted.
Consistent with the comparable cold tolerance of O. officinalis and O. sativa ssp. japonica, the dominant functional signatures (protection of photosynthesis, maintenance of intracellular CO2, oxidative defenses, osmotic adjustment) implied by the transcriptome data were also remarkably similar across the two species. While the oxidative-mediated, ABA-dependent, and DREB-mediated regulons appeared to be generally conserved, O. officinalis has a functional BES1 network for fine-tuning the integration of growth and stress-related responses. The fact that O. officinalis has an added regulatory feature that no longer exist in cultivars (i.e., BES1) reiterates its enormous value for expanding the genetic base of cultivars by wide hybridization.
In this study, we were also inspired by common observations that transgressive segregants are quite common in the progenies of crosses between cultivars and certain wild Oryza. These observations suggested that physiological complementation is possible between genetically distant genotypes although they may not exhibit clear phenotypic contrasts. We pursued our studies under the assumption that while O. officinalis and O. sativa ssp. japonica had similar cold tolerance, each may confer distinct molecular mechanisms with potential for positive complementation46. Indeed, the BES1-network that appeared to have been lost in cultivars provide a relevant proof of concept to test such hypothesis.
Methods
Plant materials, stress experiments, and RNA sampling
To substantiate the stress tolerance classification of Oryza species based on the bioclimatic models of Atwell et al.4, comparative physiological studies under cold stress were performed across a panel of accessions that included O. sativa ssp. japonica (cv. Nipponbare) as reference for AA-genome, O. officinalis IRGC100896 as reference for CC-genome, mega-variety IR64 (O. sativa ssp. indica) as sensitive check, and other representative diploid and tetraploid wild species (O. rufipogon/IRGC106424, O. longistaminata/IRGC110404, punctata/IRGC105690, O. rhizomatis/IRGC105442, O. australiensis/IRGC100882, O. brachyantha/IRGC101232, O. eichingeri/IRGC101424). Plants were established under control conditions at IRRI phytotron (25 °C to 28 °C day/22 °C to 24 °C night; 14 hr photoperiod) from germination to four-leaf stage (V4) in Yoshida hydroponics medium18. Cold stress experiments were performed in growth chamber at 4 °C day/night, 14 hr photoperiod, with tissue sampling for RNA extraction and physiological analysis at 24, 48, 72, and 168 hr. Electrolyte leakage analysis was performed after 48 hr of cold stress22,47. Post-stress plant recovery was assessed after 15 days at control conditions with three replicates. Plant recovery was assessed based on IRRI’s Standard Evaluation Score (SES) in a scale of 0 to 10 representing death to 100% recovery.
RNA-Seq library construction and sequencing
Total RNA was extracted from frozen leaf tissues with the mirVana RNA kit (Ambion-ThermoFisher Scientific, USA). Samples from each time-point (0, 24, 48, 72,168 hr at 4 °C) were used to construct the time-course RNA-Seq libraries at 150 bp paired-end using the Illumina TruSeq RNA Library Prep kit V2 with three biological replicates (Illumina Inc., USA). Indexed libraries were sequenced across ten lanes on Illumina HiSeq2500, with sequencing coverage of ~90x for Nipponbare libraries, and ~60x for IRGC100896/W0065 libraries, each with ten technical replicates.
Transcriptome data processing, assembly, and analysis
Sequence output from indexed RNA-Seq libraries were preprocessed with Cutadapt48, and mapped against the Nipponbare IRGSP1.0 reference for AA-genome (http://rapdb.dna.affrc.go.jp/download/irgsp1.html), GFF gene models, and O. officinalis-W0002 reference for CC-genome by TopHat2 and Cufflinks49,50. The draft assembly of W0002 genome (unpublished) was provided by Dr. Nori Kurata, National Institute of Genetics, Japan. Merging of sequence assemblies and test for statistical significance of mapped RNA-Seq reads were performed with Cuffdiff with default parameters (p-value = 0.05, FDR 5%).
The degree of orthologous sequence conservation is well established in the genus Oryza5. Within-genus orthologous transcripts (japonica versus officinalis) were established by searching the W0002 draft for open reading frames and using them to establish gene models by Augustus (3.2.3)51. Using blat at default parameters, orthologous transcripts established from officinalis were aligned to Nipponbare IRGSP1.0 gene models. The longest alignment was determined for each O. officinalis locus and final transcript sequences were annotated according to homologous loci in Nipponbare52. In cases where multiple O. officinalis loci corresponded to a single locus in Nipponbare, loci with the most stable mapping was used. Establishment of orthologous gene pairs for subsequent genome-wide comparison of gene expression and cis-element analysis were guided by established computational logic5,53.
Analysis of differentially expressed genes and K-means clustering
For transcription factor transcripts, maximum fold-induction were calculated based on cumulative expression for each family in order to assess saturation. Gene-by-gene expression analysis was performed to identify the specific transcription factor transcripts with significant changes in expression in either or both IRGC100896 and Nipponbare. Analysis of expression of non-transcription factor transcripts was performed by first identifying the genes according to functional annotation using the RiceXPro database54. Threshold for differential expression was set at >2 (log2 scale) and p-value <0.01. Patterns of temporal co-expression was established across IRGC100896 and Nipponbare transcriptomes by K-means clustering in R-package MBCluster version 3.3.3 using the negative binominal with eight total clusters55.
Metabolic and hormone biosynthetic pathway mapping
Transcripts relevant to metabolic and hormone biosynthetic pathways were first identified from transcriptome datasets and their abundances were compared across the two genotypes. Transcripts were mapped to the reference metabolic pathways using the KaPPA-View analysis for glycolysis, TCA cycle, starch synthesis and catabolism, triacylglyceride synthesis and catabolism, photosynthesis (chlorophyll biosynthesis, Calvin cycle), ubiquinone synthesis, ROS scavenging (catalase, peroxidase, superoxide dismutase, GSH synthesis), osmotic adjustment (trehalose, sucrose, and proline) and hormone biosynthesis (http://kpv.kazusa.or.jp)56.
Cis-element analysis
Genomic sequences corresponding to −3,000 to +2,000 regions were extracted for all loci with mapped transcripts in both W0002 and Nipponbare. These sequences were scanned for motifs corresponding to known and/or putative cis-elements by MAMA on CUDA version 7.557. Of all the motifs occurring along the −3,000 to +2,000 region, those that were specifically enriched around −500 region or −150 to +50 interval only among upregulated genes were given high MAMA scores. High MAMA-scoring motifs were used to generate spatial maps of −1,200 to +500 regions of all candidate genes for biological hypothesis testing. Cis-element spatial maps were visualized using the Matlab version R2017a (http://www.mathworks.com). Baseline annotation of all putative cis-elements in PLACE database58 was further elaborated with additional information from the literature29,45.
Transcriptional network modeling
The dataset used for modeling the brassinosteroid (BL) network in IRGC100896 included a subset of tightly co-expressed genes with either high (8 to 10 copies) or very high (at least 11 copies) density of sequence motifs for brassinosteroid response element (BRRE) across the −1,200 to +500 regions. Nipponbare loci orthologous to the BRRE-enriched and co-regulated genes in IRGC100896 were used to establish the corresponding networks for O. sativa ssp. japonica. Models of co-expression networks were visualized using Cytoscape version 3.5.1 (http://www.cytoscape.org/59. In all network models, length of the edge reflects the strength of co-expression.
Electronic supplementary material
Acknowledgements
This research was supported by the National Science Foundation (NSF), Plant Genome Research Program (Grant 1602494), Bayer Crop Science Endowed Professorship to BGDR, and NIG-JOINT Research Fund (16B2012, 47B2014) from the National Institute of Genetics of Japan to NK and BGDR. All Oryza accessions were obtained from the Wide Hybridization Program of IRRI. We acknowledge the assistance of Joie Ramos, Junrey Amas, and Boyet Manalaysay for all experiments performed at IRRI. The O. officinalis accession W0002 for the CC-genome draft was obtained from the National Institute of Genetics through the National Bioresource Project of Japan. High-throughput computing was performed at the DNA Database of Japan (DDBJ), ROIS-NIG.
Author Contributions
A.K. performed all transcriptome experiments, processed and analyzed all transcriptomic and genomic datasets, performed the KaPPA-View analysis and network modeling, and co-wrote the manuscript with B.G.d.l.R. I.C.M.P. assisted in the analysis of transcriptomic datasets and KaPPA-View pathway mapping, and helped write the manuscript. H.O. and N.K. assisted in RNA-Seq experiments, reference-guided assembly and analysis against the Nipponbare reference and officinalis W0002 draft genome. M.S., M.F., Y.N. and Y.K. assisted in all aspects of data analysis that made use of the W0002 draft genome. B.O. and Y.K. contributed to the analysis of cis-elements, assisted in network modeling, and provided technical assistance in programming. D.S.B. determined the representative Oryza accessions for this study and helped design the stress physiological experiments. B.G.d.l.R. conceptualized the project, designed all physiological experiments, integrated and interpreted all datasets, and wrote the entire manuscript.
Data Availability and Other Supplementary Information
The RNA-Seq data described in this manuscript are publicly available as DDBJ accession DRA006704. Supplementary data files 1–21 accompany this paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-34608-z.
References
- 1.Sang T, Ge S. Understanding rice domestication and implications for cultivar improvement. Curr. Opin. Plant Biol. 2013;16:139–46. doi: 10.1016/j.pbi.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Xu X, et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2012;30:105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- 3.Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–8. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atwell BJ, Wang H, Scafaro AP. Could abiotic stress tolerance in wild relatives of rice be used to improve Oryza sativa? Plant Sci. 2014;215–216:48–58. doi: 10.1016/j.plantsci.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Ammiraju JSS, et al. Dynamic evolution of Oryza genomes is revealed by comparative genomic analysis of a genus-wide vertical data set. Plant Cell. 2008;20:3191–209. doi: 10.1105/tpc.108.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurata, N. Chromosome and Genome Evolution in Rice, In Rice Biology in the Genomics Era. (eds Hirano, H.-Y., Hirai, A., Sano, Y. & Sasaki, T.) 62, 235–245 (2008).
- 7.Wang B, et al. Polyploid evolution in Oryza officinalis complex of the genus Oryza. BMC Evol. Biol. 2009;9:250. doi: 10.1186/1471-2148-9-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirabayashi H, et al. qEMF3, a novel QTL for the early-morning flowering trait from wild rice, Oryza officinalis, to mitigate heat stress damage at flowering in rice, O. sativa. J. Exp. Bot. 2015;66:1227–36. doi: 10.1093/jxb/eru474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu R, et al. Drought-tolerant rice germplasm developed from an Oryza officinalis transformation-competent artificial chromosome clone. Genet. Mol. Res. 2015;14:13667–78. doi: 10.4238/2015.October.28.29. [DOI] [PubMed] [Google Scholar]
- 10.Multani DS, Khush GS. delos Reyes, B. G. & Brar, D. S. Alien gene introgression and development of monosomic alien addition lines from Oryza latifolia Desv. to rice, Oryza sativa L. Theor. Appl. Genet. 2003;107:395–405. doi: 10.1007/s00122-003-1214-3. [DOI] [PubMed] [Google Scholar]
- 11.Khush G. S., Brar D. S. Distant Hybridization of Crop Plants. Berlin, Heidelberg: Springer Berlin Heidelberg; 1992. Overcoming the Barriers in Hybridization; pp. 47–61. [Google Scholar]
- 12.Shin Y-B, Katayama T. Cytogenetical studies on the genus. Oryza. Japanese J. Genet. 1979;54:1–10. doi: 10.1266/jjg.54.1. [DOI] [Google Scholar]
- 13.Bao W, et al. Diversity of centromeric repeats in two closely related wild rice species, Oryza officinalis and Oryza rhizomatis. Mol. Genet. Genomics. 2006;275:421–30. doi: 10.1007/s00438-006-0103-2. [DOI] [PubMed] [Google Scholar]
- 14.Jin H, et al. Molecular and cytogenetic characterization of an Oryza officinalis-O. sativa chromosome-4 addition line and its progenies. Plant Mol. Biol. 2006;62:769–77. doi: 10.1007/s11103-006-9056-4. [DOI] [PubMed] [Google Scholar]
- 15.Amante-Bordeos, A. et al. Transfer of bacterial blight and blast resistance from the tetraploid wild rice Oryza minuta to cultivated rice. Oryza sativa. Theor. Appl. Genet.84–84, 345–354 (1992). [DOI] [PubMed]
- 16.Brar DS, Khush GS. Alien introgression in rice. Plant Mol. Biol. 1997;35:35–47. doi: 10.1023/A:1005825519998. [DOI] [PubMed] [Google Scholar]
- 17.de los Reyes, B. G. et al. Cold and water deficit regulatory mechanisms in rice: Optimizing stress tolerance potential by pathway integration and netowrk engineering, In Rice Genomics, Genetics and Breeding (Sasaki T. & Ashikari M. eds), Springer-Nature Singapore, pp. 317–359 (2018).
- 18.de los Reyes BG, et al. Phenotypic, physiological, and molecular evaluation of rice chilling stress response at the vegetative stage. Methods Mol. Biol. 2013;956:227–41. doi: 10.1007/978-1-62703-194-3_16. [DOI] [PubMed] [Google Scholar]
- 19.Kurata N, Yamazaki Y. Oryzabase. An integrated biological and genome information database for rice. Plant Physiol. 2006;140:12–7. doi: 10.1104/pp.105.063008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park M-R, et al. Supra-optimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development. Plant. Cell Environ. 2010;33:2209–30. doi: 10.1111/j.1365-3040.2010.02221.x. [DOI] [PubMed] [Google Scholar]
- 21.Yun Kil-Young, Park Myoung Ryoul, Mohanty Bijayalaxmi, Herath Venura, Xu Fuyu, Mauleon Ramil, Wijaya Edward, Bajic Vladimir B, Bruskiewich Richard, de los Reyes Benildo G. Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biology. 2010;10(1):16. doi: 10.1186/1471-2229-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng C, et al. An early response regulatory cluster induced by low temperature and hydrogen peroxide in seedlings of chilling-tolerant japonica rice. BMC Genomics. 2007;8:175. doi: 10.1186/1471-2164-8-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zou Xin-Hui, Zhang Fu-Min, Zhang Jian-Guo, Zang Li-Li, Tang Liang, Wang Jun, Sang Tao, Ge Song. Analysis of 142 genes resolves the rapid diversification of the rice genus. Genome Biology. 2008;9(3):R49. doi: 10.1186/gb-2008-9-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vaughan DA, Morishima H, Kadowaki K. Diversity in the Oryza genus. Curr. Opin. Plant Biol. 2003;6:139–46. doi: 10.1016/S1369-5266(03)00009-8. [DOI] [PubMed] [Google Scholar]
- 25.Sakai H, Itoh T. Massive gene losses in Asian cultivated rice unveiled by comparative genome analysis. BMC Genomics. 2010;11:121. doi: 10.1186/1471-2164-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ammiraju JSS, et al. Evolutionary dynamics of an ancient retrotransposon family provides insights into evolution of genome size in the genus Oryza. Plant J. 2007;52:342–351. doi: 10.1111/j.1365-313X.2007.03242.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang S, et al. New insights into Oryza genome evolution: high gene colinearity and differential retrotransposon amplification. Plant Mol. Biol. 2007;64:589–600. doi: 10.1007/s11103-007-9178-3. [DOI] [PubMed] [Google Scholar]
- 28.Tanksley SD, McCouch SR. Seed banks and molecular maps: unlocking genetic potential from the wild. Science (80-.). 1997;277:1063–6. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- 29.Xu F, et al. Cis-regulatory signatures of orthologous stress-associated bZIP transcription factors from rice, sorghum and Arabidopsis based on phylogenetic footprints. BMC Genomics. 2012;13:497. doi: 10.1186/1471-2164-13-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun S, et al. Brassinosteroid signaling regulates leaf erectness in Oryza sativa via the control of a specific U-type cyclin and cell proliferation. Dev. Cell. 2015;34:220–8. doi: 10.1016/j.devcel.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 31.Zhang C, Bai M-Y, Chong K. Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 2014;33:683–96. doi: 10.1007/s00299-014-1578-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharma I, Kaur N, Pati PK. Brassinosteroids: A promising option in deciphering remedial strategies for abiotic stress tolerance in rice. Front. Plant Sci. 2017;8:2151. doi: 10.3389/fpls.2017.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng H, Tang Q, Hua X. Arabidopsis brassinosteroid mutants det2-1 and bin2-1 display altered salt tolerance. J. Plant Growth Regul. 2010;29:44–52. doi: 10.1007/s00344-009-9111-x. [DOI] [Google Scholar]
- 34.Xia X-J, et al. Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol. 2009;150:801–14. doi: 10.1104/pp.109.138230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia XJ, et al. Pesticides-induced depression of photosynthesis was alleviated by 24-epibrassinolide pretreatment in Cucumis sativus L. Pestic. Biochem. Physiol. 2006;86:42–48. doi: 10.1016/j.pestbp.2006.01.005. [DOI] [Google Scholar]
- 36.Yu X, et al. A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 2011;65:634–46. doi: 10.1111/j.1365-313X.2010.04449.x. [DOI] [PubMed] [Google Scholar]
- 37.Sun Y, et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell. 2010;19:765–77. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin Y, et al. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–59. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 39.Chen, X. et al. OsNAC2 encoding a NAC transcription factor that affects plant height through mediating the gibberellic acid pathway in rice. 1, 302–314 (2015). [DOI] [PubMed]
- 40.Chen L, et al. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta. 2012;1819:120–8. doi: 10.1016/j.bbagrm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 41.Tong H, et al. DWARF and LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009;58:803–16. doi: 10.1111/j.1365-313X.2009.03825.x. [DOI] [PubMed] [Google Scholar]
- 42.Wang Q, et al. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol. Biol. 2008;67:589–602. doi: 10.1007/s11103-008-9340-6. [DOI] [PubMed] [Google Scholar]
- 43.Huot B, Yao J, Montgomery BL, He SY. Growth-defense tradeoffs in plants: A balancing act to optimize fitness. Mol. Plant. 2014;7:1267–1287. doi: 10.1093/mp/ssu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Vleesschauwer D, et al. Brassinosteroids antagonize gibberellin- and salicylate-mediated root immunity in rice. Plant Physiol. 2012;158:1833–1846. doi: 10.1104/pp.112.193672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de los Reyes BG, Mohanty B, Yun SJ, Park M-R, Lee D-Y. Upstream regulatory architecture of rice genes: summarizing the baseline towards genus-wide comparative analysis of regulatory networks and allele mining. Rice. 2015;8:14. doi: 10.1186/s12284-015-0041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCouch SR, et al. Through the genetic bottleneck: O. rufipogon as a source of trait-enhancing alleles for O. sativa. Euphytica. 2007;154:317–339. doi: 10.1007/s10681-006-9210-8. [DOI] [Google Scholar]
- 47.Ballou SM, Yun K-Y, Cheng C. & de los Reyes, B. G. Cold sensitivity gradient in tuber-bearing Solanum based on physiological and transcript profiles. Crop Sci. 2007;47:2027. doi: 10.2135/cropsci2007.01.0039sc. [DOI] [Google Scholar]
- 48.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 49.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12:R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stanke M, Schöffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–64. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y, et al. Differentially regulated orthologs in sorghum and the subgenomes of maize. Plant Cell. 2017;29:1938–1951. doi: 10.1105/tpc.17.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sato Y, et al. RiceXPro version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013;41:D1206–13. doi: 10.1093/nar/gks1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Si Y, Liu P, Li P, Brutnell TP. Model-based clustering for RNA-seq data. Bioinformatics. 2014;30:197–205. doi: 10.1093/bioinformatics/btt632. [DOI] [PubMed] [Google Scholar]
- 56.Tokimatsu T, et al. KaPPA-view: a web-based analysis tool for integration of transcript and metabolite data on plant metabolic pathway maps. Plant Physiol. 2005;138:1289–300. doi: 10.1104/pp.105.060525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kakei Y, et al. Development of a novel prediction method of cis-elements to hypothesize collaborative functions of cis-element pairs in iron-deficient rice. Rice. 2013;6:22. doi: 10.1186/1939-8433-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higo K. PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 1998;26:358–359. doi: 10.1093/nar/26.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinformatics. 2014;47:8.13.1–8.13.24. doi: 10.1002/0471250953.bi0813s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq data described in this manuscript are publicly available as DDBJ accession DRA006704. Supplementary data files 1–21 accompany this paper.