Abstract
The razor clam genus Novaculina represents an example of a marine-derived, secondary freshwater group. It was thought to comprise three species: N. gangetica (Ganges and smaller basins in Bangladesh and northwestern Myanmar), N. siamensis (Bang Pakong and Pasak rivers in Thailand and Mekong River in Vietnam), and N. chinensis (lower Yangtze River, China). Here we describe Novaculina myanmarensis sp. nov., an additional species from the Ayeyarwady and Salween basins representing a divergent lineage that appears to be sister to N. gangetica. This new record closes a Novaculina range disjunction between northwestern Myanmar and Thailand. The populations of this novel species share a shallow molecular divergence from each other indicating potential dispersal events between the two distant freshwater basins during the Late Pleistocene. Our ancestral area modeling suggests that the MRCA of Novaculina crown group was a salt-tolerant freshwater species. The recent Novaculina species most likely originated via allopatric speciation. Our findings highlight that generalist estuarine species could have played the role as a source for bivalve expansions into freshwater and that western Indochina is a separate biogeographic subregion, which is clearly distinct from India. A new synonymy is proposed as follows: Pharellinae Stoliczka, 1870 = Novaculininae Ghosh, 1920 syn. nov.
Introduction
Freshwater bivalves are a taxonomically diverse ecological group, which includes representatives of at least 19 families1,2. Unionida is the only strictly freshwater order among Bivalvia representing a monophyletic entity with six families, i.e. Unionidae, Margaritiferidae, Etheriidae, Iridinidae, and Mulleriidae2,3. However, several other orders have small to large radiations in freshwater, e.g. Venerida, which includes families such as Cyrenidae, Dreissenidae, Sphaeriidae, and Pharidae1.
Pharidae is a primary marine family4, but it contains a single typically freshwater genus, Novaculina that was thought to include three species: N. gangetica, N. siamensis, and N. chinensis. This genus belongs to the subfamily Novaculininae, which also comprises a second genus with two species, Sinonovacula constricta5 and S. mollis. Annandale6 suggested that Novaculina is a relict marine-derived freshwater lineage, and this hypothesis has recently been supported by multi-locus phylogenetic analyses4.
Novaculina gangetica was considered an endemic species of the Ganges River system in India and Bangladesh7,8, but it was recently discovered in the Kaladan and Lemro rivers in northwestern Myanmar4. Novaculina siamensis was known from the Bang Pakong and Pasak rivers in Thailand9,10, but Sayenko et al.11 found this species in the Mekong Delta in Vietnam. Finally, Novaculina chinensis was described from the Taihu Lake, a large floodplain water body in the lower Yangtze River basin12, and later reported from two additional lakes and a river in the same region13–15.
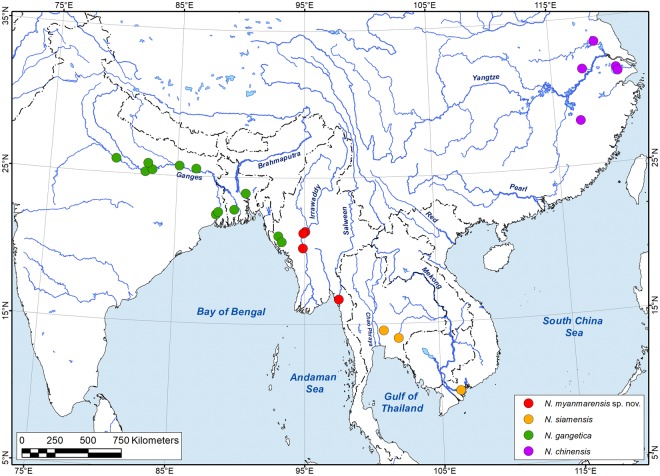
A large Novaculina range disjunction was situated in central and eastern Myanmar (Ayeyarwady and Salween river basins) (Fig. 1). However, we have discovered an additional species in this genus during a recent field trip to Myanmar. The present study aims to describe a new species, Novaculina myanmarensis sp. nov., to provide a brief taxonomic overview of all Novaculina species, and to discuss the putative origin of freshwater lineages in estuarine bivalves within a broad phylogenetic and biogeographic context.
Figure 1.
Distribution range of the genus Novaculina Benson, 1830 based on available georeferenced records (Supplementary Table 2). The map was created using ESRI ArcGIS 10 software (www.esri.com/arcgis); the topographic base of the map was created with Natural Earth Free Vector and Raster Map Data (www.naturalearthdata.com) and Global Self-consistent Hierarchical High-resolution Geography, GSHHG (http://www.soest.hawaii.edu/wessel/gshhg/). (Map: Mikhail Yu. Gofarov).
Results
Multi-locus phylogeny of the Pharidae
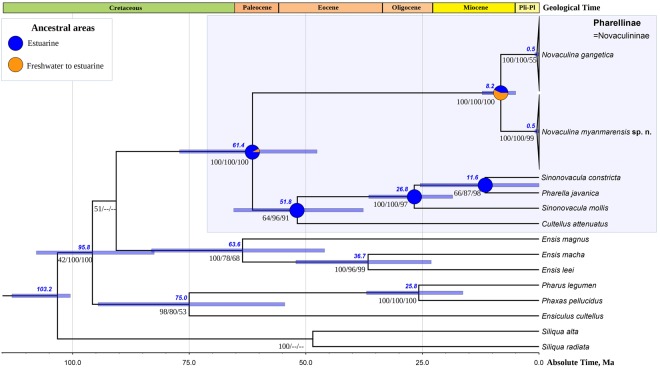
Our multi-locus phylogeny (five partitions: three codons of COI, 16S rRNA, and 28S rRNA) indicates that Novaculina myanmarensis sp. nov. and N. gangetica represent phylogenetically distant lineages belonging to a separate, fully-supported subclade (Fig. 2). The mean p-distances (±standard error estimates) between the new species and Novaculina gangetica are as follows: COI = 8.0 ± 1.0%, and 16S rRNA = 1.9 ± 0.6%. There is a single nucleotide substitution in the nuclear 28S rRNA gene between these species. The Novaculina subclade is fully supported by both Bayesian and maximum likelihood models, and it appears to be closely related to another Pharidae subclade, which includes representatives of Sinonovacula, Pharella javanica, and Cultellus attenuatus. Pharella javanica belongs to the Sinonovacula subclade, and this pattern is strongly supported by our models, indicating the synonymy of Pharellinae Stoliczka, 1870 and Novaculininae Ghosh, 1920.
Figure 2.
Fossil-calibrated chronogram of the Pharidae computed under a lognormal relaxed clock model and a Yule process speciation implemented in BEAST 1.8.4 and obtained for the complete data set of mitochondrial and nuclear sequences (five partitions: three codons of COI + 16S rRNA + 28S rRNA). Bars indicate 95% confidence intervals of the estimated divergence times between lineages (Ma). Black numbers near nodes are BPP values of BEAST model/BPP values of MrBayes model/BS of RAxML model. Blue numbers near nodes are mean ages (Ma). Stratigraphic chart according to the International Commission on Stratigraphy, 2018. The node pies indicate ancestral area reconstructions (probability of each area combination) in accordance with the combined biogeographic model (combination of the S-DIVA + DEC + S-DEC models). Age values for weakly supported nodes are not shown.
Divergence times
Our fossil-calibrated phylogeny suggests that the crown group of the Pharidae has been originated in the mid-Cretaceous (mean age = 103 Ma, 95% HPD 100–113 Ma) (Fig. 2). The Pharellinae (=Novaculininae) clade most likely originated in the Paleocene (mean age = 61 Ma, 95% HPD 48–77 Ma). The origin of the Novaculina crown group placed in the Miocene (mean age = 8 Ma, 95% HPD 5–12 Ma). Finally, the crown group of Pharella + Sinonovacula clade most likely originated in the Oligocene (mean age = 27 Ma, 95% HPD 19–36 Ma).
Ancestral areas
The ancestral area modeling indicates that the most recent common ancestor (MRCA) of the Pharellinae (=Novaculininae) clade was an estuarine species (probability 92.1% by integrative model, 100% by S-DIVA model, 87.5% by DEC model, and 88.8% by S-DEC model). The MRCA of the Novaculina crown group was most likely a salt-tolerant freshwater species like its recent descendants (probability 56.9% by integrative model, 85.0% by DEC, and 85.6% by S-DEC model), although S-DIVA model assumes that it might be an estuarine species (probability 100%).
Phylogeography
A unique COI haplotype of Novaculina myanmarensis sp. nov. has been found in the Ayeyarwady River, and four unique COI haplotypes were recorded in the Salween Basin (Table 1). The mean COI p-distance (±standard error estimates) between these groups is 0.3 ± 0.1%. Almost all specimens from both rivers share a single 16S rRNA haplotype, with exception of a specimen from the Salween Basin having another haplotype with a single nucleotide substitution (239 G). The 28S rRNA sequences were identical among the samples.
Table 1.
List of Novaculina (Bivalvia: Pharidae) sequences used in this study.
| Species | Locality | Voucher no. | COI haplotype code | Acc. numbers of reference sequences | Reference | ||
|---|---|---|---|---|---|---|---|
| COI | 16S rRNA | 28S rRNA | |||||
| N. gangetica Benson, 1830 | Myanmar: Lemro River | biv_150_1 | L1 | MF958986 | MF958997 | MF959011 | 4 |
| biv_150_2 | L2 | MF958987 | MF958998 | MF959012 | 4 | ||
| biv_150_3 | K1 | MF958988 | MF958999 | MF959013 | 4 | ||
| Myanmar: Kaladan River | biv_151_1 | K1 | MF958989 | MF959000 | MF959014 | 4 | |
| biv_151_2 | K2 | MF958990 | MF959001 | MF959015 | 4 | ||
| biv_151_3 | K3 | MF958991 | MF959002 | MF959016 | 4 | ||
| N. myanmarensis sp. nov. | Myanmar: Donthami River | biv_369_1 | D1 | MH670876 | MH670886 | MH664920 | This study |
| biv_369_2 | D2 | MH670877 | MH670887 | MH664921 | This study | ||
| biv_369_3 | D3 | MH670878 | MH670888 | MH664922 | This study | ||
| biv_369_4 | D4 | MH670879 | MH670889 | MH664923 | This study | ||
| biv_369_5 | D3 | MH670880 | MH670890 | MH664924 | This study | ||
| Myanmar: Ayeyarwady River | biv_420_1 | A1 | MH670881 | MH670891 | MH664925 | This study | |
| biv_420_3 | A1 | MH670882 | MH670892 | MH664926 | This study | ||
| biv_420_4 | A1 | MH670883 | MH670893 | MH664927 | This study | ||
| biv_420_5 | A1 | MH670884 | MH670894 | MH664928 | This study | ||
| biv_420_6 | A1 | MH670885 | MH670895 | MH664929 | This study | ||
Taxonomy
Family Pharidae H. Adams & A. Adams, 1856
Subfamily Pharellinae Stoliczka, 1870
Type genus: Pharella Gray, 1854
= Novaculininae Ghosh, 1920 syn. nov.
Type genus: Novaculina Benson, 1830
Genus Novaculina Benson, 1830
Type species: Novaculina gangetica Benson, 1830 (by monotypy).
Novaculina myanmarensis sp. nov
Figures 3A,B, 4A, 5 and 6A, Tables 1 and 2.
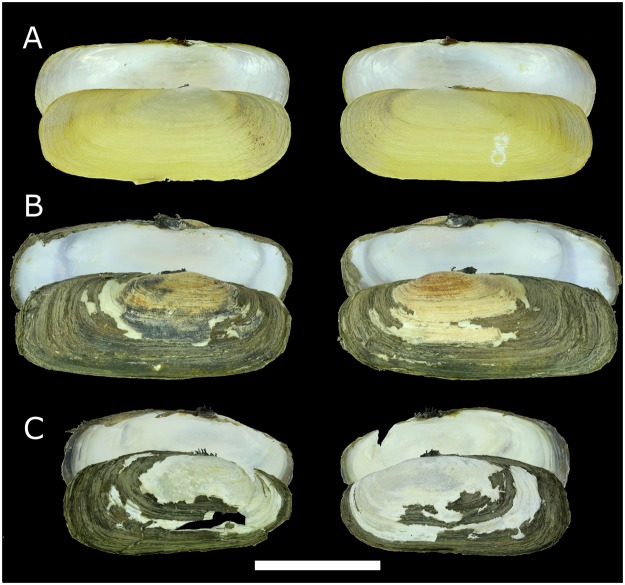
Figure 3.
Shells of Novaculina spp. (A) N. myanmarensis sp. nov., holotype RMBH no. biv0420_8, Ayeyarwady River near Thin Baw Kone village, Pakokku Region, Myanmar. (B) N. myanmarensis sp. nov., paratype RMBH no. biv0369_3, Donthami River, Salween River basin, Myanmar. (C) N. gangetica, RMBH no. biv0150_24, Lemro River, Myanmar. Scale bar = 2 cm. (Photos: Ekaterina S. Konopleva).
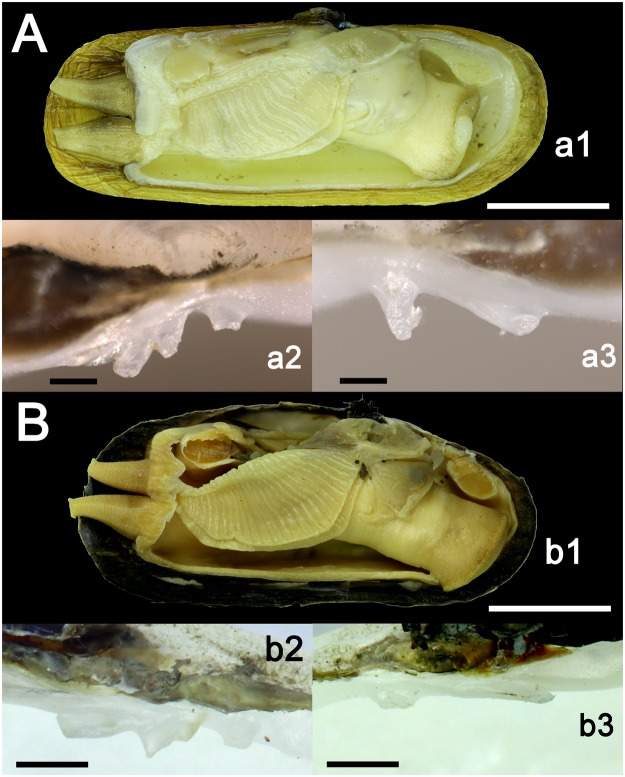
Figure 4.
Soft body morphology (right valve and corresponding mantle tissue were removed) and hinge structure of Novaculina spp. (A) N. myanmarensis sp. nov. (holotype RMBH biv0420_8), including (a1) soft body (scale bar = 10 mm), (a2) pseudocardinal teeth on the left valve, and (a3) pseudocardinal teeth on the right valve (scale bars = 0.4 mm). (B) N. gangetica (RMBH biv0150_24), including (b1) soft body (scale bar = 10 mm), (b2) pseudocardinal teeth on the left valve, and (b3) pseudocardinal teeth on the right valve (scale bars = 1 mm). (Photos: Ekaterina S. Konopleva and Ilya V. Vikhrev).
Figure 5.
Type locality and habitats of Novaculina myanmarensis sp. nov. (A) Habitat in the downstream of the Donthami River, 16.6935°N, 97.5819°E. (B) Type locality: the middle section of the Ayeyarwady River near Thin Baw Kone village (Pakokku Region), 21.3146°N, 95.0591°E. (C) Сlay bottom substrate with clam burrows, the type locality. (D) A clam in its burrow, the type locality. (Photos: Ilya V. Vikhrev).
Figure 6.
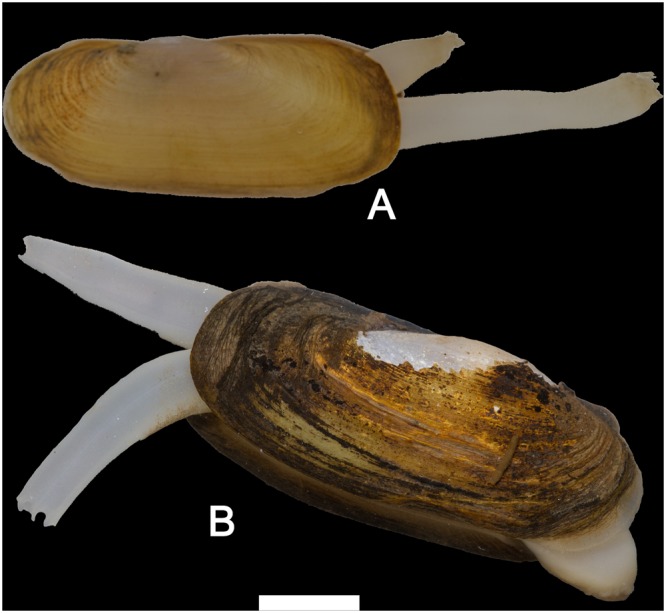
Live Novaculina clams with protruded siphons. (A) N. myanmarensis sp. nov., Donthami River. (B) N. gangetica, Lemro River. Scale bar = 10 mm. (Photos: Ilya V. Vikhrev).
Table 2.
Measurements of the type series of Novaculina myanmarensis sp. nov.
| Locality | Status of specimen | Voucher no.* | Shell length, mm | Shell height, mm | Shell width, mm |
|---|---|---|---|---|---|
| Myanmar: Ayeyarwady River, Pakokku Region, near Thin Baw Kone village | Holotype | Biv0420_8 | 40.6 | 15.2 | 9.8 |
| Paratype | Biv0420_1 | 34.5 | 12.8 | 7.9 | |
| Paratype | Biv0420_2 | 38.1 | 13.9 | 8.7 | |
| Paratype | Biv0420_3 | 36.4 | 12.9 | 8.7 | |
| Paratype | Biv0420_4 | 34.9 | 11.8 | 7.8 | |
| Paratype | Biv0420_5 | 34.1 | 12.3 | 7.7 | |
| Paratype | Biv0420_6 | 38.5 | 13.9 | 8.5 | |
| Paratype | Biv0420_7 | 33.7 | 12.2 | 7.4 | |
| Paratype | Biv0420_9 | 40.6 | 15.2 | 9.8 | |
| Paratype | Biv0420_10 | 30.3 | 11.1 | 6.4 | |
| Paratype | Biv0420_11 | 31.4 | 11.9 | 7.0 | |
| Paratype | Biv0420_12 | 35.2 | 12.4 | 8.1 | |
| Paratype | Biv0420_13 | 27.6 | 10.0 | 5.8 | |
| Paratype | Biv0420_14 | 35.4 | 12.9 | 7.9 | |
| Paratype | Biv0420_15 | 33.5 | 12.2 | 7.3 | |
| Paratype | Biv0420_16 | 27.9 | 10.9 | 6.1 | |
| Paratype | Biv0420_17 | 30.3 | 11.0 | 6.7 | |
| Paratype | Biv0420_18 | 36.3 | 12.5 | 8.0 | |
| Paratype | Biv0420_19 | 29.6 | 11.0 | 7.0 | |
| Paratype | Biv0420_20 | 36.4 | 12.2 | 7.5 | |
| Paratype | Biv0420_21 | 30.9 | 11.3 | 6.5 | |
| Paratype | Biv0420_22 | 34.1 | 12.7 | 8.0 | |
| Paratype | Biv0420_23 | 33.4 | 12.0 | 7.0 | |
| Paratype | Biv0420_24 | 33.1 | 11.4 | 7.1 | |
| Paratype | Biv0420_25 | 29.8 | 10.5 | 6.7 | |
| Paratype | Biv0420_26 | 30.2 | 11.0 | 6.5 | |
| Paratype | Biv0420_27 | 29.1 | 10.8 | 6.8 | |
| Paratype | Biv0420_28 | 32.3 | 11.8 | 7.0 | |
| Paratype | Biv0420_29 | 25.4 | 9.0 | 6.0 | |
| Paratype | Biv0420_30 | 28.3 | 10.1 | 6.3 | |
| Paratype | Biv0420_31 | 26.4 | 9.7 | 6.2 | |
| Paratype | Biv0420_32 | 27.1 | 10.5 | 6.1 | |
| Paratype | Biv0420_33 | 24.3 | 9.3 | 5.4 | |
| Paratype | Biv0420_34 | 24.1 | 9.5 | 4.9 | |
| Paratype | Biv0420_35 | 26.2 | 10.2 | 6.3 | |
| Paratype | Biv0420_36 | 21.6 | 8.1 | 4.9 | |
| Paratype | Biv0420_37 | 28.5 | 11.6 | 5.6 | |
| Paratype | Biv0420_38 | 20.5 | 7.9 | 4.5 | |
| Paratype | Biv0420_39 | 25.0 | 9.5 | 5.8 | |
| Paratype | Biv0420_40 | 38.1 | 14.2 | 9.0 | |
| Paratype | Biv0420_41 | 40.4 | 15.0 | 9.3 | |
| Paratype | Biv0420_42 | 35.6 | 12.8 | 7.9 | |
| Paratype | Biv0420_43 | 34.3 | 12.5 | 7.6 | |
| Paratype | Biv0420_44 | 33.5 | 11.4 | 7.1 | |
| Paratype | Biv0420_45 | 33.1 | 11.4 | 6.9 | |
| Paratype | Biv0420_46 | 32.3 | 12.0 | 7.5 | |
| Paratype | Biv0420_47 | 36.8 | 12.7 | 7.9 | |
| Paratype | Biv0420_48 | 26.5 | 10.0 | 5.6 | |
| Myanmar: downstream of Donthami River | Paratype | Biv0369_1 | 46.5 | 17.0 | 12.0 |
| Paratype | Biv0369_2 | 41.9 | 16.2 | 11.0 | |
| Paratype | Biv0369_3 | 43.9 | 17.0 | 13.3 | |
| Paratype | Biv0369_4 | 43.3 | 17.3 | 12.9 | |
| Paratype | Biv0369_5 | 42.6 | 17.5 | 12.3 | |
| Mean ± s.e.m. | 32.91 ± 0.81 | 12.12 ± 0.31 | 7.58 ± 0.27 |
*RMBH – Russian Museum of Biodiversity Hotspots, Federal Center for Integrated Arctic Research, Russian Academy of Sciences (Arkhangelsk, Russia).
Type locality
Myanmar: Ayeyarwady River, near Thin Baw Kone village (Pakokku Region) [21.3146°N, 95.0591°E].
Holotype RMBH Biv420_8
Myanmar: Ayeyarwady River, near Thin Baw Kone village (Pakokku Region), clay bottom near the river shore, 21.3146°N, 95.0591°E, 2 March 2018, Bolotov, Vikhrev, Zau Lunn, Nyein Chan, and locals leg.
Paratypes
Myanmar: Type locality, same label data, 47 specimens [RMBH Biv0420]; downstream of Donthami River, hard gravel-clay bottom, 16.6935°N, 97.5819°E, 11 February 2018, 5 specimens, local collector leg. [RMBH Biv0369]; Magway Division, Ayeyarwady River, large sandbar 1/2 mi SE of Nyaung-U, 21.2066°N, 94.9062°E, November 2009, 3 specimens, C. N. Piotrowski leg. [CAS 180843]; Ayeyarwady River, near Minbu, 20.1911°N, 94.8788°E, 29 April 2018, 4 specimens, Nyein Chan leg. [FFI].
Etymology
The name of this species is derived from the country of Myanmar.
Conchological diagnosis
Shell length 20.5–46.5 mm, shell height 7.9–17.5 mm, shell width 4.5–13.3 mm (N = 54, Table 2). This species has an elongated shell, and is closely related to N. gangetica and N. chinensis, but it can be distinguished from these taxa by a more rectangular shell shape with truncated posterior end (vs more oval shell shape with rounded posterior end).
Molecular diagnosis
The new species differs from N. gangetica by the fixed nucleotide substitutions: 49 substitutions in the COI gene fragment [29 G, 38 A, 53 G, 59 A, 92 A, 128 C, 134 C, 161 G, 170 T, 173 A, 182 A, 185 A, 197 A, 200 A, 212 G, 213 C, 215 T, 230 A, 243 T, 245 A, 254 A, 299 A, 302 A, 308 T, 311 C, 314 G, 329 G, 347 T, 362 G, 371 G, 377 C, 380 A, 404 G, 413 G, 464 T, 467 G, 470 G, 485 G, 491 G, 497 T, 521 G, 545 A, 551 G, 560 G, 569 A, 572 A, 590 T, 617 C, 644 A], 8 substitutions in the 16S rRNA gene fragment [24 G, 74 G, 237 A, 239 G, 242 A, 287 G, 288 T, 305 A], and one substitution in the nuclear 28S rRNA gene fragment [429 A].
Description
Shell shape from rectangular to oval-elongated, dorsal and ventral margin are almost parallel to each other (Fig. 3A,B). Shell thin or moderately thick, not inflated. Periostracum from light yellow to brown; nacre whitish, shining. Umbo more or less prominent, in the first half of the shell. Pseudocardinal teeth small and distant from each other, two on the right valve and three on the left valve. Anterior muscle scar pyriform, posterior muscle scar shallow and with rounded shape. The mantle and its edges colored in light yellow. The gills elongated and ribbed (Fig. 4A). The anterior margin of inner gills slightly longer and wider than the outer gills. Foot stumpy, slightly dilated at the end and somewhat truncated; branchial siphon stouter than the anal one, almost the same length, surface of siphon ribbed (Figs 4A and 6A).
Intraspecific conchological variability
Specimens from the Donthami and Ayeyarwady rivers are rather different from each other conchologically (compare Fig. 3A,B). The first ones are stronger and thicker, have more truncated posterior end, slightly concave dorsal margin, more developed umbo and hinge. The specimens from the Ayeyarwady River are characterized by more elliptical and very thin shell with light-yellow and smoother periostracum. At first glance, these conchological differences may reflect an environment-induced variability, because the populations were recorded from sites with different bottom substrate (i.e. soft clay substrate in the Ayeyarwady River vs hard gravel-clay substrate in the Donthami River).
Distribution
Donthami (Salween River basin) and Ayeyarwady rivers in Myanmar.
Habitat
Downstream and middle section of large rivers, in fresh water (Fig. 5A,B). This species inhabits gravel-clay and clay bottom, in which it makes deep vertical holes (Fig. 5C,D).
Comments
Local villagers harvest N. myanmarensis sp. nov. from the downstream section of the Donthami River (food for consumption). In contrast, this species seems to be unutilized in the Ayeyarwady River.
Novaculina gangetica Benson, 1830
Novaculina gangetica Benson16: p. 63; Subba Rao7: p. 223; Graf2: p. 152.
Type locality
Ganges, Calcutta [India, approximately 22.6°N, 88.3°E].
Type series
The University Museum of Zoology, Cambridge, UK [UMZC I.102125: eleven syntypes from the Robert McAndrew collection, labeled “Bens. Coll., Ganges, Calcutta”].
Conchological diagnosis
Shell length 28.1–39.7 mm, shell height 12.9–17.5 mm, shell width 8.1–12.7 mm (N = 24). This species has an elongated shell, and is closely related to N. myanmarensis sp. nov. and N. chinensis. It could be distinguished from N. myanmarensis sp. nov. by a more ovate shell shape with rounded posterior end (vs more rectangular shell shape with truncated posterior end). N. gangetica differs from N. chinensis by somewhat higher and shorter shell with slightly convex ventral margin (vs more elongated shell with straight ventral margin).
Intraspecific conchological variability
Some specimens have somewhat trapezoidal shell, with slightly expanded posterior end4.
Distribution
Ganges River and its tributaries in India and Bangladesh ranging from the delta to at least 1,500 km upstream7,8,17,18, Buriganga and Pashur river systems in Bangladesh19,20, and Kaladan and Lemro rivers in Myanmar4.
Habitat
The species inhabits downstream and middle sections of large rivers, in fresh or slightly brackish water4,8. N. gangetica prefers clay bottom, in which it makes cylindrical holes4,16,17, but it was also recorded in soft sand and silt bottom8. In the Kaladan River, this species also inhabits submerged rocks, in which it was recorded from the vacant borings of Lignopholas fluminalis, filled with clay4. Benson16 noted that this species rarely occurs from holes in rocks in the Jumna and Gumti rivers, and that the specimens from such a habitat have an asymmetrical shell.
Comments
Local villagers harvest N. gangetica from the Kaladan and Lemro rivers in Myanmar (food for consumption and local market trade), but it seems to be unutilized in India7.
Novaculina siamensis Morlet, 1889
Novaculina siamensis Morlet10: p. 172, 198; Brandt9: p. 303; Graf2: p. 152; Sayenko et al.11: p. 182.
Type locality
Marais de Chantakam (Siam)10 [M. Chantakam, rainfall station on a tributary of the Phra Prong River21,22, Thailand, approximately 14.0°N, 102.0°E].
Type series
Whereabouts unknown. Morlet’s collection of shells from Indochina went to P. Dautzenberg and is in the Royal Belgian Institute of Natural History, Brussels, Belgium. However, the type series of N. siamensis seems to be lacking in this collection (Thierry Backeljau, pers. comm., 2018).
Conchological diagnosis
Shell length 30–38 mm, shell height 13–18 mm, shell width 10–15 mm9. This species could be distinguished from all the other Novaculina taxa by its much shorter and higher shell, less prominent umbo, clear sculpture with concentric growth lines, and dark yellow periostracum.
Intraspecific conchological variability: Some shells in the Mekong Delta population are asymmetrical and torsed11.
Distribution
Bang Pakong and Pa Sak River basins in Thailand9,10, and the Mekong Delta in Vietnam11. We assume that a population of N. cf. siamensis from a tidal creek in the Trang Province of Thailand23 belongs to another species, because this creek empties into the Andaman Sea.
Habitat
Upstream section of medium-sized rivers, in fresh water, probably on clay bottom substrate. However, it was found in a slightly brackish, tidal channel in the Mekong Delta11.
Comments
This species seems to be unutilized in Thailand.
Novaculina chinensis Liu & Zhang, 1979
Novaculina chinensis Liu & Zhang12: p. 356; Qin24: p. 305; He & Zhuang25: p. 128; Graf2: p. 152; Chen et al.26: p. 4.
Type locality
Wuxi, Jiangsu Province [Lake Taihu, approximately 31.4402°N, 120.3143°E]12.
Type series
National Zoological Museum of China, Institute of Zoology, Chinese Academy of Sciences, Beijing, China [holotype NZMC KS 747703, paratypes NZMC FM00855]25.
Conchological diagnosis
Shell length 34–46 mm, shell height 11–16 mm, shell width 8–10 mm12. This species is closely related to N. myanmarensis sp. nov. and N. gangetica by an elongated shell shape, but could be distinguished from these species by more prominent, somewhat acute umbo.
Intraspecific conchological variability
Not known.
Distribution
Downstream of the Yangtze River, China, most records from Lake Taihu12,24,27, Lake Hongze14, and Lake Chaohu13.
Habitat
Large floodplain lakes, in fresh water12–14,24,27. A single record from the Shangqing River15.
Comments
This species seems to be unutilized in China. A parasitic mite species, Unionicola imamurai Hevers, 1978, has been reported from N. chinensis15.
Discussion
Taxonomic conclusions
Our results reveal that the genus Novaculina comprises four species: N. gangetica from India, Bangladesh and northwestern Myanmar, N. myanmarensis sp. nov. from central and eastern Myanmar, N. siamensis from Thailand and southern Vietnam, and N. chinensis from southeastern China (Fig. 1). An additional species, Novaculina andamanensis, was described from the Andaman Islands but without a precise locality28 (holotype no. ZSI M4060/1, paratype no. ZSI 20765/4 [Subba-Rao7 considered the latter specimen to be the holotype], malacological collection of the Zoological Survey of India, Kolkata, India29). However, this species has been considered a junior synonym of the marine bivalve species Azorinus coarctatus29,30. We agree with that taxonomic conclusion, because, according to the original description and figure of the type specimen28, this species has somewhat trapezoidal shell with concave ventral margin as seen in Azorinus coarctatus.
Records of Novaculina are still lacking from the downstream sections of several large and medium-sized Southeast and East Asian rivers such as the Pearl River in China and Red River in Vietnam. Taking into account a poor knowledge of freshwater fauna in these basins, further records of new Novaculina taxa could not be ruled out. An occasional record of Novaculina cf. siamensis from a small creek in southern Thailand23 suggests that the members of this genus could also establish permanent populations in small-sized freshwater basins, the fauna of which is almost unknown.
Two pharid genera, i.e. Sinonovacula and Novaculina, were hitherto placed in the Novaculininae5. This subfamily was established for Novaculina gangetica31, but, later, Sinonovacula constricta had also been placed within it32. However, we found that Pharella javanica belongs to a well-supported clade together with Sinonovacula and Novaculina species (Fig. 2), as it was also shown by another study4. According to this, we propose the Novaculininae as a junior synonym of the Pharellinae. A close similarity between Pharella and Sinonovacula has also been recorded by a functional morphology, particularly in the presence of crescentric anterior and posterior pedal protractor muscles in both taxa5.
Biogeographic implications
Discovery of the new Novaculina species from the Salween and Ayeyarwady river drainage basins in Myanmar indicates that the range of this genus is rather continuous and extends along the continental margin of Asia from the Ganges River to the Yangtze River (Fig. 1). Unfortunately, the phylogenetic affinities of the two eastern species, i.e. Novaculina siamensis and N. chinensis, are still unknown because of the lack of available molecular data. However, they may represent a divergent clade, because the Thai–Malay Peninsula is a significant biogeographic barrier to longitudinal dispersal of aquatic animals33. This barrier could have existed during most of the Cenozoic Epoch34, although it may have occasionally been incised, but not breached, at the Isthmus of Kra33. Our statistical biogeographic modeling strongly supports the hypothesis4,6,18 that the genus Novaculina is a relict, marine-derived freshwater clade. Similar examples of such secondary freshwater lineages are known among a variety of other taxa, e.g. in fishes, gastropods, polychaetes, and crustaceans4,35.
The high level of molecular divergence between the two western species, i.e. Novaculina gangetica and N. myanmarensis sp. nov., supports a new freshwater biogeographic division of Southeast Asia that has been developed on the basis of unionid mussel research36–38. According to this model, the drainages of the Arakan coast of Myanmar, the Ayeyarwady, Bago, Sittaung, and Bilin river basins, and east to the Salween River and drainages of southern Myanmar belong to the Western Indochina Subregion of the Oriental Region38. This subregion has high levels of faunal endemism and is separated well from the Indian and Sundaland subregions38,39. However, our new study reveals that the northern drainages of the Aracan coast such as the Kaladan and Lemro rivers seem to be a rather marginal part of the Indian Subregion that has already been shown by another research4. Anyway, the presence of sister but highly divergent species in the Ganges and Ayeyarwady rivers even in salt-tolerant freshwater taxa such as Novaculina strongly indicates that these basins were separated at least since the Miocene (Fig. 2).
In contrast, a shallow genetic divergence between populations of Novaculina myanmarensis sp. nov. from the Salween and Ayeyarwady river basins in Myanmar suggests that there were relatively recent (i.e. Late Pleistocene) dispersal events in this species among the downstream sections of these large river drainages. The phylogeography of freshwater mussels (Unionidae) partly reflects this pattern, e.g. the distribution range of Leoparreysia tavoyensis crosses numerous freshwater drainages from the Tavoy (north of the Thai–Malay Peninsula) to the Ayeyarwady36,37. However, the majority of unionid species in Myanmar appear to be restricted to certain drainage basins or their tributaries36,37.
There are several widespread salt-tolerant estuarine and freshwater species, e.g. a polychaete, Neanthes meggitti (Nereididae), and a pholadid bivalve, Lignopholas fluminalis (Pholadidae)4, the range of which encompasses the downstream sections of the Ganges and Ayeyarwady rivers. Such taxa were described from the delta of Ayeyarwady and later have been discovered from the Ganges, or vice versa, and they are ecologically associated with the typical Novaculina habitats4. The discovery of a new Novaculina species in Myanmar indicates that such taxa with broad distribution may actually represent cryptic species complexes, although this preliminary assumption is in need of future molecular research with extensive field surveys in South and Southeast Asia.
Methods
Data sampling and mapping
The samples of Novaculina myanmarensis sp. nov. were collected from two localities during a field trip to Myanmar in 2018. Additional materials were investigated in the collections of the Fauna & Flora International – Myanmar Program [FFI] (Yangon, Myanmar) and California Academy of Sciences [CAS] (San Francisco, USA). We processed new COI, 16S rRNA and 28S rRNA sequences from ten specimens of Novaculina myanmarensis sp. nov. (Table 1) using the standard approach as described in our previous work4. Sequences of Novaculina gangetica and other Pharidae taxa were obtained from GenBank (Table 1 and Supplementary Table 1). We collected a dataset of reliable georeferenced records of Novaculina species from published sources and museum specimens (Supplementary Table 2). The map was created using ESRI ArcGIS 10 software (www.esri.com/arcgis).
Morphological study
The samples were studied using a stereomicroscope (Leica M165C, Leica Microsystems, Germany). The comparative analysis of taxa was performed according to the standard conchological patterns, i.e. the shape of shell, hinge structure, muscle attachment scars, and position of umbo.
Sequence alignment, saturation analyses and congruence of phylogenetic signals
We aligned each gene data set using the MUSCLE algorithm in MEGA640. We performed the saturation test of Xia et al.41 with DAMBE v. 5.3.10842, but we found no evidence of substitution saturation (P < 0.001). A partition homogeneity test was calculated in PAUP* v. 4.0a151 to confirm the congruence of phylogenetic signals among sequence data sets43. This test revealed that the signals among the data sets are consistent (P > 0.1 in all the combinations).
Phylogenetic analyses
We computed maximum likelihood and Bayesian inference phylogenetic models with RAxML v. 8.2.6 HPC Black Box44 and MrBayes v. 3.2.645, respectively. The settings of analyses were as described in Bolotov et al.4. The best substitution models that were used for the Bayesian analyses are listed in Supplementary Table 3. The phylogenetic analysis was done at the San Diego Supercomputer Center through the CIPRES Science Gateway46.
Divergence time modeling
A time-calibrated phylogenetic model has been calculated with BEAST 1.8.447 using the same substitution models as for the MrBayes analyses (Supplementary Table 3). A lognormal relaxed clock and Yule speciation process with continuous quantile parametrization were assigned as model’s priors. To timing the phylogeny, we used the following new crown fossil calibration: †Leptosolen otterensis Cragin (1894)48. Diagnosis and phylogenetic placement: Shell thin, elongated, moderately convex, inequilateral, compressed anteriorly, with anterior fold and angular growth lines around anterior and posterior margins. This species seems to be the oldest member of the genus49, and may represent an ancestral lineage of the Pharidae. Stratigraphic horizon and locality: dark-gray shale of Kiowa Formation (Albian) in central Kansas48. Absolute age estimate: Lower Cretaceous, upper Albian boundary, 100.5 Ma, based on stratigraphy48; 95% soft upper bound 113.0 Ma (lower Albian boundary). Prior setting: exponential distribution, mean (lambda) = 3.4, MRCA: Novaculina gangetica – Siliqua radiata. Five independent runs of 30,000,000 generations were processed, with sampling every 1,000 generation. The resulting tree sets were combined using LogCombiner 1.8.447. An appropriate burn-in was chosen for each tree set with Tracer v. 1.650. A maximum clade credibility tree has been computed with TreeAnnotator 1.8.4 with an additional resampling every 10,000 generation47.
Ancestral area modeling
Ancestral area reconstruction has been performed on the basis of three algorithms, i.e., Statistical Dispersal-Vicariance Analysis (S-DIVA), Dispersal-Extinction Cladogenesis (DEC), and Statistical Dispersal-Extinction Cladogenesis (S-DEC) implemented in RASP v. 3.251 as described in Bolotov et al.4. We assigned two possible ancestral areas of the in-group species, i.e., (a) estuarine and (ab) freshwater to estuarine. The three primary models were combined into an integrative model using the Combine Results option of RASP v. 3.251.
Molecular diagnoses
To test the molecular differences between N. myanmarensis sp. nov. and N. gangetica, we used an approach of Bolotov et al.37. The mean p-distances and number of fixed nucleotide substitutions were accessed using MEGA640.
Nomenclatural acts
The electronic edition of this article conforms to the requirements of the amended International Code of Zoological Nomenclature (ICZN), and hence the new names contained herein are available under that Code from the electronic edition of this article. This published work and the nomenclatural acts it contains have been registered in ZooBank (http://zoobank.org), the online registration system for the ICZN. The LSID for this publication is: urn:lsid:zoobank.org:pub:19E34605-30C2-4DAB-B81F-53A1FDB324DB. The electronic edition of this paper was published in a journal with an ISSN, and has been archived and is available from PubMed Central.
Electronic supplementary material
Acknowledgements
We are grateful to the Editor and two anonymous reviewers for their important comments which improved an initial version of the manuscript. This work was partly funded by grants from the Russian Ministry of Education and Science (project no. 6.2343.2017/4.6), Federal Agency for Scientific Organizations (project no. 0409-2015-0143), Russian Foundation for Basic Research (project nos. 16-34-00638 and 18-44-292001), National Geographic Society (project no. NGS-274R-18), and Northern Arctic Federal University. M.L.-L. was funded by FCT – Foundation for Science and Technology under grant no. SFRH/BD/115728/2016. We are grateful to Dr. Thierry Backeljau (Royal Belgian Institute of Natural History, Brussels, Belgium), the late Dr. Tony Whitten (Fauna & Flora International – Asia-Pacific), Mr. Frank Momberg (Fauna & Flora International – Myanmar Program, Myanmar), staff of the Department of Fisheries of the Ministry of Agriculture, Livestock and Irrigation of Myanmar, and Khin Lin Lin Kyaw (Department of Zoology, Hpa-An University, Myanmar) for their great help during this study. Our research has been performed under the survey permission no. 5/6000/LFR(210/2018) dated on 23 January 2018 issued by the Ministry of Agriculture, Livestock and Irrigation of Myanmar and the export permission no. NWCD/CITES/9/5666/2018 dated on 28 June 2018 issued by the Forest Department of the Ministry of Environmental Conservation and Forestry of Myanmar.
Author Contributions
I.N.B. developed the concept of the study. I.V.V., I.N.B., M.L.-L., Z.L., N.C. and T.W. collected samples. A.V.K. designed and processed molecular analyses. E.S.K. performed morphological and anatomical research. M.Y.G. created the map. I.N.B. performed phylogenetic modeling and wrote the paper, with input from E.S.K., I.V.V., M.L.-L., Z.L., N.C., A.V.K., M.Y.G., O.V.A., S.T. and T.W. All authors discussed the manuscript.
Data Availability
The sequences used in this study are available from GenBank. Accession numbers for each specimen are presented in Table 1. The type series of the new species is available in the Russian Museum of Biodiversity Hotspots [RMBH], Federal Center for Integrated Arctic Research, Russian Academy of Sciences (Arkhangelsk, Russia), Fauna & Flora International – Myanmar Program [FFI] (Yangon, Myanmar), and California Academy of Sciences [CAS] (San Francisco, USA).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-34491-8.
References
- 1.Bogan AE. Global diversity of freshwater mussels (Mollusca, Bivalvia) in freshwater. Hydrobiologia. 2008;595:139–147. doi: 10.1007/s10750-007-9011-7. [DOI] [Google Scholar]
- 2.Graf DL. Patterns of freshwater bivalve global diversity and the state of phylogenetic studies on the Unionoida, Sphaeriidae, and Cyrenidae. American Malacological Bulletin. 2013;31:135–153. doi: 10.4003/006.031.0106. [DOI] [Google Scholar]
- 3.Carter JG, et al. A synoptical classification of the Bivalvia (Mollusca) Paleontological Contributions. 2011;4:1–47. doi: 10.17161/PC.1808.8287. [DOI] [Google Scholar]
- 4.Bolotov IN, et al. Discovery of a silicate rock-boring organism and macrobioerosion in fresh water. Nature Communications. 2018;9:2882. doi: 10.1038/s41467-018-05133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morton B. The functional morphology of Sinonovacula constricta with a discussion on the taxonomic status of the Novaculininae (Bivalvia) Journal of Zoology. 1984;202:299–325. doi: 10.1111/j.1469-7998.1984.tb05085.x. [DOI] [Google Scholar]
- 6.Annandale N. The marine element in the fauna of the Ganges. Bijdragen tot de Dierkunde. 1922;22:143–154. [Google Scholar]
- 7.Subba Rao, N. V. Handbook of freshwater molluscs of India. (Calcutta, 1989).
- 8.Nesemann HA, Sharma SU, Sharma GO, Sinha RK. Illustrated checklist of large freshwater bivalves of the Ganga river system (Mollusca: Bivalvia: Solecurtidae, Unionidae, Amblemidae) Nachrichchtenblatt der Ersten Vorarlberger Malakologischen Gesellschaft. 2005;13:1–51. [Google Scholar]
- 9.Brandt RAM. The non-marine aquatic mollusca of Thailand. Archiv für Mollusckenkunde. 1974;105:1–423. [Google Scholar]
- 10.Morlet L. Catalogue des Coquilles recueillies, par M. Pavie, dans le Cambodge et le Royaume de Siam, et description d’espèce nouvelles. Journal de Conchyliologie. 1889;37:121–199. [Google Scholar]
- 11.Sayenko, E. M., Quang, N. X. & Lutaenko, K. A. Bivalves of the Ba Lai River – one of estuary of the Mekong Delta, Vietnam. In Life-Supporting Asia-Pacific Marine Ecosystems, Biodiversity and Their Functioning (eds Dautova, T. N, Sun, X., Sun, S. & Adrianov, A.V.) 178–184 (Science Press Beijing, 2017).
- 12.Liu YY, Zhang WZ. A new species of freshwater razor clam, Novaculina chinensis, from Jiangsu Province, China. Acta Zootaxonomica Sinica. 1979;4:356–357. [Google Scholar]
- 13.Cai Y, Gong Z, Xie P. Community structure and spatiotemporal patterns of macrozoobenthos in Lake Chaohu (China) Aquatic Biology. 2012;17:35–46. doi: 10.3354/ab00455. [DOI] [Google Scholar]
- 14.Hu Z, et al. The habitat type and trophic state determine benthic macroinvertebrate communities in lowland shallow lakes of China. Journal of Limnology. 2016;75:330–339. doi: 10.4081/jlimnol.2016.1220. [DOI] [Google Scholar]
- 15.Wen C, Zhu Z. Seven species of water mites in the genus Unionicola from Jiangxi (Acari: Unionicolidae) Acta Zootaxonomica Sinica. 1999;24:30–37. [Google Scholar]
- 16.Benson WH. Description of Novaculina, a new genus of fresh-water bivalves, inhabiting the Ganges and its branches. Gleanings in Science. 1830;2:63–65. [Google Scholar]
- 17.Benson WH. Characters of Tanysiphon, a new genus of fluviatile shells, allied to Myacidae. Annals and Magazine of Natural History (3rd series) 1858;1:407–410. doi: 10.1080/00222935808696948. [DOI] [Google Scholar]
- 18.Nesemann H, Sharma G, Sinha R. Benthic macro-invertebrate fauna and “marine elements” sensu Annandale (1922) highlight the valuable dolphin habitat of river Ganga in Bihar-India. Taprobanica. 2011;3:18–30. doi: 10.4038/tapro.v3i1.3230. [DOI] [Google Scholar]
- 19.Baki MA, Hossain MM, Bhouiyan NA. Checklist of freshwater mollusca (Gastropoda and Bivalvia) recorded from the Buriganga and Turag rivers, Dhaka, Bangladesh. The Festivus. 2016;48:221–228. [Google Scholar]
- 20.Khan AN, Kamal D, Mahmud MM, Rahman MA, Hossain MA. Diversity, distribution and abundance of benthos in Mouri River, Khulna, Bangladesh. International Journal of Sustainable Crop Production. 2007;2:19–23. [Google Scholar]
- 21.Anonymous. Burma, Siam, French Indo-China, and Straits Settlements; Inset Map of Singapore and Vicinity. Printed map (London, 1907).
- 22.Anonymous. Rain-Report: 1st of April, 1906-1st of April, 1907. The Journal of the Siam Society4, 35–60 (1907).
- 23.Kon K, Kurokura H, Hayashizaki K. Role of microhabitats in food webs of benthic communities in a mangrove forest. Marine Ecology Progress Series. 2007;340:55–62. doi: 10.3354/meps340055. [DOI] [Google Scholar]
- 24.Qin, B. (Ed.). Lake Taihu, China: Dynamics and environmental change. Monographiae Biologicae87, 1–348 (2008).
- 25.He, J. & Zhuang, Z. The Freshwater Bivalves of China. (Harxheim, 2013).
- 26.Chen J, Hu D, Zhang C, Ding Z. Temporal and spatial changes of macrobenthos community in the regions frequently occurring black water aggregation in Lake Taihu. Scientific Reports. 2018;8:5712. doi: 10.1038/s41598-018-24058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ji L, Song C, Cao X, Zhou Y, Deng D. Spatial variation in nutrient excretion by macrozoobenthos in a Chinese large shallow lake (Lake Taihu) Journal of Freshwater Ecology. 2015;30:169–180. doi: 10.1080/02705060.2014.997816. [DOI] [Google Scholar]
- 28.Preston HB. XXIII. — Descriptions of new species of land, marine and freshwater shells from the Andaman Islands. Records of the Indian Museum. 1908;2:187–210. [Google Scholar]
- 29.Ramakrishna K, Dey A, Mitra SC. Catalogue of type species (Bivalvia, Scaphopoda) present in the Mollusca section of Zoological Survey of India. Records of the Zoological Survey of India, Occasional Paper. 2004;228:1–97. [Google Scholar]
- 30.Sartori, A. F. Novaculina andamanensis Preston, 1908. World Register of Marine Species http://marinespecies.org/aphia.php?p=taxdetails&id=820235 (2018).
- 31.Ghosh R. Taxonomic studies on the soft parts of the Solenidae. Records of the Indian Museum. 1920;19:47–78. [Google Scholar]
- 32.Annandale TN, Prashad B. Report on a small collection of molluscs from the Chekiang Province of China. Journal of Molluscan Studies. 1924;16:27–49. doi: 10.1093/oxfordjournals.mollus.a063832. [DOI] [Google Scholar]
- 33.Parnell J. The biogeography of the Isthmus of Kra region: a review. Nordic Journal of Botany. 2013;31:001–015. doi: 10.1111/j.1756-1051.2012.00121.x. [DOI] [Google Scholar]
- 34.Woodruff DS. Neogene marine transgressions, palaeogeography and biogeographic transitions on the Thai–Malay Peninsula. Journal of Biogeography. 2003;30:551–567. doi: 10.1046/j.1365-2699.2003.00846.x. [DOI] [Google Scholar]
- 35.Adams Nicole E., Inoue Kentaro, Seidel Richard A., Lang Brian K., Berg David J. Isolation drives increased diversification rates in freshwater amphipods. Molecular Phylogenetics and Evolution. 2018;127:746–757. doi: 10.1016/j.ympev.2018.06.022. [DOI] [PubMed] [Google Scholar]
- 36.Bolotov IN, et al. Ancient river inference explains exceptional Oriental freshwater mussel radiations. Scientific Reports. 2017;7:2135. doi: 10.1038/s41598-017-02312-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolotov IN, et al. New taxa of freshwater mussels (Unionidae) from a species-rich but overlooked evolutionary hotspot in Southeast Asia. Scientific Reports. 2017;7:11573. doi: 10.1038/s41598-017-11957-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolotov IN, et al. A new genus and tribe of freshwater mussel (Unionidae) from Southeast Asia. Scientific Reports. 2018;8:10030. doi: 10.1038/s41598-018-28385-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfeiffer, J. M., Graf, D. L., Cummings, K. S. & Page, L. M. Molecular phylogeny and taxonomic revision of two enigmatic freshwater mussel genera (Bivalvia: Unionidae incertae sedis: Harmandia and Unionetta) reveals a diverse clade of Southeast Asian Parreysiinae. Journal of Molluscan Studies, 10.1093/mollus/eyy028 (2018).
- 40.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia X, Xie Z, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution. 2003;26:1–7. doi: 10.1016/S1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- 42.Xia, X. & Lemey, P. Assessing substitution saturation with DAMBE. In The Phylogenetic Handbook: A Practical Approach to DNA and Protein Phylogeny, Second Edition (Lemey, P., Salemi, M. & Vandamme, A. eds) 615–630 (Cambridge University Press, 2009).
- 43.Swofford, D. L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0b10 (Sinauer Associates, Sunderland, Massachusetts, 2002).
- 44.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 45.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, M., Pfeiffer, W. & Schwartz, T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Gateway Computing Environments Workshop (GCE) 1–8 (IEEE, 2010).
- 47.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott RW. Paleoecology and paleontology of the Lower Cretaceous Kiowa Formation, Kansas. The University of Kansas Paleontological Contributions. 1970;52:1–94. [Google Scholar]
- 49.Stephenson LW, Stenzel HB. Larger invertebrate fossils of the Woodbine Formation (Cenomanian) of Texas with Decapod Crustaceans from the Woodbine Formation of Texas. Geological Survey Professional Paper. 1952;242:1–226. [Google Scholar]
- 50.Rambaut, A., Suchard, M. & Drummond, A. J. Tracer v1.6 at http://beast.bio.ed.ac.uk/software/tracer/ (2013).
- 51.Yu Y, Harris AJ, Blair C, He XJ. RASP (Reconstruct Ancestral State in Phylogenies): a tool for historical biogeography. Molecular Phylogenetics and Evolution. 2015;87:46–49. doi: 10.1016/j.ympev.2015.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences used in this study are available from GenBank. Accession numbers for each specimen are presented in Table 1. The type series of the new species is available in the Russian Museum of Biodiversity Hotspots [RMBH], Federal Center for Integrated Arctic Research, Russian Academy of Sciences (Arkhangelsk, Russia), Fauna & Flora International – Myanmar Program [FFI] (Yangon, Myanmar), and California Academy of Sciences [CAS] (San Francisco, USA).