Abstract
Genome-wide association studies (GWASs) have identified thousands of loci associated with hundreds of complex diseases and traits, and progress is being made toward elucidating the causal variants and genes underlying these associations. Functional characterization of mechanisms at GWAS loci is a multi-faceted challenge. Challenges include linkage disequilibrium and allelic heterogeneity at each locus, the noncoding nature of most loci, and the time and cost needed for experimentally evaluating the potential mechanistic contributions of genes and variants. As GWAS sample sizes increase, more loci are identified, and the complexities of individual loci emerge. Loci can consist of multiple association signals, each of which can reflect the influence of multiple variants, inseparable by association analyses. Each signal within a locus can influence the same or different target genes. Experimental studies of genes and variants can differ on the basis of cell type, cellular environment, or other context-specific variables. In this review, we describe the complexity of mechanisms at GWAS loci—including multiple signals, multiple variants, and/or multiple genes—and the implications these complexities hold for experimental study design and interpretation of GWAS mechanisms.
Keywords: human genetics, genome-wide association study, signal, variant, SNP, gene, mechanism, allelic heterogeneity, complex trait
Main Text
Introduction
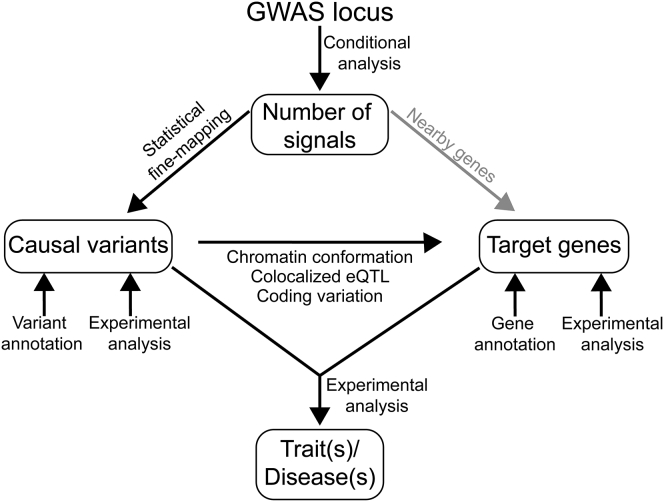
Genome-wide association studies (GWASs) have identified thousands of loci associated with complex traits and diseases.1, 2 Converting GWAS findings into trait or disease insights includes elucidating both molecular mechanisms, by which genetic variants affect gene expression or function, and biological mechanisms, by which target genes affect a trait or disease. Progress is being made to identify candidate causal variants and genes underlying these associations, and complex molecular and biological mechanisms at GWAS loci are appearing. A recent review provides an excellent framework for the functional dissection of a genetic risk locus.3 Here, we review the emerging complexities of molecular mechanisms at GWAS loci. After providing background to the challenges, we review three major questions: (1) How many association signals exist at a locus? (2) What are the candidate causal variant(s)? (3) What are the target gene(s)? In each section, we provide historical context to the question, methods available for addressing it, and evidence and observations from examples of GWAS loci that have been mechanistically characterized to date. Identifying mechanisms responsible for GWAS loci requires an accumulation of consistent evidence for the genes and variants that influence the trait or disease in humans (Figure 1). We conclude with future directions for researchers to consider in experimental design and interpretation of GWAS locus mechanisms.
Figure 1.
Process for Evaluating a GWAS Locus
Many approaches exist for identifying mechanisms at GWAS loci. In this review, we address three major questions at GWAS loci: (1) How many association signals exist at a locus? (2) What are the candidate causal variant(s)? (3) What are the target gene(s)? This flowchart shows how an accumulation of evidence can address these questions.
Background
Complex genetic traits and diseases differ from monogenic traits and diseases. Monogenic diseases are caused by variation in single genes, whereas complex genetic traits are influenced by variation in multiple genes and environmental factors. GWASs have successfully identified thousands of genomic regions associated with hundreds of complex traits and diseases. GWAS publications typically report association results as a list of loci, distinguished from one another for counting purposes and labeled with a variant and one or more gene names as signposts. The variant named is typically the most strongly associated variant and is referred to as the lead, index, sentinel, or top variant (for other terminology, see Table 1). The gene names make referring to loci easier than using genome positions or variant labels, although the genes named in GWAS reports have variable evidence supporting their role in the trait or disease. Some GWAS reports simply indicate the nearest gene; others label loci with nearby gene(s) that have some annotation or experimental support. Early GWASs were performed with less densely spaced sets of variants, so the reported variant might not have been the strongest associated variant at a locus. More recent GWASs and GWAS meta-analyses are larger with sample sizes approaching one million for some traits, and although GWASs have often been performed in a single population, a growing number of trans-ancestry studies combine data across populations. For most identified loci, the molecular and biological mechanisms remain to be determined.
Table 1.
Definitions of Terms Used in This Review
| Monogenic disease or trait | a disease caused by or a trait influenced by variation in a single gene, although variants in multiple single genes can cause the same disease |
| Complex disease or trait | a disease or trait caused by a combination of variants in many genes; it can be influenced by behavioral and environmental factors |
| Molecular mechanism | a mechanism by which a genetic variant affects gene expression or function |
| Biological mechanism | a mechanism by which a gene affects a trait or disease |
| Allelic heterogeneity | the phenomenon by which multiple variants act on one gene to influence the same phenotype |
| Locus | a genomic region associated with a complex trait or disease; it is often defined by a distance, e.g., within 500 kb or 1 Mb of a reported variant |
| Signal | within a locus, a set of variants that are in strong pairwise LD with each other and are associated with a trait or disease; multiple signals can be conditionally distinct from each other, a subset of which are independent |
| Causal variant | a variant that affects a molecular or cellular process to have an impact on a trait or disease |
| Functional variant | a variant that shows evidence of allelic differences affecting gene regulation or function; variants can be functional but not affect a trait or disease |
| Target gene | a gene affected by a functional variant; also called an effector gene |
Much of the complexity of mechanisms at GWAS loci is due to allelic heterogeneity, in which multiple alleles act through the same gene to influence the same phenotype; allelic heterogeneity is common at monogenic disease-associated loci. For example, the Cystic Fibrosis Mutation Database includes >2,000 disease-causing mutations,4 and at least 17 mutations can cause sickle cell disease.5 As GWAS sample sizes become larger and we delve further into the mechanisms at GWAS loci, we are learning that allelic heterogeneity is also prevalent in complex genetic traits, and this heterogeneity influences both the design and interpretation of experimental studies.
Allelic heterogeneity of complex traits has been identified in studies of model organisms. Initially, quantitative trait loci (QTLs) in model organisms were generally assumed to harbor one causal and multiple passenger alleles that affect a single causal gene; however, dissection of QTLs in inbred organisms has identified evidence of more than one gene in the same region.6, 7, 8 In addition to identifying multiple genes at QTLs, fine-mapping efforts in model organisms have suggested multiple causal variants at a single locus.8, 9 Thus, genetic studies in model organisms suggest complex mechanisms that can involve multiple genes and variants at a single locus.
Experimental characterization of GWAS loci has lagged behind locus discovery because each locus presents a multi-faceted challenge. The location of many GWAS variants in noncoding regions10 provides less straightforward hypotheses for mechanisms than variants within protein-coding regions. Each association signal typically consists of multiple variants in linkage disequilibrium (LD), and the sheer numbers of candidate variants can pose a challenge for interpreting annotations and performing experimental analyses; for example, 6,324 SNPs were reported to be in high LD (r2 > 0.5) with 146 lead variants in 100 regions associated with prostate cancer.11 Often, the cell type or tissue of action and the cellular state are unknown, and researchers must choose a cell type or model organism and potential stimuli to test mechanisms. It can be difficult to recapitulate the exact conditions of a trait or disease in model systems to determine the precise mechanistic effects in the human body. Identification of mechanisms, even in high throughput, requires a locus-by-locus interpretation, involving significant time and resources. Despite these challenges, significant progress has been made to identify molecular and biological mechanisms for GWAS loci across many complex diseases and traits.
How Many Association Signals Exist at a Locus?
Historical Context
Candidate-gene and early genome-wide studies identified multiple variants in the same gene in association with a complex trait. For example, four rare coding variants in IFIH1 (MIM: 606951) were associated with lower type 1 diabetes risk,12 and seven coding variants in NOD2 (MIM: 605956) were associated with Crohn disease.13, 14 These examples represent allelic heterogeneity at complex-trait loci. In each example, the coding variants showed independent evidence of association with the disease or trait in larger GWAS analyses.14
Initial GWAS analyses identified genomic regions harboring variants associated with a given trait or disease as loci and typically defined distinct loci according to distance. When trait-associated variants at a locus do not exhibit strong pairwise LD with each other, they represent distinct association “signals.” For example, Willer and colleagues15 aligned GWAS loci for cholesterol and triglyceride levels to previously reported causal variants to demonstrate that the GWAS analysis had identified additional signals of association at these loci. Early studies had limited statistical power to detect loci with two or more significant signals.
Methods
Linkage Disequilibrium. To determine the number of signals at a locus, one strategy is to evaluate pairwise LD between a lead variant and other variants at the locus. GWAS analyses can define multiple signals within a genomic region on the basis of the LD measure r2 in a selected reference population. GWASs have used different LD thresholds and different reference panels. For example, a schizophrenia GWAS used a threshold of r2 < 0.1 to identify 128 independent signals at 108 loci,16 and a coronary artery disease GWAS used a threshold of r2 < 0.2 to identify 104 independent signals at 46 loci;17 both used 1000 Genomes Project data as a reference panel.
Conditional Analysis. An additional strategy for identifying multiple signals at a locus is conditional association analysis. An initial lead associated variant is included as a covariate in association analyses testing other nearby variants; if a nearby variant remains significant, it is considered a conditionally distinct additional signal. When individual-level genotype and trait data are available, stepwise conditional analysis can be performed by including each newly identified lead variant as an additional covariate. When only summary-level association statistics from GWAS meta-analysis are available, a conditional and joint analysis option in the Genome-wide Complex Trait Analysis (GCTA) software18 uses estimated LD from a provided reference sample to identify conditionally distinct signals. Significance thresholds for additional signals at a locus are typically set to account for the number of variants tested, either within a locus or genome-wide. Limitations of defining signals on the basis of LD in a reference panel are that reference panels missing variants analyzed in the association study can fail to detect signals and that population differences between the reference and GWAS samples can cause signals to be mis-characterized.
Haplotype Association. When individual-level genotype and trait data are available, haplotype association analysis can help interpret the inheritance patterns of signals that are not fully independent from one another. Variant alleles that are present on the same or different homologous copies of a genomic region can have differing physiological consequences. Several methods are available for estimating haplotypes from genotype data.19 Haplotypes are then tested for association with a trait either via one test per haplotype or through a global test for all haplotypes in a region.20, 21 Estimates of haplotype effect sizes and directions can aide interpretation of multiple nearby signals.22, 23, 24
Multiple Signals at GWAS Loci
Loci that appear to consist of only one signal can be found to harbor multiple signals after denser genotyping, conditional analysis, or the inclusion of more samples or ancestry groups. Using 1000 Genomes instead of HapMap as an imputation reference panel to analyze the same individuals detected additional loci and different lead variants,25 and genome sequencing by the UK10K Consortium identified additional low-frequency and rare alleles that had not been detected by array-based genotyping.26 Conditional analysis in larger sample sizes of GWAS meta-analyses has detected additional signals at established loci for many phenotypes; for example, an early report identified additional signals at 26 loci for lipids,27 and a more recent study of lipids in a multi-ancestry cohort identified 121 new association signals, including 15 additional signals at established loci.28 In another example, a blood pressure analysis accounting for smoking behavior identified nine novel signals at known loci, including eight that were detected only through the inclusion of data from study subjects of all ancestries.29
As more signals are identified, the definition of a “locus” can change. Signals can span distances of hundreds of kilobases, and a single 500 kb locus containing two signals could easily be defined as two separate loci with narrower spans. A study of 36 blood cell traits reported signals without reporting loci; investigators used stepwise multiple regression to identify 3,755 conditionally distinct associations that clustered into 2,706 LD-group signals.30
Given the growing number of signals identified for some traits, determining whether association signals are new or previously reported is becoming more challenging. Some studies have aimed to identify additional new signals by simultaneously conditioning on all known signals. Such “global” conditional analyses can include hundreds of variants as covariates in a GWAS. For example, an age-related macular degeneration GWAS using a modified global conditional analysis identified 52 signals across 24 loci,31 and a GWAS of lipoprotein particle subclasses conditioned on 885 variants at 157 previously identified lipid loci and identified five novel signals.32
Identifying multiple signals at a locus can help explain some of the “missing heritability” of complex diseases and traits. A stepwise conditional analysis that identified seven signals at three loci associated with fetal hemoglobin levels increased the explained heritable variation in fetal hemoglobin from 38.6% to 49.5%.33 Re-sequencing at five low-density lipoprotein cholesterol (LDL-C) loci identified additional signals and increased the estimate of heritability from 3.1% to 6.5%,34 and including additional ancestries further increased lipid heritability estimates from 1.3- to 1.8-fold across all signals and traits.35 Similar patterns were identified in expression QTL (eQTL) associations—9% of cis-eQTL loci showed evidence of a secondary signal, resulting in a 31% average increase in explained phenotypic variance.22
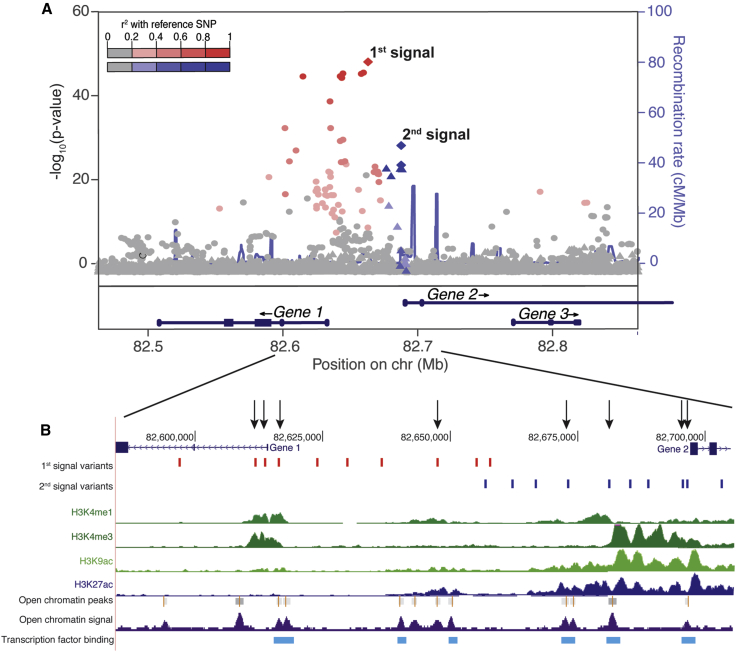
Identifying additional signals enables additional mechanisms to be characterized. For example, variants in three enhancers representing association signals bind three different transcription factors to influence expression of RET (MIM: 164761), leading to Hirschsprung disease.36 In addition, the TCF7L2 (MIM: 602228) locus for type 2 diabetes initially appeared to consist of a single signal, and early variant characterization suggested that rs7903146 affected islet enhancer activity.37, 38 Now, eight signals at the TCF7L2 locus have been reported to be associated with diabetes risk, and several do not overlap islet regulatory elements.39 One or more of the new signals could affect other mechanisms of TCF7L2 regulation, including alternative splicing, expression in other tissues, or both.40 In these examples, the additional signals could target the same candidate gene, but signals could also target different nearby genes. In a recent analysis of eQTLs at GWAS loci, Gamazon et al. observed more than one colocalized gene and one tissue at more than 50% of signals.41 Nearby signals that target different genes or transcripts, possibly with different mechanisms across cell types, could be especially common in gene-dense regions (Figure 2).
Figure 2.
Hypothetical GWAS Locus with Two Signals that Affect Two Genes
(A) Plot of association for two signals within 100 kb at a single GWAS locus. The first signal is shown by red circles, and the second is shown by blue triangles. The intensity of color corresponds to the strength of LD between the lead variant and other variants in the signal.
(B) Hypothetical regulatory marks overlapping the positions of candidate variants. Arrows point to variants that overlap predicted regulatory regions: four for signal 1 and four for signal 2. Signal 1 variants could target gene 1, and signal 2 variants could target gene 2 because variants are located in each respective promoter.
Haplotype analysis can aid the interpretation of multiple signals. Identifying shared haplotypes between alleles of multiple signals can help explain why a variant with low initial evidence of association becomes much more significant after being conditioned on a nearby variant and why a variant with strong initial evidence of association becomes less significant but still meets a significance threshold.22 Haplotype analysis can also help interpret the mechanistic consequences of regulatory and coding variants at the same locus.23, 24 In a study of G6PC2 (MIM: 612108) missense variants associated with fasting glucose, single-variant association results showed an apparent discrepancy with results of cellular functional studies. Haplotype analysis explained the discrepancy by showing that the glucose-lowering allele of the coding variant was always inherited with the glucose-raising allele of a more common noncoding GWAS signal.42 Haplotype analysis can be especially relevant to identifying the functional consequences of variants at multi-signal loci.
Conclusions
As GWAS sample sizes increase, the observable complexity of association signals at individual GWAS loci is increasing. Multiple signals exist at many GWAS loci, and a pattern is emerging whereby the strongest GWAS loci are often influenced by multiple nearby association signals. These multiple signals represent more of the disease or trait heritability than initial signals, and the additional candidate variants can have distinct mechanisms affecting the associated trait or disease, such as variants in different regulatory elements that regulate different genes. Alleles at distinct, but not completely independent, signals can act together through haplotypes. We encourage researchers to consider the possibility that more than one signal contributes to a GWAS locus as a valuable step in accurately delineating the mechanisms at GWAS loci. Considering more than one signal can be especially helpful when the direction of effect of a signal appears inconsistent with other data. When complex relationships between signals are identified, consider the potential contribution of gene regulation, and embrace the opportunity to identify potentially novel interacting genetic mechanisms.
What Are the Candidate Causal Variant(s)?
Historical Context
Identifying candidate causal variants underlying GWAS signals is valuable in helping to identify target genes for GWAS loci because the gene(s) responsible for an association are often not clear, and identifying causal links allows variant-gene links to be validated. The value of identifying candidate causal variants can be questioned; however, specific candidate causal variants provide a data-driven link to genes rather than relying on assumptions about the connections to specific genes and their directions of effect to alter the amount or function of gene products. At noncoding GWAS loci, identifying candidate causal variants further informs our understanding of gene regulation, especially distances at which variants and regulatory regions can act and mechanisms by which variants affect protein-coding RNAs directly and via antisense, long noncoding, or other molecular moieties.
For monogenic disorders, disease-associated chromosomes are typically found to contain a single causal variant. For complex traits, a single causal variant per GWAS signal is the simplest explanation, and although such mechanisms have been described,43, 44, 45, 46, 47 it is becoming increasingly evident that multiple variants can be causal at a single GWAS signal.48, 49, 50 That is, for complex traits, allelic heterogeneity exists at the level of both the locus (multiple signals) and the signal (multiple causal variants).
Edwards et al.10 noted that “… variants are unlikely to act alone, and the importance of combinatorial effects should be considered.” In one study, the authors suggested that “throughout the evolution of species it is haplotype blocks, rather than individual genes and mutations, that serve as the fundamental unit of inheritance” on the basis of highly inbred yeast strains.8 These observations further support the potential role for multiple variants at a single locus. Variants can act in concert in promoters, enhancers, repressors, and coding regions, adding to the complexity of determining mechanisms at GWAS loci.
Methods
Statistical Fine-Mapping. As a first step in identifying candidate causal variants in humans, the strongest GWAS variants and/or variants in LD with the most significantly associated variant at each signal are considered candidates. Typically, an LD r2 threshold of 0.7 or 0.8 is used for determining candidate variants at a GWAS locus; however, various studies use different, less stringent thresholds (e.g., r2 > 0.5) given the strong contribution of allele frequency to r2 estimates or population differences between samples and reference panels.
Statistical fine-mapping uses association statistics to predict which variants are more likely to be causal. Analyses fit in two broad categories: (1) prioritizing variants on the basis of association statistics and LD and (2) Bayesian methods that assign posterior probabilities of causality to each variant (Table 2), as reviewed recently.52 Methods differ in the required input (such as original genotype-trait data versus summary statistics), assumptions (such as one versus more than one potential causal variant), and output. Some methods, such as CAVIAR,53 allow for an arbitrary number of candidate causal variants and return a list, or “credible set” of variants. FINEMAP explores a set of the causal configurations of variants in a region.54 PAINTOR,55 fGWAS,56 and deterministic approximation of posteriors (DAP)57 incorporate functional annotations in the analysis. Performing fine-mapping across diverse populations (e.g., with MANTRA51) can further refine a shared signal on the basis of differing LD between the populations; however, this approach assumes a shared variant between populations, which might or might not reflect the true causal variants within each population. In most cases, statistical fine-mapping predicts a set of variants that are likely to be causal and can then be further examined for functional effects.39, 58 A notable limitation of these approaches is that they analyze only the variants provided; other variants not included as a result of failed genotyping, poor imputation, or other exclusions (e.g., indels and triallelic variants) cannot be considered candidates. The approaches can be sensitive to differences in evidence of association as a result of sample-size variation or inaccurate imputation, and some methods are limited by computation, reducing the number of variants that can be analyzed simultaneously.
Table 2.
Toolbox for Identifying and Annotating Candidate GWAS Variants
These example tools are intended as a starting point for researchers to identify available tools.
Variant Annotation. The effects of coding variants can be interpreted more directly than noncoding variants and are thus frequently the first variants considered for causality. The effects of coding variants can be predicted by computational algorithms, including SIFT,59 PolyPhen-2,60 MutationTaster2,61 CADD,62 MAPPIN,63 and others.64 These methods consider sequence conservation, protein structure, and amino acid properties to predict the effect of missense coding variants. Some methods, such as MutationTaster2 and CADD, also integrate functional genomic data (e.g., DNase I hypersensitivity). The MAPPIN algorithm additionally includes post-translational modifications of proteins, biological networks, and allele frequency to determine whether a variant is predicted to be deleterious or not. Cell or organism models can show the effect of a coding variant in a model system. Variants within or near the conserved splice site between exons can affect splicing, and deleterious effects can be predicted with tools, as reviewed by Jian et al.65
Noncoding variants can be annotated by colocalization with genomic regulatory regions in relevant cell types. Large consortium efforts, including the Encyclopedia of DNA Elements (ENCODE),66 the Roadmap Epigenomics Project,67 and the International Human Epigenome Consortium,68 have created robust datasets for many cell and tissue types to describe regions characteristic of regulatory activity. These datasets include chromatin immunoprecipitation sequencing (ChIP-seq) of histone marks often observed at enhancers, promoters, and insulators and transcription factors and open chromatin profiles generated by DNase hypersensitivity sequencing (DNase-seq), formaldehyde-assisted isolation of regulatory elements sequencing (FAIRE-seq), and the assay for transposase-accessible chromatin using sequencing (ATAC-seq). Algorithms can be used to assess which annotations are enriched in association results (e.g., GREGOR69 and GARFIELD70), to annotate variants at a signal (e.g., HaploReg and RegulomeDB), and predict functional consequence of noncoding variants (e.g., CADD and GWAVA) (Table 2).
Sequencing data generated from ChIP-seq, DNase-seq, FAIRE-seq, or ATAC-seq can be used for identifying heterozygous sites of allelic imbalance in transcription factor binding or chromatin accessibility. A site of allelic imbalance occurs when a sample has disproportionate sequencing reads for each allele in comparison with the expected 50:50. An important aspect of evaluating allelic imbalance is to align reads by using a strategy that avoids bias toward the reference genome allele at the expense of an alternate allele.71 Numerous methods have been described for the identification of allelic imbalance in high-throughput sequence data.72, 73, 74 These strategies require sufficient reads to detect imbalances and require the sequenced sample to be heterozygous at a position of interest.
Experimental Analysis. Experimental analysis can determine whether variants show allelic differences in gene function or regulation, such as allele-specific effects on gene expression levels.47, 75 Experimental analysis of coding and splicing variants requires gene-specific assays that include examining protein function and downstream phenotypes in cells and model organisms.76 Many approaches also exist for testing regulatory variants in functional experiments.77
Transcriptional reporter assays test variant alleles located in regulatory regions for differences in transcriptional activity. When an individual regulatory region is analyzed, the regions surrounding an associated variant are cloned into a vector containing a reporter gene, usually luciferase or GFP, and transfected into a cell line or transiently expressed in a model organism. The activity of reporter genes is measured, and the variant alleles are compared for the detection of any allelic differences in transcriptional activity. Transcriptional reporter assays can also be performed in a high-throughput manner with massively parallel reporter assays (MPRAs), in which hundreds of regulatory regions are tested simultaneously.78 Reporter assays require the presence of the transcription factors that drive allelic differences in activity, so selection of cell type and context, such as differentiation state and stimuli, is important. The limitations of reporter assays in low or high throughput are that (1) the size of the cloned segment and the location of the variant in the segment and in relation to the transcriptional start site can affect detection of allelic differences, leading to at least false-negative results and possibly false positives, and (2) the variant is removed from the chromosomal context; however, this latter limitation can be somewhat remedied by lentiviral transduction and recombineering technologies that allow the construct to incorporate into the genome.
Protein binding assays—including electrophoretic mobility shift assays (EMSAs), DNA-affinity pull-downs, and ChIP experiments—are used to identify variant alleles that bind transcription factors differentially. EMSAs are an in vitro approach that visualizes nuclear protein complexes that bind to ∼20–100 bp DNA probes surrounding a candidate variant. The DNA-protein complexes are visualized on a gel, and the identity of the transcription factor can be determined with antibodies to transcription factors predicted by conserved transcription factor binding motifs or identified by ChIP-seq datasets. DNA-affinity pull-downs are similar to EMSAs: all DNA-protein complexes are captured by a probe including a candidate variant allele and visualized on a gel. Proteins in allele-specific bands are identified by mass spectrometry. Allelic differences in transcription factor binding can also be evident in differences in ChIP-seq or ChIP-qPCR between samples of different genotypes.48 Protein binding assays can also be performed in high throughput.79, 80
An increasingly popular method for determining the function of regulatory variants and elements is to use CRISPR-Cas9 genome editing to delete the regulatory region, create the alternate allele of a variant, or alter the epigenome and regulatory regions with CRISPRi.81, 82 After genome editing, assays of gene expression and/or gene function are performed. Deleting a regulatory element or substituting the alternate allele allows for direct observation of phenotypic effects in the native chromatin context. Challenges that arise in optimizing CRISPR-Cas9 genome editing include the following: it is difficult to edit many disease-specific cell types, targeting can create mutations at unwanted sites, the deletion of a regulatory element might not have the same effect as that of a single-nucleotide change, and editing in model organisms can create mosaicism.
Finally, model organisms can be used to model the effect of candidate causal variants. Reporter assays can be performed in model organisms, often in mice or zebrafish, for assessing variant effects in the context of multiple cell types and controllable environmental conditions. Genome editing in organisms allows more complex drug- or chemically inducible effects to be characterized. Model organisms have the benefit of providing the context of an entire biological system but can sometimes show phenotypic effects that are not consistent in humans.
Candidate Variants at GWAS Signals
Fine-mapping and computational approaches often detect multiple candidate causal variants at individual GWAS loci. For example, in a recent GWAS meta-analysis of type 2 diabetes, only 18 of 380 signals resulted in a single-variant credible set.39 Examination of autoimmune GWAS loci suggested a “multiple enhancer variant” hypothesis, whereby multiple associated variants in clusters of enhancers work together to alter gene expression.49 CAVIAR fine-mapping of skin pigmentation GWAS loci resulted in multiple predicted causal variants at most loci.58 Fine-mapping analyses can reduce the list of candidate variants at a GWAS locus.
Statistical fine-mapping methods use different models and can generate inconsistent results. Methods make different assumptions about the contributions of underlying variants and apply different strategies incorporating genetic association, LD, and functional annotation. At the ANGPTL8 (MIM: 616223) locus, associated with high-density lipoprotein (HDL) cholesterol across populations, three fine-mapping methods (MANTRA, CAVIAR, and PAINTOR) of prioritizing candidate variants generated differing results;83 MANTRA identified a credible set of ten variants, CAVIAR identified a credible set of 24 variants by using Finnish association data and a credible set of two variants by using African American association data, and PAINTOR identified ten, seven, and five variants in Finnish, African American, and the combined studies, respectively. Of the 39 variants identified by least one method, only 12 were identified in at least two analyses, and only four were identified in all three analyses. A candidate causal variant that was identified by all three methods showed significant allelic differences in two assays of regulatory function; however, further experiments are needed to determine the full molecular mechanism.
Great progress has been made in identifying regulatory mechanisms at GWAS loci. Among numerous examples, at the PHACTR1 (MIM: 608723) locus (associated with vascular disease), rs9349379 was identified as a regulator of EDN1 (MIM: 131240) expression by evidence from enhancer signatures in heart tissue and genome editing.47 At the 6q22.1 locus (associated with prostate cancer), rs339331 risk alleles showed increased expression of RFX6 (MIM: 612659), increased binding of HOXB13, increased enhancer histone mark H3K4me2, and altered cell morphology and adhesion.84 At a PARP1 (MIM: 173870) locus associated with melanoma, the risk allele showed increased expression of PARP1 and decreased RECQL binding. Interestingly, RECQL overexpression in melanoma cells resulted in significant allelic differences in transcriptional activity, whereas basal levels of RECQL resulted in no allelic differences, suggesting that cellular context can be important for identifying functional effects of GWAS variants.75
Multiple variants within a single GWAS signal have shown evidence of functional effects. For example, at the MFSD12 (MIM: 617745) locus (associated with skin pigmentation), six variants showed allelic differences in transcriptional reporter luciferase assays.58 Similarly, at the GALNT2 (MIM: 602274) locus (associated with HDL cholesterol), at least two regulatory variants were described as having effects on transcriptional reporter luciferase assays, in vitro protein binding, and allelic imbalance in ChIP-seq and DNase reads.48 Four regulatory variants could contribute to the mechanism at the HKDC1 (MIM: 617221) locus (associated with gestational hyperglycemia).85 MPRAs identified 32 functional variants at 23 GWAS loci associated with red blood cell traits, and targeted genome editing showed that three functional variants at one locus affect transcription of SMIM1 (MIM: 615242), RBM38 (MIM: 612428), and CD164 (MIM: 603356) and that RBM38 is involved in erythropoiesis.86 Multiple variants can be tested together in transcriptional reporter assays, but the distance between variants can present a problem for cloning, and plasmids do not provide the native chromatin context that is most likely important for the regulatory regions to act together. As genome-editing methods are optimized for cell types of interest, it will be important to test multiple variants together in vivo to determine the effects of haplotypes and to truly delineate the complex mechanisms. These functional-mechanism success stories highlight the great progress in understanding genetic contribution to disease in the post-GWAS era.
The TERT (MIM: 187270) locus (associated with breast cancer, ovarian cancer, and telomere length) provides an example of both multiple signals and multiple variants. This GWAS locus consists of at least three signals; signals 2 and 3 are distinct but not completely independent (r2 = 0.33).87 At the first signal, three regulatory variants located in the TERT promoter decrease transcriptional activity in breast and ovarian cancer cell lines. At the second signal, one variant in a TERT intron increases transcriptional activity. At the third signal, a variant alters TERT splicing, resulting in a premature stop codon and truncated protein. The mechanistic connections for multiple variants provide strong evidence for TERT as a plausible target gene.
Conclusions
Multiple candidate causal variants at a single GWAS locus adds complexity to delineating the molecular mechanism at the locus. If one functional variant is identified without full evaluation of all potential candidates, additional variants could contribute, and part of the mechanistic impact on a gene or genes could be missed. The effect of individual variants could be small and might not be observed in functional experiments, perhaps because the effect is observed in only a specific cellular environment or in combination with other variants. Additionally, a variant that affects a gene in functional assays does not necessarily demonstrate that it causes trait variation or a disease; further evidence is needed to prove this connection definitively. Many new tools and methods need further vetting to determine their effectiveness for a given situation. Modeling a single variant in some assays and systems could fail to exhibit a sufficient genetic or physiological consequence if multiple candidate causal variants act together to affect the gene of its function. Identifying different variants in different assays (e.g., transcriptional activity and chromatin interaction) can lead to seemingly inconsistent evidence when assay variability or missing data are responsible. In these cases, we encourage researchers to identify a consistent direction of allelic effect across multiple experiments, which together can provide strong conclusions about candidate causal variants at GWAS loci.
What Are the Target Gene(s)?
Historical Context
Identifying the target gene(s) at a GWAS locus is a fundamental part of elucidating the molecular mechanism because these genes provide a key to understanding the pathogenic processes and provide potential new targets for drug development. However, the target genes remain largely unknown for most GWAS loci. Although some early studies of complex genetic traits were designed on the basis of expecting perhaps tens of contributing genes, GWASs have found that hundreds or thousands of genes might contribute to complex genetic traits.
A large number of genes contributing to a trait or disease is consistent with the possibility that multiple susceptibility genes are located relatively close to each other, even at a single locus (Figure 2). GWAS loci can have multiple genes that appear to be good functional candidates on the basis of gene function and expression, coding variants, chromosome interactions, and/or literature review. Although one candidate causal gene per locus is perhaps still the most likely scenario, GWAS variants can affect multiple genes, perhaps at multi-signal loci or in gene-dense regions.
Methods
Colocalized Variant Association with Gene Expression. One method of identifying target genes for GWAS signals is identifying colocalized eQTL associations. eQTL analysis identifies variants associated with the RNA levels of genes (or transcripts, isoforms, or exons), usually in a single tissue or cell type. Whereas some eQTLs are shared across tissues, others are tissue specific.88 Typically, representative GWAS variants are examined in eQTL datasets for the identification of GWAS variants that show significant association with the expression level of one or more genes. However, the GTEx Consortium reported that a remarkable 92.7% of common variants tested show nominal association (p < 0.05) with the expression level of at least one gene in at least one tissue.88 In addition, given the very strong associations that can be observed between variants and gene expression level, noncausal variants in only moderate LD with candidate causal regulatory variants can also show significant association with gene expression levels. Hence, further analyses are required to determine whether the same variant(s) underlying the GWAS trait association are also likely to be the variant(s) that affect the RNA levels of the eQTL gene.
In the simplest scenario, a lead GWAS variant is also the lead variant associated with a gene in an eQTL study using samples from the same ancestral population. In this setting, eQTL and GWAS associations are considered to be colocalized through a comparison of the lead GWAS and eQTL variants. When the lead GWAS and eQTL variants are identical or LD r2 between them is high (e.g., r2 > 0.8 or 0.9), then the signals can be colocalized. A recent analysis of 3,718 independent GWAS signals found that 58.0% were in LD (r2 > 0.8) with at least one eQTL and that 27.8% were in LD with the best eQTL variant for a gene.41 Conditional analyses in the eQTL dataset can provide further support for colocalization; if the association between the lead eQTL variant and gene expression is no longer significant after conditioning on the GWAS variant and if the association between the lead GWAS variant and gene expression is no longer significant after conditioning on the lead eQTL variant, then the signals are typically considered colocalized.
Other methods more fully assess colocalization of GWAS and eQTL signals by using additional statistical tests, and some methods can be applied to summary association statistics and/or incorporate variant annotation. For example, COLOC applies a Bayesian procedure to estimate the posterior probabilities that a variant is causal in both GWASs and eQTL studies,89 and eCAVIAR applies a Bayesian procedure that allows for more than one candidate causal variant.90 Summary Mendelian randomization in conjunction with a test for heterogeneity in dependent instruments (HEIDI) tests whether gene expression and a trait are associated because of a shared candidate causal variant and can distinguish that model from two or more distinct genetic variants that are in LD and independently affect the gene expression level and the trait.91 Limitations of using eQTLs to identify target genes include that eQTL datasets can still be underpowered to detect associations or unavailable in appropriate cell types or contexts, the LD structure might not be identical between available GWAS and eQTL datasets, and colocalization approaches can be computationally intensive and not robust to the presence of multiple eQTL signals, leading to potential false-positive and -negative colocalization. Finally, evidence of an eQTL colocalized with a GWAS signal does not necessarily mean that expression of that gene mediates the effect of the signal on the trait.
Similar to eQTL associations, allelic imbalance in gene expression can identify potential target genes. Allelic expression imbalance (AEI) analysis involves examining the cDNA or RNA-sequencing reads of genes of interest in individuals heterozygous for a transcribed variant. If the ratio of reads from each allele deviates from 1:1, the correlated alleles of a noncoding and transcribed variant can determine cis-acting variants. Advantages of this approach are that the two variant alleles are assayed in the same environment and that significant differences can be detected in smaller sample sizes than needed for eQTLs. Disadvantages of AEI are that some genes do not contain common variants in available samples and that especially in small sample sizes, a variant can exhibit AEI merely through moderate LD with a candidate causal regulatory variant.
Predicted Gene Expression Association Studies. A complementary approach to prioritizing candidate genes and direction of effect is predicted expression association studies. In this approach, GWASs and eQTL studies are integrated to identify disease associations on the basis of sets of variants that influence gene expression. The portion of gene expression due to genetic variants is estimated from reference eQTL studies, and these estimates are used for predicting gene expression in larger GWAS, where the imputed gene expression can be correlated to the trait for the identification of candidate genes.92, 93, 94 These strategies increase power to detect genes that exhibit differences in genotype-dependent expression patterns, although power is reduced by pleiotropy, and false positives can be identified as a result of LD and the complex genetic architecture of GWAS loci.57, 95 Nonetheless, these studies can be used to help identify candidate genes for complex traits.96, 97
Chromatin Conformation. Another approach to identifying target genes is identifying contact between GWAS variants and the promoters and transcription start sites of target genes by using chromatin conformation assays. Assays such as 3C, Hi-C, and Capture-C identify physical interactions and loops in the genome,98, 99 and these interactions can differ by cell and tissue type. Locus-specific assays can be performed to test interactions between GWAS variant regions and promoters of nearby target genes.100, 101, 102, 103 Genome-wide datasets can be used as a resource for detecting the physical interactions between GWAS variants and nearby gene promoters or established enhancers. Genome-wide chromatin conformation datasets have been generated in multiple cell types104, 105, 106 and can be visualized with tools such as the 3D Genome Browser,107 the Hi-C Unifying Genomic Interrogator (HUGIn),108 or Juicebox.109 Resolution of the chromosomal regions involved in the interaction depends on the assay and/or sequencing depth.105, 110 Cell type is an important consideration given that chromatin conformation assays identify active regulatory elements, which can differ between cell types. Limitations of using chromatin contacts to identify candidate genes are that significance thresholds are not well established, the strategy is uninformative when the candidate variants are located in close proximity to a target gene as a result of the large number of chance contacts between adjacent genome sites, chromatin loops can be general mechanisms of gene regulation and not relevant to GWAS mechanisms, and physical interaction of chromatin does not guarantee a consequence of that interaction.
Coding Variation. Coding variants in genes that are associated with a complex disease or trait can point to target genes at a locus. Numerous resources exist for annotating variants as nonsense, frameshift, and splice altering, all of which are expected to lead to loss of function of a gene,111 and for predicting which missense variants are most likely to alter protein function according to evolutionary, biochemical, and structural information. In addition, multiple, typically rare, coding variants can independently associate with the same phenotype as a GWAS signal. For example, at a GWAS locus for type 2 diabetes, a study of coding variants in multiple populations identified 12 rare protein-truncating variants in SLC30A8 (MIM: 611145), associated with decreased disease risk,112 and of 31 rare (MAF < 1%) coding or splice-site variants newly identified to be independently associated with hematological traits, 30 mapped to loci previously implicated in hematopoiesis by GWASs.113 However, coding variants are not necessarily causal and can be in LD with variants affecting other genes.114, 115 When mechanistic consequences are considered, coding variants located at GWAS loci can help recognize biologically relevant genes.
Functional Studies. Functional studies of genes at GWAS loci can provide insight into target genes. Studies including overexpression, knockdown, or knockout of a target gene in cells or model organisms with the use of plasmids, viruses, oligos, or CRISPR-Cas9 approaches can point to genes with relevant biology.116 For example, functional studies in mice identified a role of CDKAL1 (MIM: 611259) in insulin secretion, and variants near CDKAL1 are associated with type 2 diabetes.117 Studies of gene function can provide insight into the biochemical pathways, protein interactions, and relevant cell or tissue types. Of note, whereas recapitulating some phenotypes in model systems is straightforward, recapitulating many others, especially neurological phenotypes, is very difficult. Deletion or substitution of a noncoding region containing candidate variants and subsequent analysis of gene expression and/or function can also point to target genes.46, 47 Another approach is to epigenetically alter a regulatory element by using a “dead” Cas9 with no nuclease activity tethered to an “epigenetic switch” consisting of chromatin modifying domains to mimic either an enhancer or a repressor.103, 116 Studies in human cells are of particular interest when a gene function is cell autonomous, and experiments in model organisms are especially informative when the biological process or pathway and disease context is well represented in the organism.
Candidate Genes at GWAS Signals
Evaluation of eQTLs colocalized with GWAS signals can point to one or more than one target gene. For example, a study of seven autoimmune diseases found colocalized eQTLs in immune cell types for 91 unique GWAS loci, including more than one gene (up to as many as 12) at 31 loci.118 A study of cardiometabolic traits found colocalized subcutaneous adipose tissue eQTLs for 109 GWAS loci, including more than one gene (up to as many as five) at 25 loci.119 Analyses of GTEx data found that among the GWAS signals that co-localized with an eQTL in one or more tissues, up to 62% of the signals co-localized with more than one gene.41, 88 In addition, recent eQTL studies have identified conditionally distinct eQTL signals, including some that colocalize with GWAS signals.88, 120, 121, 122 Although the associated changes in gene expression levels might not all have an effect on phenotype, the many GWAS loci with more than one colocalized eQTL provide many examples where more than one gene can be causal.
Although chromatin interactions often suggest multiple plausible target genes, they can provide stronger evidence when observed with colocalized eQTLs or functional evidence. At the ARL15 locus (associated with insulin resistance traits and type 2 diabetes), HiC data in multiple cell types show significant interaction between the GWAS variant region and the FST (MIM: 136470) promoter,108 ∼500 kb away, and these data support evidence that the GWAS signals exhibit significant colocalized eQTLs for FST,83 although functional studies support a role for ARL15 in insulin secretion.123 At a schizophrenia-associated locus, variants were linked by both HiC and genome editing to FOXG1 (MIM: 164874) >700 kb away,124 and at the TMEM106B (MIM: 613413) GWAS locus (associated with dementia), a variant in a CTCF binding site altered chromatin architecture to change TMEM106B expression and cell toxicity.101 Recent analysis with ATAC-seq open-chromatin data identified over 15,000 interactions between distal regions of open chromatin, and allele-specific effects were inferred from the read data, resulting in improved fine-mapping of eQTL signals.125
Chromatin interactions with a GWAS variant region can also suggest multiple candidate genes. For example, at a breast-cancer-associated locus at 11q13, the GWAS variant region interacted with two long noncoding RNAs, LINC01488 (MIM: 617696) and CUPID2 (MIM: 617697), in allele-specific 3C experiments.103 At a locus associated with schizophrenia and bipolar disorder, an enhancer variant interacted with two nearby miRNAs by 3C, supported by transcriptional activity and transcription factor binding assays.100 At the FTO (MIM: 610966) locus (associated with obesity), the GWAS variants showed physical chromatin interaction with eight genes, at least two of which (IRX3 [MIM: 612985] and IRX5 [MIM: 606195]) showed genotype-expression association in preadipocytes.102 Although the chromatin interactions might not all lead to an effect on phenotype, the GWAS loci with more than one interacting promoter provide examples where more than one gene can be causal.
Coding variants in genes can point to target genes, and rare coding variants can provide evidence for the target genes of common noncoding variants at the same locus. At the fasting-glucose-associated GWAS locus near G6PC2 and ABCB11 (MIM: 603201), for which the target gene was unknown, exome array data from ∼33,000 individuals identified three rare G6PC2 coding variants associated with fasting glucose,42 suggesting that G6PC2 might also be altered by the noncoding variants. In vitro assays confirmed the effect of the coding variants and established G6PC2 as an effector gene and likely target gene of the noncoding variants at this GWAS locus.
Experiments in model organisms also identify multiple target genes at GWAS loci. At the Agtrap-Plod1 locus (associated with blood pressure), six nearby potential target genes exist. Flister et al. performed mutagenesis in mice to create mutant strains for all six of these genes. They found that five genes showed an effect on blood pressure or renal function, suggesting that there might be multiple target genes at this GWAS locus.126 One important consideration is that gene function is dependent on cell type and cellular context and might not be replicated in humans versus model organisms.
Genome and epigenome editing of regulatory regions pinpoints target genes. A noncoding region near PHACTR1 (MIM: 608723) and EDN1 (MIM: 131240) is associated with five vascular diseases—coronary artery disease, migraine, dissection, fibromuscular dysplasia, and hypertension. Deletion of 88 bp surrounding a fine-mapped SNP resulted in increased expression of EDN1, but not PHACTR1 or four other nearby genes,47 providing initial evidence of EDN1 as a target gene. EDN1 has known roles in the physiology of vasculature, suggesting a mechanism for the associated diseases.47 In a second example, at the ADCY5 (MIM: 600293) GWAS locus (associated with type 2 diabetes), deletion of the orthologous associated regulatory region in rat pancreatic islet cells resulted in decreased ADCY5 expression and reduced insulin secretion, supporting other evidence that ADCY5 is a plausible target gene at the GWAS signal.46 At the 11q13 GWAS locus (associated with breast cancer), epigenetically silencing a regulatory region with nuclease-inactivated Cas9 fused to the Kruppel-associated (KRAB) repressor reduced expression levels of three targeting genes: LINC01488, CUPID2, and CCND1 (MIM: 168461).103 These experiments provide evidence that genome and epigenome editing can validate target genes in vivo.
Emerging molecular QTL associations, such as histone, splice, methylation, metabolite, and other endophenotype QTLs, will aid in the identification of target genes and biological mechanisms. For example, an analysis that combined eQTLs, histone QTLs, splicing QTLs, and methylation QTLs to annotate 41 diseases and complex traits found that these QTLs are strongly enriched with disease heritability and provide complementary information about disease.127 Together, these molecular associations can suggest disease mechanisms.
Conclusions
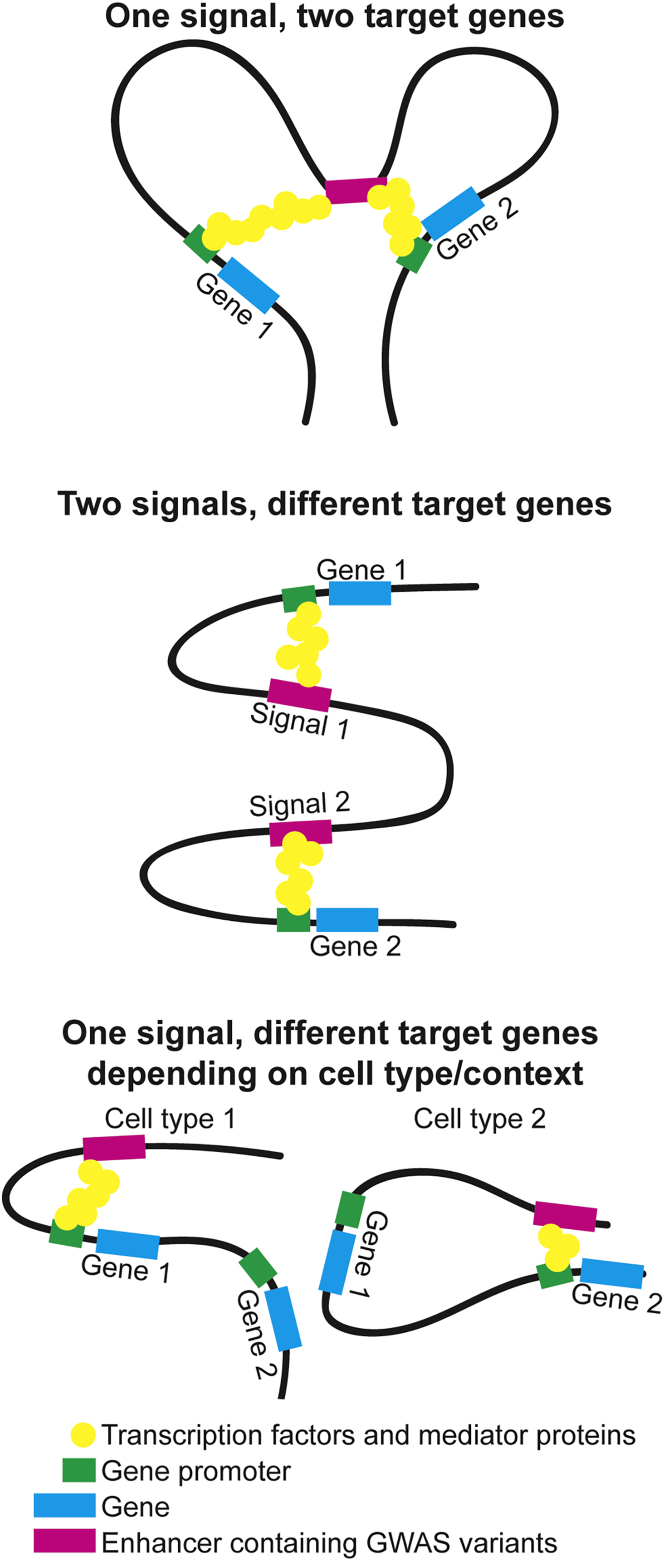
Each approach to identifying genes at a GWAS signal can provide evidence supporting a potential contribution of more than one gene. As expression datasets increase in size; the number of tissues, cell types, and contexts; and gene, isoform, and exon specificity, more colocalized eQTLs are being identified. Similarly, as chromatin-interaction datasets are generated in additional tissues, cell types, and contexts, more GWAS signals can be connected to one or more genes. Functional assays can suggest different genes depending on cell type, cellular environment, or other factors. In addition, two signals at a locus can act on the same or different genes; variants could target multiple genes via chromatin looping or different genes via tissue-specific enhancers (Figure 3). If multiple genes show strong candidacy, researchers should consider pursuing both genes in functional experiments because they could both be true target genes. Lack of support for a gene from any one approach could reflect that data are not available in the appropriate cell type or environmental state. Given limitations in concluding causality, multiple lines of genetic, bioinformatic, and experimental evidence supporting the role of a gene strengthen its candidacy.
Figure 3.
Hypotheses for Multiple Target Genes at a GWAS Locus
Multiple target genes can be present at a single GWAS locus. Three examples show how multiple genes might be affected. At a locus with one GWAS signal, an enhancer containing GWAS variants could target two genes simultaneously, or different genes could be targeted depending on cell type or cellular context. At a locus with two signals, each signal could target different genes. Other mechanisms could exist.
Concluding Remarks
In this review, we have outlined three important aspects of evaluating GWAS loci (Figure 1). Generally, multiple pieces of evidence supporting a gene or variant that affects a complex trait can show a consistent direction and a single mechanism. However, given the contributions of multiple genes and variants at complex-trait loci and the imperfect nature of experimental systems, some evidence might not fit a simple model. When interpreting the results of a computational or experimental analysis, especially unexpected results, researchers should consider that additional signals might exist at a locus and that variants not considered candidates according to LD might nonetheless contribute to the mechanism of the locus. When searching for target genes, consider that variants might act through more than one nearby gene to influence disease. When identifying a variant that exhibits allelic effects on a gene, consider that additional variants might also have functional effects. Finally, when evaluating the biological effects of genes on disease, consider that cell type, cellular context, and multiple molecular mechanisms acting together can affect disease pathogenesis.
As GWASs are performed in more samples and additional populations, more loci with multiple signals and variants will be identified. The future is bright, given that progress is being made more quickly with high-throughput assays and with genome-editing experiments in the native chromatin context. Better statistical methods are continually being developed for identifying and localizing loci, signals, genes, and variants. The field can look forward to a better understanding of gene regulation, biological mechanisms, and disease pathways by closely examining GWAS loci.
Acknowledgments
The authors thank James Davis, Cassandra Spracklen, Chelsea Raulerson, and Kevin Currin for helpful discussion and feedback. This work was supported by National Institutes of Health grants F31 HL127984, U01 DK105561, R01 DK 093757, and R01 DK072193.
References
- 1.Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J. 10 Years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallagher M.D., Chen-Plotkin A.S. The post-GWAS era: From association to function. Am. J. Hum. Genet. 2018;102:717–730. doi: 10.1016/j.ajhg.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cutting G.R. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat. Rev. Genet. 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rees D.C., Williams T.N., Gladwin M.T. Sickle-cell disease. Lancet. 2010;376:2018–2031. doi: 10.1016/S0140-6736(10)61029-X. [DOI] [PubMed] [Google Scholar]
- 6.Legare M.E., Bartlett F.S., 2nd, Frankel W.N. A major effect QTL determined by multiple genes in epileptic EL mice. Genome Res. 2000;10:42–48. [PMC free article] [PubMed] [Google Scholar]
- 7.Dixit S., Kumar Biswal A., Min A., Henry A., Oane R.H., Raorane M.L., Longkumer T., Pabuayon I.M., Mutte S.K., Vardarajan A.R. Action of multiple intra-QTL genes concerted around a co-localized transcription factor underpins a large effect QTL. Sci. Rep. 2015;5:15183. doi: 10.1038/srep15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.She R., Jarosz D.F. Mapping causal variants with single-nucleotide resolution reveals biochemical drivers of phenotypic change. Cell. 2018;172:478–490.e15. doi: 10.1016/j.cell.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Logan R.W., Robledo R.F., Recla J.M., Philip V.M., Bubier J.A., Jay J.J., Harwood C., Wilcox T., Gatti D.M., Bult C.J. High-precision genetic mapping of behavioral traits in the diversity outbred mouse population. Genes Brain Behav. 2013;12:424–437. doi: 10.1111/gbb.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards S.L., Beesley J., French J.D., Dunning A.M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thibodeau S.N., French A.J., McDonnell S.K., Cheville J., Middha S., Tillmans L., Riska S., Baheti S., Larson M.C., Fogarty Z. Identification of candidate genes for prostate cancer-risk SNPs utilizing a normal prostate tissue eQTL data set. Nat. Commun. 2015;6:8653. doi: 10.1038/ncomms9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nejentsev S., Walker N., Riches D., Egholm M., Todd J.A. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugot J.P., Chamaillard M., Zouali H., Lesage S., Cézard J.P., Belaiche J., Almer S., Tysk C., O’Morain C.A., Gassull M. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 14.Rivas M.A., Beaudoin M., Gardet A., Stevens C., Sharma Y., Zhang C.K., Boucher G., Ripke S., Ellinghaus D., Burtt N., National Institute of Diabetes and Digestive Kidney Diseases Inflammatory Bowel Disease Genetics Consortium (NIDDK IBDGC) United Kingdom Inflammatory Bowel Disease Genetics Consortium. International Inflammatory Bowel Disease Genetics Consortium Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deloukas P., Kanoni S., Willenborg C., Farrall M., Assimes T.L., Thompson J.R., Ingelsson E., Saleheen D., Erdmann J., Goldstein B.A., CARDIoGRAMplusC4D Consortium. DIAGRAM Consortium. CARDIOGENICS Consortium. MuTHER Consortium. Wellcome Trust Case Control Consortium Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tewhey R., Bansal V., Torkamani A., Topol E.J., Schork N.J. The importance of phase information for human genomics. Nat. Rev. Genet. 2011;12:215–223. doi: 10.1038/nrg2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaid D.J., Rowland C.M., Tines D.E., Jacobson R.M., Poland G.A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Datta A.S., Biswas S. Comparison of haplotype-based statistical tests for disease association with rare and common variants. Brief. Bioinform. 2016;17:657–671. doi: 10.1093/bib/bbv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wood A.R., Hernandez D.G., Nalls M.A., Yaghootkar H., Gibbs J.R., Harries L.W., Chong S., Moore M., Weedon M.N., Guralnik J.M. Allelic heterogeneity and more detailed analyses of known loci explain additional phenotypic variation and reveal complex patterns of association. Hum. Mol. Genet. 2011;20:4082–4092. doi: 10.1093/hmg/ddr328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khetarpal S.A., Edmondson A.C., Raghavan A., Neeli H., Jin W., Badellino K.O., Demissie S., Manning A.K., DerOhannessian S.L., Wolfe M.L. Mining the LIPG allelic spectrum reveals the contribution of rare and common regulatory variants to HDL cholesterol. PLoS Genet. 2011;7:e1002393. doi: 10.1371/journal.pgen.1002393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castel S.E., Cervera A., Mohammadi P., Aguet F., Reverter F., Wolman A., Guigo R., Iossifov I., Vasileva A., Lappalainen T. Modified penetrance of coding variants by cis-regulatory variation contributes to disease risk. Nat. Genet. 2018;50:1327–1334. doi: 10.1038/s41588-018-0192-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J., Ellinghaus D., Franke A., Howie B., Li Y. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 Data. Eur. J. Hum. Genet. 2012;20:801–805. doi: 10.1038/ejhg.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walter K., Min J.L., Huang J., Crooks L., Memari Y., McCarthy S., Perry J.R., Xu C., Futema M., Lawson D., UK10K Consortium The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann T.J., Theusch E., Haldar T., Ranatunga D.K., Jorgenson E., Medina M.W., Kvale M.N., Kwok P.Y., Schaefer C., Krauss R.M. A large electronic-health-record-based genome-wide study of serum lipids. Nat. Genet. 2018;50:401–413. doi: 10.1038/s41588-018-0064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sung Y.J., Winkler T.W., de Las Fuentes L., Bentley A.R., Brown M.R., Kraja A.T., Schwander K., Ntalla I., Guo X., Franceschini N., CHARGE Neurology Working Group. COGENT-Kidney Consortium. GIANT Consortium. Lifelines Cohort Study A large-scale multi-ancestry genome-wide study accounting for smoking behavior identifies multiple significant loci for blood pressure. Am. J. Hum. Genet. 2018;102:375–400. doi: 10.1016/j.ajhg.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fritsche L.G., Igl W., Bailey J.N., Grassmann F., Sengupta S., Bragg-Gresham J.L., Burdon K.P., Hebbring S.J., Wen C., Gorski M. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis J.P., Huyghe J.R., Locke A.E., Jackson A.U., Sim X., Stringham H.M., Teslovich T.M., Welch R.P., Fuchsberger C., Narisu N. Common, low-frequency, and rare genetic variants associated with lipoprotein subclasses and triglyceride measures in Finnish men from the METSIM study. PLoS Genet. 2017;13:e1007079. doi: 10.1371/journal.pgen.1007079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galarneau G., Palmer C.D., Sankaran V.G., Orkin S.H., Hirschhorn J.N., Lettre G. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat. Genet. 2010;42:1049–1051. doi: 10.1038/ng.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanna S., Li B., Mulas A., Sidore C., Kang H.M., Jackson A.U., Piras M.G., Usala G., Maninchedda G., Sassu A. Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet. 2011;7:e1002198. doi: 10.1371/journal.pgen.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y., Waite L.L., Jackson A.U., Sheu W.H., Buyske S., Absher D., Arnett D.K., Boerwinkle E., Bonnycastle L.L., Carty C.L. Trans-ethnic fine-mapping of lipid loci identifies population-specific signals and allelic heterogeneity that increases the trait variance explained. PLoS Genet. 2013;9:e1003379. doi: 10.1371/journal.pgen.1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chatterjee S., Kapoor A., Akiyama J.A., Auer D.R., Lee D., Gabriel S., Berrios C., Pennacchio L.A., Chakravarti A. Enhancer variants synergistically drive dysfunction of a gene regulatory network in hirschsprung disease. Cell. 2016;167:355–368.e10. doi: 10.1016/j.cell.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyssenko V., Lupi R., Marchetti P., Del Guerra S., Orho-Melander M., Almgren P., Sjögren M., Ling C., Eriksson K.F., Lethagen A.L. Mechanisms by which common variants in the TCF7L2 gene increase risk of type 2 diabetes. J. Clin. Invest. 2007;117:2155–2163. doi: 10.1172/JCI30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaulton K.J., Nammo T., Pasquali L., Simon J.M., Giresi P.G., Fogarty M.P., Panhuis T.M., Mieczkowski P., Secchi A., Bosco D. A map of open chromatin in human pancreatic islets. Nat. Genet. 2010;42:255–259. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahajan A., Taliun D., Thurner M., Robertson N.R., Torres J.M., Rayner N.W., Payne A.J., Steinthorsdottir V., Scott R.A., Grarup N. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 2018 doi: 10.1038/s41588-018-0241-6. Published online October 8, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mondal A.K., Das S.K., Baldini G., Chu W.S., Sharma N.K., Hackney O.G., Zhao J., Grant S.F., Elbein S.C. Genotype and tissue-specific effects on alternative splicing of the transcription factor 7-like 2 gene in humans. J. Clin. Endocrinol. Metab. 2010;95:1450–1457. doi: 10.1210/jc.2009-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gamazon E.R., Segrè A.V., van de Bunt M., Wen X., Xi H.S., Hormozdiari F., Ongen H., Konkashbaev A., Derks E.M., Aguet F., GTEx Consortium Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat. Genet. 2018;50:956–967. doi: 10.1038/s41588-018-0154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahajan A., Sim X., Ng H.J., Manning A., Rivas M.A., Highland H.M., Locke A.E., Grarup N., Im H.K., Cingolani P., T2D-GENES consortium and GoT2D consortium Identification and functional characterization of G6PC2 coding variants influencing glycemic traits define an effector transcript at the G6PC2-ABCB11 locus. PLoS Genet. 2015;11:e1004876. doi: 10.1371/journal.pgen.1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregory A.P., Dendrou C.A., Attfield K.E., Haghikia A., Xifara D.K., Butter F., Poschmann G., Kaur G., Lambert L., Leach O.A. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature. 2012;488:508–511. doi: 10.1038/nature11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen E.K., Randolph A.G., Bhangale T., Dogra P., Ohlson M., Oshansky C.M., Zamora A.E., Shannon J.P., Finkelstein D., Dressen A. SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat. Med. 2017;23:975–983. doi: 10.1038/nm.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sankaran V.G., Ludwig L.S., Sicinska E., Xu J., Bauer D.E., Eng J.C., Patterson H.C., Metcalf R.A., Natkunam Y., Orkin S.H. Cyclin D3 coordinates the cell cycle during differentiation to regulate erythrocyte size and number. Genes Dev. 2012;26:2075–2087. doi: 10.1101/gad.197020.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roman T.S., Cannon M.E., Vadlamudi S., Buchkovich M.L., Wolford B.N., Welch R.P., Morken M.A., Kwon G.J., Varshney A., Kursawe R., National Institutes of Health Intramural Sequencing Center (NISC) Comparative Sequencing Program A type 2 diabetes-associated functional regulatory variant in a pancreatic islet enhancer at the ADCY5 locus. Diabetes. 2017;66:2521–2530. doi: 10.2337/db17-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta R.M., Hadaya J., Trehan A., Zekavat S.M., Roselli C., Klarin D., Emdin C.A., Hilvering C.R.E., Bianchi V., Mueller C. A genetic variant associated with five vascular diseases is a distal regulator of endothelin-1 gene expression. Cell. 2017;170:522–533.e15. doi: 10.1016/j.cell.2017.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roman T.S., Marvelle A.F., Fogarty M.P., Vadlamudi S., Gonzalez A.J., Buchkovich M.L., Huyghe J.R., Fuchsberger C., Jackson A.U., Wu Y. Multiple hepatic regulatory variants at the GALNT2 GWAS locus associated with high-density lipoprotein cholesterol. Am. J. Hum. Genet. 2015;97:801–815. doi: 10.1016/j.ajhg.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corradin O., Saiakhova A., Akhtar-Zaidi B., Myeroff L., Willis J., Cowper-Sal lari R., Lupien M., Markowitz S., Scacheri P.C. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stadhouders R., Aktuna S., Thongjuea S., Aghajanirefah A., Pourfarzad F., van Ijcken W., Lenhard B., Rooks H., Best S., Menzel S. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J. Clin. Invest. 2014;124:1699–1710. doi: 10.1172/JCI71520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris A.P. Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol. 2011;35:809–822. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spain S.L., Barrett J.C. Strategies for fine-mapping complex traits. Hum. Mol. Genet. 2015;24(R1):R111–R119. doi: 10.1093/hmg/ddv260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hormozdiari F., Kostem E., Kang E.Y., Pasaniuc B., Eskin E. Identifying causal variants at loci with multiple signals of association. Genetics. 2014;198:497–508. doi: 10.1534/genetics.114.167908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benner C., Spencer C.C., Havulinna A.S., Salomaa V., Ripatti S., Pirinen M. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–1501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kichaev G., Yang W.Y., Lindstrom S., Hormozdiari F., Eskin E., Price A.L., Kraft P., Pasaniuc B. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10:e1004722. doi: 10.1371/journal.pgen.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pickrell J.K. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wen X., Lee Y., Luca F., Pique-Regi R. Efficient integrative multi-SNP association analysis via deterministic approximation of posteriors. Am. J. Hum. Genet. 2016;98:1114–1129. doi: 10.1016/j.ajhg.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crawford N.G., Kelly D.E., Hansen M.E.B., Beltrame M.H., Fan S., Bowman S.L., Jewett E., Ranciaro A., Thompson S., Lo Y., NISC Comparative Sequencing Program Loci associated with skin pigmentation identified in African populations. Science. 2017;358:eaan8433. doi: 10.1126/science.aan8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 60.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 62.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gosalia N., Economides A.N., Dewey F.E., Balasubramanian S. MAPPIN: a method for annotating, predicting pathogenicity and mode of inheritance for nonsynonymous variants. Nucleic Acids Res. 2017;45:10393–10402. doi: 10.1093/nar/gkx730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eilbeck K., Quinlan A., Yandell M. Settling the score: variant prioritization and Mendelian disease. Nat. Rev. Genet. 2017;18:599–612. doi: 10.1038/nrg.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jian X., Boerwinkle E., Liu X. In silico tools for splicing defect prediction: a survey from the viewpoint of end users. Genet. Med. 2014;16:497–503. doi: 10.1038/gim.2013.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanyal A., Lajoie B.R., Jain G., Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stunnenberg H.G., Hirst M., International Human Epigenome Consortium The International Human Epigenome Consortium: A blueprint for scientific collaboration and discovery. Cell. 2016;167:1145–1149. doi: 10.1016/j.cell.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 69.Schmidt E.M., Zhang J., Zhou W., Chen J., Mohlke K.L., Chen Y.E., Willer C.J. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics. 2015;31:2601–2606. doi: 10.1093/bioinformatics/btv201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iotchkova V., Ritchie G.R.S., Geihs M., Morganella S., Min J.L., Walter K., Timpson N.J., UK10K Consortium. Dunham I., Birney E. GARFIELD - GWAS analysis of regulatory or functional information enrichment with LD correction. bioRxiv. 2018 [Google Scholar]
- 71.Degner J.F., Marioni J.C., Pai A.A., Pickrell J.K., Nkadori E., Gilad Y., Pritchard J.K. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009;25:3207–3212. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reddy T.E., Gertz J., Pauli F., Kucera K.S., Varley K.E., Newberry K.M., Marinov G.K., Mortazavi A., Williams B.A., Song L. Effects of sequence variation on differential allelic transcription factor occupancy and gene expression. Genome Res. 2012;22:860–869. doi: 10.1101/gr.131201.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buchkovich M.L., Eklund K., Duan Q., Li Y., Mohlke K.L., Furey T.S. Removing reference mapping biases using limited or no genotype data identifies allelic differences in protein binding at disease-associated loci. BMC Med. Genomics. 2015;8:43. doi: 10.1186/s12920-015-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harvey C.T., Moyerbrailean G.A., Davis G.O., Wen X., Luca F., Pique-Regi R. QuASAR: quantitative allele-specific analysis of reads. Bioinformatics. 2015;31:1235–1242. doi: 10.1093/bioinformatics/btu802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi J., Xu M., Makowski M.M., Zhang T., Law M.H., Kovacs M.A., Granzhan A., Kim W.J., Parikh H., Gartside M. A common intronic variant of PARP1 confers melanoma risk and mediates melanocyte growth via regulation of MITF. Nat. Genet. 2017;49:1326–1335. doi: 10.1038/ng.3927. [DOI] [PubMed] [Google Scholar]
- 76.MacArthur D.G., Manolio T.A., Dimmock D.P., Rehm H.L., Shendure J., Abecasis G.R., Adams D.R., Altman R.B., Antonarakis S.E., Ashley E.A. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ward L.D., Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat. Biotechnol. 2012;30:1095–1106. doi: 10.1038/nbt.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Starita L.M., Ahituv N., Dunham M.J., Kitzman J.O., Roth F.P., Seelig G., Shendure J., Fowler D.M. Variant interpretation: Functional assays to the rescue. Am. J. Hum. Genet. 2017;101:315–325. doi: 10.1016/j.ajhg.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stormo G.D., Zhao Y. Determining the specificity of protein-DNA interactions. Nat. Rev. Genet. 2010;11:751–760. doi: 10.1038/nrg2845. [DOI] [PubMed] [Google Scholar]
- 80.Jolma A., Yan J., Whitington T., Toivonen J., Nitta K.R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G. DNA-binding specificities of human transcription factors. Cell. 2013;152:327–339. doi: 10.1016/j.cell.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 81.Zhang F., Wen Y., Guo X. CRISPR/Cas9 for genome editing: Progress, implications and challenges. Hum. Mol. Genet. 2014;23(R1):R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 82.Thakore P.I., D’Ippolito A.M., Song L., Safi A., Shivakumar N.K., Kabadi A.M., Reddy T.E., Crawford G.E., Gersbach C.A. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods. 2015;12:1143–1149. doi: 10.1038/nmeth.3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cannon M.E., Duan Q., Wu Y., Zeynalzadeh M., Xu Z., Kangas A.J., Soininen P., Ala-Korpela M., Civelek M., Lusis A.J. Trans-ancestry fine mapping and molecular assays identify regulatory variants at the ANGPTL8 HDL-C GWAS locus. G3 (Bethesda) 2017;7:3217–3227. doi: 10.1534/g3.117.300088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spisák S., Lawrenson K., Fu Y., Csabai I., Cottman R.T., Seo J.H., Haiman C., Han Y., Lenci R., Li Q., GAME-ON/ELLIPSE Consortium CAUSEL: an epigenome- and genome-editing pipeline for establishing function of noncoding GWAS variants. Nat. Med. 2015;21:1357–1363. doi: 10.1038/nm.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo C., Ludvik A.E., Arlotto M.E., Hayes M.G., Armstrong L.L., Scholtens D.M., Brown C.D., Newgard C.B., Becker T.C., Layden B.T. Coordinated regulatory variation associated with gestational hyperglycaemia regulates expression of the novel hexokinase HKDC1. Nat. Commun. 2015;6:6069. doi: 10.1038/ncomms7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ulirsch J.C., Nandakumar S.K., Wang L., Giani F.C., Zhang X., Rogov P., Melnikov A., McDonel P., Do R., Mikkelsen T.S., Sankaran V.G. Systematic functional dissection of common genetic variation affecting red blood cell traits. Cell. 2016;165:1530–1545. doi: 10.1016/j.cell.2016.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bojesen S.E., Pooley K.A., Johnatty S.E., Beesley J., Michailidou K., Tyrer J.P., Edwards S.L., Pickett H.A., Shen H.C., Smart C.E., Australian Cancer Study. Australian Ovarian Cancer Study. Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab) Gene Environment Interaction and Breast Cancer (GENICA) Swedish Breast Cancer Study (SWE-BRCA) Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) Epidemiological study of BRCA1 & BRCA2 Mutation Carriers (EMBRACE) Genetic Modifiers of Cancer Risk in BRCA1/2 Mutation Carriers (GEMO) Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 2013;45:371–384, e1–e2. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration & Visualization—EBI. Genome Browser Data Integration & Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis & Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 89.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hormozdiari F., van de Bunt M., Segrè A.V., Li X., Joo J.W.J., Bilow M., Sul J.H., Sankararaman S., Pasaniuc B., Eskin E. Colocalization of GWAS and eQTL signals detects target genes. Am. J. Hum. Genet. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 92.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., Nicolae D.L., Cox N.J., Im H.K., GTEx Consortium A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W., Jansen R., de Geus E.J., Boomsma D.I., Wright F.A. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Manor O., Segal E. Robust prediction of expression differences among human individuals using only genotype information. PLoS Genet. 2013;9:e1003396. doi: 10.1371/journal.pgen.1003396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Veturi Y., Ritchie M.D. How powerful are summary-based methods for identifying expression-trait associations under different genetic architectures? Pac. Symp. Biocomput. 2018;23:228–239. [PMC free article] [PubMed] [Google Scholar]
- 96.Gusev A., Mancuso N., Won H., Kousi M., Finucane H.K., Reshef Y., Song L., Safi A., McCarroll S., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 2018;50:538–548. doi: 10.1038/s41588-018-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mancuso N., Shi H., Goddard P., Kichaev G., Gusev A., Pasaniuc B. Integrating gene expression with summary association statistics to identify genes associated with 30 complex traits. Am. J. Hum. Genet. 2017;100:473–487. doi: 10.1016/j.ajhg.2017.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Belmont A.S. Large-scale chromatin organization: the good, the surprising, and the still perplexing. Curr. Opin. Cell Biol. 2014;26:69–78. doi: 10.1016/j.ceb.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Risca V.I., Greenleaf W.J. Unraveling the 3D genome: genomics tools for multiscale exploration. Trends Genet. 2015;31:357–372. doi: 10.1016/j.tig.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duan J., Shi J., Fiorentino A., Leites C., Chen X., Moy W., Chen J., Alexandrov B.S., Usheva A., He D., Molecular Genetics of Schizophrenia collaboration. Genomic Psychiatric Cohort consortium A rare functional noncoding variant at the GWAS-implicated MIR137/MIR2682 locus might confer risk to schizophrenia and bipolar disorder. Am. J. Hum. Genet. 2014;95:744–753. doi: 10.1016/j.ajhg.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gallagher M.D., Posavi M., Huang P., Unger T.L., Berlyand Y., Gruenewald A.L., Chesi A., Manduchi E., Wells A.D., Grant S.F.A. A dementia-associated risk variant near TMEM106B alters chromatin architecture and gene expression. Am. J. Hum. Genet. 2017;101:643–663. doi: 10.1016/j.ajhg.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Claussnitzer M., Dankel S.N., Kim K.H., Quon G., Meuleman W., Haugen C., Glunk V., Sousa I.S., Beaudry J.L., Puviindran V. FTO obesity variant circuitry and adipocyte browning in humans. N. Engl. J. Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Betts J.A., Moradi Marjaneh M., Al-Ejeh F., Lim Y.C., Shi W., Sivakumaran H., Tropée R., Patch A.M., Clark M.B., Bartonicek N. Long noncoding RNAs CUPID1 and CUPID2 mediate breast cancer risk at 11q13 by modulating the response to DNA damage. Am. J. Hum. Genet. 2017;101:255–266. doi: 10.1016/j.ajhg.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Montefiori L.E., Sobreira D.R., Sakabe N.J., Aneas I., Joslin A.C., Hansen G.T., Bozek G., Moskowitz I.P., McNally E.M., Nóbrega M.A. A promoter interaction map for cardiovascular disease genetics. eLife. 2018;7:e35788. doi: 10.7554/eLife.35788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., Li D., Choudhary M.N.K., Li Y., Hu M. The 3D Genome Browser: A web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19:151. doi: 10.1186/s13059-018-1519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Martin J.S., Xu Z., Reiner A.P., Mohlke K.L., Sullivan P., Ren B., Hu M., Li Y. HUGIn: Hi-C Unifying Genomic Interrogator. Bioinformatics. 2017;33:3793–3795. doi: 10.1093/bioinformatics/btx359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Durand N.C., Robinson J.T., Shamim M.S., Machol I., Mesirov J.P., Lander E.S., Aiden E.L. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016;3:99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jin F., Li Y., Dixon J.R., Selvaraj S., Ye Z., Lee A.Y., Yen C.A., Schmitt A.D., Espinoza C.A., Ren B. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cooper G.M., Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat. Rev. Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 112.Flannick J., Thorleifsson G., Beer N.L., Jacobs S.B., Grarup N., Burtt N.P., Mahajan A., Fuchsberger C., Atzmon G., Benediktsson R., Go-T2D Consortium. T2D-GENES Consortium Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat. Genet. 2014;46:357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mousas A., Ntritsos G., Chen M.H., Song C., Huffman J.E., Tzoulaki I., Elliott P., Psaty B.M., Auer P.L., Johnson A.D., Blood-Cell Consortium Rare coding variants pinpoint genes that control human hematological traits. PLoS Genet. 2017;13:e1006925. doi: 10.1371/journal.pgen.1006925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Huyghe J.R., Jackson A.U., Fogarty M.P., Buchkovich M.L., Stančáková A., Stringham H.M., Sim X., Yang L., Fuchsberger C., Cederberg H. Exome array analysis identifies new loci and low-frequency variants influencing insulin processing and secretion. Nat. Genet. 2013;45:197–201. doi: 10.1038/ng.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mahajan A., Wessel J., Willems S.M., Zhao W., Robertson N.R., Chu A.Y., Gan W., Kitajima H., Taliun D., Rayner N.W., ExomeBP Consortium. MAGIC Consortium. GIANT Consortium Refining the accuracy of validated target identification through coding variant fine-mapping in type 2 diabetes. Nat. Genet. 2018;50:559–571. doi: 10.1038/s41588-018-0084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tak Y.G., Farnham P.J. Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin. 2015;8:57. doi: 10.1186/s13072-015-0050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wei F.Y., Tomizawa K. Functional loss of Cdkal1, a novel tRNA modification enzyme, causes the development of type 2 diabetes. Endocr. J. 2011;58:819–825. doi: 10.1507/endocrj.ej11-0099. [DOI] [PubMed] [Google Scholar]
- 118.Chen L., Ge B., Casale F.P., Vasquez L., Kwan T., Garrido-Martín D., Watt S., Yan Y., Kundu K., Ecker S. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016;167:1398–1414.e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Civelek M., Wu Y., Pan C., Raulerson C.K., Ko A., He A., Tilford C., Saleem N.K., Stančáková A., Scott L.J. Genetic regulation of adipose gene expression and cardio-metabolic traits. Am. J. Hum. Genet. 2017;100:428–443. doi: 10.1016/j.ajhg.2017.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dobbyn A., Huckins L.M., Boocock J., Sloofman L.G., Glicksberg B.S., Giambartolomei C., Hoffman G.E., Perumal T.M., Girdhar K., Jiang Y., CommonMind Consortium Landscape of conditional eQTL in dorsolateral prefrontal cortex and co-localization with schizophrenia GWAS. Am. J. Hum. Genet. 2018;102:1169–1184. doi: 10.1016/j.ajhg.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jansen R., Hottenga J.J., Nivard M.G., Abdellaoui A., Laport B., de Geus E.J., Wright F.A., Penninx B.W.J.H., Boomsma D.I. Conditional eQTL analysis reveals allelic heterogeneity of gene expression. Hum. Mol. Genet. 2017;26:1444–1451. doi: 10.1093/hmg/ddx043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhernakova D.V., Deelen P., Vermaat M., van Iterson M., van Galen M., Arindrarto W., van ’t Hof P., Mei H., van Dijk F., Westra H.J. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 2017;49:139–145. doi: 10.1038/ng.3737. [DOI] [PubMed] [Google Scholar]
- 123.Thomsen S.K., Ceroni A., van de Bunt M., Burrows C., Barrett A., Scharfmann R., Ebner D., McCarthy M.I., Gloyn A.L. Systematic functional characterization of candidate causal genes for type 2 diabetes risk variants. Diabetes. 2016;65:3805–3811. doi: 10.2337/db16-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Won H., de la Torre-Ubieta L., Stein J.L., Parikshak N.N., Huang J., Opland C.K., Gandal M.J., Sutton G.J., Hormozdiari F., Lu D. Chromosome conformation elucidates regulatory relationships in developing human brain. Nature. 2016;538:523–527. doi: 10.1038/nature19847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kumasaka N., Knights A., Gaffney D. High resolution genetic mapping of causal regulatory interactions in the human genome. bioRxiv. 2018 [Google Scholar]
- 126.Flister M.J., Tsaih S.W., O’Meara C.C., Endres B., Hoffman M.J., Geurts A.M., Dwinell M.R., Lazar J., Jacob H.J., Moreno C. Identifying multiple causative genes at a single GWAS locus. Genome Res. 2013;23:1996–2002. doi: 10.1101/gr.160283.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hormozdiari F., Gazal S., van de Geijn B., Finucane H.K., Ju C.J., Loh P.R., Schoech A., Reshef Y., Liu X., O’Connor L. Leveraging molecular quantitative trait loci to understand the genetic architecture of diseases and complex traits. Nat. Genet. 2018;50:1041–1047. doi: 10.1038/s41588-018-0148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]