Abstract
The dataset presented here profiles oxidative modification of mRNAs in wheat seeds in response to after-ripening, a treatment that releases seeds from the state of dormancy. The level of dormancy in wheat seeds is closely associated with preharvest sprouting, defined as the germination of seeds while they are on the mother plant, which negatively affects wheat yield and quality. Understanding the molecular mechanisms involved in the control of seed dormancy is critical for improving the tolerance of wheat seeds to preharvest sprouting. The dataset were generated using oxidized mRNA samples derived from three independent biological replicates of dormant and after-ripened (non-dormant) wheat seeds and a microarray based experimental procedures that involved the use of Affymetrix GeneChip Wheat Genome Array. The raw and normalized data are available in NCBI׳s Gene Expression Ominbus (GEO) database with accession number GSE41949, and a related research article has been published in Plant Biotechnology Journal (Gao et al., 2013).
Keywords: After-ripening, Dormancy, Microarray, mRNA oxidation, Seed, Wheat
Specifications table
| Subject area | Biology |
|---|---|
| More specific subject area | Oxidative modification of mRNAs |
| Type of data | Data Table in Excel and CEL files |
| How data was acquired | Affymetrix GeneChip Wheat Genome Array |
| Data format | Raw and normalized data |
| Experimental factors | Dormant seeds vs. after-ripened (non-dormant) seeds |
| Experimental features | Isolation of oxidized mRNAs, large scale analysis of oxidative modification of seed stored mRNAs |
| Data source location | University of Manitoba, Winnipeg, Canada |
| Data accessibility | The raw and normalized data are available from NCBI׳s Gene Expression Ominbus (GEO) database with accession number GSE41949. |
Value of the data
-
•
Large scale analysis of oxidative modification of seed stored mRNAs in dormant and after-ripened (non-dormant) seeds of wheat.
-
•
Genes corresponding to mRNAs identified as differentially oxidized by after-ripening might be involved in the control of seed dormancy.
-
•
The data set has a paramount significance in enhancing meta-analysis and provides an important platform for comparison of similar datasets derived from seeds of other crop species.
1. Data
This study was performed to gain insights into the molecular mechanisms involved in the regulation of seed dormancy in wheat. Microarray based seed stored mRNA oxidation profiles of dry dormant and after-ripened (non-dormant) seeds of wheat were compared. The mRNA samples were extracted from three independent biological replicates of the two seed samples and then subjected to isolation of oxidized seed stored mRNAs. Using microarray experiments that involved the use of the Affymetrix GeneChip Wheat Genome Array and 100 ng of oxidized mRNA samples, the study presented a profile of oxidative modification of seed stored mRNAs in response to after-ripening, a period of dry storage that releases seeds from the state of dormancy. The raw and normalized files of the data are available at NCBI׳s Gene Expression Ominbus database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41949). Detailed description of the data can be found in Gao et al. [1].
2. Experimental design, materials and methods
2.1. Plant materials and growth conditions
Wheat plants of cv. AC Domain, a hard red spring wheat cultivar with high level of dormancy [2], were grown in a greenhouse at 18–22 °C/14–18 °C (day/night) under a 16/8 h photoperiod until harvest as described previously [1]. Mature seeds were harvested and divided into two groups. One group of the seeds was stored at −80 °C to maintain dormancy while the other group was stored at room temperature for 10 months to generate after-ripened seeds.
2.2. Isolation of total and mRNAs, and oxidized mRNAs
Isolation of total RNA was performed from three independent biological replicates of both dormant and after-ripened wheat seed samples. RNAqueous columns (Ambion, Austin, TX) were used for extraction of the total RNA samples. After checking their purity and integrity, the total RNA samples were digested with DNase (Ambion, Austin, TX, USA) for removing any genomic DNA contaminants. The total RNA samples were subsequently used for isolating mRNAs, which was conducted using PolyATract kit (Promega, Madison, WI) following the manufacturer׳s instruction. Oxidized mRNAs were then isolated from the mRNA samples using immunostaining with 8-hydroxydeoxyguanosine (OHdG) monoclonal antibody as described previously [1], [3]. Approximately 3 μg of the mRNA from each sample was used for the isolation of oxidized mRNAs.
2.3. Microarray experiment
Microarray analysis was performed with 100 ng of oxidized mRNA samples derived from dormant and after-ripened seeds. After cDNA synthesis and purification, biotinylated cRNA samples were prepared using the GeneChip IVT Labelling Kit and the GeneChip Sample Cleanup Module. Quality of the labelled cRNAs was examined using an Agilent 2000 Bioanalyzer. Following fragmentation, labelled cRNA samples were hybridized for 16 h at 45 °C on GeneChip Wheat Genome Array. GeneChips were then subjected to washing and staining procedures using the Affymetrix Fluidics Station 450. Scanning of the GeneChips was performed with Affymetrix Scanner 3000.
2.4. Data analysis
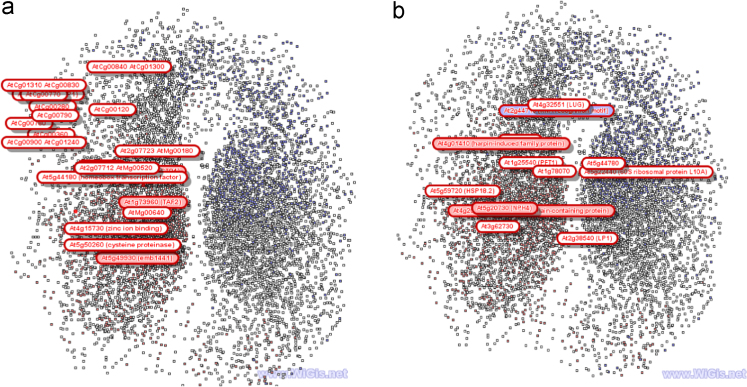
Normalization of the raw data files was carried out with Robust Multi-Array Average (RMA), and annotation of the probesets was performed using HarvEST WheatChip (http://harvest.ucr.edu/) [4]. Identification of probesets that are differentially oxidized between dormant and after-ripened seeds was performed by analysis of variance (ANOVA) using FlexArray software [5]. Probesets were considered to be differentially oxidized if they showed ≥2-fold changes at probability level of ≤0.05. Gene ontological analysis of the probesets oxidized in response to after-ripening, which was performed using the AgriGO analysis toolkit [6], indicated their distribution over different functional categories [1]. Analysis of the Arabidopsis genes corresponding to the wheat probesets representing mRNAs differentially oxidized between dormant and after-ripened seeds using SeedNet (http://vseed.nottingham.ac.uk; [7]) revealed that genes corresponding to the oxidized probesets are overrepresented in a region consisting of gene sets associated with seed dormancy (Fig. 1).
Fig. 1.
Distribution of genes highly oxidized in dry dormant (a) and after-ripened (b) wheat seeds in SeedNet topology consisting of gene sets associated with dormancy (red) and germination (purple) (http://vseed.nottingham.ac.uk; [7]).
Acknowledgements
This study was supported by a grant from the Natural Sciences and Engineering Research Council of Canada to BTA. We would like to thank Zhen Yao and Brenda Oosterveen for their technical assistance.
Footnotes
Transparency data associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.10.036.
Transparency document. Supplementary material
Supplementary material
References
- 1.Gao F., Rampitsch C., Chitnis V.R., Humphreys G.C., Jordan M.C., Ayele B.T. Integrated analysis of seed proteome and mRNA oxidation reveals distinct post-transcriptional features regulating dormancy in wheat (Triticum aestivum L.) Plant Biotechnol. J. 2013;11:921–932. doi: 10.1111/pbi.12083. [DOI] [PubMed] [Google Scholar]
- 2.Townley-Smith T.F., Czarnecki E.M. AC Domain hard red spring wheat. Can. J. Plant Sci. 2008;88:347–350. [Google Scholar]
- 3.Bazin J., Langlade N., Vincourt P., Arribat S., Balzergue S., El-Maarouf-Bouteau H., Bailly C. Targeted mRNA oxidation regulates sunflower seed dormancy alleviation during dry after-ripening. Plant Cell. 2011;23:2196–2208. doi: 10.1105/tpc.111.086694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harvest: WheatChip version 1.57. 〈http://harvest.ucr.edu〉 (Accessed 2 June 2012).
- 5.Blazejczyk M., Miron M., Nadon R. Genome Quebec; Montreal, Canada: 2007. FlexArray: A Statistical Data Analysis Software for Gene Expression Microarrays. [Google Scholar]
- 6.Du Z., Zhou X., Ling Y., Zhang Z.H., Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassel G.W., Lan H., Glaab E., Gibbs D.J., Gerjets T., Krasnogor N., Bonner A.J., Holdsworth M.J., Provart N.J. Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc. Natl. Acad. Sci. USA. 2011;108:9709–9714. doi: 10.1073/pnas.1100958108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material