Abstract
PGC-1α is a key regulator of oxidative metabolism facilitating the expression of genes critical for the function and biogenesis of the two key oxidative organelles, mitochondria and peroxisomes, in skeletal muscle (SKM) and other organs. Our recent studies have found that the transcription factor Bhlhe40 negatively regulates PGC-1α gene expression and its coactivational activity, therefore, this factor should have profound influence on the biogenesis and metabolic activity of mitochondria and peroxisomes. Here we found that both the number and activity of peroxisomes were increased upon knockdown of Bhlhe40 expression but were repressed by its over-expression. Mitochondrial efficiency was significantly reduced by Bhlhe40 knockdown, resulting in the burst of ROS. Over-expression of a constitutively active PGC-1α-interactive domain (named as VBH135) of Bhlhe40 mimicked the effects of its knockdown on peroxisomes but simultaneously reduced ROS level. Furthermore, the efficiency, but not the number, of mitochondria was also increased by VBH135, suggesting differential regulation of peroxisomes and mitochondria by Bhlhe40. Unsaturated fatty acid oxidation, insulin response, and oxidative respiration were highly enhanced in Bhlhe40 knockdown or VBH135 over-expressed cells, suggesting the importance of Bhlhe40 in the regulation of unsaturated fatty acid and glucose oxidative metabolism. Expression profiling of genes important for either organelle also supports differential regulation of peroxisomes and mitochondria by Bhlhe40. These observations have established the important role of Bhlhe40 in SKM oxidative metabolism as the critical regulator of peroxisome and mitochondrion biogenesis and functions, and thus should provide a novel route for developing drugs targeting SKM metabolic diseases.
Keywords: Skeletal muscle, Bhlhe40, PGC-1α, Oxidative metabolism, Peroxisome
Graphical abstract
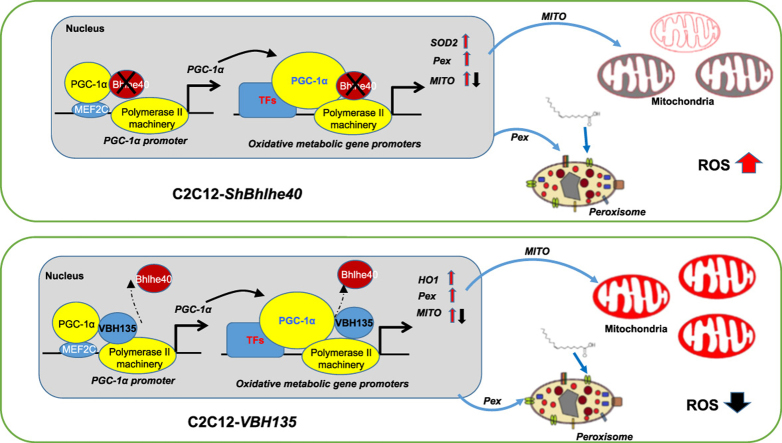
Bhlhe40 normally represses PGC-1α expression and its coactivational activity on target gene promoters. When Bhlhe40 is knockdown (as in C2C12-ShBhlhe40 cells), PGC-1α and its target genes, such as SOD2 and peroxisome related genes (Pex), is increased to enhance/increase peroxisome function and number. Mitochondria (MITO) related genes are regulated differentially, which leads to reduced MITO efficiency (in grey) and surged ROS level. In C2C12-VBH135 cells, wildtype Bhlhe40 is competed off the promoters and the expression of both Pex and HO1 genes are increased, which increased peroxisome function and number. Although MITO genes are also regulated differentially, VBH135 increased MITO efficiency (in red) and reduced ROS level.
Highlights
-
•
Knockout of Bhlhe40 increased ROS but over-expression of Bhlhe40 reduced ROS.
-
•
Peroxisome number was increased by Bhlhe40 knockout or VBH135 overexpression.
-
•
Mitochondrial efficiency was reduced by Bhlhe40 knockout but increased by VBH135.
-
•
Oxidative respiration was enhanced by Bhlhe40 knockdown or VBH135 overexpression.
-
•
Bhlhe40 repressed PGC-1α coactivation of nuclear gene expression.
1. Introduction
Skeletal muscle (SKM) relies very much on the transcriptional coactivator PGC-1α to promote oxidative metabolism, metabolic thermogenesis adaptation, biogenesis of mitochondria, and fatty acid oxidation for adapting to high energy demands [1], [2], [3]. In SKM, PGC-1α is preferentially expressed in oxidative metabolism dependent slow-twitch fibers [4] and its over-expression can convert putative fast-twitch fibers into slow-twitch fibers [4]. The expression of PGC-1α in skeletal muscle is critically regulated by transcription factors with bHLH DNA-binding motif, as it can be activated by myogenic regulatory factors (MRFs, including Myf5, MyoD, Myogenin and Mrf4) but repressed by Bhlhe40 [5]. However, this antagonism can be relieved when P/CAF, a key coactivator of MRFs, is supplied in over-dose, suggesting the sequestration of P/CAF by Bhlhe40 [6].
Bhlhe40 (also known as Stra13, Dec1, Sharp2, or BHLHB2) is ubiquitously expressed but with strong expression in skeletal muscle [7], [8], where it regulates the activation of myogenic stem cells (named as satellite cells) by antagonizing Notch signaling [8] and protects SKM from reactive oxidative species (ROS) induced damage by activating the expression of heme-oxygenase-1 (HO-1) [9]. Multiple cellular processes, including differentiation, tumorigenesis, peripheral circadian output, and response to hypoxia, have been reported to involve Bhlhe40 [7], [10], [11], [12]. Bhlhe40 can either function as a transcriptional repressor through both histone deacetylase (HDAC)-dependent and -independent mechanisms on most target genes [13] or as an activator on ∆Np63 and Survivin genes [14], [15].
Mitochondria and peroxisomes are the major organelles involved in the cellular oxidative metabolism and both are ubiquitous and highly dynamic. Mitochondria are the power houses of eukaryotic cells and they provide ATP currency through oxidative phosphorylation (OXPHOS) of reducing equivalents [16]. Peroxisomes participate in the β-oxidation of very long chain, unsaturated, and branched fatty acids [17]. Additionally, they can also metabolize carboxylates containing a 3-methyl or 2-hydroxy group via α-oxidation, whereby the one-carbon shortened products can be passed onto the β-oxidation system. Another important function of peroxisomes is removing the harmful reactive oxygen species (ROS) concomitantly generates from the oxidation of substrates in mitochondria and peroxisomes, which is performed by several antioxidant enzymes, including Catalase, SOD1, and PRDX5.
It has been shown that PGC-1α play critical roles in the regulation of the biogenesis and function of mitochondria and peroxisomes through coactivating the activity of various DNA-binding transcription factors, such as PPARγ, ERRα, and NRF1 [1], [18]. Recently we found that PGC-1α directly interacted with Bhlhe40 and they co-occupied PGC-1α targeted gene promoters/enhancers, which in turn repressed PGC-1α transactivational activity [19]. Relief of this repression by knockdown of Bhlhe40 expression increased levels of ROS, fatty acid oxidation, mitochondria DNA, and the expression of PGC-1α target genes. These observations suggest that Bhlhe40 should be an important regulator of mitochondrial and peroxisome biogenesis and functions [19]. Therefore, more endeavor should be devoted to decipher the regulation of these two organelles by Bhlhe40.
2. Materials and methods
2.1. Plasmids
The plasmids pEGFP-N1-roGFP-PTS1 and –KillerRed-PTS1 were generous gifts from Dr. Marc Fransen (Katholieke Universiteit Leuven, Belgium) and their construction can be found in Dr. Fransen's publication [20]. The expression vectors of PGC-1α and Bhlhe40 and their target promoters driven reporters have been described in our previous studies [19], [21]. A TET-off expression vector, pCEGFP-TRE, was created by inserting the SalI/MluI fragment of pBI-EGFP vector into the BglII/HindIII sites of pCDNA3 vector. The CDS of Bhlhe40-Flag or VP16-Bhlhe40-(1−135) was inserted into the EcoRV site of pCEGFP-TRE for stable cloning and doxycycline repressible expression. The basic region (ETYKLPHRLIEKKRR) of the DNA-binding motif in VBH135 was mutated into ETYALPAALIEKAAAA by site-directed PCR mutagenesis before inserted back to the EcoRV site of pCEGFP-TRE vector to generate pCEGFP-TRE -VBH135m. The coding sequence of RFP was PCR amplified from PLKO-RFP vector with primers containing PTS1 sequence in the N-terminus and inserted into the EcoRV site of the pCDNA3.0 vector. The CDSs of PPARγ, Bhlhe40, and PGC-1α carried in pCDNA3.0 vector were used for transient transfection assays (Fig. 4). The reporters driven by rUCP-1 and M-cadherin promoters were in pGL3-basic and pStable-luc vectors, respectively.
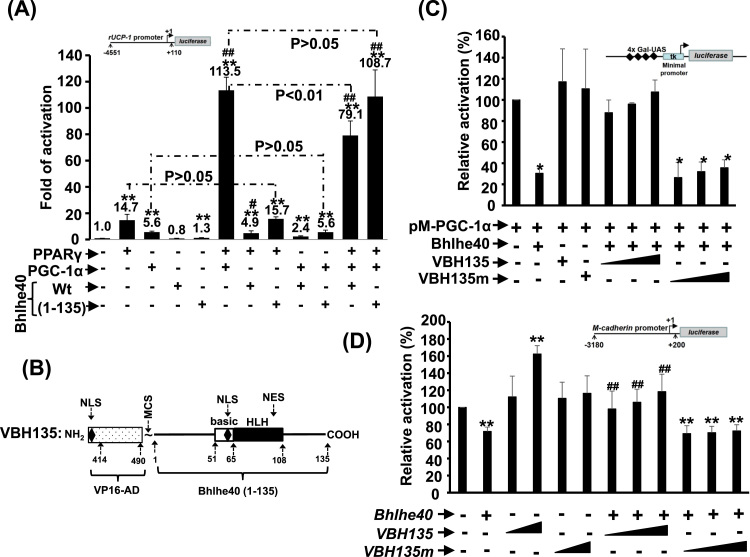
Fig. 4.
The chimeric factor VBH135 functions as a dominant and constitutive transcription activator. (A) Rat UCP-1 gene promoter driven reporter, rUCP-1-luc, was co-transfected with PPARγ, PGC-1α, and Bhlhe40 (full length or 1–135) expression vectors into C2C12 myoblasts in the presence of troglitazone (1 μM). * *: p < 0.01 vs. promoter only; # and ##: p < 0.05 and p < 0.01, respectively, vs. PPARγ activated promoter activity. (B) Schematic representations of the molecular organization of VBH135, a chimeric fusion protein of large T antigen nuclear localization signal, VP16 activation domain (414−490), and Bhlhe40 N-terminal 1–135 amino acids. (C) Gal-PGC-1α expression vector was co-transfected with MH100-luc reporter, with or without Bhlhe40 (FL or 1–135) into C2C12 myoblasts. * and * *: p < 0.05 and p < 0.01 vs. Gal-DBD only. # and ##: p < 0.05 and p < 0.01 vs. Gal-PGC-1α-(FL). $: p < 0.05 vs Gal-PGC-1α-(1−128). (D)M-cadherin-luc reporter was co-transfected with or without Bhlhe40, VBH135 or VBH135m expression vectors into C2C12 myoblasts. All transfections were initiated when myoblasts were at about 70% confluence and lasted for overnight till cells become confluent. Then, cells were kept in differentiation medium to induce myotube formation for 72 h before harvested for determining luciferase activity.
2.2. Cell culture, promoter assay, and stable transfection
The detailed protocol of cell culture and promoter assay has been reported in our previous studies [19], [21], [22]. Briefly, C2C12 myoblasts were kept in DMEM containing 20% FCS and induced to differentiate by changing to differentiation medium (DM) containing DMEM supplemented with 2% horse serum. Myotubes (MT) were examined after 4 days in DM (DM4) unless otherwise indicated. Fusion index indicates the percentage of nuclei in the myotubes among all nuclei and was calculated by the equation: nuclei in myotubes/total nuclei.
For promoter assay, reporter constructs (0.67 μg/well) and expression vectors (0.16 μg each/well) were transfected into myoblasts (70% confluence) grown in 12-well dish by T-Pro NTR-II transfection reagent (T-Pro Biotechnology) mixture in DM for overnight. Cells were harvested and assayed for luciferase activity 72 h after transfection as they differentiated into myotubes. The luciferase activity was determined using the luciferin (VivoGlo Luciferin, Promega) mixture (20 mM Tricine, 2.67 mM MgSO4, 1.07 mM (MgCO3)4. Mg (OH)2·5H2O, 0.1 mM EDTA, 33.3 mM DTT, 270 μM Coenzyme A, 530 μM ATP, 470 μM luciferin) with a Clarity 2 luminometer (BioTEK; Winooski, VM). All experiments were performed in triplicates and repeated at least 3 times.
For establishing the Tet-off stable clones, the tetracycline controlled transactivator (tTA) expression vector pEF-tTA-IRES-puro (5 μg/well) was transfected into C2C12 myoblasts and selected with puromycin (2.5 μg/ml) for 2–3 weeks. Then, pCEGFP-TRE vector (5 μg/well) carrying Bhlhe40, VBH135, or VBH135m was transfected into the tTA expressing cells and selected with G418 (800 μg/ml) for 2–3 weeks until stable clones were acquired.
2.3. Gene knockdown by lentivirus expressed shRNA
For establishing Bhlhe40 knockdown clone, pCMV-Δ8.91 (5.625 μg), pMD.G (0.625 μg), and the shRNA expressing pLKO1 vectors (6.25 μg) were transfected into 293FT cells (on a 10 cm dish) to generate infectious lentivirus, which was collected 48 h later before added to C2C12 culture medium. Infected C2C12 myoblasts were selected with puromycin (2 μg/ml) for at least 2 weeks to generate monoclonal colonies and these clones were expanded and examined for their Bhlhe40 levels with RT-PCR. Clones derived from pLKO1–218239 plasmid (clone ID TRCN0000218239) showed reduced Bhlhe40 level were used for further experiments. For transient knockdown of PGC-1α, viruses derived from pLKO1–218239 plasmid (clone ID TRCN0000218239) were pooled and used for infection. All vectors used for the knockdown experiments were purchased from the RNAi core facility of Academia Sinica.
2.4. Quantitative RT-PCR (qRT-PCR)
C2C12 cells were lysed with home-made Solution D (4 M guanidinium thiocyanate, 25 mM sodium citrate, pH7.0, 0.5% sarcosyl, 0.1% β-mecaptoethanol) and the genomic DNA and proteins were removed by repeated (x4) acid phenol/chloroform extractions and followed by ethanol precipitation of total RNA [23]. The detailed protocol for traditional and quantitative RT-PCR has been described in our previous studies [21], [22]. Briefly, the first strand of cDNA was synthesized using the Superscript III kit (Invitrogen) according to the manufacturer's protocol. All qPCR was performed in a 15 μl reaction mixture containing 5 μM forward/ reverse primers, template cDNA (equivalent to about 25, 50, or 100 ng of total RNA was used in separate tubes to ensure the linearity of PCR amplification), and 1X SYBR Green reaction mix (Power SYBR Green PCR master mix, Applied Biosystems) for detecting PCR product. Gapdh was used as internal control amplified in the same PCR assay. The expression level of each gene was normalized to that of Gapdh first, then, the normalized expression level (△Ct) in control cells was arbitrarily set as 1, and the control cell △Ct was deduced from those of the treatment groups to derived △△Ct. The relative expression level of each gene in Bhlhe40 knockdown or over-expressed cells was calculated using the equation (2-△△Ct). All reactions were performed in ABI 7300 sequence detection system. The primer sets used are listed in supplementary table I.
2.5. Determination of ROS levels
The detailed protocols for these assays have been described in our previous study [19]. Cells were grown in 6-well plates until confluent before used assays. For measuring ROS, cells were incubated in KRPH buffer (20 mM Hepes at pH 7.4, 5 mM KH2PO4, 1 mM MgSO4, 1 mM CaCl2, 136 mM NaCl, 4.7 mM KCl, and 1% BSA) containing 30 μM H2-DCFDA for 30 min. Then, cells were extensive wash with PBS before trypsinzed (with 0.25X trypsin) and re-suspended in PBS buffer. The fluorescence of H2-DCFDA in the cells was measured with a fluorescence spectrophotometer (Hitachi F-4500) and normalized with cell number.
2.6. Fatty acid oxidation
For measuring fatty acid oxidation, BSA conjugated 2 μCi 3H-labeled oleic acid (NET289005MC, Perkin Elmer) and palmitic acid (NET043001MC, Perkin Elmer) was added into cells kept in αMEM and incubated for 6 h in the presence of 100 μM cold oleic or 50 μM palmitic acid, respectively. Then, medium was collected and extracted with phenol/chloroform to remove un-metabolized 3H-palmitic acid or oleic acid as reported before [24]. The 3H activity in the aqueous phase was determined by a scintillation β-counter (LS6500, Beckman).
2.7. O2 consumption
The Winkler test for determining dissolved O2 level was performed as previously described [25]. Briefly, after one hour of incubation, 3 ml medium was transferred to a 15 ml centrifuge tube before 500 μl 1 M MnSO4 and 500 μl mixture of 1 M KI and 6 N NaOH (1:3 vol: vol) was added, then mixed well until a brown precipitate of Mn3+ appeared. The Mn3+ brown precipitate was dissolved by adding 500 μl 6 N HCl until the solution turned into a deep yellow color. Then, 500 μl 1% starch solution was added to serve as an indicator of I2 before titrating with 18 mM S2O32- to reduced I2 and I3- to I- until the solution turned clear. The amount of dissolved oxygen in the original medium can be deducted from the following equations: (1) 2 M S2O32- could reduce 1 M I2, and (2) 1 M I2 was produced from costing 0.5 M O2. Therefore, the net equation was that 1 mol dissolved oxygen was required to titrate with 4 mol S2O32-. The result was normalized to protein content and showed as nM O2/mg/h.
2.8. Glucose uptake
The assay of glucose uptake was modified from previously described protocol [26]. Serum starvation was performed in HBSS (5 mM glucose) for at least 4 h to further enhance the basal and insulin-induced glucose uptake. After starvation, the culture medium of myotubes in 6-well plates were replaced with Krebs-Ringer phosphate (KRPH) buffer containing 0.2 μCi 2-deoxy-D-[3 H]-glucose (2-DG; NET328A250UC, Perkin Elmer) with/without 100 nM insulin and incubate at 37 °C for another 2 h. One well was treated with 10 μM cytochalasin B (CB) for inhibiting actin cytoskeleton mediated glucose uptake and thus served as non-specific/negative glucose uptake control. After two times wash with cold 1x phosphate-buffered saline (PBS), cells were lysed with 500 μl radioimmunoprecipitation assay (RIPA) buffer. The radioactivity in the lysate is quantified by liquid scintillation counting. The value of the result is normalized to protein amount and showed as cpm/μg.
2.9. NAD(P)H content
This assay was performed as previously described [27]. Briefly, cells grown on 6-well plates were harvested by trypsinzation and suspended in PBS. Cell numbers were counted on hemocytometer and their emission of 460 nm light after excited with 340 nm light was determined using the F-4500 fluorescence spectrophotometer (Hitachi).
2.10. Mitotracker staining and Mitochondrial DNA level determination
Cells grown on 6-well plates were washed with PBS thoroughly before incubated with KRPH buffer containing MitoTracker Red CMXRox (50 ng/ml) for 30 min. Then cells were trypsined and re-suspended in PBS. Cell numbers were counted on hemocytometer and their emission of 595 nm light after excited with 575 nm light was determined using the F-4500 fluorescence spectrophotometer (Hitachi) normalized with cell number.
The relative amounts of mitochondria in the cells were determined by qRT-PCR using the ratio of mitochondrial genes (cytochrome b and Cox II) and nuclear genes (MyoD and Oct4) derived from total genomic DNA. The nuclear gene levels served as input control.
2.11. Catalase activity assay
The assay was modified from previous studies [28]. Briefly, C2C12 cells were harvested by trypsinzation and re-suspended in PBS. Then cells were lysed by repeated passing through gauge #27 needle and examined under microscope for cellular fragmentation. The protein concentration was determined with BCA protein assay and 0.1 μg total lysate was included in the 100 μl PBS based reaction mixture (10 mM H2O2 and with or without 20 mM catalase inhibitor 3-amino-1, 2, 4-triazole). Various times (5, 10, or 20 min) after the reaction, residual H2O2 level was determined with HRP conjugated IgG in the presence of equal amount enhanced chemilminescence (ECL) reagent (Amersham Pharmacia Biotech) in a Clarity 2 luminometer (BioTEK; Winooski, VM). For reactions containing 3-amino-1, 2, 4-triazole (3AT), lysate and 3AT were pre-incubate for 20 min to ensure the inhibition of catalase by 3-AT.
2.12. SDH assays
This assay was performed as previously described [29]. Briefly, cells were harvested by lifting in PBS containing 2 mM EDTA and washed extensively with PBS before sonicated to lyse the cells. After centrifugation, the protein concentration in the supernatant was determined by BCA reaction and equal amount (50 μg) of protein was used for enzymatic assays. For SDH assay, lysate, sodium succinate (40 mM, pH>7), sodium azide (8 mM), and 2, 6-dinitrophenolindophenol (DCPIP, 0.005%) were incubated at 37 °C for 30–60 min before SDS (2%) was added to stop the reaction. The reaction product was determined by reading absorbance at 600 nm in a spectrophotometer.
2.13. SOD assay
Samples for SOD2 assay was processed as in the SDH assay, and the SOD activity was determined using a SOD assay-WST kit (19160–1KT, Sigma) according to the manufacturer's protocol. Briefly, lysate was mixed with WST and enzyme solutions at 37 °C for 20 min. Then, the reaction product was determined by reading absorbance at 450 nm in a spectrophotometer. The SOD activity in the lysate was calculated by the percentage of reduction in the 450 nm absorbance.
2.14. Western blot
The detailed Western blot protocol has been described previously [30]. Briefly, aliquots of total lysate (50 μg) in RIPA buffer supplemented with protease inhibitors and phosphatase inhibitors were run on 10% SDS-PAGE gels before blotted onto a PVDF membrane (Pall FluoroTrans W membrane, PALL). After extensively washed with 1X PBS containing 0.5% Tween 20 (PBST), PVDF membranes were blocked by 5% skimmed milk in PBST for 30–60 min. Primary antibody was diluted 1:1000 in blocking solution and incubated with the blot at 4 °C for overnight. After several washes with PBST, HRP conjugated secondary antibody (1:10000 dilution) was added and incubated at room temperature for 1 h. After extensive wash, the HRP signal was detected by an enhanced chemiluminescence kit (Amersham Pharmacia Biotech). The Flag antibody was purchased from Sigma (M2 antibody, F3165), and the rabbit polyclonal antibody against Bhlhe40 amino acids 259–411 was generated in our laboratory [19]. The antibodies for SDHB2, Gapdh, Lamin B1, OXPHOS subunits, MTCO1, and MTCO2 were from Abcam (ab178423, ab9482-200, ab16043-25, ab110413, ab1475, and ab198286, respectively).
2.15. Statistic analyses
All experiments were performed at least 3 times with similar results observed. Results from independent experiments were pooled and their means and standard errors are shown. The difference between various treatments was examined with student's t-test. All tests were 1-sided, and a probability value of < 0.05 was considered statistically significant. For real-time PCR, each assay was performed in duplicate of two different input levels to ensure linear PCR amplification and no difference in ΔCt between input levels should be detected.
2.16. Detection and counting of peroxisomes
The peroxisome reporter plasmids (pEGFP-N1-roGFP-PTS1, -KillerRed-PTS1, and pcDNA3-RFP-PTS1) were transfected into C2C12 myoblasts (about 40–50% confluence) grown on glass cover slides held in 12-well dishes for overnight, then medium was changed and cells were harvested 48 h later. Cell were washed 3 times with PBS before fixed in in 4% paraformaldehyde for 30 min. After fixation, cells were washed 3 times with PBS and then quenched in PBS containing 50 mM NH4Cl. To visualize the nuclei, cells were incubated with DAPI (2 μg/ml in PBS) at room temperature for 10 min and were washed with PBS thoroughly afterward. All the images were observed and photographed under a Carl Zeiss Axio Observer A1 fluorescence microscope with AxioVision software. The peroxisome numbers on photographed images were counted either manually (Fig. 1, Fig. 2) or automatically using the Image J software (Fig. 6). The reliability of this assay was confirmed by the overlapped images of RFP-PTS1 and Catalase detected by immunofluorescence (Supplementary Fig. 3).
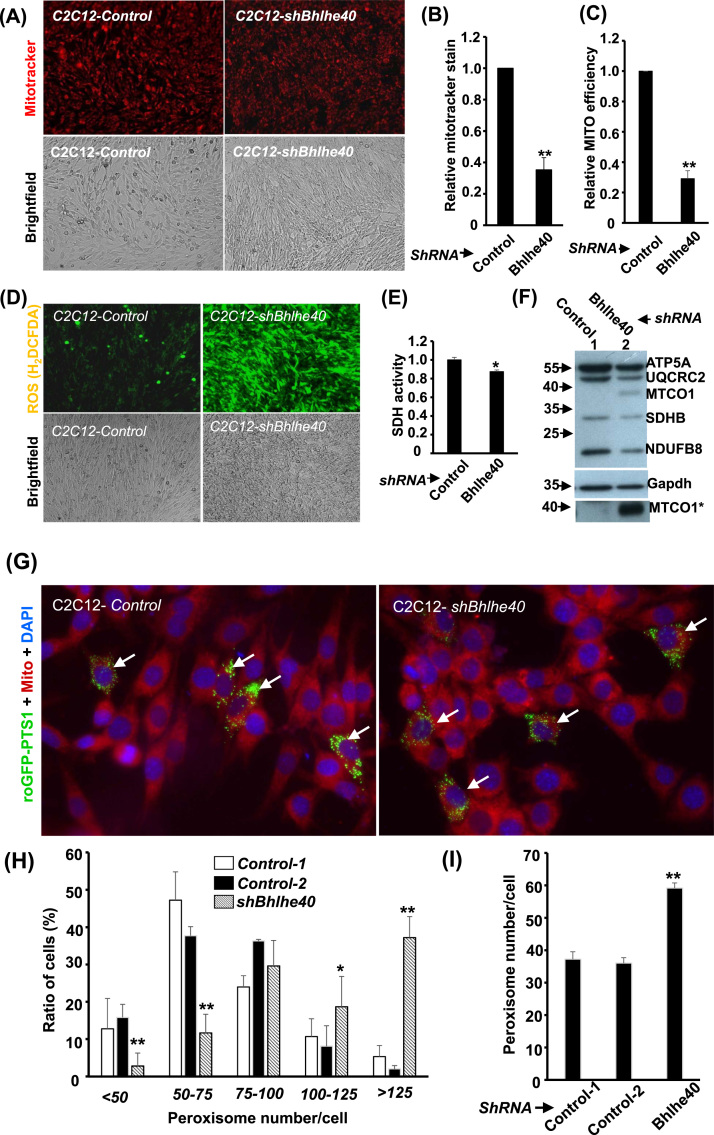
Fig. 1.
The effects of Bhlhe40 knockdown on mitochondria and peroxisomes. (A) Images of Mitotracker stained C2C12 myoblasts over-expressed with control (C2C12-Control) or Bhlhe40-targeting (C2C12-shBhlhe40) shRNA. Their relative intensity of Mitotracker stain and mitochondrial efficiency (Mitotracker stain/mtDNA) at CMB stage are shown in (B) and (C) respectively. (D) Images of C2C12-Control or -shBhlhe40 myoblasts stained with H2DCFDA. Their relative SDH activity and levels of OXPHOS subunits are shown in (E) & (F). MTCO1 * : signals of MTCO1 under longer exposure. (G) Images of C2C12-Control or -shBhlhe40 myoblasts transfected with roGFP-PTS1 expression vector and counterstained with DAPI and Mitotracker. The distribution (%) of myoblasts with different numbers of peroxisome/cell and the average peroxisome number/cell are shown in (H) and (I), respectively. Both Mitotracker and ROS levels were quantified by fluorescence spectrophotometry. * and * *: p < 0.05 and p < 0.01 vs. C2C12-Control.
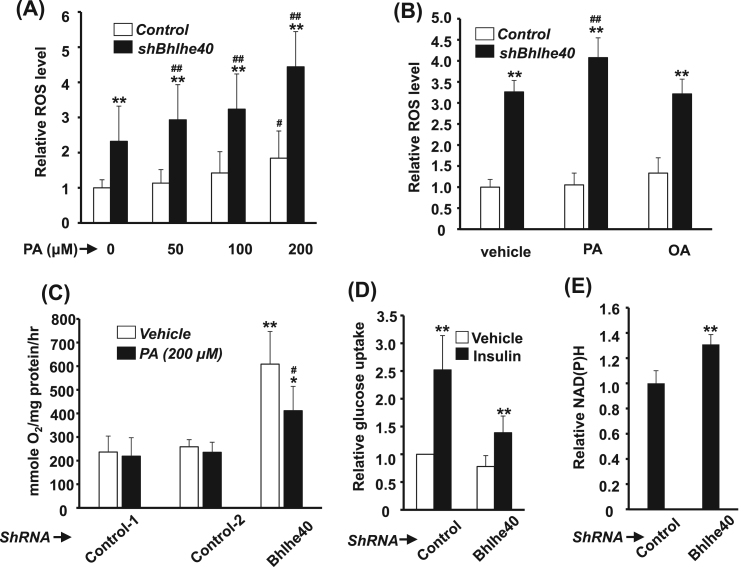
Fig. 2.
The effects of Bhlhe40 knockdown on oxidative metabolism. C2C12-Control and -shBhlhe40 myoblasts were treated with various doses of palmitic acid (A) or oleic acid (B) and then with their relative intracellular ROS levels determined. The effect of palmitic acid treatment on cellular O2 consumption is shown in (C). Basal and insulin (100 nM) stimulated glucose uptake of myoblasts is shown in (D), and their relative insulin response is shown in (E). The value in vehicle treated control cells was arbitrarily set as 1 fold. * *: p < 0.01 vs. C2C12-Control. # and ##: p < 0.05 and p < 0.01 vs. vehicle.
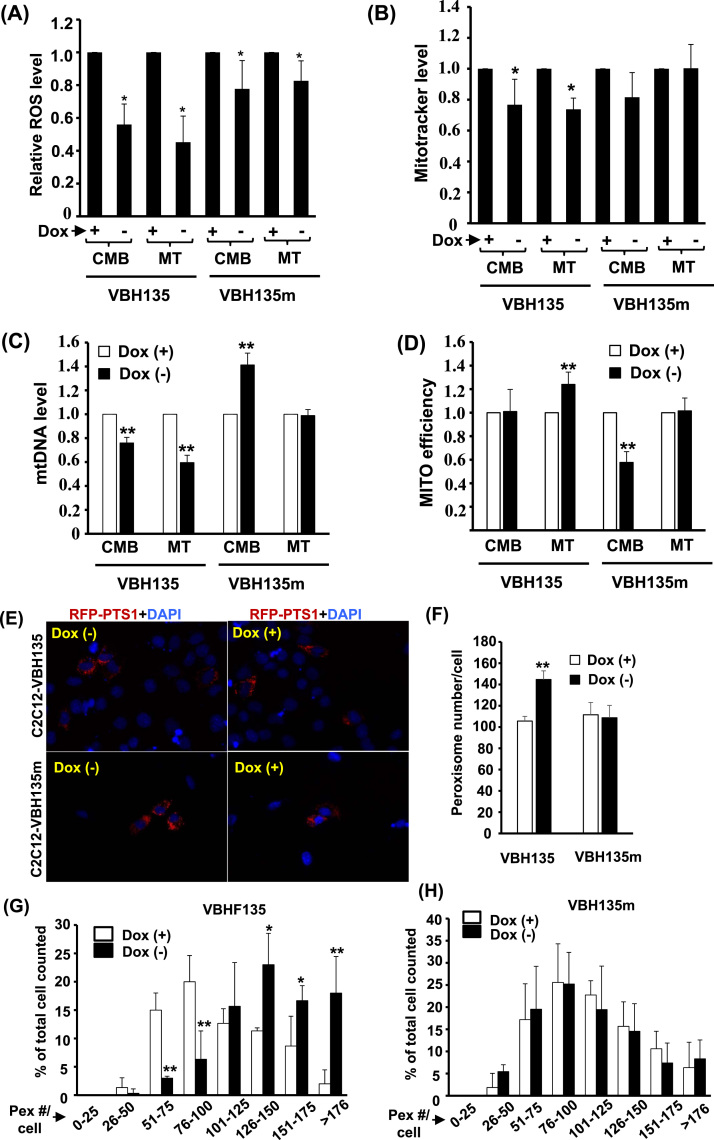
Fig. 6.
The effects of VBH135 or VBH135m over-expression on mitochondrion and peroxisome activity. The levels of ROS, mtDNA, and Mitotracker stain intensity of C2C12-VBH135 and -VBH135m cells at CMB and MT stages in the presence and absence of Dox are shown in (A), (B), and (C), respectively. The MITO efficiency (D) was calculated by Mitoctracker/mtDNA and compared with cells treated with Dox (+). (E) Images of C2C12-VBH135 and -VBH135m myoblasts transfected with RFP-PTS1 expression vector and counterstained with DAPI. The average number of peroxisomes/cell (F) and the distribution (%) of cells with different numbers of peroxisome/cell (G & H) of peroxisome number/cell are calculated 48 h after transfection. * and * *: p < 0.05 and p < 0.01, respectively, vs. Dox (+) cells.
2.17. Immunostaining
After in DM for 72–96 h, stable clone cells were washed with cold PBS before fixed in 4% paraformaldehyde for 20 min. Then, they were quenched in 50 mM NH4Cl for 15 min before permeablized in 2% Triton-X 100 overnight. Cells were incubated in blocking solution (2% BSA and 2% goat serum in PBS) for 20 min followed by incubating with MHC antibody (1: 1000 dilution; clone MY-32, Sigma) over night. After extensive wash with PBS, HRP-conjugated secondary antibody (Goat anti mouse IgG, Santa Cruze) was added and incubated for an hour. The expression of MHC was visualized with AEC substrate kit (Zymed Laboratories) and the cells were counter-stained with hematoxylin and eosin.
For immunofluorescent staining of Catalase, cells were processed as above but with the Catalase antibody (SC-271803, Santa Cruz) and FITC-conjugated secondary antibody (ab6785-1, Abcam) used. To visualize the nuclei, cells were incubated with DAPI at room temperature for 10 min after the secondary antibody incubation and were washed with PBS thoroughly afterward. All the images were observed and photographed under the Carl Zeiss Axio Observer A1 fluorescence microscope with AxioVision software.
3. Results
3.1. Bhlhe40 is a critical regulator of mitochondrion and peroxisome activity
Mitochondria (MITO) are the power houses of a cell and it is of interest to know if their functions and number are regulated by Bhlhe40; therefore, stable clones of Bhlhe40 knockdown cells, C2C12-shBhlhe40, were established (Supplementary Fig. 1A). Our previous data had shown that mitochondrial DNA (mtDNA) was increased in C2C12-shBhlhe40 cells [21]; however, here we were surprised to find that their staining intensity of Mitotracker, an inner membrane potential dependent MITO stain, was significantly reduced (Fig. 1A & B). As their relative membrane potential/efficiency (mitotracker stain normalized by mtDNA level) was also reduced (Fig. 1C and Supplementary Fig. 1B), it indicates deficiency of MITO functions amid increased mtDNA and MITO number. The deficiency of MITO functions was further demonstrated by the radical increase in cellular ROS level but reduced SDH activity and the expression levels of oxidative phosphorylation system (OXPHOS) subunits (Fig. 1D, E & F). Surprisingly, the OXPHOS complex IV subunit MTCO1 (Fig. F), but not MTCO2, was highly induced in C2C12-shBhlhe40 cells, implying unbalance of this OXPHOS complex. Whether surged MTCO1 protein was the cause of ROS increase or secondary to it needs to be further investigated.
Another important oxidative organelle and ROS contributor is the peroxisome, but whose number is difficult to quantify due to the lack of DNA in this organelle. The quantification difficulty was overcome by the use of PTS1-roGFP protein that specifically localized to peroxisomes and light up them with green light. After counting these green dots in the cells, we found the number of peroxisomes was significantly increased in the C2C12-shBhlhe40 cells (Fig. 1G~I), suggesting the shift from MITO-dependent to peroxisome-dependent oxidative metabolism. We further found that the ROS level in C2C12-shBhlhe40 could be increased by palmitic acid (PA), a MITO-dependent saturated fatty acid, in a dose-dependently manner (Fig. 2A), but not by oleic acid (OA), an unsaturated fatty acid depending on peroxisome for its metabolism (Fig. 2B). Knockdown of Bhlhle40 significantly enhanced oxidative respiration but which was compromised by PA treatment (Fig. 2C). On the contrary, insulin stimulated glucose uptake and response were reduced in C2C12-shBhlhe40 cells (Fig. 2D and Supplementary Fig. 1C), although the insulin signaling pathway was not affected (Supplementary Fig. 1D). These observations suggest the shifting from glucose-dependent metabolism to other energy sources, which was further confirmed by the discovery that the NAD(P)H, important cofactors for fatty acid metabolism, level in these cells was significantly increased (Fig. 2E), suggesting for increased fatty acid metabolism.
3.2. Over-expression of Bhlhe40 reduces peroxisome number
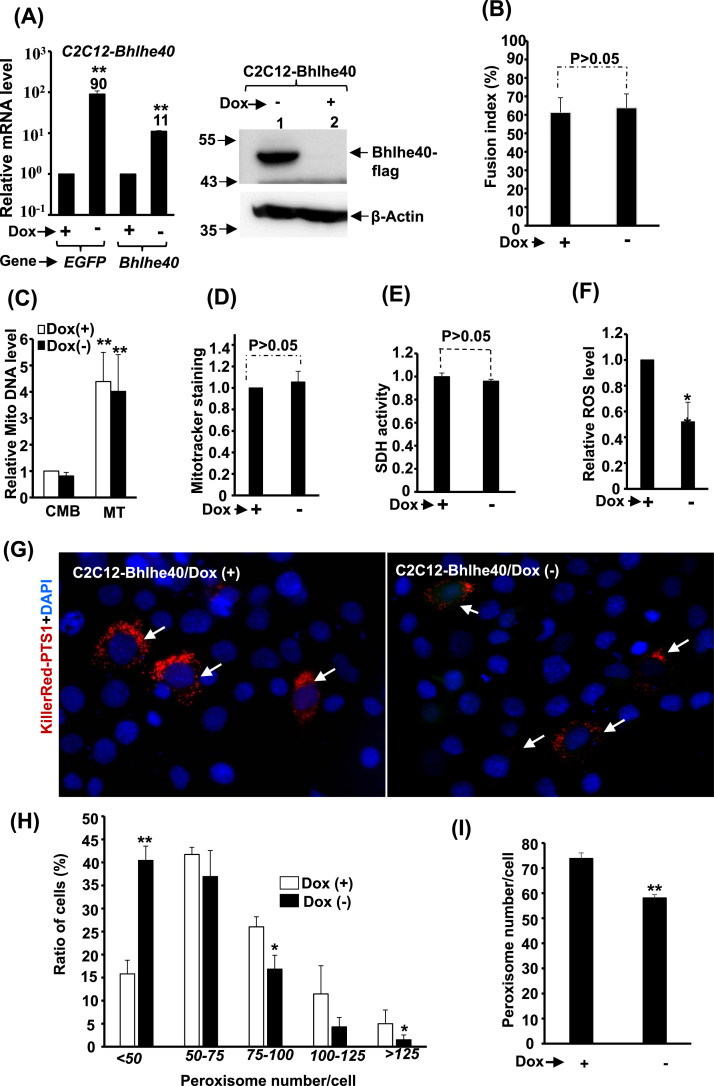
A doxycycline (Dox) regulated inducible system (Tet-off) of Bhlhe40 was established (Fig. 3A) to discuss the effects of its over-expression on MITO and peroxisome activity. We found that the C2C12-Bhlhe40 cells differentiated normally (Fig. 3B and Supplementary Fig. 2B) and neither mtDNA level nor Mitotracker stain intensity was altered in these cells (Fig. 3C, D & Supplementary Fig. 2 A), indicating no observable effect on MITO number and function, which is further demonstrated by unaltered SDH activity (Fig. 3E). However, the level of ROS and number of peroxisomes were both reduced by Bhlhe40 over-expression (Fig. 3F~I). We further examined their SOD activity and found marginal but significant increase by Bhlhe40 overexpression (Supplementary Fig. 4 A). These observations suggest that Bhlhe40 is an important negative regulator of cellular peroxisome number and ROS level.
Fig. 3.
The effects of Bhlhe40 over-expression on differentiation and metabolism. A Tet-off system was established to stably over-express Bhlhe40 and GFP simultaneously in C2C12 cells (C2C12-Bhlhe40), in which the expression is shut off in the presence of Doxycycline (Dox, 25 ng/ml). The expression of Bhlhe40 mRNA (left panel) and protein (right panel) in myoblasts is shown in (A) and its effects on myogenic differentiation (B) was represented by the fusion index (nuclei number in myotubes/total nuclei number). The effects on mtDNA levels of CMB and MT cells, and on Mitotracker stain intensity, SDH activity, and ROS levels in myotubes are shown in (C), (D), (E), and (F), respectively. (G) Images of C2C12-Bhlhe40 myoblasts transfected with KillerRed-PTS1 expression vector and counterstained with DAPI. The distribution (%) of cells with different numbers of peroxisome/cell and the average peroxisome number/cell are shown in (H) and (I), respectively. * and * *: p < 0.05 and p < 0.01 vs. Dox (+) cells.
3.3. VBH135 serves as an antagonist of Bhlhe40
One of the mechanisms employed by Bhlhe40 to regulate cellular oxidative metabolism is repressing the coactivational activity of PGC-1α via direct interaction with each other's N-terminal domains [19]. The N-terminal domain (1–135 amino acids) of Bhlhe40 mediate strong interaction with PCG-1α without affecting its coactivational activity on PPARγ activated UCP1 promoter driven luciferase reporter (Fig. 4A). Fusion of this domain with VP16 activation domain creates a constitutively active factor (VBH135, Fig. 4B) that can rescue wildtype Bhlhe40 repressed PGC-1α coactivational activity (Fig. 4C), which in turn might enhance oxidative respiration. As this domain retains the DNA-binding domain of Bhlhe40, it raises the query whether it can also bind to Bhlhe40 target genes to activate their expression? We have reported that M-cadherin promoter can be repressed by Bhlhe40 [21], and with this promoters we found strong activation by VBH135 and which can further overcomes the repression by wildtype Bhlhe40 (Fig. 4D). As the DNA-binding mutant of VBH135 (VBH135m) failed to affect M-cadherin promoter activity, suggesting that VBH135 can be used as a Bhlhe40 antagonist to rescue its repression of PGC-1α- and Bhlhe40-targeted gene expression via protein-protein interaction and direct DNA-binding.
3.4. VBH135 increases the efficiency of both MITO and peroxisomes
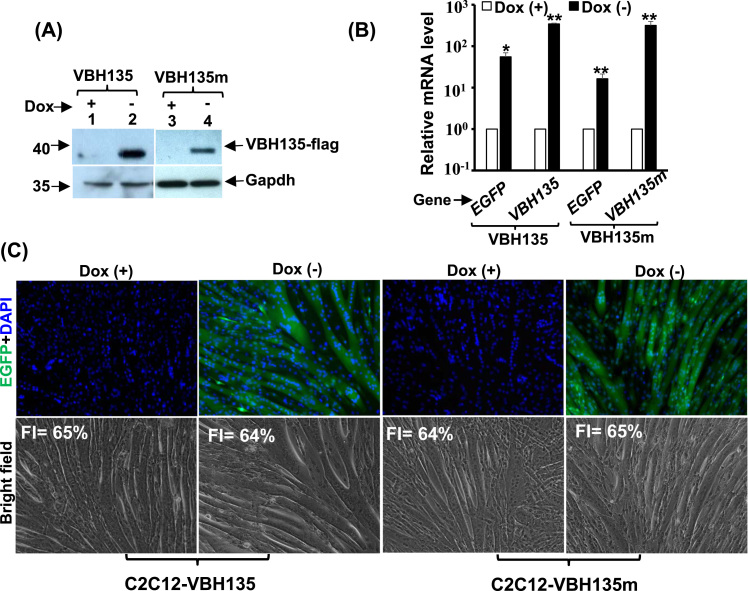
VBH135 and its DNA-binding domain mutant were stably over-expressed in C2C12 using the Tet-off system, and their mRNA and protein could be induced by removal of Dox (Fig. 5A & B, Suppl. Fig. 2C, and in [19]). We further found that their expression did not affect myogenic differentiation (Fig. 5C & Suppl. Fig. 2C), so their effects on SKM metabolism would not be complicated by that on differentiation.
Fig. 5.
Over-expression of VBH135 has no effect on myogenic differentiation. The expression levels of VBH135 and its DNA-binding mutant, VBH135m, in Tet-off regulated stable clones (C2C12-VBH135 and -VBH135m) were determined by Western blot (A) and quantitative RT-PCR (B). The morphology of these cells at MT stage (DM4) is shown in (C). FI: fusion index.
Although VBH135 over-expression had no effect on terminal differentiation, significantly reduced levels of ROS, mtDNA, and Mitotracker stain were found in C2C12-VBH135 cells (Fig. 6A~C), however, these reducing effects were compromised in cells over-expressed with VBH135m, suggesting the involvement of DNA-binding ability of VBH135 in its metabolic regulation. The MITO efficiency was found increased by VBH135 (Fig. 6D), which partially explained the cause of reduced ROS and Mitotracker stain, since only high efficient mitochondria remained. Unexpectedly, the mtDNA level was increased, but the MITO efficiency was reduced, in confluent C2C12-VBH135m myoblasts (Fig. 6B~D), implying that DNA-binding independent protein-protein competition of VBH135m with some factors, such as PGC-1α, negatively affected MITO functions in myoblasts, but not in myotubes. On the contrary, the regulation of peroxisome number by VBH135 was more DNA-binding dependent as it was significantly increased by VBH135, but not VBH135m, in confluent myoblasts (Fig. 6E~H).
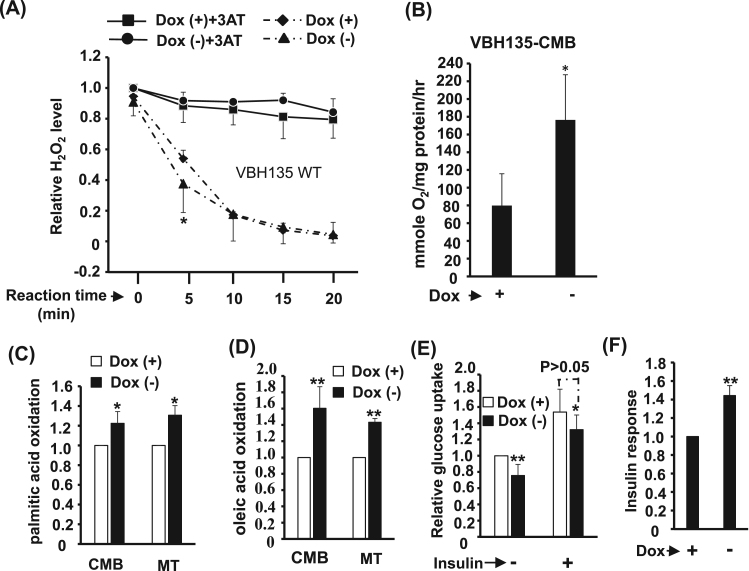
The effects of VBH135 expression on metabolism were analyzed and we found that Catalase activity was enhanced by VBH135, suggesting for better peroxisome activity/efficiency (Fig. 7A). We further found that O2 consumption was highly increased by VBH135 (Fig. 7B), and which was consistent with the increased oxidation of both PA and OA (Fig. 7C & D), implying more dependent on mitochondria and peroxisome mediated fatty acid oxidation. Surprisingly, although basal glucose uptake was slightly mitigated by VBH135, cellular response to insulin treatment was improved by it and the insulin induced glucose uptake was similar to control cells (Fig. 7E & F).
Fig. 7.
The effects of VBH135 over-expression on oxidative metabolism. (A) Catalase activity is enhanced in C2C12-VBH135 cells. Catalase activity in C2C12-VBH135 myotubes was examined in the presence/absence of 20 mM 3-amino-1, 2, 4-triazole (3AT, a catalase inhibitor) and H2O2 (10 mM) for 5–20 min by determining the residual H2O2 amount. The O2 consumption rate of CMB myoblast (B) and the oxidation of palmitic (C) and oleic acids (D) of cells at CMB and MT stages are determined. The basal and insulin (100 nM) stimulated glucose uptake of these cells at CMB stage is shown in (E), and their relative insulin responses (insulin stimulated/basal) are shown in (F). * and * *: p < 0.05 and p < 0.01, respectively, vs. Dox (+) cells.
3.5. Differential regulation of the expression of key MITO and peroxisome regulators by VBH135 and Bhlhe40
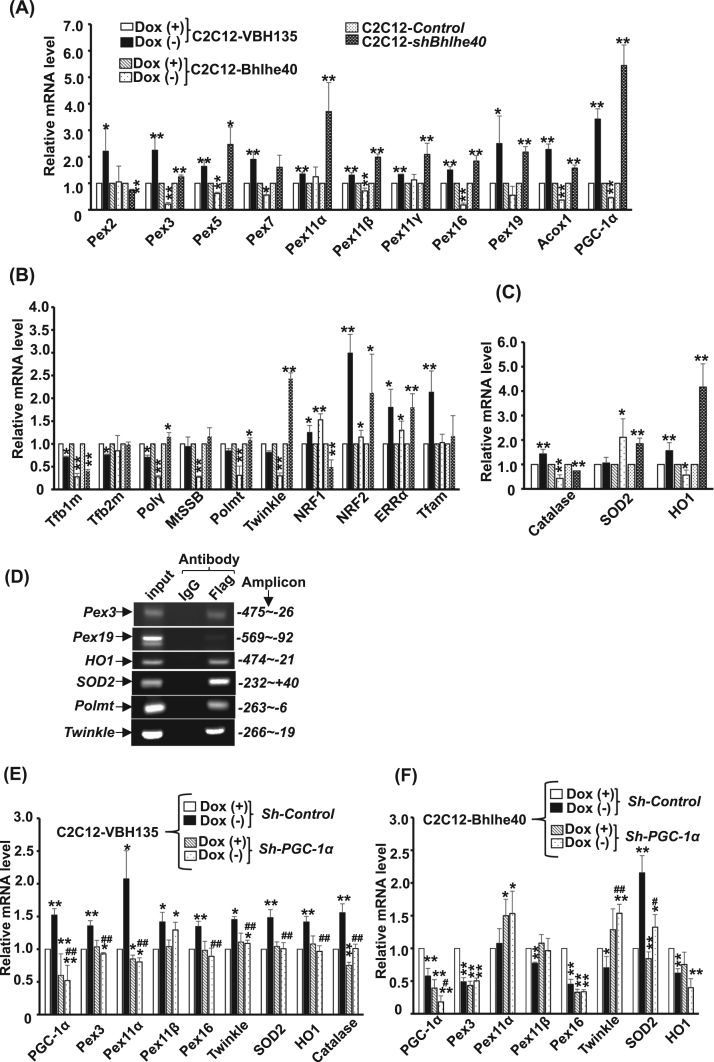
To identify factors mediating Bhlhe40 effects on MITO and peroxisomes, the expression of key factors regulating the biogenesis and functions of MITO and peroxisomes was examined by quantitative RT-PCR (qRT-PCR). We found that the mRNA levels of factors involved in peroxisome biogenesis, such as Pex3, Pex16, and Pex19, protein import, such as Pex5 and Pex7, and fission, such as Pex11β, were significantly increased by VBH135 but reduced by Bhlhe40 (Fig. 8A). Similar effects were seen on the expression of PGC-1α and the key fatty acid metabolizing enzyme Acox1 in peroxisomes. The expression of most peroxisome-related factors (except for Pex2) was also increased by Bhlhe40 knockdown.
Fig. 8.
The genes targeted by Bhlhe40 and VBH135. The genes expression patterns of C2C12-VBH135 and C2C12-Bhlhe40 myotubes and C2C12-shBhlhe40 myoblasts were examined with qRT-PCR. The relative levels of genes involved in peroxisome or mitochondrial biogenesis and functions are shown in (A) and (B) respectively. The results of genes involved in ROS scavenging are shown in (C). Binding of Bhlhe40 to target gene promoters was examined in C2C12-Bhlhe40 myotubes (Dox (-)) by ChIP assay (D). PGC-1α was knockdowned in C2C12-VBH135(E) and -Bhlhe40(F) myotubes to reveal PGC-1α-dependent gene regulation. * and * *: p < 0.05 and p < 0.01, respectively, vs. Dox (+) or control cells. # and ##: p < 0.05 and p < 0.01 vs. control ShRNA treated Dox (+)/(-) in (E) and (F).
Interestingly, the mRNA levels of most factors involved in mitochondrial replication or gene expression, such as Tfb1m, DNA polymerase γ (Polγ), and Twinkle, were repressed by both VBH135 and Bhlhe40, although other factors, such as MtSSB and RNA polymerase (Polmt), were only repressed by Bhlhe40 (Fig. 8B). NRF1, NRF2, and ERRα are transcription factors regulating MITO function and they are all upregulated by both VBH15 and Bhlhe40, implying indirect regulation. Bhlhe40 knockdown repressed Tfb1m and NRF1, but increased the expression of Twinkle, NRF2, and ERRα. Tfam expression was only activated by VBH135 but not altered by Bhlhe40 and its knowdown. The expression of ROS scavenging enzymes Catalase and HO1 was activated by VBH135 but reduced by Bhlhe40 as that of peroxisomal factors. SOD2, the major ROS scavenging enzymes in mitochondria, mRNA level was not affected by VBH135 but highly increased by Bhlhe40 (Fig. 8C). Both SOD2 and HO1 mRNA was enhanced by Bhlhe40 knockdown, and the SOD2 protein was also increased in this stable clone (Suppl. Fig. 4B). Furthermore, increased total SOD activity was found in C2C12-Bhlhe40 cells (Suppl. Fig. 4A). Taken together, the expression profiling results support the notion that VBH135 enhances peroxisomal, but slightly represses mitochondrial, biogenesis and oxidative metabolism; meanwhile, Bhlhe40 strongly represses oxidative metabolism in both organelles but enhances total ROS scavenging.
The recruitment of Bhlhe40 to these target gene promoters was also examined with ChIP assay and strong binding of Bhlhe40 to promoters of all 3 categories of genes was detected, suggesting regulation of these genes by Bhlhe40 in vivo (Fig. 8D). It was of interest to know whether Bhlhe40 directly binding to cis-elements in these genes to regulate their expression or indirectly through repressing PGC-1α expression/activity. To clarify this issue, the expression of some key factors was examined in C2C12-VBH135 and -Bhlhe40 cells after PGC-1α knockdown. We are surprised to find that, after PGC-1α knockdown, all the activation effects of VBH135 was suppressed and the repression of Pex3, Pex11, Pex16, and Twinkle by Bhlhe40 disappeared, implying a PGC-1α-dependent regulation (Fig. 8E & F). The activation of SOD2 by Bhlhe40 was also compromised by PGC-1α knockdown, demonstrating a partially PGC-1α-dependent regulation. However, the repression of HO1 by Bhlhe40 remained after PGC-1α knockdown, suggesting HO1 might be regulated by Bhlhe40 without involving PGC-1α. These observations together demonstrated the importance of Bhlhe40-mediated PGC-1α regulation in cellular oxidative respiration and ROS metabolism.
4. Discussion
Although Bhlhe40 has been implicated in many cellular processes, including differentiation, tumorigenesis, peripheral circadian output, and response to hypoxia [7], [10], [11], [12], its involvement in the regulation of mitochondrial and peroxisomal function and biogenesis has not been reported before. This study is the first one reporting its critical role in the regulation of these two organelles and should provide more support for its involvement in many physiological processes as both organelles play critical roles in energy supply and adaptation.
Our current study found that mtDNA was slightly increased in C2C12-shBhlhe40 cells (Supplementary Fig. 1B) but not in C2C12-Bhlhe40 cells (Fig. 3C), implying the minor role of Bhlhe40 in the regulation of mitochondrial number. On the other hand, ROS level was increased in C2C12-shBhlhe40 cells but reduced in C2C12-Bhlhe40 and -VBH135 cells, which strongly implicates the involvement of Bhlhe40 in the suppression of ROS in SKM and sufficient amount of Bhlhe40 is required to maintain ROS under harmful level. Furthermore, the increased mtDNA level but reduced MITO efficiency in the C2C12-shBhlhe40 cells might reflect mitochondrial dysfunction caused by surged ROS level [33]. Both MITO and peroxisomes are the major sources of ROS and they contain separate sets of enzymes for detoxification of ROS, for instance, SOD2 (Mn-SOD) is functioning in MITO but Catalase, SOD1 (Cu/Zn-SOD), and Prdx5, are functioning in the peroxisomes [32]. The previously reported Bhlhe40 target gene HO1 is actually a cytoplasmic ROS scavenging enzyme. Our results suggest that HO1 is a bona fide repressive target of Bhlhe40 containing Bhlhe40-binding sites in its regulatory cis-elements, but Catalase and SOD2 might be regulated by Bhlhe40 via antagonizing PGC-1α activity (Fig. 8D & E). These results suggest that Bhlhe40 regulates intracellular ROS level mainly by activating the expression of one set of ROS scavenging enzymes but repressing the expression of others, either directly or indirectly through PGC-1α. Under the condition of low Bhlhe40 expression, such as in C2C12-shBhlhe40, the expression of these activating targets is reduced, which in turn results in the accumulation of ROS and dysfunction of MITO.
Although it has been reported before that HO1 is activated by Bhlhe40, but evidence showing direct regulation of HO1 promoter or other cis-elements by Bhlhe40 is still lacking [9]. As the previous observation is contradictory to our current observations, further study on its promoter regulation by Bhlhe40 is required to solve this contradiction. The repressor role of Bhlhe40 has been studied in many genes, and it binds to the B class E-box (CACGTG) as homodimer instead of the N-box (CACGCG) on the promoters/enhancers of these genes [33]. Using bioinformatics tool, multiple E-boxes can be identified in the HO1 promoter region, and we are currently trying to dissect their function in the Bhlhe40 mediated repression of HO1.
It has been shown that Bhlhe40 protects muscle cells from oxidative damage and its absence leads to muscle necrosis in response to injury, a phenomenon usually associated with Duchenne Muscular Dystrophy [9]. These can be demonstrated by the Bhlhe40-/- mice, where the levels of TNFα is increased but the level of HO1 is reduced and apparent signs of oxidative stress is shown before muscle death. Its role in SKM protection can be further perceived as it is induced by genotoxic stress and hypoxia, two conditions that are associated with exercise [34]. As Bhlhe40 can function as a transcriptional activator and repressor, like the two blades of a sword, genes involved in producing genotoxic stress can be repressed by Bhlhe40 and those involved in scavenging ROS should be activated by Bhlhe40. The induction of SOD2 but repression of Catalase and HO1 by Bhlhe40 indicates that Bhlhe40 selectively activates some ROS scavenging enzymes but repress the others. Although PGC-1α level is increased in the C2C12-shBhlhe40 cells (Fig. 8), the ROS level still surged significantly (Fig. 2A & B), implying PGC-1α-independent activation of ROS scavenging enzymes by Bhlhe40 should play an important role in Bhlhe40-mediated ROS reduction. Further investigating on its regulation of other ROS scavenging enzymes, such as SOD1 and those in the peroxidase systems (including the thioredoxin/thioredoxin reductase/peroxiredoxin and the glutathione/glutathione peroxidase systems), should provide better insight into how ROS level is regulated by Bhlhe40.
Bhlhe40 has been shown to repress adipocyte differentiation via repressing the key adipogenic factor, PPARγ2, so its role in negative regulation of fatty acid oxidation in adipocytes is predictable [35]. Its negative role in fatty acid oxidation is also found in myocytes, but its mechanism remain elusive [36]. Our previous study had shown the enhanced fatty acid oxidation in C2C12-VBH135 and -shBhlhe40 cells, and the expression of factors participating in oxidation of fatty acids in mitochondria was also increased in these cells [19]. Here we further demonstrated that Bhlhe40 can also negatively regulate the expression of factors involved in peroxisomal fatty acid oxidation and in the biogenesis of peroxisomes (Fig. 8). As Bhlhe40 is ubiquitously expressed so its role in tissues with low PGC-1α expression, such as liver and spleen [37], should be further assessed to reveal the participation of its PGC-1α-independent regulation of fatty acid metabolism in these tissues.
The number of peroxisomes in a cell reflects its metabolic status and it is balanced by the biogenesis and autophagy (or pexophagy for peroxisomes) processes. The number of peroxisomes can be increased, through either fission of existing peroxisomes or de novo biogenesis from ER derived membrane, by treating cells with peroxisome proliferator, such as fibrates, or its substrates, such as oleic acid (an unsaturated fatty acid) to switch the metabolic dependence to peroxisome-dependent metabolism [17]. Our observations here suggest that Bhlhe40 targets both mitochondria and peroxisomes oxidative metabolism, but only negatively regulates the number of peroxisomes by reducing the expression of its key biogenic factors. This peroxisome-specific regulation seems to be independent of PGC-1α as PGC-1α is deeply involved in the function and biogenesis of both organelles [1], [18]. Further study on the regulation of peroxisome biogenesis genes by Bhlhe40 should shed light on this PGC-1α-independent biogenesis pathway. Besides, the fission of peroxisomes is probably mildly or not targeted by Bhlhe40 as the expression of Pex11 and factors in the fission machinery (shared with mitochondria) is only mildly affected by both VBH135 and Bhlhe40 (Fig. 8 and data not shown).
Most obsolete peroxisomes (about 80%) are degraded through pexophagy [38], in which both Pex14, a component of the Pex5 PTS-1 receptor docking complex, and ubiquitinated peroxisome membrane proteins (Ub-PMPs, such as Pex5 and PMP70), can trigger pexophagy [39], [40], [41]. The ubiquitination of PMPs is majorly mediated by Pex2, an E3 ubiquitin ligase in the Pex2/Pex10/Pex12 translocation complex, which then recruits the autophagic adaptor p62/NBR1 and receptor LC3 to initiate pexophagy [42]. Under starvation, Pex14 can directly interact with LC3-II, the phosphatidylethanolamine conjugated and active form of LC3, to activate pexophagy. As Bhlhe40 negatively regulates peroxisome number, it will be interesting to know whether pexophagy is enhanced by Bhlhe40 under normal and pathologic conditions.
Bhlhe40 is involved in many diverse physiological processes and also responses to various physiological stimuli. It can be induced by retinoic acid in p19 embryonic carcinoma stem cells, where it inhibits mesodermal, but promotes neuronal, differentiation [43]. Both parathyroid hormone and its 2nd messenger cAMP can induce Bhlhe40 mRNA in chondrocytes [44], but EGF can only accumulate Bhlhe40 protein in mammalian epithelium [45]. In breast cancer cells, Bhlhe40 is induced by TGFβ to mediate the anti-apoptotic effect [46]. It is highly induced by hypoxia and is highly augmented in most types of tumors, although reduced in some cases [7], [12], [47]. In tumor tissues examined, it is predominantly expressed in viable cells surrounding the necrosis tissues, suggesting its dual roles in response to hypoxia: induced by hypoxia to antagonize apoptosis but turns into an apoptotic factor in extreme hypoxia condition [12]. Bhlhe40 can also response to circadian rhythm, in which it is induced by Clock-Bmal1 proteins to regulate a subset of peripheral circadian output [11], [48]. It can also be induced by insulin via PI3K pathway in the liver of diabetic mice [49]. The discovery of its implication in the regulation of mitochondrial and peroxisome functions and number in this study has provided a very sound answer to why/how it is involved in such a diverse range of physiological processes, as both organelles are metabolically required in these processes. Therefore, understanding its role in the regulation of both organelles is key to further understanding the regulation of these physiological processes. In the near future, more endeavor should be devoted to unravel the mechanisms by which Bhlhe40 employed to regulate nuclear/mitochondrial genes participating in peroxisome and mitochondrial functions and regulation, so novel drugs targeting SKM metabolic diseases can be developed.
Acknowledgements
The authors like to thank Dr. Marc Fransen (Katholieke Universiteit Leuven, Belgium) for providing roGFP-PTS1 and KillerRed-PTS1 expression vectors. We are also grateful to Dr. Shuang En-Chuang (National Health Research Institutes) for help with the promoter assays. Thanks also go to Drs. Hong-Chen Chen and Won-Jin Wang (Institute of Biochemistry and Molecular Biology, National Yang Ming University, Taiwan) for their help during revision. This study was supported by funding from the National Science Council of Taiwan, ROC to Shen Liang Chen [NSC-102-2311-B-008-003 and MOST 105-2311-B-008-001]. The authors have no conflict of interest to declare.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.10.009.
Appendix A. Supplementary material
Supplementry figure 1: The differences between Control and Bhlhe40 knockdown cells. (A) The expression levels of Bhlhe40 were determined by qRT-PCR (left panel) and Western blot (right panel). Gapdh and Lamin B1 signals are used as input controls for mRNA and protein, respectively. (B) The levels of mitochondrial DNA were examined with quantitative PCR, in which nuclear genes MyoD and Oct4 were used as input control for determining the levels of mitochondrial genes cytochrome b and Cox2. (C) The relative insulin responses of control and Bhlhe40 knockdown cells. (D) Detection of Phospho- and total mTOR in vehicle and insulin treated myoblasts by Western blot. Relative levels of phospho-mTOR are shown at the bottom panel. (E) The growth curve of control and Bhlhe40 knockdown cells established during a 4 day culture on 6-well dishes. * and * *: p < 0.05 and p < 0.01, respectively, vs. control cells. Supplementry figure 2: The influences of Bhlhe40 or VBH135 on cells. (A) Images of Mitotracker and DAPI stained C2C12-Bhlhe40 myoblasts. (B, C) The protein levels of myosin heavy chain (MHC), Bhlhe40-flag, and VBH135-flag in myotubes (triplicates) of Tet-off regulated stable clones were determined by Western blot. (D) The fusion indexes of C2C12-VBH135 and -VBH135m after in DM for 4 days. Supplementry figure 3: The RFP-PTS1 specially marks peroxisomes Immunofluorescent detection of Catalase was performed on C2C12 cells stably expressing RFP-PTS1 (C2C12-RFP-PTS1). The RFP (A) and FITC-labeled Catalase (B) images were merged in (C) to demonstrate co-localization of both signals. A higher magnification image is shown in (D). All images were taken at 400X magnification. Supplementry figure 4: SOD activity and SOD2 expression. Total SOD activity in C2C12-VBH135 and -Bhlhe40 myotubes and in C2C12-Control/-shBhlhe40 myoblasts was determined (A), The SOD2 protein levels under the same condition were determined by Western blot and is shown in (B). Supplementary material
Supplementary material
Supplementary material
References
- 1.Wu Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., Spiegelman B.M. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 2.Yoon J.C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C.R., Granner D.K., Newgard C.B., Spiegelman B.M. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 3.Finck B.N., Kelly D.P. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Investig. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin J., Wu H., Tarr P.T., Zhang C.Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., Lowell B.B., Bassel-Duby R., Spiegelman B.M. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 5.Chang J.H., Lin K.H., Shih C.H., Chang Y.J., Chi H.C., Chen S.L. Myogenic basic helix-loop-helix proteins regulate the expression of peroxisomal proliferator activated receptor-gamma coactivator-1alpha. Endocrinology. 2006;147:3093–3106. doi: 10.1210/en.2005-1317. [DOI] [PubMed] [Google Scholar]
- 6.Hsiao S.P., Huang K.M., Chang H.Y., Chen S.L. P/CAF rescues the Bhlhe40-mediated repression of MyoD transactivation. Biochem. J. 2009;422:343–352. doi: 10.1042/BJ20090072. [DOI] [PubMed] [Google Scholar]
- 7.Ivanova A.V., Ivanov S.V., Danilkovitch-Miagkova A., Lerman M.I. Regulation of STRA13 by the von Hippel-Lindau tumor suppressor protein, hypoxia, and the UBC9/ubiquitin proteasome degradation pathway. J. Biol. Chem. 2001;276:15306–15315. doi: 10.1074/jbc.M010516200. [DOI] [PubMed] [Google Scholar]
- 8.Sun H., Li L., Vercherat C., Gulbagci N.T., Acharjee S., Li J., Chung T.K., Thin T.H., Taneja R. Stra13 regulates satellite cell activation by antagonizing Notch signaling. J. Cell Biol. 2007;177:647–657. doi: 10.1083/jcb.200609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vercherat C., Chung T.K., Yalcin S., Gulbagci N., Gopinadhan S., Ghaffari S., Taneja R. Stra13 regulates oxidative stress mediated skeletal muscle degeneration. Human. Mol. Genet. 2009;18:4304–4316. doi: 10.1093/hmg/ddp383. [DOI] [PubMed] [Google Scholar]
- 10.Guillaumond F., Lacoche S., Dulong S., Grechez-Cassiau A., Filipski E., Li X.M., Levi F., Berra E., Delaunay F., Teboul M. Altered Stra13 and Dec2 circadian gene expression in hypoxic cells. Biochem Biophys. Res Commun. 2008;369:1184–1189. doi: 10.1016/j.bbrc.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Grechez-Cassiau A., Panda S., Lacoche S., Teboul M., Azmi S., Laudet V., Hogenesch J.B., Taneja R., Delaunay F. The transcriptional repressor STRA13 regulates a subset of peripheral circadian outputs. J. Biol. Chem. 2004;279:1141–1150. doi: 10.1074/jbc.M305369200. [DOI] [PubMed] [Google Scholar]
- 12.Turley H., Wykoff C.C., Troup S., Watson P.H., Gatter K.C., Harris A.L. The hypoxia-regulated transcription factor DEC1 (Stra13, SHARP-2) and its expression in human tissues and tumours. J. Pathol. 2004;203:808–813. doi: 10.1002/path.1585. [DOI] [PubMed] [Google Scholar]
- 13.Sun H., Taneja R. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc. Natl. Acad. Sci. USA. 2000;97:4058–4063. doi: 10.1073/pnas.070526297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian Y., Jung Y.S., Chen X. DeltaNp63, a target of DEC1 and histone deacetylase 2, modulates the efficacy of histone deacetylase inhibitors in growth suppression and keratinocyte differentiation. J. Biol. Chem. 2011;286:12033–12041. doi: 10.1074/jbc.M110.207241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y., Xie M., Yang J., Yang D., Deng R., Wan Y., Yan B. The expression of antiapoptotic protein survivin is transcriptionally upregulated by DEC1 primarily through multiple sp1 binding sites in the proximal promoter. Oncogene. 2006;25:3296–3306. doi: 10.1038/sj.onc.1209363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ribas V., Garcia-Ruiz C., Fernandez-Checa J.C. Mitochondria, cholesterol and cancer cell metabolism. Clin. Transl. Med. 2016;5:22. doi: 10.1186/s40169-016-0106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honsho M., Yamashita S., Fujiki Y. Peroxisome homeostasis: mechanisms of division and selective degradation of peroxisomes in mammals. Biochim. Biophys. Acta. 2016;1863:984–991. doi: 10.1016/j.bbamcr.2015.09.032. [DOI] [PubMed] [Google Scholar]
- 18.Bagattin A., Hugendubler L., Mueller E. Transcriptional coactivator PGC-1alpha promotes peroxisomal remodeling and biogenesis. Proc. Natl. Acad. Sci. USA. 2010;107:20376–20381. doi: 10.1073/pnas.1009176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung S.Y., Kao C.H., Villarroya F., Chang H.Y., Chang H.C., Hsiao S.P., Liou G.G., Chen S.L. Bhlhe40 represses PGC-1alpha activity on metabolic gene promoters in myogenic cells. Mol. Cell. Biol. 2015;35:2518–2529. doi: 10.1128/MCB.00387-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ivashchenko O., Van Veldhoven P.P., Brees C., Ho Y.S., Terlecky S.R., Fransen M. Intraperoxisomal redox balance in mammalian cells: oxidative stress and interorganellar cross-talk. Mol. Biol. Cell. 2011;22:1440–1451. doi: 10.1091/mbc.E10-11-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsiao S.P., Chen S.L. Myogenic regulatory factors regulate M-cadherin expression by targeting its proximal promoter elements. Biochem. J. 2010;428:223–233. doi: 10.1042/BJ20100250. [DOI] [PubMed] [Google Scholar]
- 22.Teng H.F., Kuo Y.L., Loo M.R., Li C.L., Chu T.W., Suo H., Liu H.S., Lin K.H., Chen S.L. Valproic acid enhances Oct4 promoter activity in myogenic cells. J. Cell Biochem. 2010;110:995–1004. doi: 10.1002/jcb.22613. [DOI] [PubMed] [Google Scholar]
- 23.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 24.Belke D.D., Larsen T.S., Lopaschuk G.D., Severson D.L. Glucose and fatty acid metabolism in the isolated working mouse heart. Am. J. Physiol. 1999;277:R1210–R1217. doi: 10.1152/ajpregu.1999.277.4.R1210. [DOI] [PubMed] [Google Scholar]
- 25.Butler I.B., Schoonen M.A., Rickard D.T. Removal of dissolved oxygen from water: a comparison of four common techniques. Talanta. 1994;41:211–215. doi: 10.1016/0039-9140(94)80110-x. [DOI] [PubMed] [Google Scholar]
- 26.Nedachi T., Kanzaki M. Regulation of glucose transporters by insulin and extracellular glucose in C2C12 myotubes. Am. J. Physiol. Endocrinol. Metab. 2006;291:E817–E828. doi: 10.1152/ajpendo.00194.2006. [DOI] [PubMed] [Google Scholar]
- 27.de Andrade P.B., Casimir M., Maechler P. Mitochondrial activation and the pyruvate paradox in a human cell line. FEBS Lett. 2004;578:224–228. doi: 10.1016/j.febslet.2004.10.088. [DOI] [PubMed] [Google Scholar]
- 28.Aebi H. Catalase in vitro. Methods Enzymol. 1984;105:121–126. doi: 10.1016/s0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- 29.Jardim-Messeder D., Caverzan A., Rauber R., de Souza Ferreira E., Margis-Pinheiro M., Galina A. Succinate dehydrogenase (mitochondrial complex II) is a source of reactive oxygen species in plants and regulates development and stress responses. New Phytol. 2015;208:776–789. doi: 10.1111/nph.13515. [DOI] [PubMed] [Google Scholar]
- 30.Lang K.C., Lin I.H., Teng H.F., Huang Y.C., Li C.L., Tang K.T., Chen S.L. Simultaneous overexpression of Oct4 and Nanog abrogates terminal myogenesis. Am. J. Physiol. Cell Physiol. 2009;297:C43–C54. doi: 10.1152/ajpcell.00468.2008. [DOI] [PubMed] [Google Scholar]
- 31.Theilen N.T., Kunkel G.H., Tyagi S.C. The role of exercise and TFAM in Preventing skeletal muscle atrophy. J. Cell Physiol. 2016 doi: 10.1002/jcp.25737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antonenkov V.D., Grunau S., Ohlmeier S., Hiltunen J.K. Peroxisomes are oxidative organelles. Antioxid. Redox Signal. 2010;13:525–537. doi: 10.1089/ars.2009.2996. [DOI] [PubMed] [Google Scholar]
- 33.St-Pierre B., Flock G., Zacksenhaus E., Egan S.E. Stra13 homodimers repress transcription through class B E-box elements. J. Biol. Chem. 2002;277:46544–46551. doi: 10.1074/jbc.M111652200. [DOI] [PubMed] [Google Scholar]
- 34.Thin T.H., Li L., Chung T.K., Sun H., Taneja R. Stra13 is induced by genotoxic stress and regulates ionizing-radiation-induced apoptosis. EMBO Rep. 2007;8:401–407. doi: 10.1038/sj.embor.7400912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yun Z., Maecker H.L., Johnson R.S., Giaccia A.J. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev. Cell. 2002;2:331–341. doi: 10.1016/s1534-5807(02)00131-4. [DOI] [PubMed] [Google Scholar]
- 36.Takeshita S., Suzuki T., Kitayama S., Moritani M., Inoue H., Itakura M. Bhlhe40, a potential diabetic modifier gene on Dbm1 locus, negatively controls myocyte fatty acid oxidation. Genes Genet. Syst. 2012;87:253–264. doi: 10.1266/ggs.87.253. [DOI] [PubMed] [Google Scholar]
- 37.Chung S.Y., Huang W.C., Su C.W., Lee K.W., Chi H.C., Lin C.T., Chen S.T., Huang K.M., Tsai M.S., Yu H.P., Chen S.L. FoxO6 and PGC-1alpha form a regulatory loop in myogenic cells. Biosci. Rep. 2013;33 doi: 10.1042/BSR20130031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim P.K., Hailey D.W., Mullen R.T., Lippincott-Schwartz J. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. USA. 2008;105:20567–20574. doi: 10.1073/pnas.0810611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hara-Kuge S., Fujiki Y. The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Exp. Cell Res. 2008;314:3531–3541. doi: 10.1016/j.yexcr.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J., Tripathi D.N., Jing J., Alexander A., Kim J., Powell R.T., Dere R., Tait-Mulder J., Lee J.H., Paull T.T., Pandita R.K., Charaka V.K., Pandita T.K., Kastan M.B., Walker C.L. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 2015;17:1259–1269. doi: 10.1038/ncb3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sargent G., van Zutphen T., Shatseva T., Zhang L., Di Giovanni V., Bandsma R., Kim P.K. PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J. Cell Biol. 2016;214:677–690. doi: 10.1083/jcb.201511034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boudjelal M., Taneja R., Matsubara S., Bouillet P., Dolle P., Chambon P. Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev. 1997;11:2052–2065. doi: 10.1101/gad.11.16.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen M., Kawamoto T., Teramoto M., Makihira S., Fujimoto K., Yan W., Noshiro M., Kato Y. Induction of basic helix-loop-helix protein DEC1 (BHLHB2)/Stra13/Sharp2 in response to the cyclic adenosine monophosphate pathway. Eur. J. Cell Biol. 2001;80:329–334. doi: 10.1078/0171-9335-00167. [DOI] [PubMed] [Google Scholar]
- 45.St-Pierre B., Cooper M., Jiang Z., Zacksenhaus E., Egan S.E. Dynamic regulation of the Stra13/Sharp/Dec bHLH repressors in mammary epithelium. Dev. Dyn. 2004;230:124–130. doi: 10.1002/dvdy.20013. [DOI] [PubMed] [Google Scholar]
- 46.Ehata S., Hanyu A., Hayashi M., Aburatani H., Kato Y., Fujime M., Saitoh M., Miyazawa K., Imamura T., Miyazono K. Transforming growth factor-beta promotes survival of mammary carcinoma cells through induction of antiapoptotic transcription factor DEC1. Cancer Res. 2007;67:9694–9703. doi: 10.1158/0008-5472.CAN-07-1522. [DOI] [PubMed] [Google Scholar]
- 47.Miyazaki K., Kawamoto T., Tanimoto K., Nishiyama M., Honda H., Kato Y. Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J. Biol. Chem. 2002;277:47014–47021. doi: 10.1074/jbc.M204938200. [DOI] [PubMed] [Google Scholar]
- 48.Honma S., Kawamoto T., Takagi Y., Fujimoto K., Sato F., Noshiro M., Kato Y., Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 49.Yamada K., Kawata H., Shou Z., Mizutani T., Noguchi T., Miyamoto K. Insulin induces the expression of the SHARP-2/Stra13/DEC1 gene via a phosphoinositide 3-kinase pathway. J. Biol. Chem. 2003;278:30719–30724. doi: 10.1074/jbc.M301597200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementry figure 1: The differences between Control and Bhlhe40 knockdown cells. (A) The expression levels of Bhlhe40 were determined by qRT-PCR (left panel) and Western blot (right panel). Gapdh and Lamin B1 signals are used as input controls for mRNA and protein, respectively. (B) The levels of mitochondrial DNA were examined with quantitative PCR, in which nuclear genes MyoD and Oct4 were used as input control for determining the levels of mitochondrial genes cytochrome b and Cox2. (C) The relative insulin responses of control and Bhlhe40 knockdown cells. (D) Detection of Phospho- and total mTOR in vehicle and insulin treated myoblasts by Western blot. Relative levels of phospho-mTOR are shown at the bottom panel. (E) The growth curve of control and Bhlhe40 knockdown cells established during a 4 day culture on 6-well dishes. * and * *: p < 0.05 and p < 0.01, respectively, vs. control cells. Supplementry figure 2: The influences of Bhlhe40 or VBH135 on cells. (A) Images of Mitotracker and DAPI stained C2C12-Bhlhe40 myoblasts. (B, C) The protein levels of myosin heavy chain (MHC), Bhlhe40-flag, and VBH135-flag in myotubes (triplicates) of Tet-off regulated stable clones were determined by Western blot. (D) The fusion indexes of C2C12-VBH135 and -VBH135m after in DM for 4 days. Supplementry figure 3: The RFP-PTS1 specially marks peroxisomes Immunofluorescent detection of Catalase was performed on C2C12 cells stably expressing RFP-PTS1 (C2C12-RFP-PTS1). The RFP (A) and FITC-labeled Catalase (B) images were merged in (C) to demonstrate co-localization of both signals. A higher magnification image is shown in (D). All images were taken at 400X magnification. Supplementry figure 4: SOD activity and SOD2 expression. Total SOD activity in C2C12-VBH135 and -Bhlhe40 myotubes and in C2C12-Control/-shBhlhe40 myoblasts was determined (A), The SOD2 protein levels under the same condition were determined by Western blot and is shown in (B). Supplementary material
Supplementary material
Supplementary material