Abstract
ADP-ribosylation is a reversible posttranslational modification used to regulate protein function. ADP-ribosyltransferases transfer ADP-ribose from NAD+ to the target protein, and ADP-ribosylhydrolases, such as ADPRHL2, reverse the reaction. We used exome sequencing to identify five different bi-allelic pathogenic ADPRHL2 variants in 12 individuals from 8 families affected by a neurodegenerative disorder manifesting in childhood or adolescence with key clinical features including developmental delay or regression, seizures, ataxia, and axonal (sensori-)motor neuropathy. ADPRHL2 was virtually absent in available affected individuals’ fibroblasts, and cell viability was reduced upon hydrogen peroxide exposure, although it was rescued by expression of wild-type ADPRHL2 mRNA as well as treatment with a PARP1 inhibitor. Our findings suggest impaired protein ribosylation as another pathway that, if disturbed, causes neurodegenerative diseases.
Keywords: ribosylation, posttranslational modification, ADPRHL2, ARH3, PARP, neurodegeneration, seizure, ataxia, neuropathy, cerebellar atrophy
Main Text
Pediatric neurodegenerative diseases are progressive conditions typically associated with severe disability or even death in early infancy. Identification of the underlying genetic defects is a prerequisite for a better understanding and eventually prevention of the pathophysiological cascades causing pathology. Despite the implementation of unbiased genome-wide sequencing in clinical routine, it is estimated that at least half of the affected individuals and their families remain without a definitive molecular diagnosis. One explanation among many others is that for many loci, a putative disease association remains to be established, and although a specific gene defect might be an obvious candidate in a single affected individual, it could take years to collectively observe enough additional individuals with the same rare condition.
Here, we report on the results of an exome sequencing study in eight families with individuals clinically diagnosed with a neurodegenerative syndrome but without a molecular diagnosis. Informed consent was obtained from all affected individuals or their guardians. The study was approved by the local ethics committees.
Individual F1:II.3, a female, was born after an uneventful pregnancy. Her early development was normal. At 1 year, 5 months of age, she was able to walk independently, but a delay of speech development was noticed. From the age of 3 years on, she had recurrent episodes of diplopia and right-sided ataxic-dystonic posturing possibly triggered by exercise. These signs progressed over several years, and she developed tics such as facial grimacing and throat clearing. Initial brain MRI at age 3 years was without obvious pathological findings. At the age of 12 years, her condition worsened with more frequent episodes of ataxic-dystonic posturing, ataxia, hypokinetic movements, intermittent hypesthesia, pain, and fatigue. Although cognitive function was reported to be unaffected, brain MRI showed bilateral hippocampal diffusion restriction. Laboratory investigations of blood and cerebrospinal fluid, as well as extensive metabolic investigations, were noncontributory. At the age of 12 years, 2 months, she experienced a drowning event necessitating resuscitation and hypothermia therapy. She completely recovered, but brain MRI at age 12 years, 4 months showed novel pathologies (putatively secondary to hypoxia) involving bilateral basal ganglia, cerebellar, and parietal white matter, as well as the central cortex. Disease progressed with onset of pain in the lumbar area, increasing trunk inclination, ataxia, inability to climb stairs, unsteady gait, and impaired cognition followed by unprovoked pain in the upper limbs and tetraplegia. Respiratory insufficiency necessitated mechanically assisted ventilation from the age of 12 years, 11 months on. Her clinical state further deteriorated, and she deceased at the age of 14 years.
Individual F2:II.2, a female, was born to healthy consanguineous parents from Lebanon. Her two siblings are healthy. Pregnancy, delivery, and postnatal adaption were reportedly normal. Cognitive and motor developmental milestones were reached during infancy. However, from the age of 4 years on, febrile convulsions occurred, and she had mild developmental delay in fine motor skills. At the age of 8 years, muscular weakness, slowed movements, and a progressive nodding of the head triggered by, e.g., cold water were noticed. She developed a progressive ataxia and weakness leading to gait abnormalities and loss of independent ambulation. Brain and spinal MRI at the age of 12 years revealed atrophy of the cerebellum and spinal cord. At the age of 13 years, an acute life-threatening event occurred with unclear acute respiratory insufficiency possibly in the context of a seizure. She was subsequently resuscitated twice because of suspected neurogenic asystole. Prolonged episodes of respiratory insufficiency required mechanical ventilation. A tracheostomy was performed, and a percutaneous endoscopic gastrostomy tube was placed because of dysphagia. Her condition slightly improved to intermediate usage of ventilator support and independent eating. Over the course of the disease, she developed a neurogenic bladder-voiding disorder. At the age of 14 years, 9 months, her condition deteriorated during an infectious episode with respiratory insufficiency requiring mechanical ventilation. Feeding problems increased, and she developed a paralytic ileus. Furthermore, increasing sleep disturbances were reported, and she displayed extensive facial myoclonia. A clinically suspected axonal sensorimotor peripheral neuropathy was supported by absent responses in nerve conduction studies. There was no reliable response to acoustic or optical stimuli, and she lost her ability to speak. Follow-up brain MRI showed progressive atrophy of the cerebellum, edema of the cortex with especially right-sided volume increase, and signal alterations in putamina, caudate nuclei, and the white matter of the corpus callosum. We assume that the neuroimaging findings that evolved in addition to the cerebellar atrophy are likely to represent secondary findings resulting from episodes of respiratory insufficiency with hypoxia. Extensive laboratory testing was noncontributory. Electroencephalography showed a burst suppression pattern, and she died at 17 years of age.
Clinical and genetic findings are summarized in Table 1, pedigrees are shown in Figure 1, and neuroimaging findings are shown in Figure 2. Clinical descriptions of the remaining individuals are provided in the Supplemental Note.
Table 1.
Clinical and Genetic Findings in Individuals with Confirmed Bi-allelic ADPRHL2 Variants
| F1:II.2 | F1:II.3 | F2:II.2 | F3:II.1 | F4:II.3 | F4:II.4 | F5:II.2 | F6:II.1 | F7:II.1 | F7:II.2 | F8:II.1 | F8:II.3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | male | female | female | female | male | female | male | female | female | female | male | female |
| Ethnicity | German | German | Lebanese | ND | ND | ND | Kosovan | Polish | Chinese | Chinese | Turkish | Turkish |
| Identified homozygous changes (cDNA [GenBank: NM_017825.2] and protein GenBank: NP_060295) | c.1004T>G (p.Val335Gly) | c.1004T>G (p.Val335Gly) | c.744_746del p.(Lys248_Ile249delinsAsn) |
c.1038C>G (p.Tyr346∗) | c.1004T>G (p.Val335Gly) | c.1004T>G (p.Val335Gly) | c.1004T>G (p.Val335Gly) | c.1004T>G (p.Val335Gly) | c.309−1G>T (p.?) | c.309−1G>T (p.?) | c.292delG (p.Val98Trpfs∗23) | c.292delG (p.Val98Trpfs∗23) |
| Phenotypic Features | ||||||||||||
| Age of onset (current age or age of death) | 1 y (27 y) | 17 m (Ɨ 14 y) | 4 y (Ɨ 17 y) | 2 y (12 y) | 13 y (32 y) | 11 y (Ɨ 30 y) | 3 y (7 y) | 2 y (Ɨ 11 y) | 1 y (Ɨ 12 y, 10 m) | 2 y (Ɨ 5 y) | 15 m (Ɨ 4 y, 4 m) | 14 m (22 m) |
| Developmental delay or intellectual impairment | + | + | + | + | − | − | + | + | + | + | + | + |
| Gait abnormalities | + | + | + | + | + | + | + | + | + | + | + | + |
| Ataxia | ND | + | + | + | + | + | + | + | + | + | + | + |
| Seizures | ND | − | + | + | − | − | − | + | + | − | + | + |
| Neuropathy | ND | + | + | + | ND | ND | + | + | + | + | − | ND |
| Facial myoclonia | − | + | + | − | − | − | − | − | − | − | − | − |
| Sensorineural hearing loss | − | − | − | + | − | − | − | − | ND | − | ND | ND |
| Ophthalmologic features | − | diplopia | nystagmus | strabismus | nystagmus, strabismus | − | putative external ophthalmoplegia with ptosis, impaired saccades and upward gaze, and nystagmus; putative retinal pigment epithelium anomalies | − | ND | − | − | − |
| Microcephaly | ND | ND | ND | + | ND | ND | + | − | ND | ND | − | + |
| Respiratory insufficiency | − | + | + | − | − | + | − | − | + | + | + | − |
| cMRI Findings (Affected Regions) | ||||||||||||
| Basal ganglia | ND | + | + | − | − | − | − | − | − | − | + | − |
| Cortex | ND | + | + | − | − | − | − | − | − | − | − | − |
| Corpus callosum | ND | − | + | + | − | − | − | − | − | − | − | − |
| Cerebellum | ND | + | + | + | + | + | + | − | + | + | − | − |
| Other | ||||||||||||
| Nerve conduction studies (biopsy) | ND | axonal sensorimotor neuropathy | no signal | axonal sensorimotor neuropathy | ND | ND | axonal motor neuropathy | axonal motor neuropathy | ND | axonal sensorimotor neuropathy (decreased number of nerve fibers) | reportedly normal | ND |
| Respiratory chain activities (muscle) | ND | unremarkable | unremarkable | ND | ND | unremarkable | ND | unremarkable | ND | unremarkable | unremarkable | unremarkable |
Abbreviations are as follows: y, years; m, months; ND, not determined; +, present; −, absent, and Ɨ, individual deceased.
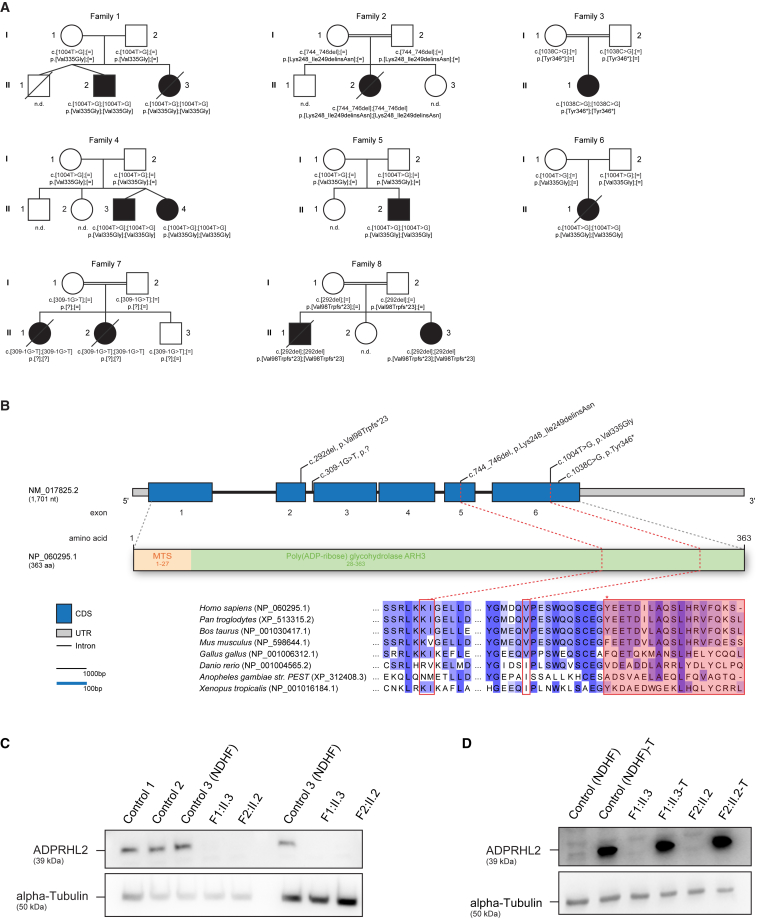
Figure 1.
Pedigrees of Investigated Families, Structure of ADPRHL2, and Investigation of ADPRHL2 Amounts
(A) Pedigrees of eight families carrying pathogenic variants in ADPRHL2 illustrate the mutation-carrier status of affected (closed symbols) and healthy (open symbols) family members. ND, not determined.
(B) Structure of ADPRHL2 (top) and ADPRHL2 (bottom) with known protein domains and motifs of the gene product and position of the identified variants. Intronic regions are not drawn to scale.
(C and D) Immunoblot studies on ADPRHL2-mutant fibroblast cell lines (C) and transduced cell lines (D) indicated that the homozygous variants c.1004T>G (p.Val335Gly) and c.744_746del (p.Lys248_Ile249delinsAsn) demonstrate a severe reduction of ADPRHL2. Transduction with wild-type ADPRHL2 led to increased amounts of the protein. Immunoblotting was performed on whole-cell lysates with anti-ADPRHL2 antibody (Sigma-Aldrich, HPA027141, dilution 1:200). An anti-alpha-tubulin antibody (Sigma-Aldrich, T5168, dilution 1:20,000) was used as a loading control.
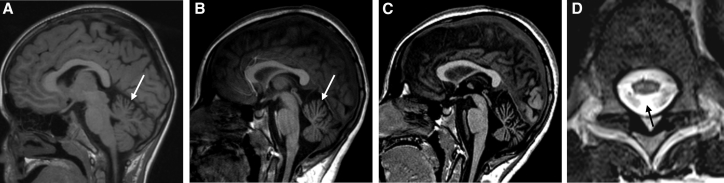
Figure 2.
Neuroimaging Findings in Individuals with ADPRHL2 Variants
(A) Brain MRI (T1-weighted image, sagittal view) of individual F5:II.2 at the age of 7 years demonstrates mild upper vermian atrophy (arrow).
(B and C) Brain MRI (T1-weighted image, sagittal view) of individual F2:II.2 at the ages of 12 years (B) and 14 years (C) demonstrates progressive cerebellar atrophy (arrow).
(D) MRI of the myelon (T2-weighted image, axial view) of individual F2:II.2 at the age of 12 years shows atrophy of the spinal cord and bilateral cord T2 hyperintensities (arrow).
Pregnancy, postnatal adaption, and perinatal development were reportedly normal in all individuals, and all but individual F1:II.2 were born at term. Neurodevelopmental problems involving a delay in speech and psychomotor development were noted in 10 of 11 individuals within the first years of life, and five individuals presented with infection-associated episodes of ataxia or dystonic posturing. Over the course of the disease, gait abnormalities were present in all individuals. 10 of 11 developed ataxia, and individual F1:II.2 showed a spastic diplegia, which could have resulted from perinatal hypoxic brain damage. Seizures and corresponding electroencephalography abnormalities were documented in six individuals. Peripheral axonal isolated motor or sensorimotor neuropathy, as shown by decreased amplitudes with normal latencies in nerve conduction studies, was present in six of eight individuals. Facial myoclonia, a possible sign of developing bulbar palsy, was present in two of the affected individuals. Visual impairment manifesting as diplopia (1/5), nystagmus (3/5), strabismus (2/5), and impaired upward gaze and saccadic movements and ptosis (1/5) was reported in 5 of 11 affected individuals. Additional findings included acquired microcephaly in individuals F5:II.2, F3:II.2, and F8:II.3, and F3:II.2 also showed sensorineural hearing loss. Disease progress was variable but frequently associated with periods of increased stress, such as infections. Three individuals died in childhood, whereas in another five individuals, disease progressed into their teens, and two had life-threatening events requiring resuscitation and mechanically assisted ventilation for respiratory insufficiency.
Extensive laboratory testing and metabolic investigations were not contributory in any affected individuals. Biochemical analysis was performed on skeletal muscle specimen of four individuals and showed normal activity of mitochondrial respiratory-chain enzymes. Histological examinations showed evidence of neurogenic muscle atrophy.
Neuroimaging data were available for all but one individual and were considered unremarkable at an early disease stage. However, over the course of disease, eight of ten individuals developed cerebellar atrophy (Figure 2). At a late stage of the disease, putatively secondary additional abnormalities affecting the central cortical region (2/10), basal ganglia (3/10), and corpus callosum (2/10) were observed.
Exome sequencing was performed at four centers—Munich (families 1, 2, 4, 5, and 8), Baylor Genetics (family 3), Warsaw (family 6), and Beijing (family 7)—on genomic DNA from affected individuals F1:II.3, F2:II.2, F3:II.1, F4:II.3, F5:II.2, F6:II.1, F7:II.2, and F8:II.3, as well as the parents from families 4 and 5, as described previously.1, 2, 3 In individual F1:II.3, prioritization of potentially pathogenic variants included a search for recessive-type non-synonymous variants with a minor allele frequency (MAF) < 0.1% in both an in-house database containing ∼3,000 control exomes and the Exome Aggregation Consortium (ExAC) Browser (accessed January 2018). This search failed to identify pathogenic or likely pathogenic variants in established disease-related genes associated with clinical features of the affected individuals. Given the proposed role of the encoded protein in posttranslational protein modification and in silico prediction, the homozygous missense change c.1004T>G (p.Val335Gly) (GenBank: NM_017825.2) in ADPRHL2 (MIM: 610624), coding for ADP-ribosylhydrolase like 2, was initially considered a promising candidate in individual F1:II.3. The same homozygous missense variant was subsequently identified in the similarly affected individuals of families F4–F6. In families F1, F4, and F5, a comparison of the sequence variation observed in the ∼2 Mb region surrounding the variant identified ten homozygous rare (MAF < 1% in public databases) variants. This finding is in line with shared ancestry. Furthermore, an additional four different homozygous ADPRHL2 variants—an in-frame deletion (c.744_746del [p.Lys248_Ile259delinsAsn]), a predicted truncating variant (c.1038C>G [p.Tyr346∗]), a canonical splice-site variant (c.309−1G>T [p.?]), and a frameshift variant (c.292del [p.Val98Trpfs∗23])—were identified in four additional unrelated individuals with a similar clinical phenotype. The results of carrier testing performed on available family members were in line with autosomal-recessive inheritance. The change c.1004T>G (p.Val335Gly) (rs201735454) is observed 27 times in a heterozygous state in 277,240 alleles of the gnomAD browser, the change c.1038C>G (p.Tyr346∗) (rs531916765) is present three times in 246,234 alleles, and the variants c.744_746del (p.Lys248_Ile259delinsAsn), c.292del (p.Val98Trpfs∗23), and c.309−1G>T (p.?) have not been observed in at least 227,988 alleles. None of the variants have been reported in a homozygous state in public databases (ExAC Brower or gnomAD) or an in-house database containing >3,000 exome datasets of individuals with unrelated phenotypes. No bi-allelic loss-of-function variants were observed in ∼120,000 alleles of the ExAC Browser. Immunoblot studies in primary fibroblasts available from individuals F1:II.3 and F2:II.2 showed an absence of ADPRHL2 (Figure 1). In summary, the identification of five different bi-allelic functionally relevant ADPRHL2 variants in eight unrelated families establishes ADPRHL2 as a gene confidently implicated in this neurodegenerative disease that includes developmental delay or regression, seizures, ataxia, and neuropathy as key clinical features.
ADPRHL2 is thought to function in the pathway of ADP-ribosylation, which is a reversible posttranslational modification used to regulate key cellular processes such as transcription, DNA repair, translation, and apoptosis.4 In humans, several ADP-ribose transferases transfer ADP-ribose from nicotinamide adenine dinucleotide (NAD+) to target proteins. Among different protein families putatively catalyzing this reaction, the probably best-characterized enzymes are so-called poly(ADP-ribose) polymerases (PARPs).5 Some members of the PARP family are able to transfer only ADP-ribose monomers, whereas others produce poly(ADP-ribose) chains.5 A well-characterized member is PARP1. Once activated by genotoxic stress-induced single-strand DNA breaks, it produces poly(ADP)-ribosylation associated with depletion of cellular NAD+ and translocation of mitochondrial proapoptotic factors to the nucleus.6 Given that the ultimate consequence of persistent ADP-ribosylation is parthanatos, these dynamic processes require precise regulation, and ADP-ribosylation has to be reversible.7, 8 Poly(ADP-ribose) (PAR) hydrolysis is catalyzed by several enzymes. The function of PAR glycohydrolase (PARG) is well studied: it cleaves the ribose-ribose bonds between ADP-ribose subunits of the poly(ADP-ribose) chains.9 Another enzyme able to reverse protein poly(ADP)-ribosylation is ADPRHL2. It hydrolyses PAR chains on proteins, albeit less efficiently than PARG.10 So far, ADPRHL2 is the only known poly(ADP-ribose)-hydrolyzing enzyme in mitochondria.11 Given that PARG and ADPRHL2 exclusively hydrolyze poly(ADP-ribose) chains, a number of additional factors—including OARD1 (also called TARG1), MACROD1, MACROD2, and ARH—are necessary to remove the last ADP-ribose subunit attached to a protein.12, 13, 14
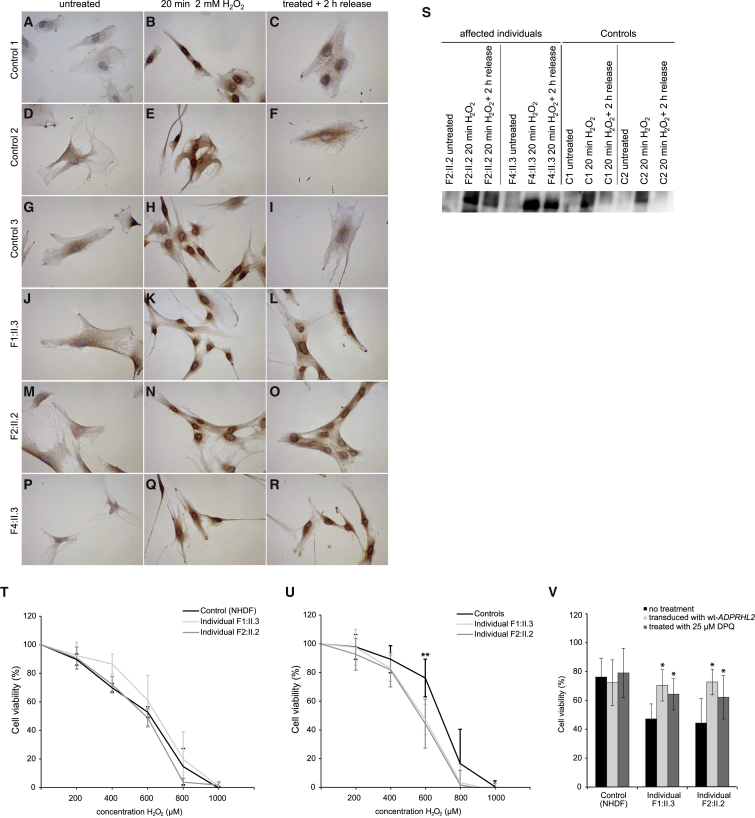
We used fibroblast cell lines of individuals F1:II.3 and F2:II.2 to study the cellular consequences of ADPRHL2 deficiency. PARP1 is the main polymerase contributing to the intracellular PAR pool.15, 16 Hydrogen peroxide (H2O2) stimulates PARP1 via oxidative DNA damage and thereby leads to an increase in intracellular PAR.17 Given the proposed role of ADPRHL2 in reversing poly(ADP)-ribosylation by hydrolyzing PAR into mono(ADP-ribose), we postulated ADPRHL2 deficiency to promote H2O2-mediated accumulation of PAR. To test this hypothesis, we performed immunohistochemical staining of ADP-ribosylation in fibroblasts of affected and control individuals. Treatment of fibroblasts with 2 mM H2O2 for 20 min resulted in a marked ring-shaped accumulation of ADP-ribose in the perinuclear and nuclear region in both control and affected individuals’ cell lines (Figure 3). Amounts of ADP-ribose normalized 2 hr after H2O2 removal in control fibroblasts, whereas a ring-shaped signal remained in ADPRHL2-mutant fibroblasts (Figure 3). This observation is in line with impaired cellular removal of ADP-ribose as a consequence of ADPRHL2 deficiency. PAR accumulation in ADPRHL2 fibroblasts was also documented by immunoblotting (Figure 3).
Figure 3.
Investigation of ADP-Ribosylation and Cell Viability upon H2O2 Treatment
(A–R) Control (A–I) and mutant (J–R) primary fibroblasts were seeded on chamber slides and allowed to attach overnight. Treatment with 2 mM H2O2 for 20 min increased ADP ribose in both control (B, E, and H) and mutant (K, N, and Q) cell lines, as shown by immunohistochemistry using the anti-pan-ADP-ribose binding reagent (MABE1016, Merck Millipore, diluted 1:1,000 in 1× blocking solution [Roche] in PBS and 0.5% Tween-20). Subsequent incubation for 2 hr in normal cell-culture medium led to a marked decrease in staining intensity in control cell lines (C, F, and I), whereas the signal intensity remained high in fibroblasts from ADPRHL2-deficient individuals (L, O, and R).
(S) Immunoblot analysis of ADP ribose in fibroblasts of affected and control individuals. Fibroblasts were untreated, treated with 2 mM H2O2 for 20 min, or treated with H2O2 for 20 min and allowed to recover for 2 hr (release). Staining was performed with anti-pan-ADP-ribose binding reagent (MABE1016, Merck-Millipore).
(T and U) Cell-viability analysis using alamarBlue Cell Viability Reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. 2,500–3,000 cells were plated per well on 96-well plates 24 hr before treatment. Cells were cultured either in high-glucose (T) or low-glucose (U) DMEM before and during treatment with H2O2 at concentrations from 0 to 1,000 μM for 48 hr (T) or 24 hr (U).
(V) Cell-viability analysis using alamarBlue Cell Viability Reagent after treatment with 600 μM H2O2 for 24 hr in low-glucose DMEM after transduction and treatment with 25 μM DPQ.
The data represent at least five experiments for each cell line grown and treated in parallel. Error bars indicate 1 SD from the mean. ∗p < 0.01, ∗∗p < 0.001, two-tailed unpaired t test.
PAR accumulation has been associated with increased cell death.7 To assess this hypothesis, we exposed ADPRHL2-mutant and control fibroblast cell lines cultured in high-glucose DMEM (4.5 g/L; Thermo Fisher Scientific) to H2O2 concentrations ranging from 0 to 1,000 μM for 48 hr. We assessed cell viability by quantification with alamarBlue Cell Viability Reagent (Thermo Fisher Scientific) as a readout. We did not detect a significant difference in cell viability between affected individuals’ fibroblasts and control cells (Figure 3). A potential explanation for this observation could be that in this setting, alternative cytosolic PAR-hydrolyzing enzymes can compensate for defective ADPRHL2 function.
As mentioned previously, ADPRHL2 is the only characterized PAR-hydrolyzing enzyme to date in mitochondria.11 Furthermore, an increase in PARP1 activity has been linked to impaired mitochondrial metabolism.18 In order to more specifically challenge stress ADPRHL2-dependent PAR hydrolysis, we next cultured the cell lines in low-glucose DMEM (1 g/L; Thermo Fisher Scientific) to promote mitochondrial respiration as a source of cellular energy supply. Cell viability was determined after 24 hr treatment with H2O2 at concentrations of 0 to 1,000 μM. At a concentration of 600 μM, we observed significantly lower cell viability in ADPRHL2-mutant cell lines than in control cells (Figure 3).
To confirm that this effect was indeed caused by a deficiency of ADPHRL2, we performed a rescue experiment. We transduced ADPRHL2-deficient and control fibroblasts with wild-type ADPRHL2 cDNA by using a feline-immunodeficiency-virus-based lentiviral expression vector (GeneCopoeia) as described previously.19 Increased amounts of ADPHRL2 were confirmed by immunoblot analysis (Figure 3). Transduced fibroblasts grown on low-glucose DMEM with 600 μM H2O2 showed a significant higher cell viability than naive ADPRHL2-mutant cell lines (Figure 3).
To further corroborate the hypothesis that PAR accumulation is indeed the mechanism mediating increased H2O2 sensitivity in ADPRHL2-deficient cell lines, we treated the cells with the PARP1 inhibitor 3,4-dihydro-5-[4-(1-piperidinyl)butoxyl]-1(2H)-isoquinolinone (DPQ, Sigma-Aldrich). Viability of ADPRHL2-mutant fibroblasts cultured in low-glucose DMEM with 600 μM H2O2 for 24 hr was significantly increased upon treatment with 25 μM DPQ. Our findings provide additional evidence for the functional relevance of the investigated ADPRHL2 variants, supporting PAR accumulation as a pathomechanism mediating cell death from increased oxidative stress.
In conclusion, we identified bi-allelic ADPRHL2 variants as the disease-causing molecular defects underlying a progressive neurodegenerative disorder in nine affected individuals from seven families. Although larger studies are needed to fully define the phenotypic spectrum associated with ADPRHL2 deficiency, overlapping findings in several individuals suggest that disease manifestation with episodic movement disorders at the beginning with periods of partial recovery might constitute clinical hallmarks that deserve special notion in this disease. Additional suggestive features are development of ataxia and peripheral (sensori-)motor axonal neuropathy over the course of the disease. Similar to murine fibroblasts deficient of the mouse ortholog (Arh3−/−), affected individuals’ fibroblasts grown on respiratory medium were less viable upon H2O2 stress than control cells.20 This cellular phenotype was rescued by expression of wild-type ADPRHL2 mRNA as well as treatment with a PARP1 inhibitor, the latter of which supports the hypothesis that PAR accumulation is an important factor in the pathophysiology of this disease. PAR accumulation by PARP1 hyperactivation has been suggested as a mechanism involved in the pathogenesis of the autosomal-recessive disorders spinocerebellar ataxia 26 (SCAR26 [MIM: 617633]) and ataxia oculomotor apraxia 4 (AOA4 [MIM: 616267]), caused by mutations in XRCC1 and its’ interaction partner PNKP, respectively. Notably, the phenotypic spectrum associated with SCAR26 and especially AOA4 dysfunction shows remarkable overlap with ADPRHL2 deficiency, including progressive ataxia, oculomotor abnormalities, and peripheral sensorimotor neuropathy.21 More generally, an increase in intracellular PAR has been associated with more common neurological disorders, such as Alzheimer disease,22 Parkinson disease, and amyotrophic lateral sclerosis.23 Although the above-mentioned changes are likely to be secondary phenomena, in addition to our report, currently only one report directly implicates impaired recycling of ADP-ribosylation in neurodegeneration. In 2013, Sharifi et al. described several affected individuals from a single large family affected by childhood-onset neurodegeneration manifesting as severe neurodevelopmental delay, seizures, and peripheral neuropathy leading to death by the age of 10–11 years. Functional studies showed that the identified mutations in the candidate gene OARD1 (MIM: 614393; also known as TARG1 or C6orf130) result in defective degradation of ADP-ribosylation.14 Our findings further support the concept of disturbed posttranslational protein-modification pathways, such as ADP-ribosylation, in neurodegenerative diseases.
In both C. elegans and mouse neurons, inhibition of poly(ADP-ribosylation) leads to improved neuronal regeneration after axonal injury.24 Speculatively, the prevention of excessive PAR accumulation with its detrimental downstream effects culminating in cell death could evolve as a potential therapeutic target in selected neurodegenerative disease entities in humans.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank all the families for their participation. This study was supported by the German Bundesministerium für Bildung und Forschung (BMBF) through the Juniorverbund in der Systemmedizin “mitOmics” (FKZ 01ZX1405C to T.B.H.). H.P. was supported by the E-Rare project GENOMIT (01GM1603 and 01GM1207), by the EU FP7 Mitochondrial European Educational Training Project (317433), and by the EU Horizon2020 Collaborative Research Project SOUND (633974). D.P. was supported by the intramural grants S148/16 and S145/16. R.P. was supported by the National Science Centre (NCN) Poland grant 2013/11/B/NZ7/04944. Research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number R01NS08372 to P.E.B., and F.F. and M.X. were supported by the Key Special Project of National Key Research and Development Program “Research on prevention and control of major chronic non-communicable diseases: Early identification and comprehensive intervention of brain development disorders in children.”
Published: October 25, 2018
Footnotes
Supplemental Data include a Supplemental Note and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.10.005.
Web Resources
ExAC Browser, http://exac.broadinstitute.org
OMIM, http://www.omim.org
Combined Annotation Dependent Depletion (CADD), http://cadd.gs.washington.edu/
Supplemental Data
References
- 1.Haack T.B., Hogarth P., Kruer M.C., Gregory A., Wieland T., Schwarzmayr T., Graf E., Sanford L., Meyer E., Kara E. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am. J. Hum. Genet. 2012;91:1144–1149. doi: 10.1016/j.ajhg.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ploski R., Pollak A., Müller S., Franaszczyk M., Michalak E., Kosinska J., Stawinski P., Spiewak M., Seggewiss H., Bilinska Z.T. Does p.Q247X in TRIM63 cause human hypertrophic cardiomyopathy? Circ. Res. 2014;114:e2–e5. doi: 10.1161/CIRCRESAHA.114.302662. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y., Muzny D.M., Xia F., Niu Z., Person R., Ding Y., Ward P., Braxton A., Wang M., Buhay C. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibson B.A., Kraus W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 5.Vyas S., Chesarone-Cataldo M., Todorova T., Huang Y.H., Chang P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat. Commun. 2013;4:2240. doi: 10.1038/ncomms3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiber V., Dantzer F., Ame J.C., de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 7.Andrabi S.A., Kim N.S., Yu S.W., Wang H., Koh D.W., Sasaki M., Klaus J.A., Otsuka T., Zhang Z., Koehler R.C. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc. Natl. Acad. Sci. USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Dawson V.L., Dawson T.M. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp. Neurol. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slade D., Dunstan M.S., Barkauskaite E., Weston R., Lafite P., Dixon N., Ahel M., Leys D., Ahel I. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477:616–620. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oka S., Kato J., Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J. Biol. Chem. 2006;281:705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 11.Niere M., Mashimo M., Agledal L., Dölle C., Kasamatsu A., Kato J., Moss J., Ziegler M. ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose) J. Biol. Chem. 2012;287:16088–16102. doi: 10.1074/jbc.M112.349183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mashimo M., Kato J., Moss J. Structure and function of the ARH family of ADP-ribosyl-acceptor hydrolases. DNA Repair (Amst.) 2014;23:88–94. doi: 10.1016/j.dnarep.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barkauskaite E., Jankevicius G., Ahel I. Structures and mechanisms of enzymes employed in the synthesis and degradation of PARP-dependent protein ADP-ribosylation. Mol. Cell. 2015;58:935–946. doi: 10.1016/j.molcel.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Sharifi R., Morra R., Appel C.D., Tallis M., Chioza B., Jankevicius G., Simpson M.A., Matic I., Ozkan E., Golia B. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32:1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai P., Cantó C. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell Metab. 2012;16:290–295. doi: 10.1016/j.cmet.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 16.Krishnakumar R., Kraus W.L. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol. Cell. 2010;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang S., Lin Y., Kim Y.S., Hande M.P., Liu Z.G., Shen H.M. c-Jun N-terminal kinase mediates hydrogen peroxide-induced cell death via sustained poly(ADP-ribose) polymerase-1 activation. Cell Death Differ. 2007;14:1001–1010. doi: 10.1038/sj.cdd.4402088. [DOI] [PubMed] [Google Scholar]
- 18.Brunyanszki A., Szczesny B., Virág L., Szabo C. Mitochondrial poly(ADP-ribose) polymerase: The Wizard of Oz at work. Free Radic. Biol. Med. 2016;100:257–270. doi: 10.1016/j.freeradbiomed.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danhauser K., Iuso A., Haack T.B., Freisinger P., Brockmann K., Mayr J.A., Meitinger T., Prokisch H. Cellular rescue-assay aids verification of causative DNA-variants in mitochondrial complex I deficiency. Mol. Genet. Metab. 2011;103:161–166. doi: 10.1016/j.ymgme.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Mashimo M., Kato J., Moss J. ADP-ribosyl-acceptor hydrolase 3 regulates poly (ADP-ribose) degradation and cell death during oxidative stress. Proc. Natl. Acad. Sci. USA. 2013;110:18964–18969. doi: 10.1073/pnas.1312783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoch N.C., Hanzlikova H., Rulten S.L., Tétreault M., Komulainen E., Ju L., Hornyak P., Zeng Z., Gittens W., Rey S.A., Care4Rare Canada Consortium XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature. 2017;541:87–91. doi: 10.1038/nature20790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love S., Barber R., Wilcock G.K. Increased poly(ADP-ribosyl)ation of nuclear proteins in Alzheimer’s disease. Brain. 1999;122:247–253. doi: 10.1093/brain/122.2.247. [DOI] [PubMed] [Google Scholar]
- 23.Lee Y., Kang H.C., Lee B.D., Lee Y.I., Kim Y.P., Shin J.H. Poly (ADP-ribose) in the pathogenesis of Parkinson’s disease. BMB Rep. 2014;47:424–432. doi: 10.5483/BMBRep.2014.47.8.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byrne A.B., McWhirter R.D., Sekine Y., Strittmatter S.M., Miller D.M., Hammarlund M. Inhibiting poly(ADP-ribosylation) improves axon regeneration. eLife. 2016;5:e12734. doi: 10.7554/eLife.12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.