Abstract
During development, transcriptional properties of progenitor cells are stably propagated across multiple cellular divisions. Yet, at each division, chromatin faces structural constraints imposed by the important nuclear re-organization operating during mitosis. It is now clear that not all transcriptional regulators are ejected during mitosis, but rather that a subset of transcription factors, chromatin regulators and epigenetic histone marks are able to ‘bookmark’ specific loci, thereby providing a mitotic memory. Here we review mechanisms of mitotic bookmarking and discuss their impact on transcriptional dynamics in the context of multicellular developing embryos. We document recent discoveries and technological advances, and present current mathematical models of short-term transcriptional memory.
Keywords: Mitotic bookmarking, Epigenetics, Transcription factors, Development, Memory, Histone modifications, Mathematical modeling, Transcription
Graphical abstract
Highlights
-
•
Mitotically retained factors are able to ‘bookmark’ specific loci during embryogenesis.
-
•
Mitotic bookmarking can elicit rapid post-mitotic transcriptional re-activation.
-
•
Mathematical models relating transcriptional memory predict that efficient memory requires slow dynamics.
-
•
Mitotic memory leads to a spectrum of consequences: stability, flexibility or plasticity.
Introduction
During development of multicellular organisms, transcriptional programs must be faithfully transmitted during each cellular division to ensure cell fate maintenance. Two steps of the cell cycle, mitosis and replication exhibit particular topological constraints, hindering stable inheritance of transcriptional repertoires. However it is now well established that mitosis is not always accompanied by a total erasure of past chromatin states, from mother to daughter cells [1]. On the contrary, memory of active and repressed chromatin states exists, both at the short-term between successive mitoses and in longer time scale through multiple generations [2].
In this review, we focus on recent advances in understanding how active states are transmitted through cellular divisions in vivo. We will discuss potential mechanisms and possible mathematical models of mitotic bookmarking and present their consequences in developing multicellular embryos.
Mechanisms of short-term memory have also been analyzed in the context of cultured cell lines and recent reviews summarize these findings 1, 3. Similarly, long-term memory of repressed states mediated by Polycomb family has been intensively reviewed and will not be discussed [4].
Mitosis, a challenging chromatin environment?
During mitosis, genome undergoes a dramatic metamorphose, resulting in clear morphological features of mitotic cells. This chromatin landscape was long thought to represent a hostile environment, incompatible with transcription and transcription factor binding [5].
However, recent studies using sensitive techniques nuance this view. While chromatin folding and compartmentalization are wiped out during mitosis 3, 6, local accessibility can be maintained. ATAC-Seq experiments on mitotic Drosophila embryos revealed that patterns of accessibility are largely maintained through mitosis for several regulatory elements [7]. One may think that this is peculiar to the early fly embryo, where divisions are particularly fast. However genome accessibility is also widely preserved during mitosis in murine erythroblast cells, yet with local loci specific modulations [8]. Mitotic chromatin landscape has not yet been examined in vertebrate embryos, probably because of the relatively rapid loss in synchrony of divisions, operating as early as the 4-cell stage in zebrafish embryos [9]. This technical issue should be bypassed by recent developments of single cell technologies. Indeed, single cell Hi-C experiments were performed in Embryonic Stem Cells (ESCs) throughout the cell cycle, thus revealing a much more dynamic picture of chromosome organization than previous bulk experiments with unsynchronized cells [10].
Mitotic bookmarking and development
Transmission of chromatin states from mother to daughter cells can be achieved passively or via active mechanisms. Among known active supports of mitotic memory, stand three classes of regulators: epigenetic histone marks/chromatin regulators, general transcription factors (GTF) and sequence specific transcription factors (TF) (Figure 1).
Figure 1.
Putative supports of memory of active genes/of transcriptional memory. Mutually non-exclusive mechanisms of mitotic bookmarking occurring at promoters and/or enhancers. A. Previously transcribing locus remains partially accessible during mitosis, thus facilitating post-mitotic re-activation. B. Histone marks and chromatin regulators (readers, writers) of an active chromatin state bind mitotic chromatin, thus ‘bookmarking’ particular loci for subsequent transcriptional activation. C. Transcription factors such as pioneer factors can associate to a subset of their targets during mitosis, consequently favoring their activation at mitotic exit. D. Through enhancer/promoter priming, all mechanisms illustrated in (A–C) lead to a rapid post-mitotic transcriptional activation.
The vast majority of experiments aiming to assess mitotic retention of these particular factors has been performed in cultured cells with more or less conflicting conclusions, partly due to the level of purity of the mitotic population, cross linking conditions and to the methods of detection (global decoration by imaging versus specific binding assessed by ChIP). In this section, rather than providing an exhaustive survey of potential mitotic bookmarkers [1], we focus on potential supports for transmission of active chromatin states in the context of multicellular developing embryos (Figure 2).
Figure 2.
Known mitotic chromatin landscapes. Potential supports of memory are illustrated by examples reported in embryos from different model organisms. A. Example of an accessible chromatin in mitotic Drosophila embryos, as shown by ATAC-seq experiments by Ref. [7]. B. Example of a histone modification mark, present in zebrafish embryonic dividing cells, revealed by fluorescent immunostaining (Dapi in blue, acetylated histone 3 and 4 in red) [23]. C. Representative example of mitotic chromosomal decoration by a transcription factor (Essrb), in mouse embryo at morula stage [30]. D. Illustration of mitotic retention of a chromatin regulator, the histone methyltransferase Ash1 in living Drosophila embryos. Snapshot images from a time-lapse movie [20].
Passive mechanisms of mitotic bookmarking
So far, clear evidences of passive supports of memory are lacking. In a recent work, by monitoring transcriptional activation in living early Drosophila embryos, it was shown that experiencing transcription prior to mitosis does not always lead to a rapid post-mitotic activation [11]. With a mesodermal enhancer, memory of active transcriptional states is unequivocally occurring [12], while with a dorsal ectoderm enhancer, memory is not detected [11].
However, given the widespread maintenance of chromatin accessibility during mitosis in the fly embryo, a passive mechanism is plausible (Figure 1).
In interphase, gene expression seems to be partially regulated by local permissive or repressive environments, triggered by nuclear compartmentalization resulting from protein liquid–liquid demixing through phase separation [13] (e.g. HP1 in embryos [14]). The impact of phase separation during mitosis remains elusive, however mitotic structures can be assembled through phase separation in early Caenorhabditis elegans embryos [15]. Therefore, it is tempting to speculate that part of the mitotic maintenance of regulatory region accessibility could be due to local biochemical nuclear compartmentalization via phase separation. Whether this phenomenon stands as a passive or active support of mitotic memory is an open question.
Epigenetic marks, their readers and writers
Owing to decades of genetic and biochemical studies, it is well established that antagonistic actions of Polycomb group proteins (PcG) and Trithorax group proteins (TrxG) allow for dynamic regulation of developmental genes, yet with a cellular memory 4, 16. Epigenetic transmission of active transcriptional states is supported by conserved multifaceted TrxG complexes 4, 16. Among the best-characterized TrxG function is its histone methyltransferase activity leading to the trimethylation of lysine 4 of histone H3 tails (H3K4me3).
Whether H3K4me3 qualifies as an epigenetic mark during development is debated. Indeed, using a sensitive imaging technique (proximal ligation assay) in Drosophila embryos, H3K4me3 does not appear to be stable through replication [17], whereas it is clearly retained at discrete loci during mitosis [18]. In another model organism, Xenopus embryos, H3K4me3 has been functionally associated to memory of active transcriptional states using nuclear transfer experiments [19]. Contrary to the myriad of available data concerning PcG complexes, little is known concerning the mechanisms of TrxG recruitment and particularly during mitosis in vivo. Generally speaking, we can distinguish two non-mutually exclusive mechanisms of mitotic epigenetic bookmarking: either through histone-modified tails (epigenetic mark), protected or not by their ‘reader’ enzymes or through their re-establishment after mitosis by chromatin ‘writer’ enzymes (Figure 1).
In Drosophila embryos, Ash1, a ‘writer’ member of TrxG, decorates mitotic chromosomes [20] (Figure 2). It is not known whether the mark deposited by this enzyme, H3K36me2 is also retained during mitosis. Interestingly, Ash1 enhances the recruitment of other histone methyltransferases like Trx and its mammalian homolog MLL [21], which in turn triggers H3K4 trimethylation at promoters 16, 22.
Another mitotically retained member of TrxG family is the bromodomain-containing protein 4 (BRD4), shown to coat mitotic chromosomes of zebrafish embryos [23], even before zygotic transcriptional activation. Although with little validation in embryos, extensive work in mammalian cultured cells shows that BRD4 is a pleiotropic transcriptional and epigenetic regulator [24]. Interestingly H4K5ac, a mark recognized by BRD4 is detectable at particular promoters during mitosis in mammalian cells [25] as well as acetylated H4 in zebrafish embryos [23] and could thus be considered as a true ‘epigenetic’ bookmark (Figure 2).
Mitotic bookmarking by RNA Pol II machinery
Very few studies directly examined the localization of Pol II machinery during mitosis in embryos. The early fly embryo exhibits synchronous mitotic waves, whereby various steps of mitosis can be visualized in a single embryo. Using this ideal context, it was recently shown that active Pol II (Pol II-Ser5P) is present during prophase but is largely evicted at metaphase [7]. During mitosis, the clear majority of transcription ceases. However recent studies that employ sensitive techniques (for example pulse-labeling nascent RNA) nuance this statement and reveal low-level mitotic transcription in cell culture [26]. These paradigm-shifting findings would need to be confirmed in physiological conditions in vivo. However, they are in agreement with mitotic retention of GTF as the TATA binding protein, TBP, shown to bind mitotic chromosomes in ES [27] cells and to decorate chromatin in dividing mouse blastocysts [28].
Mitotic bookmarking by transcription factors
In developing embryos, very few sequence specific transcription factors have been shown to remain associated to their targets during mitosis. However recent whole-genome profiling and live imaging revealed that several pluripotency TFs have the ability to bind mitotic chromosomes in cultured dividing ES cells 1, 29. This mitotic retention might also occur in mouse embryos but remains to be demonstrated. So far, among the few reported mitotically bound TF during development, stand Essrb (Figure 2) and Klf4 in dividing mouse blastomeres •30, 31 and HNF1beta, retained during renal development in mice (Pontoglio lab, personal communication). Future investigations regarding TF retention during mitosis in embryos would require testing multiple fixation procedures and cross-validations with live imaging approaches. Indeed, formaldehyde fixation can lead to the artifactual displacement of some TFs from mitotic chromosomes •32, 33.
Interestingly, the majority of the known mitotically retained TFs, for example some pluripotency factors (e.g. Oct4/Sox2/Klf4) function as pioneer factors [34]. Pioneer factors are a particular class of TFs, able to engage their target DNA at compacted nucleosomal regions, thereby fostering subsequent access to classical non-pioneer TF to their targets [35]. It is thus legitimate to ask whether pioneer factors might have intrinsic properties to bind chromatin during mitosis. However, little is known regarding the functional importance of pioneer factors for mitotic memory in developing multicellular organisms. Intriguingly, the pioneer factor Zelda, an essential activator of the early zygotic genome in Drosophila embryos, is not retained during mitosis and does not contribute to mitotic memory [11]. This could reflect that mitotic bookmarking is not a general feature of all pioneer factors, or alternatively, that Zelda is not a canonical pioneer factor.
Mitotic retention: stable versus dynamic bookmarking
Whole-genome kinetics of nucleosome turnover is much more dynamic that what conceptual ideas of stable epigenetic inheritance of chromatin states may provide. In cultured cells, histone modifications can be often erased and re-established several time during the cell cycle [36]. This finding may not be contradictory with mitotic bookmarking, when the binding properties of chromatin regulators are examined with advanced fluorescent microscopy methods (FCS, FRAP or SPT [37]).
In early Drosophila embryos, quantitative in vivo analysis of Ash1 dynamic properties demonstrate that this mitotically retained histone methyltransferase engages chromatin dynamically with estimated residence times in the order of seconds [20].
Similarly, in ES cells, pluripotency transcription factors bind mitotic chromosomes dynamically (e.g. residence time of Sox2 ∼10 s) 1, 29, •38.
Of note, dynamic binding could be attributed to either sequence specific binding events, or to non sequence specific DNA binding [38]. The long-lived interactions, in the order of seconds generally reflect sequence specific bindings [29]. However, evidence for non-specific DNA binding during mitosis exists (e.g. FoxA) [39].
In order to clearly distinguish specific from non-specific binding events, one should ideally use mitotic Chip-seq experiments with quantitative imaging of DNA binding mutant versions of the TF assessed [37].
In sharp contrast to the dynamic mode of binding described earlier, some factors can bind to mitotic chromosomes in a stable manner. This is exemplified with TBP, which exhibits an average residence time in the order of minutes, in living dividing ES cells [27]. In conclusion, mitotic retention can occur through dynamic or stable binding, via sequence specific binding and non-specific binding.
Consequences of mitotic binding on transcriptional dynamics
Mitotic retention does not directly bestow a bookmarking function. To firmly prove that a mitotically retained factor is involved in ‘memory’, one need to examine the consequences of its transient mitotic-specific depletion on dynamics of post-mitotic transcriptional activation.
These experiments are challenging and have thus far never been performed in embryos. However knockdown experiments of candidate bookmarking factors have been performed in synchronized mitotic cells 26, 27, •38 and in Dictyostelium [40]. Generally two types of outputs are assessed: whole-genome approaches to examine bulk transcriptional levels or live imaging following transcriptional reporters in single cells.
Mitotic-specific depletion of the hematopoietic transcription factor GATA1 in erythroid precursors delays the reactivation of key lineage specifying mitotically bookmarked genes [41]. Similarly, TBP mitotic transient depletion in ES cells delays the reactivation of the global ES cell transcriptional programme [27]. While displaying an overall genome-wide picture, these ensemble approaches lack temporal resolution. The MS2/MCP system allows to directly monitor actively transcribing loci (tagged with MS2 repeats) in living cells [42]. This technique not only measures the dynamics of transcriptional activation but also provides access to the lineage and thus to mitotic memory of active genes.
Using such approaches, memory of active genes has been first visualized in Dictyostellium [40], in mammalian cells [25] and more recently in Drosophila [12]. Using a stochastically expressed transgene, Ferraro et al. [12] could distinguish transcriptionally active mother nuclei from their inactive neighbors. Quantitative analyses reveal that there is a higher probability for rapid reactivation after mitosis, when the mother was active. This bias corresponds to transcriptional mitotic memory. In the future, generalization of live imaging of transcription methods should allow for memory detection in other model organisms.
Mathematical modeling of memory
A quantitative measure of mitotic memory is the difference in the timing of post-mitotic re-activation between descendants of active and inactive mothers. This difference can be explained by the fact that these two populations 'travel' through different states prior to activating transcription after mitosis. We can distinguish one active (ON) state and various inactive states (OFF), interpreted as more or less favorable chromatin landscapes [43]. The number of rate limiting kinetic steps leading from an inactive to an active state and vice versa can be higher than one, with lifetimes of inactive states ranging from one to several hundreds of minutes [43]. By stabilizing a competent yet inactive state (OFF1), mitotic bookmarking could prevent the decline of a mother cell to a less permissive state (OFF2), allowing a faster transcriptional recovery of the daughters (Figure 3).
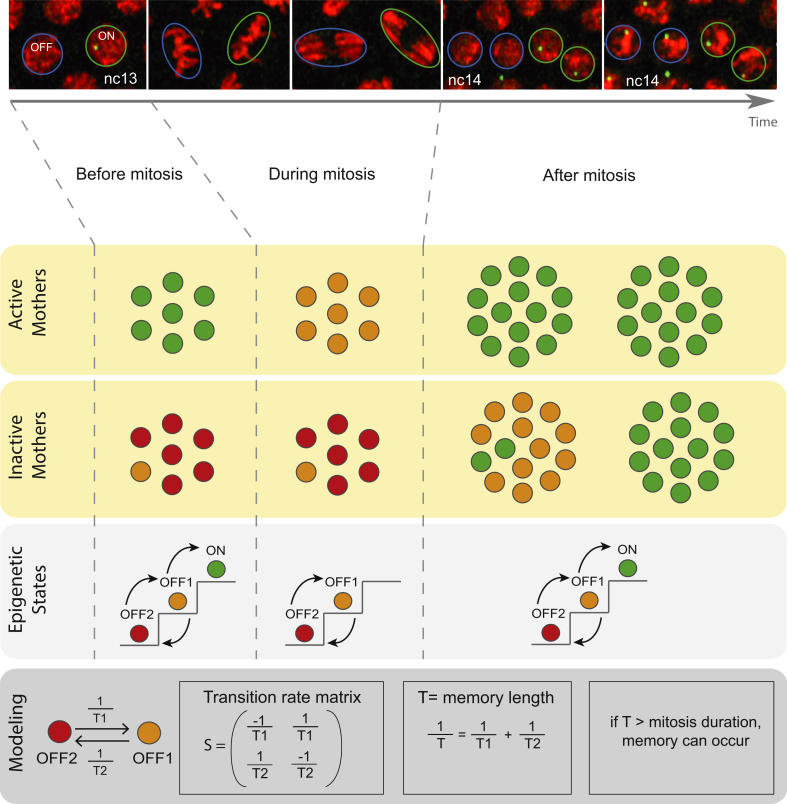
Figure 3.
Modeling memory of active states in an embryo. A. Snapshots of live imaging of an MS2-reporter transgene in a Drosophila embryo at nuclear cycle (nc) 13, during mitosis and at nc14. Nuclei are labeled with a Histone-RFP transgene and nascent MS2-mRNAs with MCP-GFP in green spots (Images from M. Lagha laboratory). B. Model of memory with two inactive states OFF1 (inactive but competent) and OFF2 (inactive and repressed). A third state ON is transcribing. The lifetimes of states OFF1, OFF2 are T1, T2, and correspond to the transition time from OFF1 to OFF2, and from OFF2 to OFF1, respectively. T3 is the time to go from the state OFF1 to the active state. The transition rates 1/T1, 1/T2, 1/T3 are reciprocals of the transition times. The memory length T, computed from the eigenvalues of the transition rate matrix, is T1T2/(T1+T2). Prior to mitosis, active mother nuclei (bookmarked) will be in state ON, whereas the population of inactive mothers will be distributed among states OFF1 and OFF2. During mitosis, the ON state is no longer accessible (transcription ceases). Therefore, previously active nuclei will be simply downgraded to state OFF1 while, provided that mitosis length is short compared to T, inactive nuclei will keep their states. After mitosis, daughters will be able to switch from OFF1 to ON or travel progressively from OFF2 to OFF1 to ON. The path to activation is shorter and consequently, the probability per unit of time to activate transcription after mitosis is increased for descendants of active mothers. Within a developmental pattern, this memory bias could favor the temporal coordination in gene activation (i.e. synchrony), with consequences in terms of cell fate.
A rather general class of mathematical models of memory can be based on Markov chains, used to explain transcriptional memory in Drosophila embryos [11] (Figure 3). Originally, Markov chains were used to describe memoryless processes. However, these models can acquire memory by an operation called “projection”. By projection, instead of following the dynamics of all the states of the system, one observes only the result of this “hidden” dynamics, namely the mRNA production. The projected dynamics exhibit memory in the sense that the future of the observed variable (mRNA production) depends not only on its present, but also on all past events that occurred prior to a time T (T is called memory length). This method, known as Mori-Zwanzig projection operator technique [44], has been recently applied to analyze propagation of signals in gene networks, as those occurring during vertebrate neural tube patterning [45]. In the model presented here (Figure 3), memory results from hidden states preservation, whereas memory length corresponds to the required time to forget differences between initial values of these states. For a finite-state, continuous-time Markov chain model, this time can be easily computed as the inverse of the smallest, non-zero eigenvalue of the transition rate matrix, in absolute value [46]. In the case of two inactive states, characterized by lifetimes T1 and T2, respectively (Figure 3), memory length (T) is the harmonic mean of the states lifetimes, which is long when both lifetimes are long. Thus, this model suggests that efficient memory requires slow dynamics. Slow dynamics could in principle be favored by the presence of mitotic bookmarking, but this remains to be demonstrated.
Consequences of memory during development
In the context of a developing multicellular embryo, mitotic memory can lead to a multitude of consequences, sometimes with opposite outcomes.
Inheritance of active transcriptional states ensures fidelity of transcriptional programs during their propagations through mitosis. Combined with other priming mechanisms during interphase, such as local chromatin opening at enhancers and promoter poising with paused polymerase, mitotic memory results in augmented transcriptional precision with less inter-cellular variability in levels of expression [47]. In sharp contrast, by allowing some cells to re-activate transcription faster than others, mitotic memory can enhance transcriptional noise [48]. From a theoretical point of view, memory can be seen as a ‘low pass filter’, buffering fast fluctuations, while allowing longer-lived fluctuations. Depending on memory length relative to mitosis duration and the architecture of the gene regulatory network (e.g existence of feedback loops), mitotic memory can thus create precision and increase noise, but both are valuable during development [47]. For example, temporal precision in gene expression has been shown to be essential to gastrulation in the fast developing Drosophila embryo [49]. However, in slower developing embryos such as the mouse blastocyst, heterogeneity in gene expression has been proposed as advantageous since it allows for mis-patterning corrections [50].
Mitotic memory thus allows for a spectrum of consequences ranging from stability to flexibility and plasticity. However, for the particular developmental contexts of trans-differentiation or reprogramming, transcriptional states must be erased rather than memorized. Accordingly, demethylation of H3K4 facilitates reprogramming of Xenopus embryos [19] and mouse epiblast cells [22].
Prospects
The vast majority of our knowledge regarding memory stems from studies performed in drug-synchronized cells in culture and has not yet been mirrored in multicellular developing embryos. This is mainly attributed to the dual technical challenge of profiling small number of cells/embryos and imaging living organisms.
With the recent technological advances in gene editing, signal amplification, microscopy combined to the possibility to probe the entire genome of limited material [51], developmental biology is embracing a new exciting era. Synergizing quantitative biological data with synthetic biology frameworks [52] and mathematical modeling opens promising avenues for a better understanding of mechanisms and functional relevance of memory.
Recent combination of optogenetics with concomitant detection of transcriptional readouts in living embryos [53] should greatly facilitate testing the direct impact of candidate bookmarking factors on kinetics of transcriptional post-mitotic reactivation.
Decoding the role of mitotic bookmarking in developing model organisms endows two obvious consequences for human therapy. Indeed defective mitotic bookmarking has been associated to human pathological conditions, as exemplified by HNF1beta [54]. Moreover understanding how cells remember their past should greatly facilitate the design of reprogramming strategies for gene therapy.
Conflict of interest statement
Nothing declared.
Acknowledgements
We thank Bernd Schuttengruber, Jeremy Dufourt, Yousra Yahia and Cyril Esnault for comments on the manuscript.
Research in the M.L laboratory is supported by the Centre National de la Recherche Scientifique, the European Research Council (Sync_DEV). O.R thanks CNRS and LABEX Epigenmed for support.
We apologize to colleagues whose work could not be cited due to space constraints.
This review comes from a themed issue on Development and differentiation
Edited by Stanislav Shvartsman and Robert Zinzen
References
- 1.Festuccia N., Gonzalez I., Owens N., Navarro P. Mitotic bookmarking in development and stem cells. Development. 2017;144:3633–3645. doi: 10.1242/dev.146522. [DOI] [PubMed] [Google Scholar]
- 2.Zenk F. Germ line–inherited H3K27me3 restricts enhancer function during maternal-to-zygotic transition. Science (80-.) 2017;357:212–216. doi: 10.1126/science.aam5339. [DOI] [PubMed] [Google Scholar]
- 3.Oomen M.E., Dekker J. Epigenetic characteristics of the mitotic chromosome in 1D and 3D. Crit Rev Biochem Mol Biol. 2017;52:185–204. doi: 10.1080/10409238.2017.1287160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piunti A., Shilatifard A. Epigenetic balance of gene expression by polycomb and compass families. Science (80-.) 2016;352 doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- 5.de Castro I.J., Gokhan E., Vagnarelli P. Resetting a functional G1 nucleus after mitosis. Chromosoma. 2016;125:607–619. doi: 10.1007/s00412-015-0561-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hug C.B., Grimaldi A.G., Kruse K., Vaquerizas J.M. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell. 2017;169 doi: 10.1016/j.cell.2017.03.024. 216–228.e19. [DOI] [PubMed] [Google Scholar]
- ••7.Blythe S.A., Wieschaus E.F. Establishment and maintenance of heritable chromatin structure during early drosophila embryogenesis. Elife. 2016;5:1–21. doi: 10.7554/eLife.20148. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using ATAC-Seq in single Drosophila embryos, the authors revealed that patterns of chromatin accessibility are maintained through mitosis.
- 8.Hsiung C.C.S. Genome accessibility is widely preserved and locally modulated during mitosis. Genome Res. 2015;25:213–225. doi: 10.1101/gr.180646.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olivier N. Cell lineage reconstruction of early. Science (80-.) 2010;70:967–971. doi: 10.1126/science.1189428. [DOI] [PubMed] [Google Scholar]
- 10.Nagano T. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature. 2017;547:61–67. doi: 10.1038/nature23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dufourt J. Temporal control of transcription by Zelda in living Drosophila embryos. bioRxiv. 2018 [Google Scholar]
- ••12.Ferraro T. Transcriptional memory in the Drosophila embryo. Curr Biol. 2016;26:212–218. doi: 10.1016/j.cub.2015.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using MS2-MCP system in the early Drosophila embryo, the authors visualized transcriptional memory for the first time in a living multicellular embryo.
- 13.Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strom A.R. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodruff J.B. The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell. 2017;169 doi: 10.1016/j.cell.2017.05.028. 1066–1077.e10. [DOI] [PubMed] [Google Scholar]
- 16.Schuettengruber B., Bourbon H.M., Di Croce L., Cavalli G. Genome regulation by polycomb and trithorax: 70 years and counting. Cell. 2017;171:34–57. doi: 10.1016/j.cell.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Petruk S. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 2012;150:922–933. doi: 10.1016/j.cell.2012.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Black K.L. Chromatin proteins and RNA are associated with DNA during all phases of mitosis. Cell Discov. 2016;2 doi: 10.1038/celldisc.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hörmanseder E. H3K4 methylation-dependent memory of somatic cell identity inhibits reprogramming and development of nuclear transfer embryos. Cell Stem Cell. 2017;21 doi: 10.1016/j.stem.2017.03.003. 135–143.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••20.Steffen P.A. Quantitative in vivo analysis of chromatin binding of polycomb and trithorax group proteins reveals retention of ASH1 on mitotic chromatin. Nucleic Acids Res. 2013;41:5235–5250. doi: 10.1093/nar/gkt217. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using time-lapse microscopy on living EGFP tagged Ash1 transgenic Drosophila embryos, the authors showed that Ash1 decorates mitotic chromosomes. Moreover, they employed quantitative imaging methods to reveal that Ash1 binds to chromatin dynamically.
- •21.Blobel G.A. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell. 2009;36:970–983. doi: 10.1016/j.molcel.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper was among the firsts to show that mitotic binding by MLL at specific loci accelerates their post-mitotic transcriptional reactivation.
- 22.Zhang H. MLL1 inhibition reprograms epiblast stem cells to naive pluripotency. Cell Stem Cell. 2016;18:481–494. doi: 10.1016/j.stem.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyama R., Rebbert M.L., Dey A., Ozato K., Dawid I.B. Brd4 associates with mitotic chromosomes throughout early zebrafish embryogenesis. Dev Dynam. 2008;237:1636–1644. doi: 10.1002/dvdy.21576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Devaiah B.N., Gegonne A., Singer D.S. Bromodomain 4: a cellular Swiss army knife. J Leukoc Biol. 2016;100:679–686. doi: 10.1189/jlb.2RI0616-250R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •25.Zhao R., Nakamura T., Fu Y., Lazar Z., Spector D.L. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat Cell Biol. 2011;13:1295–1304. doi: 10.1038/ncb2341. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrated the consequences of mitotic bookmarking (by BRD4 and H4K5 acetylation) on post-mitotic transcriptional dynamics in mammalian cells.
- 26.Palozola K.C. Mitotic transcription and waves of gene reactivation during mitotic exit. Science (80-.) 2017;358:119–122. doi: 10.1126/science.aal4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teves S.S., An L., Bhargava-Shah A., Xie L., Darzacq X., Tjian R. A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. Elife. 2018;7:1–22. doi: 10.7554/eLife.35621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun S.C. TBP dynamics during mouse oocyte meiotic maturation and early embryo development. PLoS One. 2013;8:1–6. doi: 10.1371/journal.pone.0055425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raccaud M., Suter D.M. Transcription factor retention on mitotic chromosomes: regulatory mechanisms and impact on cell fate decisions. FEBS Lett. 2017;592:878–887. doi: 10.1002/1873-3468.12828. [DOI] [PubMed] [Google Scholar]
- •30.Festuccia N. Mitotic binding of Esrrb marks key regulatory regions of the pluripotency network. Nat Cell Biol. 2016;18:1139–1148. doi: 10.1038/ncb3418. [DOI] [PubMed] [Google Scholar]; By using live imaging and ChIP experiments in ES cells, this paper identified Essrb as a potential mitotic bookmarker. A subset of interphase-bound targets, remain bound by Essrb during mitosis and correspond to key regulatory regions (super-enhancers).
- 31.Liu Y. Widespread mitotic bookmarking by histone marks and transcription factors in pluripotent stem cells. Cell Rep. 2017;19:1283–1293. doi: 10.1016/j.celrep.2017.04.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •32.Teves S.S. A dynamic mode of mitotic bookmarking by transcription factors. Elife. 2016;5 doi: 10.7554/eLife.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper demonstrated that the majority of transcription factors examined remain associated to mitotic chromosomes in ES cells. A quantitative study on Sox2 revealed that this pluripotency factor exhibits a dynamic mode of mitotic binding. This paper points to the artifactual mitotic eviction of some transcription factors triggered by formaldehyde fixation.
- 33.Pallier C. Association of chromatin proteins high mobility group box (HMGB) 1 and HMGB2 with mitotic chromosomes. Mol Biol Cell. 2003;14:2372–2384. doi: 10.1091/mbc.E02-09-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soufi A. Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell. 2015;161:555–568. doi: 10.1016/j.cell.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwafuchi-Doi M., Zaret K.S. Cell fate control by pioneer transcription factors. Development. 2016;143:1833–1837. doi: 10.1242/dev.133900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deal R.B., Henikoff J.G., Henikoff S. Genome-wide kinetics of nucleosome. Science (80-.) 2010;328:1161–1165. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller F., Stasevich T.J., Mazza D., McNally J.G. Quantifying transcription factor kinetics: at work or at play? Crit Rev Biochem Mol Biol. 2013;48:492–514. doi: 10.3109/10409238.2013.833891. [DOI] [PubMed] [Google Scholar]
- •38.Deluz C. A role for mitotic bookmarking of SOX2 in pluripotency and differentiation. Genes Dev. 2016;30:2538–2550. doi: 10.1101/gad.289256.116. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this paper, the authors profiled the mitotic binding for Sox2 and Oct4 and quantified their binding dynamics with quantitative imaging methods. By analyzing the consequences of Sox2 mitotic depletion, they demonstrated Sox2 bookmarking activity.
- •39.Caravaca J.M. Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes Dev. 2013;27:251–260. doi: 10.1101/gad.206458.112. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified two modes of mitotic bookmarking for the pioneer factor FoxA: via sequence specific binding and via nonspecific binding.
- ••40.Muramoto T., Müller I., Thomas G., Melvin A., Chubb J.R. Methylation of H3K4 is required for inheritance of active transcriptional states. Curr Biol. 2010;20:397–406. doi: 10.1016/j.cub.2010.01.017. [DOI] [PubMed] [Google Scholar]; This paper was the first to directly visualize the inheritance of transcriptionally active states down cell lineages in Dictyostelium. By using various genetic mutants, the authors demonstrated that this memory can be partially attributed to H3K4 methylation.
- 41.Kadauke S. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell. 2012;150:725–737. doi: 10.1016/j.cell.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertrand E. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 43.Lionnet T., Singer R.H. Transcription goes digital. EMBO Rep. 2012;13:313–321. doi: 10.1038/embor.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Forster D. Hydrodynamic fluctuations, broken symmetry, and correlation functions. (Frontiers in Physics 47, XIX, 326 S, 1975).
- 45.Herrera-Delgado E., Perez-Carrasco R., Briscoe J., Sollich P. Memory functions reveal structural properties of gene regulatory networks. PLoS Comput Biol. 2018;14 doi: 10.1371/journal.pcbi.1006003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karlin S., Taylor H.E. 2nd ed. 2012. A first course in stochastic processes. [Google Scholar]
- 47.Bentovim L., Harden T.T., DePace A.H. Transcriptional precision and accuracy in development: from measurements to models and mechanisms. Development. 2017;144:3855–3866. doi: 10.1242/dev.146563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chubb J.R. Gene regulation: stable noise. Curr Biol. 2016;26:R61–R64. doi: 10.1016/j.cub.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Lagha M. XPaused Pol II coordinates tissue morphogenesis in the drosophila embryo. Cell. 2013;153:976–987. doi: 10.1016/j.cell.2013.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon C.S., Hadjantonakis A.-K., Schröter C. Making lineage decisions with biological noise: lessons from the early mouse embryo. Wiley Interdiscip Rev Dev Biol. 2018;e319 doi: 10.1002/wdev.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skene P.J., Henikoff J.G., Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat Protoc. 2018;13:1006–1019. doi: 10.1038/nprot.2018.015. [DOI] [PubMed] [Google Scholar]
- 52.Bintu L. Dynamics of epigenetic regulation at the single cell level. Science (80-.) 2016;351:720–724. doi: 10.1126/science.aab2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang A., Amourda C., Zhang S., Tolwinski N.S., Saunders T.E. Decoding temporal interpretation of the morphogen bicoid in the early drosophila embryo. Elife. 2017;6 doi: 10.7554/eLife.26258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lerner J. Human mutations affect the epigenetic/bookmarking function of HNF1B. Nucleic Acids Res. 2016;44:8097–8111. doi: 10.1093/nar/gkw467. [DOI] [PMC free article] [PubMed] [Google Scholar]